Abstract
Alternative polyadenylation (APA) is a major mechanism of post-transcriptional regulation in various cellular processes including cell proliferation and differentiation, but the APA heterogeneity among single cells remains largely unknown. Single-cell RNA sequencing (scRNA-seq) has been extensively used to define cell subpopulations at the transcription level. Yet, most scRNA-seq data have not been analyzed in an “APA-aware” manner. Here, we introduce dynamic analysis of APA from single-cell RNA-seq (scDaPars), a bioinformatics algorithm to accurately quantify APA events at both single-cell and single-gene resolution using either 3′-end (10x Chromium) or full-length (Smart-seq2) scRNA-seq data. Validations in both real and simulated data indicate that scDaPars can robustly recover missing APA events caused by the low amounts of mRNA sequenced in single cells. When applied to cancer and human endoderm differentiation data, scDaPars not only revealed cell-type-specific APA regulation but also identified cell subpopulations that are otherwise invisible to conventional gene expression analysis. Thus, scDaPars will enable us to understand cellular heterogeneity at the post-transcriptional APA level.
Alternative polyadenylation (APA) is a major mechanism of post-transcriptional regulation under diverse physiological and pathological conditions (Elkon et al. 2013; Tian and Manley 2017). The process of polyadenylation involves endonucleolytic cleavage of the nascent RNA followed by synthesis of a poly(A) tail on the 3′ terminus (Tian and Manley 2017). By using different polyadenylation sites [poly(A) sites], which are defined by flanking RNA sequence motifs, APA can generate mRNA isoforms with various 3′ untranslated regions (3′ UTRs) in the majority of human genes (Derti et al. 2012; Tian and Manley 2017). Although APA in most cases does not alter the protein-coding regions in those mRNA isoforms, it disrupts important cis-regulatory elements located in the 3′ UTRs, including adenylate-uridylate-rich elements (AREs) and binding sites of miRNAs and RNA-binding proteins, resulting in altered mRNA stability, localization, and translation efficiency (Garneau et al. 2007; An et al. 2008; Hoffman et al. 2016).
High-throughput sequencing technologies have revolutionized our understanding of APA over the last decade, illustrating both the pervasiveness of dynamic APA events and complexity of the APA regulatory processes. Recently, multiple studies have shed light on the global regulation of APA in response to changes in cell proliferation and cell differentiation in human diseases, including cancer (Tian and Manley 2017; Gruber and Zavolan 2019). Both proliferating cells and transformed cells often express a multitude of alternative mRNA isoforms with shortened 3′ UTRs through APA (Sandberg et al. 2008), leading to the activation of several proto-oncogenes such as CCND1, by escaping miRNA-mediated repression (Mayr and Bartel 2009). On the other hand, 3′ UTR lengthening is more prevalent in cell differentiation (Ji et al. 2009; Ji and Tian 2009). For example, progressive 3′ UTR lengthening is observed during mouse embryonic development (Ji et al. 2009), and the generation of induced pluripotent stem cells (iPSCs; dedifferentiation) is accompanied by global 3′ UTR shortening (Ji and Tian 2009). Besides regulating cognate transcripts in cis, APA-induced 3′ UTR changes can also disrupt competing endogenous RNA (ceRNA) regulation in trans, thus repressing several crucial tumor suppressors such as PTEN in breast cancer (Park et al. 2018). Although these observations imply a possible cell-state- or cell-type-dependent manner of APA regulation, the variability of APA among individual cells and the utility of APA in revealing novel cell subpopulations remain largely unknown.
Single-cell RNA sequencing (scRNA-seq) has become one of the most widely used technologies in biomedical research by providing an unprecedented opportunity to quantify the abundance of diverse transcript isoforms among individual cells (Shapiro et al. 2013; Saliba et al. 2014). However, methods to quantify relative APA usage across single cells remain underdeveloped. Recently, Velten et al. (2015) developed an experimental protocol, BATseq, to quantify various 3′UTR isoforms at the single-cell resolution. By integrating the standard scRNA-seq protocol and the 3′ enriched bulk RNA-seq protocol, Velten et al. (2015) found that cell types can be well separated based exclusively on their 3′ UTR isoform usage, indicating that APA is a molecular feature intrinsic to cell states. Although a compelling method, BATseq is hampered by its low sensitivity (∼5%) and high procedural complexity (Chen et al. 2017), thereby not being widely adopted in practice. In contrast, standard scRNA-seq data are widely available, yet most of the scRNA-seq data have not been analyzed in an “APA-aware” manner. Because scRNA-seq only captures a small fraction (typically 5%–15%) of the total mRNAs in each cell (Stegle et al. 2015), it can falsely quantify genes, especially lowly expressed ones, as unexpressed; this phenomenon is termed as “dropout.” Existing bulk RNA-seq-based APA methods such as DaPars (Xia et al. 2014) cannot overcome this vexing challenge when applied directly to scRNA-seq data, as they would lead to a high degree of sparsity in the resulting APA profiles. To address this sparsity, recently published computational approaches such as scDAPA (Ye et al. 2020) and scAPA (Shulman and Elkon 2019) extract and combine reads from cells aggregated based on predefined cell types. Alternatively, another study (Kim et al. 2019) aggregates individual genes into “metagenes” with reference to common functionality. Although these strategies cope with the problem of sparsity to some extent, they fail to retain the single-cell or single-gene resolution (Supplemental Table S1).
To fill this knowledge gap, we developed dynamic analysis of alternative polyadenylation from scRNA-seq (scDaPars), a bioinformatics algorithm for quantifying and recovering APA usage at the single-cell and single-gene resolution using standard scRNA-seq data. Because APA is reported to be regulated in a cell-state- or cell-type-specific manner, scDaPars uses a regression model that enables sharing of APA information across related cells to tackle the sparsity, achieving considerable robustness when applied to noisy scRNA-seq data. In addition, unlike scDAPA and scAPA, which are only applicable to 3′-end scRNA-seq data sets, scDaPars can be applied to both 3′-end and full-length scRNA-seq data. To the best of our knowledge, scDaPars is the first single-cell- and single-gene-level APA quantification method for analyzing standard scRNA-seq data.
