Abstract
Single-cell genomics is rapidly advancing our knowledge of the diversity of cell phenotypes, including both cell types and cell states. Driven by single-cell/-nucleus RNA sequencing (scRNA-seq), comprehensive cell atlas projects characterizing a wide range of organisms and tissues are currently underway. As a result, it is critical that the transcriptional phenotypes discovered are defined and disseminated in a consistent and concise manner. Molecular biomarkers have historically played an important role in biological research, from defining immune cell types by surface protein expression to defining diseases by their molecular drivers. Here, we describe a machine learning-based marker gene selection algorithm, NS-Forest version 2.0, which leverages the nonlinear attributes of random forest feature selection and a binary expression scoring approach to discover the minimal marker gene expression combinations that optimally capture the cell type identity represented in complete scRNA-seq transcriptional profiles. The marker genes selected provide an expression barcode that serves as both a useful tool for downstream biological investigation and the necessary and sufficient characteristics for semantic cell type definition. The use of NS-Forest to identify marker genes for human brain middle temporal gyrus cell types reveals the importance of cell signaling and noncoding RNAs in neuronal cell type identity.
Cells are the fundamental functional units of life. In multicellular organisms, different cell types play different physiological roles in the body. The identity and function of a cell—the cell phenotype—is dictated by the subset of genes/proteins expressed in that cell at any given point in time. Abnormalities in this expressed genome are disorders that form the physical basis of disease (Scheuermann et al. 2009). Thus, understanding normal and abnormal cellular phenotypes is key for diagnosing disease and identifying therapeutic targets.
Single-cell transcriptomic technologies that measure cell transcriptional phenotypes using single-cell/single-nucleus RNA sequencing (scRNA-seq) are revolutionizing cell biology. The expression profiles produced by these technologies can be used to define cell types and their states based on the genes they express. For simplicity, throughout the text we will use the term “cell type” to refer to these distinct cell phenotypes that include discrete canonical cell types and distinct cell states. Numerous atlas projects designed to provide a comprehensive enumeration of normal cell types and states are currently underway, including the Human Cell Atlas (Regev et al. 2017), California Institute for Regenerative Medicine (CIRM) (Darmanis et al. 2015; Enge et al. 2017; Nowakowski et al. 2017), LungMAP (Schiller et al. 2019), Pancreas atlas (Muraro et al. 2016), Heart atlas (Asp et al. 2019), and NIH Brain Initiative (Mott et al. 2018). By leveraging these atlases of normal cell types defined using specimens from healthy patients as references, the role of expression deviations in disease are now being investigated (Levitin et al. 2018; Chaudhry et al. 2019; Al-Dalahmah et al. 2020).
Despite the incredible promise of single-cell transcriptomic analysis, representations of these cell type clusters and their transcriptional phenotypes have not been adequately formalized in a standardized way to ensure effective dissemination in accordance with FAIR principles (Wilkinson et al. 2016). One approach for formalizing this type of knowledge representation and dissemination is to use the semantic framework provided by biomedical ontologies. For cell types defined by single-cell transcriptomics, the Cell Ontology (CL) is an established biomedical ontology that could be used to address FAIR-compliant cell phenotype dissemination (Bard et al. 2005; Diehl et al. 2011; Meehan et al. 2011; Bakken et al. 2017). With the rapid expansion in both data sets and cell types being defined using scRNA-seq, the challenge will be to make the generation of these semantic knowledge representations scalable.
Toward a scalable dissemination solution, we previously proposed to define cell types based on the minimum combination of necessary and sufficient features that capture cell type identity and uniquely characterize a discrete cell phenotype (Bakken et al. 2017). In the case of cell types identified by scRNA-seq experiments, these features would correspond to the combination of marker genes unique to a given gene expression cluster that provides high sensitivity and high specificity for cell type classification.
In this regard, determining marker gene combinations for cell type clusters is different from differential expression analysis (DE). Commonly used scRNA-seq analysis tools—Seurat (Stuart et al. 2019) and SCANPY (Wolf et al. 2018)—are often used for differential gene analysis. After cluster analysis, genes are evaluated by comparing expression in cells in a target cluster versus expression in all other cells using, for example, the Wilcoxon rank-sum test. However, the resulting ranked set of genes cannot be used to determine the best individual marker or the best marker combinations from either the P-value rank or fold difference in expression. In contrast, marker gene determination should explicitly test for classification power and ability to discriminate a gene expression cluster of interest.
The ideal marker gene would show a “binary expression” pattern. These are markers that are expressed at high levels in all individual cells of a given cell type and not expressed in the cells of any other cell type. These binary expression markers are particularly useful in many downstream assays such as RT-PCR (Aevermann et al. 2021) or spatial transcriptomics where low level expression in nontarget cells could be problematic. However, candidate marker genes identified by traditional differential expression analysis do not necessarily enrich for binary expression. Candidate marker genes produced by these approaches are often expressed at high levels in the target cluster and lower but measurable levels in off-target clusters. We refer to these markers as quantitative markers as their discriminatory power is derived from specific expression level thresholds, and so their utility would be dependent on the analytical sensitivity of the assay performed. In other cases, a single binary marker may not be available for the cell type cluster in question, requiring the identification of marker combinations for optimal classification.
Here, we describe Necessary and Sufficient Forest (NS-Forest) version 2.0, which improves on the simple approach to feature selection implemented in the initial version of NS-Forest (Aevermann et al. 2018). By leveraging the nonlinear attributes of random forest feature selection, NS-Forest v2.0 identifies optimal combinations of markers for classification while simultaneously enriching for genes with binary expression patterns.
Results
User driven development of NS-Forest
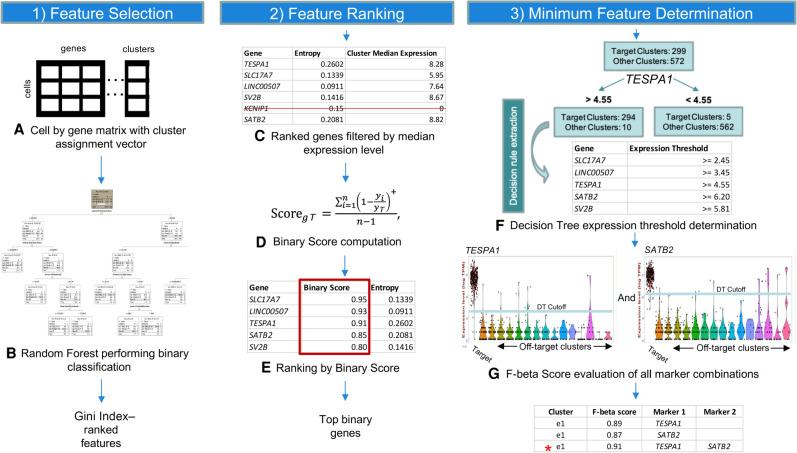
NS-Forest v2.0 was developed in close collaboration with the neuroscience user community. The primary goal was to further optimize the NS-Forest method in order to discover marker genes that can be better used for both unique cell type definition and downstream experimental investigation (Fig. 1). In order to accomplish this, several major changes were made to NS-Forest v1.3 (Table 1). First, negative markers were removed by implementing a positive expression level filter (Fig. 1C). A negative marker is defined as a gene that is not expressed in the target cluster while having expression in off-target clusters. These markers are not optimal for many downstream assays or definitional purposes. These genes are now filtered out by applying a cluster median expression threshold, with a default setting of zero.
Figure 1.
NS-Forest version 2.0 workflow. The method begins with a cell-by-gene expression matrix with cluster assignments for each cell (A). This clustered expression matrix is used to generate binary classification models for each cell cluster using the random forest machine learning method. Features are extracted from the model and ranked by Gini Index (B). Top features are filtered by expression level to remove negative markers (C) before being reranked by Binary Expression Score (D,E). Decision branch expression level cutoffs are derived from decision tree analysis for the most binary features (F) and F-beta score used as an objective function to evaluate the discriminatory power of all permutations of selected markers (G).
Table 1.
