A transcriptional regulator Foxq2 controls blue-type cone opsin gene in vertebrate species having tetrachromatic visual system.
Abstract
Most vertebrate lineages retain a tetrachromatic visual system, which is supported by a functional combination of spectrally distinct multiple cone photoreceptors, ultraviolet (UV), blue, green, and red cones. The blue cone identity is ensured by selective expression of blue (sws2) opsin, and the mechanism is poorly understood because sws2 gene has been lost in mammalian species such as mouse, whose visual system has been extensively studied. Here, we pursued loss-of-function studies on transcription factors expressed predominantly in zebrafish cone photoreceptors and identified Foxq2 as a blue cone–specific factor driving sws2 gene expression. Foxq2 has dual functions acting as an activator of sws2 transcription and as a suppressor of UV (sws1) opsin transcription in blue cones. A wide range of vertebrate species retain both foxq2 and sws2 genes. We propose that Foxq2-dependent sws2 expression is a prevalent regulatory mechanism that was acquired at the early stage of vertebrate evolution.
INTRODUCTION
Most vertebrates have highly developed camera-type eyes with duplex retinas equipped with rod and cone photoreceptor cells (1–3). Rods with a higher light-sensitivity respond to single photons and mediate scotopic vision under twilight conditions at night. In contrast, cones show a relatively lower sensitivity without saturating in brighter light and mediate photopic vision under a daylight condition. Color discrimination is established by a combination of spectrally distinct cone subtypes, each expressing a single cone opsin out of four subfamilies: ultraviolet-sensitive [SWS1, wavelength of maximum sensitivity (λmax): 360 to 420 nm], blue-sensitive (SWS2, λmax: 400 to 470 nm), green-sensitive (RH2, λmax: 460 to 510 nm), and red-sensitive opsins (LWS, λmax: 510 to 560 nm) (4, 5). Most vertebrates retain the tetrachromatic visual system organized by the four cone opsin subfamilies. A full set of genes encoding the four cone opsins is present in the Southern Hemisphere lamprey, a jawless vertebrate belonging to the earliest-branching vertebrate group (6). This fact supports the idea that the last common ancestor of vertebrates should have had color vision based on the four cone opsin subfamilies (4, 7).
Retinal progenitor cells differentiate into all types of retinal neurons in a temporal order, conserved among many species (8, 9). In the later process, transcription factors regulate photoreceptor-specific gene expression. Cone-rod homeobox (Crx) is an upstream transcriptional regulator for both rod and cone photoreceptors (10). A rod master regulator, neural retina leucine zipper (NRL), and its downstream factor, nuclear receptor 2E3 (Nr2e3), enhance rod-specific gene expression and repress sws1 expression (11, 12). With regard to cone subtypes, thyroid hormone receptor beta (Thrb) is a master transcriptional regulator for expression of lws opsin and responsible for differential expression between lws and sws opsins in mice (13), zebrafish (14), and human (15). Another transcription factor, T-box 2b (Tbx2b), plays an essential role in sws1 opsin expression in zebrafish (16). On the other hand, much less is known about a regulatory network governing expression of the middle-wavelength–sensitive opsin genes, sws2 and rh2, which have been lost in most mammalian species.
In zebrafish, a tetrachromatic freshwater fish, we found that sine oculis homeobox 7 (Six7) is required for expression of all the four subclasses of rh2 genes (rh2-1, rh2-2, rh2-3, and rh2-4) (17), which are tandemly arrayed, expressed in different cone cells, and spectrally distinct from each other (18, 19). Six7 and its homolog Six6b control sws2 expression as well (20). The cone-enriched transcription factors, Six7 and Six6b, share common DNA binding sites in both rh2 and sws2 gene loci, and Six6b overexpression rescues the reduced level of rh2 expression in six7 deficient fish (20). The overlapping functions between Six6b and Six7 for sws2 and rh2 expression imply that an additional factor(s) directs selective expression of either sws2 or rh2 gene and determines the identities of the middle-wavelength–sensitive cone subtypes. In the present study, gene expression profiling with isolated rods and cones enabled us to identify a list of cone-enriched transcription factors. Our in vivo functional analyses revealed a core transcriptional network, in which Foxq2 acts as a downstream regulator of Six7 and regulates sws2 expression. We demonstrate that Foxq2 is a terminal selector determining SWS2 cone identity during development of the middle-wavelength–sensitive cone subtypes.