Results
Overview of the scDaPars algorithm
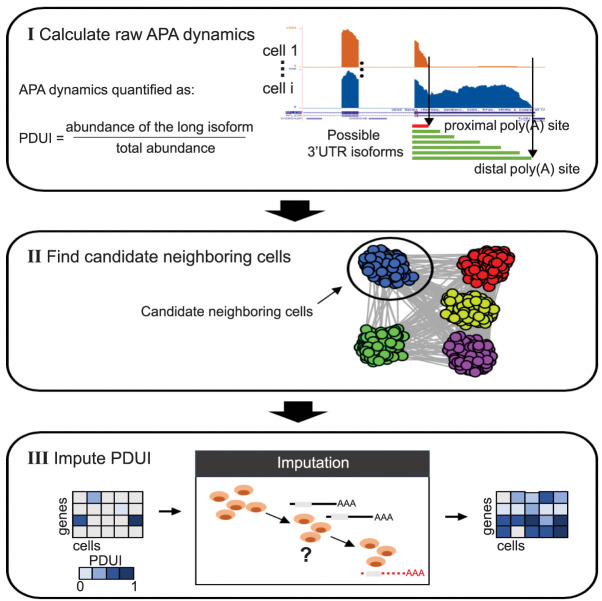
Figure 1 presents a schematic illustration of the scDaPars algorithm (for detailed definition and computational procedures, see Methods). Given a scRNA-seq data set, scDaPars first calculates raw relative APA usage, measured by the percentage of distal poly(A) site usage index (PDUI), based on the two-poly(A)-site model introduced in DaPars (Xia et al. 2014). scDaPars takes scRNA-seq genome coverage data as input and forms a linear regression model to jointly infer the exact location of proximal poly(A) sites by minimizing the deviation between the observed read density and the expected read density in all single cells. The relative APA usage is then quantified as the proportion of the estimated abundances of transcripts with distal poly(A) sites (longer 3′ UTRs) out of all transcripts (longer and shorter 3′ UTRs), and therefore, genes favoring distal poly(A) site usage (long 3′ UTRs) will have PDUI values near one, whereas genes favoring proximal poly(A) site usage (short 3′ UTRs) will have PDUI values near zero. This step (step I) will generate a PDUI matrix with rows representing genes and columns representing single cells. Of note, the raw PDUI values can only be estimated for genes with sufficient read coverages (default coverage of five reads per base), which automatically separates genes into robust genes (genes unaffected by dropout events) and dropout genes for further analysis. Because of the intrinsically low coverage of scRNA-seq data (Brennecke et al. 2013), the resulting PDUI matrix from step I is overly sparse with widespread missing data. To further recover the complete PDUI matrix independent of gene expression, we develop a new imputation method by sharing APA information across different cells. For a given cell, scDaPars begins by constructing a nearest neighbor graph based on the sparse PDUI matrix generated in step I (Fig. 1) to identify a pool of candidate neighboring cells that have similar APA profiles (step II). Finally, scDaPars uses a nonnegative least square (NNLS) regression model to refine neighboring cells based on robust genes and then borrow APA information in these neighboring cells to impute PDUIs of dropout genes in each cell (step III).
Figure 1.
A schematic illustration of the scDaPars algorithm. (I) scDaPars predicts both distal and proximal poly(A) sites by joint analysis of all single-cell samples and quantifies the raw relative APA usage by the proportion of estimated abundances of transcripts with distal poly(A) sites (long isoform). (II) scDaPars determines potential neighboring cells by applying community detection methods in APA profiles generated in step I. (III) scDaPars uses the NNLS regression model to refine neighboring cells and impute missing values by borrowing APA information from neighboring cells.
Evaluation of the accuracy and robustness of scDaPars
To quantitatively evaluate the accuracy of imputed APA usage by scDaPars, we used 384 scRNA-seq libraries of individual human peripheral blood cells (PBMCs) sequenced by a Smart-seq2 (Picelli et al. 2013) protocol and a matched bulk RNA-seq library from a benchmark study by Ding et al. (2020). Because we can estimate poly(A) sites and quantify differential poly(A) sites usage with high sensitivity and specificity in bulk RNA-seq data sets (Xia et al. 2014), we treated the results from the matched bulk sample as the pseudo-gold-standard for the following evaluation.
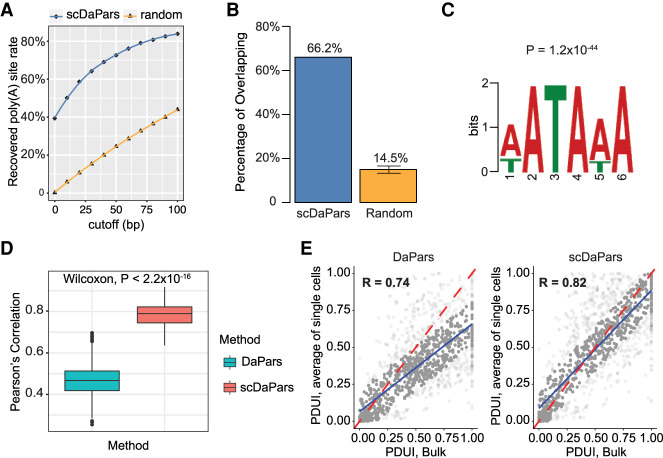
First, we showed that scDaPars reliably identified the location of proximal poly(A) sites in single cells. We found that ∼84% of poly(A) sites predicted from scRNA-seq data are within 100 bp of those predicted in bulk, whereas only ∼44% of randomly selected sites from 3′ UTR regions are within 100 bp of bulk predictions (Fig. 2A). We found that ∼66.2% of poly(A) sites predicted from scRNA-seq data also overlapped with annotated poly(A) sites complied from RefSeq, Ensembl, UCSC gene models, and poly(A)_DB (Wang et al. 2018) within 100 bp, and this overlap showed an approximately fivefold enrichment compared with random sites (Fig. 2B). In addition, canonical poly(A) signal (PAS) AATAAA was successfully identified by de novo motif analysis (Bailey 2011) within the upstream (−100 bp) sequence of single-cell predicted poly(A) sites with a P-value (P = 1.2 × 10−44) similar to that of bulk samples (P = 5.4 × 10−48) (Fig. 2C; Supplemental Fig. S1), supporting the validity of scDaPars's prediction of poly(A) sites.
Figure 2.
Evaluation of APA detection accuracy of scDaPars using human PBMCs data sets. (A) Fraction of poly(A) sites predicted in matched bulk RNA-seq data recovered in single cells using scDaPars or random control. Poly(A) sites predicted in scRNA-seq are considered true if they are located within cutoff distance from the bulk results. The cutoffs range from 0 to 100 bp with 10-bp increments. (B) Percentage of scDaPars predicted poly(A) sites or random control overlapped with annotated poly(A) sites from RefSeq, Ensembl, UCSC gene models, and poly(A)_DB. The confidence interval was derived by taking random sites 10 times. (C) The top-scoring signal identified by de novo motif analysis (DREME) from the upstream (−100 bp) of scDaPars predicted poly(A) sites from single cells. (D) Box plot showing Pearson's correlations between PDUI values of B cell pairs estimated by DaPars and scDaPars (Wilcoxon test P < 2.2 × 10−16). (E) Scatter plots of PDUI values between the average of all single cells and bulk results estimated by DaPars (left) and scDaPars (right). Red line represents the theoretical linear relationships between bulk and average of all single-cell PDUIs, and blue represents the actual linear relationships estimated from data.
