A metal-independent glutaminyl cyclase from the black-legged tick I. scapularis is reported. The metal-independent inhibition of glutaminyl cyclases by imidazole-derived inhibitors is described for the first time.
Keywords: glutaminyl cyclases, Ixodes scapularis, pyroglutamate, zinc ion, posttranslational modification
Abstract
Glutaminyl cyclases (QCs) from mammals and Drosophila are zinc-dependent enzymes that catalyze N-terminal pyroglutamate formation of numerous proteins and peptides. These enzymes have been found to be critical for the oviposition and embryogenesis of ticks, implying that they are possible physiological targets for tick control. Here, 1.10–1.15 Å resolution structures of a metal-independent QC from the black-legged tick Ixodes scapularis (Is-QC) are reported. The structures exhibit the typical scaffold of mammalian QCs but have two extra disulfide bridges that stabilize the central β-sheet, resulting in an increased thermal stability. Is-QC contains ∼0.5 stoichiometric zinc ions, which could be removed by 1 mM EDTA. Compared with the Zn-bound form, apo-Is-QC has a nearly identical active-site structure and stability, but unexpectedly possesses significantly increased QC activities towards both synthetic and physiological substrates. Enzyme-kinetic analysis revealed that apo-Is-QC has a stronger substrate-binding affinity, suggesting that bound zinc interferes with substrate binding during catalysis. The structures of Is-QC bound to the inhibitor PBD150 revealed similar binding modes to both forms of Is-QC, with the exception of the inhibitor imidazole ring, which is consistent with the comparable inhibition activities of the inhibitor towards both forms of Is-QC. These findings have implications for the design of new QC inhibitors.
1. Introduction
Glutaminyl cyclases (QCs; EC 2.3.2.5) catalyze the cyclization of glutaminyl or glutamyl residues to form pyroglutamate (5-oxoproline; pGlu) at the N-terminus of numerous hormones, cytokines and other secretory peptides (Fischer & Spiess, 1987 ▸; Busby et al., 1987 ▸; Schilling et al., 2004 ▸). The formation of pGlu is believed to protect these bioactive proteins and peptides from exopeptidase degradation and, in numerous cases, such as thyrotropin-releasing hormone and some cytotoxic ribonucleases, to endow them with a functional conformation (Goren et al., 1977 ▸; Lou et al., 2006 ▸). QCs can be classified into two types on the basis of their distinct molecular structures and protein stabilities: type I QCs have been identified in plants and some bacteria (Wintjens et al., 2006 ▸; Huang et al., 2010 ▸), while type II QCs have so far been found in humans, some mammals and insects (Busby et al., 1987 ▸; Huang, Liu, Cheng et al., 2005 ▸; Schilling et al., 2007 ▸). Recent studies have indicated that QCs are emerging drug targets for the treatment of Alzheimer’s disease and some inflammatory disorders, such as septic arthritis (Schilling et al., 2008 ▸; Nussbaum et al., 2012 ▸; Hellvard et al., 2013 ▸). These studies were motivated by the findings that human secretory QC is able to catalyze N-terminal pGlu formation of several amyloid β-peptides, leading to enhanced aggregation and neurotoxicity of these peptides (Schilling et al., 2008 ▸; Cynis, Scheel et al., 2008 ▸; Schlenzig et al., 2009 ▸), and that the Golgi-resident QC is able to catalyze pGlu formation of monocyte chemoattractant proteins, a crucial step for the migration and infiltration of monocytes in some inflammatory conditions (Cynis, Rahfeld et al., 2008 ▸; Cynis et al., 2011 ▸; Chen et al., 2012 ▸).
Schilling, Niestroj et al. (2003 ▸) have proposed that human secretory QC is a metalloenzyme based on its time-dependent inhibition by several metal-chelating agents and imidazole derivatives as well as the reactivation of the inactive ‘apoenzyme’ by the addition of metal. A couple of years later, the recombinant human and mouse QCs were shown to contain stoichiometric amounts of zinc (Huang, Liu & Wang, 2005 ▸; Schilling et al., 2005 ▸). Moreover, the crystal structures of human QCs have revealed a bound zinc ion coordinated by three conserved residues (Huang, Liu, Cheng et al., 2005 ▸; Huang et al., 2011 ▸). Therefore, type II QCs are commonly believed to be zinc-dependent enzymes. Along these lines, a series of compounds derived from imidazole have been developed as QC inhibitors, such as PBD150 (Buchholz et al., 2006 ▸, 2009 ▸), which is a potent inhibitor of human QCs, with a K i value of ∼0.1 µM towards the secretory form. The structures of human QCs in complex with imidazole derivatives have confirmed the binding between the inhibitor imidazole ring and the active-site zinc of QCs (Huang et al., 2011 ▸). Furthermore, oral administration of PBD150 in transgenic mouse models of Alzheimer’s disease resulted in significantly reduced depositions of Aβ3(pGlu)-40/42 in the brain (Schilling et al., 2008 ▸), leading to improved learning and memory in the mice. Recently, Hellvard et al. (2013 ▸) reported that some QC inhibitors were able to significantly delay the onset of clinical signs of arthritis in a septic arthritis mouse model. These findings imply that QC inhibitors may be potential drugs for the treatment of Alzheimer’s disease and some inflammatory disorders.
To date, however, several lines of results argue against the requirement for a metal in QCs. For example, (i) recombinant human QC expressed in Drosophila S2 cells showed ∼0.3 moles of zinc per mole of enzyme, but possessed an enzymatic activity that was comparable to those of QC samples produced by Pichia and Escherichia coli cells, which were shown to contain stoichiometric amounts of zinc (Huang, Liu & Wang, 2005 ▸; Booth et al., 2004 ▸; Schilling et al., 2002 ▸); (ii) the recombinant human and mouse QCs appeared to be refractory to the strong metal chelator EDTA even at concentrations above 20 mM (Schilling, Niestroj et al., 2003 ▸; Schilling et al., 2005 ▸); and (iii) the enzyme activity of porcine QC partially purified from pituitary extracts was shown to be enhanced around twofold by the addition of 10 mM EDTA (Busby et al., 1987 ▸). These phenomena are very unusual in the families of zinc-dependent enzymes (Auld, 1995 ▸). Because no reports have described the exact quantity of zinc from QCs treated with EDTA and other metal-chelating agents, and the structure of an inactive ‘apo form’ of a QC is not yet available, the role of zinc in QC catalysis still remains controversial.
More recently, Adamson et al. (2013 ▸) reported that QCs are functionally linked to the bloodmeal uptake of ticks. Knockdown of tick QC by RNA interference resulted in remarkably attenuated oviposition and embryogenesis, which is most likely to be attributable to improper pGlu formation and thus a shorter half-life of several pGlu-modified neuropeptides, such as corazonin, sulfakinin and periviscerokinin, in the ticks (Neupert et al., 2009 ▸). Since ticks, especially the black-legged tick Ixodes scapularis, are important vectors of some human pathogens such as Borrelia burgdorferi, Anaplasma phagocytophilum and Babesia microti (Burgdorfer et al., 1982 ▸; Telford et al., 1996 ▸; Rodgers & Mather, 2007 ▸), this finding implies that QCs are potential physiological targets for tick control to prevent the spread of tick-borne diseases. In the present study, we report the production and atomic resolution crystal structures of a metal-independent QC from I. scapularis (Is-QC). Structural and ICP-MS analyses revealed that Is-QC contains ∼0.5 stoichiometric zinc ions, which could readily be removed by EDTA at low-millimolar concentrations. Unexpectedly, apo-Is-QC was shown to possess a significantly increased QC activity compared with the Zn-bound form, despite having a nearly identical active-site structure. The enzyme-kinetic properties and thermal stabilities of both zinc-bound and apo-Is-QC were analyzed, which suggest that the presence of zinc was owing to nonspecific binding. We further analyzed the inhibition potency of imidazole derivatives towards both forms of Is-QC and determined the structures of both forms of Is-QC bound to PBD150.
2. Materials and methods
2.1. Materials
Chemicals were purchased as follows: l-glutaminyl 2-naphthylamide from Bachem (Weil am Rhein, Germany); 2-naphthylamide, N-ω-acetylhistamine, 1,10-phenanthroline and dipicolinic acid from Sigma–Aldrich (St Louis, Missouri, USA); 1-benzylimidazole from Fluka (St Louis, Missouri, USA). Human pyroglutamyl aminopeptidase I (PAP I) was prepared as described previously (Huang et al., 2008 ▸). The peptides [Gln1]-corazonin, [Gln1]-sulfakinin and [Gln1]-periviscerokinin were synthesized by standard FMOC peptide chemistry (Huang, Liu & Wang, 2005 ▸). The QC inhibitor PBD150 was chemically synthesized according to a previous report (Buchholz et al., 2006 ▸). All other chemicals were of analytical or HPLC grade.
