Abstract
After five patients were diagnosed with nosocomial invasive aspergillosis caused by Aspergillus fumigatus and A. flavus, a 14-month surveillance program for pathogenic and nonpathogenic fungal conidia in the air within and outside the University Hospital in Rotterdam (The Netherlands) was begun. A. fumigatus isolates obtained from the Department of Hematology were studied for genetic relatedness by randomly amplified polymorphic DNA (RAPD) analysis. This was repeated with A. fumigatus isolates contaminating culture media in the microbiology laboratory. The density of the conidia of nonpathogenic fungi in the outside air showed a seasonal variation: higher densities were measured during the summer, while lower densities were determined during the fall and winter. Hardly any variation was found in the numbers of Aspergillus conidia. We found decreasing numbers of conidia when comparing air from outside the hospital to that inside the hospital and when comparing open areas within the hospital to the closed department of hematology. The increase in the number of patients with invasive aspergillosis could not be explained by an increase in the number of Aspergillus conidia in the outside air. The short-term presence of A. flavus can only be explained by the presence of a point source, which was probably patient related. Genotyping A. fumigatus isolates from the department of hematology showed that clonally related isolates were persistently present for more than 1 year. Clinical isolates of A. fumigatus obtained during the outbreak period were different from these persistent clones. A. fumigatus isolates contaminating culture media were all genotypically identical, indicating a causative point source. Knowledge of the epidemiology of Aspergillus species is necessary for the development of strategies to prevent invasive aspergillosis. RAPD fingerprinting of Aspergillus isolates can help to determine the cause of an outbreak of invasive aspergillosis.
Aspergillus species are widely distributed fungi whose conidia are present in the outside air in a year-round fashion; relatively little seasonal variation has been documented (11). After inhalation of airborne conidia, Aspergillus species can cause various forms of disease, invasive infections in immunocompromised patients being the most serious (17). Despite early treatment with high dosages of amphotericin B, these infections remain associated with high morbidity and mortality (2). Aspergillus fumigatus is by far the most prevalent species in cases of invasive disease. Small outbreaks of aspergillosis have been reported, as have been pseudo-outbreaks due to contamination of culture media (1, 7, 8, 10, 12). Elucidation of the complex epidemiology in such cases requires detailed molecular typing studies. Randomly amplified polymorphic DNA (RAPD) analysis is a PCR fingerprinting procedure that can be applied to the typing of fungal isolates (13, 15, 19, 20).
After an outbreak of nosocomial invasive aspergillosis (caused by A. fumigatus and A. flavus) (10), a surveillance program was started for the detection of fungal conidia in the air within and in the area immediately surrounding the University Hospital in Rotterdam (The Netherlands). Isolates of A. fumigatus obtained from the department of hematology were studied for genetic relatedness, as were A. fumigatus isolates contaminating culture media in the microbiology laboratory. We started this study to address three different questions. First, we wished to improve our knowledge on the aerobiology of fungi in the air inside and outside our hospital. Second, we wanted to investigate whether the density of the conidia of Aspergillus species as higher than usual at the time of the outbreak of invasive aspergillosis. Finally, we wanted to investigate the role of genotyping fungal isolates in elucidating the epidemiology of Aspergillus species.
MATERIALS AND METHODS
Air sampling and fungal isolation.
Over a 62-week period (July 1994 to September 1995) serial air samples of 1 cubic meter each were taken with a Surface Air System (SAS) sampler (PBI International, Milan, Italy) containing Sabouraud agar plates. On each occasion, four air samples were taken from the high-efficiency particulate air (HEPA)-filtered rooms. Eight samples were taken from other sites within the hematology ward, which is separated from the rest of the hospital by closed doors. In addition, four samples were taken from generally accessible sites within the hospital, and four samples came from locations outside the hospital. Separate air samples were taken to detect Aspergillus species and nonpathogenic fungi. The Sabouraud agar plates recovered from the SAS samplers were incubated for 120 h at 22°C to examine the growth of nonpathogenic fungi and for 48 h at 37°C to examine the growth pathogenic fungi. Initially, air samples were taken twice a week. After 2 months, further samples were taken once a week. Air sampling of the microbiology laboratory was done in several places during and at 1 month after an episode of increased fungal contamination of culture media.
