High-quality genome assembly of ancient asexual rotifer reveals homologous chromosomes that frequently recombine.
Abstract
Bdelloid rotifers are notorious as a speciose ancient clade comprising only asexual lineages. Thanks to their ability to repair highly fragmented DNA, most bdelloid species also withstand complete desiccation and ionizing radiation. Producing a well-assembled reference genome is a critical step to developing an understanding of the effects of long-term asexuality and DNA breakage on genome evolution. To this end, we present the first high-quality chromosome-level genome assemblies for the bdelloid Adineta vaga, composed of six pairs of homologous (diploid) chromosomes with a footprint of paleotetraploidy. The observed large-scale losses of heterozygosity are signatures of recombination between homologous chromosomes, either during mitotic DNA double-strand break repair or when resolving programmed DNA breaks during a modified meiosis. Dynamic subtelomeric regions harbor more structural diversity (e.g., chromosome rearrangements, transposable elements, and haplotypic divergence). Our results trigger the reappraisal of potential meiotic processes in bdelloid rotifers and help unravel the factors underlying their long-term asexual evolutionary success.
INTRODUCTION
Sexual reproduction and recombination are prevalent throughout eukaryotes, despite the substantial evolutionary costs such as the twofold cost of males or the cost of recombination that breaks up coadapted gene combinations (1, 2). Several eukaryotic species appear to have evolved adaptations that reduce these costs of males, for example, by producing males only facultatively as in cyclical parthenogens [e.g., Brachionus plicatilis (3)] or by retaining a modified meiosis rescuing diploidy without fertilization by males [e.g., Diploscapter pachys (4)]. Very few, however, appear to have renounced sex and recombination completely by abolishing males and meiosis. Theory predicts that, in the absence of recombination during meiosis, physical linkage among loci reduces the effectiveness of selection upon individual loci, resulting in a decreased rate of adaptation and the accumulation of mildly deleterious mutations (5). Obligate asexuals are therefore suitable model systems to gain general insights into the long-term consequences of the lack of recombination and sexual reproduction.
Bdelloid rotifers are notorious ancient asexual animals. The longevity [>60 million years (Ma)] of the bdelloid rotifer clade and their diversity (>400 morphospecies) challenge the expectation that obligatory asexual animal lineages, in which recombination and outcrossing are absent, are evolutionary dead-ends. Historical observations (or lack thereof) had yielded a consensus that bdelloid rotifers do not produce male or hermaphrodite individuals (6) and that they are strictly parthenogenetic without any meiosis (7, 8). Moreover, the initial description of the structure of Adineta vaga genome, lacking colinear homologous scaffolds, was irreconcilable with meiosis (9). A draft genome assembly of the closely related bdelloid species Adineta ricciae found colinearity between homologous regions but could not verify it at chromosome scale (10), which was also the case for previous studies based on a handful of genomic regions (11–13). The presence or the absence of an ameiotic genome structure in bdelloids therefore remained unresolved, and a chromosome-scale assembly appeared critical.
Besides its asexual evolution, the bdelloid rotifer A. vaga also became a model species for its extreme resistance to desiccation, freezing, and ionizing radiation, with implications for space research (14, 15). Both prolonged desiccation, encountered in their ephemeral limno-terrestrial habitats, and ionizing radiation induce oxidative stress and massive genome breakage that A. vaga seems able to cope with, retaining high survival and fecundity rates while efficiently repairing DNA damage (15–17). Maintaining such long-term survival and genome stability following DNA fragmentation likely requires the use of homologous recombination (HR), at least in the germ cells. Given the supposed absence of homologous chromosomes in A. vaga (9), the exact nature of their double-strand break (DSB) repair mechanism remains elusive.
Recent studies have provided evidence for recombination in bdelloid rotifers. These include a drop of linkage disequilibrium (LD) with increasing distance between genomic loci in A. vaga (13), signatures of gene conversion (9, 12), heterozygosity levels within the range of those reported for sexual metazoans (9, 10, 18), and reports of allele sharing between bdelloid individuals from the wild (13, 19–21). While recombination likely takes place in bdelloid rotifers, its underlying mechanisms remain unknown. Recombination might theoretically occur in a mitotic or meiotic cellular context, involve short genomic regions or canonical chromosome pairing, and take place between homologous and nonhomologous (i.e., ectopic) loci. The interpretation of these recombination events has yet to be reconciled with the long-standing apparent absence of males and meiosis in bdelloid rotifers (6) and to account for their ubiquity in semiterrestrial habitats where frequent desiccation occurs, inducing DNA DSBs (14, 15, 17).
Here, we present a high-quality chromosome-level genome assembly of A. vaga. This is pivotal to tackle the contradictions between its putative ameiotic structure and the footprints of recombination, possibly associated to DSB repair and desiccation. We combined the use of short reads (Illumina), long reads (ONT and PacBio), and chromosome conformation capture data (Hi-C) with three assembly methods to successfully assemble the A. vaga genome. We provide the first telomere-to-telomere haploid and phased assemblies of a parthenogenetic lineage, paving the way to study genome evolution in an asexual clade. Using a newly developed and publicly available tool to detect horizontal gene transfers (HGTs), Alienomics, we annotated HGT candidates (HGTc) and confirmed that A. vaga has the highest number of HGTs across all animals. A. vaga has a diploid genome made of six pairs of homologous chromosomes, refuting the ameiotic structure previously described for this genome (9) and challenging the complete absence of meiosis in one of the most notable asexual animal clade. In addition, by observing large tracks of losses of heterozygosity (LOHs), we show that large-scale recombination between homologous chromosomes occurs in A. vaga. The possibility of chromosome pairing in A. vaga, during either a meiotic-like or mitotic process, allows a reinterpretation of the signatures of LD decay and allele sharing. Until now, the lack of chromosome-scale assemblies of parthenogenetic genomes hampered the investigation of the impact of meiosis, recombination, outcrossing, or their absence on entire genomes. Moreover, characterizing homologous chromosomes as potential templates for DNA repair through HR in A. vaga is an important landmark in understanding of bdelloid extreme resistance. This high-quality genome assembly of A. vaga (AV20) is also timely for comparative biology within rotifers and protostomians, extending the list of chromosome-level genomes in overlooked phyla.