2.2. Protein expression and purification
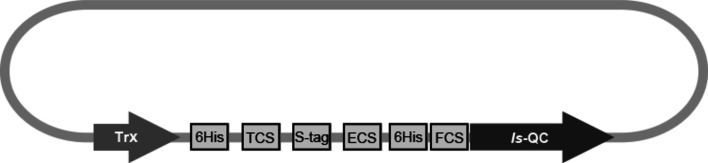
The cDNA encoding I. scapularis QC (nucleotide entry XM_002415508.1) was obtained previously (Adamson et al., 2013 ▸). The cDNA fragment covering the putative catalytic domain (Trp25–Leu353) of the enzyme was amplified and inserted into a thioredoxin-fusion expression vector via HindIII and XhoI cloning sites as described previously (Huang, Liu & Wang, 2005 ▸). The resulting construct contained a thioredoxin-fusion protein, two 6×His tags and a factor Xa cleavage site upstream of the Is-QC coding region (Fig. 1 ▸). The vector was transformed into E. coli BL21(DE3) CodonPlus-RIL cells (Stratagene, La Jolla, California, USA). The bacteria were grown in Luria–Bertani medium containing ampicillin (100 µg ml−1) and chloramphenicol (34 µg ml−1) at 37°C until the cell density reached an A 600 of ∼0.8. The cultures were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 18–20 h at 20°C. The cells were then harvested by centrifugation (8983g for 30 min at 4°C) followed by freezing at −80°C. Frozen bacterial pellets were resuspended in lysis buffer (50 mM Tris–HCl pH 7.8 containing 150 mM NaCl and 10 mM imidazole) and the cells were lysed using a cell disruptor (Constant Systems, Kennesaw, Georgia, USA). The cell lysate was clarified by centrifugation (104 630g for 60 min at 4°C) and the supernatant was loaded onto a nickel–nitrilotriacetic acid column (Amersham Biosciences, Piscataway, New Jersey, USA) pre-equilibrated with buffer A (50 mM Tris–HCl pH 7.8, 150 mM NaCl, 10 mM imidazole, 5% glycerol). The column was washed with the same buffer and the bound materials were eluted with a linear gradient of 0–100% buffer B (50 mM Tris–HCl pH 7.8, 150 mM NaCl, 300 mM imidazole, 5% glycerol). The fractions containing thioredoxin-fusion Is-QC were pooled and were digested with factor Xa (0.3 units ml−1; Novagen, Darmstadt, Germany). To minimize interference from the imidazole present in the protein solution, the digestion reaction was carried out in a dialysis bag and the imidazole was removed by dialysis against buffer C (50 mM Tris–HCl pH 7.8, 150 mM NaCl, 5% glycerol) at 25°C for 18–20 h. Subsequently, because of the relatively high content (4.9%) of histidine residues in Is-QC, the cleaved Is-QC products were isolated from the digest on a second nickel–nitrilotriacetic acid column pre-equilibrated with buffer C. The bound Is-QC was eluted with a linear gradient of 0–100% buffer D (50 mM Tris–HCl pH 7.8, 150 mM NaCl, 40 mM imidazole, 5% glycerol). The protein was concentrated and further desalted on a Sephacryl S-100 column (Amersham Biosciences) eluted with buffer E (50 mM Tris–HCl pH 7.8 containing 5% glycerol). The purified Is-QC was shown to be homogeneous as judged by SDS–PAGE with Coomassie Blue staining. The D144A and D238A mutants were constructed using KOD Hot Start DNA Polymerase (Novagen). In addition, the expression and purification of human secretory QC (sQC) was performed according to a previous protocol (Huang, Liu & Wang, 2005 ▸).
Figure 1.
Schematic representation of the expression construct for Is-QC. Trx, thioredoxin-fusion protein; TCS, thrombin cleavage site; ECS, enterokinase cleavage site; FCS, factor Xa cleavage site.
2.3. Enzyme-activity assay
The enzymatic activity of recombinant Is-QC was evaluated at 25°C using the fluorescent substrate l-glutaminyl 2-naphthylamide (Gln-βNA) as described previously (Huang et al., 2008 ▸). The 100 µl reaction mixture contained 300 µM fluorogenic substrate, ∼0.2 units of the auxiliary enzyme human PAP I (where one unit is defined as the amount that hydrolyzes 1 µmol of pGlu-βNA per minute under the same conditions) and an appropriately diluted aliquot of recombinant Is-QC in 50 mM Tris–HCl pH 8.0 with 5% glycerol. The excitation and emission wavelengths were set to 320 and 410 nm, respectively. The reaction was initiated by the addition of Is-QC. Enzymatic activity was determined from the amount of released βNA and was calculated using a standard curve for βNA under the same assay conditions. The measurements were made using a Synergy H4 microplate reader (BioTek, Winooski, Vermont, USA).
The activity of QC towards the physiological substrates of I. scapularis was analyzed based on the time-dependent change of the elution profile from reversed-phase HPLC as described previously (Huang, Liu & Wang, 2005 ▸). Briefly, for each individual substrate, a 30 µl reaction mixture containing ∼0.1 µM Is-QC and 1 mM substrate in 50 mM Tris–HCl pH 8.0 was incubated at 25°C for 5–180 min in the absence or presence of 20 mM EDTA. Subsequently, a 20 µl aliquot of the mixture was subjected to HPLC analysis on a Thermo UltiMate 3000 liquid chromatograph using a C18 column (Thermo Fisher, Waltham, Massachusetts, USA). The bound materials were eluted with a linear gradient of 0–100% acetonitrile containing 0.1% TFA. The elution of the peptides was monitored by the absorbance at 214 nm and the pGlu-containing product was identified by ESI-LC-MS/MS analysis.
2.4. Enzyme-kinetic assay
The kinetic constants were determined at pH 8.0 at 25°C using the substrate Gln-βNA as described previously (Huang et al., 2008 ▸). The reaction was initiated by adding Is-QC (final concentration of ∼43 nM) to the 100 µl reaction mixture. The initial rate was measured with less than 10% substrate depletion for the first 2–12 min. Since a weak substrate inhibition was observed, the kinetic parameters K m, V and K i were evaluated by fitting the equation v 0 = V max[S]/(K m + [S] + [S]2/K i) to the initial velocity data by nonlinear regression using the KaleidaGraph software (Synergy Software, Reading, Pennsylvania, USA), where v 0 is the initial velocity, V max is the limiting rate, [S] is the substrate concentration, K m is the Michaelis constant and K i is the inhibition constant. Correlation coefficients of better than 0.995 were obtained throughout the fittings. The substrate concentrations used were in the range 1–240 µM. The k cat values were calculated from V max/[E] according to the enzyme concentration determined from the UV absorbance at 280 nm (ɛ = 46 785 M −1 cm−1) in the presence of 6.0 M guanidinium chloride. Therefore, the k cat values reported below represent minimal estimates based on the assumption that the enzyme active-site concentration is equivalent to the concentration of the protein.
2.5. Inhibitor assay
The inhibition activity of metal-chelating agents and imidazole derivatives was evaluated according to a previous report (Huang et al., 2011 ▸). Briefly, the reaction mixture contained 300 µM Gln-βNA and ∼0.2 units of human PAP I as described above. Is-QC was first incubated with the inhibitor at 25°C for 30 min and the enzyme–inhibitor mixture was then added to the reaction mixture to initiate the cyclization reaction. The final concentration of Is-QC was ∼45 nM. The IC50 values were obtained by fitting the initial reaction rate versus the inhibitor concentrations using KaleidaGraph. K i values were calculated according to the equation IC50 = K i(1 + [S]/K m) (Segel, 1993 ▸), where [S] is the substrate concentration and K m is the Michaelis–Menten constant.
2.6. Metal analysis
The metal contents of the Is-QCs and human sQC were determined by inductively coupled plasma mass spectrometry (ICP-MS). For each analysis, 100 µl sample was pipetted into 3 ml ultrapure nitric acid in a microwave tube. The solution was heated in a microwave oven for 15 min to reach 200°C. The sample was held at 200°C for 15 min and then cooled to room temperature. Subsequently, the digested sample was diluted with 2%(v/v) nitric acid to 25 ml and subjected to ICP-MS analysis. Measurement of the amount of metal was made using an ICP-MS X series II (Thermo Fisher, Waltham, Massachusetts, USA), with direct nebulization, a normal mode of operation and an optimized condition consisting of an X-series skimmer cone with a Platinum sampler cone. The extraction voltage was −35 V with an RF power of 510 W, a focus voltage of 4.5 V and a nebulizer gas-flow rate of 1.04 l min−1. The dwell times were 10 ms, and 100 sweeps were required for each replicate, with five replicates every sample.
2.7. Thermal stability
The thermal stability was evaluated using SYPRO Orange dye (Invitrogen, Karlsruhe, Germany). Samples were prepared in 96-well plates and the experiments were conducted in 50 mM Tris–HCl pH 8.0. For each well in the plates, the 100 µl solution contained 0.20–0.23 mg ml−1 protein sample with SYPRO Orange dye added at a concentration of 5×. The dye was originally delivered as a 5000× solution in 100%(v/v) DMSO and was first diluted into a 500× solution using 50 mM Tris–HCl pH 8.0 before adding the sample. The fluorescence was recorded on a CFX Connect real-time PCR system (Bio-Rad, Hercules, California, USA) and was analyzed using the CFX Manager software. Data were collected at 1°C intervals ranging from 25 to 95°C. The melting temperature (T m) was determined as the maximum of the first derivative of the melting curves. Furthermore, to measure the residual enzymatic activity at different thermal conditions, QC samples in 50 mM Tris–HCl pH 8.0 were incubated at 25, 30, 35, 40, 45, 50, 55 or 60°C for 10 min and then cooled to room temperature. The QC activities were determined as described above, and the percentage residual activity at each temperature was calculated based on the activity at 25°C as 100%.
2.8. Crystallization and X-ray data collection
Purified Is-QC in 50 mM Tris–HCl with 5% glycerol pH 7.8 was concentrated to ∼8.5 mg ml−1 using an Ultracel 10K centrifugal filter (Millipore, Billerica, Massachusetts, USA). An initial crystallization screening of ∼900 conditions was performed at the Core Facilities for Protein Structural Analysis in Academia Sinica, Taipei, Taiwan. The crystals were grown at 20°C by the sitting-drop vapour-diffusion method. The final crystallization condition was 10%(w/v) PEG 8000, 8%(v/v) ethylene glycol, 0.1 M HEPES pH 7.5. Is-QC was mixed with an equal volume of the crystallization solution, and rod-like crystals with dimensions reaching 0.1 × 0.1 × 0.2 mm appeared within 5–8 d. High-resolution X-ray diffraction data were collected on beamline 13C1 of the National Synchrotron Radiation Research Center, Hsinchu, Taiwan and on beamline 4.2.2 of the Advanced Light Source at Lawrence Berkeley National Laboratory, Berkeley, California, USA. Before being mounted on the goniometer, the crystals were briefly soaked in reservoir solution containing 15%(v/v) glycerol as a cryoprotectant. All diffraction data were processed and scaled with the HKL-2000 package (Otwinowski & Minor, 1997 ▸). The data-collection statistics are listed in Table 1 ▸. For the structures of Is-QCs bound to PBD150, the crystals were soaked in mother liquor containing 5 mM PBD150 at 20°C for 3–5 d.