Next, we showed that scDaPars was able to recover APA usage for genes affected by dropouts in scRNA-seq data. APA is found to be uniquely regulated in distinct immune cell types in PBMCs (Kim et al. 2019). Yet the median Pearson's correlation between APA (PDUI values) of single-cell pairs in the same B cell cluster is only 0.46 when PDUI values were calculated by DaPars (our previous method for bulk RNA-seq) owing to dropout effects (Fig. 2D). In contrast, scDaPars successfully recovered PDUI values for most of the affected dropout genes (Supplemental Fig. S2) and increased the median cell–cell correlation by a large margin (0.79; P < 2.2 × 10−16) (Fig. 2D). We further compared the average APA usage of all single cells with the bulk results. The Pearson's correlation between the average PDUI values of single cells and those of the bulk increased from 0.74 to 0.82 after scDaPars imputation (Fig. 2E). Notably, even though the correlation increase was not large, the regression slope increased significantly from 0.59 (DaPars) to 0.8 (scDaPars; P = 4.89 × 10−26), indicating APA usage quantified by scDaPars better represents the linear relationship between the average of single cells and the corresponding bulk.
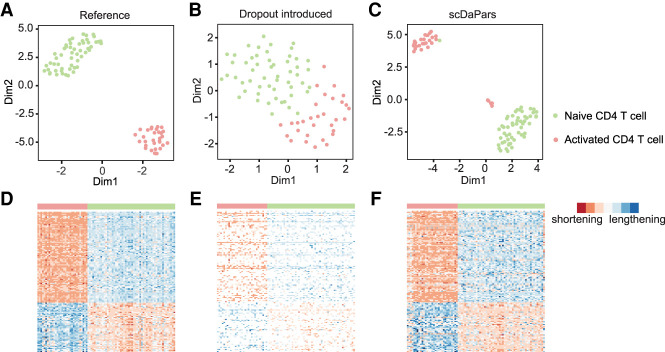
Finally, we used a simulation study to illustrate scDaPars's ability to identify dynamic APA events (see Methods) between two cell types. We created a synthetic PDUI matrix of naive and activated CD4 T cells based on bulk RNA-seq data from the DICE project (see Methods; Schmiedel et al. 2018). The naive and activated CD4 T cells are clearly distinguishable using the reference APA profiles estimated from bulk samples (Fig. 3A). Additionally, the reference data showed a strong inclination of 3′ UTR shortening in activated CD4 T cells (P = 3.8 × 10−4) (Fig. 3D), in line with previous reports that 3′ UTR shortening is widely observed upon activation of T cells (Sandberg et al. 2008). However, manually introduced dropout events obscured this differential 3′ UTR pattern, in which only ∼38% of differential APA genes remained, and the two cell types became less separated by their APA profiles (Fig. 3B,E). After we applied the imputation steps of scDaPars, ∼79% of differential APA genes are recovered, and the clear separation of these two cell types was restored (Fig. 3C,F). We further examined the robustness of scDaPars against varying dropout rates. Even though the accuracy of dynamic APA events identified by scDaPars decreased as the dropout rate increased, scDaPars could still achieve a >0.75 area under the receiver operating characteristics (ROC) curve when the proportion of dropout events was as high as 70% (Supplemental Fig. S3).
Figure 3.
Evaluation of scDaPars in identifying dynamic APA events between two cell types using naive and activated CD4 T cells. (A–C) Scatter plots showing UMAP results of 54 naive CD4 T cells and 31 activated CD4 T cells based on reference APA profiles (A), dropout events introduced APA profiles (B), or scDaPars corrected APA profiles (C). (D–F) Heat maps showing APA profiles of 136 differential APA genes (FDR ≤ 0.05 and PDUI differences ≥0.2) in the reference data (D), dropout events introduced data (E), and scDaPars corrected data (F). Rows represent differential APA genes and columns represent cells. Eighty-eight out of 136 differential APA genes have shorter 3′ UTRs in activated CD4 T cells in the reference data.
scDaPars outperforms existing methods by providing single-cell-resolution APA quantification applicable to both 3′-end and full-length scRNA-seq data
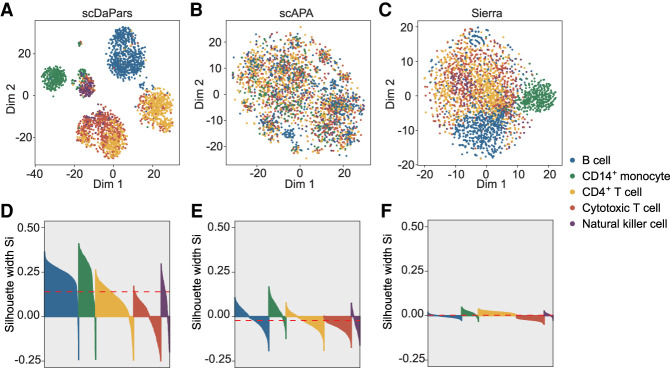
Several bioinformatics tools have been developed to analyze APA usage using scRNA-seq data (i.e., scDAPA [Ye et al. 2020] and scAPA [Shulman and Elkon 2019]), yet, unlike scDaPars, they were not designed to quantify APA usage at the single-cell resolution. During the preparation of this paper, we noticed another method, Sierra (Patrick et al. 2020), that detects differential transcript usage in scRNA-seq data may also be used for quantifying dynamic APA events. To illustrate the superiority of scDaPars over these existing methods, we applied scDaPars, scAPA, and Sierra to a benchmark 10x Chromium data set containing 902 single cells from three lung adenocarcinoma cell lines (see Methods; Tian et al. 2019). scDAPA was excluded from this study because it identifies APA events by pair-wise comparison without quantifying APA usage. scDaPars outperformed both scAPA and Sierra by generating clear and compact cell clusters according to annotated cell lines (UMAP) for visualization, see Supplemental Fig. S4A–C; McInnes et al. 2018). We used silhouette analysis to quantitatively assess the resulting clusters. Compared with scAPA and Sierra, scDaPars showed higher silhouette coefficients, which indicated the clustering results from scDaPars are more congruent with the true cell-line labels (Supplemental Fig. S4D–F). To further benchmark scDaPars in more complex biological systems, we applied scDaPars, scAPA, and Sierra to an immune data set containing 3362 PBMCs (see Methods; Ding et al. 2020). Again, the APA usage quantified by scDaPars generated compact and accurate immune cell clusters (Fig. 4A,D). In contrast, although Sierra outperformed scAPA and was able to separate B cell and CD14+ monocytes (Fig. 4B,C), both Sierra and scAPA failed to accurately distinguish the five immune cell types (Fig. 4E,F). Besides generating accurate cell clusters, scDaPars also identified 169 dynamic APA genes (genes with differential poly(A) site usage) among the five immune cell types, most of which (96%) were unseen by existing methods. For example, scDaPars identified EIF1 as a dynamic APA gene between B cells and CD14+ monocytes. Both cluster- and single-cell-level coverage plots corroborated that EIF1 shows 3′ UTR lengthening in B cells compared with CD14+ monocytes (Supplemental Fig. S5). However, EIF1 was not captured by previous methods (i.e., scAPA), indicating the advantage of scDaPars. More importantly, scDAPA, scAPA, and Sierra rely on peak calling using 3′-end enriched reads in 10x Chromium to quantify APA usage and thus are not applicable to data generated by full-length sequencing protocols like Smart-seq2 that do not contain enriched peaks in the 3′ UTR regions (Picelli et al. 2013).