RESULTS AND DISCUSSION
A diploid genome with a tetraploid past
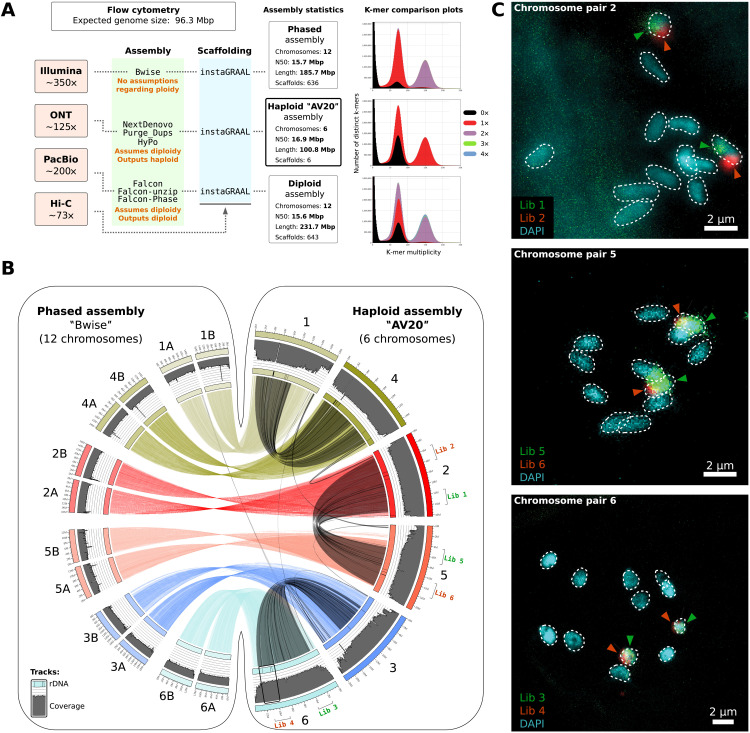
Distinct independent genome assembly procedures, relying on different assumptions regarding ploidy levels [Bwise (22), NextDenovo (23), and Falcon (24)], were first used on a combination of short and long sequencing reads. These assemblies were then scaffolded using Hi-C data and instaGRAAL (25), revealing similar chromosome-level assemblies and genome size estimations consistent with flow cytometry measurements (Fig. 1A and figs. S1 and S2). All pairwise alignments of the three independent assemblies (referred to as “phased” without ploidy assumption, “haploid,” and phased “diploid”; see Fig. 1A) confirmed chromosome-level synteny and converged toward an identical genome structure, with the six longest scaffolds from the haploid assembly (hereafter named “AV20”) being each colinear to exactly two long scaffolds from the phased assembly (Fig. 1B; see also figs. S3 to S5). To validate these assemblies, we performed fluorescence in situ hybridization (FISH) analyses with three pairs of fluorescent probe libraries complementary to separate parts of chromosomes 2, 5, and 6 from the AV20 assembly (Fig. 1B, right). For each pair of probes (one green and one red), two individual chromosomes were labeled with little or no overlap between both signals (Fig. 1C). Chromosome painting on the karyotype of 12 chromosomes of A. vaga (26) was consistent with our chromosome-scale assemblies, showing that the A. vaga genome is diploid, being composed of six pairs of colinear homologous chromosomes.
Fig. 1. The genome structure of A. vaga is diploid.
(A) Outline of the three genome assembly approaches underlined by different assumptions on genome ploidy, with median read coverage for all sequencing technologies indicated on the left and estimated with respect to the AV20 haploid genome assembly. The haploid genome size estimate of A. vaga obtained by flow cytometry (under the assumption that the genome is diploid), as well as the summary statistics of the genome assemblies, is given. The number of chromosomes corresponds to the number of scaffolds longer than 10 Mbp. Ploidy levels of assemblies are indicated by the KAT plots of k-mers distribution (first and second peaks correspond to heterozygous and homozygous k-mers, respectively; red and purple indicate haploidy and diploidy, respectively). (B) Circos plot of the pairwise colinearity between the haploid AV20 and the phased Bwise genome assemblies, depicted by colored links and obtained using nucmer. Synteny blocks within AV20 genome (between homoeologous copies) are depicted as gray links and were obtained using MCScanX. Coverage along scaffolds of both AV20 and the phased assembly is depicted as gray histograms and was computed on the basis of Illumina reads from sample GC047403. Thin black bars on the scaffold ideograms correspond to ribosomal RNA (rRNA) genes. Schematic position of the FISH probe libraries on chromosome pairs 2, 5, and 6 is indicated on the corresponding AV20 chromosomes. (C) Fluorescence microscopy images of the 12 chromosomes of A. vaga [4′,6-diamidino-2-phenylindole (DAPI) staining] with chromosome pairs 2, 5, and 6 highlighted by oligo painting using the FISH probe libraries depicted in (B).
We compared our new AV20 assembly to the previously published draft genome assembly [hereafter named “AV13” (9)]. None of the previously described colinearity break points and palindromes were retrieved in the new AV20 genome, indicating that these were likely assembly artifacts resulting from erroneous scaffolding of phased haplotypes (figs. S6 and S7). Chromosome-level colinearity, albeit weaker than between homologous chromosomes, was also observed between pairs of homoeologous (or ohnologous) chromosomes in the AV20 genome, a signature confirming the previously reported paleotetraploidy of A. vaga (9, 10, 12) (gray links in Fig. 1B). The three chromosome pairs 1, 2, and 3 are homoeologous to the three pairs 4, 5, and 6, respectively. A. vaga is thus a diploid, paleotetraploid species in which the level of synteny between homoeologous chromosomes is high. Notably, 30.8% of the genes have homoeologous copies within conserved synteny blocks (see Materials and Methods) and with an average nucleotide divergence of about 13% (fig. S8).
Recombination between homologous chromosomes causes loss of heterozygosity
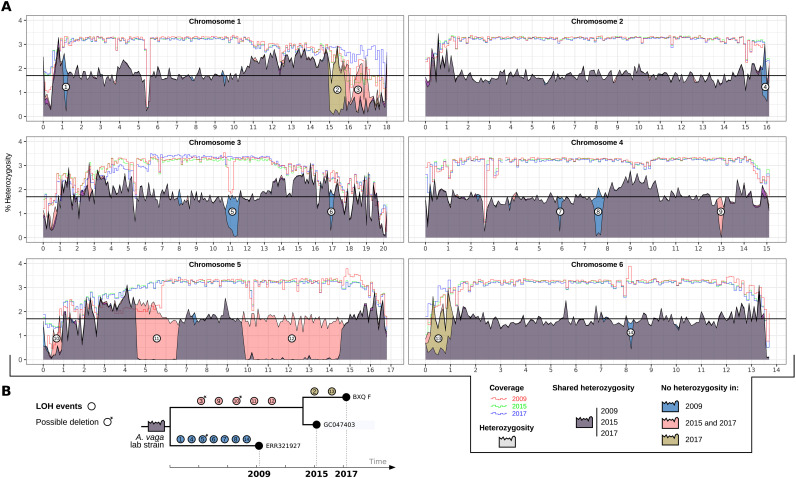
The discovery of homologous chromosomes in the oldest known asexual animal clade represents a major shift for studies of ancient asexuals and leads us to reconsider the possibility of HR in A. vaga. One potential genetic consequence of recombination between homologous chromosomes is loss of heterozygosity (LOH). We measured and compared heterozygosity along the chromosomes of three A. vaga samples cultured from the same ancestral laboratory strain that never underwent stresses causing recombinogenic DSBs and that were sequenced at three distinct time points (2009, 2015, and 2017; Fig. 2A and table S1). Mean single-nucleotide polymorphism (SNP) heterozygosity (i.e., divergence between homologous chromosomes) was around 1.7% [horizontal line in Fig. 2A; similar to previous reports (9, 10)]. We observed large regions (from 100 kb to 4.5 Mb) that were fully homozygous, except for a few SNPs, in specific isolates, while heterozygous in others (numbered tokens in Fig. 2A). Note that a few homozygous tracks were associated with coverage variation and could have been caused by a hemizygous deletion (when coverage drops by approximately 50%, e.g., event 5 in Fig. 2A) or by the high density of repeated sequences (e.g., event 13 in Fig. 2A). Given the genealogy of these laboratory lines, we argue that the large homozygous tracks that are associated with homogeneous median coverage are signatures of allelic recombination events causing LOH (Fig. 2B).
Fig. 2. Heterozygosity dynamics in A. vaga.
(A) Heterozygosity and coverage distributions of three independent A. vaga samples from the same laboratory strain along the six chromosomes. Samples are labeled by the date of the extraction of their DNA (i.e., 2009, 2015, and 2017). Data from 2009 were used to assemble the previous version of A. vaga genome (9). Lines indicate short read coverage (normalized), and filled areas indicate the percentage of heterozygosity (y axis). Chromosome lengths (x axis) are in Mb. Mean SNP heterozygosity (1.7%) is depicted by the horizontal black line. (B) Schematic reconstruction of heterozygosity evolution among three samples from the same initial A. vaga laboratory strain. Note that each sample had its own independent evolution, and the exact sequence and timing of loss of heterozygosity (LOH) events is unknown. LOH events noted with a small asterisk might correspond to deletions given the drop of coverage associated with the absence of heterozygosity.
