Abstract
A major challenge for droplet-based single-cell sequencing technologies is distinguishing true cells from uninformative barcodes in data sets with disparate library sizes confounded by high technical noise (i.e., batch-specific ambient RNA). We present dropkick, a fully automated software tool for quality control and filtering of single-cell RNA sequencing (scRNA-seq) data with a focus on excluding ambient barcodes and recovering real cells bordering the quality threshold. By automatically determining data set–specific training labels based on predictive global heuristics, dropkick learns a gene-based representation of real cells and ambient noise, calculating a cell probability score for each barcode. Using simulated and real-world scRNA-seq data, we benchmarked dropkick against conventional thresholding approaches and EmptyDrops, a popular computational method, showing greater recovery of rare cell types and exclusion of empty droplets and noisy, uninformative barcodes. We show for both low- and high-background data sets that dropkick's weakly supervised model reliably learns which genes are enriched in ambient barcodes and draws a multidimensional boundary that is more robust to data set–specific variation than existing filtering approaches. dropkick provides a fast, automated tool for reproducible cell identification from scRNA-seq data that is critical to downstream analysis and compatible with popular single-cell Python packages.
Single-cell RNA sequencing (scRNA-seq) allows for untargeted profiling of genome-scale expression in thousands of individual cells, providing insights into tissue heterogeneity and population dynamics. Droplet-based platforms that involve microfluidic encapsulation of cells in water–oil emulsions (Klein et al. 2015; Macosko et al. 2015; Zheng et al. 2017) have grown widely popular for their robustness and throughput. The use of barcoded poly-thymidine capture oligonucleotides provides information for assigning eventual sequencing reads to each droplet downstream from bulk library preparation. Because of the low cellular density required to avoid doublets (i.e., two or more cells captured in the same droplet), the vast majority of droplets are empty, ideally containing only tissue dissociation buffer and a barcoded RNA-capture bead with no cellular RNA. However, during tissue dissociation, cells may die and lyse, shedding ambient mRNA into the supernatant that is then captured as background in droplets containing cells and so-called “empty droplet” reactions. Ultimately, a droplet-based scRNA-seq data set contains up to hundreds of thousands of barcodes that correspond to these “empty droplets,” which include sequenced material from ambient RNA alone.
To prepare these data for downstream analysis, empty droplets and other uninformative barcodes with little to no molecular information must be removed. Often, computational biologists will define manual thresholds on global heuristics such as total counts of unique molecular identifiers (UMIs) or the total number of genes detected in each barcode in order to isolate high-quality cells. Although these hard cutoffs may generally yield expected cell populations and remove the bulk of populational noise in low-background samples, they are highly arbitrary, batch specific, and generally biased against cell types with low RNA content or genetic diversity (Lun et al. 2019). Furthermore, lenient thresholds often yield filtered data sets with populations of dead and dying cells or empty droplets with high ambient RNA content, especially in encapsulations with high background resulting from tissue-specific cell viability and dissociation protocols. These cell clusters may be gated out manually by the experienced single-cell biologist, but they will distort dimension-reduced embeddings and alter statistical testing for differential gene expression if left unchecked.
Here we introduce dropkick, a fully automated machine learning software tool for data-driven filtering of droplet-based scRNA-seq data. dropkick provides a quality-control (QC) module for initial evaluation of global distributions that define barcode populations (real cells vs. empty droplets) and quantifies the batch-specific ambient gene profile. The dropkick filtering module establishes initial thresholds on predictive global heuristics using an automated gradient-descent method and then trains a gene-based logistic regression model to assign confidence scores to all barcodes in the data set. dropkick model coefficients are sparse and biologically informative, identifying a minimal number of gene features associated with empty droplets and low-quality cells in a weakly supervised fashion. The following study aims to show how dropkick outperforms basic threshold-based filtering and a similar data-driven model (Lun et al. 2019) in recovery of expected cell types and exclusion of empty droplets, with robustness and reproducibility across encapsulation platforms, samples, and varying degrees of noise from ambient RNA.
Results
Evaluating data set quality with the dropkick QC module
Global data quality and predominance of ambient RNA affect both reliable cell identification as well as downstream analyses including clustering, cell type annotation, and trajectory inference in scRNA-seq data (Fleming et al. 2019; Yang et al. 2020; Young and Behjati 2020). Single-cell data with a low signal-to-noise ratio owing to high ambient background can result in information loss that may ultimately confound cell type and cell state identification and related statistical analyses (Zhang et al. 2019). For instance, an scRNA-seq encapsulation with a high degree of cell lysis can cause marker genes from abundant cell types to be present in the ambient RNA profile that contaminates all cell barcodes. In this scenario, global differences between cell populations would be diminished by the common detection of ambient noise, leading to loss of resolution in inference of cell identity and state.
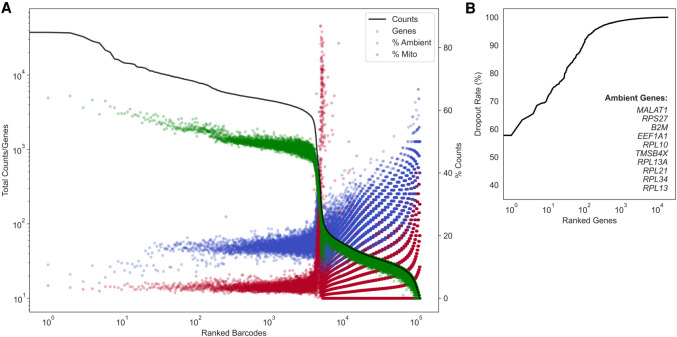
To quantify ambient contamination that reduces this batch-specific signal-to-noise ratio, we have developed a comprehensive QC report for unfiltered, postalignment UMI count matrices. Figure 1 provides an example dropkick QC report for a human T cell data set encapsulated using the 10x Genomics Chromium platform (Zheng et al. 2017). This sample is exemplary of a low-background data set, as the cells isolated from human blood do not require dissociation that causes cell stress and lysis in other tissues (Supplemental Fig. 1). Barcodes are ranked by total counts to yield a profile that describes the expected number of high-quality cells, empty droplets, and uninformative barcodes (Fig. 1A; Fleming et al. 2019). The number of genes detected per barcode follows a similar distribution to total counts, which informs our choice of dropkick training thresholds in the following sections. The first plateau in the total count profile of the T cell data set indicates approximately 4000 high-quality cells, followed by a sharp drop in the distribution (Fig. 1A). This drop-off in total UMI content signifies an estimated location for a manual cutoff as seen in the 10x CellRanger version 2 analysis software (Lun et al. 2019).
Figure 1.