Figure 4.
scDaPars outperforms existing methods by quantifying APA usage at single-cell resolution. (A–C) Scatter plots showing UMAP results of 3362 PBMCs based on scDaPars quantified APA usage (A), scAPA quantified APA usage (B) or Sierra quantified APA usage (C). (D–F) Silhouette plots for clustering results from scDaPars (D), scAPA (E), and Sierra (F). The x-axis represents cells, and y-axis is the corresponding silhouette coefficient Si for each cell. The silhouette coefficient measures how similar a cell is to its own cluster compared with other clusters; therefore, a higher silhouette coefficient indicates a better clustering result, and a negative coefficient may suggest the cell is assigned to the wrong cluster. The red dashed line is the average Si for all cells.
scDaPars revealed intrinsic tumor APA variations and immune cell subpopulations in primary breast cancer
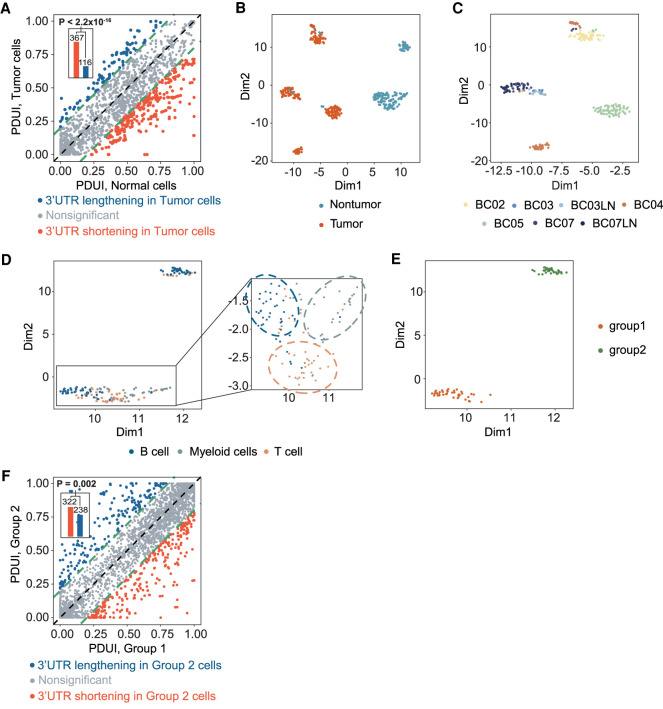
Global-scale coordinated APA events are commonly observed in cancers (Xia et al. 2014), and APA-induced 3′ UTR shortening was shown to be associated with tumor aggressiveness and poor survival of cancer patients (Lembo et al. 2012; Xia et al. 2014). However, knowledge of APA regulations in cancer has been largely derived from bulk RNA-seq studies. Therefore, although global APA variations between tumor and normal cells have been well characterized, little is known about the intertumoral APA heterogeneity at the single-cell resolution. To illustrate scDaPars’ capacity of characterizing single-cell APA variations in cancers, we applied scDaPars to a Smart-seq2 (Picelli et al. 2013) scRNA-seq data set containing 563 single cells from 11 breast cancer patients (Chung et al. 2017). In consistent with bulk results, 3′ UTRs were shortened in tumor cells compared with normal cells (P < 2.2 × 10−16) (Fig. 5A). Even PDUI values before scDaPars imputation could separate tumor cells from nontumor cells with an effectiveness comparable with that of gene expression values (Supplemental Fig. S6A), suggesting an important role of dynamic APA events in breast cancer progression. As expected, scDaPars-imputed APA profiles showed a better separation between tumor and nontumor groups (Fig. 5B; Supplemental Fig.S7).
Figure 5.
scDaPars reveals tumor-specific and immune-cell-type-specific APA landscape in primary breast cancer. (A) Scatter plot of PDUI values in tumor and normal cells. For each gene, the mean PDUI values in tumor cells (y-axis) are plotted against that in normal cells (x-axis). Genes with shortened or lengthened 3′ UTR (FDR ≤ 0.05 and PDUI difference ≥0.2) in tumor cells are shown in red and blue. Bar plot shows the number of shortening genes or lengthening genes in tumor cells, and P-value is calculated using a single-tailed binomial test. (B) Scatter plot gives UMAP results calculated from scDaPars-restored APA profiles. Each dot represents a cell, and cells are labeled based on cell index provided in the original publication. (C) Scatter plot of UMAP results of tumor cells. Cells are labeled by patient information. (D) Scatter plot of UMAP results of immune cells. Cells are labeled by cell type information. (E) Scatter plot of UMAP results of B cells based on scDaPars results. (F) Scatter plot of PDUI values in group 1 B cells and group 2 B cells. For each gene, the mean PDUI values in group 2 B cells (y-axis) are plotted against that in group 1 B cells (x-axis). Genes with shortened or lengthened 3′ UTR (FDR ≤ 0.05 and PDUI difference ≥0.2) in group 2 B cells are shown in red and blue. Bar plot shows the number of shortening genes or lengthening genes in group 2 cells.
To further elucidate APA variations among cell subgroups, we analyzed APA profiles of tumor and nontumor cells separately. On the one hand, contrary to a previous single-cell APA analysis performed on aggregated “metagenes” in the same breast cancer data set (Chung et al. 2017), which showed that no differences in APA were associated with cancer subtypes or patients (Kim et al. 2019), we found that tumor cells were not only separated into patient-specific clusters based on scDaPars-imputed APA profiles (Fig. 5C) but also further classified into different molecular subtypes (Supplemental Fig. S8), showing evidence of both intertumoral and cancer-subtype-specific APA heterogeneity, as well as scDaPars's advantage over the existing method. On the other hand, nontumor cells, which were derived from the same group of patients as tumor cells, were clustered mainly according to their cell types (B cells, myeloid cells, and T cells) instead of patients (Fig. 5D; Supplemental Fig. S6B). This result not only reaffirmed that dynamic APA events are cell-type-specific characteristics of immune cells but also indicated that the patient-specific APA profiles observed in tumor cells were unlikely owing to batch effects in patient samples but rather reflected true intertumoral variations in APA.
