Abstract
The tumor immune microenvironment is a main contributor to cancer progression and a promising therapeutic target for oncology. However, immune microenvironments vary profoundly between patients, and biomarkers for prognosis and treatment response lack precision. A comprehensive compendium of tumor immune cells is required to pinpoint predictive cellular states and their spatial localization. We generated a single-cell tumor immune atlas, jointly analyzing published data sets of >500,000 cells from 217 patients and 13 cancer types, providing the basis for a patient stratification based on immune cell compositions. Projecting immune cells from external tumors onto the atlas facilitated an automated cell annotation system. To enable in situ mapping of immune populations for digital pathology, we applied SPOTlight, combining single-cell and spatial transcriptomics data and identifying colocalization patterns of immune, stromal, and cancer cells in tumor sections. We expect the tumor immune cell atlas, together with our versatile toolbox for precision oncology, to advance currently applied stratification approaches for prognosis and immunotherapy.
Single-cell RNA sequencing (scRNA-seq) techniques are powerful tools for the unbiased charting of cellular phenotypes (Lafzi et al. 2018). Analyzing transcriptome profiles of individual cells enables the fine-grained annotation of cell types and cellular states, as well as charting the composition of complex samples. Interrogating healthy tissues at single-cell resolution provides a reference atlas of normal tissue organization (Regev et al. 2017) and defines variability across individuals (van der Wijst et al. 2018) or during development (Asp et al. 2019; Popescu et al. 2019; Park et al. 2020) and aging (Salzer et al. 2018; The Tabula Muris Consortium 2020). Diseased tissues display an additional layer of complexity, presenting cell type composition shifts and newly emerging disease-specific phenotypes (Ramachandran et al. 2019; Vieira Braga et al. 2019; Chua et al. 2020). In cancer, in addition to diverse neoplastic cell states (Patel et al. 2014; Tirosh et al. 2016b), the remodeling of the host tissue microenvironment has been characterized using scRNA-seq (Tirosh et al. 2016a; Puram et al. 2017). Phenotyping single cells from the tumor microenvironment (TME) has led to the identification of cancer-specific stromal cell states and supported their contribution to tumor progression. Functional and integrative analysis further support dependencies of stromal and cancer cells and their predictive value for patient outcome. In particular, cancer-associated fibroblasts (CAFs) (Merlos-Suárez et al. 2011; Calon et al. 2015) and tumor-resident immune cells (Fridman et al. 2012; Yofe et al. 2020) have been identified as biomarkers for patient stratification and, importantly, as an effective target for therapeutic intervention in oncology (Tumeh et al. 2014; Tauriello et al. 2018). Clinically most advanced, immune cells are now the target of immunotherapy (e.g., immune checkpoint inhibitors, ICI), stimulating the immune system to recognize and destroy cancer cells.
Single-cell transcriptomics has critically advanced our understanding of immune cell diversity in the TME by generating high-resolution landscapes of different cancer types. Combined with T cell receptor (TCR) genotyping and receptor-ligand screening (Mimitou et al. 2019), scRNA-seq identified transient immune cell states and dynamic tissue remodelling. Seminal studies include the scRNA-seq-based immunophenotyping of breast cancer, describing a tumor-specific heterogeneity and expansion of T cell states, supporting a continuous cell activation toward terminal exhaustion, rather than discrete cellular states (Azizi et al. 2018). In melanoma, similar dynamics were shown to activate transitional and dysfunctional T cell states (Li et al. 2019). Moreover, single-cell immunophenotyping identified tumor-specific T cell states, which are predictive for ICI-based therapy outcome (Sade-Feldman et al. 2018). A lung cancer study identified an immune activation module characterized through high frequencies of PDCD1+ CXCL13+ activated T cells, IgG+ plasma cells, and SPP1+ macrophages, introducing the concept of an immune cell composition-based patient stratification to complement current genomic (e.g., mutational load) or biomarker (e.g., CD274 or CTLA4) strategies (Leader et al. 2020).
In more general terms, single-cell sequencing-based immunophenotyping identified cancer-specific states and composition biases across all major immune cell types that colocalize with cancer cells (Qian et al. 2020). Major alterations have been described for T cells (Yost et al. 2019), B cells (Helmink et al. 2020), tumor-associated macrophages (TAMs) (Lee et al. 2020), and dendritic cells (Lavin et al. 2017), suggesting a global perturbation of the immune system in cancer. Comprehensively understanding the causes and consequences of perturbed immune cell function across cancer types could provide the basis to identify novel therapeutic targets and could lay the groundwork for an immune-based patient stratification in precision oncology. Current ICI stratification strategies involve the assessment of the tumor mutational burden or inflammation signatures. However, despite having favorable immune profiles, many tumors do not respond to treatment, suggesting additional mechanisms that confer resistance to therapy. In this regard, the spatial distribution of immune cells has proven to be important for ICI response, with excluded tumors blocking effective immune cell action, despite the presence of favorable cell types at their boundaries (Chen and Mellman 2017). In contrast, tumors invaded by reactive immune cells that clonally expand have shown increased response rates. However, current spatial immune profiling approaches enable only targeted profiling of mRNAs or proteins. On the other hand, the latest spatial transcriptomics (ST) techniques (Ståhl et al. 2016; Rodriques et al. 2019) provide unbiased transcriptome-wide profiles but average gene expression profiles of multiple cells.
More broadly, it remains challenging to generalize findings across cancer types. There are no standardized analysis pipelines and annotation systems, resulting in data sets characterized by varying granularity and nomenclature. Cell annotation is especially challenging when describing novel phenotypes (e.g., cancer-specific cell states), leading to discrepancies in cell labels between studies. In this work, we generate a consensus atlas of the tumor immune microenvironment through the integration of published scRNA-seq data sets from 13 cancer types for a harmonized and automated cell annotation, as an input for patient stratification and to spatially localize immune cell populations in tumor sections.
Results
Generating a tumor immune cell atlas
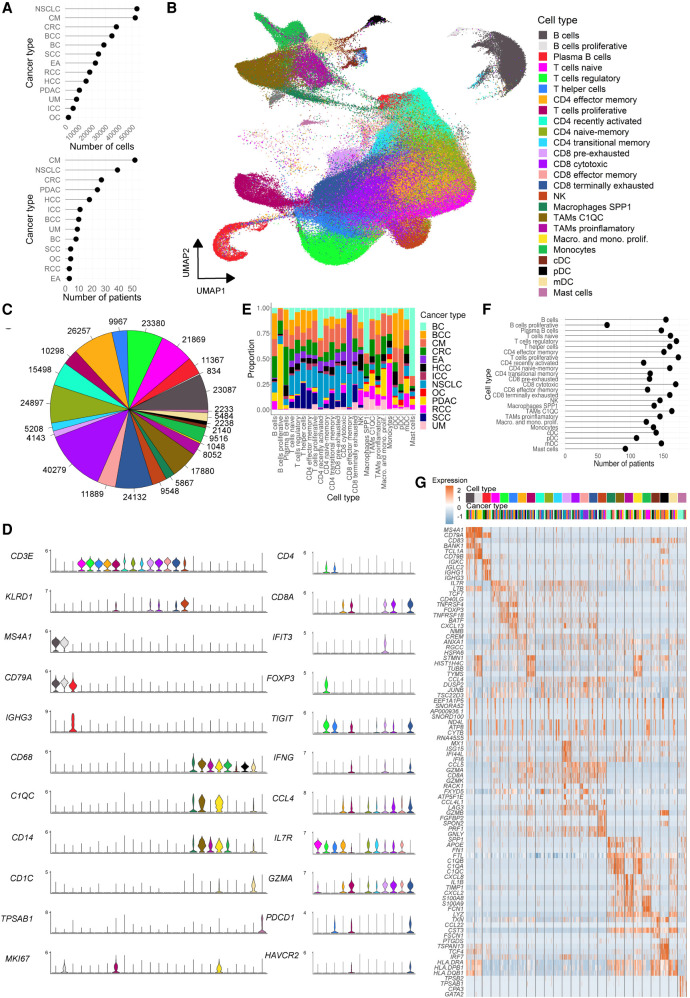
Single-cell transcriptome profiling of tumors provides an unbiased overview of the heterogeneity of cancer cells and their microenvironment. Following sample dissociation, single cells enter scRNA-seq processes either directly or following the enrichment of specific cell types (e.g., tumor or PTPRC+ immune cells). To generate a comprehensive tumor immune cell atlas of human cancers, we collected scRNA-seq data sets from 13 different cancer types, 217 patients, and 526,261 cells. In detail, we processed data from breast carcinomas (BC) (Azizi et al. 2018), basal cell and squamous cell carcinomas (BCC) (Yost et al. 2019), endometrial adeno- (EA) and renal cell carcinomas (RCC) (Wu et al. 2020), intrahepatic cholangio- (ICC) and hepatocellular carcinomas (HCC) (Ma et al. 2019; Zhang et al. 2019), colorectal cancers (CRC) (Lee et al. 2020; Wu et al. 2020), pancreatic ductal adenocarcinomas (PDAC) (Peng et al. 2019), ovarian cancers (OC) (Schelker et al. 2017), non-small-cell lung cancers (NSCLC) (Lavin et al. 2017; Lambrechts et al. 2018; Wu et al. 2020), and cutaneous (CM) and uveal (UM) melanomas (Supplemental Table 1; Fig. 1A; Sade-Feldman et al. 2018; Li et al. 2019; Durante et al. 2020). For a cell type stratification and consistent annotation, the data sets were analyzed separately before joining immune cells into a pan-cancer tumor immune reference atlas (Supplemental Figs. 1, 2). In line with previous findings, immune cells clustered by cell identity rather than patient origin, allowing the straightforward subsetting of the immune compartment.