RESULTS
A severe reduction of sws2 expression in foxq2 mutant zebrafish
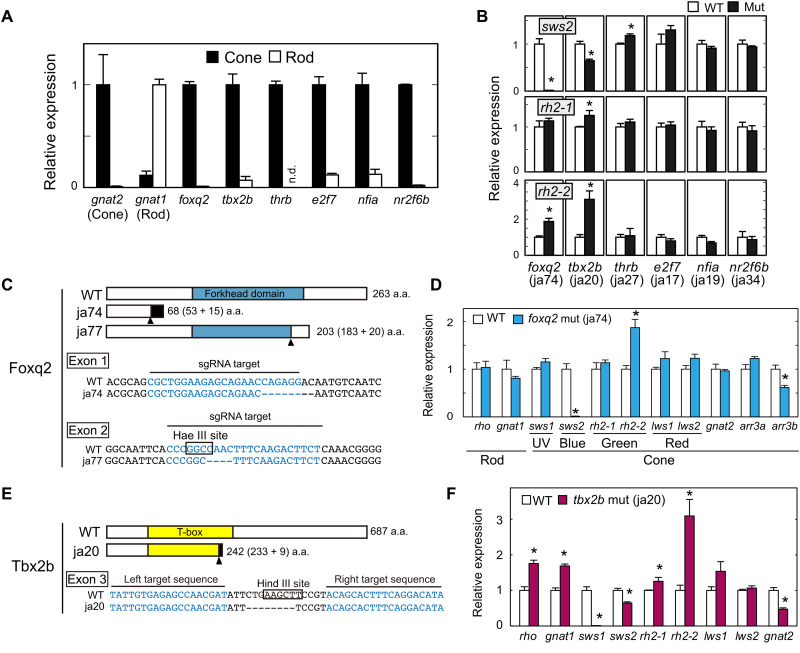
Six6b and Six7 are predominantly expressed in zebrafish cone photoreceptors and responsible for expression of the middle-wavelength–sensitive opsin genes, sws2 (blue) and rh2 (green) (20). To identify a transcription factor(s) that governs differentiation between SWS2 and RH2 cone subtypes, we searched for cone-specific genes by comparing gene expression profiles between cones and rods. These photoreceptor cells were purified from the retinas of transgenic adult zebrafish, each of which express enhanced green fluorescent protein (EGFP) in all cone subtypes [Tg(gnat2:egfp)] or in rods [Tg(rho:egfp)] (17). The cone or rod enrichment in these purified samples was validated by reverse transcription quantitative polymerase chain reaction (RT-qPCR) analyses of cone- and rod-specific transducin alpha-subunit genes, i.e., gnat2 and gnat1, respectively (Fig. 1A) (17). A subsequent microarray analysis revealed approximately 500 genes showing more than 10-fold higher expression in cones than in rods (data file S1). These cone-enriched genes included four transcription factors, foxq2, E2F transcription factor 7 (e2f7), nuclear factor 1A (nfia), and nuclear receptor 2F6b (nr2f6b), the roles of which in photoreceptor development were not known. Genomic loci of these genes harbor Six6b- and Six7-binding sites as revealed by our chromatin immunoprecipitation sequencing analysis (fig. S1) (20), implying that some of these cone-enriched transcription factors mediate a regulatory function(s) downstream of Six6b and Six7. In addition, we paid attention to two cone-enriched transcription factors, tbx2b and thrb. They are known to be required for expression of sws1 (16) and lws (14), respectively, but their contributions to the sws2 and rh2 gene expression have not been well characterized. Cone-enriched expression of these six transcription factors was verified by RT-qPCR analyses with purified cone and rod cells (Fig. 1A).
Fig. 1. The loss-of-function analysis for cone-enriched transcription factors.