Identification of fungal isolates.
Species growing at 22°C (nonpathogenic fungi) were counted and not identified further. Species growing at 37°C (pathogenic fungi) were identified by using culture characteristics and morphology of conidiophores and conidia. Air samples from the microbiology laboratory were only examined for the growth of Aspergillus species.
Collection of fungal isolates.
Aspergillus isolates obtained from the department of hematology and the HEPA-filtered rooms were propagated on fresh Sabouraud agar plates and subsequently stored at −70°C in brain heart infusion broth containing 10% glycerol. Aspergillus isolates recovered from other places were counted and identified but not stored.
Fungi contaminating culture media in the microbiology laboratory that were macroscopically suspected to be A. fumigatus were collected, as were isolates from air samples taken in the laboratory. Species were identified as described above. Isolates of A. fumigatus of patients with invasive aspergillosis in the hematology and surgical wards of the Erasmus University Medical Center Rotterdam were used to monitor the discriminative power of the RAPD assay, as published previously, and included as RAPD quality and reproducibility controls (10).
Fungal DNA isolation.
Strains were inoculated in 25 ml of Sabouraud maltose medium containing 4 mg of gentamicin per kg and incubated at 37°C for 72 h until abundant mycelial growth was observed. The entire thallus was collected in a porcelain bowl, frozen under liquid nitrogen, and grounded with a pestle; this work was done in a safety cabinet. Between 10 and 25 ml of lysis buffer (0.1 M Tris-HCl, pH 6.4; 40 mM EDTA, pH 8.0; 1% Triton X-100, 4 M guanidium isothiocyanate) was added, and the suspension was put on ice. Then 1 ml of the suspension was centrifuged for 5 min (15,000 rpm). Next, 100 μl of a Celite suspension (200 mg/ml) (Aoroa Organics, Grel, Belgium) was added to the supernatant, and this suspension was shaken vigorously by hand. The sediment was washed in a second lysis buffer (0.1 M Tris-HCl, pH 6.4; 4 M guanidinium isothiocyanate), 70% ethanol, and acetone in succession (3). After being dried, the pellet was resuspended in 150 μl of 10 mM Tris-HCl (pH 8.0) and incubated at 56°C for 10 min. Approximately 125 μl of the supernatant was collected. The DNA concentration was estimated by electrophoresis of DNA-containing aliquots through 1% agarose gels, run in 0.5× Tris-borate-EDTA buffer in the presence of ethidium bromide, and compared with the staining intensities of known amounts of bacteriophage lambda DNA.
PCR-mediated DNA fingerprinting.
DNA typing by RAPD assay was performed exactly as described previously (21, 22). The enterobacterial repetitive intergenic consensus primers ERIC-1 and ERIC-2 were employed, as they had been shown to discriminate well between epidemiologically nonrelated isolates in earlier assays (10). The resulting banding patterns were indexed by capital lettering, and even a single band difference led to a different letter code. Differences found in ethidium bromide staining intensities were ignored. Banding patterns were interpreted visually by two persons working independently. The intralaboratory reproducibility of the RAPD tests has been demonstrated on previous occasions (10, 20–22). Moreover, the well-documented reproducibility problems with RAPD were documented to be of interlaboratory nature primarily (23); although data sets produced in different laboratories differed, the individual data corroborated the epidemiological relatedness between the various strains of microorganisms. To prevent problems with day-to-day differences, all assays were performed batchwise by a single technician. Standardized assays were performed in order to achieve maximum reproducibility. Normalization included the use of identical brands and batches of thermostable polymerase and nucleotide triphosphatases, the use of commercially available PCR buffers, and the application of standardized electrophoresis conditions. Furthermore, exactly the same laboratory equipment was used for all experiments described here.