Figure 1.
Characterization of the tumor immune cell atlas. (A) Number of cells (top) and patients (bottom) per cancer type included in the atlas. (B) UMAP of 317,111 immune cells from 13 cancer types colored by annotated cell type. (C) Total number of cells of each immune cell type/state; color code as in A. (D) Marker gene expression levels for broader cell types (left) and only T cells states (right); color code as in A. (E) Cancer type proportions for each annotated cell type/state. (F) Number of unique patients representing each cell type/state in the atlas. (G) Expression of the top four differentially expressed genes per cell type/state; colored by cell type (as in A) and cancer type (as in E).
We hypothesize that joining cells from different cancer types into a single reference data set may define commonalities and harmonize annotations between studies. To this end, we integrated 317,111 immune cells using canonical correlation analysis (Fig. 1B; Butler et al. 2018). This approach identifies common cellular phenotypes and allows merging data sets from different studies and technologies for joint analyses. Data integration with alternative tools resulted in comparable results, though with reduced iLISI integration score (Korsunsky et al. 2019) and data set-specific technical clusters (Supplemental Fig. 3). Following integration, cells separated into 25 clusters representing major immune cell types, including 12 T cell, five macrophage/monocyte, three dendritic cell (DC), three B- and plasma B cell, one natural killer (NK), and one Mast cell cluster (Fig. 1B,C). To test the robustness of the clusters and their associated signatures, we trained a random forest (RF) classifier to predict cell annotation based on the 25 signatures and performed a fivefold cross-validation to assess biases and variance. The mean accuracy and kappa statistic across folds were 0.76 (sd = 0.0048) and 0.75 (sd = 0.0050), respectively, a 3.0-fold and 4.2-fold increase with respect to random signatures and comparable to values obtained in other high-quality atlases (Supplemental Fig. 4A,B; Zeisel et al. 2018). In addition, both the interquartile range (IQR) and the range of accuracies were lower when using cell type–specific signatures, suggesting a low variance across test sets. Finally, stratifying the accuracies by cell type revealed that the low degree of misclassification corresponded to cell types within the same lineage, which share several markers (Supplemental Fig. 4C). Taken together this suggests that the clustering of the atlas displays an optimal balance between granularity and robustness. Of note, deeper clustering (31 clusters) resulted in a more fine-grained annotation of mainly the B cells, with subpopulations representing naive, activated, memory, unswitched memory, and proliferative subtypes (Supplemental Fig. 5).
Clusters were manually annotated using canonical markers and curated gene signatures that defined their identities (Supplemental Tables 2, 3). Exemplarily, CD4 T cells representing regulatory, helper, and naive states were defined by FOXP3, CXCL13, and SELL marker gene expression, respectively (Fig. 1D; Supplemental Fig. 6). Cytotoxic CD8 T cells expressed high levels of GZMA, whereas their pre- and terminally exhausted subtypes expressed IFIT3 and HAVCR2, respectively. Macrophages split in different subtypes, such as C1QC, SPP1, and pro-inflammatory states, with C1QC, SPP1, and CXCL8 as respective marker genes. We further observed actively dividing lymphoid and myeloid cell types with MKI67 and STMN1 as common markers for proliferation. Plasma B cells specifically expressed IGKC, which distinguished them from naive/memory B cells expressing MS4A1. The large number of immune cells enabled the identification of rare cell states, such as proliferative B cells (834 cells) and macrophages (1048 cells) as well as plasmacytoid and conventional DC populations (2140 and 2238) (Fig. 1C). All cell types were present in multiple cancer types and patients (Fig. 1E,F), which supports the robustness of the integration approach. Our analyses revealed that several cell states such as naive, proliferative, transitional memory and terminally exhausted T cell subtypes, and all macrophage states were abundant in all tumors, highlighting common mechanisms that operate in the TME independently of the tissue of origin and mutational background. Yet, we also found cell states enriched in specific cancer types, suggesting cancer-specific immune environments (Fig. 1E). In particular, effector memory CD8 T cells were frequently found in EA and NSCLC, proliferating B cells were very abundant in PDAC, whereas Mast cells were mainly detected in BC. In summary, we built a catalog of immune cell types and state markers present in the TME of multiple cancer types (Fig. 1G; Supplemental Table 4). This resource may help annotate future single-cell tumor data sets.
Tumor stratification by immune cell composition
Tumors of the same cancer type have been described to be heterogeneous, presenting distinct genetic and epigenetic alterations as well as gene expression signatures, allowing their stratification into subtypes. Cancer subtypes have a clear impact on clinical management, being predictive for patient prognosis and therapy response (e.g., consensus molecular subtypes in CRC [CMS]) (Guinney et al. 2015). To date, gene expression subtypes are defined through the analysis of bulk tumor samples and are frequently characterized by gene signatures of stromal or immune cell types (Calon et al. 2015). Single-cell resolved tumor maps confirmed this contribution of the TME to the subtype classification and allowed an even more fine-grained interpretation of subtype composition (Lee et al. 2020).
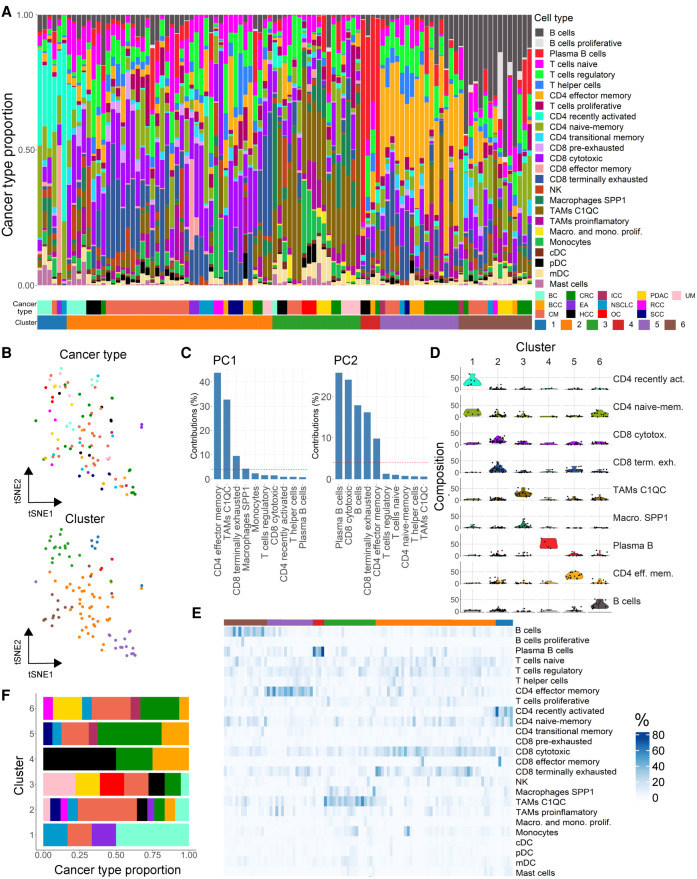
Detecting varying immune cell compositions within cancer types but conserved profiles across cancers, we sought to establish a pan-cancer immune classification system. We used immune cell type and state frequencies of the reference atlas as input for similarity assessment across the 13 cancer types (Fig. 2A). Samples of different tumor types intermixed, and cancer type was not a main source of variance as shown in a t-distributed stochastic neighbor embedding (t-SNE) of immune cell type proportions (Fig. 2B). In contrast, abundance of CD4 effector memory T cells explained most variance in the data set (principal component 1, 43.7%), followed by the relative frequencies of C1QC TAMs and terminally exhausted CD8 T cells (32.6% and 9.5%, respectively) (Fig. 2C).