Evaluating data set quality with the dropkick QC module. (A) Profile of total counts (black trace) and genes (green points) detected per ranked barcode in the 4000 pan–T cell data set (10x Genomics). Percentage of mitochondrial (red) and ambient (blue) reads for each barcode included to denote quality along data set profile. (B) Profile of dropout rate per ranked gene. Ambient genes are identified by dropkick and used to calculate ambient percentage in A.
dropkick next defines a subset of ambient genes using the dropout rate, or the fraction of barcodes in which each gene is not detected. Ranking genes in ascending order by dropout rate (Fig. 1B), dropkick labels those with dropout rates lower than the top 10 as “ambient.” High-background data sets may have many (more than 10) genes that are detected in nearly every barcode (dropout rate ≈ 0) (Supplemental Fig. 1). The dropkick definition of an ambient profile thus ensures that all relevant genes are included. The contribution of this ambient subset to the total counts of each barcode can then be calculated, shown as blue points in the dropkick QC report (Fig. 1A). Similarly, an overlay of mitochondrial read percentage indicates dead or dying cells undergoing apoptosis (Tait and Green 2010). Indeed, the ambient and mitochondrial contributions to the empty droplets in the second plateau of the total count log-rank curve are markedly higher than those in the first plateau (Fig. 1A). Another noteworthy observation is that dropkick defines an ambient profile that is distinct from the subset of mitochondrial genes. This is important for assessing cell quality in downstream clustering and dimension reduction, as any empty droplets that remain in the data set after filtering often cluster together in low-dimensional embeddings and can be highlighted by their enrichment in ambient genes. As stated previously, marker genes from abundant cell types may show up in the ambient gene set owing to excessive lysis of these common cells during tissue preparation (Supplemental Fig. 1; Fleming et al. 2019; Yang et al. 2020; Young and Behjati 2020). Accordingly, analysts should be cognizant of background expression levels that contaminate adjacent cell populations and confound cell type identification during subsequent analysis.
As each scRNA-seq data set has unique, batch-specific ambient RNA profiles and barcode distributions, the dropkick QC module allows for estimation of global data quality. The mouse colonic mucosa dissociated and encapsulated in parallel using inDrop and 10x Genomics platforms (Supplemental Fig. 1) exemplifies high-background scRNA-seq data, as indicated by elevated RNA levels in the second plateau of the total counts and genes curves. Moreover, the marker genes Car1 and Muc2 from abundant colonocytes and goblet cells, respectively, are identified by dropkick as ambient genes for these data. This signifies lysis of common epithelial cell populations during tissue preparation and dissociation. Given the dropkick QC report, the user should thus expect background expression across all barcodes, which could prove pivotal to downstream processing and biological interpretation. Taken together, dropkick can estimate the number of high-quality cells in our data set, determine average background noise from ambient RNA, and thus predict performance of filtering and ensuing analysis based on global data quality.
Description of dropkick filtering method
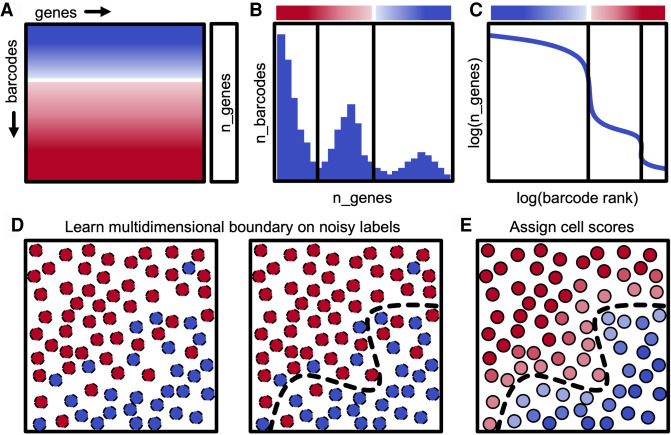
dropkick uses weakly supervised machine learning to build a model of single-cell gene expression in order to score and classify barcodes as real cells or empty droplets within individual scRNA-seq data sets. To construct a training set for this model, dropkick begins by calculating batch-specific global metrics that are generally predictive of barcode quality, such as the total number of genes detected (Fig. 2A, n_genes), which was chosen as the default training heuristic for dropkick by testing concordance with three alternative cell labels across 46 scRNA-seq samples (Supplemental Fig. 2). A data set similar to the 10x Genomics human T cell encapsulation (Fig. 1) will show a multimodal distribution of n_genes across all barcodes (Fig. 2B), where the peaks of the distribution match the plateaus seen in the log-rank representation (Fig. 2C). Next, dropkick performs multilevel thresholding on the n_genes histogram using Otsu's method (Fig. 2B,C; Otsu 1979). This automated gradient-descent technique divides the barcode distribution into three levels in this “heuristic space”: a lower level containing uninformative barcodes (which are thrown away), an upper level containing barcodes with very high cell probability based on n_genes, and an intermediate level that consists of both high-RNA empty droplets and relatively low-RNA cells. The upper and intermediate barcode populations are labeled as real cells and putative empty droplets, respectively, for initial dropkick model training. These weakly self-supervised labels based on threshold cutoffs in “heuristic space” are expected to be noisy, and the goal of the next step in the dropkick pipeline is to redraw these rough boundaries in “gene space” using logistic regression in order to recover real cells from the intermediate barcode cohort while removing ambient barcodes from the upper plateau (Fig. 2D,E).
Figure 2.
Description of dropkick filtering method. (A) Diagram of scRNA-seq counts matrix with initial cell confidence for each barcode based solely on total genes detected (n_genes), depicted by color (red, empty droplet; blue, real cell). (B) Histogram showing the distribution of barcodes by their n_genes value. Black lines indicate automated thresholds for training the dropkick model. (C) log(n_genes) versus log(rank) representation of barcode distribution as in dropkick QC report (Fig. 1A). Thresholds from B are superimposed. (D) Thresholds in heuristic space (B,C) are used to define initial training labels for logistic regression. (E) dropkick chooses an optimal regularization strength through cross-validation and then assigns cell probabilities and labels to all barcodes using the trained model in gene space.
The logistic regression model used by dropkick uses elastic net regularization (Zou and Hastie 2005), which balances feature selection and grouping by preserving or removing correlated genes from the model in concert. The motivation for choosing this regularization method is twofold. First, the resulting model exists in “gene space,” maintaining the relative dimensionality of the data set and providing biologically interpretable coefficients that describe barcode quality. Second, the model is penalized for complexity, which yields the simplest model (sparse coefficients) that adjusts the noisy initial labels while compensating for expected collinearities and errors in measurement.
