In this study, Kawasumi et al. investigated the establishment of sister chromatid cohesion in vertebrate cells. Using genetically amenable chicken DT40 B-cell lines to establish conditional knockout and point mutations in cohesin-related genes and regulators, the authors report an unknown joint essential role for vertebrate CTF18 and DDX11 in sister chromatid cohesion and cellular proliferation.
Keywords: CTF18, DDX11, sister chromatid cohesion, WAPL, cohesin balance on chromatin, ESCO1/2, cohesin acetylation
Abstract
The alternative PCNA loader containing CTF18-DCC1-CTF8 facilitates sister chromatid cohesion (SCC) by poorly defined mechanisms. Here we found that in DT40 cells, CTF18 acts complementarily with the Warsaw breakage syndrome DDX11 helicase in mediating SCC and proliferation. We uncover that the lethality and cohesion defects of ctf18 ddx11 mutants are associated with reduced levels of chromatin-bound cohesin and rescued by depletion of WAPL, a cohesin-removal factor. On the contrary, high levels of ESCO1/2 acetyltransferases that acetylate cohesin to establish SCC do not rescue ctf18 ddx11 phenotypes. Notably, the tight proximity of sister centromeres and increased anaphase bridges characteristic of WAPL-depleted cells are abrogated by loss of both CTF18 and DDX11. The results reveal that vertebrate CTF18 and DDX11 collaborate to provide sufficient amounts of chromatin-loaded cohesin available for SCC generation in the presence of WAPL-mediated cohesin-unloading activity. This process modulates chromosome structure and is essential for cellular proliferation in vertebrates.
Organizing chromosomal DNA is a fundamental but challenging task in all domains of life. The cohesin complex plays important roles in several aspects of chromosome dynamics (Yatskevich et al. 2019). Structurally, cohesin is a molecular ring composed of several essential subunits: SMC3, SMC1, RAD21, and SA1/SA2. Cohesin further associates with the WAPL and PDS5A/B subunits, which control opening and closure of the cohesin ring (Peters and Nishiyama 2012). Cohesin links DNA double helices but can also interact nontopologically with DNA. Cohesin-mediated in cis interactions of DNA within the same chromatid mediate the formation of chromosomal domains and the establishment of transcriptional programs. Cohesin-mediated in trans interactions between the sister chromatids leads to sister chromatid cohesion (SCC), which is essential for faithful chromosome segregation and facilitates error-free DNA repair by recombination (Peters and Nishiyama 2012; Tittel-Elmer et al. 2012; Fumasoni et al. 2015; Yatskevich et al. 2019). How these different interactions of cohesin with DNA are modulated is not well understood, but cohesin regulators may provide insights into these processes (Dauban et al. 2020).
Cohesin binds dynamically to DNA, and this interaction is converted to a more stable one during replication via SMC3 acetylation at conserved lysine residues (Ben-Shahar et al. 2008; Unal et al. 2008; Zhang et al. 2008; Onn et al. 2009). Cohesin acetylation is mediated by Eco1 in budding yeast and its ESCO1 and ESCO2 orthologs in vertebrates and mammalian cells. SMC3 acetylation is thought to stabilize cohesin on DNA by inactivating or preventing the access of the cohesin release factor WAPL (Rowland et al. 2009; Sutani et al. 2009; Nishiyama et al. 2010; Chan et al. 2012; Beckouët et al. 2016; Elbatsh et al. 2016; Ouyang and Yu 2017). Although ESCO1 functions throughout the cell cycle (Minamino et al. 2015), replication-associated cohesin acetylation is mediated primarily by ESCO2, via temporal regulation of ESCO2 degradation and its interactions with the MCM2-7 helicase and/or the polymerase clamp PCNA (Higashi et al. 2012; Ivanov et al. 2018; Minamino et al. 2018; Bender et al. 2020).
A number of other replisome-associated factors facilitate SCC in budding yeast by mechanisms not fully understood. One such factor is Ctf18-RFC, composed of Ctf18-Dcc1-Ctf8 (CTF18-DSCC1-CTF8 in mammalian cells), which associates with Rfc2-5 (Hanna et al. 2001; Mayer et al. 2001) and polymerase ε (García-Rodríguez et al. 2015; Stokes et al. 2020). The Ctf18-RFC complex represents an alternative PCNA clamp loader, besides the Rfc1 canonical clamp loader and the Elg1 alternative clamp loader (Lee and Park 2020). While the Elg1-RFC unloads PCNA, Ctf18 and Rfc1 facilitate PCNA loading to leading and lagging strands (Liu et al. 2020). The roles of the budding yeast Ctf18 clamp loader in SCC are controversial. One view holds that Ctf18-RFC function is linked to cohesin acetylation by supplying additional PCNA at replication forks to facilitate recruitment of Eco1 (Liu et al. 2020). The other model holds that Ctf18-RFC mediates de novo loading of cohesin at replication forks (Srinivasan et al. 2020).
The above models of Ctf18 function were developed in budding yeast, in which nonessential SCC regulators are associated with the replisome (Lengronne et al. 2006) and are divided into two SCC groups (Xu et al. 2007). Recent work provided evidence that factors in the two groups have different modes of action. Ctf18-RFC functions together with Scc2 to mediate de novo cohesin loading, whereas Ctf4 (AND-1 in vertebrates), Chl1 (DDX11 in vertebrates), and Csm3-Tof1 (TIMELESS-TIPIN in vertebrates) convert cohesin associated with unreplicated DNA into cohesive structures, independently of Scc2 (Srinivasan et al. 2020).
Of note, with the exception of DDX11 whose mutations in humans cause a cohesinopathy known as Warsaw breakage syndrome (van der Lelij et al. 2010; Pisani et al. 2018) and whose contributions to SCC are studied in cell lines and mouse models also in relation to its physical interaction with TIMELESS-TIPIN (Inoue et al. 2007; de Lange et al. 2015; Abe et al. 2016; Cortone et al. 2018; Faramarz et al. 2020; van Schie et al. 2020), little is known about the role of other replisome-associated SCC regulators in vertebrate and mammalian cells. Importantly, the role of the vertebrate CTF18 complex, belonging to a SCC group different from Chl1/DDX11 in budding yeast, where it contributes to checkpoint activation (Pan et al. 2006; Crabbé et al. 2010; Kubota et al. 2011; García-Rodríguez et al. 2015) and prevents fragility at repeat elements (Gellon et al. 2011), is currently unknown. Because CTF18 is mutated in cancers (Price et al. 2013) and required for fertility (Berkowitz et al. 2012; Holton et al. 2020), understanding its molecular function and identifying functionally related pathways in vertebrates is important and may provide medical value.
To model the consequence of CTF18 inactivation in vertebrate cells, we established CTF18 knockout and conditional depletion in avian DT40 cells defective in the p53 apoptotic pathway (Abe and Branzei 2014), often inactivated in cancers (Negrini et al. 2010). We found that ctf18 mutant cell lines proliferate normally in spite of a mild reduction in fork speed but are fully dependent on the DDX11 helicase for proliferation and normal chromosome segregation. We uncover that the essential joint role of vertebrate CTF18 and DDX11 is linked to SCC and is no longer required in the absence of the cohesin-releaser WAPL, in contrast to the situation in budding yeast. Notably, the cohesin-mediated apparent overcondensation state of chromosomes in WAPL-defective cells is attenuated by concomitant CTF18 and DDX11 inactivation. Chromatin fractionation and fluorescence recovery after photobleaching (FRAP) experiments further revealed reduced stable binding of cohesin to chromatin in ctf18 ddx11 mutant cell lines and restoration of this pool upon WAPL depletion. Altogether, the results highlight vertebrate DDX11 and CTF18 as critical complementary regulators of SCC and show that they act to provide sufficient amounts of chromatin-loaded cohesin available for SCC generation in the presence of WAPL-mediated cohesin-unloading activity.
Results
CTF18 inactivation causes replication fork slow-down and sensitivity to mitotic poisons
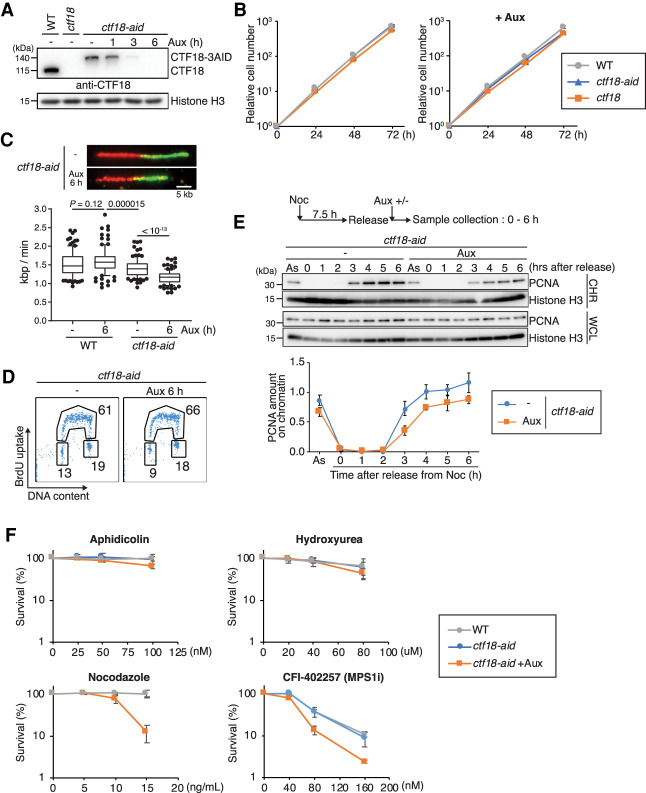
To establish CTF18 inactivation in DT40 cells, we applied the auxin-induced degron (AID) system, which enables rapid degradation of target proteins by the proteasome (Nishimura et al. 2009; Kawasumi et al. 2017; Abe et al. 2018a). Specifically, we used the flip-in system (Kobayashi et al. 2015) to C-terminally tag the endogenous CTF18 alleles with 3AID-6FLAG and 3AID-6HA, respectively (Supplemental Fig. S1A). After sequential transfection, we obtained CTF183AID6FLAG/3AID6HA cells expressing TIR1-9myc (referred to here as ctf18-aid cells). The correct insertions of the AID tags to CTF18 and auxin-induced depletion of CTF18-AID proteins were confirmed by Western blotting (Fig. 1A). After auxin addition, CTF18-AID proteins largely disappeared within 3 h (Fig. 1A) without detrimental effects on cellular proliferation (Fig. 1B). Moreover, we established CTF18 knockout DT40 cells (Supplemental Fig. S1B), referred to as ctf18, and found that they supported normal proliferation, similarly with WT and CTF18-depleted DT40 cells (Fig. 1B).