These LOHs appear to accumulate through time as some are shared by two samples (e.g., event 12 in Fig. 2), while others appeared in only one of these two samples (e.g., event 2 in Fig. 2). Note that no ancestral LOH was found that would be shared by all of the strains. This is likely because large LOH events increase the chance of exposing recessive deleterious mutations and are thus likely to be selectively eliminated in nature, maintaining the relatively homogeneous heterozygosity level in the ancestral laboratory strain (Fig. 2A). The presence of LOH tracks on all six chromosome pairs in the three laboratory samples over a relatively short period of time (i.e., several years; Fig. 2B) might be due to the culturing conditions, allowing possible bottlenecks and relaxed selection. Recombination occurring along the entire chromosomes, instead of being restricted to the telomeres only (27), invalidates the hypothesis that an Oenothera-like meiosis underlies their reproductive mode [in agreement with a recent study (13)]. Overall, these LOH tracks combined with the recently reported LD decay (13) represent a clear footprint that molecular processes involving recombination between homologous chromosomes occur in the germ line of A. vaga.
Recombination could be accidental or programmed
Theoretically, recombination between homologous chromosomes resulting in inheritable LOH could occur in the germ line during mitotic repair of accidental DSBs or when handling programmed DSBs during meiosis [potentially induced by the Spo11 protein (9, 28)]. DSBs can be repaired by different recombination pathways, but LOH of large chromosome regions without coverage reduction (e.g., events 1, 6 to 9, 11, and 12; Fig. 2) can primarily arise from two processes, break-induced replication (BIR) and the formation of crossing-over (CO). BIR is a mechanism of mitotic recombination characterized by replication fork progression over hundreds of kilobases on the repair template (29). When involving allelic loci, it causes LOH in the segment, extending from the break point site until the end of the chromosome. If a double BIR occurs, switching templates from the homologous chromosome back to the sister or the original chromatid, an LOH tract, possibly long, that does not encompass the telomere is produced (30). Such LOH could also be generated by the recombinational repair of one or two DSBs, respectively, leading to CO (i.e., a reciprocal genetic exchange between chromosomes). Compared to BIR, CO is, however, a minor pathway in mitotically cycling cells (31) that preferentially takes place between sister chromatids and therefore remains genetically silent (32).
Alternatively, programmed DSBs during meiosis can produce large LOH tracks by favoring CO formation between homologous chromosomes (31). LOH signatures in the A. vaga genome could therefore be acquired through meiotically induced recombination instead of during mitosis. Several mechanisms of meiotic parthenogenesis, globally referred to as automixis, have been described in various species such as in Daphnia pulex (33), Artemia parthenogenetica (34), or Apis mellifera capensis (35). If automixis occurs in A. vaga, the heterozygosity patterns observed here (Fig. 2), in which maternal heterozygosity is conserved along chromosomes due to the nonsegregation of homologous chromosomes, while large LOH tracks (likely counterselected in nature) could result from their CO recombination, are genetically equivalent to what is referred to as central fusion automixis (34). Nevertheless, no cytological evidence of any meiotic process has been described so far in bdelloid rotifers. Whether recombination is a key feature of the reproductive mode of bdelloids (through programmed DSBs during a modified meiosis) or whether it is mainly driven by desiccation resistance mechanisms (through accidental DSB repair in the germ line), or both, remains an open question. Whichever mechanism is involved, recombination likely plays a major role in the long-term evolution of the A. vaga genome.
Dynamic subtelomeric regions
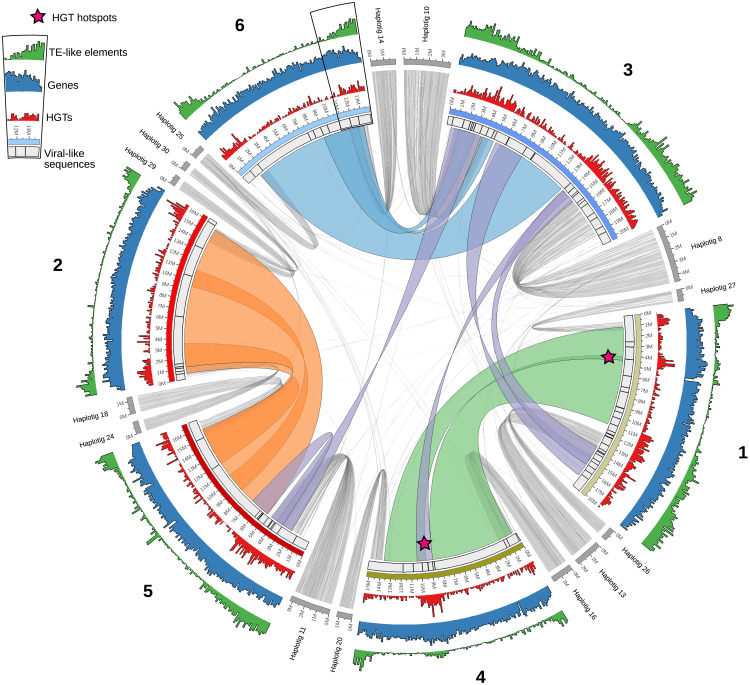
We found a low amount of transposable elements (TEs) in A. vaga (Fig. 3 and fig. S9). By combining two approaches to annotate both TE-like elements, including repeated sequences, as well as canonical TEs (i.e., the EDTA and REPET pipelines), we detected 6.6% of TE-like elements and 1.9% of canonical TEs, predominantly located in subtelomeric regions (Fig. 3). In addition, rotifer-specific telomeric repeats [i.e., (TGTGGG)n (36)] were detected at the extremities of every scaffold of the AV20 assembly, indicating that they correspond to telomeric and subtelomeric regions and that AV20 reached a chromosome-level assembly (fig. S10). Most consensus TE sequences were found at low copy numbers (i.e., 96% of canonical TE consensus sequences are present in less than six copies in AV20; see fig. S9). Notably, terminal inverted repeat (TIR) DNA transposons (i.e., Class-II TIRs) were quantitatively dominant (48% of all TEs) among the low amount of TEs in A. vaga genome (figs. S9 and S11). These results are in line with previous studies of TEs in bdelloids (9, 10, 37, 38). Using sequence similarity between a TE copy and its consensus as a proxy for how recent this copy is, we found that Class-II TIRs and Class-I LINEs (long interspersed nuclear elements) and LTRs (long terminal repeats) had high average similarity to their consensus sequences, suggesting that they have been at least recently active in A. vaga genome (fig. 12). Investigating putative endogenous viral elements (EVEs) in A. vaga revealed very few viral-like sequences (i.e., 94 loci scattered along the six chromosomes; Fig. 3), with potential donor candidates belonging to the group of large double-stranded DNA viruses. However, none of these EVE candidates had definitive viral origins, as their similarity was not restricted to viral sequences and there was no conservation of viral gene synteny.
Fig. 3. DNA content of the haploid AV20 genome assembly.
Conservation of synteny of HGTc is depicted by colored links between the six chromosome pairs. Violet links correspond to the synteny block of HGTc between nonhomoeologous chromosomes. Localization of alternative haplotigs, removed before genome scaffolding, is depicted by gray links. Distribution of repeated elements, genes, HGTc, and viral-like sequences are depicted in green, blue, red, and black bars, respectively. Ancient HGTc hotspots are indicated by pink stars.