(A) Relative expression levels of phototransduction genes and transcription factors in isolated rods and cones at the adult stage (means ± SD, n = 2). n.d., not detected. The expression levels of gnat1 and gnat2 genes are reproduced from our previous paper (17). (B) Relative expression levels of sws2 opsin in the larval eyes at 5 days postfertilization (dpf). Means ± SEM. *P < 0.05 by Student’s t test. The number of fish used was as follows: n = 5 [foxq2 wild type (WT)], n = 5 (foxq2 mut); n = 5 (tbx2b WT), n = 5 (tbx2b mut); n = 3 (thrb WT), n = 4 (thrb mut); n = 4 (e2f7 WT), n = 4 (e2f7 mut); n = 3 (nfia WT), n = 4 (nfia mut); n = 4 (nr2f6b WT), n = 4 (nr2f6b mut). See also fig. S3. (C and E) Schematic representation of Foxq2 and Tbx2b and their partial nucleotide sequences. The frameshift site is indicated by an arrowhead. Nucleotide deletions are indicated by dashes. The nucleotide sequences (letters in blue) indicate the target sequences of TAL effector nucleases or Cas9–single-guide RNA (sgRNA) complexes. The recognition sites of the restriction endonucleases, Hae III and Hind III, are surrounded by black lines. The ja74, ja77, and ja20 mutations caused a frameshift of the amino acid (a.a.) sequence of Foxq2 or Tbx2b by 8-, 4-, and 8-bp loss, respectively. (D and F) Expression profiles of phototransduction genes in the 5-dpf larval eyes. Means ± SEM. *P < 0.05 by Student’s t test. The number of fish used was as follows: n = 5 (foxq2 WT), n = 5 (foxq2 mut); n = 5 (tbx2b WT), n = 5 (tbx2b mut). The expression levels of sws2 and rh2 genes are reproduced in (B). UV, ultraviolet.
We generated loss-of-function mutants of zebrafish for each of the six transcription factors by introducing a frameshift mutation. In these mutants, ocular transcript levels of the middle-wavelength–sensitive opsin genes were examined by RT-qPCR analysis (Fig. 1B). Among the mutant larvae, a foxq2 mutant (Fig. 1C, ja74) displayed the most notable reduction in sws2 expression as compared to the wild-type (WT) siblings (Fig. 1B). Another mutant line of foxq2 (Fig. 1C, ja77) similarly showed a severe reduction of sws2 expression in the larvae (fig. S2A). The decrease of sws2 expression in the mutant larvae (Fig. 1D and fig. S2A) was not due to delayed development of SWS2 cones because the adult foxq2 mutant (ja77) also exhibited minimal expression of sws2 gene (fig. S2B). In the foxq2 mutant retinas, in situ hybridization signals of sws2 transcripts were undetectable with no apparent change in retinal morphology at the larval and adult stages (figs. S2C and S3A). These results demonstrate that foxq2 is indispensable for sws2 expression in SWS2 cone subtype.
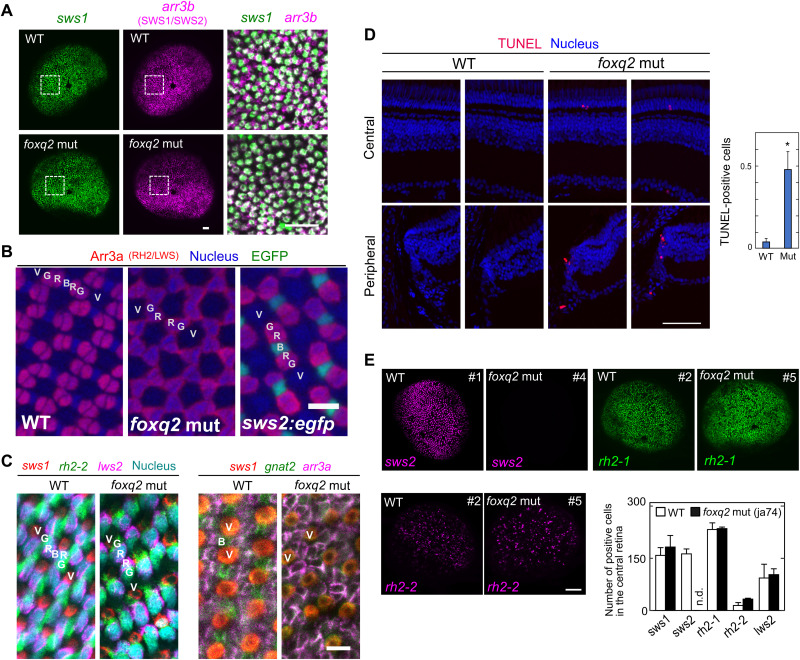
The foxq2 mutants (ja74 and ja77) were not only deficient in sws2 expression but also characterized by significant reduction in mRNA level of arrestin 3b (arr3b), a cone phototransduction gene selectively expressed in SWS1 and SWS2 cone subtypes (Fig. 1D and fig. S2) (21). sws1- and arr3b-expressing cone cells in the larval eye were visualized by in situ hybridization chain reaction (HCR) (Fig. 2A). Almost all arr3b-positive cells in the foxq2 mutant coexpress sws1, suggesting the absence of arr3b-positive and sws1-negative cells, i.e., SWS2 cones, in the foxq2 mutant. We then assessed mosaic arrays of cone photoreceptor cells in the adult retina, in which a SWS2 cone (B in Fig. 2B) is flanked by RH2/LWS double cones (G/R) in each mosaic unit (G/R/B/R/G/V; see Fig. 2B and fig. S3B) (22). Immunohistochemistry and in situ HCR labeling in the flat-mounted retina revealed that SWS2 cone is selectively lost in each mosaic unit of the foxq2 mutant retina (G/R/R/G/V; Fig. 2, B and C, and fig. S3, B and C). The expression of cone-specific phototransduction gene, gnat2 (in SWS1/SWS2/RH2/LWS cones), was colocalized with mRNA expression of either sws1 (in SWS1 cones) or arr3a (in RH2/LWS cones) in the mutant. These observations support that the mutant retina contains SWS1, RH2, and LWS cones and has no “opsin-empty” cones (Fig. 2C and fig. S3C). In the foxq2 mutant retina, the number of apoptotic cells [terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL)–positive cells] was increased in the outer nuclear layer of the central region and in the ciliary marginal zone of the peripheral region (Fig. 2D and fig. S3D). These observations suggest that foxq2 deficiency impairs formation and/or maturation of SWS2 cones.