Figure 1.
CTF18 inactivation causes replication fork slow-down and sensitivity to mitotic poisons. (A) Total cell lysates were prepared from cells of the indicated genotypes and analyzed by Western blotting using a CTF18 antibody. Cells were treated with auxin (Aux) when indicated. (B) Growth curves of WT, ctf18, and ctf18-aid cells. The left panel shows growth curves without auxin, and the right panel shows the ones with auxin treatment. (C) Replication fork velocity was determined by DNA fiber assay for WT and ctf18-aid cells with or without auxin treatment. Scale bar on the representative fiber image, 5 kb. More than 50 molecules were analyzed for each condition. (Middle line) Median, (box) 25th and 75th percentiles, (bars) 10th and 90th percentiles. P-values were calculated by Student's t-test, and indicated. (D) Cell cycle distribution of CTF18-depleted cells using BrdU FACS. After 6 h of auxin treatment, ctf18-aid cells were pulse-labeled with BrdU for 15 min and then stained with anti-BrdU and FITC-conjugated antimouse antibodies sequentially. DNA content was determined by propidium iodide (PI) staining. Percentages of cells in each cell cycle are indicated. (E) Chromatin association of PCNA was monitored by chromatin fractionation. Nocodazole-arrested cells were released into fresh medium with or without auxin. Whole-cell lysate (WCL) and chromatin fraction (CHR) samples were prepared every hour after the release and analyzed by Western blotting. PCNA amount on chromatin was calculated by normalizing PCNA amount of CHR to that of WCL. Mean of four independent experiments was plotted. Error bars represent the SD. (F) Sensitivity assays to indicated drugs. Cells were treated with auxin for 6 h prior to exposure to drugs as indicated. After 48 h of incubation at 39.5°C, cell density was determined by CellTiter-Glo viability assay. Each line and error bar represents the mean value and SD from three independent experiments, respectively.
Because the CTF18 complex affects replication fork speed, either decreasing it in budding yeast (Lopez-Serra et al. 2013) or accelerating it in human cells (Terret et al. 2009), we performed DNA fiber analysis, which revealed a mild but significant reduction in fork speed upon CTF18 depletion (Fig. 1C), similar in trend to the situation in human cells (Terret et al. 2009). We noted a modest decrease in fork speed in ctf18-aid cells even in the absence of auxin (Fig. 1C), suggesting a mild dysfunction in CTF18 caused by the C-terminal tagging. The defect was further aggravated by induced CTF18 depletion with auxin. Moreover, we observed similar reduction in fork speed in ctf18 cells (Supplemental Fig. S1C).
Next, we examined BrdU incorporation by flow cytometry and found that CTF18-depleted cells had a modest increase in the S-phase population (Fig. 1D). Because the alternative CTF18-RFC clamp loader affects PCNA loading and stimulates Polε-mediated replication (García-Rodríguez et al. 2015; Stokes et al. 2020), we monitored the levels of PCNA associated with chromatin in normal conditions and upon CTF18 depletion. By analyzing chromatin-bound versus total PCNA levels detected in whole-cell extracts upon release of cells from nocodazole-induced metaphase arrest, we found a reduction in chromatin-bound PCNA in CTF18-depleted cells (Fig. 1E).
To address whether changes in chromatin-associated PCNA levels, which correlate with a reduction in fork speed, affect the ability of cells to deal with replication stress cues and mitotic poisons, we examined the viability of CTF18-depleted cells in various conditions. We found no significant viability drop in response to aphidicolin and hydroxyurea, which cause replication slow-down by Polα uncoupling and reduction of dNTP pools, respectively, but increased sensitivity to nocodazole that causes microtubule depolymerization and to MPS1 (mitotic kinase Monopolar Spindle 1) inhibitor CFI-402257 that causes premature mitotic entry (Fig. 1F).
Sensitivity to microtubule depolymerizing drugs can be associated and caused by defective SCC, phenotypes reported for budding yeast CTF18 deletion mutants (Mayer et al. 2001). To examine whether this is the case in DT40 cells depleted for CTF18, we scored SCC defects on metaphase chromosomes of ctf18-aid cells. In line with previous reports (Abe et al. 2016; Kawasumi et al. 2017), in control cells, a high fraction of DT40 cell metaphases has well-cohered tight chromosomes, with only 5%–25% of metaphase cells having chromosomes with arms slightly apart, a phenotype scored as a modest chromosome arm SCC defect (Supplemental Fig. S1D). The SCC status was not aggravated by CTF18 depletion, indicating that CTF18 does not have a major impact on SCC (Supplemental Fig. S1D). We further analyzed whether CTF18 loss causes a defect in SMC3 acetylation as reported in budding yeast (Borges et al. 2013; Liu et al. 2020). We observed only a mild decrease in SMC3 acetylation in ctf18-aid cells, which was not further decreased by addition of auxin and was not statistically significant (Supplemental Fig. S1E). This mild reduction in SMC3 acetylation may be related to CTF18 dysfunction caused by tagging. Thus, vertebrate CTF18 is not critical for SCC, although it facilitates cellular viability in the presence of mitotic poisons.
CTF18 acts in parallel with DDX11 to mediate cellular proliferation
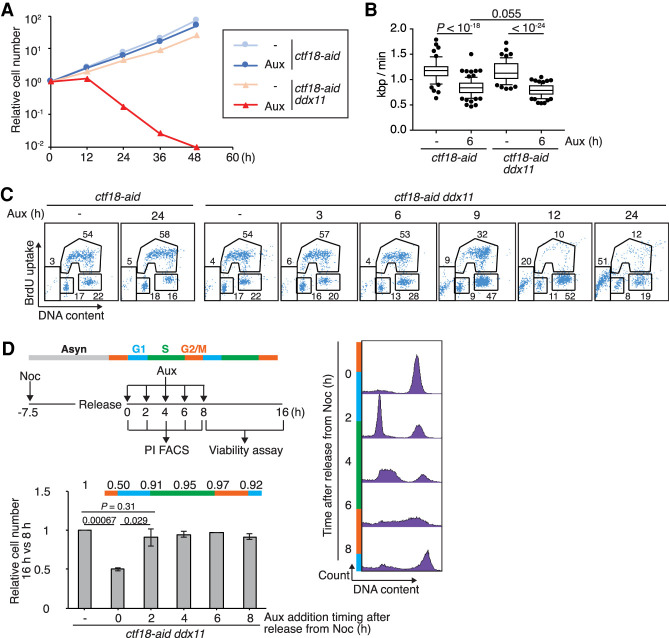
DDX11 encodes a DNA helicase that interacts physically with CTF18 (Farina et al. 2008) and contributes to SCC in budding yeast (Mayer et al. 2004; Xu et al. 2007) and in vertebrates (van der Lelij et al. 2010; de Lange et al. 2015; Abe et al. 2016; Cortone et al. 2018; Faramarz et al. 2020). To address whether DDX11 mediates proliferation in CTF18-depleted cells, in analogy to the situation reported in budding yeast (Xu et al. 2007; Srinivasan et al. 2020), we knocked out DDX11 in the ctf18-aid background to establish CTF183AID6FLAG/3AID6HA DDX11−/− cells (referred to here as ctf18-aid ddx11 cells). Differently from single ctf18 and ddx11 mutants that proliferate normally, ctf18-aid ddx11 cells are severely impaired in proliferation upon auxin addition, indicating synthetic lethality (Fig. 2A).
Figure 2.
CTF18 contributes in parallel with DDX11 to cellular proliferation. (A) Growth curves of ctf18-aid and ctf18-aid ddx11 cells with or without auxin treatment. (B) Replication fork velocity was determined by DNA fiber assay for ctf18-aid and ctf18-aid ddx11 cells with or without auxin treatment as in Figure 1C. P-values were calculated by Student's t-test and indicated. (C) Cell cycle distribution using BrdU FACS. ctf18-aid and ctf18-aid ddx11 cells were incubated with auxin for indicated times, pulse-labeled with BrdU for 15 min, and analyzed as in Figure 1D. (D) Viability of ctf18-aid ddx11 cells after depletion of CTF18-AID at different times in the cell cycle. (Top left panel) Experimental scheme. (Right panel) Cell cycle progression after release from nocodazole monitored by PI staining. (Bottom left panel) Cell number was determined by CellTiter-Glo viability assay 16 h after release from nocodazole; the results were normalized to those of 8 h. Mean values of three independent experiments are plotted. P-values were calculated by Student's t-test and indicated.