Major changes between NS-Forest v1.3 and v2.0
Next, the way genes are ranked after random forest selection was refined. Genes selected by random forest have an expression level threshold that is optimized to distinguish between target and off-target clusters. Often the genes selected discriminate based on a specific expression value resulting in quantitative expression markers. Although these quantitative markers may be good for classification, they are less useful in many downstream biological assays. To address this issue, we modified NS-Forest v2.0 to enrich for selection of binary expression markers. Binary expression markers are characterized by having expression within the target cell type while being expressed at low or negligible levels in other cell types. We accomplished this by developing a new Binary Expression Score metric with subsequent reranking of the candidate markers produced by random forest feature selection based on this score (Fig. 1D,E).
Last, the marker gene evaluation framework was redesigned. In the initial NS-Forest version, top-ranked genes were evaluated using an unweighted F1 score in an additive fashion. Candidate genes produced by random forest were ranked by unweighted F1 and the top gene selected. Next, the second-ranked gene was added to the top-ranked gene to determine if an improvement in the F-score was obtained. This stepwise additive process continued until the F-score plateaued or the selected number of top-rank genes were all tested.
In NS-Forest v2.0, all permutations of the selected top-ranked genes are tested and their performance assessed using the weighted F-beta score. The F-beta score contains a weighting term, beta, that allows for emphasizing either precision or recall. By weighting for precision (the contributions of false positives) versus recall (the contributions of false negatives), we limit the impact of zero inflation (or drop-out), a known technical artifact with scRNA-seq data, on marker gene assessment. In addition, by testing all permutations of candidate marker genes, local optima resulting from gene ranking can be avoided. These adjustments result in better final marker gene combinations given the known limitations of scRNA-seq analysis (Fig. 1F,G).
Performance testing of the Binary Expression Score approach
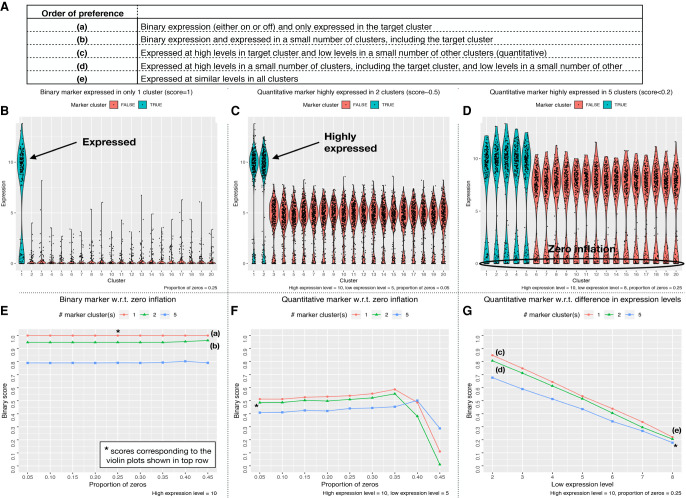
Simulation testing of the NS-Forest Binary Expression Score was performed to evaluate the impact of different data characteristics on reranking behavior. First, anticipated marker gene expression patterns were themselves ranked in order of theoretical preference (Fig. 2A). The highest preference was given to a marker gene that shows a binary expression pattern and is only expressed in the target cluster (Fig. 2A[a] ,B). The next highest preference is given to a marker gene that shows binary expression and is only expressed in the target cluster and a limited number of off-target clusters (Fig. 2A[b]). This is followed by quantitative markers that have high expression in the target cluster and lower expression in off-target clusters (Fig. 2A[c],C) or high expression in the target cluster and a limited number of off-target clusters (Fig. 2A[d]). The least preferred pattern is when the marker is expressed at only slightly different levels between the target and off-target clusters (Fig. 2A[e],D). The Binary Expression Score developed (see Methods section) was designed to quantify this order of expression pattern preference, with a range of 0 (least desirable) to 1 (most desirable).
Figure 2.
Performance testing of Binary Expression Score. Gene expression data were simulated as described in the Methods section for different expression scenarios. (A) Possible marker gene expression patterns were ranked by order of preference. Panels B–D show violin plots for three different expression scenarios: (B) binary expression only in the target cluster; (C) quantitative expression with high expression in the target cluster and one other cluster and large differences in expression in the other off-target clusters; and (D) quantitative expression with high expression in the target cluster and four other clusters, small differences in expression in the other off-target clusters, and higher levels of zero inflation. Panels E–G show line graphs of the full range of tested simulations from three defined test cases: one cluster with high expression of the marker gene (red); two clusters with high expression of the marker gene (green); and five clusters with high expression of the marker gene (blue). (E) Proportion of zeros was increased while maintaining off-target expression at zero. (F) Off-target clusters were given moderate levels of expression while the proportion of zeros was increased. (G) Expression levels were varied in all off-target clusters from low (2) to high expression (8).
Simulations varying the binary expression pattern and level of zero inflation (Fig. 2E) were then generated to test the performance of the Binary Expression Score developed. First, the ideal scenario of binary expression only in the target cluster produced a simulated Binary Expression Score of 1 (Fig. 2E, red). When the candidate marker gene was expressed in one (Fig. 2E, green) or four (Fig. 2E, blue) off-target clusters, the Binary Expression Score decreased to 0.95 and 0.80, respectively. These scores were robust to high zero inflation proportions, demonstrating no decrease in Binary Expression Score up to 45% zero values.
Next, quantitative marker expression patterns were added to the simulation (Fig. 2F,G) by varying the number of off-target clusters with high expression levels and adding moderate expression to other off-target clusters. In all cases in which quantitative differences in expression were simulated, the Binary Expression Scores was reduced accordingly (Fig. 2F). In the best case, where only the target cluster had high expression and the off-target clusters have moderate expression, the Binary Expression Score was 0.52. Further Binary Expression Score reductions were found when the high expression levels were present in additional off-target clusters. Adjusting the level of zero inflation for these scenarios showed that these Binary Expression Scores were also robust to increasing zero inflation levels until they dropped dramatically above 35% zero values.
Finally, simulations were performed to again test how a high-expressing marker is affected by the addition of 1 or 4 high-expressing off-target clusters together with increasing expression levels in the remaining off-target clusters from low (2) to high (8) expression (Fig. 2G). With the remaining off-target clusters held at low expression levels, these three scenarios returned high Binary Expression Scores (0.7–0.85), but these Binary Expression Scores quickly decreased with increasing levels of off-target expression. For example, when the off-target expression level was set to 6, all three high-expressing off-target scenarios returned Binary Expression Scores below 0.5. In the worst case, where the candidate marker had relatively high expression in all off-target clusters, the Binary Expression Score was less than 0.2.
These simulations demonstrate that the Binary Expression Score value produced by the algorithm recapitulates the preferred expression pattern ranking order (Fig. 2A). In all simulations tested, the Binary Expression Scores decreased with the addition of marker expression in off-target clusters and were robust to zero inflation.
Marker gene comparison between NS-Forest versions
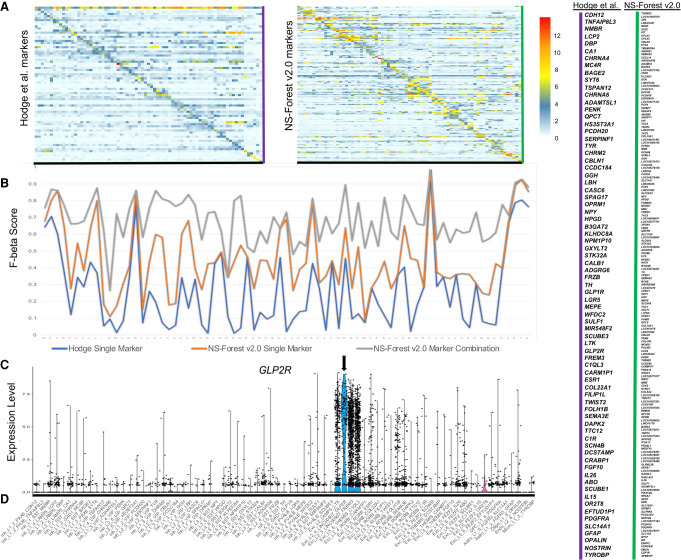
To evaluate the differences in results between NS-Forest v1.3 and v2.0, we analyzed marker genes selected for cell type clusters generated from single-nuclei transcriptomes prepared from all cortical layers (1–6) of the human middle temporal gyrus (MTG) obtained from postmortem and surgically resected samples. For this data set, three broad classes of cells were initially identified: excitatory neurons (10,708 nuclei), inhibitory neurons (4297 nuclei), and nonneuronal cells (923 nuclei). The median depth of sequencing was 2.6 ± 0.5 million reads per nucleus, with a median gene detection of 9046 for neurons and 6432 for nonneuronal cells. These nuclei were clustered iteratively by first clustering into the larger groups, followed by subsequent reclustering within each group until 75 putative cell types were found (see Hodge et al. 2019 for more details on the data set and the iterative clustering methodology). From left to right of the hierarchical clustering of clusters shown at the top of both heat maps, there are 46 inhibitory, 23 excitatory, and six nonneuronal cell types identified (Fig. 3). Subsequent figures investigating these cell type clusters are ordered by these taxonomic relationships (Fig. 3C).