Fig. 2. The loss of SWS2 cones in the foxq2 mutant.
(A) Expression patterns of sws1 and arr3b (SWS1 and SWS2 cones) in 5-dpf larval eyes of the foxq2 mut (ja74) examined by in situ HCR. Magnified view (a box surrounded with white lines) is indicated in the right side of each panel. Scale bars, 20 μm. (B) Fluorescent images of the flat-mounted retinas prepared from the adult WT, the foxq2 mut (ja74), and Tg(-3.5opn1sw2:EGFP)kj11Tg (sws2:egfp), where EGFP is expressed in SWS2 cones (green). The retinas were immunostained with zpr1 antibody (arr3a, red) and also stained with DRAQ5 to highlight cell nuclei (blue). V, SWS1 cone; B, SWS2 cone; G, RH2 cone; R, LWS cone. Scale bar, 10 μm. See also fig. S3B. (C) Expression patterns of phototransduction genes in the flat-mounted retinas examined by in situ HCR. See also fig. S3C. (D) Left: Fluorescent images in retinal cryosections from the adult fish labeled for terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) (red). The cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 50 μm. Right: Quantification of TUNEL-positive cells in the central and peripheral retina. The numbers of TUNEL-positive cells were counted for each cryosection and averaged (means ± SEM, n = 80 for WT, n = 63 for the foxq2 mut; *P < 0.05, Student’s t test). See also fig. S3D. (E) Expression patterns of sws2, rh2-1, and rh2-2 in 5-dpf larval eyes of WT and foxq2 mut (ja74) examined by in situ HCR. The number of opsin gene–positive cells in the central region of the retina is indicated in a bar graph. The number in the upper-right corner for each panel represents the unique identity of the eye. Data are represented by means ± SEM (n = 3). Scale bar, 50 μm. See also fig. S3E.
In parallel, the foxq2 mutation caused significant increase in mRNA levels of two major rh2 subclasses, rh2-1 in the larvae (fig. S2A, ja77) and rh2-2 in the adult (fig. S2B, ja77), as well as up-regulation of rh2-2 in the larvae (Fig. 1D, ja74, and fig. S2A, ja77) and rh2-4 in the adult (fig. S2B, ja77). In contrast, the numbers of rh2-1– and rh2-2–expressing cone cells in the foxq2 mutant larvae were similar to those in WT as revealed by in situ HCR analysis (Fig. 2E and fig. S3E). The number of RH2 cones also seemed normal in the adult foxq2 mutant retina (Fig. 2, B and C), in which each cone mosaic unit contains two RH2/LWS double cones as observed in WT. In addition, we observed no substantial expression of rh2-1 and rh2-2 outside the photoreceptor layer in the mutant retina (figs. S2C and S3A). It is most probable that rh2 expression is up-regulated in the individual RH2 cones of the foxq2 mutant retina.