Syntenic HGTc among nonhomoeologous chromosomes are visible, mainly in subtelomeric regions (violet links on Fig. 3) and may suggest chromosomal rearrangements. Subtelomeric regions are also the regions on which almost all divergent haplotypes (i.e., haplotigs corresponding to uncollapsed haplotypes during genome assembly process) were located (gray links on Fig. 3). Overall, these subtelomeric regions in A. vaga not only are enriched in canonical TEs, TE-like elements, HGTc, and viral-like sequences but also retain a higher haplotypic divergence (i.e., uncollapsed haplotigs) and most chromosomal rearrangements. When accounting for coding sequences only, no distinct increase or decrease of heterozygosity could, however, be observed in subtelomeric regions (fig. S13). At this stage, it is therefore unclear whether HR rate covaries with distance from telomeres in A. vaga. Nevertheless, our results suggest that subtelomeric regions seem more prone to chromosomal rearrangements, incorporation of foreign DNA (TEs and HGTs), and structural variations such as putative allelic deletions (see LOH events 3 and 10 in Fig. 2), evolving faster than the rest of the genome.
HGTs in A. vaga genome
The acquisition of foreign DNA has been hypothesized to play an important role in bdelloid evolution (20). HGTs could circumvent some deleterious effects of the lack of sexual outcrossing, and the occasional integration of foreign DNA could trigger adaptation (10, 17, 20, 39, 40). No automated tool existed to detect HGTs; therefore, we developed Alienomics, an innovative pipeline to detect both HGTs and contaminants in a genome assembly. Alienomics combines several genomic parameters such as gene taxonomy, GC (guanine-cytosine) content, and sequencing depth, also taking into account gene integration into the genome using synteny and expression data, to detect HGTs from nonmetazoan species. In contrast with the overall low amount of TEs, many HGTc (2679, about 8.3% of all genes) were detected in the A. vaga AV20 genome assembly, confirming previous reports of the highest HGT content among metazoans (9, 10, 41, 42). HGTc were enriched in subtelomeric regions as previously reported (41), although many HGTc were distributed along the chromosomes and two visible local hotspots were detected outside of the subtelomeric regions (pink stars in Fig. 3). One HGT hotspot was associated with a slight increase of interstitial telomeric repeats (fig. S10) that could represent a signature of an ancient chromosome fusion. Overall, the heterogeneity in HGTc density between subtelomeric regions and the rest of the genome could be explained either by varying rates of HGT incorporation along the chromosomes or by varying successful integration of HGTs within the genome through selection.
Using both MCScanX and Alienomics outputs, we determined that 257 foreign genes (9.6% of all HGTc) had conserved their synteny across homoeologous chromosomes, including the HGTc hotspots notably visible on homoeologous chromosomes 1 and 4 (stars on Fig. 3). These horizontal transfer events therefore occurred before the ancestral tetraploidization of modern bdelloids. This amount of ancient HGTc is, however, likely underestimated as any loss or translocation of an ancient HGTc copy would break the ancestral synteny. When looking specifically at HGTc that occurred before the tetraploidization, we observed an enrichment of genes involved in DNA recombination and DNA ligation being part of the DNA DSB repair pathways, among other enriched functional categories (see table S2). These HGTc might have allowed bdelloids to resist and overcome massive DNA breakage. When analyzing all HGTc, we found that they are enriched in genes involved in oxidation reduction and carbohydrate metabolic processes (9) as well as in the response to nitrosative stress (see table S2). Acquisition of HGTs might therefore be central in their resistance to extreme desiccation and toward more efficient homeostasis. Overall, these results are in line with previous studies, suggesting that HGTs have been continuously acquired within bdelloid rotifers, even before their tetraploidization (10, 42). However, if bdelloids have the same low rate of HGT acquisition from other individuals of the same species than from nonmetazoans (12.8 HGT/Ma), HGT is possibly insufficient to compensate for the plausible lack of outcrossing in bdelloid rotifers (40). Nevertheless, a high rate of HGT acquisition from distinct species might be deleterious for A. vaga, because recombination between homologous chromosomes appears to be central to the maintenance of heterozygosity and/or genome structure in this species.
Reasoning on bdelloid rotifers’ reproductive mode
Bdelloid rotifer species are both supposedly devoid of males and prone to integrate foreign DNA (through HGTs) into their genome. In this context, several reports of allele sharing between bdelloid individuals sampled from the wild triggered a debate whether they could exchange genetic content at all and whether this might be done through HGT or through sexual reproduction (13, 19–21, 43–45). At a first glance, showing that homologous chromosomes exist and recombine in A. vaga could be viewed as a support to the hypothesis that bdelloids might undergo meiotic sexual reproduction (13). However, this hypothesis has yet to be reconciled with the absence of both males and canonical meiosis in bdelloid rotifers, and here, we speculate on the mechanisms of HR in A. vaga. The three A. vaga lineages analyzed here (Fig. 2) were kept in hydrated conditions, leaving few opportunities for desiccation-induced, accidental DNA DSBs. Moreover, a much lower heterozygosity than for A. vaga has been observed in two obligate aquatic bdelloid rotifer species (i.e., in the genus Rotaria in which the upper limit of homologous divergence ranged between 0.033 and 0.075), also described as asexual and never experiencing desiccation. Both these observations favor the hypothesis that HR in bdelloids could be caused by programmed DNA DSBs during a meiotic-like process. Frequent and programmed recombination would cause LOH in hydrated A. vaga (Fig. 2) and would have lowered heterozygosity in the obligately aquatic Rotaria species.
Whatever the underlying mechanism, the observed recombination signatures in bdelloid rotifers are compatible with the three hypotheses proposed to explain the previous reports of allele-sharing patterns in bdelloid rotifers: (i) allele sharing may be an artifact due to undetected contamination between cultures, either during colony culture itself or during sample preparation for sequencing (46–50); (ii) allele sharing is the result of horizontal genetic transfers between bdelloid individuals through unknown molecular mechanisms, possibly associated with desiccation (but not for the nondesiccating species) and potentially linked to the high propensity of bdelloids to retain nonmetazoan genes into their genomes (13, 17, 20, 40, 43, 51); and (iii) allele sharing is caused by cryptic sexual reproduction (52), with sex events being rare enough that males, sperm, fertilization, and meiosis were never observed, but sufficiently frequent to leave a distinctive footprint in every population sample studied so far (13, 19, 21, 45). The mechanism behind the observed signatures of genetic exchanges between bdelloid individuals remains puzzling, and therefore, the significance of outcrossing in this asexual lineage is still unclear. We anticipate that the chromosome-level genome assembly of A. vaga presented here will stimulate future population genomic studies that will help to determine the cause of these allele-sharing patterns.
Long-term asexual evolution
This high-quality telomere-to-telomere assembly firmly establishes A. vaga as a model system to study long-term asexual evolution. Homologous chromosomes are present in the bdelloid species A. vaga and might well occur in all bdelloid rotifers, as colinear pairs of sequenced fosmids were found in the two distinct bdelloid species A. vaga and Philodina roseola, with each colinear pair in one species resembling a colinear pair in the other species (12). The observed long LOH tracks indicate the existence of long-range HR (Fig. 4). Whether this occurs during a meiotic-like parthenogenetic mode of reproduction or in a mitotic context during frequent repair of accidental DNA DSBs remains speculative. Recombination (mitotic or meiotic) could increase the rate of gene conversion in asexual lineages, a signature previously observed in A. vaga (9). Gene conversion, particularly when slightly biased, could correct deleterious mutations and reduce the rate of clonal deterioration (53) or even speed up the fixation of beneficial mutations (54). However, besides signatures of LOH via allelic recombination, we also observed LOH via deletions in the genome of A. vaga. The random accumulation of LOH events could expose deleterious recessive mutations in asexuals through loss of complementation (55). Our new chromosome-level genome assembly of asexual A. vaga therefore provides a critical tool to be able to evaluate the relative benefits of these recombination events on their long-term evolution and paves the way for studies on genome dynamics in A. vaga.