Evaluating dropkick filtering performance with synthetic data
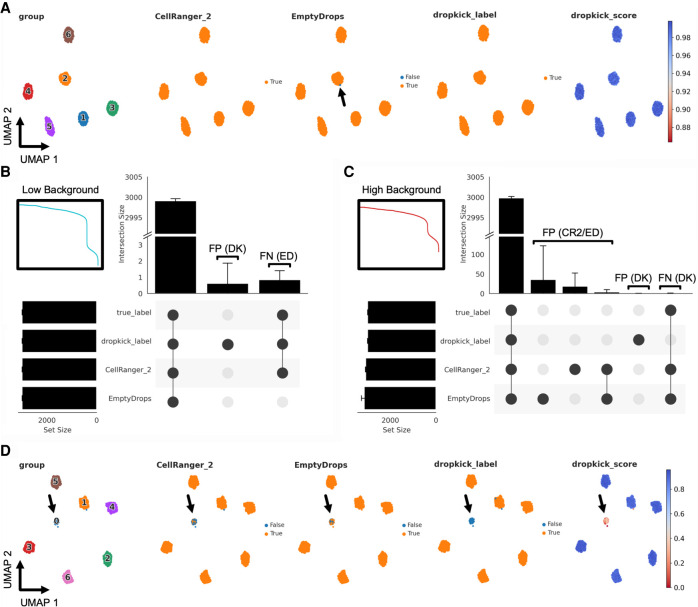
We tested dropkick filtering on single-cell data simulations that define both empty droplets and real cells, providing ground-truth labels for comparison to dropkick outputs (Fleming et al. 2019). These synthetic data sets modeled ambient RNA noise in the cell populations to confound filtering, as seen in real-world data sets. We simulated both low-background (Fig. 3A,B) and high-background (Fig. 3C,D) scenarios (see Methods: Synthetic scRNA-seq data simulation).
Figure 3.
Evaluating dropkick filtering performance with synthetic data. (A) UMAP embedding of all barcodes kept by dropkick_label, CellRanger_2, and EmptyDrops for an example low-background simulation. Points colored by each of the three filtering labels, as well as ground-truth clusters determined by the simulation and dropkick score (cell probability). Arrow highlights a single false-negative (FN) barcode in the EmptyDrops label set for this replicate. (B) UpSet plot showing mean size of shared barcode sets across dropkick_label, CellRanger_2, EmptyDrops, and true labels for 10 simulations. Error bars, SD. Unique sets show false-positive (FP) barcodes labeled by dropkick and FN barcodes excluded by EmptyDrops. Inset shows log-rank representation of the low-background simulation in A. (C) Same as in B, for 10 high-background simulations. Inset shows log-rank representation of the high-background simulation in D. (D) Same as in A, for an example high-background simulation. Arrow highlights cluster 0, designated as “empty droplets” by simulation (see Methods: Synthetic scRNA-seq data simulation).
To show the utility of the dropkick model over one-dimensional thresholding and an analogous data-driven filtering model, we ran dropkick, 10x Genomics CellRanger version 2 (CellRanger_2), and the EmptyDrops R package (Lun et al. 2019) on 10 iterations of low- and high-background simulations. An example UMAP embedding of all barcodes kept by dropkick_label (dropkick score ≥ 0.5) and the two analogous methods shows that all three methods excluded empty droplets (assigned cluster 0 from the simulation), with a single false-negative (FN) barcode highlighted in the EmptyDrops label set (Fig. 3A). An UpSet plot (Fig. 3B; Lex et al. 2014) tabulating shared barcode sets across 10 low-background simulations reveals nearly perfect specificity, sensitivity, and area under the receiver operating characteristic curve (AUROC) for all three methods in the low-background scenario (Supplemental Fig. 3A,B,D; Supplemental Tables 1, 2).
Conversely, the high-background simulations produced a large number of false positives (FPs) in the CellRanger_2 and EmptyDrops labels (Fig. 3C), as ambient barcodes with high-RNA content lie above the total count threshold identified by CellRanger and the inflection point used as a testing cutoff by EmptyDrops (Lun et al. 2019). A UMAP embedding of an example high-background simulation reveals a large population of empty droplets (assigned cluster 0 by the simulation) that dropkick_label removes from the final data set (Fig. 3D). Accordingly, dropkick displayed overall specificity and AUROC of 0.9999 ± 0.0002 and 0.9998 ± 0.0002 for the high-background simulations compared with 0.9910 ± 0.0018 and 0.9955 ± 0.0009 for CellRanger_2 and 0.9838 ± 0.0133 and 0.9917 ± 0.0071 for EmptyDrops, respectively (Supplemental Fig. 3E,F,H; Supplemental Tables 1, 2).
We also compared outputs from the trained model (dropkick_label) to automated dropkick training labels (thresholding on n_genes) in both low- and high-background scenarios to further show the utility of dropkick's machine learning model over heuristic cutoffs alone. Similar to CellRanger_2, the dropkick threshold performed favorably for the low background simulation, in which real cells are separated distinctly from empty droplets in heuristic space, indicated by a sharp drop-off in total counts and genes in the dropkick QC log-rank plot (Fig. 3B, inset). This one-dimensional thresholding resulted in sensitivity, specificity, and AUROC of 0.9986 ± 0.0007, 0.997 ± 0.0006, and 0.9978 ± 0.0005, respectively, for 10 low-background simulations (Supplemental Fig. 3C; Supplemental Table 1). The trained dropkick model, on the other hand, recovered all real cells (sensitivity 1.0), with a perfect average AUROC of 1.0 ± 0.0 (Supplemental Fig. 3D; Supplemental Table 1). This modest improvement indicates the utility of the dropkick model for sensitively discerning real cells from ambient barcodes over simple heuristic thresholding, even in a relatively low-background sample. In the high-background simulations, sensitivity of dropkick training labels fell to 0.8762 ± 0.0092, with an average AUROC of 0.9074 ± 0.0043 (Supplemental Fig. 3G; Supplemental Table 1). Following model training, dropkick's sensitivity and AUROC once again improved to 0.9995 ± 0.0004 and 0.9998 ± 0.0002, respectively (Supplemental Fig. 3H; Supplemental Table 1). These data further signify that the dropkick logistic regression model results in enhanced performance over one-dimensional heuristic thresholding, especially in the presence of high ambient noise in the training set.