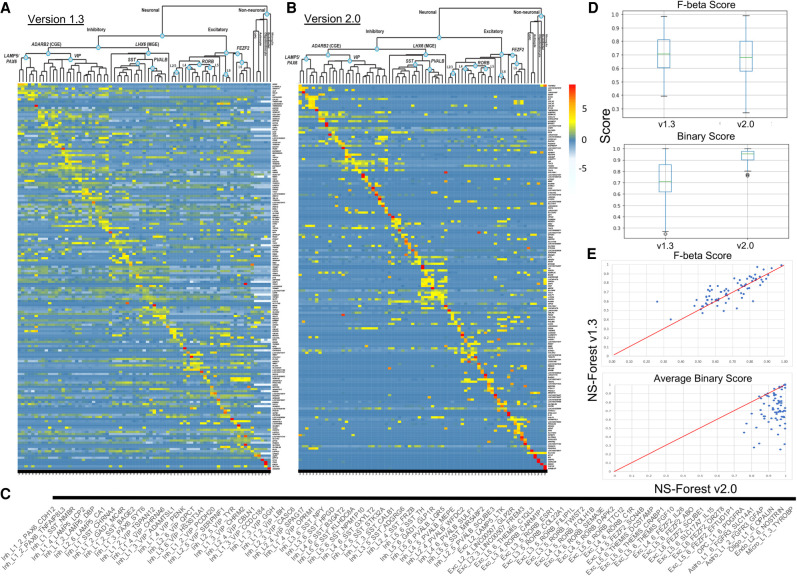
Figure 3.
Comparing NS-Forest v1.3 and v2.0 marker gene sets. Heat maps of NS-Forest v1.3 (A) and v2.0 (B) markers from human middle temporal gyrus. The taxonomy along the top of each heat map is based upon the hierarchical clustering result described in Bakken et al. (2018) and Hodge et al. (2019). Expression values are log2 CPM cluster medians normalized by row. The colors correspond to the normalized median expression level for the marker gene (rows) for a given cell type cluster (columns), with high expression (greater than five) in red and low expression (zero to negative five) in blue/white. (C) A blowup of the cell type labels corresponding to the heat map columns in parts A and B. (D) Box plots of F-beta and Binary Scores produced by NS-Forest v1.3 and v2.0 for all 75 cell type marker gene combinations. (E) Correlation of F-beta and Binary Scores between NS-Forest v1.3 and v2.0.
In total, 155 and 157 marker genes to optimally distinguish between these 75 different cell type classes were identified by NS-Forest v1.3 and v2.0, respectively (Supplemental Tables S1–S3). Of these two unique sets of markers, 51 were common to both sets (∼30%). For each method, the average number of markers per cell type was just above 2 (2.4 and 2.3, respectively). This trend of cell types requiring a combination of an average of 2–3 markers has been seen in other data sets and other tissue types (Aevermann et al. 2018, 2021), with cell types requiring only one marker reflecting very distinct types, such as the nonneuronal types found in this brain region. From the heat maps (Fig.3A,B), it is clear that the selection of genes with binary expression patterns has dramatically improved between NS-Forest v1.3 and v2.0. The diagonal for NS-Forest v2.0 contains more genes with high expression levels and, importantly, the off-diagonal expression levels are closer to zero, which demonstrates binary marker expression on a global level. Cluster median expression for markers genes are provided in Supplemental Tables S4 and S5.
Given the objective of the Binary Expression Score ranking step to preferentially find marker genes with binary expression, there are tradeoffs in both the number of genes required and the classification power when compared to markers ranked strictly by importance from the random forest model in NS-Forest v1. In general, NS-Forest v2.0 requires more unique genes for a given data set. In the case of the full MTG data set, the increase is marginal, requiring only two additional unique genes (155 vs. 157 genes); similar differences in the number of marker genes required have been observed in other data sets (Aevermann et al. 2021). Furthermore, the genes that have a high Binary Expression Score are usually not the same genes that were ranked highest by Gini Index in the random forest models. This suggests that, in terms of pure classification, the markers identified by v2.0 might be expected to underperform as compared to NS-Forest v1.3. To directly compare the F scores between these two versions of NS-Forest, an additional analysis was run setting the beta weight of the F score to 0.5 in v1.3, thereby making it directly comparable to v2.0. As expected, the median F-beta score for v2.0 (0.68) was slightly lower than for v1.3 (0.71) (Fig. 3D) and also slightly lower on a cluster-by-cluster basis (Fig. 3E). However, the Binary Expression Scores for the v1.3 markers were significantly lower—mean of 0.72 for v1.3 versus 0.94 for v2.0 (Fig. 3D,E). These results show that, although adding the Binary Expression Score criteria does slightly decrease the overall classification power of the markers selected, it dramatically increases the binary expression pattern, making the markers more useful for many downstream experimental applications.
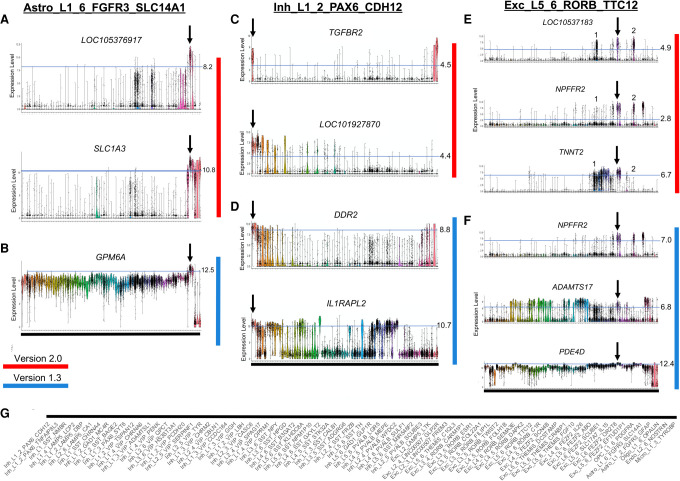
To demonstrate more clearly the differences between markers determined either by NS-Forest v1.3 or NS-Forest v2.0, we looked at one cell type cluster from each major group (nonneuronal, inhibitory neuron, and excitatory neuron) in the taxonomy (Fig. 4). For clarity, cluster labels are given along the bottom (Fig. 4G). The expression patterns for the astrocyte cell type Astro_L1_6_FGFR3_SLC14A1 illustrates the differences in marker gene characteristics broadly (Fig. 4A,B). NS-Forest v1.3 selects a single marker gene to best discriminate this cluster, whereas v2.0 selects two. NS-Forest v1.3 selects only the GPM6A gene, which performs well at classifying this cell type along a quantitative boundary at the high log2 expression level of 12.5 but also shows intermediate expression centered around 10 in many off-target clusters (Fig. 4A). Consequently, this quantitative marker is good for classification only when this small window of expression difference is discernible. In contrast, version 2.0 selects LOC105376917 and SLC1A3, both of which have binary expression patterns across clusters (Fig. 4B). LOC105376917 is highly expressed only in the target cluster and one additional closely related off-target cluster. Adding SLC1A3 further improves classification by discarding cells from this off-target cluster.
Figure 4.
Marker gene expression for representative cell type clusters of the three major taxonomy classes: nonneuronal, inhibitory neurons, and excitatory neurons. Panels A, C, and E (red) show markers determined by NS-Forest v2.0; panels B, D, and F (blue) show markers from NS-Forest v1.3. Expression level violin plots are log2 CPMs with cell types enumerated along the x-axis in taxonomic order. Expression thresholds are demarcated by light blue lines and cutoff values are given on the right. Thresholds for NS-Forest v2.0 were determined by decision tree split points, whereas, for NS-Forest v1.3, they were fixed for a given gene at the expression level where 75% of cells had expression within the target cluster. (G) Taxonomy ordered labels corresponding to the x-axis of all violin plots.