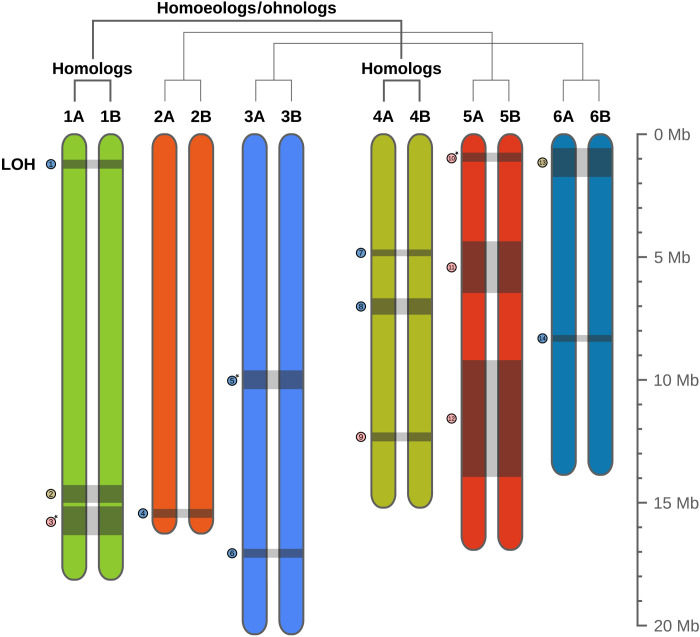
Fig. 4. Schematic representation of the karyotype of A. vaga.
The 12 chromosomes correspond to six pairs of homologous chromosomes (i.e., a diploid genome structure) sharing the same color. Ancestral genome hybridization (or whole-genome duplication) led to the existence of pairs of homoeologs (or ohnologs), represented in different but similar colors. Gray blocks linking homologous chromosomes and their corresponding numbered tokens depict LOH events produced by HR (see also Fig. 2). The length of chromosomes (in Mb) is indicated by the scale on the right.
In general, asexual populations suffer from the absence of gene shuffling with other individuals, and the long-term evolutionary success of bdelloid rotifers in the absence of outcrossing therefore remains puzzling. It is important to try to discriminate between the consequences of the two aspects underlying sex: recombination and outcrossing. Theoretical work on population genetics showed that selection could be at least as efficient in automictic lineages than in sexuals under certain circumstances [e.g., effective population size and recombination rates (56)]. It is therefore conceivable that the combination of potentially large populations, a relatively high level of heterozygosity (or mutation rate), and specific recombination rates might explain how A. vaga maintains a delicate balance between losing and accumulating heterozygosity and how it adapts and persists in the long term. Unfortunately, critical knowledge about bdelloid biology (e.g., quantitative estimates of mutation and recombination rates) is still lacking to determine whether they might circumvent the lack of outcrossing through a genetic equivalent of automixis. Outcrossing through sexual reproduction might speed up adaptation by allowing the combination of independently evolved alleles within the same individual, but might not be essential for bdelloid rotifers, especially if a high frequency of HGT is also taking place. Despite presenting the highest amount of HGTs among animals, our results also suggest that bdelloid rotifers might have to balance the acquisition rate of HGTs, a source of functional novelties, with the maintenance of faithful homology between chromosomes for HR. Overall, our work reinforces the hypothesis that recombination is critical for lineage longevity. Ancient asexual animals without a minimal rate of recombination, programmed through meiotic processes and/or accidental through their lifestyle, might not exist at all.
MATERIALS AND METHODS
Complete description of Materials and Methods can be found in the Supplementary Materials.
Genome sequencing and assembly
Cultures of the A. vaga AD008 laboratory strain were processed to obtain the following sequencing data: about 350× coverage of WGS 250–base pair (bp) paired-end Illumina reads, 200× coverage of PacBio RSII long reads, 125× coverage of ONT long reads, and 75-bp paired-end Illumina reads of Hi-C libraries. Three independent genomes were assembled using Bwise (22) (on Illumina short reads), NextDenovo (23) (on ONT long reads), and Falcon (24) (on PacBio long reads). Uncollapsed haplotypes in the ONT-based assembly were detected and discarded using purge_dups. This NextDenovo assembly was then polished on the basis of Illumina short reads and PacBio long reads using HyPo and referred to as AV20. All assemblies were scaffolded with instaGRAAL (25) using Hi-C data. Ploidy level and genome size were confirmed through interpretation of k-mers spectra using KAT (57) as well as by synteny analyses between the three genome assemblies presented here and the previously published AV13 genome assembly using MCScanX (58) (synteny blocks detection), nucmer (59) (genome alignment), and D-GENIES (60) (dot plot visualization). Flow cytometry measurements further confirmed this genome size, and FISH was used to confirm the ploidy level. For this, we designed three pairs of oligos on three different chromosomes using the OligoMiner pipeline (61), and embryos fixed at one-cell stage in methanol were permeabilized (i.e., mechanical squash, Triton, and saponin). Oligo probes were added to embryos followed by DNA denaturation (at 92°C) before hybridization of probes and DNA (at 37°C). Karyotype images were obtained using confocal imaging.
Genome annotation
TE-like elements and canonical TE consensus were predicted from the AV20 genome assembly using EDTA (62) and TEdenovo pipeline (63, 64) (part of the REPET pipeline). TE-like elements were then annotated using TEannot. Genes were annotated using funannotate (65) as described below. First, repeated elements were masked using BEDTools (66). Second, part of the available RNA sequencing (RNA-seq) reads were mapped onto the genome, while the other part of RNA-seq reads were used to produce a de novo assembled transcriptome that was subsequently aligned onto the genome as part of the PASA pipeline (67). Third, PASA annotations, the de novo assembled transcriptome, the metazoan BUSCO database, and the protein UniProt database were used as inputs within funannotate predict function. This function first produced ab initio gene annotations using GeneMark-ES, which were then used along with transcripts and protein data to train Augustus to generate a second set of gene annotations. Fourth, funannotate used EVidenceModeler as a weighted approach to combine gene annotations from PASA, GeneMark, and high-quality annotations from Augustus into a single, integrated, gene annotation set. Last, InterProScan 5 (68) was used to produce functional annotations to these genes. These functional annotations, along with the BUSCO metazoan database, were then used as input for the funannotate annotate function with default parameters to produce the final functional annotation for the predicted genes. Alienomics, a newly designed pipeline, was used to detect HGTs. This approach combines GC content, coverage, sequence similarity, taxonomic information, expression level, and synteny information to detect both HGTc (i.e., alien genes integrated into host scaffolds) and potential contaminants (i.e., alien genes present on alien scaffolds). Note that our approach can only detect transfers from alien source outside of a given clade (here, metazoans). Viral-like genes were detected by performing a DIAMOND BLASTX search on AV20 scaffolds using all viral proteins extracted from the nr database of the National Center for Biotechnology Information (NCBI) (February 2020) to the exception of Retroviridae and Hepadnaviridae.
Genome analyses
Coverage along AV20 scaffolds was computed using read mapping with BWA-MEM. These mappings were used for measuring heterozygosity in three samples (i.e., GC047403, BXQF, and ERR321927) by genotyping them using GATK (HaplotypeCaller function with -ERC GVCF option). The resulting gvcf files were combined (CombineGVCFs function) and were then jointly genotyped (GenotypeGVCFs function). The variants were then filtered to only retain SNPs using custom bash and perl scripts. Divergence between homoeologous chromosomes was assessed by producing a self-alignment of AV20 genome using nucmer, which was then filtered to only retain genomic alignments between homoeologous regions ranging from 500 to 10,000 bp. MCScanX was used to detect synteny among HGTc, and custom scripts were used to detect strictly homoeologous HGTc synteny blocks stemming from the paleotetraploidization of bdelloids (i.e., ancient HGTc). Gene Ontology (GO) terms from the functional annotation were extracted for the 2422 recent HGTc; the 257 ancient HGTc were then compared to the entire gene set of the AV20 genome containing 32,378 proteins. Enrichment analyses were performed using the topGO package with a Fisher test and the “elim” algorithm (69).