In addition, consistent with prior knowledge of two B cell subclasses (proliferating and naive/memory B cells) in this data set, we observed that B cells were classified into two cell subgroups based on scDaPars-imputed APA profiles (Fig. 5E) with group 2 B cells showing global 3′ UTR shortening compared with group 1 B cells (P = 2 × 10−3) (Fig. 5F). We found that most B cell proliferation signature genes from the literature (Chung et al. 2017) were up-regulated in group 2 B cells compared with group 1 B cells (Supplemental Fig. S9; Supplemental Table S2), suggesting that group 2 B cells may represent proliferating B cells. Indeed, the proliferating and naive/memory B cells determined by the expression of B cell proliferating marker genes are highly congruent with scDaPars-derived cell subgroups (Supplemental Fig. S10A,B). These results are also in line with previous reports that proliferating cells (i.e., group 2 cells) express more isoforms with shortened 3′ UTRs through APA (Sandberg et al. 2008). However, expression analysis of all genes failed to identify these B cell subgroups (Supplemental Fig. S10C), revealing the potential benefits of APA analysis in delineating cell subpopulations. In summary, scDaPars improves the characterization of APA variations and cell subpopulations in single cells.
scDaPars enables identification of novel cell subpopulations invisible to conventional gene expression analysis in endoderm differentiation
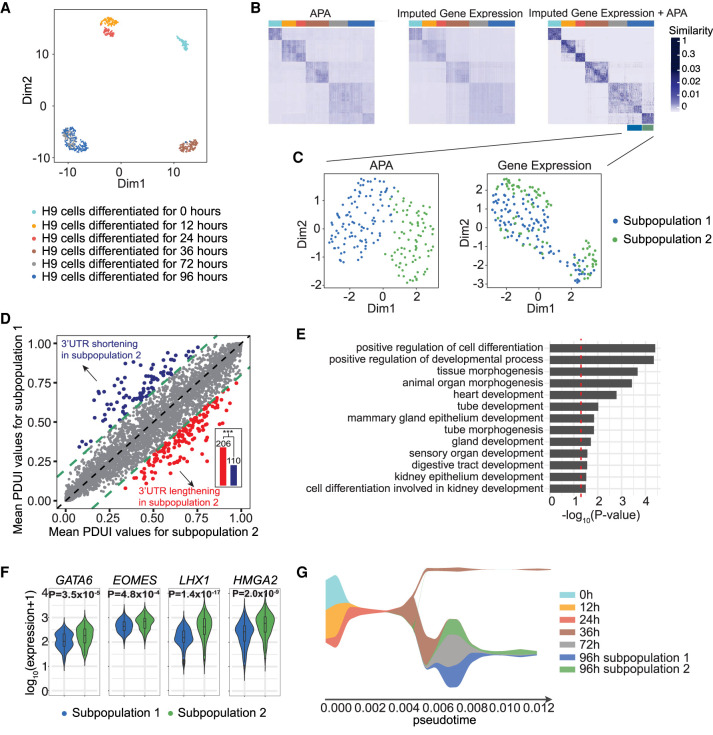
As APA patterns appear to be globally regulated in cell differentiation (i.e., decreased proximal poly(A) site usage in more differentiated states of embryonic development) (Ji et al. 2009; Tian and Manley 2017), we hypothesized that they could provide a new aspect to identify cell subpopulations during differentiation. To test this hypothesis, we applied scDaPars to a time course Smart-seq2 (Picelli et al. 2013) scRNA-seq data set containing 758 cells sequenced at 0, 12, 24, 36, 72, and 96 h of differentiation during human definitive endoderm (DE) emergence (Chu et al. 2016). scDaPars revealed clear and accurate cell clusters from each time point along the differentiation process (Fig. 6A). Dimension 2 of the UMAP projection of raw PDUI values reconstructed single-cell orders matching the true differentiation time points, reflecting the global APA dynamics during cell differentiation (Supplemental Fig. S11).
Figure 6.
scDaPars helps identify novel cell subpopulations during human embryonic development. (A) Scatter plot shows UMAP results of single cells based on scDaPars-recovered APA profiles. Cells are labeled based on cell differentiation time points given in the original publication. (B) Cell-by-cell similarities represented by similarity matrices generated by R package SNFtool. (C) Scatter plots of UMAP results of cells in 96 h of differentiation based on scDaPars results (left) and imputed gene expression (right). Cells are labeled by results from B. (D) Scatter plot shows mean PDUI values of genes in subpopulation 2 (x-axis) and subpopulation 1 (y-axis). Genes with 3′ UTR shortening and lengthening (FDR ≤ 0.05 and PDUI differences≥0.2) in subpopulation 2 are labeled in blue and red, respectively. Bar plot shows the number of genes with shortening or lengthening in subpopulation 2, and P-value is calculated using single-tailed binomial test. (E) Selected GO terms enriched in the up-regulated genes in subpopulation 2. (F) Example gene expression levels in two subpopulations. (G) Stream plot from STREAM that shows cell density along different trajectories at a given pseudotime.
Next, we investigated whether APA could help delineate novel cell subpopulations invisible to gene expression analysis alone. Imputation based on observed gene expression has been shown to enhance the identification of cell subpopulations (Li and Li 2018). Therefore, to ensure APA is providing additional information beyond expression, we first recovered plausible single-cell gene expression data using scImpute (Li and Li 2018), a state-of-the-art gene expression imputation method. Notably, although the imputed gene expression profile outputs more compact clusters than the raw-expression, single cells collected from 72 and 96 h of differentiation were still largely overlapped (Supplemental Fig. S12). To characterize additional cellular heterogeneity, we integrated APA information with imputed gene expression using similarity network fusion (SNF) (Wang et al. 2014). By creating and converging separate similarity networks for APA and gene expression, SNF reduced noisy intercluster similarities among cells in 12 and 24 h of differentiation and enhanced intracluster similarities observed in one or both similarity networks (Fig. 6B). We then quantitatively compared the clustering results by using a spectral clustering algorithm (Ng et al. 2002) on different similarity networks with the number of clusters k = 6. The clustering results are evaluated by normalized mutual information (NMI) (Witten et al. 2016), where NMI = 1 indicates a perfect match between the clustering results and the known differentiation time points. Although gene expression imputation increased NMI from 0.76 to 0.85, integration of APA usages with imputed gene expression further increased NMI from 0.85 to 0.89, suggesting the benefits of adding APA information.
Besides unifying the clustering results of APA and gene expression, the fused similarity network also revealed novel and potentially meaningful subpopulations. For example, cells at 96 h of differentiation were divided into two previously unidentified subpopulations (Fig. 6B). Through analyzing APA and gene expression between the two subpopulations, we found that APA usage alone can accurately separate the two subpopulations (Fig. 6C; Supplemental Fig. S13), and subpopulation 2, which was more distinct from cells in 72 h of differentiation than subpopulation 1, showed global 3′ UTR lengthening compared with subpopulation 1 (P = 3.64 × 10−8) (Fig. 6D), whereas the imputed gene expression profile alone failed to distinguish the two subpopulations (Fig. 6C). The APA profile quantified by DaPars also failed to identify the two subgroups (Supplemental Fig. S14), indicating the superiority of scDaPars.