In the case of the inhibitory neuron Inh_L1_2_PAX6_CDH12, both v1.3 and v2.0 select two marker genes; however, their characteristics are very different (Fig. 4C,D). NS-Forest v1.3 again found markers that classified along quantitative boundaries. DDR2 is expressed in all the related clusters in the taxonomy and in some glial clusters at the far end of the taxonomy. The addition of IL1RAPL2 removes the glial clusters and improves the classification; however, IL1RAPL2 is another example of a quantitative marker, as it separates the target cluster from the related cluster by narrow differences in expression. NS-Forest v2.0 selected two highly binary markers: TGFBR2, which is very specific to only two clusters, the target cluster and a nonneuronal type at the other end of the taxonomy; the addition of the LOC101927870 gene eliminates cells in the nonneuronal cluster to refine the classification.
Lastly, the excitatory neuron Exc_L5_6_RORB_TTC12 required three markers by both NS-Forest versions to optimize classification (Fig. 4E,F). Again, NS-Forest v1.3 identified genes that used a quantitative boundary for classification, whereas NS-Forest v2.0 discovered binary markers. A more detailed look at these binary markers provides a clear demonstration of the combinatorics captured by NS-Forest v2.0. Within the target cluster, demarcated by the arrow, all three markers have high expression; however, the off-target excitatory clusters marked as 1 and 2 also express some but not all these markers. By leveraging the combinatorics of the three-marker combination, a highly discriminative solution is obtained. Gene LOC105371833 is the most binary marker; however, it has high expression in a number of off-target cells in clusters 1 and 2. The addition of the NPFFR2 gene removes most of the false positives in cluster 1, whereas adding the TNNT2 gene removes the false positives from cluster 2. Together, this combination of three marker genes discriminates Exc_L5_6_RORB_TTC12 from other excitatory cell types.
Comparison with previous MTG marker genes
To understand how the NS-Forest marker genes compare to previously published markers for the human middle temporal gyrus, we compared the NS-Forest markers to those reported in Hodge et al. (2019) using a different binary expression approach used for cell cluster naming. In addition to a broad marker determined by the taxonomy and prior knowledge (such as GAD1 or SST), a single marker gene per cell type cluster was assigned in Hodge et al. Sixteen of the 75 Hodge markers overlapped with the NS-Forest markers [BAGE2, GGH, CASC6, NPY, HPGD, STK32A, ADGRG6, TH, MEPE, PENK, CARM1P1, TWIST2, IL26, SULF1, ADAMTSL1, PDGFRA]. These 16 were spread across the taxonomy, representing cell type clusters from all three major cell type lineages. Unscaled heat maps of mean gene expression per cluster for both the Hodge and NS-Forest marker sets (Fig. 5A) demonstrate that both are characterized by largely binary expression patterns, having a higher expression along the diagonal versus off-diagonal. However, the Hodge markers have an overall lower mean expression level of 4.8 log2 CPM in comparison with the mean expression for the NS-Forest markers of 7.0 log2 CPM.
Figure 5.
Comparison of Hodge et al. (2019) markers with NS-Forest v2.0 for the full MTG. (A) Unscaled heat map for both sets of markers where the values are the mean expression per gene. (B) F-beta scores (y-axis) for the single Hodge marker gene (blue), the best NS-Forest single marker gene (orange), and the combination of marker genes found by NS-Forest (gray). (C) An example violin plot of a binary expression pattern selected by the method used by Hodge et al. (2019) for cluster Exc_L2_4_LINC00507_GLP2R, with expression given as log2 CPMs. For all panels, cell type clusters are listed along the x-axis in taxonomic order. (D) Taxonomy ordered labels corresponding to the x-axis of the heat maps in A and also the violin plot in C.
One major difference between these two approaches is that the Hodge marker set contains a single marker per cluster, selected to label a distinct cluster phenotype, whereas NS-Forest selects combinations of markers that optimize classification power. By running the Hodge markers through NS-forest v2.0, we estimated F-beta scores for the single Hodge markers in order to compare their classification power to the best single NS-Forest markers and the NS-Forest marker combinations (Fig. 5B). Overall, the trend lines show that the F-beta scores for single markers (blue and orange lines) follow a similar trajectory, with some clusters being more difficult to classify than others, that is, having lower F-beta scores. However, the NS-Forest marker combinations (gray line) provide a uniformly higher power of discrimination over either single marker, regardless of how the single best marker is chosen.
When evaluating the F-beta scores for the Hodge markers, it became clear that many had elevated false positive rates. To directly compare the two sets of markers, we computed the false discovery rate (FDR = FP/FP + TP) for each cell type and averaged across the entire set. The Hodge markers had an average FDR of 0.7 versus 0.14 for the NS-Forest markers. GLP2R, which is a marker for Exc_L2_4_LINC00507_GLP2R, offers a good visual example (Fig. 5C). This gene is expressed in the target cluster but also the nearest cell types within the LINC00507 group. NS-Forest also has difficulty finding markers for this cell cluster phenotype, requiring three markers in total; however, in combination these markers helped reduce the FDR rate from 0.89 to 0.11. For clarity, cluster labels for the x-axis are given along the bottom (Fig. 5D).
NS-Forest markers as cell type barcode
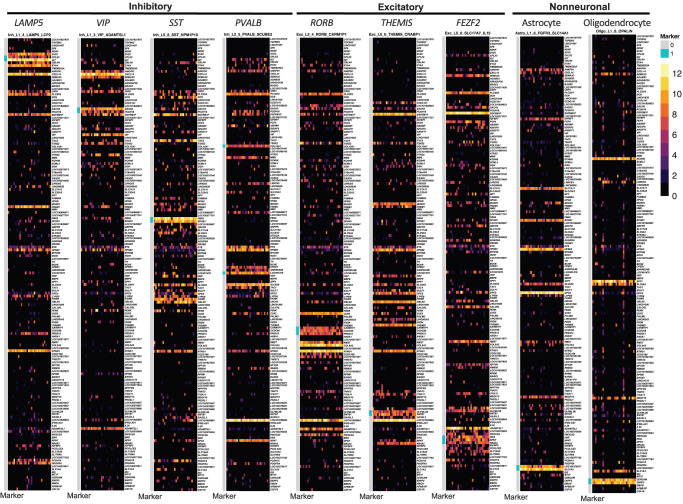
From the complete panel of NS-Forest marker genes for a given data set, it is possible to generate a “transcriptional barcode” for each cell type. As an illustration, barcodes randomly selected nuclei from nine different cell types representing each major subclass in the taxonomy, with the 157 NS-Forest v2.0 markers displayed as rows and individual nuclei as columns, as shown (Fig. 6). The markers that are specific for the given cluster are demarcated in pink within the blue bar along the left side of the barcode. The distinct patterns of these transcriptional barcodes are clearly apparent and include not only distinct expression of the specific marker genes in the target cells but also variable but distinct expression patterns of marker genes from other clusters. Barcodes for all cell types within the human MTG are provided in Supplemental Figures S1–S9. These barcodes can be used as a clear visualization of a given cell type within the context of its data set or projected onto new data sets to demonstrate cell type similarity.
Figure 6.
Molecular barcode examples for representative cell types. A representative cell type was selected from each of the major cell type subclasses in the taxonomy from left to right: LAMP5, VIP, SST, PVALB, RORB, THEMIS, FEZF2, astrocytes, and oligodendrocytes. Each cell type is represented by 30 individual cells selected at random (columns) with the heat map color-coded by log2 CPM expression values for each marker gene (rows).