Reappraisal of the AV13 genome assembly
ONT reads were trimmed using porechop (70) and were mapped onto the previous AV13 genome assembly using NGMLR (71). The AV20 and AV13 genome assemblies were aligned together using Sibelia (72). This genome alignment was used to localize putative break points, which were then inspected using Tablet (73) to evaluate whether ONT reads confirmed previously reported break points in the AV13 genome assembly. Synteny blocks from the Sibelia alignment between AV20 and AV13 were analyzed using a custom perl script (available at https://github.com/jnarayan81/huntPalindrome) to localize putative palindromes, and circos plots of these locations were inspected to evaluate the existence of palindromes in the AV20 genome.
Acknowledgments
We thank A. Mayer for help with score computation and transformation within Alienomics, M. Colinet for help with FISH experiments, and N. Verbruggen for providing Arabidopsis plantlets for genome size estimation. The Morphim imaging platform of UNamur is acknowledged for technical help with the FISH image analyses. Funding: This project received funding from the Horizon 2020 research and innovation program under European Research Council (ERC) grant agreement 725998 (RHEA) and from BELSPO PRODEX for the ESA selected ILSRA-2014-0106 Project to K.V.D., from the Fédération Wallonie-Bruxelles via an “Action de Recherche Concertée” (ARC) grant to K.V.D. and B.Ha., from the European Union’s Horizon 2020 research and innovation programme under Marie Skłodowska-Curie grant agreement 764840 to J.-F.F. (ITN IGNITE; www.itn-ignite.eu), from the Fédération Wallonie-Bruxelles via an Action de Recherche Concertée (ARC) grant to J.-F.F., from the European Research Council under the Horizon 2020 Program (ERC grant agreement 260822) and JPI-EC-AMR STARCS ANR-16-JPEC-0003-05 grant to R.K., and through funding by the French Government “Investissement d’Avenir” program FRANCE GENOMIQUE (ANR-10-INBS-09). E.N. obtained an FSR UNamur fund. A.H., A.D., and R.A. are Research Fellows of the Fonds de la Recherche Scientifique—FNRS. Author contributions: Conceptualization: P.S., J.N., T.L., E.N., A.H., A.D., M.C., R.K., A.L., M.T., J.-F.F., and K.V.D. Data curation: P.S., J.N., A.H., A.D., and R.A. Formal analysis: P.S., J.N., T.L., J.-F.F., E.N., A.H., C.G., F.R.G., A.D., M.C., L.B., and D.K.L.K. Funding acquisition: K.V.D. Investigation: P.S., J.N., T.L., A.H., E.N., M.C., M.L., A.D., L.B., R.K., E.G.J.D., D.K.L.K., R.A., A.L., J.-F.F., and K.V.D. Methodology: P.S., J.N., T.L., J.-F.F., E.N., A.H., C.G., R.C., M.L., A.D., M.C., L.B., R.K., D.K.L.K., M.M., N.G., and A.L. Project administration: J.-F.F. and K.V.D. Resources: L.B., C.C., M.M., B.He., K.L., J.V., J.-F.F., and K.V.D. Software: J.N., P.S., J.-F.F., A.H., A.D., M.C., D.K.L.K., N.G., and A.L. Supervision: T.L., R.K., E.G.J.D., R.C., B.Ha., J.-F.F., and K.V.D. Validation: J.N., P.S., A.H., J.-F.F., and K.V.D. Visualization: P.S., J.N., J.-F.F., E.N., A.H., D.K.L.K., and K.V.D. Writing—parts of the original draft: P.S., J.N., J.-F.F., A.H., M.T., E.G.J.D., T.L., and K.V.D. Writing—review and editing: All authors. Supervising the entire article writing: P.S. and K.V.D. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Haploid genome assembly, gene annotation, and sequencing reads are available under the project accession number PRJNA680543. Phased and diploid genome assemblies (i.e., “Bwise” and “Falcon”) as well as annotation of HGTc (gff format) and TE-like elements (bed format) for the AV20 genome assembly are available on Zenodo: 10.5281/zenodo.5126644.
Supplementary Materials
This PDF file includes:
Supplementary Materials
Figs. S1 to S13
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.J. Maynard Smith, The Evolution of Sex (Cambridge Univ. Press, 1978). [Google Scholar]
- 2.Lehtonen J., Jennions M. D., Kokko H., The many costs of sex. Trends Ecol. Evol. 27, 172–178 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Seudre O., Vanhoenacker E., Mauger S., Coudret J., Roze D., Genetic variability and transgenerational regulation of investment in sex in the monogonont rotifer Brachionus plicatilis. J. Evol. Biol. 33, 112–120 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Fradin H., Kiontke K., Zegar C., Gutwein M., Lucas J., Kovtun M., Corcoran D. L., Baugh L. R., Fitch D. H. A., Piano F., Gunsalus K. C., Genome architecture and evolution of a unichromosomal asexual nematode. Curr. Biol. 27, 2928–2939.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill W. G., Robertson A., The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294 (1966). [PubMed] [Google Scholar]
- 6.Birky C. W., Positively negative evidence for asexuality. J. Hered. 101, S42–S45 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Hsu W. S., Oogenesis in the bdelloidea rotifer Philodina roseola Ehr. La Cellule 57, 283–296 (1956). [Google Scholar]
- 8.Hsu W. S., Oogenesis In Habrotrocha tridens (Milne). Biol. Bull. 111, 364–374 (1956). [Google Scholar]
- 9.Flot J.-F., Hespeels B., Li X., Noel B., Arkhipova I., Danchin E. G. J., Hejnol A., Henrissat B., Koszul R., Aury J. M., Barbe V., Barthélémy R. M., Bast J., Bazykin G. A., Chabrol O., Couloux A., da Rocha M., da Silva C., Gladyshev E., Gouret P., Hallatschek O., Hecox-Lea B., Labadie K., Lejeune B., Piskurek O., Poulain J., Rodriguez F., Ryan J. F., Vakhrusheva O. A., Wajnberg E., Wirth B., Yushenova I., Kellis M., Kondrashov A. S., Mark Welch D. B., Pontarotti P., Weissenbach J., Wincker P., Jaillon O., van Doninck K., Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500, 453–457 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Nowell R. W., Almeida P., Wilson C. G., Smith T. P., Fontaneto D., Crisp A., Micklem G., Tunnacliffe A., Boschetti C., Barraclough T. G., Comparative genomics of bdelloid rotifers: Insights from desiccating and nondesiccating species. PLOS Biol. 16, e2004830 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welch D. B. M., Welch J. L. M., Meselson M., Evidence for degenerate tetraploidy in bdelloid rotifers. Proc. Natl. Acad. Sci. U.S.A. 105, 5145–5149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hur J. H., Van Doninck K., Mandigo M. L., Meselson M., Degenerate tetraploidy was established before bdelloid rotifer families diverged. Mol. Biol. Evol. 26, 375–383 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Vakhrusheva O. A., Mnatsakanova E. A., Galimov Y. R., Neretina T. V., Gerasimov E. S., Naumenko S. A., Ozerova S. G., Zalevsky A. O., Yushenova I. A., Rodriguez F., Arkhipova I. R., Penin A. A., Logacheva M. D., Bazykin G. A., Kondrashov A. S., Genomic signatures of recombination in a natural population of the bdelloid rotifer Adineta vaga. Nat. Commun. 11, 6421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontaneto D., Bunnefeld N., Westberg M., Long-term survival of microscopic animals under desiccation is not so long. Astrobiology 12, 863–869 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Hespeels B., Penninckx S., Cornet V., Bruneau L., Bopp C., Baumlé V., Redivo B., Heuskin A. C., Moeller R., Fujimori A., Lucas S., van Doninck K., Iron ladies—How desiccated asexual rotifer Adineta vaga deal with x-rays and heavy ions? Front. Microbiol. 11, 1792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gladyshev E., Meselson M., Extreme resistance of bdelloid rotifers to ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 105, 5139–5144 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hespeels B., Knapen M., Hanot-Mambres D., Heuskin A. C., Pineux F., Lucas S., Koszul R., van Doninck K., Gateway to genetic exchange? DNA double-strand breaks in the bdelloid rotifer Adineta vaga submitted to desiccation. J. Evol. Biol. 27, 1334–1345 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Romiguier J., Gayral P., Ballenghien M., Bernard A., Cahais V., Chenuil A., Chiari Y., Dernat R., Duret L., Faivre N., Loire E., Lourenco J. M., Nabholz B., Roux C., Tsagkogeorga G., Weber A. A. T., Weinert L. A., Belkhir K., Bierne N., Glémin S., Galtier N., Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515, 261–263 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Signorovitch A., Hur J., Gladyshev E., Meselson M., Allele sharing and evidence for sexuality in a mitochondrial clade of bdelloid rotifers. Genetics 200, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debortoli N., Li X., Eyres I., Fontaneto D., Hespeels B., Tang C. Q., Flot J.-F., Van Doninck K., Genetic exchange among bdelloid rotifers is more likely due to horizontal gene transfer than to meiotic sex. Curr. Biol. 26, 723–732 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Signorovitch A., Hur J., Gladyshev E., Meselson M., Evidence for meiotic sex in bdelloid rotifers. Curr. Biol. 26, R754–R755 (2016). [DOI] [PubMed] [Google Scholar]
- 22.A. Limasset, “Novel approaches for the exploitation of high throughput sequencing data”, PhD thesis, Université Rennes 1 (2017). [Google Scholar]
- 23.NextDenovo (2020); https://github.com/Nextomics/NextDenovo.