Benchmarking dropkick performance on simulated high-background data
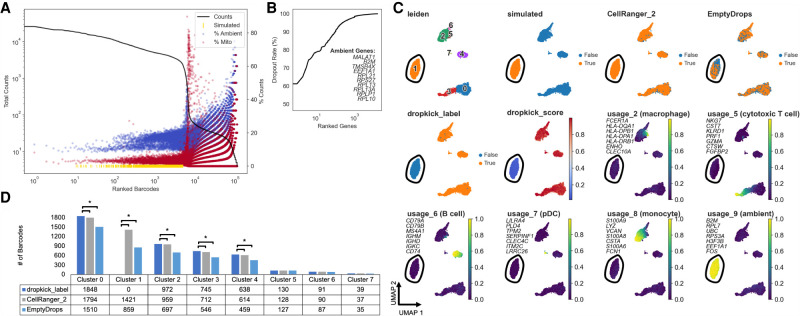
Next, we aimed to further confirm dropkick's utility in filtering high-background data by simulating extremely high ambient droplets to overlay on the 10x Genomics human PBMC data set. These data are particularly clean and easy to filter in its raw state, as the suspended cells from human blood were minimally agitated before encapsulation. To imitate empty droplets with high mRNA content, we combined all reads in barcodes with less than 100 total UMI counts and used the resulting pseudobulk as weightings for a random generation of count vectors from a multinomial distribution with UMI sums between 10 and 5000 total counts. We added 2000 of these count vectors back to the original matrix, modeling high-background empty droplets (Fig. 4A). Upon filtering with dropkick, CellRanger version 2, and EmptyDrops, a large subset of the simulated ambient barcodes remained in the latter two label sets but was discarded entirely by dropkick (Fig. 4C,D). We jointly processed all barcodes kept by the three filtering tools using nonnegative matrix factorization (NMF) (Kotliar et al. 2019) to define cell clusters and corresponding cell type metagene scores (Fig. 4C; Supplemental Fig. 4). dropkick recovered significantly more lymphoid progenitors, monocytes, and T and B cells than both EmptyDrops and CellRanger according to sc-UniFrac (Liu et al. 2018) analysis, indicating that it successfully parsed the noise introduced by the simulated droplets (Fig. 4D). dropkick also completely excluded Leiden cluster 1, the simulated barcodes with high NMF scores for usage 9, which contained high loadings for several ambient genes (Fig. 4B,C; Supplemental Fig. 4B). This result both confirmed the effectiveness of the pseudobulk multinomial simulation and further established dropkick's robustness in filtering high-background data.
Figure 4.
Benchmarking dropkick performance on simulated high-background data. (A) Log-rank total counts curve for the high-background PBMC simulation. The horizontal dashed line indicates the threshold below which ground-truth empty droplets were used to build simulated barcodes from a multinomial distribution (100 total counts). Gold rug plot indicates the location along the total counts curve of 2000 simulated high-UMI droplets (see Methods: High-Background PBMC Simulation). (B) Genes in PBMC simulation ranked by dropout rate. Top 10 ambient genes are listed, defining ambient profile used to calculate percentage in A. (C) UMAP embedding of all barcodes kept by dropkick_label, CellRanger_2, and EmptyDrops. Points colored by each of the three filtering labels, Leiden clusters determined by NMF analysis, dropkick score (cell probability), and select cell type metagene usages from NMF. Top seven gene loadings for each NMF factor are printed on their respective plots, in axis order from top to bottom. Circled area shows independent cluster of simulated empty droplets. (D) Table and bar graph enumerating the total number of barcodes detected by each algorithm in all NMF clusters. Significant cluster enrichment as determined by sc-UniFrac is denoted by brackets.
Dropkick recovers expected cell populations and eliminates low-quality barcodes in experimental data
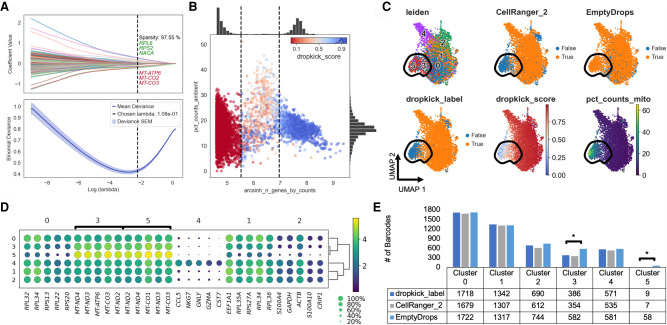
To evaluate dropkick's performance against existing scRNA-seq filtering algorithms with real-world data, we processed a human T cell data set from 10x Genomics (Fig. 1) and again compared default dropkick results (dropkick_label) to CellRanger version 2 and EmptyDrops. The final dropkick coefficients and chosen regularization strength (lambda; Fig. 5A) reveal that the model is sparse—with nearly 98% of all coefficient values equal to zero—offering an interpretable gene-based output. Without prior training or supervision, dropkick identified higher counts of mitochondrial genes, which are markers of cell death and poor barcode quality (Tait and Green 2010), as predictive of empty droplets (Fig. 5A). To visualize heuristic distributions within the T cell data set, the number of detected genes and the percentage of ambient counts per barcode are shown along with dropkick's automatic training thresholds (Fig. 5B). Uninformative barcodes below the lower n_genes threshold were discarded before model training and assigned a dropkick score of zero. Barcodes between the two thresholds were initially assigned a label indicating putative empty droplets, whereas those above the upper threshold were labeled as real cells for model training. The dropkick score overlay illustrates how dropkick redrew label boundaries in gene space (Fig. 5B). dropkick scores are noticeably lower for barcodes with high ambient RNA content, whereas some putative empty droplets with lower background are “rescued” and labeled as real cells by the trained dropkick model. It is important to note that this high-dimensional boundary was learned by dropkick with no prior labeling of “ambient” transcripts. Rather, dropkick's weakly supervised algorithm excluded barcodes with high ambient content based solely on their transcriptional similarity to the least informative barcodes (lower n_genes) in the training set.
Figure 5.
dropkick recovers expected cell populations and eliminates low-quality barcodes in experimental data. (A) Plot of coefficient values for 2000 highly variable genes (top) and mean binomial deviance ± SEM (bottom) for fivefold cross-validation along the lambda regularization path defined by dropkick. The top and bottom three coefficients are shown, in axis order, along with total model sparsity representing the percentage of coefficients with values of zero (top). Chosen lambda value indicated by dashed vertical line. (B) Joint plot showing scatter of percentage of ambient counts versus arcsinh-transformed genes detected per barcode, with histogram distributions plotted on margins. Initial dropkick thresholds defining the training set are shown as dashed vertical lines. Each point (barcode) is colored by its final dropkick score after model fitting. (C) UMAP embedding of all barcodes kept by dropkick_label, CellRanger_2, and EmptyDrops. Points colored by each of the three filtering labels, as well as Leiden clusters determined by NMF analysis, dropkick score (cell probability), and percentage counts mitochondrial. Circled area shows high mitochondrial enrichment in a population discarded by dropkick. (D) Dot plot showing top differentially expressed genes for each NMF cluster. The size of each dot indicates the percentage of cells in the population with nonzero expression for the given gene, and the color indicates the average normalized expression value in that population. Bracketed genes indicate significantly enriched populations in EmptyDrops compared with dropkick_label as shown in E. (E) Table and bar graph enumerating the total number of barcodes detected by each algorithm in all NMF clusters. Significant cluster enrichment as determined by sc-UniFrac is denoted by brackets.