Comparison with other marker gene selection approaches
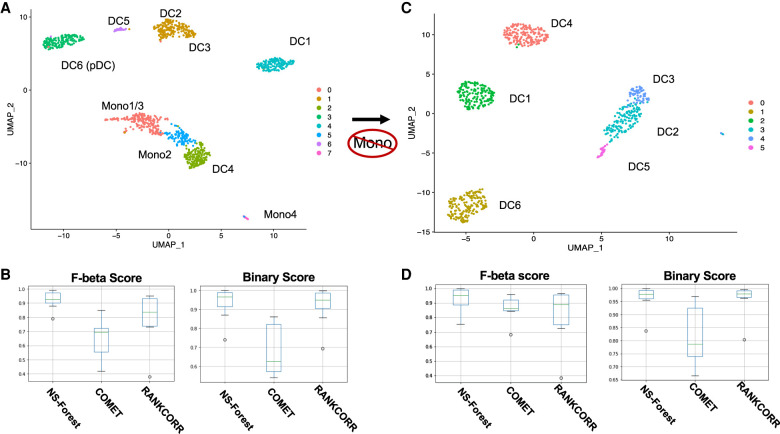
In order to assess the performance of NS-Forest v2.0 for marker gene selection, we compared it to two other marker gene selection tools—COMET (Delaney et al. 2019) and RANKCORR (Vargo and Gilbert 2020). These three tools were evaluated using an independent monocyte/dendritic cell data set produced by Villani et al. (2017). For the eight clusters produced by reprocessing these data (Fig. 7A), NS-Forest v2.0, COMET, and RANKCORR required 17, 16, and 28 markers, respectively, to produce optimal classification results. When comparing F-beta scores and Binary Scores (Fig. 7B), NS-Forest v2.0 was found to outperform both COMET and RANKCORR. Although there was a significant overlap of 10 genes between NS-Forest v2.0 and RANKCORR, none were shared with COMET. Given the overlap, it is not surprising to find that both the F-beta scores and Binary Scores are close between NS-Forest v2.0 and RANKCORR. The median F-beta scores were [0.92 > 0.84 > 0.69] for NS-Forest v2.0 > RANKCORR > COMET, and the median Binary Scores were [0.97 > 0.95 > 0.62] for NS-Forest v2.0 > RANKCORR > COMET.
Figure 7.
Results from marker gene set determination for monocyte and dendritic cell types described in Villani et al. (2017). (A) Louvain clustering result for all monocytes and dendritic cell types with labels indicating the cell types defined in Villani et al. (2017). In comparison with the original result derived from iterative clustering, monocyte 1 and 3 and dendritic type DC2 and DC3 have been merged in this clustering result. (B) Box plots showing F-beta scores and Binary Scores for markers determined for the clusters in panel A by NS-Forest v2.0, COMET, and RANKCORR. (C) Louvain clustering results of dendritic cells only with labels indicating the cell type defined in Villani et al. (2017). (D) Box plots showing F-beta scores and Binary Scores for markers determined for the cell type clusters in panel C by NS-Forest v2.0, COMET, and RANKCORR.
Clustering was also performed after removal of the monocytes, which resulted in six clusters corresponding to the DC1-DC6 types as characterized in the original study (Fig. 7C). For the six clusters produced by reprocessing these data, NS-Forest v2.0, COMET, and RANKCORR required 9, 12, and 19 markers, respectively, to produce optimal classification results (Fig. 7D). Three markers were shared by all methods and four markers shared between NS-Forest v2.0 and COMET and between NS-Forest v2.0 and RANKCORR. Again, NS-Forest v2.0 outperformed both COMET and RANKCORR, but the F-beta score results were more comparable with these clusters. The median F-beta scores were [0.95 > 0.89 > 0.86] for NS-Forest v2.0 > RANKCORR > COMET, and the average Binary Scores were [0.979 > 0.978 > 0.78] for NS-Forest v2.0 > RANKCORR > COMET.
In general, these results show that NS-Forest v2.0 outperforms these other marker gene identification methods. However, it should be noted that these other methods were not designed for the purpose of selecting the minimum set of marker genes. COMET may have underperformed because its XL-mHG framework, that uses the X and L parameters, optimizes for the true positives and false positives, whereas NS-Forest v2.0 and its associated metrics are more focused on false positives only. In addition, although RANKCORR performance is comparable to NS-Forest v2.0, it required substantially more marker genes for optimal performance. Thus, NS-Forest v2.0 appears to be optimal for the specific use case of finding the minimal set of markers for maximal classification accuracy. Marker genes identified by each method are given in Supplemental Figure S10.
Of the three methods, NS-Forest v2.0 was the most time-intensive. Both COMET and RANKCORR completed in under 3 min when analyzing both the monocyte/dendritic cell and dendritic cells-only data set, whereas NS-Forest v2.0 took 45 min at the default settings. To investigate the performance of NS-Forest v2.0 further, the dendritic cell data set was run while varying the parameters (Supplemental Fig. S11). Whereas the number of trees used during the random forest modeling step did not have a significant impact on the run time, the number of genes tested for all permutations was the limiting step. We found that running 1–4 genes resulted in run times under 4 min, using five genes increased this to 7 min, and using the default of six genes resulted in a 45-min run time. Looking at the resulting F-beta scores for each run, we can see clear improvement in marker determination up to the five-gene selection and only a slight improvement thereafter. Consequently, it may be advisable to change the default to five genes in situations where time is a limiting factor.
Validation of human MTG NS-Forest v2.0 markers
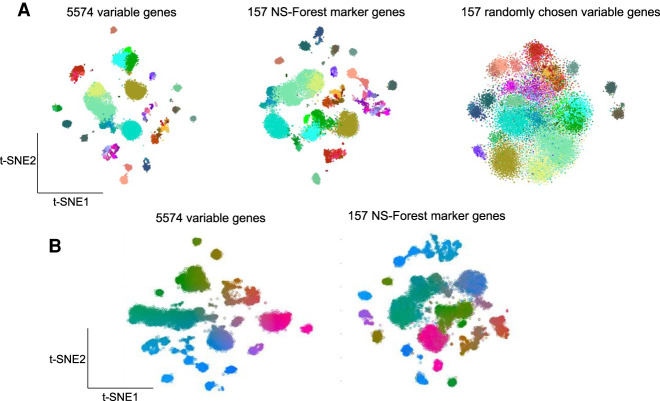
The ground truth for neuron types and their marker genes in human MTG is not known as this is currently an active area of investigation. Consequently, a true biological validation of the marker genes is not possible. As an alternative, we asked the question, does the minimum set of marker genes selected by NS-Forest capture the underlying diversity of cell type identity reflected in the entire expressed transcriptome? To do this, we generated t-SNE plots using the complete set of 5574 variable genes used for the original MTG clustering, the minimum set of 157 NS-Forest v2.0 marker genes, and sets of 157 genes randomly selected from the complete variable genes list. These embeddings were then painted using the cell type assignments from the MTG taxonomy. From the t-SNE plots, it is clear that the NS-Forest markers closely recapitulate the clustering and embedding structure of the complete variable genes set, much better than the randomly selected genes (Fig. 8A). For example, in the bottom of the complete variable genes t-SNE, there are light salmon- and dark salmon-colored groups of clusters; these two clusters are nicely preserved in the right-hand side of the NS-Forest marker t-SNE, whereas, in the t-SNE from the randomly selected variable genes, these two clusters are spread out and a third brown cluster is now merged with the light salmon cluster. Examples like this can be seen throughout the three embeddings. A more quantitative analysis of these t-SNE embeddings using the Nearest-Neighbor Preservation metric showed that both the precision and recall are higher using the 157 NS-Forest markers compared with 50 samplings of 157 genes randomly selected from the variable gene set (Supplemental Fig. S12).
Figure 8.
Validation of NS-Forest v2.0 MTG marker genes. (A) t-SNE plots generated using the full 5574 variable gene list, the 157 NS-Forest v2.0 markers, and 157 genes randomly selected from the variable gene list painted by taxonomic assignment. (B) t-SNE map generated from the full 5574 variable gene list was painted by CIELAB color space using coordinate position for each nucleus (left). t-SNE map generated using the 157 NS-Forest markers was then painted according to the CIELAB color space established in the complete variable genes t-SNE (right).
In addition, the local embedding structures as reflected by expression gradients within a given t-SNE cluster also appear to be well preserved (Fig. 8B). The complete variable genes t-SNE map was painted using coordinate positioning. This yields a visual way of comparing where individual nuclei are located within the full t-SNE embedding versus other t-SNE embeddings. The NS-Forest marker t-SNE was then painted using the colors derived from the complete variable genes t-SNE. The fact that the same color gradients are observed in the NS-Forest embedding demonstrates that the positional gradients, and thus the nuclei-to-nuclei relationships, in the NS-Forest embedding closely reflect the positional gradients in the complete variable genes t-SNE embedding. For example, in the full t-SNE, there is a long cluster of nuclei beginning on the left in green that extends toward the middle, moving into bluish green, and ending in purplish blue. This same cluster, with the same color gradient, is preserved within the center left cluster of the NS-Forest t-SNE.