We further assessed whether the cause of the observed synthetic lethality relates to replication problems. DDX11 knockout did not affect fork speed in either wild-type (ctf18-aid without auxin) or CTF18-depleted backgrounds (Fig. 2B), suggesting that DNA replication defects are unlikely the source of the observed synthetic lethality. Consistent with this notion, cell cycle analysis by BrdU uptake revealed that BrdU incorporation is normal but the mitosis is severely impaired in ctf18 ddx11 cells (Fig. 2C).
To examine the time in the cell cycle in which CTF18 is essential for proliferation in DDX11 knockout cells, we synchronized ctf18-aid ddx11 cells in metaphase with nocodazole and added auxin at different times, up to 8 h of release from nocodazole, scoring the cell cycle profile of the cultures. We then measured cellular viability at 16 h after release from nocodazole arrest and compared it with the one measured at 8 h and different auxin treatments to evaluate whether one cell division has occurred upon CTF18 depletion in DDX11 knockout cells. The results revealed that only when auxin was added early enough to achieve CTF18 depletion prior to S phase, the viability of ddx11 cells was impaired (Fig. 2D). Thus, CTF18 acts prior to or early during replication to facilitate proliferation in DDX11 knockout cells but without impairing the ability of cells to complete bulk replication.
CTF18-depleted cells rely on the DDX11 helicase and TIPIN-mediated SCC axis for proliferation
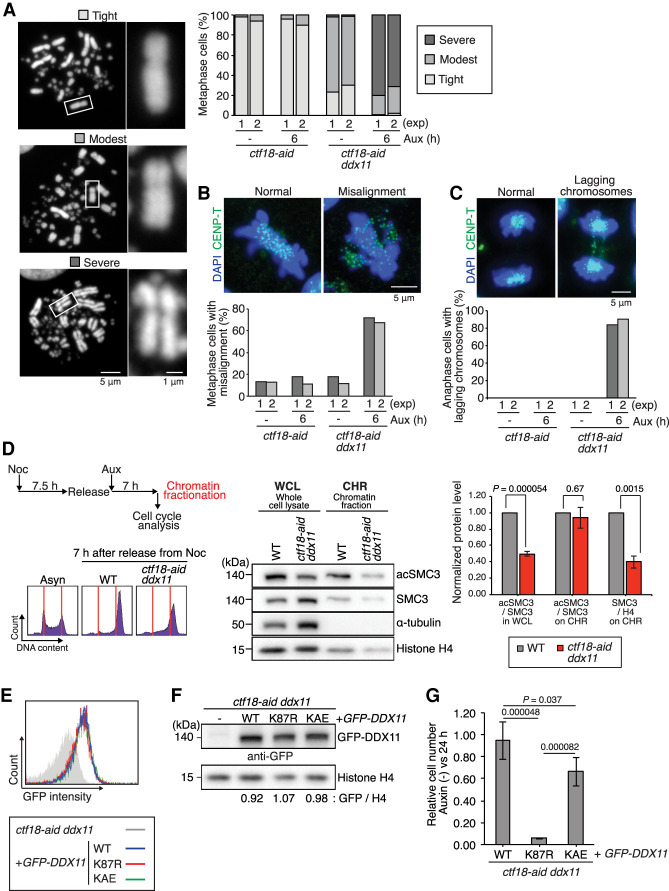
One hypothesis that could explain the above results is that CTF18 supports SCC generation in parallel with DDX11. Supporting this notion, ctf18-aid ddx11 cells treated with auxin showed a strong increase in metaphases exhibiting centromere separation defects (severe type of SCC defect), not observed in ctf18 and ddx11 single mutants (Fig. 3A). Moreover, ctf18-aid ddx11 cells have a high percentage of misaligned chromosomes in metaphase (Fig. 3B) and lagging chromosomes in anaphase (Fig. 3C), common phenotypes of SCC-defective mutants (Sonoda et al. 2001; Abe et al. 2016; Kawasumi et al. 2017). Thus, vertebrate CTF18 and DDX11 compensate for each other in SCC.
Figure 3.
CTF18-depleted cells rely on the DDX11 helicase-mediated SCC axis. (A) Sister chromatid cohesion status was assessed for the cells of indicated genotypes. Metaphase chromosomes were classified in three groups depending on the level of cohesion, and >100 metaphase cells were analyzed for each genotype with or without auxin treatment. The results of two independent experiments are plotted. Representative images of each class are shown. (B,C) Metaphase cells with chromosome misalignment (B) and anaphase cells with lagging chromosomes (C) were scored. CENP-T was used to visualize small chromosomes that are barely visible by DAPI staining. At least 50 cells were analyzed for each condition. The results of two independent experiments are plotted. Representative images of misalignment and lagging chromosomes are shown. (D) Analysis of SMC3 and its acetylated form in whole-cell lysates and chromatin fractions of WT and ctf18-aid ddx11 cells, released for 7 h in the presence of auxin from nocodazole arrest. Cell cycle profile was analyzed by PI staining. Mean values and SEs of three independent experiments showing ratios of acSMC3 versus SMC3 in whole-cell lysates, chromatin, and the ratio of SMC3 versus histone H4 on chromatin are plotted. P-values were calculated by Student's t-test and indicated. (E) FACS profile showing GFP-DDX11 expression levels for various DDX11 variants. The Y-axis represents cell count, and the X-axis represents GFP intensity in log scale. (F) Western blotting showing GFP-DDX11 expression levels for various DDX11 variants normalized versus histone H4. (G) Relative cell number after 24 h of auxin treatment determined by CellTiter-Glo viability assay for indicated genotypes. Results of four independent clones for each genotype were combined. P-values were calculated by Student's t-test and plotted. Error bars represent SD obtained from the result of four independent clones.
To examine whether the SCC defects and mitotic lethality in ctf18-aid ddx11 cells relate to aberrant distribution of cohesin or its acetylation, we first analyzed the relative levels of SMC3 and its acetylation in whole-cell lysates (Supplemental Fig. S2A). The ratio of acetylated-SMC3 versus total SMC3 was reduced in whole-cell extracts of double mutants compared with WT, although this decrease was observed even in the absence of auxin, reminiscent of the defect observed in ctf18-aid cells (Supplemental Fig. 1E). Next, we investigated the levels of cohesin and its acetylation in whole-cell extracts and chromatin fractions from WT and ctf18-aid ddx11 cells that reached mitosis after release from nocodazole arrest in the presence of auxin (Fig. 3D). Importantly, while we recapitulated the reduction in acetylated SMC3 versus SMC3 in whole-cell extracts of double mutants, this pattern does not apply to the chromatin fraction, in which the ratio is similar to what is observed in WT cells (Fig. 3D). This phenotype of double mutants is likely due to reduced levels of SMC3 on chromatin as observed from quantifying SMC3 versus histone H4 in the chromatin fraction (Fig. 3D).
Previous work indicated that DDX11 facilitates cohesion in cooperation with TIPIN-TIMELESS with which it physically interacts (Leman et al. 2010; Abe et al. 2016; Calì et al. 2016; Cortone et al. 2018). To inquire on the DDX11 functions required for viability in CTF18-depleted cells, we transfected ctf18-aid ddx11 cells with GFP-tagged WT DDX11, the helicase-dead K87R DDX11 variant, mutated in the Walker A domain, or the KAE DDX11 variant defective in interaction with TIM/TIMELESS (Cortone et al. 2018), selecting clones expressing these variants at similar levels, as assessed by FACS (Fig. 3E) and then confirmed by Western blotting (Fig. 3F). Proliferation experiments measuring cell number after 24 h upon auxin addition revealed that both the helicase activity of DDX11 and the interaction with TIMELESS contributed to normal proliferation, with the helicase activity being essential in this regard (Fig. 3G).
To further inquire whether DDX11 compensates for CTF18 loss jointly with TIPIN-TIMELESS, we established combinations between the CTF18 KO and TIPIN conditional mutant in which TIPIN depletion is induced by doxycycline (Dox) (Supplemental Fig. S2B; Hosono et al. 2014). Tet-tipin ctf18 cells stopped proliferation after Dox-induced TIPIN depletion (Supplemental Fig. S2C) and displayed a strong increase in anaphase lagging chromosomes (Supplemental Fig. S2D), resembling the lethality of ctf18 ddx11 cells (see Fig. 3C). Thus, vertebrate CTF18 compensates for DDX11-TIPIN regarding SCC and their concomitant loss leads to cell death due to massive chromosome missegregation.