RESULTS
The results section is divided into two parts. The first part describes the results of the 14-month surveillance of fungi in air samples and the molecular analysis of the Aspergillus strains thus isolated. The second part describes the isolation and molecular analysis of the Aspergillus strains recovered from contaminated microbiological culture media.
Surveillance of fungi in air.
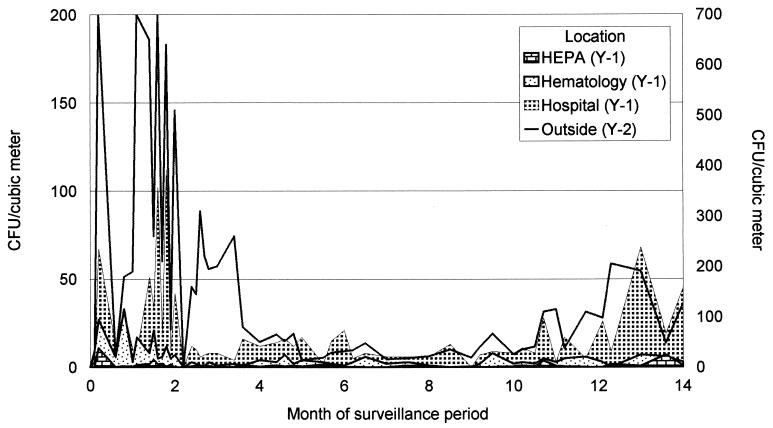
The results of the surveillance for nonpathogenic fungi in the air outside and at several locations inside the hospital are shown in Fig. 1. During the first 2 months of the surveillance period, a median of more than 400 CFU per cubic meter was counted in the outside air. In the air samples collected from within the hospital, but outside the hematology ward, this number was significantly lower (32 CFU/m3), while samples from the hematology ward itself showed a median of 7 CFU/m3. Air samples from HEPA-filtered rooms showed very low fungal densities (<2 CFU/m3). During the next 2 months (fall to early winter), the density of nonpathogenic fungal CFU declined outside as well as inside the hospital, but the same decreasing gradient between the clinical locations could still be observed. The numbers of CFU during the winter and spring were low. During the following summer, the numbers of CFU increased again, but not to the levels observed during the preceding summer. Fungi obtained during the first sampling weeks were identified. This analysis showed that the majority (>80%) of the airborne species were Cladosporium species. Alternaria and Botrytis species formed the majority of the remainder of isolates.
FIG. 1.
Results of a 14-month surveillance period of the conidia of nonpathogenic fungi present in air samples from within the hospital outside the department of hematology, air samples within the hematology ward, and air samples in HEPA-filtered rooms. The numbers of conidia outside are depicted in the line (enumeration on the y2 axis). The surveillance began in July 1994 (0 on the x axis) and ended in September 1995 (14 on the x axis).
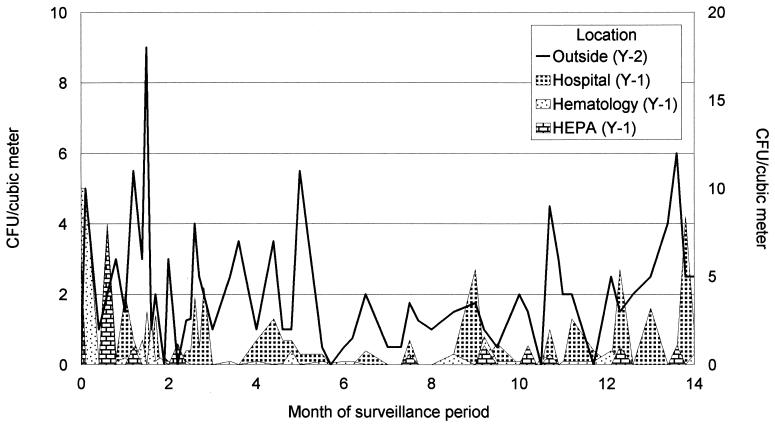
The results of the surveillance of Aspergillus species at several locations inside the hospital are presented in Fig. 2. During the surveillance period the density of aspergilli in the outside air was relatively constant; only from January to April was the number somewhat lower. The same was true for the number of CFU within the hospital outside the department of hematology. Within the confinement of the department of hematology and in the HEPA-filtered rooms, very few conidia of Aspergillus (<1 CFU/m3) were found, except at the very beginning of the surveillance period.