Figure 2.
Patient stratification based on the tumor immune cell composition. (A) Cell type composition of patients colored by cell type/state frequencies. Patients are clustered (C1–6) into groups of similar cell type composition. Cancer and cluster identities are indicated below. (B) Dimensionality reduction representation (t-SNE distribution) of cell type frequencies in cancer patients colored by cancer type (top) and cluster identity (bottom). (C) Variance contribution to the two first principal components (PCs) of the top variable cell types. (D) Frequencies (% total cells) of cell types representative for cluster 1–6. (E) Heat map representation of cell type frequencies within each cluster. (F) Cancer type contribution to the six immune clusters; color code as cancer types in A.
Correlating immune cell population frequencies identified an intriguing pattern of tumors being mutually exclusive for the presence of lymphoid or myeloid cell types (Supplemental Figs. 7, 8A). Moreover, lymphoid-enriched tumors presented either naive-memory, effector-memory and recently activated profiles, or more differentiated (regulatory, cytotoxic, and exhausted) phenotypes. Myeloid-enriched tumors further split into pro-inflammatory and inhibitory (C1QC and SPP1 TAMs) subtypes, the former correlating with the presence of plasmacytoid and myeloid DCs. A hierarchical k-means clustering using immune cell proportions as features defined six clusters with largely different compositions (Fig. 2A,B,D,E). In spite of the different sizes of the clusters, almost all cancer types were presented in each cluster, confirming large commonalities of immune cell compositions between them (Fig. 2F). Tumors in cluster 1 (C1) showed a high proportion of recently activated and naive-memory CD4 T cells, whereas C2 had high amounts of cytotoxic and terminally exhausted CD8 cells (Fig. 2D,E). C3 displayed exceptionally high levels of macrophages (C1QC and SPP1), C4 was driven by increased frequencies of plasma B cells, C5 contained high proportions of CD4 effector memory T cells, and C6 was very high in B cells. To confirm the proportional differences of cell types between clusters, we applied scCODA, confirming the enrichment of cluster-specific cell states and the largely mutually exclusive distribution of myeloid and lymphoid cell types in tumors (Supplemental Fig. 7). The cell-type enrichment was further confirmed by generalized linear models interrogating the differences in cell abundance across experimental conditions and cancer types, also accounting for batch effects. We observed the cluster-defining cell types to be statistically enriched within their respective clusters (Supplemental Table 5). Consistent with the distinct cell type proportions, the clusters presented specific gene expression signatures (Supplemental Fig. 8B; Supplemental Table 6). Intriguing from a therapeutic perspective, C5 showed a striking increase of exhaustion markers on CD8 cells with significantly higher levels of LAG3, PDCD1, and CTLA4 (ANOVA, P < 0.01) (Supplemental Fig. 8C,D). The fact that these markers are also target for immune-therapy suggests C5 tumors to be more susceptible to ICI treatment but could also shed light on the role of CD4 effector memory cells, strongly enriched in these patients (Fig. 2D,E). In line with this, the presence of specific immune cell states, such as regulatory T cells or anti-inflammatory TAMs, has been linked to immune-therapy efficacy. Thus, it is tempting to speculate that the here-identified patient clusters differ in their response to ICIs.
To facilitate the classification of immune profiles of future data sets, we trained a RF classifier with the 25 immune cell population achieving a highly accurate classification (accuracy: 0.8, 95% CI: [0.593, 0.932]; P-value: 5.36 × 10−5) (Supplemental Methods). Of note, the cluster-specific cell types were also the most important variables when training the RF classifier using cell type proportions as features to predict cluster identities (Supplemental Fig. 9). To integrate our patient clusters with large clinical RNA sequencing cohorts, we used immune cell types defining our single-cell derived clusters (Fig. 2C) to deconvolute bulk RNA sequencing data sets from the Tumor Cancer Genome Atlas (TCGA) program. Specifically, we inferred tumor immune cell proportions for 1222 breast invasive carcinoma (BRCA), 594 lung adenocarcinoma (LUAD), 472 skin cutaneous melanoma (SKCM), and 521 colon adenocarcinoma (COAD) samples. Following deconvolution, we applied the aforementioned RF classifier to assign TCGA samples to the six single-cell derived patient immune subtypes. Here, we could assign patients from all cancer types to the single-cell derived clusters, with the exception of our smallest cluster enriched in plasma B cells (Supplemental Fig. 10).
A resource for immune cell annotation
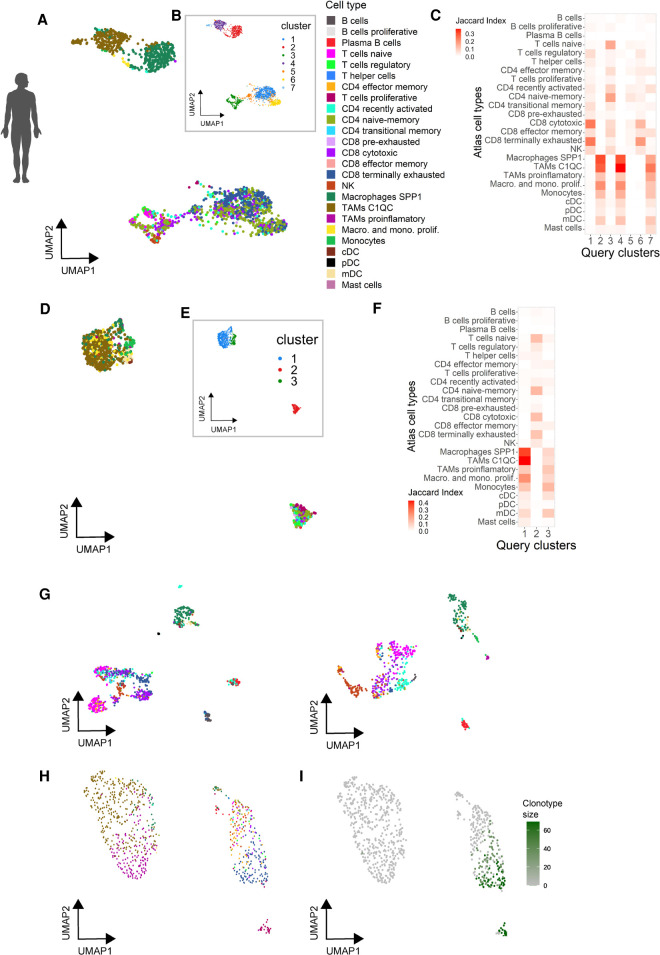
To demonstrate the predictive value of the atlas, we generated query data sets from different cancer types and varying experimental designs. Following clustering of the query data sets, we projected either single cells or clusters onto the atlas using a reference-based projection (Butler et al. 2018) or cluster matching (Mereu et al. 2020) tools, respectively. As proof-of-concept, we performed scRNA-seq for human primary OC and UM, liver metastases (from primary UM), as well as a brain metastasis (from primary CM).