Table 1. Data-collection and refinement statistics.
These crystals belonged to space group P212121 and the asymmetric unit comprises one QC molecule. Values in parentheses are for the highest resolution shell.
| Zn-Is-QC | Apo-Is-QC | Zn-Is-QC–PBD150 | Apo-Is-QC–PBD150 | |
|---|---|---|---|---|
| Data collection | ||||
| Resolution (Å) | 30–1.15 (1.19–1.15) | 50–1.10 (1.14–1.10) | 30–1.38 (1.43–1.38) | 50–1.95 (2.02–1.95) |
| Unit-cell parameters (Å) | ||||
| a | 55.28 | 55.39 | 54.98 | 55.26 |
| b | 71.38 | 71.15 | 71.08 | 71.44 |
| c | 80.08 | 80.12 | 80.07 | 80.25 |
| Total observations | 793696 | 759300 | 359056 | 83869 |
| Unique reflections | 112442 (11084) | 128122 (12774) | 64300 (6353) | 21418 (2102) |
| Multiplicity | 7.1 (5.8) | 5.9 (5.7) | 5.6 (5.5) | 3.9 (3.7) |
| Completeness (%) | 99.6 (99.3) | 99.5 (100.0) | 99.1 (99.5) | 89.9 (90.4) |
| 〈I/σ(I)〉 | 43.9 (2.6) | 38.8 (2.8) | 31.3 (2.8) | 9.5 (1.9) |
| R merge (%) | 5.6 (79.7) | 5.4 (77.7) | 7.1 (73.2) | 13.2 (75.4) |
| Refinement | ||||
| Resolution (Å) | 30–1.15 | 30–1.10 | 30–1.38 | 50–1.95 |
| Reflections [>0σ(F)] (working/test) | 101091/5632 | 115164/6434 | 57726/3259 | 14875/843 |
| R factor/R free | 0.162/0.181 | 0.165/0.186 | 0.153/0.184 | 0.166/0.221 |
| R.m.s.d. | ||||
| Bond lengths (Å) | 0.005 | 0.005 | 0.006 | 0.012 |
| Angles (°) | 1.125 | 1.131 | 1.169 | 1.530 |
| No. of atoms | ||||
| Protein | 2665 | 2665 | 2650 | 2656 |
| Inhibitor | 22 | 22 | ||
| Zinc ion | 1 | 1 | ||
| Water | 381 | 427 | 334 | 262 |
| Average B factor (Å2) | ||||
| Protein | 16.1 | 14.4 | 16.1 | 20.5 |
| Inhibitor | 19.5 | 26.6 | ||
| Zinc ion | 14.8 | 12.2 | ||
| Water | 27.0 | 25.9 | 27.7 | 30.0 |
| Ramachandran plot, residues in (%) | ||||
| Most favoured regions | 90.1 | 90.1 | 91.0 | 89.0 |
| Additionally allowed regions | 9.6 | 9.6 | 8.7 | 10.7 |
| Generously allowed regions | 0.3 | 0.3 | 0.3 | 0.3 |
| Disallowed regions | 0 | 0 | 0 | 0 |
| PDB code | 4mhn | 4mhp | 4mhy | 4mhz |
2.9. Structure determination and refinement
The crystal structure of Is-QC was solved by the molecular-replacement method with the program MOLREP (Vagin & Teplyakov, 2010 ▸), using the published structure (PDB entry 2afm; Huang, Liu, Cheng et al., 2005 ▸) of human sQC as a search model and a 2.2 Å resolution data set for recombinant Is-QC (Zn-Is-QC) which was collected using an FR-E+ generator equipped with an R-AXIS HTC image-plate detector at the Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan. Only one clear solution for the translation function indicated the presence of one Is-QC molecule in the asymmetric unit. On the basis of the initial phase obtained from the molecular-replacement search, ∼90% of the model was automatically traced into the electron-density map with ARP/wARP (Perrakis et al., 1999 ▸) and the remainder was manually built with Coot (Emsley & Cowtan, 2004 ▸). The resulting model was subjected to computational refinement with the program REFMAC5 (Murshudov et al., 2011 ▸). Throughout refinement, a randomly selected 5% of the data was set aside as a free data set, and the model was refined against the remaining data with F > 0 as a working data set. The parameters for ideal protein geometry of Engh and Huber were used during refinement (Engh & Huber, 1991 ▸). Subsequently, several rounds of model adjustment with Coot and refinement with REFMAC5 were performed using a 1.15 Å resolution data set to improve the quality and completeness of the structure. The well ordered water molecules and one zinc ion were located with Coot. Finally, the refinement converged at a final R factor and R free of 0.162 and 0.181, respectively, in the 30–1.15 Å resolution range. The stereochemical quality of the refined structure was checked with the program PROCHECK (Laskowski et al., 1993 ▸). The final refinement statistics are listed in Table 1 ▸. The initial difference Fourier maps for the structures of apo-Is-QC and the PBD150-bound Is-QCs were obtained employing the refined structure of Zn-Is-QC. The molecular figures were produced with PyMOL (Schrödinger, New York, USA).
3. Results
3.1. Protein production and enzymatic activity
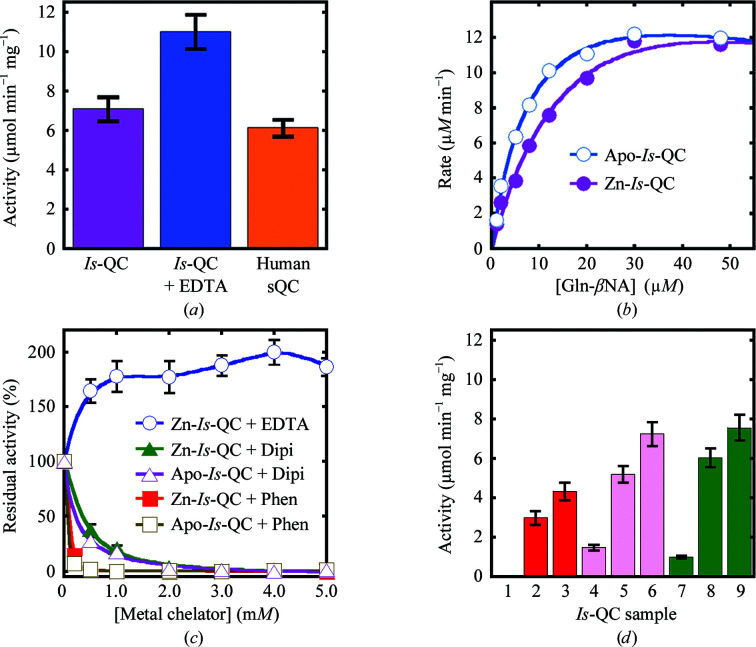
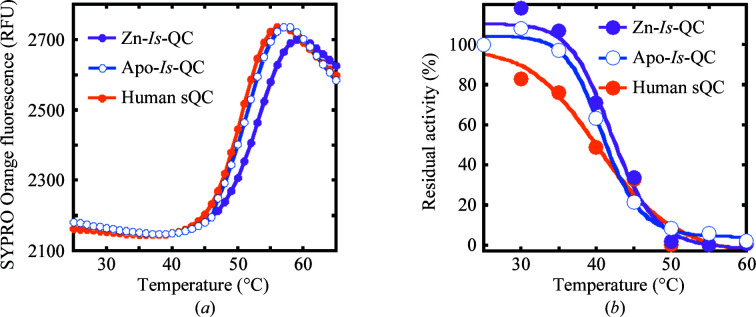
The segment Trp25–Leu353 of I. scapularis QC (Is-QC), which contains the putative catalytic domain of the enzyme, was overexpressed in E. coli cells and purified to near-homogeneity. The purified enzyme showed a QC activity that was ∼1.2-fold stronger than the activity of human secretory QC (sQC; Fig. 2 ▸ a). Unexpectedly, when 1 mM EDTA was added during the last purification step by size-exclusion chromatography the enzymatic activity increased by ∼55%, indicating that EDTA could stimulate the activity of Is-QC. Similar results were obtained when the enzyme and relevant materials in the QC activity assay were treated with the metal-chelating resin Chelex-100 at a concentration of 100 g per litre of sample (data not shown). ICP-MS analysis revealed a stoichiometry of ∼0.5 mol of zinc per mole of enzyme for recombinant Is-QC (Zn-Is-QC); the zinc content was reduced to ∼0.1 mol per mole of enzyme on the addition of 1–10 mM EDTA (apo-Is-QC) (Table 2 ▸). In contrast, human sQC showed a nearly equimolar amount of zinc, consistent with our previous data determined by the atomic absorption method (Huang, Liu & Wang, 2005 ▸). Indeed, the crystal structure of apo-Is-QC, as described below, demonstrated that the zinc was effectively removed by 1 mM EDTA. Enzyme-kinetic analysis revealed that Zn-Is-QC has k cat and K m values of 8.8 ± 0.8 s−1 and 21.8 ± 3.5 µM, respectively, while those for apo-Is-QC were 6.9 ± 0.2 s−1 and 8.8 ± 0.6 µM, respectively (Fig. 2 ▸ b), implying that the bound zinc interfered with substrate binding during catalysis.