Characterization of NS-Forest v2.0 markers
Overall, the results from NS-Forest v2.0 reflect the high quality of the data and clustering analysis; as a supervised machine learning method, NS-Forest v2.0 is reliant on the quality of the clustering results. The median number of markers required for optimal classification was two, with only two clusters needing four markers, producing a mean F-beta score of 0.69. Overall, the 75 clusters required 157 unique genes to achieve optimal classification. Occasionally, marker genes are shared between clusters, with 11 genes that were not unique (MOXD1, MME, LOC101928196, SULF1, NPFFR2, LINC01583, TAC1, COL15A1, LOC401478, CPED1, TAC3).
Out of the 157 NS-Forest v2.0 marker genes, 37 (24%) were long noncoding RNAs (lncRNAs) or uncharacterized loci (LOCs). Noncoding RNAs have been previously found to be prevalent when analyzing RNA-seq data from single neuronal cells or nuclei, and, surprisingly, these noncoding RNAs had higher specificity as markers when compared to coding genes (Bakken et al. 2018). In particular, lncRNAs are known to show cell line–specific expression (Djebali et al. 2012). In contrast, little is known about the LOC genes. These genes are particularly intriguing as they are highly specific to individual cell types and are likely important for their function. As such, they represent areas of unknown biology discovered by scRNA-seq and NS-Forest machine learning that warrant further investigation.
For the characterized marker genes, the most enriched annotations both by adjusted P-value and number of genes involved are for signaling (signal peptide, signal, secreted), including neuropeptide signaling (GO:0007218∼neuropeptide) and calcium, and extracellular matrix (glycoprotein, extracellular matrix, GO:0005615∼extracellular space, GO:0005578∼proteinaceous extracellular matrix, GO:0030198∼extracellular matrix organization, GO:0005576∼extracellular region, GO:0031012∼extracellular matrix), and calcium (Supplemental Table S6; Huang et al. 2009). There are fewer genes annotated with specific neurological functions in the marker gene list, as molecular neuroscience is a relatively nascent field. However, many of genes assessed here are known signaling peptides in other contexts and would benefit from further characterization in a neurological context. Taken together, these results suggest that specific signaling pathways and extracellular signaling molecules are key to neuronal cell type identity.
Discussion
Here, we describe the development and performance of NS-Forest version 2.0, a method for the identification of cell type–specific gene expression markers from scRNA-seq data. Development was driven by user community requirements for data-driven cell type definitions that are testable in future investigations. To this end, a number of changes were made after the random forest feature selection step. In earlier versions of NS-Forest, negative markers were occasionally found. These are marker genes that are expressed in many off-target clusters but not the target cluster. Given that experimental testing for a gene that is not expressed is methodologically difficult, NS-Forest v2.0 was designed to avoid this category of markers. By implementing a median expression level cutoff greater than zero for the target cluster, all possible negative marker genes were removed. In addition, this cutoff also defines another core characteristic of NS-Forest Markers: selected marker genes must be expressed in greater than half of the individual cells within the cell type cluster.
In addition to negative markers, the standard random forest feature selection approach used in early NS-Forest versions discovered quantitative markers that were good for classification but problematic for further biological investigation. This limitation of random forest feature selection could be shared with other machine learning methods. Consequently, a ranking step to select marker genes with binary expression patterns was incorporated. Simulation testing performed to assess this Binary Expression Score ranking step demonstrated that marker genes with binary expression patterns were preferentially selected and accurately ranked according to the levels of binary expression. As a result, NS-Forest v2.0 demonstrated clear improvement in the enrichment for binary expression patterns, with a nominal impact on the overall classification power and number of marker genes necessary. Consequently, if a user prefers the highest level of classification accuracy without the practical constraint imposed by many types of downstream investigations, NS-Forest v1.3 might be preferred. However, if binary expression for downstream application is important, NF-Forest v2.0 would be the best choice. Both versions are available as official GitHub releases.
Beyond their use for defining and investigating cell types, necessary and sufficient marker genes also offer a dimensionality reduction with limited loss of fidelity to the originally clustering solution. This dimensionality reduction offers a feasible way of representing the clustering solution with a minimal amount of information, which is ideal for data dissemination. These marker genes can then be used to generate a reference knowledgebase for cell types, generating expression barcodes that can be used to identify these cell types within new data sets. Indeed, NS-Forest marker genes have been used to facilitate reference cell type matching in the FR-Match algorithm (Zhang et al. 2020).
As mentioned above, NS-Forest markers are optimized for downstream experimental investigation. There are a number of assays for which known markers could facilitate biological investigation, such as qPCR and the burgeoning field of spatial transcriptomics based on multiplex FISH. To date, a number of projects have used NS-Forest markers for these purposes. For example, qPCR probes based on NS-Forest markers were made to detect genes in scRNA-seq libraries from myeloid dendritic cells (mDCs) FACS sorted from peripheral blood in patients treated with the hepatitis B vaccine (Aevermann et al. 2021). In a similar fashion, gene probes were designed based on NS-Forest markers for cell type detection using a number of spatial transcriptomic technologies. These technologies aim to resolve the location of cell types derived from scRNA-seq generated taxonomies within intact tissue specimens (Perkel 2019).
Another possible application of NS-Forest is to utilize selected gene sets of particular interest as input to produce marker gene sets designed to capture specific cell type properties. For example, the input of gene sets composed of transcription factors could reveal master regulators of developmental programs (Cui et al. 2019). Input gene sets composed of neuropeptides and neurotransmitters could be used to shed new light on the specific signaling properties of different neuronal cell subsets (Smith et al. 2019). Input gene sets composed of cell surface markers could be used to identify markers for use in fluorescence-activated cell sorting.
As the number of experiments performed and data sets made publicly available dramatically increase, the greater biological community is left with the monumental task of integrating these data into a consensus of canonical cell types. With cell types defined by NS-Forest marker genes, we can move ahead with the creation of a dissemination framework that defines ontological classes based upon these molecular markers as the necessary and sufficient criteria in an axiomatic semantic representation compliant with FAIR principles. Ontological representations have numerous advantages over simple vocabularies, including the structuring of knowledge in a computationally readable format so that findings from many experiments can be easily accessible and “reasoning” can be performed to ensure the consistency of the representation as the knowledge rapidly grows. These instances of “cell type clusters” defined by NS-Forest markers can form the basis for the instantiation of an ontology class for adoption into the official Cell Ontology. Progress is already underway in developing programmatic and scalable methods to handle the volumes of single-cell data being generated. This ontological representation can address several pressing needs of the wider biological research community, producing an easy, visually accessible overview of the results of many single-cell experiments in a traversable structure while preserving the hierarchical relationships inherent in a taxonomy of cell types. In addition, this ontology will provide a platform for integration with other data modalities, such as cell morphology, electrophysiology, and cell–cell interactions. A Provisional Cell Ontology (pCL) generated in this manner for human middle temporal gyrus and human, mouse, and marmoset primary motor cortex is available for exploration at https://bioportal.bioontology.org/ontologies/PCL.
Development of NS-Forest is ongoing; a number of functionalities are planned for near term release. One major update to NS-Forest v2.0 will be to add the option to run marker determination within a hierarchical framework, for example, to determine markers for a series of cluster labels that reflect a relational structure such as a taxonomy dendrogram. Another key aspect will be to include cross-validation or some other methodology to estimate the reliability of a given marker gene for a given cell type cluster. On a broader level, incorporating NS-Forest into the library of easily available SCANPY plugins is a high priority. Last, we will be increasing the number of output reports to facilitate the generation of ontological type artifacts, including OWL and RDF representations.
Methods
NS-Forest version 2.0
Initial feature selection
The NS-Forest v2.0 workflow (Fig. 1A,B) begins with a cell-by-gene expression matrix, with an additional column containing cluster membership labels, produced by any expression data clustering method applied to single-cell/-nucleus RNA sequencing data sets. This cluster-labeled expression matrix is then used to generate random forest classification models distinguishing each target cluster from all other clusters (binary classification) using RandomForestClassifier scikit. RandomForestClassifier hyperparameters were left at default except that the number of trees was set at 10,000 to give sufficient coverage of the sample and gene expression feature space; necessary coverage for a given feature space is estimated as the square root of the number of samples (∼10,000 cells) times the square root of the number of features (∼10,000 genes). From the resulting random forest model, the average Gini Index value is used to initially rank genes based on their feature importance. The output from the random forest model is a ranked list of all the input features from most informative to least informative.