FIG. 2.
Results of a 14-month surveillance period of the conidia of Aspergillus spp. present in air samples within the hospital outside the department of hematology, air samples within the hematology ward, and air samples in HEPA-filtered rooms. The numbers of conidia outside are depicted in the line (enumeration on the y2 axis). The surveillance began in July 1994 (0 on the x axis) and ended in September 1995 (14 on the x axis).
Most of the isolates obtained from air samples obtained outside were identified as A. fumigatus (>90%) and A. niger (5%). The same species were also found in the hospital. However, in the first 2 months, A. flavus was isolated in high numbers, especially in samples from the department of hematology. Since other Aspergillus species were only rarely isolated in this environment, A. flavus turned out to be the most frequently clinically isolated species.
A. fumigatus isolates from the department of hematology (including the HEPA-filtered rooms) were further characterized by means of RAPD (Table 1). The number of different genotypes in air samples was limited to seven during the entire 14-month period. Only three genotypes were found during the first 4 months; a fourth genotype was introduced in the fourth month and remained present during the remainder of the surveillance period. In the last 3 months, the number of A. fumigatus clones increased, with three new genotypes being introduced. No correlation between rooms and genotypes was found. All clinical isolates of A. fumigatus were different from the environmental isolates. Also, isolates from different patients could be discriminated from each other.
TABLE 1.
A. fumigatus clones isolated from various locations and sources in the hematology department over a 14-month perioda
| Date of isolation | Location or source | Genotype with PCR primer:
|
Overall genotype | |
|---|---|---|---|---|
| ERIC-1 | ERIC-2 | |||
| 1994 | ||||
| June | ||||
| 4 | HEPA 87 | A | A | I |
| 8 | Room 92 | B | B | II |
| 19 | Room 96 | B | B | II |
| 24 | Room 75 | C | A | III |
| August | ||||
| 3 | Room 72 | A | A | I |
| 8 | Room 73 | A | A | I |
| 15 | Room 96 | B | B | II |
| 22 | Corridor | A | A | I |
| 29 | Room 96 | A | A | I |
| 31 | Room 96 | A | A | I |
| September | ||||
| 9 | Room 88 | C | A | III |
| 12 | HEPA 91 | B | B | II |
| 12 | HEPA 89 | A | A | I |
| 22 | HEPA 93 | C | A | III |
| October | ||||
| 26 | Corridor | B | A | IV |
| 31 | Room 88 | A | A | I |
| 31 | Room 74 | C | A | III |
| November | ||||
| 9 | Examination room | B | A | IV |
| 16 | Room 73 | B | A | IV |
| 23 | Room 96 | C | A | III |
| 1995 | ||||
| January | ||||
| 4 | Room 73 | A | A | I |
| 20 | Room 88 | C | A | III |
| 20 | Room 93 | B | B | II |
| March | ||||
| 10 | Room 88 | A | A | I |
| 10 | HEPA 87 | B | A | IV |
| April | ||||
| 20 | Room 96 | A | A | I |
| 25 | HEPA 89 | C | A | III |
| May | ||||
| 3 | Room 96 | B | B | II |
| 10 | Room 74 | A | A | I |
| 10 | Room 84 | B | B | II |
| 16 | Room 84 | B | A | IV |
| 16 | Room 82 | B | B | II |
| 30 | Room 88 | A | A | I |
| 30 | Room 96 | C | A | III |
| 30 | HEPA 89 | A | A | I |
| June | ||||
| 7 | Room 84 | D | C | V |
| 14 | Room 75 | A | A | I |
| 21 | Room 75 | A | D | VI |
| 27 | Room 95 | A | A | I |
| July 26 | HEPA 93 | B | A | IV |
| August | ||||
| 9 | Room 75 | A | A | I |
| 23 | Room 84 | B | B | II |
| September | ||||
| 8 | HEPA 91 | E | E | VII |
| 20 | Room 95 | B | A | IV |
| 1994 | Clinical patient 1 | F | F | VIII |
| 1994 | Clinical patient 2 | G | G | IX |
| 1994 | Clinical patient 3 | H | H | X |
| 1994 | Clinical patient 4 | I | I | XI |
| 1994 | Clinical patient 5 | I | I | XI |
| 1994 | Clinical patient 6 | J | G | XII |
The overall PCR type was based on the number of different banding patterns when both assays were combined. HEPA, HEPA-filtered room.