- 24.Chin C.-S., Peluso P., Sedlazeck F. J., Nattestad M., Concepcion G. T., Clum A., Dunn C., O’Malley R., Figueroa-Balderas R., Morales-Cruz A., Cramer G. R., Delledonne M., Luo C., Ecker J. R., Cantu D., Rank D. R., Schatz M. C., Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baudry L., Guiglielmoni N., Marie-Nelly H., Cormier A., Marbouty M., Avia K., Mie Y. L., Godfroy O., Sterck L., Cock J. M., Zimmer C., Coelho S. M., Koszul R., instaGRAAL: chromosome-level quality scaffolding of genomes using a proximity ligation-based scaffolder. Genome Biol. 21, 148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mark Welch J. L., Meselson M., Karyotypes of bdelloid rotifers from three families. Hydrobiologia 387, 403 (1998). [Google Scholar]
- 27.Golczyk H., Massouh A., Greiner S., Translocations of chromosome end-segments and facultative heterochromatin promote meiotic ring formation in evening primroses. Plant Cell 26, 1280–1293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecox-Lea B. J., Mark Welch D. B., Evolutionary diversity and novelty of DNA repair genes in asexual Bdelloid rotifers. BMC Evol. Biol. 18, 177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakofsky C. J., Malkova A., Break induced replication in eukaryotes: Mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 52, 395–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yim E., O’Connell K. E., Charles J. S., Petes T. D., High-resolution mapping of two types of spontaneous mitotic gene conversion events in Saccharomyces cerevisiae. Genetics 198, 181–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bzymek M., Thayer N. H., Oh S. D., Kleckner N., Hunter N., Double Holliday junctions are intermediates of DNA break repair. Nature 464, 937–941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadyk L. C., Hartwell L. H., Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiruta C., Nishida C., Tochinai S., Abortive meiosis in the oogenesis of parthenogenetic Daphnia pulex. Chromosome Res. 18, 833–840 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Nougué O., Rode N. O., Jabbour-zahab R., Ségard A., Chevin L.-M., Haag C. R., Lenormand T., Automixis in Artemia: Solving a century-old controversy. J. Evol. Biol. 28, 2337–2348 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Goudie F., Oldroyd B. P., Thelytoky in the honey bee. Apidologie 45, 306–326 (2014). [Google Scholar]
- 36.Mason J. M., Randall T. A., Capkova Frydrychova R., Telomerase lost? Chromosoma 125, 65–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arkhipova I., Meselson M., Deleterious transposable elements and the extinction of asexuals. BioEssays 27, 76–85 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Nowell R. W., Wilson C. G., Almeida P., Schiffer P. H., Fontaneto D., Becks L., Rodriguez F., Arkhipova I. R., Barraclough T. G., Evolutionary dynamics of transposable elements in bdelloid rotifers. eLife 10, e63194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boschetti C., Carr A., Crisp A., Eyres I., Wang-Koh Y., Lubzens E., Barraclough T. G., Micklem G., Tunnacliffe A., Biochemical diversification through foreign gene expression in bdelloid rotifers. PLOS Genet. 8, e1003035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eyres I., Boschetti C., Crisp A., Smith T. P., Fontaneto D., Tunnacliffe A., Barraclough T. G., Horizontal gene transfer in bdelloid rotifers is ancient, ongoing and more frequent in species from desiccating habitats. BMC Biol. 13, 90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gladyshev E. A., Meselson M., Arkhipova I. R., Massive horizontal gene transfer in bdelloid rotifers. Science 320, 1210–1213 (2008). [DOI] [PubMed] [Google Scholar]
- 42.B. Hespeels, J.-F. Flot, A. Derzelle, K. Van Doninck, in Evolutionary Biology: Genome Evolution, Speciation, Coevolution and Origin of Life, P. Pontarotti, Ed. (Springer International Publishing, 2014), pp. 207–225. [Google Scholar]
- 43.Flot J.-F., Debortoli N., Hallet B., Doninck K. V., Response to Signorovitch et al. Curr. Biol. 26, R755 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Schwander T., Evolution: The end of an ancient asexual scandal. Curr. Biol. 26, R233–R235 (2016). [DOI] [PubMed] [Google Scholar]
- 45.V. N. Laine, T. Sackton, M. Meselson, Sexual reproduction in bdelloid rotifers. bioRxiv 2020.08.06.239590 [Preprint], 16 April 2021; 10.1101/2020.08.06.239590. [DOI]
- 46.Ballenghien M., Faivre N., Galtier N., Patterns of cross-contamination in a multispecies population genomic project: Detection, quantification, impact, and solutions. BMC Biol. 15, 25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simion P., Belkhir K., François C., Veyssier J., Rink J. C., Manuel M., Philippe H., Telford M. J., A software tool ‘CroCo’ detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 16, 28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson C. G., Nowell R. W., Barraclough T. G., Cross-contamination explains “inter and intraspecific horizontal genetic transfers” between asexual bdelloid rotifers. Curr. Biol. 28, 2436–2444.e14 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Prous M., Lee K. M., Mutanen M., Cross-contamination and strong mitonuclear discordance in Empria sawflies (Hymenoptera, Tenthredinidae) in the light of phylogenomic data. Mol. Phylogenet. Evol. 143, 106670 (2020). [DOI] [PubMed] [Google Scholar]
- 50.P. Simion, F. Delsuc, H. Philippe, in Phylogenetics in the Genomic Era, C. Scornavacca, F. Delsuc, N. Galtier, Eds. (2020), pp. 2.1:1–2.1:34; https://hal.inria.fr/PGE/.