Because subpopulation 2 showed global 3′ UTR lengthening, we hypothesized it may represent a more differentiated cell subgroup. To test our hypothesis, we performed differential gene expression analysis between subpopulation 1 and 2 using DESeq2 (Love et al. 2014). As a result, subpopulation 2 was characterized by higher expression of endoderm development marker genes, including GATA6, EOMES, and SOX17 (Fig. 6F; Supplemental Table S3; Chu et al. 2016). In addition, the transcriptional profile of subpopulation 2 also included significantly up-regulated endoderm-development-related genes like LHX1, which is important for renal development (Reidy and Rosenblum 2009), and HMGA2, which is required for epithelium differentiation during embryonic lung development (Singh et al. 2014), suggesting subpopulation 2 has a more differentiated phenotype than subpopulation 1. To further elucidate the global biological differences between the two subpopulations, we performed Gene Ontology (GO) analysis (Luo et al. 2009). We found that several endoderm-development-related GO terms were highly enriched in the up-regulated genes in subpopulation 2 (Fig. 6E). Furthermore, using the expression of differential APA genes, we were able to separate the two subpopulations (Supplemental Fig. S15), indicating that some biologically meaningful subpopulations were masked by overall gene expression analysis. Finally, we conducted a trajectory analysis by STREAM (Chen et al. 2019) to independently show the validity of the identified subpopulations. Using cells at 0 h of differentiation as a natural starting point (root), we found that most cells are projected onto the inferred branches according to their corresponding differentiation time points (Supplemental Fig. S16A,B), and the derived pseudotime progression corroborated that cells in subpopulation 2 are more differentiated than those in subpopulation 1 (Fig. 6G; Supplemental Fig. S16C). Considered collectively, scDaPars-calculated APA usage offered an additional layer of information in characterizing cellular heterogeneity that was otherwise invisible in gene expression analysis.
Discussion
Here, we developed scDaPars, a novel bioinformatics algorithm to de novo identify and quantify single-cell dynamic APA events using standard scRNA-seq data. Many methods have been developed to measure the relative APA usages in RNA-seq data from bulk samples (Xia et al. 2014). However, the widespread dropout events in scRNA-seq data impede these bulk-sample-based methods to quantify APA usage among single cells (Fig. 2D,E). To address this technical challenge in scRNA-seq, scDaPars first quantifies raw APA usage based on the two-poly(A)-site model introduced in DaPars (Xia et al. 2014). Because APA shows a cell-type-specific pattern (Velten et al. 2015; Kim et al. 2019), scDaPars then clusters cells into different cell neighbors based on their calculated raw APA profiles. Next, scDaPars imputes missing APA usage by borrowing APA information of the same gene from neighboring cells. Benchmarking on both real and simulated data shows the accuracy of scDaPars in predicting poly(A) sites, the ability in recovering missing APA usages, and the robustness in identifying dynamic APA events across different cell types (Figs. 2, 3).
Previously, methods for analyzing APA usage using scRNA-seq data mostly address the high technical noise in scRNA-seq by creating pseudobulk RNA-seq data (i.e., pooled reads from cells that are assigned to the same cell cluster) (Shulman and Elkon 2019; Ye et al. 2020). Unlike scDaPars, even though these methods perform on scRNA-seq data, they do not quantify APA usage at the single-cell resolution but rather measure cell-cluster APA usage, which contradicts the purpose of single-cell sequencing (Supplemental Table S1). Additionally, previous methods are confined by cell cluster assignments determined by conventional gene expression analysis. In contrast, scDaPars quantifies single-cell APA usage independent of gene expression, which provides an additional layer of APA information that helps identify hidden cell states. (Fig. 6C).
Finally, unlike existing methods, we expect scDaPars to be widely applicable to any scRNA-seq data sets. Although the main analysis presented in this article builds on scRNA-seq data generated by low-throughput Smart-seq2 (Picelli et al. 2013) protocol and the accuracy of scDaPars decreases as the dropout rate increases (Supplemental Fig. S3), scDaPars can also be applied to data sets generated by high-throughput high-dropout-rate droplet-based methods; for example, 10x Chromium (Zheng et al. 2017). For example, scDaPars successfully revealed cell-type-specific APA patterns in 3362 PBMCs sequenced by 10x Chromium (Fig. 4A; Ding et al. 2020). Together, scDaPars provides an additional layer of APA information that helps identify cell subpopulations invisible to conventional gene expression analysis.
Methods
De novo quantification of dynamic APA events
scDaPars first performs de novo identification and quantification of dynamic APA events based on the two-poly(A)-site model introduced in DaPars. The bedGraph files for each single cell were used as input and jointly analyzed to calculate the APA usage measured as the PDUI. For each gene, the distal poly(A) site was identified as the end point of the longest 3′ UTR among all scRNA-seq samples, and the proximal poly(A) site was inferred by optimizing the following linear regression model:
| (1) |
where and are the abundances of transcripts with distal and proximal poly(A) sites for cell i, Ci is the read coverage of cell i normalized by total sequencing depth, L is the length of the longest 3′ UTR, P is the length of the alternative proximal 3′ UTR to be inferred, and IL and IP are two indicator functions for long and short 3′ UTRs such that and . The optimal proximal poly(A) site is selected by minimizing the deviation between the observed read density Ci and the expected read density in all single cells. The APA usage is then quantified as PDUI for each gene in each single cell, with PDUI defined as
| (2) |
where and are the optimal expression levels of transcripts with the distal and proximal poly(A) site for cell i. The smaller the PDUI is, the less distal poly(A) site is used and the shorter the 3′ UTRs. The final output is a PDUI matrix in which rows represent genes and columns represent cells. Additionally, PDUIs can only be calculated in this step for genes with sufficient read coverage (default coverage of five reads per base), which automatically separates genes into robust genes and dropout genes for future analysis. On average, 50% of the genes in a cell are robust genes after quality control, and if the dropout rate in the data set is higher (e.g., in 10x Chromium data sets), the average number of robust genes in the data will decrease. There are overlaps between robust genes of different cells: In the benchmark data set in Figure 2, the overlap of robust genes between any two cells is ∼40%.
Detection of potential neighboring cells and outliers
Because APA shows alterations in different cell types and cell states in a global scale, scDaPars recovers missing single-cell-level APA usage by borrowing APA information of the same gene from neighboring cells. A critical step here is to determine which cells are from the same cell subpopulation and therefore are neighboring cells. Instead of using observed gene expression, scDaPars uses raw APA usage for this task because (1) APA is a feature intrinsic to cell types or cell states, and (2) scDaPars quantifies APA usage independent of gene expression. We first performed a quantitative comparison of clustering using raw APA usage and observed gene expression from the hESC data set in Figure 6 (Supplemental Fig. S17). We found that clustering of raw APA usage outperformed that of observed gene expression (Supplemental Fig. S17C,D) partly because differentiation is one of the biological processes with the most dramatic APA changes. To further illustrate the benefits of quantifying APA independent of gene expression, we modified our original scDaPars algorithm so that the initial clustering is performed using observed gene expression instead of raw APA usage and requantified the APA usage of cells from the hESC data set in Figure 6. We found that the two subpopulations identified by original scDaPars were obscured by the modified version (Supplemental Fig. S18), indicating the advantage of quantifying APA independent of gene expression.
Because of the technical limitation of scRNA-seq data, it is unlikely to completely cluster cells into true subpopulations based on the sparse PDUI matrix generated in last step. Instead, the goal of this step is to determine a set of potential neighboring cells that scDaPars will fine-tune in the following imputation step.