We again jointly processed all barcodes kept by dropkick_label (dropkick score ≥ 0.5), CellRanger_2, and EmptyDrops using NMF (Kotliar et al. 2019) to define cell clusters, as well as sc-UniFrac (Liu et al. 2018) to determine population differences across labeled barcode sets. A UMAP embedding of these barcodes reveals a population of cells with high mitochondrial content that is mostly excluded by dropkick (Fig. 5C). This area is enriched in clusters 3 and 5 from NMF analysis, which carry exclusively mitochondrial genes as their top differentially expressed features (Fig. 5D). Based on sc-UniFrac, these two clusters constitute the only statistically significant differences between EmptyDrops and dropkick (Fig. 5E). These data indicate that dropkick recovers as many or more real cells in expected populations than previous algorithms while also identifying and excluding low-quality dead or dying cells with high mitochondrial RNA content.
Dropkick outperforms analogous methods on challenging data sets
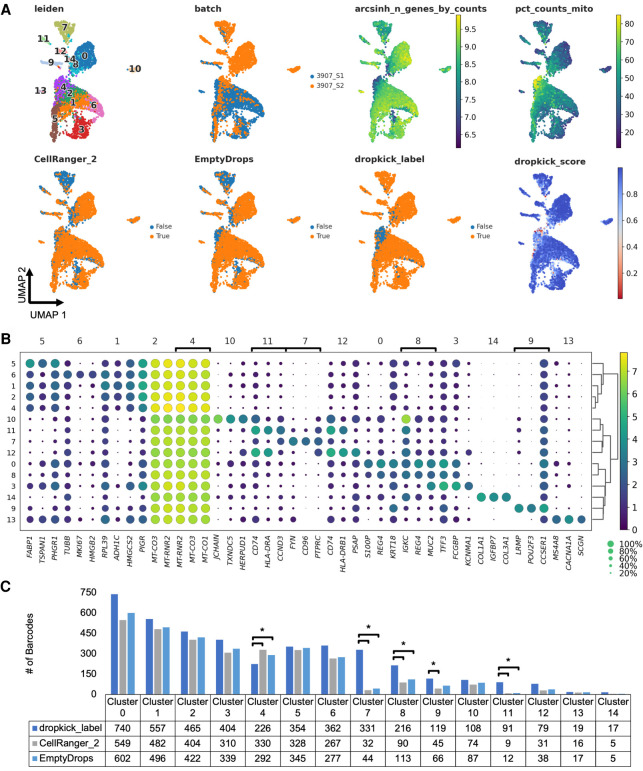
To challenge the robustness of the model, we next used dropkick to filter real-world samples with more complex cell types and higher noise. Human colorectal carcinoma (3907_S2) and adjacent normal colonic mucosa (3907_S1) samples were dissociated and encapsulated using the inDrop scRNA-seq platform (Klein et al. 2015). In contrast to the 10x Genomics pan–T cell data set (Figs. 1, 5), these samples showed high levels of background, containing empty droplets with thousands of UMI counts detected per barcode and up to 40% ambient RNA in expected cell barcodes (Supplemental Fig. 6A,D). Because of this dominant ambient profile, infiltrating immune populations with lower mRNA content than epithelial cells can be lost among empty droplets. Indeed, CellRanger_2 and EmptyDrops show depletion in T cells (cluster 7) and macrophages (cluster 11) compared with dropkick (Fig. 6A,B). Prevalence of high-RNA empty droplets also yields a population with low genetic diversity and mitochondrial gene enrichment (Fig. 6A, cluster 4) that is kept by the one-dimensional thresholding of CellRanger_2 but discarded by dropkick. sc-UniFrac analysis confirmed that dropkick recovers significantly more cells from rare populations than both CellRanger_2 and EmptyDrops in this pair of high-background data sets dominated by ambient RNA from dead and dying colonic epithelial cells (Fig. 6C; Supplemental Fig. 6). Meanwhile, dropkick also identified and removed significantly more dead cells (cluster 4) than both CellRanger_2 and EmptyDrops (Fig. 6C) by designating mitochondrial and ambient genes as negative coefficients (Supplemental Fig. 6B,E).
Figure 6.
dropkick outperforms analogous methods on challenging data sets. (A) UMAP embedding of all barcodes kept by dropkick_label (dropkick score ≥ 0.5), CellRanger_2, and EmptyDrops for human colorectal carcinoma inDrop samples. Points colored by each of the three filtering labels, as well as clusters determined by NMF analysis, dropkick score (cell probability), arcsinh-transformed total genes detected, percentage counts mitochondrial, and original batch. 3907_S1 is normal human colonic mucosa, and 3907_S2 is colorectal carcinoma from the same patient. (B) Dot plot showing top differentially expressed genes for each NMF cluster. The size of each dot indicates the percentage of cells in the population with nonzero expression for the given gene, and the color indicates the average expression value in that population. Bracketed genes indicate significantly enriched or depleted populations in dropkick compared with CellRanger_2 and/or EmptyDrops labels as shown in C. (C) Table and bar graph enumerating the total number of barcodes detected by each algorithm in all NMF clusters for the combined data set. Significant cluster enrichment as determined by sc-UniFrac is denoted by brackets.
Dropkick filters reproducibly across scRNA-seq batches
We also applied dropkick to a combined human placenta data set from six patients to show robustness of the model to batch-specific variation. dropkick learned the distribution of genes and ambient RNA specific to each data set and filtered them accordingly (Supplemental Fig. 7A), with a resulting AUROC of 0.9956 ± 0.0051 across all six replicates compared to EmptyDrops labels (Supplemental Table 3). We also performed two types of manual cell labeling as well as the CellBender remove-background model (Fleming et al. 2019) to provide additional alternative filtering labels to compare with dropkick (see Methods: CellRanger 2, EmptyDrops, CellBender, and manual filtering of real-world scRNA-seq data sets) (Supplemental Fig. 7B,D–H,J). The CellBender remove-background package primarily aims to subtract ambient background from single-cell expression data sets rather than filter alone. This resulted in the addition of a large population of high ambient barcodes unique from those labeled by dropkick, EmptyDrops, and CellRanger 2, warranting further assessment of the efficacy of background-removal methods in the context of consensus cell labels beyond the scope of this paper (Supplemental Fig. 7B–E).