Tbx2b is also a cone-specific transcription factor (Fig. 1A) and is required for expression of sws1 gene (16). In the present study, tbx2bja20 mutant was designed to encode C-terminally truncated Tbx2b protein (Fig. 1E), which lacked two DNA recognition helices of T-box domain (23) in a manner similar to tbx2bfby mutant (24). As reported in tbx2bfby (24), tbx2bja20 larvae exhibited a severe decrease of sws1 and a parallel increase of rhodopsin (rho) expression (Fig. 1F). Notably, we noticed that tbx2bja20 mutation caused 40% reduction of sws2 expression level from that in the WT siblings and substantial increase in mRNA levels of rh2-1 and rh2-2 (Fig. 1, B and F). Although the effect of tbx2b mutation on sws2 expression was weaker than that of foxq2 mutation, tbx2b appears to play a supportive role to foxq2 on sws2 expression in addition to the essential role on sws1 expression.
Thrb is another cone-specific transcription factor (Fig. 1A), and its mutant thrbja27 (fig. S4A) manifested a massive reduction of both lws1 and lws2 expression and a concomitant increase in sws1 expression in the larval eyes (Fig. 1B and fig. S4B) as reported in previous studies (14, 15, 25). We noticed that thrbja27 mutant exhibited a moderate but significant increase in sws2 gene expression (Fig. 1B and fig. S4B). The sws2 up-regulation accompanied no significant reduction of rh2 expression level (Fig. 1B and fig. S4B). Accordingly, Thrb contributes to fine-tuning of sws2 expression together with a dominant role in regulation of lws expression. In contrast, nfia, nr2f6b, and e2f7 mutant fish showed no noticeable change in sws2 or rh2 expression (Fig. 1B and fig. S4, C to H). These three transcription factors are dispensable for middle-wavelength cone opsin gene expression.
Collectively, our mutant analysis demonstrates that sws2 expression is predominantly regulated by a cone-specific transcription factor, Foxq2, with the aid of regulation by Tbx2b and Thrb. It is most probable that Foxq2 is also required for formation and/or maturation of SWS2 cones.
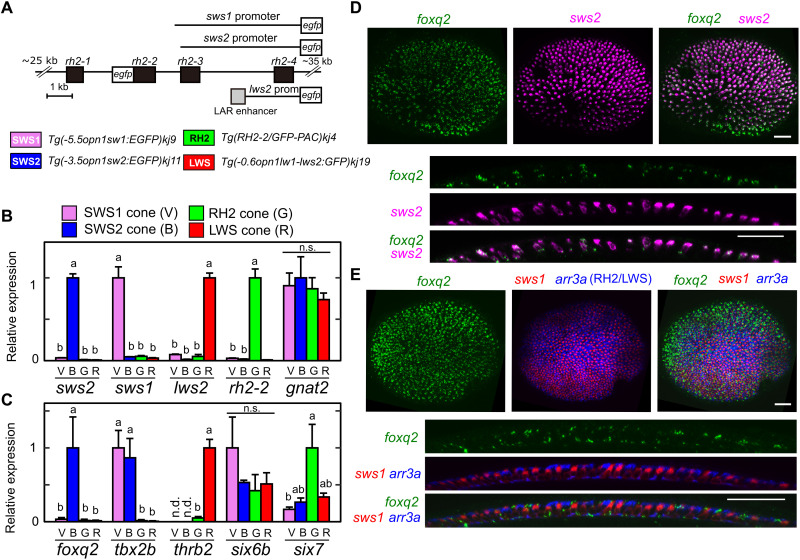
SWS2 cone subtype–specific expression of foxq2
To gain insights into how foxq2, tbx2b, and thrb regulate sws2 and rh2 expression, we investigated gene expression profiles of foxq2, tbx2b, and thrb among the four cone subtypes: SWS1, SWS2, RH2, and LWS. These four cone cells were isolated from four different lines of transgenic fish, each of which expresses EGFP in one of the four cone subtypes (Fig. 3A; see Materials and Methods for details). The cone subtype enrichment in these purified samples was validated by RT-qPCR analyses of cone opsin genes (Fig. 3B). Subsequent analysis of foxq2, tbx2b, thrb, six6b, and six7 in the four samples revealed that foxq2 was specifically expressed in SWS2 cone subtype (Fig. 3C). Another SWS2 regulator, tbx2b, was expressed in both SWS1 and SWS2 cone subtypes, whereas thrb was expressed only in the LWS cone subtype (Fig. 3C). In contrast, SWS2/RH2 regulators, six6b and six7, were expressed in all the cone subtypes (Fig. 3C). The SWS2 cone subtype–enriched expression of foxq2 and tbx2b suggests that these two factors coordinately regulate the cell type–specific expression of sws2.
Fig. 3. SWS2 cone–enriched expression of foxq2.