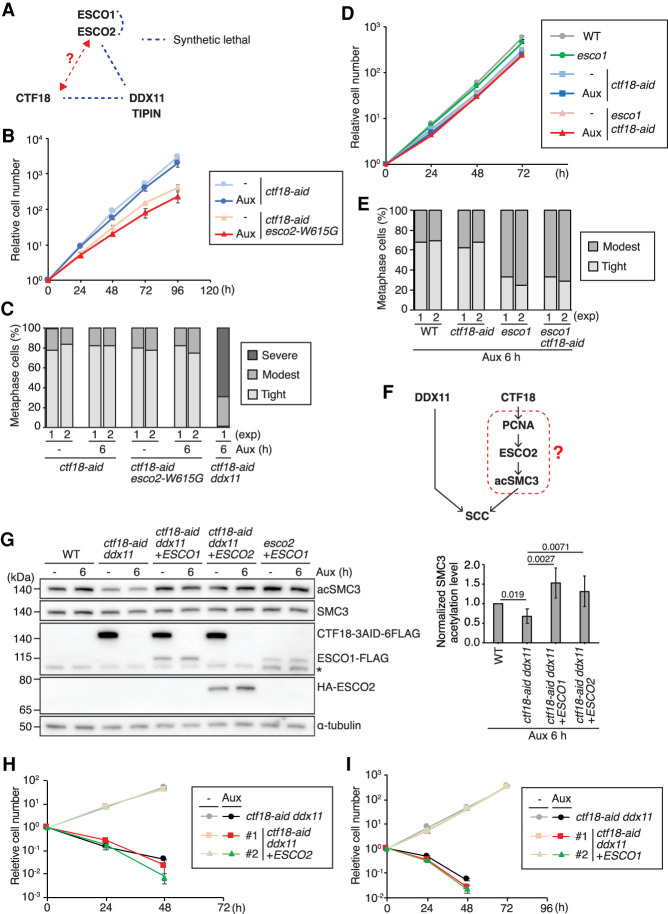
CTF18 loss does not synergize with ESCO1 and ESCO2 loss in SCC defects
In DT40 cells, individual DDX11 and TIPIN deficiency leads to synthetic lethality in combination with the esco2-W615G mutation (Abe et al. 2016), which mimics the relatively common human esco2-W539G mutation found in Roberts syndrome patients (Fig. 4A; Gordillo et al. 2008). A similar genetic relationship between DDX11 and ESCO2 toward SCC has been reported in human cells (Faramarz et al. 2020). To investigate the connections between CTF18 and ESCO2, we established ESCO2-/W615G CTF183AID6FLAG/3AID6HA (referred to here as ctf18-aid esco2-W615G cells). The proliferation of ctf18-aid esco2-W615G cells was not affected by the presence of auxin (Fig. 4B) and did not result in severe SCC defects (Fig. 4C) or lagging chromosomes in anaphase (Supplemental Fig. S3A), differently from ctf18 ddx11 mutants. We also examined the CTF18 genetic interaction with ESCO1, which shows compensatory functions with ESCO2 in centromeric cohesion (Kawasumi et al. 2017). ctf18-aid esco1 cells proliferated similarly in the presence as in the absence of auxin (Fig. 4D) and had similar levels of mild cohesion defects with esco1 cells (Fig. 4E). Thus, CTF18 functions genetically with ESCO1/2 and in complementary fashion with DDX11.
Figure 4.
CTF18 and DDX11 participate in different SCC branches, and loss of their joint essential function is not compensated by ESCO1/2 overexpression. (A) Genetic interactions of SCC-related genes. Genetic interaction data inferred from Abe et al. (2016) and Kawasumi et al. (2017). (B,D) Growth curves of the cells of indicated genotypes as in Figure 1B. (C,E) Chromosomes from metaphase spreads were analyzed for sister chromatid cohesion as in Figure 3A. (F) Hypothetical scheme of functional interactions among SCC-related factors, including DDX11, CTF18, ESCO2, and SMC3 acetylation. (G) SMC3 acetylation levels in the cells of the indicated genotypes were monitored by Western blotting, normalized to SMC3, and plotted. Each bar and error bar represents the mean value and SD from three independent experiments, respectively. (H,I) Growth curves of cells of indicated genotypes with or without auxin treatment as in B.
ESCO1/2 overexpression does not rescue ctf18 ddx11 synthetic lethality
Because the level of acSMC3 was reduced in ctf18-aid ddx11 cells, we addressed whether forced recovery of acSMC3 level by ESCO1/2 overexpression restores the SCC defects and viability of ctf18-aid ddx11 cells. We transfected ESCO2 and ESCO1 expression constructs in ctf18-aid ddx11 cells and assessed their expression levels in independent clones, using as reference points esco2 cells overexpressing ESCO1 at levels that suppressed their cohesion defects (Kawasumi et al. 2017) and ESCO2 knockout. We isolated ctf18-aid ddx11 clones expressing about 20-fold more ESCO1 or ESCO2 compared with WT levels (Supplemental Fig. S3B,C). We verified the high levels of ESCO1 and ESCO2 by western blotting and found that they fully compensated the cohesin acetylation defect observed in ctf18-aid ddx11 cells in whole-cell lysates (Fig. 4G; Supplemental Fig. S3D) but did not rescue the lethality of ctf18 ddx11 cells (Fig. 4H,I).
We further attempted to analyze whether in DDX11 KO cells ESCO1/2-mediated acetylation of cohesin at conserved lysine residues K105 and K106 accounts for the observed lethality with ctf18. To this end, we established DDX11 knockout in smc3-aid cells (Kawasumi et al. 2017) and expressed the SMC3 WT or the SMC3-AA variant containing the HA tag (Supplemental Fig. S3E). Upon auxin addition, ddx11 smc3-aid cells expressing SMC3 variants degrade endogenous SMC3-AID and rely on exogenously introduced SMC3 and SMC3-AA variants, expressed at similar levels (Supplemental Fig. S3E). Both SMC3 variants allowed proliferation of ddx11 smc3-aid cells, with only mild impairment observed upon expression of the smc3-AA variant (Supplemental Fig. S3F). We note that previous biochemical work suggested that all SMC3 variants with mutations in the acetylation sites, including SMC3-AA, may function as bypass mutants (Nishiyama et al. 2010). Although we cannot exclude the possibility that SMC3-AA is an acetylation bypass mutant, collectively, the results suggest that the CTF18 function required for proliferation in the absence of DDX11 may not be simply linked to defective cohesin acetylation.
WAPL loss suppresses the synthetic lethality of ctf18 ddx11 in vertebrate cells but not in budding yeast
How CTF18, DDX11 and other nonessential SCC regulators are involved in SCC is not fully elucidated. Yeast Chl1 (vertebrate DDX11) and Ctf18 were proposed to support SCC by providing sufficient cohesin amounts during replication through cohesin conversion and de novo loading, respectively (Srinivasan et al. 2020). As double mutants ctf18-aid ddx11 have low levels of cohesin on chromatin (Fig. 3D), we asked whether increasing chromatin-bound cohesin levels by preventing its WAPL-mediated unloading would rescue ctf18 ddx11 cohesion defects and synthetic lethality in vertebrate cells. To this end, we established ctf18-aid ddx11 wapl-aid cells after sequential transfections of the WAPL KO construct and the WAPL-3AID-6FLAG vector (Kawasumi et al. 2017) to ctf18-aid ddx11 cells. Strikingly, the lethality of ctf18-aid ddx11 cells in the presence of auxin was rescued by concomitant removal of WAPL (Fig. 5A; see below). We note that this is different from the situation of esco1 esco2 lethality, which was instead aggravated by WAPL depletion (Kawasumi et al. 2017).
Figure 5.
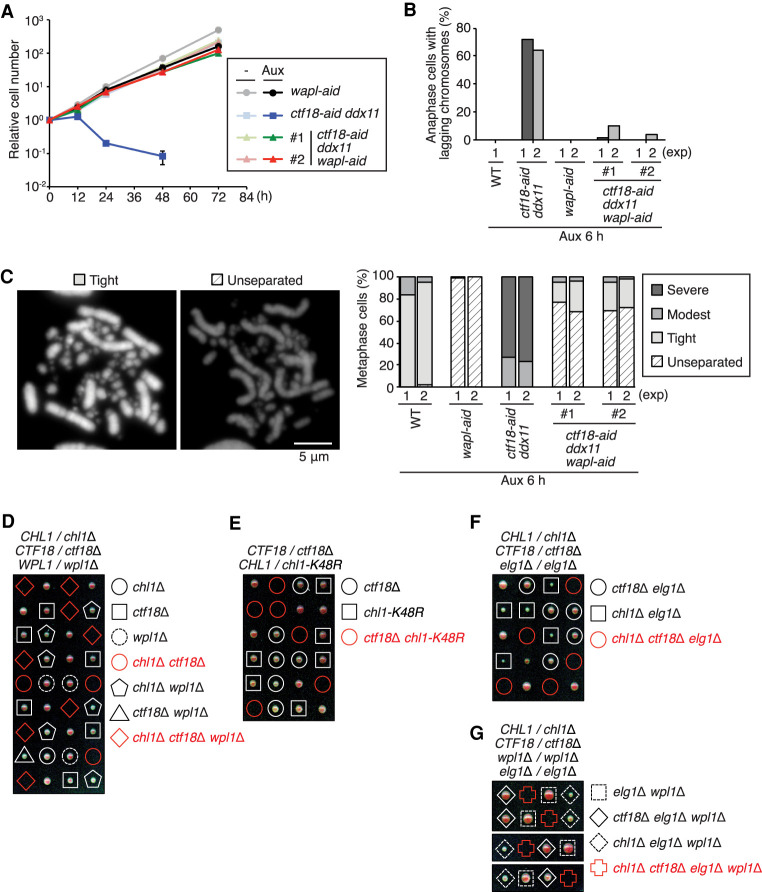
The synthetic lethality of ctf18 ddx11 is rescued by WAPL depletion in vertebrate cells but not in budding yeast. (A) Growth curves of cells of indicated genotypes with or without auxin treatment as in Figure 2A. (B) Frequency of lagging chromosomes at anaphase as in Figure 3C. The results of two independent experiments (except for WT cells) are plotted. (C) Metaphase chromosomes were analyzed for sister chromatid cohesion as in Figure 3A, with an additional class termed “unseparated type” typical of the wapl cell shown. (D–G) Genetic interactions between ctf18Δ, chl1Δ, chl1-K48R, wpl1Δ and elg1Δ were assessed by tetrad dissection analysis of the indicated diploid budding yeast strains.
Characterizing the conditional ctf18-aid ddx11 wapl-aid cells, we found that the severe SCC defects and the high frequency of lagging chromosomes observed in ctf18-aid ddx11 cells in the presence of auxin were also rescued by WAPL depletion (Fig. 5B,C). Moreover, double mutants ctf18-aid ddx11 accumulate γH2AX and mitotic phosphorylation at H3-S10, and these defects were rescued by WAPL depletion (Supplemental Fig. S4A). Thus, the severe cohesion defects and mitotic catastrophe in the absence of both CTF18 and DDX11 are linked to low levels of cohesin available for SCC generation due to the cohesin-unloading activity of WAPL.