To increase the robustness and reliability of the clustering results and to find more plausible neighboring cells, scDaPars applies principal component analysis (PCA) to the raw PDUI matrix. Although the PDUI matrix is sparse, the modularity of dynamic APA provides redundancy in gene dimensions, which can be exploited. Therefore, scDaPars selects principal components (PCs) that can together explain at least 40% of the variance in the data. Note that the neighboring cells are identified in these PCA dimensions, whereas the imputation is performed on the full PDUI matrix:
| (3) |
Next, scDaPars identifies and removes outlier cells from the analysis. The outlier cells may be the result of technical errors or may represent true rare biological variations; in either case, scDaPars will not use these outlier cells to impute missing APA usage in other cells. We calculate the distance matrix DN×N between cells based on the PCA-transformed data PDUIpca. For each cell m, we define the Euclidean distance of cell m to its nearest neighbor as dm, resulting in a set d = {d1, …, dN}. We denote the first quantile of d as Q1 and its third quantile as Q3 and the distance between Q1 and Q3 as interquartile range IQR. The outlier cells are defined as cells that are separated by more than 1.5 IQR to the third quantile Q3:
| (4) |
The remaining nonoutlier cells are then clustered into subpopulations using a graph-based community detection algorithm. The single cells are the vertices in the graph, and community detection in graphs will identify groups of vertices with high probability of being connected to each other than to members of other groups. We use R package RANN with default parameters to first identify the approximate nearest neighbors and convert neighbor relation matrix into an adjacency matrix. We then use igraph (Csardi and Nepusz 2006) to represent the resulting adjacency matrix as a graph and apply the walkstrap (Pons and Latapy 2005) algorithm to identify communities of vertices (cells) that are densely connected. Suppose scDaPars divides cells into K subpopulations in this step, for each cell m, its potential neighboring cells Nm are the other cells in the same cell subpopulation k:
| (5) |
Imputation of missing APA usage
After potential neighboring cells Nm for each cell are determined, we impute APA usage cell by cell. Recall that PDUIs can only be estimated for genes with sufficient read coverage; scDaPars thereby automatically separates genes into robust genes and dropout genes when calculating the PDUI matrix. Here, we denote the set of robust genes for cell m as Rm and the set of dropout genes that will be imputed in this step as Dm. scDaPars then learns the cells’ similarities through the robust gene set GRobust,m and impute the APA usage of Dm by borrowing information from the same gene's APA usage in other neighboring cells learned from Rm. To fine-tune the grouping of neighboring cells from Nm, we use NNLS regression:
| (6) |
where Nm represents the indices of cells that are potential neighboring cells of cell m, is a vector of response variables representing Rm rows in the mth column (cell m) of the original PDUI matrix, and is a submatrix of the original PDUI matrix with dimensions |Rm| × |Nm|. The goal is to find the optimal coefficients of length |Nm| that can minimize the deviation between APA usage of Rm in cell m and those in potential neighboring cells. The advantage of using NNLS is that it has the property of leading to a sparse estimate of θm, whose components may have exact zeros, so that true neighboring cells of cell m are conveniently selected from Nm. Once is computed, we have a vector of weighted neighbors associated with each cell in our data. scDaPars use this coefficient estimated from the set Rm to impute the APA usage of genes in the set Dm in cell m. All of the above analyses are conducted in R (R Core Team 2020).
| (7) |
Differential PDUI
We used the following two criteria to define the significant differential PDUI (dynamic APA events): First, given the PDUI values for cells in two cell types, the Benjamini–Hochberg corrected Mann–Whitney U P-value between two cell types (FDR) is less than 0.05; second, the absolute difference of mean PDUIs in cell type 1 and cell type 2 is greater than 0.2.
| (8) |
Preprocessing of scRNA-seq data
The scRNA-seq data sets used in this manuscript are all publicly available and are summarized in Supplemental Table S4. The two single-cell PBMC data are available at the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE132044. The breast cancer data are available at GEO under accession number GSE75688. The time course DE data are available at GEO under accession number GSE75748. The lung adenocarcinoma cell line data are available at GEO under accession number GSE118767. The DICE immune data used to generate synthetic data set were obtained from the NCBI database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) under study accession number phs001703.v1.p1. For low-throughput data sets generated by the Smart-seq2 (Picelli et al. 2013) protocol, we downloaded the publicly available FASTQ files from GEO database and aligned the reads using STAR 2.5.2 (Dobin et al. 2013) with default parameters, generating one BAM file for each single cell. For high-throughput data sets generated by 10x Chromium (Zheng et al. 2017), we downloaded the FASTQ files and aligned the reads using Cell Ranger 3.0.2. We then selected reads with correct unique molecular identifier (UMI) using Drop-seq tool FilterBAM (Macosko et al. 2015) and remove reads with duplicated UMIs using UMI-tool dedup (Smith et al. 2017). We next merged reads originated from same cells together and generated one BAM file for each single cell. The BAM files are used as inputs for subsequent scDaPars analysis. The average dropout rate (percentage of missing data) for Smart-seq2 data sets is ∼50% in our study. The 10x Chromium data set in our study has a dropout rate of ∼65%.
Generation of the synthetic data set
The synthetic data set was created based on bulk RNA-seq data generated from 13 immune cell types (Schmiedel et al. 2018). The different immune cell types are isolated so that each sample only contains cells from one cell type. We used DaPars to estimate the APA usage in these bulk samples and generated an APA matrix, in which rows represent genes and columns represent samples. Because widespread dynamic APA events were reported in naïve and activated CD4 T cells, we selected only samples that belong to these two cell types for the following simulation.
We down-sampled the resulting bulk APA matrix to emulate the APA profiles generated from single-cell data. We first calculated the dropout rate for each gene in the benchmark immune data set (Ding et al. 2020). Next, for each gene in the bulk APA matrix, the dropout rate is randomly selected from the set of real dropout rates with replacement. Finally, we used Bernoulli distribution with P equals to the selected dropout rate and n equals to the number of samples to introduce dropouts into the synthetic data set. The final dropout introduced data has a ∼50% dropout rate, which is similar to the dropout rate of the real data sets. Notice that the generation of the synthetic data set is independent from the models of scDaPars, so that it can be used to evaluate scDaPars in a fair way.
Benchmark comparison of scDaPars
To illustrate the advantage of scDaPars, we applied scDaPars, scAPA, and Sierra to two benchmark 10x Chromium data sets. scAPA measures differential usage of poly(A) sites between different cell types by the proximal poly(A) site usage index (proximal PUI). Because we want to test scAPA's ability for quantifying single-cell-level APA usage, we input single-cell coverage into scAPA to generate a cell-by-transcript-proximal-PUI matrix to perform the clustering analysis. The Sierra pipeline does not yield PDUI-like measurements. Instead, it generates a peak count matrix in which peak coordinates are annotated according to the genomic features they fall on, including UTRs, exons, or introns. To calculate APA usage from the peak count matrix, we first selected peaks falling on the 3′ UTRs and only kept transcripts with more than one peak. We then transferred the peak count matrix into an APA matrix by calculating the relative usage of the most distal peak. The resulting APA matrix were used for the clustering analysis. Finally, we performed silhouette analysis by silhouette () in R package cluster v2.1.0. to quantitatively evaluate the clustering accuracy of the three methods.