Figure 2.
Enzymatic activity of Is-QC and its modulation by metal-chelating agents. (a) Activities of Zn-Is-QC (purple) and apo-Is-QC (blue) and comparison with the activity of human sQC (orange). The activities ± SD were calculated from three independent experiments. (b) Enzyme-kinetic analysis of Zn-Is-QC and apo-Is-QC. The typical result of three experiments is presented. (c) Activities of Zn-Is-QC and apo-Is-QC in the presence of EDTA, dipicolinic acid (Dipi) or 1,10-phenanthroline (Phen). Before addition to the reaction mixture, the QC samples were pre-incubated with the metal-chelating agents at the indicated concentrations for 30 min. The results were calculated as residual activities ± SD from three independent experiments. (d) Restoration of Is-QC activity. Columns 1, 4 and 7: Zn-Is-QC was passed through a Sephacryl S-100 size-exclusion column in the presence of 5 and 1 mM 1,10-phenanthroline and 5 mM dipicolinic acid, respectively, and the QC activities of the resulting Is-QC samples were analyzed. Columns 2, 5 and 8: the Is-QC samples in columns 1, 4 and 7, respectively, were re-passed through a second Sephacryl S-100 column to remove the metal-chelating agents and the QC activities of the resulting samples were analyzed. Columns 3, 6 and 9: the Is-QC samples in columns 2, 5 and 8 were analyzed for QC activity in the presence of 1 mM EDTA. All enzymatic activity assays were performed at 25°C in 50 mM Tris–HCl pH 8.0 using the fluorescent substrate Gln-βNA.
Table 2. Metal contents of Is-QC and human sQC analyzed by ICP-MS.
The results are means ± SD from three experiments. ND, not detectable. —, not determined.
| Metal-to-enzyme molar ratio × 100 | ||||||||
|---|---|---|---|---|---|---|---|---|
| QC sample | Zn | Ca | Mn | Fe | Co | Cu | Ni | Cd |
| Is-QC | 51.6 ± 1.4 | 8.6 ± 0.3 | 0.15 ± 0.01 | 2.3 ± 0.1 | ND | 8.3 ± 0.4 | 5.4 ± 0.2 | ND |
| Is-QC + 1 mM EDTA | 14.2 ± 0.7 | 4.2 ± 0.2 | 0.58 ± 0.02 | 2.7 ± 0.1 | 0.12 ± 0.01 | ND | ND | ND |
| Is-QC + 10 mM EDTA | 8.5 ± 0.3 | — | — | — | — | — | — | — |
| Human sQC | 98.2 ± 3.4 | 3.2 ± 0.2 | 1.2 ± 0.1 | 1.9 ± 0.1 | ND | ND | 3.8 ± 0.1 | ND |
Moreover, although EDTA displayed a stimulatory effect, the heterocyclic metal-chelating agents 1,10-phenanthroline and dipicolinic acid showed strong inhibitory activities towards Zn-Is-QC, with K i values in the low- and medium-micromolar ranges, respectively (Fig. 2 ▸ c and Table 3 ▸). Interestingly, similar inhibition effects were also observed towards apo-Is-QC. In addition, once the metal-chelating agents had been removed by size-exclusion columns, a large portion of the QC activity could be restored (Fig. 2 ▸ d, columns 2, 5 and 8) and the activities were further enhanced by adding 1 mM EDTA (Fig. 2 ▸ d, columns 3, 6 and 9). These findings strongly suggest that the inhibition by the heterocyclic metal-chelating agents is owing to reversible binding of the chelators to Is-QC rather than zinc depletion.
Table 3. Inhibition of zinc-bound and apo-Is-QCs by heterocyclic metal-chelating agents and imidazole derivatives.
Assays were performed at 25°C in 50 mM Tris–HCl pH 8.0 using L-glutaminyl 2-naphthylamide as the substrate. The results are means ± SD from three experiments.
| K i value (µM) | |||
|---|---|---|---|
| Compound | Zn-Is-QC | Apo-Is-QC | Human sQC† |
| 1,10-Phenanthroline | 3.45 ± 0.59 | 2.88 ± 0.36 | |
| Dipicolinic acid | 36.8 ± 4.3 | 32.4 ± 4.9 | |
| PBD150 | 0.342 ± 0.038 | 0.134 ± 0.005 | 0.095 |
| 1-Benzylimidazole | 10.2 ± 0.9 | 1.48 ± 0.16 | 0.607 |
| N-ω-Acetylhistamine | 14.2 ± 1.7 | 4.53 ± 1.22 | 1.698 |
Data are taken from the previous report (Huang et al., 2011 ▸).
3.2. Efficacy towards the physiological substrates of I. scapularis
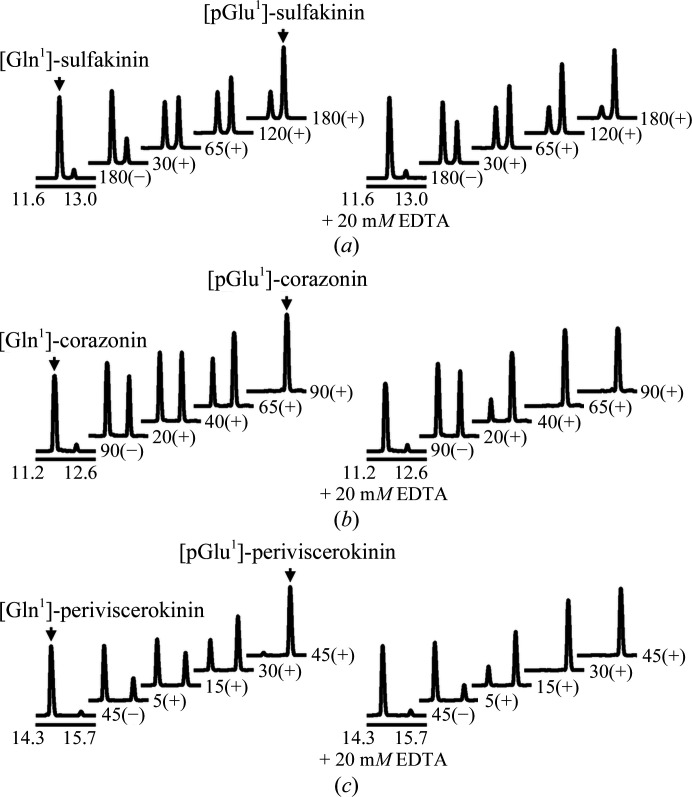
Apparently, as revealed by the time-dependent change in the HPLC elution profiles (Fig. 3 ▸), Is-QC was able to convert the three physiological substrates [Gln1]-sulfakinin, [Gln1]-corazonin and [Gln1]-periviscerokinin to their expected pGlu-containing products. Furthermore, the enzymatic activity of Is-QC was significantly increased by the addition of 20 mM EDTA (Fig. 3 ▸, right panels). On close analysis of the elution profiles, it is noteworthy that the cyclization rates of [Gln1]-corazonin and [Gln1]-periviscerokinin were significantly faster than that of [Gln1]-sulfakinin. In addition, a slightly faster rate was observed for [Gln1]-periviscerokinin than for [Gln1]-corazonin. These results suggest that Is-QC prefers substrates with neutral or hydrophobic amino-acid residues at the second and third positions, rather than negatively charged residues. Indeed, similar substrate specificities were observed for human, mouse and Drosophila QCs (Schilling, Manhart et al., 2003 ▸; Schilling et al., 2005 ▸, 2007 ▸), presumably reflecting the hydrophobic substrate-binding pocket near the catalytic centres of these enzymes.
Figure 3.
Efficacy of Is-QC towards the physiological substrates of I. scapularis. Characterization of the cyclization rates of (a) [Gln1]-sulfakinin (sequence QDDDYGHMRF), (b) [Gln1]-corazonin (QTFQYSRGWTN) and (c) [Gln1]-periviscerokinin (QGLIPFPRV). All reactions were carried out at 25°C in 50 mM Tris–HCl pH 8.0 in the presence (+) or absence (−) of Zn-Is-QC, as described in §2. Progress at the indicated time intervals was monitored by reverse-phase HPLC, and the glutaminyl peptides and the resulting pyroglutamyl [pGlu1] products were confirmed by ESI-LC-MS/MS analysis. The numbers below the lines indicate the elution time (in minutes), whereas the other numbers indicate incubation times (in minutes). In the right panels of the figure, EDTA was added to the reaction mixtures to a final concentration of 20 mM to remove the bound zinc in Is-QC.
3.3. Crystal structures of Is-QC
The crystals of Zn-Is-QC and apo-Is-QC grown at pH 7.5 as described in §2 belonged to space group P212121, with one Is-QC molecule in the asymmetric unit. The crystal structures were solved by the molecular-replacement method and were refined to 1.10–1.15 Å resolution (Table 1 ▸). The final structures comprise Leu28–Leu353, ∼400 water molecules and a zinc ion in Zn-Is-QC, with the first three N-terminal residues being excluded from the models owing to high flexibility.