The synthetic lethality between ctf18Δ and chl1Δ is also observed in budding yeast (Fig. 5D; Xu et al. 2007; Stokes et al. 2020). Similar to the situation in vertebrates, we found that the helicase activity of Chl1, disrupted in the chl1-K48R allele, is essential in the absence of Ctf18 (Fig. 5E). However, in budding yeast, the chl1Δ ctf18Δ lethality is not suppressed by deletion of the WAPL ortholog WPL1 (Fig. 5D). This difference may be owing to a paradoxical positive role for budding yeast Wpl1 toward efficient establishment of cohesion (Rowland et al. 2009; Sutani et al. 2009; Guacci and Koshland 2011). We further investigated whether increased levels of chromatin-bound PCNA caused by deletion of the PCNA unloader Elg1, reported to rescue phenotypes of ctf18Δ cells in SCC (Liu et al. 2020), can relieve the lethality of ctf18Δ chl1Δ cells. However, neither elg1Δ alone nor wpl1Δ elg1Δ combination could suppress the observed synthetic lethality (Fig. 5F,G). Moreover, higher levels of the cohesin loader Scc2, two copies of which suppress the cohesion defect of ctf4Δ cells (Srinivasan et al. 2020), expressed here from an ADH1 promoter, either alone or in combination with wpl1Δ, did not suppress the chl1Δ ctf18Δ lethality (Supplemental Fig. S4B,C). Thus, ctf18 ddx11 synthetic lethality in vertebrates is strongly linked to SCC generation defects, whereas in budding yeast, it may result from other causes or be compounded by other defects.
WAPL and DDX11-CTF18 have opposite effects on mitotic chromosome structure and sister centromere proximity
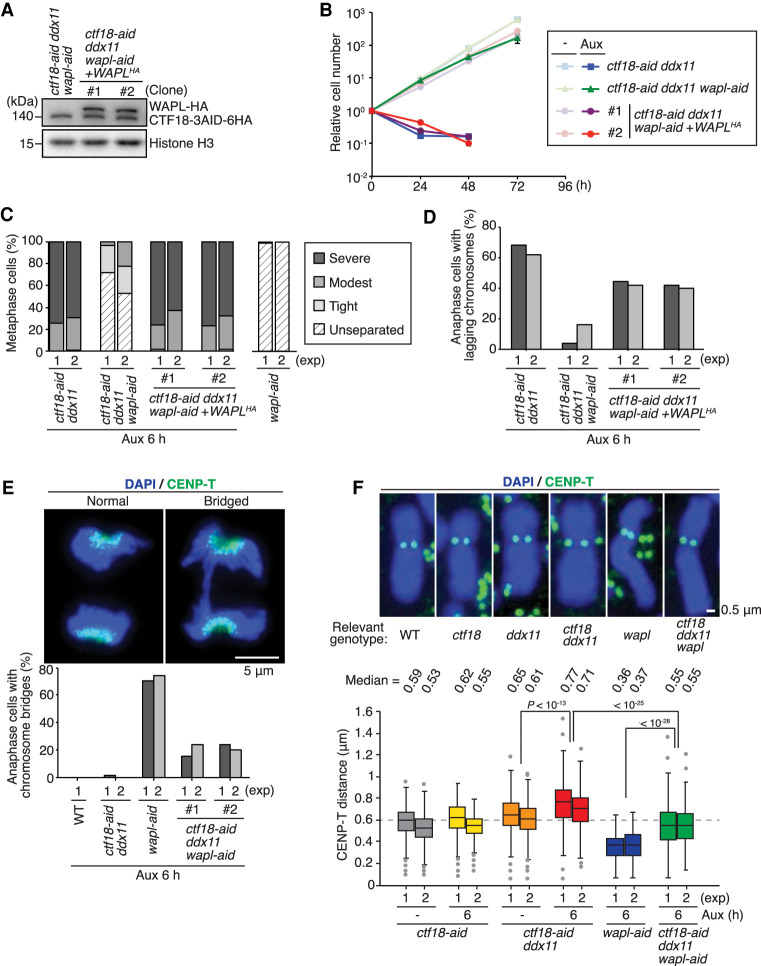
To validate the effects observed upon WAPL depletion in conditional CTF18 DDX11 mutants, we reintroduced HA-tagged WAPL to ctf18-aid ddx11 wapl-aid cells (Fig. 6A). Re-expressing WAPL-HA in ctf18-aid ddx11 wapl-aid cells caused defects in proliferation (Fig. 6B) along cohesion defects and increased lagging chromosomes similar to the situation observed in ctf18-aid ddx11 cells after auxin addition (Fig. 6C,D).
Figure 6.
WAPL and DDX11-CTF18 have opposite effects on mitotic chromosome structure and sister centromere proximity. (A) Expression of complemented WAPL-HA was monitored by Western blotting as in Figure 1A. (B) Growth curves of cells of indicated genotypes with or without auxin treatment as in Figure 2A. (C) Metaphase chromosomes were analyzed for sister chromatid cohesion as in Figure 5C. (D) Frequency of lagging chromosomes at anaphase was measured as in Figure 5B. (E) Anaphase cells with bridged chromosomes were scored. At least 50 cells for the anaphase cells were analyzed. Auxin was added 6 h before cell collection. (F) Sister centromere distance was measured for the cells of indicated genotypes. CENP-T was used to mark centromeres, and the distance between paired CENP-T signals was measured for at least 200 of chromosomes 1 and 2. (Middle line) Median, (box) 25th and 75th percentiles, (bars) 5th and 95th percentiles. P-values were calculated by Student's t-test and plotted.
We further noted that CTF18 and DDX11 deficiency in WAPL-depleted cells decreases the frequency of tight and twisted, visually “unseparated” chromosomes characteristic of WAPL-depleted cells (Fig. 6C; see Fig. 5C; Kawasumi et al. 2017). We thus asked whether such mutual suppression between wapl and ctf18 ddx11 is relevant for the mitotic bridges that are strongly increased in wapl cells. This was indeed the case (Fig. 6E). Next, we examined whether the extreme tightness of sister chromatids in WAPL-depleted cells is improved by loss of CTF18 and DDX11. To this end, we measured the sister centromere proximity in different genotypes by assessing the CENP-T distance. ctf18 ddx11 cells show increased sister centromere distance in comparison with control cells, in line with centromeric SCC defects, whereas WAPL-depleted cells have sister centromeres in very tight proximity (Fig. 6F). Strikingly, ctf18 ddx11 wapl mutants had sister centromere distances similar to control cells, indicating that CTF18-DDX11 and WAPL act in opposing manners on chromosome structure. The results thus indicate that chromosome structure is regulated through an exquisite balance between positive and negative regulation of sister chromatid cohesion and resolution.
DDX11-CTF18 and WAPL have opposite effects on the stable association of cohesin with chromatin
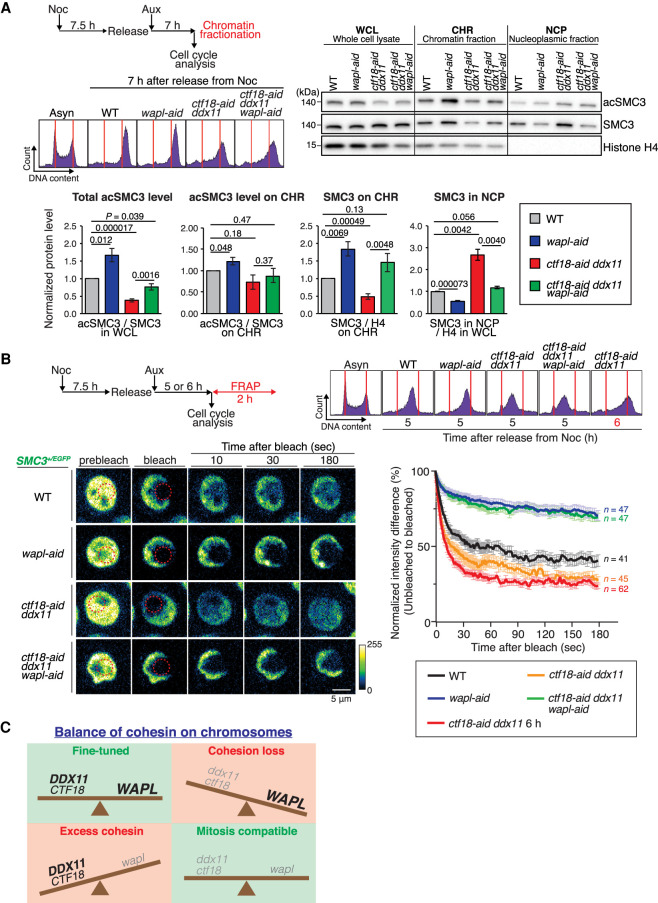
The above results could be most simply explained by CTF18 and DDX11 functioning in two different pathways that positively influence the levels of cohesin on chromatin, in manners opposite from WAPL. We probed this hypothesis by analyzing the relative levels of cohesin and its acetylated form in chromatin and nucleoplasmic fractions versus whole-cell lysates. We used mitotic cells synchronously released from nocodazole arrest in the presence of auxin to manifest different genotypes and analyzed the relative levels of SMC3 and its acetylation in different fractions (Fig. 7A). The relative acSMC3 level versus SMC3 in whole-cell lysates was decreased in ctf18-aid ddx11 cells (see also Fig. 3D) and rescued by WAPL depletion. However, the relative ratio of acSMC3 on chromatin was not significantly altered across the mutants (Fig. 7A). Instead, we observed striking differences on the levels of SMC3 associated with chromatin, which was increased in wapl-aid, decreased in ctf18-aid ddx11, and rescued in ctf18-aid ddx11 wapl-aid to levels comparable with WT cells (Fig. 7A). As expected, the relative amount of cohesin on chromatin showed an inverse pattern with the one in nucleoplasmic fractions in all mutants, being increased in ctf18-aid ddx11 and decreased in wapl-aid, whereas ctf18-aid ddx11 wapl-aid cells showed levels comparable with WT cells (Fig. 7A).