Supplementary Material
Acknowledgments
We thank Yikai Luo, Dr. Joel Neilson at Baylor College of Medicine, members of the Li laboratory at University of California at Irvine, and Dr. Jingyi Jessica Li at University of California at Los Angeles for insightful discussions. We thank Dr. Chen Chao at Baylor College of Medicine for his suggestions on this manuscript. This work is supported by US National Institutes of Health (NIH) grants R01HG007538, R01CA193466, and R01CA228140 to W.L. and the Cancer Prevention Research Institute of Texas (CPRIT) grant RR170048 to C.I.A. C.I.A. is a CPRIT research scholar.
Author contributions: W.L. conceived and supervised the project. Y.G. performed the data analysis. Y.G., L.L., and W.L. interpreted the data. Y.G., L.L., W.L., and C.I.A. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.271346.120.
Software availability
The source codes and the R package scDaPars are available as Supplemental Code. scDaPars is also freely available at GitHub (https://github.com/YiPeng-Gao/scDaPars).
Competing interest statement
The authors declare no competing financial interests.
References
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. 2008. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134: 175–187. 10.1016/j.cell.2008.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. 10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, et al. 2013. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 10: 1093–1095. 10.1038/nmeth.2645 [DOI] [PubMed] [Google Scholar]
- Chen W, Jia Q, Song Y, Fu H, Wei G, Ni T. 2017. Alternative polyadenylation: methods, findings, and impacts. Genom Proteom Bioinform 15: 287–300. 10.1016/j.gpb.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Albergante L, Hsu JY, Lareau CA, Lo Bosco G, Guan J, Zhou S, Gorban AN, Bauer DE, Aryee MJ, et al. 2019. Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nat Commun 10: 1903. 10.1038/s41467-019-09670-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L-F, Leng N, Zhang J, Hou Z, Mamott D, Vereide DT, Choi J, Kendziorski C, Stewart R, Thomson JA. 2016. Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol 17: 173. 10.1186/s13059-016-1033-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T, Ryu HS, Kim S, Lee JE, Park YH, et al. 2017. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun 8: 15081. 10.1038/ncomms15081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. 2006. The igraph software package for complex network research. Int J Complex Syst 1695: 1–9. [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. 10.1101/gr.132563.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND, Hughes TK, Wadsworth MH, Burks T, Nguyen LT, et al. 2020. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 38: 737–746. 10.1038/s41587-020-0465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506. 10.1038/nrg3482 [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126. 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- Gruber AJ, Zavolan M. 2019. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet 20: 599–614. 10.1038/s41576-019-0145-z [DOI] [PubMed] [Google Scholar]
- Hoffman Y, Bublik DR, Ugalde AP, Elkon R, Biniashvili T, Agami R, Oren M, Pilpel Y. 2016. 3′UTR shortening potentiates microRNA-based repression of pro-differentiation genes in proliferating human cells. PLoS Genet 12: e1005879. 10.1371/journal.pgen.1005879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B. 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 4: e8419. 10.1371/journal.pone.0008419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033. 10.1073/pnas.0900028106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Chung W, Eum HH, Lee H-O, Park W-Y. 2019. Alternative polyadenylation of single cells delineates cell types and serves as a prognostic marker in early stage breast cancer. PLoS One 14: e0217196. 10.1371/journal.pone.0217196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo A, Di Cunto F, Provero P. 2012. Shortening of 3′ UTRs correlates with poor prognosis in breast and lung cancer. PLoS One 7: e31129. 10.1371/journal.pone.0031129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WV, Li JJ. 2018. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun 9: 997. 10.1038/s41467-018-03405-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. 2009. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10: 161. 10.1186/1471-2105-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J. 2018. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arXiv:1802.03426 [stat.ML]. [Google Scholar]
- Ng AY, Jordan MI, Weiss Y. 2002. On spectral clustering: analysis and an algorithm. Adv Neural inform Process Syst: 849–856. [Google Scholar]
- Park HJ, Ji P, Kim S, Xia Z, Rodriguez B, Li L, Su J, Chen K, Masamha CP, Baillat D, et al. 2018. 3′ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet 50: 783–789. 10.1038/s41588-018-0118-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick R, Humphreys DT, Janbandhu V, Oshlack A, Ho JW, Harvey RP, Lo KK. 2020. Sierra: discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. Genome Biol 21: 167. 10.1186/s13059-020-02071-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10: 1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- Pons P, Latapy M. 2005. Computing communities in large networks using random walks. In International Symposium on Computer and Information Sciences, pp. 284–293. Springer, Berlin, Heidelberg. [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Reidy KJ, Rosenblum ND. 2009. Cell and molecular biology of kidney development. In Seminars in nephrology, Vol. 29, pp. 321–337. Elsevier, Philadelphia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba AE, Westermann AJ, Gorski SA, Vogel J. 2014. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res 42: 8845–8860. 10.1093/nar/gku555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, et al. 2018. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175: 1701–1715.e16. 10.1016/j.cell.2018.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. 2013. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14: 618–630. 10.1038/nrg3542 [DOI] [PubMed] [Google Scholar]
- Shulman ED, Elkon R. 2019. Cell-type-specific analysis of alternative polyadenylation using single-cell transcriptomics data. Nucleic Acids Res 47: 10027–10039. 10.1093/nar/gkz781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Mehta A, Contreras A, Boettger T, Carraro G, Wheeler M, Cabrera-Fuentes HA, Bellusci S, Seeger W, Braun T, et al. 2014. Hmga2 is required for canonical WNT signaling during lung development. BMC Biol 12: 21. 10.1186/1741-7007-12-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Heger A, Sudbery I. 2017. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res 27: 491–499. 10.1101/gr.209601.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Teichmann SA, Marioni JC. 2015. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet 16: 133–145. 10.1038/nrg3833 [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Dong X, Freytag S, Lê Cao K-A, Su S, JalalAbadi A, Amann-Zalcenstein D, Weber TS, Seidi A, Jabbari JS, et al. 2019. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat Methods 16: 479–487. 10.1038/s41592-019-0425-8 [DOI] [PubMed] [Google Scholar]
- Velten L, Anders S, Pekowska A, Järvelin AI, Huber W, Pelechano V, Steinmetz LM. 2015. Single-cell polyadenylation site mapping reveals 3′ isoform choice variability. Mol Syst Biol 11: 812. 10.15252/msb.20156198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, Goldenberg A. 2014. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 11: 333–337. 10.1038/nmeth.2810 [DOI] [PubMed] [Google Scholar]
- Wang R, Nambiar R, Zheng D, Tian B. 2018. Polya_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res 46: D315–D319. 10.1093/nar/gkx1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten IH, Frank E, Hall MA, Pal CJ. 2016. Data mining: practical machine learning tools and techniques. Morgan Kaufmann, Burlington, MA. [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. 2014. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun 5: 5274. 10.1038/ncomms6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Zhou Q, Wu X, Yu C, Ji G, Saban DR, Li QQ. 2020. scDAPA: detection and visualization of dynamic alternative polyadenylation from single cell RNA-seq data. Bioinformatics 36: 1262–1264. 10.1093/bioinformatics/btz701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.