Using cell-by-cell projection, we matched UM and OC cells to specific cell types and states of the atlas reference (Fig. 3A,D). Query cells projected to macrophage and T cells, with a large variety of cell states being detected. The UM harbored all macrophage subtypes, including abundant SPP1 and C1QC TAMs and a small fraction of proliferative macrophages (Fig. 3A). In addition, the entire spectrum of T cell phenotypes could be assigned, with a high fraction of cells projecting to the terminally exhausted CD8 T cell state. The OC data set contained less SPP1 TAMs and exhibited an increased proportion of proliferative macrophages (Fig. 3D). We also detected a small number of conventional DC and monocytes. We next used clustering analyses on the query data sets and subsequently matched the identified clusters to the atlas reference. There were seven and three clusters for UM and OC, respectively (Fig. 3B,E). Matching the query and reference atlas clusters resulted in a clear separation between the lymphoid and myeloid populations (Fig. 3C,F). In UM, cytotoxic and terminally exhausted cells were enriched in cluster 1 and cluster 6, in line with their distribution in the dimensionality reduction representation (Fig. 3A). SPP1 TAMs showed higher scores in cluster 2, whereas C1QC TAMs fell into cluster 4, an assignment consistent with the cell-by-cell projection. Cluster 7 matched to both lineages, likely depicting cell doublets.
Figure 3.
Automated annotation of external human tumor-derived immune cells using the tumor immune atlas as reference. (A,B) UMAP representation of immune cell transcriptomes from a primary uveal melanoma colored by their predicted cell type/state based on the tumor immune reference (A) or using unsupervised clustering (B). (C) Marker correspondence (Jaccard index) between uveal melanoma clusters (B) and the cell type clusters of the reference atlas. (D,E) UMAP representation of immune cell transcriptomes from a primary ovarian carcinoma colored by predicted cell type/state (D, color code as in A) and after clustering (E). (F) Marker correspondence (Jaccard index) between the ovarian cancer clusters and the cell type clusters of the reference atlas. (G) UMAP representation of immune cells from two uveal melanoma liver metastasis colored by their predicted cell type (color code as in A). (H,I) UMAP representation of T cells isolated from a brain metastasis colored by their predicted cell type (H, color code as in A) and clonal expansion profiled through TCR genotyping (I).
The projection of cells from UM liver metastases rendered a precise separation of T cell states (Fig. 3G), which split into naive, recently activated, cytotoxic, and proliferative subpopulations. We found a distinct frequency of exhausted cells between both UM metastases. Although much less abundant, all DC and B cell states could also be assigned in both samples. Cell-by-cell projection of T cells from a NSCLC brain metastasis also identified most cell states, with a high proportion of terminally exhausted CD8 T cells (Fig. 3H). Combined scRNA-seq and TCR genotyping assigned clonally expanded T cells as being either actively proliferating or terminally exhausted, in line with the expected history of tumor reactivity and proliferation of exhausted cells (Fig. 3H,I; Azizi et al. 2018; Li et al. 2019).
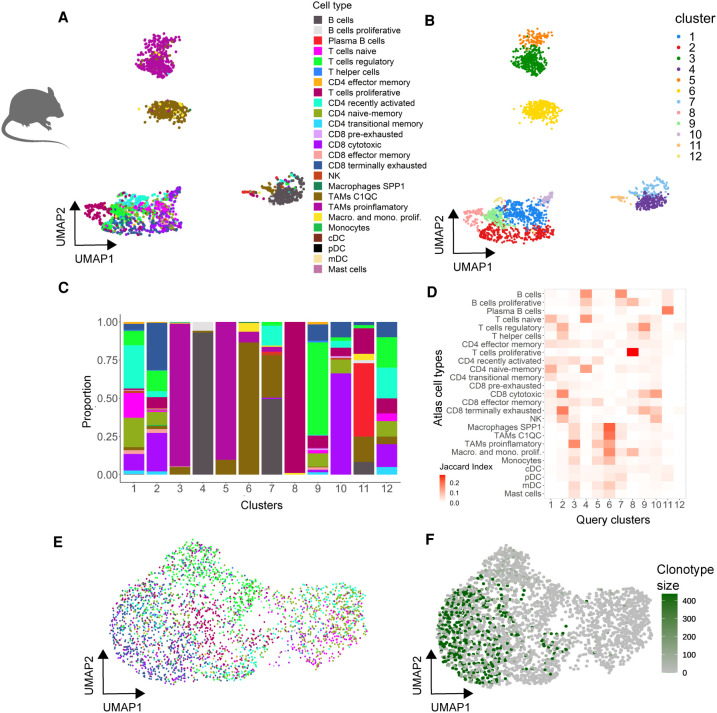
We further wondered about the applicability of the atlas as a reference across species. To tackle this question, we generated scRNA-seq data sets for two liver metastases derived from mouse CRC organoids (Tauriello et al. 2018), one PTPRC selected and the other enriched in T cells, and projected individual cells onto the human reference. The mouse data sets could be projected with high confidence (∼70% of cells with matching probabilities > 0.5). Mouse clusters of main subtypes could be readily annotated, and specific subpopulations could also be assigned to distinct T/B cell or macrophage cell states using the human reference (Fig. 4A–D). Again, distinct T cell states emerged from the dimensional reduction plots or after clustering analysis (Fig. 4A–C). Consistently, cell projection of the experimentally enriched T cell fraction to the reference atlas identified defined T cell states (Fig. 4E). Clonal expansion according to TCR sequencing could only be detected in cells assigned to be proliferative, cytotoxic, or exhausted, whereas naive, naive-memory, and recently activated cells were not of clonal origin (Fig. 4E,F).
Figure 4.
Projecting mouse tumor immune cells onto the human reference atlas. (A,B) UMAP single-cell transcriptome representations of mouse T cells isolated from tumor organoids. Cells are color-coded by their predicted cell type based on the human reference atlas (A) or by cluster identity (B). (C) Cell type composition of each cluster (color code as in A). (D) Marker gene correspondence (Jaccard index) between the mouse immune clusters and the cell type clusters of the reference atlas. (E,F) UMAP representation of mouse T cells isolated from tumor organoids colored by predicted cell type (E, color code as in A) and clonality based on expanded TCR clonotypes (F).
Spatial localization of immune cells in tumor sections
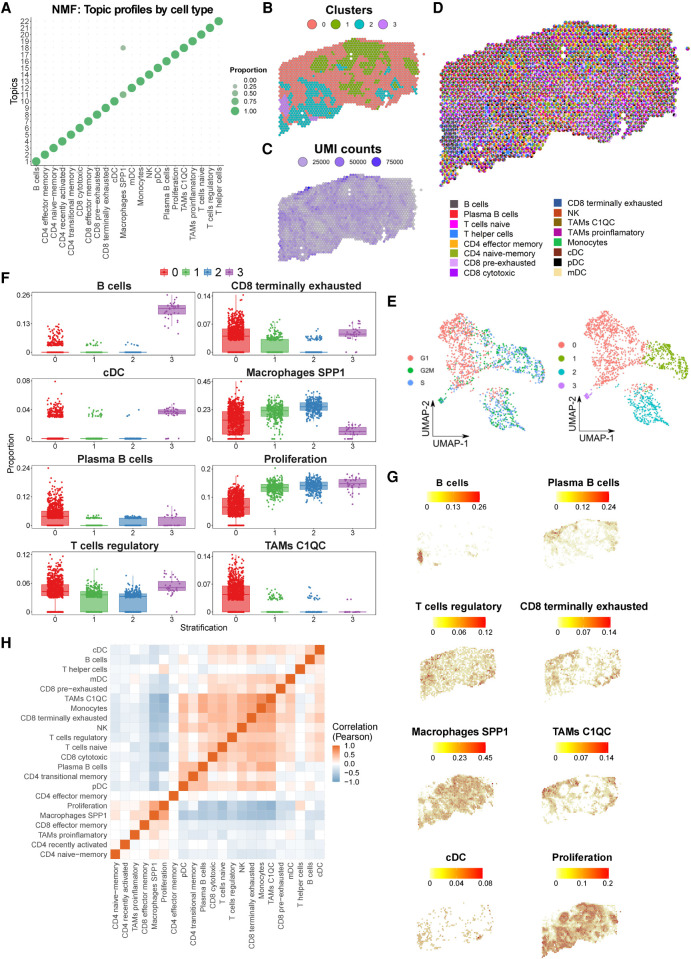
Widespread evidence indicates that the spatial distribution of immune cells is important for ICI response (Chen and Mellman 2017). To explore this issue, we combine single-cell reference atlas immune profiles with ST data from tumor sections. The aim of this approach is to provide spatial tumor maps that precisely delineate colocalization of immune cells in tumors. To integrate both data modalities, we applied SPOTlight, a nonnegative matrix factorization (NMF)-based spatial deconvolution framework (Elosua-Bayes et al. 2021). SPOTlight identifies cell type–specific topic profiles (gene expression signatures) from scRNA-seq data, which are subsequently used to deconvolute ST spots. To predict and quantify the location of the 25 cell types and states of the tumor immune reference, gene expression profiles were translated into SPOTlight topic profiles. Topics were highly specific to clusters, resulting in high specificities to localize immune cell types (Fig. 5A). To avoid conflicting signals and inaccurate predictions related to the presence of strong cell cycle signatures from cancer cells, we removed proliferating immune cells from the deconvolution step and included a general proliferation signature to identify areas of dividing cells. To map immune cells within tumor sections, we generated ST data sets for an oropharyngeal squamous cell carcinoma (SCC) metastasis and analyzed two sequential sections of an invasive ductal BC (Visium platform, 10x Genomics).