Laboratory contamination.
During renovation of the air-conditioning system of the corridor and the rooms directly opposite the microbiology laboratory, many culture media became contaminated with A. fumigatus, despite several precautions (closed doors, air corridors, and intensified cleaning). Fungal growth was detected on and in all kinds of media (including blood culture media). Within a period of 4 days, over 200 agar plates became contaminated. From these plates, 27 isolates of A. fumigatus were collected and further analyzed by RAPD assay.
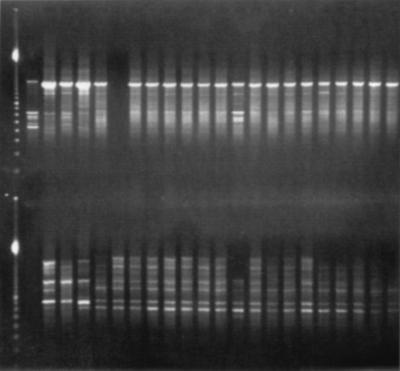
Four days after the first contamination of culture media, air samples were taken. All samples (n = 11) showed more than 10 CFU of A. fumigatus per cubic meter of air. From each of these samples one colony of A. fumigatus was genotyped by RAPD. New air samples, obtained 1 month later, still showed A. fumigatus, although in lower quantities (0 to 5 CFU/m3). From these samples, five isolates were further analyzed. All isolates obtained from contaminated culture media displayed the same genotype. Six of these isolates were randomly chosen to be compared with isolates obtained by air sampling. The results of this analysis are shown in Fig. 3 and in Table 2. The genotypes of all isolates obtained from the culture media and all except one isolate from air samples taken during the contamination period were identical. Three of the five isolates obtained 1 month later still had the same genotype.
FIG. 3.
PCR typing of A. fumigatus isolates. The first lane contains the molecular mass marker. The following banding patterns from left to right correspond to the strains given in Table 2. The top panel shows results of arbitrarily primed PCR performed with ERIC-2 primer; the bottom panel shows the ERIC-1 data. The interpretation of the picture is given in Table 2.
TABLE 2.
Clones of A. fumigatus clinical isolates and isolates contaminating culture media and air in the medical microbiology laboratorya
| Date of isolation | Source | Genotype with PCR primer:
|
Overall genotype | |
|---|---|---|---|---|
| ERIC-1 | ERIC-2 | |||
| May 1994 | Clinical patient | F | VIII | |
| June 1994 | Clinical patient | G | G | IX |
| June 1994 | Clinical patient | H | H | X |
| June 1994 | Clinical patient | I | I | XI |
| 29 Oct. 1995 | Agar plate (MML) | K | J | XIII |
| 29 Oct. 1995 | Agar plate (MML) | K | XIII | |
| 30 Oct. 1995 | Agar plate (MML) | K | J | XIII |
| 30 Oct. 1995 | Agar plate (MML) | K | J | XIII |
| 31 Oct. 1995 | Agar plate (MML) | K | J | XIII |
| 1 Nov. 1995 | Agar plate (MML) | K | J | XIII |
| 1 Nov. 1995 | Air sample (MML) | K | J | XIII |
| 1 Nov. 1995 | Air sample (MML) | K | J | XIII |
| 1 Nov. 1995 | Air sample (MML) | L | K | XIV |
| 1 Nov. 1995 | Air sample (MML) | K | J | XIII |
| 1 Nov. 1995 | Air sample (MML) | K | J | XIII |
| 1 Nov. 1995 | Air sample (MML) | K | J | XIII |
| 1 Nov. 1995 | Air sample (MML) | K | J | XIII |
| 6 Dec. 1995 | Air sample (MML) | K | K | XV |
| 6 Dec. 1995 | Air sample (MML) | K | K | XV |
| 6 Dec. 1995 | Air sample (MML) | K | J | XIII |
| 6 Dec. 1995 | Air sample (MML) | K | J | XIII |
| 6 Dec. 1995 | Air sample (MML) | K | J | XIII |
The overall PCR type was based on the number of different banding patterns when both assays were combined. MML, Medical Microbiology Laboratory.