Figure 7.
WAPL and DDX11-CTF18 have opposite effects on cohesin dynamics and cohesin levels on chromatin. (A) Analysis of relative levels of SMC3 and its acetylated form in whole-cell lysates and chromatin and nucleoplasmic fractions of WT, wapl-aid, ctf18-aid ddx11, and ctf18-aid ddx11 wapl-aid cells. Scheme of experimental flow and representative cell cycle profiles are shown. Mean values and SDs of three independent experiments showing ratios of acSMC3 versus SMC3 in whole-cell lysates, chromatin fractions, and the ratio of SMC3 versus histone H4 on chromatin and nucleoplasmic fractions. P-values were calculated by Student's t-test and indicated. (B) Graph plotting the difference in fluorescence intensity between unbleached and bleached regions against time (mean values and SEMs). Cells of the indicated genotypes, expressing SMC3-EGFP from the SMC3 promoter, were synchronized as indicated in the scheme, and cells were collected for FRAP and cell cycle analysis. The number (n) of cells analyzed for each genotype and condition is indicated. (C) Schematic model of CTF18 and DDX11 action in counterbalancing WAPL-mediated cohesin release to allow for proper sister chromatid cohesion and mitotic chromosome structure.
We next examined the cohesin dynamics using fluorescence recovery after photobleaching (FRAP) to have insight into the kinetics of cohesin diffusion on chromatin as a measure of the strength of the interaction. To this purpose, we tagged one SMC3 allele with C-terminal EGFP in all relevant backgrounds and used a similar synchronization procedure of cells to collect cells in late S–G2 for FRAP analysis (Fig. 7B). The FRAP results confirmed very stable chromatin association of cohesin in wapl cells and revealed less stable chromatin binding of cohesin to chromatin in the double mutant ctf18-aid ddx11 in comparison with WT cells. This trend was observed at different times upon release when ctf18-aid ddx11 cells were enriched in S phase (5 h) or strongly enriched at S–G2 (6 h). Notably, concomitant depletion of WAPL stabilized cohesin on chromatin in ctf18-aid ddx11 mutant cells to the levels observed in wapl single mutant (Fig. 7B).
Altogether, the results confirmed that CTF18 and DDX11 function in two different pathways that control the levels of cohesin on chromatin.
Discussion
Establishment of sister chromatid cohesion is linked in space and time to replication fork progression (Terret et al. 2009). Cohesin acetylation by Eco1 and ESCO1/2 acetyltransferases is important for cohesion establishment, but other transactions may be required. SCC establishment has been modeled in vitro using recombinant fission yeast cohesin, and it appears to critically require the cohesin loader, ssDNA, and DNA synthesis (Murayama et al. 2018). Several SCC regulators that associate with the replisome have been identified in budding yeast (Lengronne et al. 2006), and further genetic dissection separated these factors in two different classes based on their genetic interactions with the cohesin loader Scc2 and with each other (Xu et al. 2007; Srinivasan et al. 2020). The replication fork mediators have multiple roles in replication-associated processes (Crabbé et al. 2010; García-Rodríguez et al. 2015; Abe et al. 2018a), and the importance of their function in cohesion toward cellular proliferation and genome integrity remains unclear (Stokes et al. 2020).
Here we started out by investigating the functions of CTF18-RFC versus DDX11, both associated with DNA repair and implicated in human disease (Gellon et al. 2011; Price et al. 2013; Abe et al. 2018b; van Schie et al. 2020; Jegadesan and Branzei 2021). We found that in avian DT40 cells, CTF18 loss causes negligible defects, but combined deficiency in CTF18 and DDX11 genes causes synthetic lethality due to severely compromised SCC and chromosome missegregation. Of interest, chromosome replication is not affected, indicating that their joint SCC function is not required for bulk chromosome replication. Although underreplication of centromeric regions cannot be formally excluded, the suppression of ctf18 ddx11 lethality by the removal of WAPL indicates that the core problem in ctf18 ddx11 cells is related to cohesin levels on chromatin and cohesion deficiency (Fig. 7C).
While most models of SCC establishment invoke a critical role for cohesin acetylation (Borges et al. 2013; Liu et al. 2020), our analyses with ESCO1/2 overexpression, cohesin dynamics, and assessment of cohesin and its acetylated form in chromatin fractions indicate that CTF18 and DDX11 engender cohesion in a manner largely uncoupled from this modification. Importantly, the essential function of CTF18 and DDX11 in cohesion is linked to low levels of chromatin-bound cohesin in their absence that can be bypassed by WAPL depletion in vertebrate cells, indicating that their joint primary role is to allow sufficient chromatin-loaded cohesin amounts necessary for cohesion in the presence of WAPL-mediated release (Fig. 7C).
Interestingly, we found that the cohesion build-up function of CTF18 and DDX11 also yields the increased apparent condensation status and twisted aspect of chromosomes in wapl mutants. Recent work has shown that cohesin can have a negative impact by generating DNA topological stress in specific chromosomal locations, most prominently at centromeres in budding yeast (Minchell et al. 2020). In mitosis, Top2 associates with chromosomes to facilitate chromosome disentanglement of catenanes, which tend to accumulate at centromeric and pericentromeric regions (Bachant et al. 2002). The presence of high levels of cohesin on chromosomes in wapl cells (Fig. 7C) will probably cause Top2 (TOP2α in vertebrate cells) to catenate and entangle the chromosomes (Sen et al. 2016), rather than facilitate their disentanglement, being likely responsible, together with cohesin (Tedeschi et al. 2013; Kawasumi et al. 2017), for the twisted aspect of chromosomes of WAPL-depleted cells.
We propose that fine-tuning the levels of chromatin-bound cohesin is critical for allowing the right balance between cohesion and mitotic condensation of vertebrate chromosomes (Fig. 7C). We identified that CTF18 and DDX11 are essential to engender cohesion by counterbalancing WAPL and allowing optimal levels of chromatin-associated cohesin on chromosomes. While cohesin acetylation counteracts WAPL-mediated cohesin release (Nishiyama et al. 2010; Peters and Nishiyama 2012; Elbatsh et al. 2016), CTF18 and DDX11 complementary function is not essential for directing cohesin acetylation but to provide sufficient chromatin-bound cohesin amounts in the presence of WAPL-mediated cohesin-unloading activity.
Materials and methods
The cell lines are listed in the Supplemental Table S1. The constructs for knock-in and knockout are described in the Supplemental Material. Total RNA isolation, RT-PCR, immunofluorescence analysis, and sister chromatid cohesion analysis were performed as described (Abe et al. 2016). DNA fiber assay was performed as described by Abe et al. (2018a). Yeast tetrad dissection was performed following standard procedures (Sherman 2002). Yeast strains used in this study are described in the Supplemental Table S2.
Cell culture techniques and cell viability/drug sensitivity assays
DT40 cells were cultured at 39.5°C in D-MEM/F-12 medium (Gibco 31331093) supplemented with 10% fetal bovine serum (Life Technologies 10270-106), 2% chicken serum (Euroclone ECS0050D), penicillin/streptomycin mix (Euroclone ECB3001), and 2-mercaptoethanol (Life Technologies 31350-010). Medium was treated with 500 µM auxin (Sigma I5148), or 1 µg/mL doxycycline (Sigma D9891), when required. Nocodazole (Sigma M1404) was used at 100 ng/mL for metaphase arrest. Drugs used for sensitivity assay were hydroxyurea (Sigma H8627), aphidicolin (Sigma A0781), nocodazole (Sigma M1404), and CFI-402257 (Focus Bioscience HY-101340-1MG). Growth curves and drug sensitivity assays are described in the Supplemental Material.
Western blotting
Whole-cell extracts were prepared by lysing cells directly with 1× Laemmli buffer (50 mM Tris-HCl, 2% SDS, 10% glycerol, 100 mM dithiothreitol, 2.5 mg/mL bromophenol blue) and boiled for 10 min. Protein samples were separated by electrophoresis on Bolt 4%–12% gradient precast gel (Invitrogen NW04125BOX) with MOPS SDS running buffer (Invitrogen NP0001). Proteins were then transferred to PVDF membrane (Amersham GE10600023) using 1× transfer buffer (25 mM Tris-HCl, 192 mM glycine, 20% methanol, 0.01% SDS) at 100 V for 90 min on ice. After transfer, membranes were saturated by PBS-T (0.05% Tween 20) containing 5% skim milk for 30 min. After washing by PBS-T, membranes were then incubated with primary antibodies overnight at 4°C. Membranes were washed by PBS-T and then incubated with secondary antibodies for 40 min at room temperature. After washing by PBS-T, protein signals were detected by either SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific 34095) or SuperSignal West Dura extended duration substrate (Thermo Scientific 34076) using the Chemidoc XRS+ system (Bio-Rad). Primary antibodies used were as follows: Myc (1:3000; homemade), HA (1:500; Roche 11867423001), miniAID (1:1000; MBL M214-3), α-tubulin (1:1000; Santa Cruz Biotechnology sc-5286), FLAG (1:3000; Sigma F3165), PCNA (1:1000; Santa Cruz Biotechnology sc-56), SMC3 (1:1000; a gift from Dr. Losada), Ac-SMC3 (1:1000; a gift from Dr. Shirahige), and CTF18 (1:1000; Santa Cruz Biotechnology sc-374632). Secondary antibodies used were as follows: horseradish peroxidase–conjugated antirabbit IgG (1:3000; Cell Signaling 7074S), antirat IgG (1:3000; Cell Signaling 7077S), and antimouse IgG (1:3000; Cell Signaling 7076S).