Figure 5.
Spatial mapping of the reference immune cell types using ST oropharyngeal squamous cell carcinoma (SCC) sections. (A) Cell type–specific topic profiles presenting a high topic/cell type specificity. (B) ST profiled section of a SCC primary tumor. Tissue stratification according to unsupervised clustering. (C) The number of unique molecular identifiers (UMI) recovered from each spot indicate the areas transcriptionally most active. (D) Pie chart representation showing proportions (per ST spot) of SPOTlight-predicted immune cells based on the single-cell immune reference atlas. To visualize spatially variable cell types, only immune cell types present in <75% of the spots are displayed. (E) UMAP embedding of ST spots presenting the cell cycle phase (left) and cluster identity (right) for each spot. (F) Box plots displaying significant differences in cell type proportion of clusters (ANOVA test). (G) Location and proportion of significantly differentially located cell types in the SCC section. (H) Clustered correlation matrix between the predicted cell type proportions identifying colocalization (red) and exclusive (blue) immune distribution patterns.
Clustering the spatial SCC data identified four distinct transcriptional and proliferative active areas containing cancer cells (cluster 1/2), surrounded by stroma (cluster 0) and an area enriched in immune cells (cluster 3) (Fig. 5B–E). Mapping the immune cell distribution in the sections using SPOTlight returned proportions for each cell within a spot, predictions that were validated using an alternative spot deconvolution tool (Supplemental Fig. 11; Cable et al. 2021). This resulted in a clear regionalization of distinct immune cell types and states (Fig. 5D; Supplemental Fig. 12). The cancer areas cluster 1/2 presented similar patterns, with an enrichment of the proliferation signature and SPP1 macrophages (Fig. 5F,G; Supplemental Fig. 13). Cluster 3, in turn, presented a specific immune infiltration pattern dominated by an enriched presence of B cells and cDCs. The stroma section, cluster 0, harbored regulatory T cells and terminally exhausted CD8 T cells and was specifically enriched in C1QC TAMs. An interaction matrix computed based on the predicted proportions on the spatial distribution of cell states in the section highlighted colocalization and mutual exclusive patterns (Fig. 5H; Supplemental Fig. 14). Here, spatial colocalization provided further insights into immune-modulating mechanisms of cancer and immune-suppressive stromal cells (e.g., regulatory T cells). Exemplarily, recently activated T cells colocalized with effector/naive memory T cells, whereas they were excluded from areas with regulatory and exhausted T cells as well as C1QC TAMs and plasma B cells. Accordingly, regulatory T cells showed a positive correlation with cytotoxic cells (including NK), pre- and terminally exhausted CD8 T cells, and C1QC TAMs, but a negative correlation with recently activated and effector/naive memory T cells. It has been shown that regulatory T cells inhibit effector T cells by releasing ENTPD1 (previously known as CD39) and NT5E (also known as CD73) that bind to the A2A receptor (Allard et al. 2020). In line with this finding, and in addition to the exclusive pattern of regulatory and activated and effector T cells, we detected a positive expression correlation of ENTPD1 and NT5E with the regulatory T cell predicted proportions (P < 2.2 × 10−16 and P = 2.5 × 10−6, respectively). Along with the detected spatial organization with other immune cell types, regulatory T cells illustrate the value of spatial immune mapping to advance our understanding of tumor immunology. Analysis of a second SCC case confirmed the regional restriction of tumor immune cells and confirmed the previously described colocalization and mutually exclusive patterns (Supplemental Figs. 15–17).
In order to validate the colocalization of abovementioned cell type and states, we performed a digital in situ hybridization (digital ISH) experiment by correlating the presence of canonical expression markers of these cells. Although being less sensitive, ST generates accurate molecule counts, thus returning an adequate signal for targeted colocalization (same spot) analysis. To enhance the signal, we denoised the data (van Dijk et al. 2018) before computing a clustered correlation matrix of marker genes (Supplemental Fig. 18A). We observed strong correspondence of cell type marker gene colocalization with the predicted proximity of respective cell types. Specifically, the localization of GNLY and NKG7 (cytotoxic T cells) correlated significantly with FOXP3 and CTLA4 (regulatory T cells) (Supplemental Fig. 18B–D). On the other hand, the correlation between SPP1, CLEC7A (SPP1 TAMs), and cancer (EPCAM), as well as proliferation markers (STMN1, MKI67, CDK1) confirmed the colocalization of SPP1 TAMs with proliferating cancer cells (Supplemental Fig. 18E–G). Finally, our digital ISH experiment confirmed the colocalization of stromal cells (ACTA2, PDGFRA) with regulatory T cells (FOXP3, CTLA4), terminally exhausted T cells (PDCD1, LAG3, HAVCR2), and C1QC macrophages (MRC1, CD163) (Supplemental Fig. 18A).
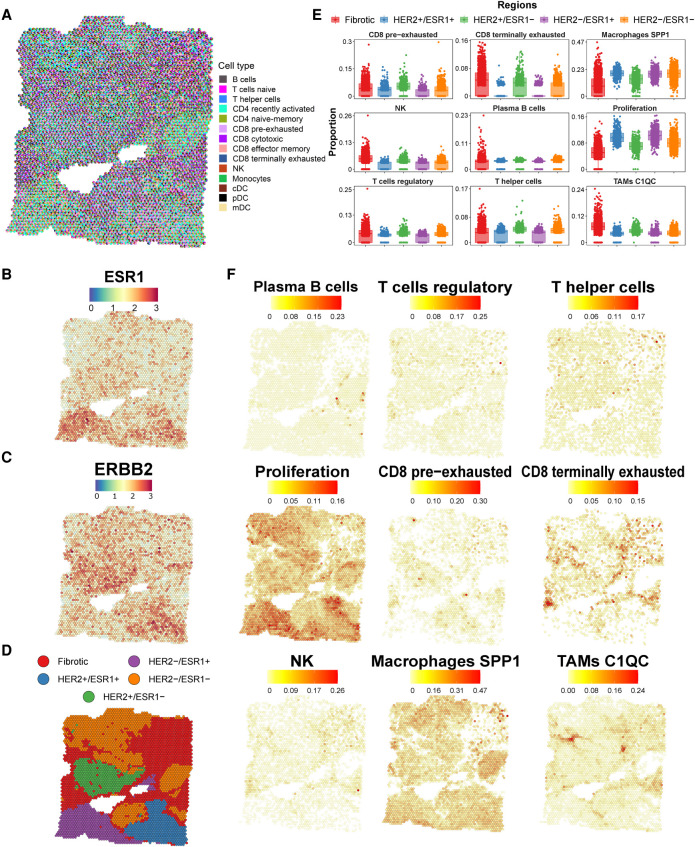
A cancer-specific regional distribution was also observed in the invasive ductal BC and replicated over serial sections of the tumor (Fig. 6A–C; Supplemental Fig. 19). The gene expression profiles of cancer cells pointed to a strong subclonal structure, with mutually exclusive localizations of BC subtype-specific ERBB2 (HER2+) and ESR1 (ESR1+) positive clones. This subclonal architecture was directly associated with local enrichment of distinct cell states (Fig. 6D; Supplemental Figs. 20, 21). The ESR1+/HER2− region showed the increased presence of proliferating cells and SPP1 macrophages (Fig. 6E,F; Supplemental Figs. 22, 23). In contrast, the HER2+/ESR1− region showed a higher proportion of regulatory and cytotoxic T cells, C1QC TAMs, and NK cells. In this region, coupled to the higher proportion of regulatory T cells, we also observed an enrichment of pre-exhausted and terminally exhausted CD8 T cells, different inactivation stages of the immune cell response. Lastly, we observed an enrichment of plasma B cells and C1QC TAMs lining the cancer areas in the fibrotic tissue. The importance of a quantitative assessment of cell types within ST spots is displayed by the ubiquitous presence of SPP1 macrophages across all areas but the significant proportional enrichment in specific tumor fractions (Fig. 6E,F). Similar to the SCC, spatial correlation analysis in BC, among other features, identified the colocalization of proliferating cells and SPP1 macrophages along with the anticorrelation with regulatory and cytotoxic cells and C1QC TAMs (Supplemental Fig. 14B,C).