- 51.J.-F. Flot, N. Debortoli, B. Hallet, J. Narayan, K. Van Doninck, Reply to Cross-contamination explains “inter and intraspecific horizontal genetic transfers” between asexual bdelloid rotifers (Wilson, Nowell & Barraclough, 2018). bioRxiv 368209 [Preprint], 15 July 2018; 10.1101/368209. [DOI] [PubMed]
- 52.Boyer L., Jabbour-Zahab R., Mosna M., Haag C. R., Lenormand T., Not so clonal asexuals: Unraveling the secret sex life of Artemia parthenogenetica. Evol. Lett. 5, 164–174 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khakhlova O., Bock R., Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 46, 85–94 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Mandegar M. A., Otto S. P., Mitotic recombination counteracts the benefits of genetic segregation. Proc. R. Soc. B Biol. Sci. 274, 1301 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Archetti M., Recombination and loss of complementation: A more than two-fold cost for parthenogenesis. J. Evol. Biol. 17, 1084–1097 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Engelstädter J., Asexual but not clonal: Evolutionary processes in automictic populations. Genetics 206, 993–1009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mapleson D., Garcia Accinelli G., Kettleborough G., Wright J., Clavijo B. J., KAT: A K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 33, 574–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Tang H., DeBarry J. D., Tan X., Li J., Wang X., Lee T. H., Jin H., Marler B., Guo H., Kissinger J. C., Paterson A. H., MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delcher A. L., Salzberg S. L., Phillippy A. M., Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics Chapter 10, Unit 10.3 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Cabanettes F., Klopp C., D-GENIES: Dot plot large genomes in an interactive, efficient and simple way. PeerJ 6, e4958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beliveau B. J., Kishi J. Y., Nir G., Sasaki H. M., Saka S. K., Nguyen S. C., Wu C. T., Yin P., OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl. Acad. Sci. U.S.A. 115, E2183–E2192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ou S., Su W., Liao Y., Chougule K., Agda J. R. A., Hellinga A. J., Lugo C. S. B., Elliott T. A., Ware D., Peterson T., Jiang N., Hirsch C. N., Hufford M. B., Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20, 275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flutre T., Duprat E., Feuillet C., Quesneville H., Considering transposable element diversification in de novo annotation approaches. PLOS ONE 6, e16526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quesneville H., Bergman C. M., Andrieu O., Autard D., Nouaud D., Ashburner M., Anxolabehere D., Combined evidence annotation of transposable elements in genome sequences. PLOS Comput. Biol. 1, e22 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.J. Palmer, J. Stajich, nextgenusfs/funannotate: funannotate v1.5.3 (Zenodo, 2019).
- 66.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haas B. J., Delcher A. L., Mount S. M., Wortman J. R., Smith R. K. Jr., Hannick L. I., Maiti R., Ronning C. M., Rusch D. B., Town C. D., Salzberg S. L., White O., Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones P., Binns D., Chang H. Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A. F., Sangrador-Vegas A., Scheremetjew M., Yong S. Y., Lopez R., Hunter S., InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alexa A., Rahnenführer J., Lengauer T., Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Wick R. R., Judd L. M., Gorrie C. L., Holt K. E., Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3, e000132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sedlazeck F. J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M. C., Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.I. Minkin, A. Patel, M. Kolmogorov, N. Vyahhi, S. Pham, in Algorithms in Bioinformatics, A. Darling, J. Stoye, Eds. (Springer, 2013), pp. 215–229. [Google Scholar]
- 73.Milne I., Bayer M., Cardle L., Shaw P., Stephen G., Wright F., Marshall D., Tablet—Next generation sequence assembly visualization. Bioinformatics 26, 401–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lazar-Stefanita L., Scolari V. F., Mercy G., Muller H., Guérin T. M., Thierry A., Mozziconacci J., Koszul R., Cohesins and condensins orchestrate the 4D dynamics of yeast chromosomes during the cell cycle. EMBO J. 36, 2684–2697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., Dekker J., Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mark Welch D. B., Meselson M., Oocyte nuclear DNA content and GC proportion in rotifers of the anciently asexual Class Bdelloidea. Biol. J. Linn. Soc. 79, 85–91 (2003). [Google Scholar]
- 77.Bennett M. D., Leitch I. J., Price H. J., Johnston J. S., Comparisons with Caenorhabditis (~100 Mb) and drosophila (~175 Mb) using flow cytometry show genome size in arabidopsis to be ~157 Mb and thus ~25% larger than the Arabidopsis genome initiative estimate of ~125 Mb. Ann. Bot. 91, 547–557 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mark Welch J. L., Mark Welch D. B., Meselson M., Cytogenetic evidence for asexual evolution of bdelloid rotifers. Proc. Natl. Acad. Sci. U.S.A. 101, 1618–1621 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beliveau B. J., Joyce E. F., Apostolopoulos N., Yilmaz F., Fonseka C. Y., McCole R. B., Chang Y., Li J. B., Senaratne T. N., Williams B. R., Rouillard J. M., Wu C. T., Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. U.S.A. 109, 21301–21306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beliveau B. J., Boettiger A. N., Avendaño M. S., Jungmann R., McCole R. B., Joyce E. F., Kim-Kiselak C., Bantignies F., Fonseka C. Y., Erceg J., Hannan M. A., Hoang H. G., Colognori D., Lee J. T., Shih W. M., Yin P., Zhuang X., Wu C. T., Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fields B. D., Nguyen S. C., Nir G., Kennedy S., A multiplexed DNA FISH strategy for assessing genome architecture in Caenorhabditis elegans. eLife 8, e42823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chikhi R., Limasset A., Medvedev P., Compacting de Bruijn graphs from sequencing data quickly and in low memory. Bioinformatics 32, i201–i208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Limasset A., Flot J.-F., Peterlongo P., Toward perfect reads: Self-correction of short reads via mapping on de Bruijn graphs. Bioinformatics 36, 1374–1381 (2020). [DOI] [PubMed] [Google Scholar]
- 84.Limasset A., Cazaux B., Rivals E., Peterlongo P., Read mapping on de Bruijn graphs. BMC Bioinformatics 17, 237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zimin A. V., Marçais G., Puiu D., Roberts M., Salzberg S. L., Yorke J. A., The MaSuRCA genome assembler. Bioinformatics 29, 2669–2677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guan D., McCarthy S. A., Wood J., Howe K., Wang Y., Durbin R., Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36, 2896–2898 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.R. Kundu, J. Casey, W.-K. Sung, HyPo: Super fast & accurate polisher for long read genome assemblies. bioRxiv 2019.12.19.882506 [Preprint]. 20 December 2019; 10.1101/2019.12.19.882506. [DOI]
- 89.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN] (16 March 2013).
- 90.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tarasov A., Vilella A. J., Cuppen E., Nijman I. J., Prins P., Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., Phillippy A. M., Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.C. Matthey-Doret, L. Baudry, A. Bignaud, A. Cournac, Remi-Montagne, N. Guiglielmoni, T. F. Rodier, V. F. Scolari, hicstuff (2020); https://github.com/koszullab/hicstuff.
- 94.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lam K.-K., LaButti K., Khalak A., Tse D., FinisherSC: A repeat-aware tool for upgrading de novo assembly using long reads. Bioinformatics 31, 3207–3209 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lomsadze A., Ter-Hovhannisyan V., Chernoff Y. O., Borodovsky M., Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33, 6494–6506 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.T. Seemann, Barrnap 0.9 (2018); https://github.com/Tseemann/Barrnap.
- 100.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., Antonescu C., Salzberg S. L., Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.R. Poplin, V. Ruano-Rubio, M. A. De Pristo, T. J. Fennell, M. O. Carneiro, G. A. Van der Auwera, D. E. Kling, L. D. Gauthier, A. Levy-Moonshine, D. Roazen, K. Shakir, J. Thibault, S. Chandran, C. Whelan, M. Lek, S. Gabriel, M. J. Daly, B. Neale, D. G. MacArthur, E. Banks, Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178 [Preprint], 14 November 2017; 10.1101/201178. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
Figs. S1 to S13
References
Tables S1 and S2