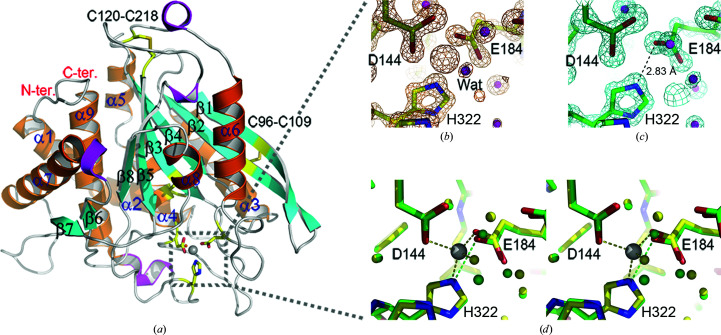
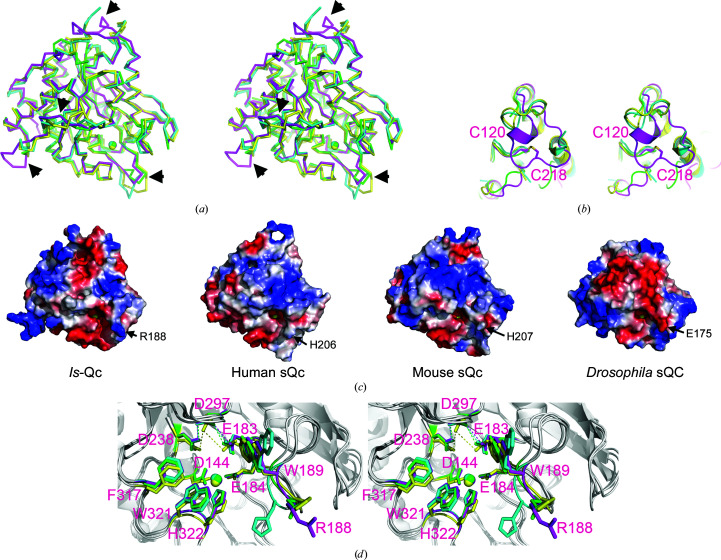
The Is-QC structure exhibits the typically globular and mixed α/β scaffold of type II QCs (Fig. 4 ▸ a). The structure displays an open sandwich topology, established by a central six-stranded (β1–β5 and β8) β-sheet flanked by two (α6 and α8) and six (α1–α5 and α9) α-helices on the concave and convex sides, respectively, with one edge of the β-sheet being sealed by an α-helix (α7) and two antiparallel β-strands (β6 and β7). The central β-sheet is twisted, consisting of five parallel (β1, β3–β5 and β8) and one antiparallel (β2) strands. Notably, the two β-strands β6 and β7 could only be identified in the structure of Drosophila mitochondria-resident QC and not in other structures of type II QCs (Koch et al., 2012 ▸). In addition, there are four short 310-helices adjacent to the N-terminus of β5, α7 and α9 and the C-terminus of α6 (Figs. 4 ▸ a and 5 ▸). Surprisingly, in addition to the conserved disulfide bridge Cys127–Cys149 near the zinc-binding site, Is-QC is unique in that it possesses two extra disulfide bonds: Cys96–Cys109 and Cys120–Cys218 (Fig. 4 ▸ a). Although the Cys127–Cys149 bond was not formed in the present structure since the protein was produced in an E. coli expression system, we noticed that two extra disulfide bridges Cys96–Cys109 and Cys120–Cys218 were formed, as shown by clear electron densities (Supplementary Fig. S11). Cys96–Cys109 connects strands β1 and β2, while Cys120–Cys218 shields the vertex of the central β-sheet; thus, it is reasonable to assume that these two disulfide bridges help to stabilize the central β-sheet of Is-QC.
Figure 4.
Crystal structures of Is-QC. (a) Overall structure of Zn-Is-QC shown as a ribbon diagram. The amino-acid sequences corresponding to the nine α-helices (α1–α9), eight β-strands (β1–β8) and four 310-helices can be seen in Fig. 5 ▸. The bound zinc ion is shown as a grey ball. The zinc-coordinated amino-acid residues (Asp144, Glu184 and His322) and water molecule are shown as yellow sticks and a red ball, respectively. The two extra disulfide bridges, as described in the text, are also shown as yellow sticks. (b) A close-up view of the zinc-binding environment of Zn-Is-QC. The 2.5σ 2F o − F c electron-density map, calculated with a zinc-free model of Is-QC, is overlaid on the final refined structure. The zinc-coordinated residues and water molecule are labelled. (c) A close-up view of the metal-binding site of apo-Is-QC. The 2F o − F c electron-density map, contoured at the 2.5σ level, is also shown. Note that electron density for zinc is not visible, and a new hydrogen bond between Glu184 Oɛ2 and His322 Nɛ2 is formed with the distance indicated. (d) Superimposition of the metal-binding sites in Zn-Is-QC (yellow) and apo-Is-QC (green). The zinc ion in Zn-Is-QC is shown as a grey ball.
Figure 5.
Structure-based sequence alignment of Is-QC with secreted QCs from human, mouse and Drosophila. The secondary-structural elements are illustrated on the basis of the refined structure of Is-QC. The completely conserved residues are shaded in black. The metal-coordinated residues are marked with grey balls. The three disulfide bridges Cys96–Cys109, Cys120–Cys218 and Cys127–Cys149 are marked with green, red and grey triangles, respectively. The segments with notable conformation change between these QC structures, as described in the text, are boxed in blue, and the segment with an eight-residue insertion in Is-QC is boxed in red. The PDB codes of these structures are as follows: Is-QC, 4mhn; human sQC, 2afm; mouse sQC, 3si1; Drosophila sQC, 4f9u.
3.4. Structures of the zinc-binding site
The putative catalytic centre and the conserved zinc-binding residues Asp144, Glu184 and His322 are located in the unstructured loops near the C-terminal edge of the central parallel strands β3–β5 (Figs. 4 ▸ a and 5 ▸). In Zn-Is-QC, the bound zinc forms a tetrahedral coordination geometry with Asp144 Oδ2 (2.03 Å), Glu184 Oɛ2 (1.84 Å), His322 Nɛ2 (1.98 Å) and a water molecule (1.90 Å) (Fig. 4 ▸ b and Supplementary Fig. S2). In apo-Is-QC, the electron density for zinc disappeared and a new hydrogen bond was formed between Glu184 Oɛ2 and His322 Nɛ2 (2.83 Å; Fig. 4 ▸ c). Consistently, when Zn-Is-QC was co-crystallized with 1 mM EDTA or when the crystals of Zn-Is-QC were soaked in buffer with 1 mM EDTA, the electron density for zinc clearly disappeared (Supplementary Fig. S3), indicating that the bound zinc in Is-QC was removed by EDTA. On superimposing the structures of Zn-Is-QC and apo-Is-QC, no significant conformational change was observed (r.m.s. deviation of 0.184 Å for all Cα atoms), apart from a slight movement of the side-chain carboxylic group of Glu184 (Fig. 4 ▸ d). Furthermore, we produced the D144A mutant of Is-QC and found that ∼11% of the enzymatic activity was retained, although a significant conformational change was detected by circular-dichroism spectroscopy (Supplementary Fig. S4), strengthening our finding that the reaction catalyzed by Is-QC is metal-independent.
3.5. Comparison with the reported structures of mammalian and Drosophila sQCs
In spite of limited sequence identity (40–43%), the Is-QC structure shares a highly conserved scaffold with the reported structures of human, mouse and Drosophila sQCs (Huang, Liu, Cheng et al., 2005 ▸; Koch et al., 2012 ▸; Ruiz-Carrillo et al., 2011 ▸), as shown by the small r.m.s. deviations of 1.118, 1.269 and 1.025 Å for all Cα atoms, respectively. Notable conformational changes were observed in four segments: Leu132–Thr138 (referring to the sequence of Is-QC), Asp208–Gln227, Leu276–Asn284 and Ser290–Ile295 (Figs. 5 ▸ and 6 ▸ a). Of these segments, the largest loop movement was found at Asp208–Gln227 owing to an eight-residue insertion in Is-QC (Fig. 5 ▸, red box). The insert forms a 310-helix at the vertex of the structure and introduces an extra disulfide bridge, Cys120–Cys218, in the molecule to stabilize the central β-sheet (Figs. 4 ▸ a and 6 ▸ b). As a consequence, several loops adjacent to the disulfide bridge, such as Thr168–Gly172 and Ser211–Ile224, became ordered and visible, whereas the corresponding loops in human (Thr183–Lys188), mouse (Ser186–Lys189) and Drosophila (Gly198–Ala204) sQCs were flexible and were not observed (Huang, Liu, Cheng et al., 2005 ▸; Koch et al., 2012 ▸; Ruiz-Carrillo et al., 2011 ▸). Moreover, the structures of human and mouse sQCs possess an additional N-terminal α-helix near the vertex, but this α-helix is not present in Is-QC; instead, the N-terminus of Is-QC is directed outwards, likely because of the narrower space caused by the eight-residue insertion (Supplementary Fig. S5a). In contrast, the first α-helix of Is-QC is not found in Drosophila sQC because of the shorter N-terminal sequence (Fig. 5 ▸ and Supplementary Fig. S5b).
Figure 6.
Structural comparison. (a) Superimposition (in stereoview) of the structures of Is-QC (magenta), human sQC (cyan), mouse sQC (yellow) and Drosophila sQC (green). The segments with notable conformational change between these QC structures are marked with arrowheads. (b) A close-up view of the structures around the disulfide bridge Cys120–Cys218 of Is-QC. (c) Comparison of the surface-charge distribution on these QC structures. The charge potentials were calculated with PyMOL (Schrödinger) and are coloured from −68.4 K B T (red) to 68.4 (blue). A notable residue substitution between these QCs is labelled. (d) A close-up view of the active-site structures of Is-QC (magenta), human sQC (cyan), mouse sQC (yellow) and Drosophila sQC (green). The catalytically important residues are shown as stick models with numbering referring to the sequence of Is-QC. Note that the catalytic glutamate residue (Glu183 in Is-QC) and the critical hydrogen-bond network (Glu183⋯Asp297⋯Asp238 in Is-QC) are superimposed well in these QC structures.