Feature reranking based on positive binary expression
Reranking the features after initial random forest ranking begins with selecting the top 15 genes ranked by Gini index. It is critical to limit the number of genes before reranking by binary expression, as the Binary Expression Score does not necessarily correlate to their importance in the classification context. As such, increasing the number of genes for reranking would potentially lower the overall classification power. Positive expression filtering (Fig. 1C) is then performed by removing genes with a median cluster expression of 0 in order to exclude genes that are not expressed in the relevant cluster, which we refer to as negative markers, or show high zero inflation. The “Median_Expression_Level” parameter, default value of 0, is tunable and can be adjusted according to the data set.
Next, genes are reranked to enrich for genes with binary expression patterns (Fig. 1D). A “Binary Expression Score” was developed to enrich for genes that show all-or-none expression patterns, with expression in the target cluster and as few other cell type clusters as possible. The Binary Expression Score is calculated for each gene in the initial random forest feature list according to the equation
where yi is the median gene expression level for each cluster i, yT is the median expression in the target cluster, and n is the number of clusters, whereas (·)+ denotes the nonnegative value of a real number. This results in a Binary Expression Score in the range of 0–1, with a Binary Expression Score of 1 being the ideal case where the gene is only expressed in the target cluster (Fig. 1E). The final list of 15 genes is ranked first on the Binary Expression Score and then by the Gini Index value. This guarantees that any genes with Binary Expression Score ties are ranked by informativeness rather than lexicographically.
Estimation of expression thresholds for evaluation
After the top genes are reranked based on positive binary expression, they are then tested for their classification power individually and in combination. First, the top M genes, a tunable parameter “Genes_to_testing,” set to six genes by default, are used to generate individual decision trees to determine the optimal expression level cut-off value for each gene (Fig. 1F). The maximum leaf nodes parameter is set at two, thereby ensuring a single split point per tree. From these trees, the optimal gene expression threshold at the split point is extracted.
Minimum feature combination determination
To evaluate the discriminative power of a given combination of candidate marker genes, we use the F-beta score as an objective function
The F-score is the harmonic mean of precision and recall providing equal weight for these two classification measures. The F-beta score includes a beta term that allows for the weighting of the function toward either precision (beta < 1) or recall (beta > 1) (Fig. 1G). The beta for the analysis described here was estimated empirically at 0.5. In brief, the empirical selection of 0.5 was based on a balance of the average values for the confusion matrix across all cell type clusters while varying the beta parameter. At a beta of 0.5, there was an optimum reached in the confusion matrix while averaging approximately two markers per cell type cluster (Supplemental Fig. S13). This parameter should be evaluated for each data set, as it adjusts for the amount of zero inflation within the data. Here, we are analyzing Smart-Seq data which are known to have comparatively lower zero inflation versus droplet-based methodologies.
Finally, all permutations of the top-ranked genes (six genes by default) are then evaluated at the expression levels determined earlier by decision tree analysis. The F-beta scores for all permutations are written to a complete results file and the gene feature combination producing the best F-beta score result selected per cluster.
Simulation testing of the Binary Expression Score
Simulation studies were conducted to investigate the properties of the Binary Expression Score weighting using a three-component mixture model to reflect the zero-inflation technical artifact and the background and positive expression signals in real data distributions. Denoting X as the gene expression value, our simulated data follow a mixture distribution
where δ0 (x) is the probability density function of the degenerate distribution at 0 for the zero-inflation technical artifact, fGamma (x) is the probability density function of a Gamma distribution (with hyperparameters α and β) for low level background expression from off-target cells or on-target cells with low expression, and fNormal (x) is the probability density function of a Normal distribution (with hyperparameters μ and σ2) for positive expression signals; parameters π1, π2, and π3 are the corresponding mixture weights for each component such that π1, π2, π3 > 0 and π1 + π2 + π3 = 1. In our simulations, we generated 20 clusters with 300 cells in each cluster. We designed cases where the simulated gene is expressed at high levels in one, two, or five clusters. Both binary and quantitative markers were simulated for on-target and off-target clusters by setting different parameters and hyperparameters in the mixture model.
scRNA-seq data
The scRNA-seq data evaluated here were obtained from the Allen Institute for Brain Science (https://portal.brain-map.org/atlases-and-data/rnaseq). The experimental design, including tissue sampling and data processing, can be found in Krishnaswami et al. (2016) and Hodge et al. (2019). In brief, layers 1–6 of the human middle temporal gyrus) were vibratome-sectioned, and nuclei were extracted and labeled for NeuN expression. Nuclei were then FACS-sorted and libraries were generated using the Smart-Seq v4 and Nextera XT chemistries. Data processing and clustering were then performed as detailed in Bakken et al. (2018).
NS-Forest v2.0 was run using the cluster assignments given in Hodge et al. (2019). Nuclei not assigned to a cluster were removed from the analysis. CPM expression values were log2 (x + 1) transformed and genes with a sum of zero median expression across all clusters were removed. After filtering, 15,928 nuclei and 13,946 genes remained. Given the size of the input matrix, we increased the number of trees in the random forest model from the default of 10,000 to 50,000.
Marker validation
In order to demonstrate the preservation of the cell type clustering characteristics using NS-Forest marker genes, t-SNE embeddings were generated using Cytosplore (Höllt et al. 2016; van Unen et al. 2017). The original clustering solution is represented by an embedding generated from the 5574 variable genes used for the iterative clustering originally performed (Hodge et al. 2019). Additional embeddings were made using the combined set of 157 marker genes for all cell type clusters determined by NS-Forest v2.0 and a set of 157 genes chosen at random from the original 5574 genes. Figures were generated using two different painting strategies. The first painted cells based upon the cluster assignment given in the taxonomy. The second painted using a CIELAB color space on the coordinate positioning, giving a visual way of comparing the relative location of individual nuclei between the full t-SNE embedding and other t-SNE embeddings.
In addition, a more quantitative analysis of these t-SNE embeddings using the Nearest-Neighbor Preservation metric was performed. In brief, this is computed as follows: for each data point, the K-Nearest-Neighborhood (KNN) in the high-dimensional space is compared with the KNN in the reduced-dimensional space. Average precision/recall curves are generated by taking into account high-dimensional neighborhoods of increasing size up to Kmax = 50. The True-Positive (TP) number is the intersection between the high-dimensional and low-dimensional neighborhoods based on the 157 selected genes. The precision is computed as TP/K and the recall as TP/Kmax (Venna et al. 2010; Ingram and Munzner 2015; Pezzotti et al. 2020).
Comparison to other marker gene methodologies
Comparisons of marker gene methodologies was performed using the monocyte/dendritic cell data set detailed in Villani et al. (2017). This data set was chosen because it is well characterized in the associated publication and offers a range of defined cell types that vary in their difficulty to classify. Raw data were obtained from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) using accession number GSE94820 and then processed using a standard Seurat analysis (Stuart et al. 2019) in two ways: first, the entire data set was processed and clustered; and second the monocytes were removed followed by processing and clustering of the dendritic cell populations only. These analyses were independent and not iterative. For both analyses, cells were filtered that had less than 1000 genes and the top 2500 variable genes were selected. The complete data set had a total of 1103 cells whereas the dendritic cell data set had 750 cells. After processing, the resulting data sets were analyzed by Louvain clustering and visualized by UMAP embedding.
Clustering assignments and expression matrices containing the top 10,000 variable genes were used to perform marker determination using NS-Forest v2.0, COMET (Delaney et al. 2019), and RANKCORR (Vargo and Gilbert 2020). All three methods were run using default parameters, with COMET being run using http://www.cometsc.com/comet web submission. To compare the resulting marker gene sets, NS-Forest v2.0 was used to compute the Binary Score and F-beta score for all results.
To benchmark NS-Forest v2.0, the dendritic-only data set was used to estimate the computations time. Two different parameters were tested: the number of trees used in the random forest model generation and the number of top genes for which all permutations were tested.