For the chromatin binding assay performed in Figure 1E, cells were lysed with CSK buffer (0.3% Triton X-100, 10 mM PIPES, 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 1 mM EDTA, 1× Complete) for 10 min on ice, and then the chromatin fraction was isolated by centrifugation at 500g for 5 min at 4°C. For the chromatin fractionation assay performed in Figures 3D and 7A and Supplemental Figure S3D for SMC3, a subcellular protein fractionation kit for cultured cells (Thermo Scientific 78840) was used following the manufacturer's protocol.
Cell cycle analysis by flow cytometry
Cells were cultured in the presence of BrdU (final: 20 µM; Sigma B5002) for 15 min and fixed in ice-cold 70% EtOH for >1 h. Cells were then treated with 2.5 N HCl to denature DNA for 30 min. After washing with PBS containing 1% BSA, cells were incubated with anti-BrdU antibody (1:5; BD Biosciences 347580) and stained with FITC-conjugated antimouse IgG antibody (1:50; Jackson ImmunoResearch 115-095-003) and propidium iodide (final: 5 µg/mL; Sigma P4170) in the presence of RNase A (final: 0.1 mg/mL; Sigma R5503). For PI single-staining analysis without BrdU incorporation, cells were fixed in 70% ethanol and stained with propidium iodide in the presence of RNase A.
FRAP assay
The FRAP assay was performed with adaptations from Huis in ’t Veld et al. (2014) and Chan et al. (2012) and are described in the Supplemental Material. Cells were synchronized by 100 ng/mL nocodazole for 7.5 h and then released into medium containing 0.5 mM of auxin. Five hours or 6 h after release, 40 µL of cells was transferred into 15 µ-slide angiogenesis ibiTreat chamber slides (ibidi 81506), and remaining cells were fixed by 70% EtOH for cell cycle analysis by propidium iodide staining. During imaging, cells were cultured in normal medium at 39.5°C and 5% CO2.
Data sets
The research data set files are available at Mendeley Data (doi:10.17632/fd89yy4ms5.1).
Supplementary Material
Acknowledgments
We thank A. Losada, T. Fukagawa, and K. Shirahige for antibodies; B. Szakal, C. Reyes, and other Branzei team members for sharing unpublished reagents; and Rika Rifana Sari (Tokyo Metropolitan University), Francesca Casagrande, and International Foundation of Medicine Imaging facility for technical help. This work was supported by the Italian Association for Cancer Research (AIRC; IG 18976 and IG 23710) and European Research Council (consolidator grant 682190) grants to D.B. and Japanese Society for the Promotion of Science KAKENHI (17K17986) to T.A. T.A. was partly supported by the Structured International Post Doc Program (SIPOD) fellowship cofunded in the context of the FP7 Marie Curie Actions-People. R.K. was partly supported by an AIRC 3-yr fellowship Mario e Valeria Rindi (Rif.22403). I.P. was partly supported by EMBO long-term fellowship ALTF 561-2014 and an AIRC/Marie Curie Actions-COFUND iCARE fellowship.
Authors contributions: D.B., R.K., and T.A. conceived the study. R.K., T.A., I.P., and K.M. performed experiments. R.K., T.A., I.P., K.H., and D.B. analyzed the data. R.K. and T.A. constructed the figures. D.B., R.K., T.A., and I.P. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348581.121.
Competing interest statement
The authors declare no competing interest.
References
- Abe T, Branzei D. 2014. High levels of BRC4 induced by a Tet-On 3G system suppress DNA repair and impair cell proliferation in vertebrate cells. DNA Repair 22: 153–164. 10.1016/j.dnarep.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T, Kawasumi R, Arakawa H, Hori T, Shirahige K, Losada A, Fukagawa T, Branzei D. 2016. Chromatin determinants of the inner-centromere rely on replication factors with functions that impart cohesion. Oncotarget 7: 67934–67947. 10.18632/oncotarget.11982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T, Kawasumi R, Giannattasio M, Dusi S, Yoshimoto Y, Miyata K, Umemura K, Hirota K, Branzei D. 2018a. AND-1 fork protection function prevents fork resection and is essential for proliferation. Nat Commun 9: 3091. 10.1038/s41467-018-05586-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T, Ooka M, Kawasumi R, Miyata K, Takata M, Hirota K, Branzei D. 2018b. Warsaw breakage syndrome DDX11 helicase acts jointly with RAD17 in the repair of bulky lesions and replication through abasic sites. Proc Natl Acad Sci 115: 8412–8417. 10.1073/pnas.1803110115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell 9: 1169–1182. 10.1016/S1097-2765(02)00543-9 [DOI] [PubMed] [Google Scholar]
- Beckouët F, Srinivasan M, Roig MB, Chan KL, Scheinost JC, Batty P, Hu B, Petela N, Gligoris T, Smith AC, et al. 2016. Releasing activity disengages cohesin's Smc3/Scc1 interface in a process blocked by acetylation. Mol Cell 61: 563–574. 10.1016/j.molcel.2016.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender D, Da Silva EML, Chen J, Poss A, Gawey L, Rulon Z, Rankin S. 2020. Multivalent interaction of ESCO2 with the replication machinery is required for sister chromatid cohesion in vertebrates. Proc Natl Acad Sci 117: 1081–1089. 10.1073/pnas.1911936117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. 2008. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566. 10.1126/science.1157774 [DOI] [PubMed] [Google Scholar]
- Berkowitz KM, Sowash AR, Koenig LR, Urcuyo D, Khan F, Yang F, Wang PJ, Jongens TA, Kaestner KH. 2012. Disruption of CHTF18 causes defective meiotic recombination in male mice. PLoS Genet 8: e1002996. 10.1371/journal.pgen.1002996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V, Smith DJ, Whitehouse I, Uhlmann F. 2013. An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae. Chromosoma 122: 121–134. 10.1007/s00412-013-0396-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calì F, Bharti SK, Di Perna R, Brosh RM Jr., Pisani FM. 2016. Tim/Timeless, a member of the replication fork protection complex, operates with the Warsaw breakage syndrome DNA helicase DDX11 in the same fork recovery pathway. Nucleic Acids Res 44: 705–717. 10.1093/nar/gkv1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Roig MB, Hu B, Beckouët F, Metson J, Nasmyth K. 2012. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell 150: 961–974. 10.1016/j.cell.2012.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortone G, Zheng G, Pensieri P, Chiappetta V, Tatè R, Malacaria E, Pichierri P, Yu H, Pisani FM. 2018. Interaction of the Warsaw breakage syndrome DNA helicase DDX11 with the replication fork-protection factor timeless promotes sister chromatid cohesion. PLoS Genet 14: e1007622. 10.1371/journal.pgen.1007622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbé L, Thomas A, Pantesco V, De Vos J, Pasero P, Lengronne A. 2010. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol 17: 1391–1397. 10.1038/nsmb.1932 [DOI] [PubMed] [Google Scholar]
- Dauban L, Montagne R, Thierry A, Lazar-Stefanita L, Bastié N, Gadal O, Cournac A, Koszul R, Beckouët F. 2020. Regulation of cohesin-mediated chromosome folding by Eco1 and other partners. Mol Cell 77: 1279–1293.e4. 10.1016/j.molcel.2020.01.019 [DOI] [PubMed] [Google Scholar]
- de Lange J, Faramarz A, Oostra AB, de Menezes RX, van der Meulen IH, Rooimans MA, Rockx DA, Brakenhoff RH, van Beusechem VW, King RW, et al. 2015. Defective sister chromatid cohesion is synthetically lethal with impaired APC/C function. Nat Commun 6: 8399. 10.1038/ncomms9399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbatsh AMO, Haarhuis JHI, Petela N, Chapard C, Fish A, Celie PH, Stadnik M, Ristic D, Wyman C, Medema RH, et al. 2016. Cohesin releases DNA through asymmetric ATPase-driven ring opening. Mol Cell 61: 575–588. 10.1016/j.molcel.2016.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faramarz A, Balk JA, van Schie JJM, Oostra AB, Ghandour CA, Rooimans MA, Wolthuis RMF, de Lange J. 2020. Non-redundant roles in sister chromatid cohesion of the DNA helicase DDX11 and the SMC3 acetyl transferases ESCO1 and ESCO2. PLoS One 15: e0220348. 10.1371/journal.pone.0220348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Shin JH, Kim DH, Bermudez VP, Kelman Z, Seo YS, Hurwitz J. 2008. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J Biol Chem 283: 20925–20936. 10.1074/jbc.M802696200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumasoni M, Zwicky K, Vanoli F, Lopes M, Branzei D. 2015. Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the polα/primase/Ctf4 complex. Mol Cell 57: 812–823. 10.1016/j.molcel.2014.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodríguez LJ, De Piccoli G, Marchesi V, Jones RC, Edmondson RD, Labib K. 2015. A conserved polɛ binding module in Ctf18-RFC is required for S-phase checkpoint activation downstream of Mec1. Nucleic Acids Res 43: 8830–8838. 10.1093/nar/gkv799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon L, Razidlo DF, Gleeson O, Verra L, Schulz D, Lahue RS, Freudenreich CH. 2011. New functions of Ctf18-RFC in preserving genome stability outside its role in sister chromatid cohesion. PLoS Genet 7: e1001298. 10.1371/journal.pgen.1001298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo M, Vega H, Trainer AH, Hou F, Sakai N, Luque R, Kayserili H, Basaran S, Skovby F, Hennekam RC, et al. 2008. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum Mol Genet 17: 2172–2180. 10.1093/hmg/ddn116 [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D. 2011. Cohesin-independent segregation of sister chromatids in budding yeast. Mol Biol Cell 23: 729–739. 10.1091/mbc.E11-08-0696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA. 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21: 3144–3158. 10.1128/MCB.21.9.3144-3158.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi TL, Ikeda M, Tanaka H, Nakagawa T, Bando M, Shirahige K, Kubota Y, Takisawa H, Masukata H, Takahashi TS. 2012. The prereplication complex recruits XEco2 to chromatin to promote cohesin acetylation in xenopus egg extracts. Curr Biol 22: 977–988. 10.1016/j.cub.2012.04.013 [DOI] [PubMed] [Google Scholar]