Similar to human, mouse and Drosophila sQCs, the active-site pocket of Is-QC is narrow but is accessible to solvent (Fig. 6 ▸ c). A notable difference between these QCs was observed in the surface charges around the active-site pocket owing to residue substitutions; for example, Arg188 in Is-QC corresponds to His206, His207 and Glu175 in human, mouse and Drosophila sQCs, respectively. In addition to the zinc-binding residues, the catalytic Glu residue (Glu183 in Is-QC) and the essential hydrogen-bond network (Glu183⋯Asp297⋯Asp238) are completely conserved in Is-QC (Fig. 6 ▸ d), suggesting a conserved catalytic mechanism between these QCs. We further produced the D238A mutant of Is-QC, which might lead to disruption of the hydrogen-bond network, resulting in a complete loss of enzymatic activity (Supplementary Fig. S4). This confirms our previous finding regarding the catalytic role of the hydrogen-bond network (Huang et al., 2008 ▸). The putative hydrophobic substrate-binding pocket is primarily defined by the residues Trp189, Phe317 and Trp321 (Fig. 6 ▸ d), reflecting the preference of Is-QC for hydrophobic and uncharged residues in the second and the third positions of the substrates, as mentioned above. Another conserved feature in the active site is a nonproline cis-peptide bond between Asp144 and Ser145 stabilized by several hydrogen bonds.
3.6. Thermal stability
Since Is-QC contains two extra disulfide bridges compared with the known structures of type II QCs, we analyzed the thermal stability of Is-QC using SYPRO Orange dye and compared it with the stability of human sQC. As shown in Fig. 7( ▸ a), the thermal unfolding transition displayed a sigmoidal curve. The T m value of Zn-Is-QC was measured to be 53°C, slightly higher than that of human sQC (51°C), implying structural stabilization by the extra disulfide bridges. Furthermore, apo-Is-QC has a T m value of ∼51.5°C, indicating a little reduction in structural stability upon zinc depletion. Moreover, the enzymatic activities of these QC samples were reduced in a temperature-dependent manner and the curves displayed a sigmoidal shape (Fig. 6 ▸ b). About 50% of the activity was retained at 42.6, 41.4 and 40.0°C for Zn-Is-QC, apo-Is-QC and human sQC, respectively, consistent with the results obtained from the SYPRO Orange fluorescence assay.
Figure 7.
Thermal stability. (a) Thermal stabilities of Zn-Is-QC, apo-Is-QC and human sQC analyzed using SYPRO Orange dye (Invitrogen). (b) Enzyme activities of Is-QCs and human sQC at various temperatures. The percentage residual activity at each temperature was calculated on the basis of the activity at 25°C as 100%. In both (a) and (b), a typical result from three experiments is presented.
3.7. Metal-independent inhibition by imidazole derivatives
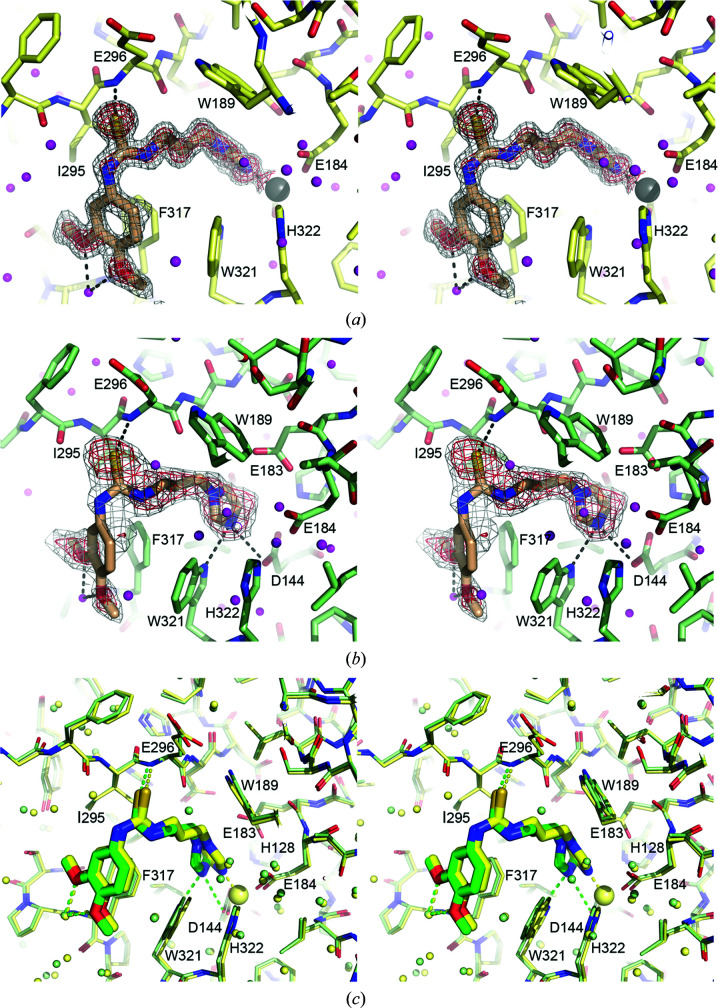
The three imidazole derivatives PBD150, 1-benzylimidazole and N-ω-acetylhistamine were shown to possess remarkable inhibitory activities towards Zn-Is-QC, with K i values in the sub- or mid-micromolar ranges (Table 3 ▸), although the effects are significantly weaker than those towards human sQC (Huang et al., 2011 ▸). Surprisingly, these inhibitors also showed comparable inhibitory activities towards apo-Is-QC (Table 3 ▸), implying metal-independent binding of the inhibitors to Is-QC. To elucidate the inhibition mechanism, we determined the structures of both Zn-Is-QC and apo-Is-QC bound to PBD150, which were refined to 1.38 and 1.95 Å resolution, respectively (Table 1 ▸). The bound PBD150 in Zn-Is-QC adopts a similar binding mode as observed in human sQC and the Drosophila mitochondria-resident QC (Fig. 8 ▸ a and Supplementary Fig. S6) (Huang et al., 2011 ▸; Koch et al., 2012 ▸). No significant conformational change (r.m.s. deviation of 0.129 Å for all Cα atoms) was found upon inhibitor binding. The imidazole ring of PBD150 ligates to the zinc ion and makes van der Waals contacts with Leu239. The thiourea S atom forms a hydrogen bond (3.20 Å) to the backbone amide of Glu296, while the propyl moiety is held in position by the hydrophobic Trp189, Ile295, Phe317 and Trp321. Moreover, the dimethoxyphenyl ring is partially stacked with Phe317 and makes van der Waals contacts with Ile295 and Trp321. The two methoxy groups are in contact with the backbone amide of Phe317 via water-mediated hydrogen bonds.
Figure 8.
Structures of Is-QC bound to PBD150. (a) A stereoview of the active-site structure of PBD150-bound Zn-Is-QC. The bound PBD150 is shown as a stick model overlaid with a simulated-annealing F o − F c OMIT map contoured at 2.0σ (grey) and 3.0σ (red) levels. The hydrogen bonds and the zinc coordination bonds are shown as dashed lines. (b) A stereoview of the active-site structure of PBD150-bound apo-Is-QC. The model of PBD150 is overlaid with a simulated-annealing F o − F c OMIT map contoured at 1.0σ (grey) and 1.5σ (red) levels. The hydrogen bonds and a possible electrostatic interaction are shown as dashed lines. (c) A stereoview of superimposition of the PBD150-bound Zn-Is-QC (yellow) and apo-Is-QC (green) structures.
In contrast, the electron density for the bound PBD150 in apo-Is-QC was broken in some places but was interpretable (Fig. 8 ▸ b), probably owing to the lower quality of the data (Table 1 ▸). Similar to that in Zn-Is-QC, the density for the inhibitor phenyl ring is relatively weak (compare the densities coloured in red in Figs. 8 ▸ a and 8 ▸ b), suggesting that the interaction between the inhibitor phenyl ring and Is-QC is relatively weak. Based on the density that is present, the PBD150 imidazole ring displays a different orientation compared with that in Zn-Is-QC (Fig. 8 ▸ c), resulting in the formation of a new hydrogen bond (2.98 Å) to the indole ring of Trp321 and a possible electrostatic interaction (3.36 Å) with the carboxylic group of Asp144 (Fig. 8 ▸ b). The positioning of PBD150 is slightly rotated in an anticlockwise direction (Fig. 8 ▸ c); as a consequence, the thiourea S atom is closer (2.97 Å) to the backbone amide of Glu296. Therefore, in spite of the higher flexibility in the phenyl ring, the electron densities for the inhibitor thiourea, propyl and imidazole moieties are visible (Fig. 8 ▸ b). In addition, the two methoxy groups appeared to also be involved in water-mediated hydrogen bonds to the backbone amide of Phe317, as shown by clear electron density.