Extending this analysis to a larger cohort of scRNA-seq samples from both 10x Genomics (n = 13) and inDrop (n = 33) encapsulation platforms, we see that dropkick is highly concordant with CellRanger version 2 (AUROC 0.9656 ± 0.0271) and EmptyDrops (AUROC 0.9817 ± 0.012), suggesting global recovery of major cell populations (Supplemental Fig. 8A,B,E,F; Supplemental Tables 3, 4). dropkick filtering for 33 inDrop samples yielded an AUROC of 0.9729 ± 0.0335 compared with manually curated labels using an inflection point cutoff followed by dimension-reduced cluster gating (Supplemental Fig. 8C; Supplemental Table 6; for review, see Chen et al. 2021). For all 46 scRNA-seq samples, we also performed bivariate thresholding on total UMI counts and percentage of mitochondrial transcripts per droplet, mimicking another popular preprocessing technique. Again, dropkick's AUROC averaged 0.9805 ± 0.0194, confirming the model's utility for robust filtering across several unique data sets (Supplemental Fig. 8D,H; Supplemental Tables 5, 6). Finally, we measured the total run time of dropkick, which was appreciably faster than both CellBender remove-background and the EmptyDrops R package on average, running to completion in 40.56 ± 25.97 sec across 10 replicates of all 46 samples when using five CPUs with dropkick's built-in parallelization (Supplemental Fig. 8J).
Discussion
Barcode filtering is a key preprocessing step in analyzing droplet-based single-cell expression data. Reliable filtering is confounded by distributions of global heuristics such as total UMI counts, total genes, and ambient RNA that can be highly variable across batches and encapsulation platforms. We have developed dropkick, a fully automated machine learning software tool that assigns confidence scores and labels to barcodes from unfiltered scRNA-seq counts matrices. By automatically curating a training set using predictive heuristics and training a gene-based logistic regression model, dropkick ensures that ambient barcodes (“empty droplets”) are removed from the filtered data set while recovering rare, low-RNA cell types that may be lost in ambient noise. We showed that unlike previous filtering approaches including one-dimensional thresholding (CellRanger 2) and a Dirichlet-multinomial model (EmptyDrops), dropkick is robust to the level of ambient RNA, performing favorably in both low- and high-background scenarios across simulated and real-world data sets.
Although we have shown that dropkick is more robust to varying degrees of ambient background than existing filtering methods, the dropkick model is still limited by the input data set. As stated previously (see Results: Evaluating data set quality with the dropkick QC module), the profile of ranked total counts/genes and the global contribution of ambient reads are vital to analysis of single-cell sequencing data, including cell filtering. Data with weak separation between high-quality cells and empty droplets (i.e., a unimodal distribution of n_genes lacking distinct plateaus in the log-rank curve) will perform poorly in inflection-point thresholding as well as data-driven models such as EmptyDrops and dropkick owing to the similarity between theoretically “high-confidence” barcodes and ambient background droplets. Moreover, data sets dominated by expression of ambient genes (>40% average ambient counts across all barcodes) will also perform poorly in automated filtering. Although such data artifacts may be handled by dropkick's heavy feature selection conferred by HVG calculation and elastic net regularization, there will also be circumstances that cause dropkick—as well as CellRanger and EmptyDrops—to return an over- or underfiltered data set. Scenarios such as those described should be considered QC failures, and further analysis should not be performed. For this reason, the dropkick QC module is extremely beneficial in postalignment evaluation of scRNA-seq data quality and should be applied to all data sets before filtering. The dropkick Python package provides a fast, user-friendly interface that integrates seamlessly with the SCANPY (Wolf et al. 2018) single-cell analysis suite for ease of workflow implementation.
Methods
Indrop data generation
The human colorectal carcinoma inDrop data were generated according to published protocols (Banerjee et al. 2020; Southard-Smith et al. 2020).
QC and ambient RNA quantification with the dropkick QC module
The dropkick QC module begins by calculating global heuristics per barcode (observation) and gene (variable) using the SCANPY (Wolf et al. 2018) pp.calculate_qc_metrics function. These metrics are used to order barcodes by decreasing total counts (black curve in Fig. 1A) and order genes by increasing dropout rate (Fig. 1B). The nth gene ranked by dropout rate determines the cutoff for calling “ambient” genes, with n determined by the n_ambient parameter in the dropkick.qc_summary function. All genes with dropout rates less than or equal to this threshold are labeled “ambient.” In a sample with many (>n) genes detected in all barcodes, this ensures that the entire ambient profile is identified. Through observation of samples used in this study, we set the default n_ambient = 10. To compile the dropkick QC summary report, the log-total counts versus log-ranked barcodes (Fig. 1A, black curve) are plotted along with total genes detected for each barcode (Fig. 1A, green points), percentage counts from “ambient” genes in each barcode (Fig. 1A, blue points), and percentage counts from mitochondrial genes in each barcode (Fig. 1A, red points).
Labeling training set with the dropkick filtering module
The dropkick filtering module also begins by calculating global heuristics per barcode (observation) and gene (variable) using the SCANPY (Wolf et al. 2018) pp.calculate_qc_metrics function. Next, training thresholds are calculated on the histogram of the chosen heuristic(s); arcsinh-transformed n_genes by default. dropkick then uses the scikit-image function filters.threshold_multiotsu to identify two local minima in the n_genes histogram that represent the transitions from uninformative barcodes to “empty droplets” and from “empty droplets” to real cells. These locations are also characterized by the two expected drop-offs in the total counts/genes profiles as shown in the dropkick QC report (Fig. 1; Supplemental Fig. 1). To label barcodes for dropkick model training, barcodes with fewer genes detected than the first multi-Otsu threshold are discarded owing to their lack of molecular information. dropkick then labels barcodes below the second threshold as “empty” and remaining barcodes above the second threshold as real cells for initial training. These inputs to the dropkick logistic regression model represent the “noisy” boundary in heuristic space that is to be replaced with a learned cell boundary in gene space.
Training and optimizing the dropkick filtering model
The dropkick filtering model uses logistic regression with elastic net regularization (Zou and Hastie 2005) and is fit as described by Friedman et al. (2010). The elastic net combines ridge and lasso (least absolute shrinkage and selection operator) penalties for optimal regularization of model coefficients. The ridge regression penalty pushes all coefficients toward zero while allowing multiple correlated predictors to borrow strength from one another, ideal for a scenario like scRNA-seq with several expected collinearities (Hoerl and Kennard 1970). The lasso penalty, on the other hand, favors model sparsity, driving coefficients to zero and thus selecting informative features (Tibshirani 1996). The combined elastic net balances feature selection and grouping by preserving or removing correlated features from the model in concert (Zou and Hastie 2005).