DISCUSSION
The numbers of conidia of nonpathogenic fungi detected in this study from samples obtained outside showed a variation that is compatible with surveillance results described before: higher numbers during the summer and lower numbers during the fall and winter (9, 11). There was a clear difference between the peak numbers of conidia during the two summers that were included in the surveillance period. This difference might be due to differences in weather conditions. A very wet period followed by hot dry weather proceeded the period of high conidial numbers in 1994. There was almost no seasonal influence on the number of aspergillus conidia (5).
We found decreasing numbers of conidia when we compared air from outside the hospital to air inside the hospital and when we compared air from open areas within the hospital to air from the closed department of hematology. The lower density of fungal conidia in the department of hematology may be the result of the increased level of environmental isolation (closed entrance doors, windows that cannot be opened, no plants or flowers, etc.). An even higher level of isolation exists within the HEPA-filtered rooms, which resulted in very low numbers of conidia, usually below the recommended threshold for such rooms (18). Only during summer months did the air samples from HEPA-filtered rooms show more conidia of nonpathogenic fungi, probably as a result of a higher density outside. Because conidia of nonpathogenic fungi are present in much higher densities than conidia of pathogenic fungi (ca. 10 to 20 times higher), enumeration of the former type of conidia can be conveniently used to control the effectiveness of barrier and filtration systems. During the surveillance period, the number of conidia of nonpathogenic fungi indicated that the barrier and filtration systems provided in the HEPA-filtered rooms worked properly.
The gradient found for the conidia of nonpathogenic fungi was also present when only conidia of Aspergillus species were counted. On two occasions, during and just after the outbreak of invasive aspergillosis, however, the numbers of these conidia within the hematology ward and the HEPA-filtered rooms were higher than usual (10). From the subsequent surveillance it became clear that during these occasions the density of nonpathogenic fungal conidia in the outside air also had peak values. However, since the density of Aspergillus conidia did not show such a pattern, the outbreak of invasive aspergillosis was probably not due to the introduction of outside air carrying higher numbers of Aspergillus conidia into the department of hematology. Furthermore, on both occasions, the majority of these conidia were identified as conidia of A. flavus, a species that was never found outside the hematology ward. This would indicate the presence of a point source within the department. Because A. flavus was never found after the outbreak of invasive aspergillosis, we speculate that a patient or patient materials may have introduced the conidia. However, at that time the numbers of A. fumigatus conidia were also higher in the department of hematology and in the HEPA-filtered rooms. An extensive search did not reveal a common source. The higher numbers of A. fumigatus conidia may have resulted from the windows being opened more frequently in other departments at the beginning of the summer. After the beginning of the outbreak, all departments were ordered to keep the windows closed, which may explain the sudden decrease in the numbers of conidia.
When A. fumigatus isolates from the department of hematology were genotyped, it was shown that in this department clonally related isolates were present. This finding does not support the hypothesis that conidia were being introduced from outside the department. Furthermore, we showed that these clonally related isolates were persistently present for over 1 year and that a newly introduced clone was able to establish itself for almost a year. This suggests that, despite our intensive search, one or more common sources of A. fumigatus may have been present. Another explanation is that personnel of the hematology department served as a carrier for conidia from a common source outside the hospital. Unfortunately, the personnel were not screened. When we investigated more than a year later whether these isolates were still present (January to May 1997), air samples taken at the same locations in the hematology department only once showed growth of A. fumigatus. This isolate was genotypically different from the clones present before (results not shown). Persistence of identical genotypes of A. fumigatus was described earlier by Girardin et al., who took samples over a period of 6 months (4). They also occasionally found identical genotypes, although the majority of their isolates could be discriminated. This suggests that in their situation there was a constant introduction of conidia from outside.