4. Discussion
Recent studies, particularly those on transgenic mouse models (Schilling et al., 2008 ▸; Nussbaum et al., 2012 ▸; Hellvard et al., 2013 ▸; Cynis et al., 2011 ▸), have indicated that QCs are potential drug targets for the treatment of Alzheimer’s disease and some inflammatory disorders; thus, compounds with the ability to ligate the active-site zinc of QCs, such as imidazole derivatives, are currently under intense development as QC inhibitors. However, the exact metal content of QCs under physiological conditions is still unclear, although it has been more than twenty-five years since the first mammalian QCs were partially purified from bovine and porcine pituitary extracts (Fischer & Spiess, 1987 ▸; Busby et al., 1987 ▸). Previously, we proposed a ‘zinc-dependent’ catalytic mechanism for mammalian QCs based on the following findings (Huang, Liu, Cheng et al., 2005 ▸; Huang et al., 2008 ▸). (i) Mammalian QCs share a highly conserved scaffold and active-site structure with some binuclear aminopeptidases, such as Aeromonas proteolytica aminopeptidase (ApAP) and Streptomyces griseus aminopeptidase (SgAP) (Bzymek & Holz, 2004 ▸; Fundoiano-Hershcovitz et al., 2004 ▸). These proteases utilize bound zinc ions, usually the tightly bound zinc ion, to polarize the carbonyl O atom of the scissile peptide bond of substrates and to stabilize the tetrahedral intermediate formed during the catalytic process (Lowther & Matthews, 2002 ▸). Therefore, it is reasonable to assume that the zinc in QCs acts by polarizing the γ-amide group of the first Gln residue of the substrate and simultaneously stabilizing the oxyanion formed by nucleophilic attack of the α-nitrogen on the γ-carbonyl carbon. This mechanism is further supported by the fact that the catalytic Glu residue and the zinc-coordinated water molecule are completely conserved in the structures of QCs and aminopeptidases (Huang, Liu, Cheng et al., 2005 ▸) and cooperatively trigger the formation of the tetrahedral intermediate during catalysis. (ii) Human and mouse QCs were shown to be susceptible to inhibition by several heterocyclic metal-chelating agents, imidazole derivatives and some cysteamine derivatives (Schilling, Niestroj et al., 2003 ▸; Schilling et al., 2005 ▸). The enzymatic activity of the ‘inactivated’ mammalian QCs, as established by dialysis against dipicolinic acid or 8-hydroxyquinoline, could be restored by adding ZnSO4 (Schilling, Niestroj et al., 2003 ▸; Schilling et al., 2005 ▸). (iii) A mutation (H330Q) at one of the zinc-binding residues of human sQC resulted in a nearly complete loss of QC activity (Bateman et al., 2001 ▸).
However, there is no structural evidence to support loss of zinc in the ‘inactivated’ mammalian QCs, and no reports have described the exact quantities of zinc in QCs treated with EDTA or other metal-chelating agents. Notably, ApAP and SgAP were highly susceptible to EDTA inhibition; for example, only 80 µM EDTA could nearly abolish the enzymatic activity of SgAP (Spungin & Blumberg, 1989 ▸), which is in sharp contrast to the refractory properties of QCs towards EDTA inhibition. In the present study, we report the first case of an active QC ‘apoenzyme’, as confirmed by ICP-MS, high-resolution crystal structure and enzyme-kinetic analyses. This finding strongly contradicts the previous conclusions about the role of zinc in QCs. Furthermore, unlike the previous observation for human sQC (Bateman et al., 2001 ▸), a mutation (D144A) at one of the zinc-binding residues of Is-QC did not abolish the enzyme activity; instead, partial activity was retained, although the mutant showed a significant structural change compared with the wild-type enzyme. These results indicate that the zinc does not seem to play a catalytic role in Is-QC. Along these lines, the issue of whether the zinc has a structural role in Is-QC is raised. Previous results regarding the structural role of zinc in QCs are diverse; Ruiz-Carrillo et al. (2011 ▸) reported that the ‘apo-form’ of human sQC displayed a remarkably decreased thermal stability, whereas no significant change in stability was observed for the mitochondria-resident form of Drosophila QC after the zinc was removed (Koch et al., 2012 ▸). In the present study, the zinc in Is-QC apparently has no structural role, suggesting that the presence of zinc in Is-QC was owing to nonspecific binding.
It has been well established that the tightly bound zinc in the active site of ApAP, SgAP and other di-zinc aminopeptidases is essential for catalysis of these enzymes, while the weakly bound zinc can modulate the enzymatic activity and substrate specificity (Lowther & Matthews, 2002 ▸). To date, however, no reports have described a structural role for the zinc ions in aminopeptidases. We noticed that the zinc ions in aminopeptidases can easily be removed and substituted with other metals by dialysis methods and that high levels of enzyme activity can be restored (Lowther & Matthews, 2002 ▸), indicating that there is no significant structural change upon zinc depletion in aminopeptidases. Since QCs and the di-zinc aminopeptidases share a common scaffold and active-site structure, it is reasonable to assume that the zinc in most QCs has no or little structural role. Moreover, no bacterial type II QCs have been found thus far in searches of the deposited genomes of bacteria, and the closest bacterial homologues are the di-zinc aminopeptidases. This implies that mammalian QCs and other animal QCs have evolved from the bacterial zinc aminopeptidases, with the two metal-binding sites being conserved during the course of evolution. Nevertheless, some unique structural elements in the active site of QCs have been created, particularly the catalytically essential hydrogen-bond network (Huang et al., 2008 ▸), which is absolutely conserved from yeast to human QCs. In this regard, to gain more insights into the catalysis mechanism of QCs, we should pay more attention to this unusual hydrogen-bond network. Undoubtedly, this hydrogen-bond network is a good target for QC inhibitors.
To date, although recombinant human QCs with varied zinc contents have been reported (Booth et al., 2004 ▸; Huang, Liu & Wang, 2005 ▸), the QC activities of these enzymes were comparable. In the present study, the zinc-bound Is-QC still showed ∼65% QC activity when compared with the activity of apo-Is-QC. In this regard, we cannot rule out the possibility that both zinc-dependent and zinc-independent catalytic mechanisms co-exist in the QC family. This is indeed not surprising since it has been reported that a lysine residue in plant and bacterial QCs or the zinc ions in some binuclear aminopeptidases can function as a polarizing group to activate the susceptible carbonyl group of the substrate during the catalysis process (Lowther & Matthews, 2002 ▸; Wintjens et al., 2006 ▸; Huang et al., 2010 ▸). In mammalian QCs, a lysine residue adjacent to the zinc-binding site, such as Lys144 in human sQC (Huang, Liu, Cheng et al., 2005 ▸), could be a good candidate for the polarizing group in the absence of bound zinc. However, this lysine residue is not found in the active site of tick QC, and mutation of human sQC at this lysine residue did not show a significant reduction in enzymatic activity (Huang, Liu, Cheng et al., 2005 ▸). Alternatively, two highly conserved histidine residues, such as His140 and His330 in human sQC and His128 and His322 in Is-QC, could also be candidates, since mutations of human sQC at each of these two histidine residues inactivated the enzyme (Bateman et al., 2001 ▸). The microenvironment of the QC active site is highly acidic, as supported by the finding that the side-chain carboxylic groups of several aspartic acid residues can form hydrogen bonds (Huang et al., 2008 ▸). This suggests that the side-chain imidazole ring of the histidine residues should be protonated, which eventually gives them sufficient electrophilicity to activate the susceptible carbonyl group of the substrate during catalysis.
With regard to the inhibition of QCs by some heterocyclic metal-chelating agents, Schilling and coworkers reported that human sQC could be inactivated by dialysis against 5 mM 1,10-phenanthroline or dipicolinic acid, but that 50–60% QC activity could be restored after repeated dialysis against chelator-free buffers (Schilling, Niestroj et al., 2003 ▸). However, when the QC sample was dialyzed against buffers containing 1 mM EDTA, no reactivation was observed. In the present study, a similar reactivation of the QC activity of Is-QC was observed after the metal-chelating agents were removed using a size-exclusion column, but the activity could be further enhanced by adding 1 mM EDTA. These differing observations between human and tick QCs give the zinc an enigmatic role in this enzyme family. Nevertheless, the strong inhibitory activities of 1,10-phenanthroline and dipicolinic acid towards apo-Is-QC may preclude the possibility that the inhibition of Is-QC is owing to zinc depletion. In fact, the efficacies of 1,10-phenanthroline and dipicolinic acid towards QCs are significantly weaker than those towards the zinc-dependent ApAP and SgAP. For instance, 0.1 mM 1,10-phenanthroline caused a nearly complete loss of aminopeptidase activity in ApAP and SgAP (Spungin & Blumberg, 1989 ▸; Prescott et al., 1983 ▸), whereas greater than 50% activity was retained by Is-QC and human sQC (Schilling, Niestroj et al., 2003 ▸). Based on these findings, 1,10-phenanthroline and dipicolinic acid might be able to bind QCs in a reversible manner, probably binding at the hydrophobic substrate-binding pocket, since the pocket can accommodate the hydrophobic methoxyphenyl ring of PBD150 well. This may be supported by the fact that the tricyclic 1,10-phenanthroline has an effect stronger by one order of magnitude than the monocyclic dipicolinic acid towards QCs.
Finally, we report the zinc-independent inhibition of QCs by imidazole-derived inhibitors for the first time, although these inhibitors were shown to be able to bind the active-site zinc of QCs. The inhibitor PBD150 bound to Zn-Is-QC exhibited a binding mode similar to that in human sQC (Huang et al., 2011 ▸), in spite of a weaker inhibitory activity towards Is-QC. Surprisingly, in the absence of zinc the inhibitor showed a similar orientation except for its imidazole ring, which forms a new hydrogen bond and a possible electrostatic interaction with Is-QC. This finding indicates that the zinc coordination of the inhibitor imidazole ring can be replaced by other interactions. In addition, we noticed that the electron densities at the inhibitor thiourea, propyl and imidazole moieties are stronger in both the Zn-bound and apo Is-QC structures, implying that these groups contribute tighter interactions with Is-QC which are critical for the inhibitory potency of the inhibitor.
In conclusion, we present the first case of a metal-independent QC and describe the metal-independent inhibition of QCs by imidazole-derived inhibitors for the first time. Because the black-legged tick is an important vector of human pathogens and the more active ‘apo-form’ of Is-QC is a good target of QC inhibitors, our results not only provide a new way of thinking about QC catalysis but also suggest new designs of QC inhibitors for tick control.