Figure 6.
Tumor immune reference mapping ST section from a ductal breast carcinoma. (A) Pie chart representation showing proportions (per ST spot) of SPOTlight-predicted immune cells based on the single-cell immune reference atlas. To visualize spatially variable cell types, only immune cell types present in <75% of the spots are displayed. (B,C) Estrogen receptor 1 (ESR1, B) and erb-b2 receptor tyrosine kinase 2 (ERBB2, also known as HER2, C) gene expression levels on the ST section, indicating profound regionality of the expression. (D) Tissue stratification and labeling according to unsupervised clustering. (E) Box plots of significantly differentially localized cell type proportions between the clusters (ANOVA test). Differences between tumor areas (i.e., HER2+ and ESR1+) are observed, suggesting differential tumor microenvironments of the tumor subclones. (F) Location and proportion of immune cell types with local enrichment.
Discussion
We jointly analyzed 317,111 immune cells from 217 patients, including major (e.g., T cells, B cells, and macrophages) and rare (e.g., proliferating and dendritic cells) cell types and states infiltrating human tumors. We stratified cell types into subpopulations and annotated a fine-grained map of immune cell states. We envision this atlas to serve as a reference for a harmonized annotation of future data sets, which we tested by projecting single cells from primary human tumors and metastases as well as mouse immunotherapy models onto the atlas. The here-identified 25 cell types/states present a compendium of immune cell populations shared across cancer types to enable personalized medicine applications. Whereas the RF classifier pointed to the robustness of the clusters, increasing the cluster number could identify additional biological relevant and functional cell states. This was exemplified by the identification of additional B cell states through deeper clustering, and we expect the large cell number of this atlas to allow an even deeper annotation.
Observing high cell state heterogeneity across patients and cancer types enabled a patient stratification based on the immune composition. This immune-based classification identified pan-cancer subtypes exclusively enriched in recently activated CD4 T cells, cytotoxic/terminally exhausted CD8 T cells, CD4 effector memory, C1QC tumor-associated macrophages, or B cells and plasma B cells. We believe pan-cancer immune classifications to be highly informative and clinically predictive when integrating tumor samples from single-cell studies with patient survival and response to therapy metadata. We thus envision our immune-based patient classifier to be a useful tool for future patient stratification, predicting prognosis and immunotherapy response. Moreover, using the classifier, the pan-cancer immune classification system could be extended to additional cancer types and drive the design of basket clinical trials in which a common immune stratification and recruitment framework is applied across cancer types.
Detecting regional immune cell enrichments in tumor clones and adjacent areas, we foresee an application of the atlas in digital pathology to guide immunotherapy decisions. The tumor immune cell reference, in combination with SPOTlight-based spatial mapping of ST data, enables harmonization of the spatial assessment of tumor infiltrating immune cells, a first step toward an automated digital pathology framework. We foresee the regional distribution of immune cell types becoming an important feature for the prediction of immunotherapy outcome. Moreover, longitudinal sampling for spatial immune cell mapping throughout ICI treatment could aid in explaining therapy action and point to further weaknesses approachable through combinatorial therapy strategies. However, it is of note that predicting the presence of immune cell types from a reference atlas cannot comprehensively take into account cell type– or patient-specific peculiarities. Overlapping gene expression signatures, as shown for the proliferation-related transcriptional program, can challenge distinguishing the presence of immune cells from cell types that are not present in the training set. Consequently, additional analysis, such as the here-applied digital ISH validation, or experimental validation should be considered.
In conclusion, our analysis allowed us to catalog tumor-resident immune cells across a large patient cohort and to draw a comprehensive tumor immune cell atlas of human cancers. We envision the application of the atlas in multiple areas of precision oncology. Therefore, we provide a framework for an immune-based patient stratification, the feasibility to integrate newly generated patient single-cell data, and a toolbox to map immune cells directly in tumor sections. Following the integration of clinical cohort single-cell studies with patient outcome and response metadata, we expect the atlas to be predictive for patient prognosis and immunotherapy response to a level that greatly exceeds currently applied stratification strategies.
Methods
Data preprocessing
We initially preprocessed each scRNA-seq data set independently: breast carcinomas (obtained from the NCBI Gene Expression Omnibus [GEO; https://www.ncbi.nlm.nih.gov/geo/] accession numbers GSE114727 and GSE114725), basal cell and squamous cell carcinomas (GEO GSE123814), endometrial adeno- and renal cell carcinomas (GEO GSE99254), intrahepatic cholangio- and hepatocellular carcinomas (GEO GSE125449, GSE140228), colorectal cancers (GEO GSE132465, GSE99254), pancreatic ductal adenocarcinomas (Genome Sequence Archive [GSA; https://ngdc.cncb.ac.cn/gsa/] accession number CRA001160), ovarian cancers (https://figshare.com/s/711d3fb2bd3288c8483a), non-small-cell lung cancers (ArrayExpress [https://www.ebi.ac.uk/arrayexpress] accession numbers E-MTAB-6149, E-MTAB-6653 and GEO GSE97168, GSE99254), and cutaneous (GEO GSE120575, GSE123139) and uveal (GEO GSE139829) melanomas.
From the original count matrices, we filtered the low-quality cells based on mitochondrial RNA percentage, number of UMIs (unique molecular identifiers), and number of different genes (thresholds adjusted separately for each data set). Then, we obtained the highly variable genes, corrected, logarithmically normalized, and scaled the counts (default parameters) of each data set using Seurat (version 3.2.0) (Stuart et al. 2019). Principal component analysis (PCA) was performed on all individual data sets. The optimal number of principal components ranged between 25 and 40, depending on data set size. Dimensionality reduction was performed by applying the Uniform Manifold Approximation and Projection (UMAP) algorithm, which also served as a two-dimensional embedding for data visualization. Cluster analysis was performed using the Louvain clustering algorithm, a method separating different communities inside the data by optimizing the modularity measure. The clusters were annotated by expression of canonical markers of major cell types to consistently identify and extract immune cells. Before data set integration, the major cell types were identified, that is, T, B, myeloid, and dendritic cells, and separated from the tumor cells. Note that, for some data sets, prior information for cell-level annotation was also available.
Tumor immune cell atlas construction
To construct the atlas, we used the raw counts from the previously annotated immune cells of each data set and followed Seurat's standard data integration pipeline. This method is based on the identification of anchor cells between pairs of data sets, which are used to harmonize the integrated data sets. To do this, canonical correlation analysis (CCA) is applied, for which we set the number of anchors to 3000. Prior to integration, we obtained the data set-specific genes in order to remove them from the integration anchors, thereby reducing noise. To do this, we joined all the data sets and, with the FindAllMarkers function, calculated the genes specific for each data set. Then, we removed these genes from the integration features.