The fraction α ∈ [0, 1] (alpha) represents the balance between the lasso and ridge penalties for the elastic net model. If α = 0, the regularization would be entirely ridge, whereas if α = 1, it would be entirely lasso. By default, dropkick fixes this alpha value at 0.1, but the user may alter this parameter or provide multiple alpha values to optimize through cross-validation (with lambda; explained below) at the expense of slightly longer computational time. All default dropkick results in this paper used α = 0.1, and we also ran dropkick on all 46 samples with given alpha values [0.1, 0.25, 0.5, 0.75, 0.9]. Optimal values chosen by dropkick cross-validation for this set of runs are shown in Supplemental Table 7. Only nine of 46 models chose a value other than α = 0.1.
For a desired length of “lambda path,” n (default n = 100 for dropkick), the model is fit n + 1 times, where the first pass determines the values of lambda (regularization strength) to test and subsequent fits determine model performance using cross-validation (CV; default fivefold for dropkick). Each fit involves selection of highly-variable genes (HVGs; SCANPY pp.highly_variable_genes; default 2000 for dropkick) from the training set. For both the first pass and the final model, the training set consists of all available barcodes, whereas training the model along the lambda path uses only the current training fold as to not bias model fitting with information from the test set. The lambda path is scored using mean deviance from the training labels for all cross-validation folds. The largest value of lambda such that its mean CV deviance is less than or equal to one standard error above the minimum deviance is chosen as the final regularization strength for the model in order to further minimize overfitting. Finally, dropkick fits a logistic regression model using all training labels and the chosen lambda value and assigns cell probability (dropkick_score) to all barcodes. By default, the resulting dropkick_label is positive (one; real cell) for barcodes with dropkick_score ≥ 0.5, but the user may define a stricter or more lenient threshold for particular applications.
Synthetic scRNA-seq data simulation
We used CellBender (Fleming et al. 2019) to build synthetic single-cell data sets. We generated a basic count matrix with 30,000 features (n_genes), 12,000 total droplets (including 3000 n_cells and 9000 n_empty), and six clusters. The default ratio between the cell-size scale factor and the empty droplet–size scale factor—d_cell at 10,000 and d_empty at 200—created an unrealistic gap between the empty droplets and the real cells but built a foundation on which to produce more realistic simulations. By adjusting these parameters, we simulated two different scenarios with the number of features, total droplets, and clusters held constant. The first scenario modeled a “low background” data set, with a realistic n_genes and total count profile and relatively low ambient RNA. We set the cell-size scale factor (d_cell) to 10,000, and the empty droplet–size scale factor (d_empty) to 1000. These settings produced a small gap between the real cells and the empty droplets, yet still mimicked a low background droplet profile. We then modeled a “high background” scenario, which had much higher ambient RNA content. For this simulation, we set d_cell to 10,000 and d_empty to 2000. This simulation mimicked a real scRNA-seq data set with a high ambient profile, as it had a smaller gap between real cells and empty droplets. Taken together, these simulations recapitulate real-world single-cell data and were tested by dropkick to compare their ground-truth labels to those determined by dropkick filtering.
High-background PBMC simulation
To imitate empty droplets with high mRNA content over a relatively low-background sample, we used the 10x Genomics 4000 human PBMC data set. Because this encapsulation was derived from suspended blood cells, there was negligible lysis and ambient contamination, and empty droplets are very clearly distinguished from real cells based on their mRNA content alone. Combining reads from the bottom 1000 genes by dropout rate across all barcodes with less than 100 total UMIs, we normalized this pseudobulk as probabilistic weightings for a random generation of count vectors. We drew 2000 random integers between 10 and 5000 to determine the total number of counts for each simulated barcode and then drew that number of random integers from a multinomial distribution using the random.default_rng.multinomial function from the numpy Python package, with pvals equal to the weightings determined from the true empty droplet pseudobulk. We then added these 2000 count vectors back to the original matrix, labeling them as “simulated” for downstream comparison (Fig. 4A).
CellRanger 2, EmptyDrops, CellBender, and manual filtering of real-world scRNA-seq data sets
CellRanger and EmptyDrops filtering algorithms were derived from Lun et al. (2019), with CellRanger 2 described by the function DefaultDrops (from the repository https://github.com/MarioniLab/EmptyDrops2017) and EmptyDrops by the EmptyDrops function within the DropletUtils R package (v1.8.0). All 10x data sets were processed as by Lun et al. (2019; https://github.com/MarioniLab/EmptyDrops2017). EmptyDrops was run for all inDrop data sets using the “inflection point” from CellRanger 2 analysis as the minimum non-ambient UMI threshold as by Lun et al. (2019; https://github.com/MarioniLab/EmptyDrops2017).
Further investigation of user-defined parameters for both methods was performed by titrating the “lower” parameter, which describes the lower proportion of total barcodes to ignore when calculating the inflection point for CellRanger 2, as well as the maximum total UMI counts under which all barcodes are considered ground-truth empty droplets for EmptyDrops. We compared dropkick scores to the resulting labels (Supplemental Fig. 9; Supplemental Tables 8–11), noting that suboptimal parameter values led to lower concordance than those in Supplemental Tables 3 and 4.
CellBender remove-background, although primarily an ambient RNA subtraction model, also provides cell labels from raw scRNA-seq counts matrices (Fleming et al. 2019). With the caveat that CellBender likely retains more previously high-background droplets after regressing out ambient reads, CellBender was performed on 10x Genomics samples using the same expected cell number used for EmptyDrops by Lun et al. (2019), and concordance was tested with dropkick labels as before, showing a slightly lower average AUROC of 0.9585 ± 0.0596 for 13 10x Genomics samples (Supplemental Fig. 8G; Supplemental Table 5).
Manual filtering was performed for each inDrop sample by initial thresholding beyond the inflection point detected in the first curve of the ranked barcodes profile (as in Fig. 1A). Then, following standard dimension reduction and high-resolution Leiden clustering, clusters with low-quality cells (high mitochondrial/ambient percentage, low total counts/genes) were manually gated out of the final data set. These manually curated labels were used as an orthogonal gold standard for benchmarking automated thresholding methods (Supplemental Fig. 2A) and final AUROC (Supplemental Fig. 8D). For further description of this manual filtering method, see Chen et al. (2021).
Bivariate thresholding was performed for all samples using total UMI counts and percentage mitochondrial counts, keeping barcodes that have greater than or equal to the minimum total count threshold and <40% mitochondrial reads.