- Holton RA, Harris AM, Mukerji B, Singh T, Dia F, Berkowitz KM. 2020. CHTF18 ensures the quantity and quality of the ovarian reserve. Biol Reprod 103: 24–35. 10.1093/biolre/ioaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono Y, Abe T, Higuchi M, Kajii K, Sakuraba S, Tada S, Enomoto T, Seki M. 2014. Tipin functions in the protection against topoisomerase I inhibitor. J Biol Chem 289: 11374–11384. 10.1074/jbc.M113.531707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in ’t Veld PJ, Herzog F, Ladurner R, Davidson IF, Piric S, Kreidl E, Bhaskara V, Aebersold R, Peters JM. 2014. Characterization of a DNA exit gate in the human cohesin ring. Science 346: 968–972. 10.1126/science.1256904 [DOI] [PubMed] [Google Scholar]
- Inoue A, Li T, Roby SK, Valentine MB, Inoue M, Boyd K, Kidd VJ, Lahti JM. 2007. Loss of ChlR1 helicase in mouse causes lethality due to the accumulation of aneuploid cells generated by cohesion defects and placental malformation. Cell Cycle 6: 1646–1654. 10.4161/cc.6.13.4411 [DOI] [PubMed] [Google Scholar]
- Ivanov MP, Ladurner R, Poser I, Beveridge R, Rampler E, Hudecz O, Novatchkova M, Hériché JK, Wutz G, van der Lelij P, et al. 2018. The replicative helicase MCM recruits cohesin acetyltransferase ESCO2 to mediate centromeric sister chromatid cohesion. EMBO J 37: e97150. 10.15252/embj.201797150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegadesan NK, Branzei D. 2021. DDX11 loss causes replication stress and pharmacologically exploitable DNA repair defects. Proc Natl Acad Sci 118: e2024258118. 10.1073/pnas.2024258118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasumi R, Abe T, Arakawa H, Garre M, Hirota K, Branzei D. 2017. ESCO1/2's roles in chromosome structure and interphase chromatin organization. Genes Dev 31: 2136–2150. 10.1101/gad.306084.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Fujii T, Asada R, Ooka M, Hirota K. 2015. Development of a targeted flip-in system in avian DT40 cells. PLoS One 10: e0122006. 10.1371/journal.pone.0122006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Hiraga S, Yamada K, Lamond AI, Donaldson AD. 2011. Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol Cell Proteomics 10: M110.005561. 10.1074/mcp.M110.005561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Park SH. 2020. Eukaryotic clamp loaders and unloaders in the maintenance of genome stability. Exp Mol Med 52: 1948–1958. 10.1038/s12276-020-00533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman AR, Noguchi C, Lee CY, Noguchi E. 2010. Human timeless and tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci 123: 660–670. 10.1242/jcs.057984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. 2006. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23: 787–799. 10.1016/j.molcel.2006.08.018 [DOI] [PubMed] [Google Scholar]
- Liu HW, Bouchoux C, Panarotto M, Kakui Y, Patel H, Uhlmann F. 2020. Division of labor between PCNA loaders in DNA replication and sister chromatid cohesion establishment. Mol Cell 78: 725–738.e4. 10.1016/j.molcel.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F. 2013. Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol 23: 64–69. 10.1016/j.cub.2012.11.030 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P. 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell 7: 959–970. 10.1016/S1097-2765(01)00254-4 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, et al. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell 15: 1736–1745. 10.1091/mbc.e03-08-0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino M, Ishibashi M, Nakato R, Akiyama K, Tanaka H, Kato Y, Negishi L, Hirota T, Sutani T, Bando M, et al. 2015. Esco1 acetylates cohesin via a mechanism different from that of Esco2. Curr Biol 25: 1694–1706. 10.1016/j.cub.2015.05.017 [DOI] [PubMed] [Google Scholar]
- Minamino M, Tei S, Negishi L, Kanemaki MT, Yoshimura A, Sutani T, Bando M, Shirahige K. 2018. Temporal regulation of ESCO2 degradation by the MCM complex, the CUL4-DDB1-VPRBP complex, and the anaphase-promoting complex. Curr Biol 28: 2665–2672.e5. 10.1016/j.cub.2018.06.037 [DOI] [PubMed] [Google Scholar]
- Minchell NE, Keszthelyi A, Baxter J. 2020. Cohesin causes replicative DNA damage by trapping DNA topological stress. Mol Cell 78: 739–751.e8. 10.1016/j.molcel.2020.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Samora CP, Kurokawa Y, Iwasaki H, Uhlmann F. 2018. Establishment of DNA–DNA interactions by the cohesin ring. Cell 172: 465–477.e15. 10.1016/j.cell.2017.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. 2010. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228. 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, et al. 2010. Sororin mediates sister chromatid cohesion by antagonizing wapl. Cell 143: 737–749. 10.1016/j.cell.2010.10.031 [DOI] [PubMed] [Google Scholar]
- Onn I, Guacci V, Koshland DE. 2009. The zinc finger of Eco1 enhances its acetyltransferase activity during sister chromatid cohesion. Nucleic Acids Res 37: 6126–6134. 10.1093/nar/gkp656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Yu H. 2017. Releasing the cohesin ring: a rigid scaffold model for opening the DNA exit gate by Pds5 and Wapl. Bioessays 39: 1600207. 10.1002/bies.201600207 [DOI] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081. 10.1016/j.cell.2005.12.036 [DOI] [PubMed] [Google Scholar]
- Peters JM, Nishiyama T. 2012. Sister chromatid cohesion. Cold Spring Harbor Perspect Biol 4: a011130. 10.1101/cshperspect.a011130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani FM, Napolitano E, Napolitano LMR, Onesti S. 2018. Molecular and cellular functions of the Warsaw breakage syndrome DNA helicase DDX11. Genes (Basel) 9: 564. 10.3390/genes9110564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Pollock LM, Rudd ML, Fogoros SK, Mohamed H, Hanigan CL, Le Gallo M, NIH Intramural Sequencing Center (NISC) Comparative Sequencing Program, Zhang S, Cruz P, et al. 2013. Sequencing of candidate chromosome instability genes in endometrial cancers reveals somatic mutations in ESCO1, CHTF18, and MRE11A. PLoS One 8: e63313. 10.1371/journal.pone.0063313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, et al. 2009. Building sister chromatid cohesion: Smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33: 763–774. 10.1016/j.molcel.2009.02.028 [DOI] [PubMed] [Google Scholar]
- Sen N, Leonard J, Torres R, Garcia-Luis J, Palou-Marin G, Aragón L. 2016. Physical proximity of sister chromatids promotes Top2-dependent intertwining. Mol Cell 64: 134–147. 10.1016/j.molcel.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. 2002. Getting started with yeast. Methods Enzymol 350: 3–41. 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger IC, Peters JM, et al. 2001. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell 1: 759–770. 10.1016/S1534-5807(01)00088-0 [DOI] [PubMed] [Google Scholar]
- Srinivasan M, Fumasoni M, Petela NJ, Murray A, Nasmyth KA. 2020. Cohesion is established during DNA replication utilising chromosome associated cohesin rings as well as those loaded de novo onto nascent DNAs. Elife 9: e56611. 10.7554/eLife.56611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes K, Winczura A, Song B, De Piccoli G, Grabarczyk DB. 2020. Ctf18-RFC and DNA Polɛ form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication. Nucleic Acids Res 48: 8128–8145. 10.1093/nar/gkaa541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. 2009. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Cur Biol 19: 492–497. 10.1016/j.cub.2009.01.062 [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. 2013. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568. 10.1038/nature12471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. 2009. Cohesin acetylation speeds the replication fork. Nature 462: 231–234. 10.1038/nature08550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M, Lengronne A, Davidson MB, Bacal J, François P, Hohl M, Petrini JH, Pasero P, Cobb JA. 2012. Cohesin association to replication sites depends on Rad50 and promotes fork restart. Mol Cell 48: 98–108. 10.1016/j.molcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569. 10.1126/science.1157880 [DOI] [PubMed] [Google Scholar]
- van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, Zdzienicka MZ, Joenje H, de Winter JP. 2010. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J H Genet 86: 262–266. 10.1016/j.ajhg.2010.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie JJM, Faramarz A, Balk JA, Stewart GS, Cantelli E, Oostra AB, Rooimans MA, Parish JL, de Almeida Estéves C, Dumic K, et al. 2020. Warsaw breakage syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nat Commun 11: 4287. 10.1038/s41467-020-18066-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Boone C, Brown GW. 2007. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics 176: 1417–1429. 10.1534/genetics.107.072876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatskevich S, Rhodes J, Nasmyth K. 2019. Organization of chromosomal DNA by SMC complexes. Annu Rev Genet 53: 445–482. 10.1146/annurev-genet-112618-043633 [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. 2008. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31: 143–151. 10.1016/j.molcel.2008.06.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.