Integrating multiple data sets is a computationally intensive procedure. Due to the large number of atlas cells, we used the alternative reciprocal PCA (RPCA) algorithm for anchor identification, as proposed by Stuart et al. (2019). This method is notably faster and performs considerably similarly to the standard CCA method. With this procedure, we built a tumor immune cell atlas of over 300,000 immune cells across 13 cancer types and 217 patients. After integration, we computed the clusters again to obtain a fine-grained resolution of cell types and states. We increased the resolution parameter in Seurat's FindClusters up to 1.2, to retain more subtle differences between subpopulations.
We performed differential expression analysis (DEA) for all clusters to determine their marker genes. For the DEA, we used the normalized RNA counts, instead of using the integrated data, to increase the accuracy identifying significant cluster makers. To further refine the cluster signatures, we computed the clusters using both the Wilcoxon rank-sum test and MAST (Finak et al. 2015) methods and kept only genes identified with both approaches. We annotated the refined clusters using a curated gene signature of immune cell subtypes and compared it with the results of the DEA analysis (using the matchSCore package [Mereu et al. 2020]). Here, not only the major immune cell types but also different immune cell states were identified.
Clustering and classification of patients based on immune cell composition
In order to find similarities in patients’ immune microenvironment across cancer types, we applied patient clustering using the proportion of 25 immune cell states of their TME. For this, we filtered out patients with less than 500 cells and applied hierarchical k-means (HKM) clustering (Chen et al. 2005; Alashwal et al. 2019) using the cell types as variables and their proportion in the TME as values. We set the number of clusters to k = 6 (Supplemental Fig. 9B) and used the agglomerative Ward 2 method with Euclidean distance metric. This algorithm iteratively joins instances in clusters, reducing intra-cluster variance. Further, we built a RF classifier to predict the assignment of each patient to one of the six immune subtype clusters. Therefore, we split the data set into training and test sets (75% and 25% of the patients, respectively) and grew 100 trees to train the classifier, while automatically assessing predictor's importance and calculating the proximity between patients for a better model. To evaluate the variable importance obtained from the RF classifier, we assessed how the accuracy of the model decreased when a variable is excluded from the model (mean decrease accuracy) and how each variable affects the homogeneity of the trees in the forest (mean decrease Gini). We then used scCODA to statistically confirm proportional differences in cell type abundance in the six patient clusters. Therefore, we included all cell types/states of the atlas, and let the algorithm automatically select a reference cell type for scCODA's model (transitional memory CD4 T cells). Of note, using proliferative T cells as a reference cell type produced similar results. We then tested each of the clusters against the rest, setting the patient and the cluster as covariates. We used scCODA's default model and algorithm and gathered the results of the estimated parameter and log2-fold change for each of the six contrasts (one per cluster). These results further confirmed and provided statistical support for the differences in patient cluster cell type composition (Supplemental Fig. 7). To further assess the robustness of the patient clusters and to identify significantly enriched cell types, we used generalized linear models (GLMs). Specifically, we used the R package “emmeans” to perform the contrasts between the clusters and included the batch (data set of origin), cancer type, and sequencing technologies, as covariates of the models (Supplemental Table 6). Both scCODA and GLM analysis confirm our observations.
Data set projection onto the tumor immune cell atlas
For the projection and annotation of external data sets onto the atlas, we used Seurat's anchor-transferring method. This algorithm uses the PCA structure of the reference atlas and projects it on the query. Then, it finds pairs of anchor genes between data sets that enable the projection. These anchors allow the transfer of annotations (labels) between data sets. Here, we used an unsupervised anchoring based on the first 30 principal components of the data sets and the RNA assays with a LogNormalize normalization method. We obtained the cell-type assignment of each cell in the query data sets plus the corresponding prediction probability. We also used matchSCore2's function to correlate the markers of query clusters with the markers of the 25 cell types/states of the atlas. Therefore, we used the top 100 markers (average log-fold change) for each of the clusters.
Spatial transcriptomics analysis
Clusters were annotated according to pathologists’ assessment, their transcriptional activity, and differentially expressed genes. A subset of the single-cell atlas was used to train the SPOTlight model; we selected up to 100 cells coming from melanoma cancer for each cell type/state. Selecting cells from one of the cancer types allowed us to reduce data set-specific noise, which could confound the model. The gene set used to train the model was the union between the marker genes of the cell types along with the top 3000 variable genes. Marker genes for each cell type were obatined with Seurat's function FindAllMarkers, considering only positive markers and setting the logFC and min.pct to 0 to include all genes. All markers were used to initialize the model basis, and unit variance normalization was carried out. Nonsmooth, nonnegative matrix factorization was the method used to carry out the factorization. Cell types contributing <3% to the spot's predicted composition were considered fitting noise and were set as 0. We then used SPOTlight to map the atlas cell types to the spatial spots. For the digital ISH, we reduced data sparsity and enhanced gene expression signals with MAGIC (van Dijk et al. 2018), applied on the normalized gene expression data. We ran MAGIC with default parameters, except for knn set to 2, to ensure that the information used to recover the data structure is based on the most similar ST spots.
Data access
The Tumor Immune Cell Atlas count matrix and metadata are deposited at Zenodo (https://zenodo.org/record/5186413#.YRqbJC1h2v6). The analysis code is available at GitHub (https://github.com/Single-Cell-Genomics-Group-CNAG-CRG/Tumor-Immune-Cell-Atlas) and as Supplemental Code. Raw data are available as stated in the Supplemental Methods. To explore the atlas, project external data, and apply the immune classifier, we are currently developing a user-friendly ShinyApp accessible at https://paula-nieto-garcia.shinyapps.io/TICA/. Raw data for the projected single-cell and ST data sets generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE158803.
Supplementary Material
Acknowledgments
This publication is part of a project (BCLLATLAS) that has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No. 810287). This work has received funding from the Ministerio de Ciencia, Innovación y Universidades (SAF2017-89109-P; AEI/FEDER, UE) and the Fundació La Marató de TV3 (201903-30-31-32). We further acknowledge funding from the St. Vincent's Clinic Foundation (V.T.C.) and the National Health and Medical Research Council Investigator Grant (APP1175781, J.E.P.), the Fundación Asociación Española contra el Cáncer (AECC), FERO (EDM), Ramón Areces Foundation, Cellex Foundation, BBVA (CAIMI), the ISCIII, FIS (PI16/01278), Juan de la Cierva formación fellowship (C.R.-P.) and Sara Borrell fellowship (E.P.-R.). Core funding is from the ISCIII and the Generalitat de Catalunya. We acknowledge support of the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) to the EMBL partnership, the Centro de Excelencia Severo Ochoa, the CERCA Programme / Generalitat de Catalunya, the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) through the Instituto de Salud Carlos III and the Generalitat de Catalunya through Departament de Salut and Departament d'Empresa i Coneixement. We also acknowledge the cofinancing by the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) with funds from the European Regional Development Fund (ERDF) corresponding to the 2014–2020 Smart Growth Operating Program.
Author contributions: H.H. designed the study. P.N. and M.E.-B. performed all data analyses. J.L.T. analyzed TCR data. E.M., R.M.-B., S.A.-F., and C.R.-P. supported the data analysis. D.M., C.M., S.R., P.L., and E.P.-R. generated single-cell data sets. D.K. and C.C. generated ST data sets. M.S., A.H., and J.N. supported data interpretation. V.T.C., R.G., I.G., J.M.P., J.S., J.E.P., and E.B. provided samples and technical and sequencing support. H.H., P.N., and M.E.-B. wrote the manuscript with contributions from the co-authors. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.273300.120.
Freely available online through the Genome Research Open Access option.
Competing interest statement
H.H. is cofounder of Omniscope. J.S. is cofounder of Mosaic Biomedicals, board member of Northern-Mosaic partnership, and has ownership interests from Mosaic Biomedicals and Northern Biologics. J.S. received grant/research support from Mosaic Biomedicals, Northern Biologics, Roche/Glycart, and Hoffmann la Roche. J.S. reports a scientific consultancy role for Merck Serono, GSK, and Eli Lilly. All other authors declare no competing interests.