Clinical isolates of A. fumigatus obtained during the outbreak could easily be distinguished from the strains present in the department of hematology. The fact that patients become infected with other strains than those present in the hospital air strongly suggests that patients already carry these strains when admitted to the hospital. If this is true, the outbreak of invasive aspergillosis and the high number of Aspergillus conidia occurring at the same time was merely a coincidence. This underscores the potential value of the use of prophylactic antifungal regimens in addition to environmental isolation. However, effective prophylactic antifungal regimens still need to be determined (1, 6, 12, 16).
Genotyping of A. fumigatus isolates obtained during the laboratory contamination showed that these isolates were all identical. Whether this contamination was caused directly by the renovation activities opposite the laboratory, with subsequent introduction of genotypically identical isolates of A. fumigatus, was not proven. Because the contamination was so extensive, the “pseudo” character of the outbreak was immediately clear, and no action was taken towards patients. However, it is not unreasonable to assume that during an episode of laboratory contamination, materials of patients prone to develop invasive aspergillosis become contaminated and show growth of Aspergillus species. We demonstrated that in such cases, RAPD analysis might be a very useful method for determining the clinical relevance of the cultured isolate. For bacterial pathogens, genotyping to determine the clinical relevance of an isolate has been described previously (14).
We conclude that knowledge of the epidemiology of Aspergillus species is important for the development of strategies to prevent invasive aspergillosis. Both seasonal variation in the density of aerial spores and construction activities can contribute to increased aerial spore concentrations. As such, both phenomena may be causally related to increased infection risks for susceptible patients or for pseudo-outbreaks. Genotyping by means of RAPD assay is helpful in elucidating the complex epidemiology of Aspergillus infections.
ACKNOWLEDGMENT
We are indebted to Marian Humphries for critically reading the manuscript.
REFERENCES
- 1.Anderson K, Morris G, Kennedy H, Croall J, Michie J, Richardson M D, Gibson B. Aspergillosis in immunocompromised paediatric patients: associations with building hygiene, design, and indoor air. Thorax. 1989;51:256–261. doi: 10.1136/thx.51.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andriole V T. Infections with Aspergillus species. Clin Infect Dis. 1993;17(Suppl. 2):S481–S486. doi: 10.1093/clinids/17.supplement_2.s481. [DOI] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girardin H, Sarfat J, Traore F, Camet J D, Derouin F, Latge J P. Molecular epidemiology of nosocomial invasive aspergillosis. J Clin Microbiol. 1994;32:684–690. doi: 10.1128/jcm.32.3.684-690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodley J M, Clayton Y M, Hay R J. Environmental sampling for aspergilli during building construction on a hospital site. J Hosp Infect. 1994;26:27–35. doi: 10.1016/0195-6701(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 6.Hay R J, Clayton Y M, Goodley J M. Fungal aerobiology: how, when and where? J Hosp Infect. 1995;30(Suppl.):352–357. doi: 10.1016/0195-6701(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 7.Hruszkewycz V, Ruben B, Hypes C, Bostic G D, Staszkiewicz J, Band J D. A cluster of pseudofungemia associated with hospital renovation adjacent to the microbiology laboratory. Infect Control Hosp Epidemiol. 1992;13:147–150. doi: 10.1086/646498. [DOI] [PubMed] [Google Scholar]