Supplementary Material
PDB reference: Zn-Is-QC, 4mhn
PDB reference: apo-Is-QC, 4mhp
PDB reference: Zn-Is-QC–PDB150, 4mhy
PDB reference: Zn-Is-QC–PDB150, 4mhz
Supporting Information. DOI: 10.1107/S1399004713033488/dw5081sup1.pdf
Acknowledgments
We thank Miss Hui-Ling Shr and Dr Ch-Chi Chou of the Core Facilities for Protein Structural Analysis, Academia Sinica, Taipei, Taiwan for assistance in crystallization screening and ESI-LC-MS/MS analysis. We are grateful to Dr Jay Nix at beamline 4.2.2 of the Advanced Light Source, Berkeley, California, USA and the staff of beamline 13C1 of the National Synchrotron Radiation Research Center (Hsinchu, Taiwan) for assistance in X-ray data collection and access to the synchrotron-radiation centres. We are also grateful to Chun-Chi Tsai of the Incubation Center of Genomic Research Center in Academia Sinica for assistance in ICP-MS analysis. This work was supported by grants from Academia Sinica and the National Science Council, Taiwan (NSC-101-2319-B-001-003 to AH-JW for the National Core Facilities for Protein Structural Analysis).
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: DW5081).
References
- Adamson, S. W., Browning, R. E., Chao, C.-C., Bateman, R. C. Jr, Ching, W. -M. & Karim, S. (2013). Insect Biochem. Mol. Biol. 43, 781–793. [DOI] [PMC free article] [PubMed]
- Auld, D. S. (1995). Methods Enzymol. 248, 228–242. [DOI] [PubMed]
- Bateman, R. C. Jr, Temple, J. S., Misquitta, S. A. & Booth, R. E. (2001). Biochemistry, 40, 11246–11250. [DOI] [PubMed]
- Booth, R. E., Lovell, S. C., Misquitta, S. A. & Bateman, R. C. Jr (2004). BMC Biol. 2, 2. [DOI] [PMC free article] [PubMed]
- Buchholz, M., Hamann, A., Aust, S., Brandt, W., Böhme, L., Hoffmann, T., Schilling, S., Demuth, H.-U. & Heiser, U. (2009). J. Med. Chem. 52, 7069–7080. [DOI] [PubMed]
- Buchholz, M., Heiser, U., Schilling, S., Niestroj, A. J., Zunkel, K. & Demuth, H.-U. (2006). J. Med. Chem. 49, 664–677. [DOI] [PubMed]
- Burgdorfer, W., Barbour, A. G., Hayes, S. F., Benach, J. L., Grunwaldt, E. & Davis, J. P. (1982). Science, 216, 1317–1319. [DOI] [PubMed]
- Busby, W. H. Jr, Quackenbush, G. E., Humm, J., Youngblood, W. W. & Kizer, J. S. (1987). J. Biol. Chem. 262, 8532–8536. [PubMed]
- Bzymek, K. P. & Holz, R. C. (2004). J. Biol. Chem. 279, 31018–31025. [DOI] [PubMed]
- Chen, Y.-L., Huang, K.-F., Kuo, W.-C., Lo, Y.-C., Lee, Y.-M. & Wang, A. H.-J. (2012). Biochem. J. 442, 403–412. [DOI] [PubMed]
- Cynis, H. et al. (2011). EMBO Mol. Med. 3, 545–558. [DOI] [PMC free article] [PubMed]
- Cynis, H., Rahfeld, J.-U., Stephan, A., Kehlen, A., Koch, B., Wermann, M., Demuth, H.-U. & Schilling, S. (2008). J. Mol. Biol. 379, 966–980. [DOI] [PubMed]
- Cynis, H., Scheel, E., Saido, T. C., Schilling, S. & Demuth, H.-U. (2008). Biochemistry, 47, 7405–7413. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Fischer, W. H. & Spiess, J. (1987). Proc. Natl Acad. Sci. USA, 84, 3628–3632. [DOI] [PMC free article] [PubMed]
- Fundoiano-Hershcovitz, Y., Rabinovitch, L., Langut, Y., Reiland, V., Shoham, G. & Shoham, Y. (2004). FEBS Lett. 571, 192–196. [DOI] [PubMed]
- Goren, H. J., Bauce, L. G. & Vale, W. (1977). Mol. Pharmacol. 13, 606–614. [PubMed]
- Hellvard, A., Maresz, K., Schilling, S., Graubner, S., Heiser, U., Jonsson, R., Cynis, H., Demuth, H.-U., Potempa, J. & Mydel, P. (2013). J. Infect. Dis. 207, 768–777. [DOI] [PMC free article] [PubMed]
- Huang, K.-F., Liaw, S.-S., Huang, W.-L., Chia, C.-Y., Lo, Y.-C., Chen, Y.-L. & Wang, A. H.-J. (2011). J. Biol. Chem. 286, 12439–12449. [DOI] [PMC free article] [PubMed]
- Huang, K.-F., Liu, Y.-L., Cheng, W.-J., Ko, T.-P. & Wang, A. H.-J. (2005). Proc. Natl Acad. Sci. USA, 102, 13117–13122. [DOI] [PMC free article] [PubMed]
- Huang, K.-F., Liu, Y.-L. & Wang, A. H.-J. (2005). Protein Expr. Purif. 43, 65–72. [DOI] [PubMed]
- Huang, K.-F., Wang, Y.-R., Chang, E.-C., Chou, T.-L. & Wang, A. H.-J. (2008). Biochem. J. 411, 181–190. [DOI] [PubMed]
- Huang, W.-L., Wang, Y.-R., Ko, T.-P., Chia, C.-Y., Huang, K.-F. & Wang, A. H.-J. (2010). J. Mol. Biol. 401, 374–388. [DOI] [PubMed]
- Koch, B., Kolenko, P., Buchholz, M., Carrillo, D. R., Parthier, C., Wermann, M., Rahfeld, J.-U., Reuter, G., Schilling, S., Stubbs, M. T. & Demuth, H.-U. (2012). Biochemistry, 51, 7383–7392. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Lou, Y.-C., Huang, Y.-C., Pan, Y.-R., Chen, C. & Liao, Y.-D. (2006). J. Mol. Biol. 355, 409–421. [DOI] [PubMed]
- Lowther, W. T. & Matthews, B. W. (2002). Chem. Rev. 102, 4581–4608. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Neupert, S., Russell, W. K., Predel, R., Russell, D. H., Strey, O. F., Teel, P. D. & Nachman, R. J. (2009). J. Proteomics, 72, 1040–1045. [DOI] [PubMed]
- Nussbaum, J. M. et al. (2012). Nature (London), 485, 651–655.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol. 6, 458–463. [DOI] [PubMed]
- Prescott, J. M., Wagner, F. W., Holmquist, B. & Vallee, B. L. (1983). Biochem. Biophys. Res. Commun. 114, 646–652. [DOI] [PubMed]
- Rodgers, S. E. & Mather, T. N. (2007). Emerg. Infect. Dis. 13, 633–635. [DOI] [PMC free article] [PubMed]
- Ruiz-Carrillo, D., Koch, B., Parthier, C., Wermann, M., Dambe, T., Buchholz, M., Ludwig, H.-H., Heiser, U., Rahfeld, J.-U., Stubbs, M. T., Schilling, S. & Demuth, H.-U. (2011). Biochemistry, 50, 6280–6288. [DOI] [PubMed]
- Schilling, S., Cynis, H., von Bohlen, A., Hoffmann, T., Wermann, M., Heiser, U., Buchholz, M., Zunkel, K. & Demuth, H.-U. (2005). Biochemistry, 44, 13415–13424. [DOI] [PubMed]
- Schilling, S., Hoffmann, T., Manhart, S., Hoffmann, M. & Demuth, H.-U. (2004). FEBS Lett. 563, 191–196. [DOI] [PubMed]
- Schilling, S., Hoffmann, T., Rosche, F., Manhart, S., Wasternack, C. & Demuth, H.-U. (2002). Biochemistry, 41, 10849–10857. [DOI] [PubMed]
- Schilling, S., Lindner, C., Koch, B., Wermann, M., Rahfeld, J.-U., von Bohlen, A., Rudolph, T., Reuter, G. & Demuth, H.-U. (2007). Biochemistry, 46, 10921–10930. [DOI] [PubMed]
- Schilling, S., Manhart, S., Hoffmann, T., Ludwig, H.-H., Wasternack, C. & Demuth, H.-U. (2003). Biol. Chem. 384, 1583–1592. [DOI] [PubMed]
- Schilling, S., Niestroj, A. J., Rahfeld, J.-U., Hoffmann, T., Wermann, M., Zunkel, K., Wasternack, C. & Demuth, H.-U. (2003). J. Biol. Chem. 278, 49773–49779. [DOI] [PubMed]
- Schilling, S. et al. (2008). Nature Med. 14, 1106–1111. [DOI] [PubMed]
- Schlenzig, D., Manhart, S., Cinar, Y., Kleinschmidt, M., Hause, G., Willbold, D., Funke, S. A., Schilling, S. & Demuth, H.-U. (2009). Biochemistry, 48, 7072–7078. [DOI] [PubMed]
- Segel, I. H. (1993). Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems, pp. 100–118. New York: John Wiley & Sons.
- Spungin, A. & Blumberg, S. (1989). Eur. J. Biochem. 183, 471–477. [DOI] [PubMed]
- Telford, S. R. III, Dawson, J. E., Katavolos, P., Warner, C. K., Kolbert, C. P. & Persing, D. H. (1996). Proc. Natl Acad. Sci. USA, 93, 6209–6214. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wintjens, R., Belrhali, H., Clantin, B., Azarkan, M., Bompard, C., Baeyens-Volant, D., Looze, Y. & Villeret, V. (2006). J. Mol. Biol. 357, 457–470. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Zn-Is-QC, 4mhn
PDB reference: apo-Is-QC, 4mhp
PDB reference: Zn-Is-QC–PDB150, 4mhy
PDB reference: Zn-Is-QC–PDB150, 4mhz
Supporting Information. DOI: 10.1107/S1399004713033488/dw5081sup1.pdf