References
- Alashwal H, El Halaby M, Crouse JJ, Abdalla A, Moustafa AA. 2019. The application of unsupervised clustering methods to Alzheimer's disease. Front Comput Neurosci 13: 31. 10.3389/fncom.2019.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard D, Allard B, Stagg J. 2020. On the mechanism of anti-CD39 immune checkpoint therapy. J Immunother Cancer 8: e000186. 10.1136/jitc-2019-000186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp M, Giacomello S, Larsson L, Wu C, Fürth D, Qian X, Wärdell E, Custodio J, Reimegård J, Salmén F, et al. 2019. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 179: 1647–1660.e19. 10.1016/j.cell.2019.11.025 [DOI] [PubMed] [Google Scholar]
- Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, et al. 2018. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174: 1293–1308.e36. 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cable DM, Murray E, Zou LS, Goeva A, Macosko EZ, Chen F, Irizarry RA. 2021. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat Biotechnol 10.1038/s41587-021-00830-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, Sevillano M, Palomo-Ponce S, Tauriello DVF, Byrom D, et al. 2015. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 47: 320–329. 10.1038/ng.3225 [DOI] [PubMed] [Google Scholar]
- Chen DS, Mellman I. 2017. Elements of cancer immunity and the cancer–immune set point. Nature 541: 321–330. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- Chen B, Harrison R, Pan Y, Tai PC. 2005. Novel hybrid hierarchical-K-means clustering method (H-K-means) for microarray analysis. In Proceedings of the 2005 IEEE Computational Systems Bioinformatics Conference - Workshops(CSBW'05), pp. 105–108. Stanford, CA. IEEE Computer Society, Washington, DC. [Google Scholar]
- Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, et al. 2020. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat Biotechnol 38: 970–979. 10.1038/s41587-020-0602-4 [DOI] [PubMed] [Google Scholar]
- Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, Snyder H, Feun LG, Livingstone AS, Harbour JW. 2020. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun 11: 496. 10.1038/s41467-019-14256-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosua-Bayes M, Nieto P, Mereu E, Gut I, Heyn H. 2021. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res 49: e50. 10.1093/nar/gkab043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. 2015. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16: 278. 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman WH, Pagès F, Sautès-Fridman C, Galon J. 2012. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 12: 298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. 2015. The consensus molecular subtypes of colorectal cancer. Nat Med 21: 1350–1356. 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, Yizhak K, Sade-Feldman M, Blando J, Han G, et al. 2020. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577: 549–555. 10.1038/s41586-019-1922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P, Raychaudhuri S. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16: 1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafzi A, Moutinho C, Picelli S, Heyn H. 2018. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat Protoc 13: 2742–2757. 10.1038/s41596-018-0073-y [DOI] [PubMed] [Google Scholar]
- Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwé H, Pircher A, Van den Eynde K, et al. 2018. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 24: 1277–1289. 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. 2017. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169: 750–765.e17. 10.1016/j.cell.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader AM, Grout JA, Chang C, Maier B, Tabachnikova A, Walker L, Lansky A, LeBerichel J, Malissen N, Davila M, et al. 2020. CITEseq analysis of non-small-cell lung cancer lesions reveals an axis of immune cell activation associated with tumor antigen load and TP53 mutations. bioRxiv 10.1101/2020.07.16.207605 [DOI]
- Lee H-O, Hong Y, Etlioglu HE, Cho YB, Pomella V, Van den Bosch B, Vanhecke J, Verbandt S, Hong H, Min J-W, et al. 2020. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 52: 594–603. 10.1038/s41588-020-0636-z [DOI] [PubMed] [Google Scholar]
- Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, Haanen JBAG, Blank CU, et al. 2019. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 176: 775–789.e18. 10.1016/j.cell.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, Rae Z, Hernandez JM, Davis JL, Martin SP, et al. 2019. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell 36: 418–430.e6. 10.1016/j.ccell.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereu E, Lafzi A, Moutinho C, Ziegenhain C, McCarthy DJ, Álvarez-Varela A, Batlle E, Sagar, Grün D, Lau JK, et al. 2020. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol 38: 747–755. 10.1038/s41587-020-0469-4 [DOI] [PubMed] [Google Scholar]
- Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Muñoz P, et al. 2011. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8: 511–524. 10.1016/j.stem.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Cheng A, Montalbano A, Hao S, Stoeckius M, Legut M, Roush T, Herrera A, Papalexi E, Ouyang Z, et al. 2019. Expanding the CITE-seq tool-kit: detection of proteins, transcriptomes, clonotypes and CRISPR perturbations with multiplexing, in a single assay. Nat Methods 16: 409–412. 10.1038/s41592-019-0392-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-E, Botting RA, Domínguez Conde C, Popescu D-M, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, et al. 2020. A cell atlas of human thymic development defines T cell repertoire formation. Science 367: eaay3224. 10.1126/science.aay3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Sun B-F, Chen C-Y, Zhou J-Y, Chen Y-S, Chen H, Liu L, Huang D, Jiang J, Cui G-S, et al. 2019. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 29: 725–738. 10.1038/s41422-019-0195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu D-M, Botting RA, Stephenson E, Green K, Webb S, Jardine L, Calderbank EF, Polanski K, Goh I, Efremova M, et al. 2019. Decoding human fetal liver haematopoiesis. Nature 574: 365–371. 10.1038/s41586-019-1652-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, et al. 2017. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171: 1611–1624.e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Olbrecht S, Boeckx B, Vos H, Laoui D, Etlioglu E, Wauters E, Pomella V, Verbandt S, Busschaert P, et al. 2020. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res 30: 745–762. 10.1038/s41422-020-0355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, Matchett KP, Brice M, Marwick JA, et al. 2019. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575: 512–518. 10.1038/s41586-019-1631-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell PJ, Carninci P, Clatworthy M, et al. 2017. Science Forum: The Human Cell Atlas. eLife 6: e27041. 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ. 2019. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363: 1463–1467. 10.1126/science.aaw1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. 2018. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175: 998–1013.e20. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer MC, Lafzi A, Berenguer-Llergo A, Youssif C, Castellanos A, Solanas G, Peixoto FO, Attolini CS-O, Prats N, Aguilera M, et al. 2018. Identity noise and adipogenic traits characterize dermal fibroblast aging. Cell 175: 1575–1590.e22. 10.1016/j.cell.2018.10.012 [DOI] [PubMed] [Google Scholar]
- Schelker M, Feau S, Du J, Ranu N, Klipp E, MacBeath G, Schoeberl B, Raue A. 2017. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun 8: 2032. 10.1038/s41467-017-02289-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, et al. 2016. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353: 78–82. 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tabula Muris Consortium. 2020. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583: 590–595. 10.1038/s41586-020-2496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Cañellas A, Hernando-Momblona X, et al. 2018. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554: 538–543. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. 2016a. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352: 189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. 2016b. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539: 309–313. 10.1038/nature20123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. 2014. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515: 568–571. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijst MGP, Brugge H, de Vries DH, Deelen P, Swertz MA, LifeLines Cohort Study, BIOS Consortium, Franke L. 2018. Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat Genet 50: 493–497. 10.1038/s41588-018-0089-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. 2018. Recovering gene interactions from single-cell data using data diffusion. Cell 174: 716–729.e27. 10.1016/j.cell.2018.05.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, Brouwer S, Gomes T, Hesse L, Jiang J, et al. 2019. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med 25: 1153–1163. 10.1038/s41591-019-0468-5 [DOI] [PubMed] [Google Scholar]
- Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen Y-JJ, Chitre AS, Chiang EY, Iftikhar H, O'Gorman WE, Au-Yeung A, et al. 2020. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579: 274–278. 10.1038/s41586-020-2056-8 [DOI] [PubMed] [Google Scholar]
- Yofe I, Dahan R, Amit I. 2020. Single-cell genomic approaches for developing the next generation of immunotherapies. Nat Med 26: 171–177. 10.1038/s41591-019-0736-4 [DOI] [PubMed] [Google Scholar]
- Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, et al. 2019. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 25: 1251–1259. 10.1038/s41591-019-0522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. 2018. Molecular architecture of the mouse nervous system. Cell 174: 999–1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, Modak M, Carotta S, Haslinger C, Kind D, et al. 2019. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 179: 829–845.e20. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.