- 8.Iwen P C, Davis J C, Reed E C, Winfield B A, Hinrichs S H. Airborne fungal spore monitoring in a protective environment during hospital construction, and correlation with an outbreak of invasive aspergillosis. Infect Control Hosp Epidemiol. 1994;15:303–306. doi: 10.1086/646916. [DOI] [PubMed] [Google Scholar]
- 9.Kersten W, von Wahl P G. Three-year survey of the mould spore flight in the air over Moers/Niederrhein (1985–1987) Allergologie. 1989;12:66–76. [Google Scholar]
- 10.Leenders A C A P, van Belkum A, Janssen S, de Marie S, Kluytmans J, Wielenga J, Löwenberg B, Verbrugh H A. Molecular epidemiology of an apparent outbreak of invasive aspergillosis in a hematology ward. J Clin Microbiol. 1996;34:345–352. doi: 10.1128/jcm.34.2.345-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D W, Kendrick B. A year-round study on functional relationships of airborne fungi with meteorological factors. Int J Biometeorol. 1995;39:74–80. doi: 10.1007/BF01212584. [DOI] [PubMed] [Google Scholar]
- 12.Loo V G, Bertrand C, Dixon C, Vitye D, Desalis B, McLean A P, Brox A, Robson H G. Control of construction-associated aspergillosis in an antiguated hematology unit. Infect Control Hosp Epidemiol. 1996;17:360–364. doi: 10.1086/647317. [DOI] [PubMed] [Google Scholar]
- 13.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 14.Morris T, Brechner S M, Fitzsimmons D, Durbin A, Arbet R D, Maslow J N. A pseudoepidemic due to laboratory contamination deciphered by molecular analysis. Infect Control Hosp Epidemiol. 1995;16:82–87. doi: 10.1086/647061. [DOI] [PubMed] [Google Scholar]
- 15.Pfaller M A. Epidemiological typing methods for mycoses. Clin Infect Dis. 1992;14(Suppl. 1):S4–S10. doi: 10.1093/clinids/14.supplement_1.s4. [DOI] [PubMed] [Google Scholar]
- 16.Philpott-Howard J. Prevention of fungal infections in hematology patients. Infect Control Hosp Epidemiol. 1996;17:545–551. doi: 10.1086/647369. [DOI] [PubMed] [Google Scholar]
- 17.Seeliger H P R, Tintelnot K. Epidemiology of aspergillosis. In: Van den Bossche E, MacKenzie D W R, Cauwenbergh G, editors. Aspergillus and aspergillosis. New York, N.Y: Plenum Press, Inc.; 1988. pp. 23–25. [Google Scholar]
- 18.Streifel A J. Air cultures for fungi. In: Isenberg H D, editor. Clinical microbiology procedures handbook. Washington, D.C: American Society for Microbiology; 1992. pp. 11.8.1–11.8.7. [Google Scholar]
- 19.Tompkins L S, Tenover F C, Arvin A. New technology in the clinical microbiology laboratory: what you always wanted to know but were afraid to ask. J Infect Dis. 1994;170:1068–1074. doi: 10.1093/infdis/170.5.1068. [DOI] [PubMed] [Google Scholar]
- 20.Van Belkum A. DNA fingerprinting of medically important microorganisms by use of PCR. Clin Microbiol Rev. 1994;7:174–184. doi: 10.1128/cmr.7.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Belkum A, Melchers W, de Pauw B E, Scherer S, Quint W, Meis J F G M. Genotypic characterization of sequential Candida albicans isolates from fluconazole-treated neutropenic patients. J Infect Dis. 1994;169:1062–1070. doi: 10.1093/infdis/169.5.1062. [DOI] [PubMed] [Google Scholar]
- 22.Van Belkum A, Mol W, van Saene R, Ball L M, van Velzen D, Quint W G V. PCR-mediated genotyping of Candida albicans strains from bone marrow transplant patients. Bone Marrow Transplant. 1994;13:811–815. [PubMed] [Google Scholar]
- 23.Van Belkum A, Kluytmans J, Van Leeuwen W, Bax R, Quint W, Peters E, Fluit A, Vandenbroucke-Grauls C, Van den Brule A, Koeleman H, Melchers W, Meis J, Elaichouni A, Vaneechoutte M, Moonens F, Maes N, Struelens M, Tenover F, Verbrugh H. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J Clin Microbiol. 1995;33:1537–1547. doi: 10.1128/jcm.33.6.1537-1547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]