Supplemental Table 7 contains parameters used for each of the above methods on all 46 data sets.
sc-UniFrac analysis of shared populations between dropkick, CellRanger 2, and EmptyDrops labels
To evaluate the preservation of expected cell clusters between dropkick and alternative labels, we used sc-UniFrac (Liu et al. 2018) to determine the global and populational differences between the label sets. We used NMF to analyze the union of barcodes kept by dropkick_label, CellRanger_2, and EmptyDrops in order to reduce dimensions into cell identity and activity “metagenes” (Kotliar et al. 2019). We then clustered this low-dimensional space using the Leiden algorithm (Traag et al. 2019) to define consensus cell populations for sc-UniFrac analysis. We then ran sc-UniFrac (v0.9.6) to evaluate statistically significant cluster differences based on both cluster membership and gene expression hierarchies between clusters. The global sc-UniFrac distance quantified the overall similarity of hierarchical trees across barcode label sets.
Dimension reduction, clustering, projection, and differential expression analysis
We used consensus nonnegative matrix factorization (cNMF) (Kotliar et al. 2019) for initial dimension reduction. The optimal number of factors, k, was determined by maximizing stability and minimizing errors across all tested values after 30 iterations of each. We then built a nearest-neighbors graph in SCANPY (pp.neighbors function) from the NMF usage scores for consensus factors in all cells, where we set n_neighbors to the square root of the total number of cells in the data set. We then clustered cells with the Leiden algorithm (SCANPY tl.leiden function) (Traag et al. 2019) applied to this graph. Resulting clusters were used in sc-UniFrac analysis, differential expression, and visualization. We performed differential expression analysis using a Student's t-test with Benjamini–Hochberg P-value correction for multiple testing (SCANPY tl.rank_genes_groups). To visualize data sets in 2D space, we ran partition-based graph abstraction (PAGA; SCANPY tl.paga) (Wolf et al. 2019) on this nearest-neighbors graph and associated Leiden clustering in order to create a simple representation of cluster similarity. Finally, a UMAP projection (McInnes et al. 2018) seeded with these PAGA positions provided a 2D embedding of all cells in the data set (SCANPY tl.umap with init_pos = ”paga”).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE158636. All publicly available data sets are listed in Supplemental Table 12. The dropkick Python package is available for download via “pip” from the Python Package Index (PyPI) at https://pypi.org/project/dropkick/. Source code for the package is available as Supplemental Code and at GitHub (https://github.com/KenLauLab/dropkick). Scripts for reproducing the analyses in this manuscript are available as Supplemental Scripts and at GitHub (https://github.com/codyheiser/dropkick-manuscript).
Supplementary Material
Acknowledgments
We thank the Epithelial Biology Center and Vanderbilt Quantitative Systems Biology Center for helpful discussions, and Paige Vega from the Lau Laboratory for assistance in tailoring the sc-UniFrac analysis pipeline. K.S.L. is funded by the National Institutes of Health (NIH, grants R01DK103831 [National Institute of Diabetes and Digestive and Kidney Diseases], P50CA236733, U01CA215798, and U54CA217450 [National Cancer Institute]), J.J.H. is funded by NIH grant R35GM124685 (National Institute of General Medical Sciences), and C.N.H. is funded by NIH grant U2CCA233291 (National Cancer Institute).
Author contributions: B.C. and C.N.H. conceived of the quality-control and filtering methodology. J.J.H. assisted in design and interpretation of the statistical model and analysis. C.N.H. developed the dropkick software package and analyzed the data. V.M.W. performed simulations and sc-UniFrac analyses. C.N.H. and V.M.W. wrote the manuscript. K.S.L. supervised the study, secured funding, and participated in writing the manuscript and interpreting results.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.271908.120.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Banerjee A, Herring CA, Chen B, Kim H, Simmons AJ, Southard-Smith AN, Allaman MM, White JR, Macedonia MC, McKinley ET, et al. 2020. Succinate produced by intestinal microbes promotes specification of tuft cells to suppress ileal inflammation. Gastroenterology 159: 2101–2115.e5. 10.1053/j.gastro.2020.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Ramirez-Solano MA, Heiser CN, Liu Q, Lau KS. 2021. Processing single-cell RNA-seq data for dimension reduction-based analyses using open-source tools. STAR Protoc 2: 100450. 10.1016/j.xpro.2021.100450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming SJ, Marioni JC, Babadi M. 2019. CellBender remove-background: a deep generative model for unsupervised removal of background noise from scRNA-seq datasets. bioRxiv 10.1101/791699 [DOI]
- Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Soft 33: 1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerl AE, Kennard RW. 1970. Ridge regression: biased estimation for nonorthogonal problems. Technometrics 12: 55–67. 10.1080/00401706.1970.10488634 [DOI] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161: 1187–1201. 10.1016/j.cell.2015.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotliar D, Veres A, Aurel Nagy M, Tabrizi S, Hodis E, Melton DA, Sabeti PC. 2019. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. eLife 8: e43803. 10.7554/eLife.43803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. 2014. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph 20: 1983–1992. 10.1109/TVCG.2014.2346248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Herring CA, Sheng Q, Ping J, Simmons AJ, Chen B, Banerjee A, Li W, Gu G, Coffey RJ, et al. 2018. Quantitative assessment of cell population diversity in single-cell landscapes. PLoS Biol 16: e2006687. 10.1371/journal.pbio.2006687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, Marioni JC. 2019. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol 20: 63. 10.1186/s13059-019-1662-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J. 2018. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arXiv:1802.03426 [stat.ML].
- Otsu N. 1979. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9: 62–66. 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Southard-Smith AN, Simmons AJ, Chen B, Jones AL, Ramirez Solano MA, Vega PN, Scurrah CR, Zhao Y, Brenan MJ, Xuan J, et al. 2020. Dual indexed library design enables compatibility of in-Drop single-cell RNA-sequencing with exAMP chemistry sequencing platforms. BMC Genomics 21: 456. 10.1186/s12864-020-06843-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait SWG, Green DR. 2010. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11: 621–632. 10.1038/nrm2952 [DOI] [PubMed] [Google Scholar]
- Tibshirani R. 1996. Regression shrinkage and selection via the lasso. J R Stat Soc B 58: 267–288. 10.1111/j.2517-6161.1996.tb02080.x [DOI] [Google Scholar]
- Traag VA, Waltman L, van Eck NJ. 2019. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9: 5233. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ. 2018. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19: 15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, Theis FJ. 2019. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 20: 59. 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Corbett SE, Koga Y, Wang Z, Johnson WE, Yajima M, Campbell JD. 2020. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol 21: 57. 10.1186/s13059-020-1950-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Behjati S. 2020. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9: 12. 10.1093/gigascience/giaa151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Kamath GM, Tse DN. 2019. Valid post-clustering differential analysis for single-cell RNA-Seq. Cell Syst 9: 383–392.e6. 10.1016/j.cels.2019.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nature Commun 8: 14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. 2005. Regularization and variable selection via the elastic net. J R Stat Soc B 67: 301–320. 10.1111/j.1467-9868.2005.00503.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.