Software availability
NS-Forest version 2.0 is available at GitHub (https://github.com/JCVenterInstitute/NSForest) under an open-source MIT license. Source code is also available with this manuscript labeled “NS_Forest_v2.ipynb”. Protocol is available at protocols.io: dx.doi.org/10.17504/protocols.io.un7evhn.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institutes of Health (R21-AI122100, U19-AI118626, and RF1-MH123220), the California Institute for Regenerative Medicine (GC1R-06673-B), the Wellcome Trust 208379/Z/17/Z, the Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation (2018-182730), the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) Gravitation 2019 grant: BRAINSCAPES: A Roadmap from Neurogenetics to Neurobiology (NWO: 024.004.012), and NWO TTW project 3DOMICS (NWO: 17126).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275569.121.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Aevermann BD, Novotny M, Bakken T, Miller JA, Diehl AD, Osumi-Sutherland D, Lasken RS, Lein ES, Scheuermann RH. 2018. Cell type discovery using single-cell transcriptomics: implications for ontological representation. Hum Mol Genet 27: R40–R47. 10.1093/hmg/ddy100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aevermann BD, Shannon CP, Novotny M, Ben-Othman R, Cai B, Zhang Y, Ye JC, Kobor MS, Gladish N, Lee A, et al. 2021. Machine learning-based single cell and integrative analysis reveals that baseline mDC predisposition predicts protective hepatitis B vaccine response. medRxiv 10.1101/2021.02.22.21251864 [DOI] [PMC free article] [PubMed]
- Al-Dalahmah O, Sosunov AA, Shaik A, Ofori K, Liu Y, Vonsattel JP, Adorjan I, Menon V, Goldman JE. 2020. Single-nucleus RNA-seq identifies Huntington disease astrocyte states. Acta Neuropathol Commun 8: 19. 10.1186/s40478-020-0880-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp M, Giacomello S, Larsson L, Wu C, Fürth D, Qian X, Wärdell E, Custodio J, Reimegård J, Salmén F, et al. 2019. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 179: 1647–1660.e19. 10.1016/j.cell.2019.11.025 [DOI] [PubMed] [Google Scholar]
- Bakken T, Cowell L, Aevermann BD, Novotny M, Hodge R, Miller JA, Lee A, Chang I, McCorrison J, Pulendran B, et al. 2017. Cell type discovery and representation in the era of high-content single cell phenotyping. BMC Bioinformatics 18: 559. 10.1186/s12859-017-1977-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Hodge RD, Miller JA, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, et al. 2018. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One 13: e0209648. 10.1371/journal.pone.0209648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard J, Rhee SY, Ashburner M. 2005. An ontology for cell types. Genome Biol 6: R21. 10.1186/gb-2005-6-2-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry F, Isherwood J, Bawa T, Patel D, Gurdziel K, Lanfear DE, Ruden DM, Levy PD. 2019. Single-cell RNA sequencing of the cardiovascular system: new looks for old diseases. Front Cardiovasc Med 6: 173. 10.3389/fcvm.2019.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, et al. 2019. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep 26: 1934–1950.e5. 10.1016/j.celrep.2019.01.079 [DOI] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. 2015. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci 112: 7285–7290. 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney C, Schnell A, Cammarata LV, Yao-Smith A, Regev A, Kuchroo VK, Singer M. 2019. Combinatorial prediction of marker panels from single-cell transcriptomic data. Mol Syst Biol 15: e9005. 10.15252/msb.20199005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl AD, Augustine AD, Blake JA, Cowell LG, Gold ES, Gondré-Lewis TA, Masci AM, Meehan TF, Morel PA, Nijnik A, et al. 2011. Hematopoietic cell types: prototype for a revised cell ontology. J Biomed Inform 44: 75–79. 10.1016/j.jbi.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enge M, Arda HE, Mignardi M, Beausang J, Bottino R, Kim SK, Quake SR. 2017. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171: 321–330.e14. 10.1016/j.cell.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. 2019. Conserved cell types with divergent features in human versus mouse cortex. Nature 573: 61–68. 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höllt T, Pezzotti N, van Unen V, Koning F, Eisemann E, Lelieveldt B, Vilanova A. 2016. Cytosplore: interactive immune cell phenotyping for large single-cell datasets. Computer Graphics Forum 35: 171–180. 10.1111/cgf.12893 [DOI] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram S, Munzner T. 2015. Dimensionality reduction for documents with nearest neighbor queries. Neurocomputing 150: 557–569. 10.1016/j.neucom.2014.07.073 [DOI] [Google Scholar]
- Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, Linker SB, Pham S, Erwin JA, Miller JA, et al. 2016. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc 11: 499–524. 10.1038/nprot.2016.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuby J, Kindt TJ, Goldsby RA, Osborne BA. 2007. Kuby immunology. W.H. Freeman, San Francisco. [Google Scholar]
- Levitin HM, Yuan J, Sims PA. 2018. Single-cell transcriptomic analysis of tumor heterogeneity. Trends Cancer 4: 264–268. 10.1016/j.trecan.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan TF, Masci AM, Abdulla A, Cowell LG, Blake JA, Mungall CJ, Diehl AD. 2011. Logical development of the cell ontology. BMC Bioinformatics 12: 6. 10.1186/1471-2105-12-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott MC, Gordon JA, Koroshetz WJ. 2018. The NIH BRAIN Initiative: advancing neurotechnologies, integrating disciplines. PLoS Biol 16: e3000066. 10.1371/journal.pbio.3000066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJ, et al. 2016. A single-cell transcriptome atlas of the human pancreas. Cell Syst 3: 385–394.e3. 10.1016/j.cels.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. 2017. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358: 1318–1323. 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel JM. 2019. Starfish enterprise: finding RNA patterns in single cells. Nature 572: 549–551. 10.1038/d41586-019-02477-9 [DOI] [PubMed] [Google Scholar]
- Pezzotti N, Thijssen J, Mordvintsev A, Höllt T, Lew BV, Lelieveldt BPF, Eisemnann E, Vilanova A. 2020. GPGPU linear complexity t-SNE optimization. IEEE Trans Vis Comput Graph 26: 1172–1181. 10.1109/TVCG.2019.2934307 [DOI] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. 2017. The Human Cell Atlas. eLife 6: e27041. 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann RH, Ceusters W, Smith B. 2009. Toward an ontological treatment of disease and diagnosis. In Proceedings of the 2009 AMIA Summit on Translational Bioinformatics, pp. 116–120. San Francisco. [PMC free article] [PubMed] [Google Scholar]
- Schiller HB, Montoro DT, Simon LM, Rawlins EL, Meyer KB, Strunz M, Vieira Braga FA, Timens W, Koppelman GH, Budinger GRS, et al. 2019. The Human Lung Cell Atlas: a high-resolution reference map of the human lung in health and disease. Am J Respir Cell Mol Biol 61: 31–41. 10.1165/rcmb.2018-0416TR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Sümbül U, Graybuck LT, Collman F, Seshamani S, Gala R, Gliko O, Elabbady L, Miller JA, Bakken TE, et al. 2019. Single-cell transcriptomic evidence for dense intracortical neuropeptide networks. eLife 8: e47889. 10.7554/eLife.47889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Unen V, Höllt T, Pezzotti N, Li N, Reinders MJT, Eisemann E, Koning F, Vilanova A, Lelieveldt BPF. 2017. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat Commun 8: 1740. 10.1038/s41467-017-01689-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargo AHS, Gilbert AC. 2020. A rank-based marker selection method for high throughput scRNA-seq data. BMC Bioinformatics 21: 477. 10.1186/s12859-020-03641-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venna J, Peltonen J, Nybo K, Aidos H, Kaski S. 2010. Information retrieval perspective to nonlinear dimensionality reduction for data visualization. J Mach Learn Res 11: 451–490. 10.5555/1756006.1756019 [DOI] [Google Scholar]
- Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, et al. 2017. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356: eaah4573. 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, et al. 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3: 160018. 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F, Angerer P, Theis F. 2018. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19: 15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Aevermann BD, Bakken TE, Miller JA, Hodge RD, Lein ES, Scheuermann RH. 2020. FR-Match: robust matching of cell type clusters from single cell RNA sequencing data using the Friedman–Rafsky non-parametric test. Brief Bioinform 22: bbaa339. 10.1093/bib/bbaa339 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.