(A) Schematic drawing of transgenes for the four transgenic lines each expressing EGFP in SWS1 (V), SWS2 (B), RH2 (G), or LWS (R) cone subtype. See also Materials and Methods. (B and C) Relative expression levels of cone opsin genes (B) and transcription factors (C) among isolated cone subtypes at the adult stage. Mean expression values with SEM (n = 3) are indicated as bars. Distinct letters indicate statistically significant differences (P < 0.05 by Tukey’s multiple comparisons test). n.s., not significant. (D and E) Expression pattern of foxq2 in 3-dpf larval eyes examined by in situ HCR. The foxq2 expression is covisualized with sws2 (D) or with sws1 and arr3a (E). The bottom image is an orthogonal view of the top z-projected image. Scale bars, 25 μm.
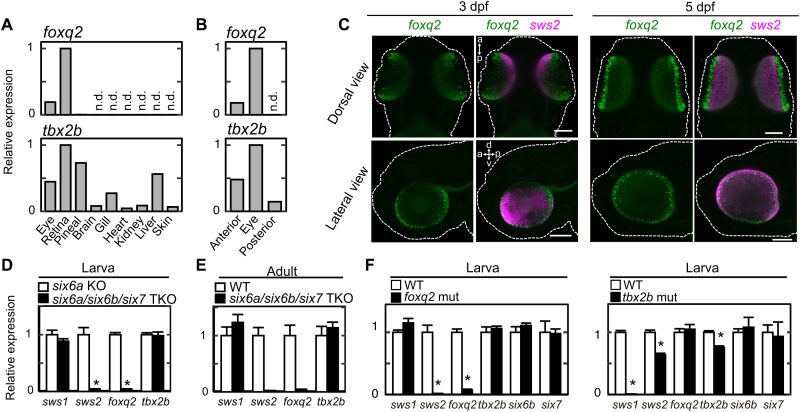
The expression of foxq2 gene in the larval stage was visualized by in situ HCR. The foxq2 transcript signals were colocalized with those of sws2 (in SWS2 cones; Fig. 3D) but not with those of sws1 (in SWS1 cones) or arr3a (in RH2/LWS cones) (Fig. 3E). foxq2 expression is thus specific to SWS2 cones among the cone subtypes at both larval (Fig. 3D) and adult (Fig. 3C) stages. RT-qPCR analysis revealed that foxq2 expression was selectively expressed in the retina among various adult tissues (Fig. 4A), which is consistent with the publicly available RNA sequencing data (fig. S5). In the larvae, foxq2 expression was detected only in the anterior half of the body (including the eyes) but not in the posterior half (Fig. 4B). The eye-specific foxq2 expression in the larvae was visualized by in situ HCR (Fig. 4C). Notably, the foxq2 signals were enriched in ciliary marginal zone (Fig. 4C), which contains a population of actively proliferating progenitor cells (26). This expression pattern suggests that foxq2 expression precedes sws2 expression in differentiating cone cells. The mRNA level of foxq2 was severely reduced in six6a/six6b/six7 triple knockout (TKO) lacking sws2 expression, i.e., loss of SWS2 cone identity from the retina (Fig. 4, D and E) (20). foxq2 expression was also abrogated in the larval eyes of foxq2 mutant (ja74) as compared to that of WT sibling (Fig. 4F). The close correlation between foxq2 and sws2 expressions implies that Foxq2 is responsible for establishing SWS2 cone identity. Meanwhile, tbx2b was expressed widely in various tissues at the larval (Fig. 4B) and adult stage (Fig. 4A and fig. S5), and the ocular tbx2b expression level was unaffected in the foxq2 mutant (Fig. 4F) and six6a/six6b/six7 TKO (Fig. 4, D and E), both of which were deficient in sws2 expression. These observations indicate dominant expression of tbx2b outside of SWS2 cones and imply that Tbx2b may have pleiotropic roles for eye development (27) including the cone identity determination.
Fig. 4. Gene expression profiles of foxq2 and tbx2b.
(A and B) Relative expression levels of foxq2 and tbx2b in adult tissues (A), and 4-dpf larval anterior segments, posterior segments, and eyes (B). (C) Expression pattern of foxq2 and sws2 in 3-dpf and 5-dpf larvae examined by in situ HCR. The larval zebrafish are outlined with white dotted lines. d-v, dorsal-ventral axis; a-p, anterior-posterior axis. Scale bars, 100 μm. (D) Relative expression levels of tbx2b and foxq2 in the 5-dpf larval eyes of the six6a/six6b/six7 triple knockout (TKO). Means ± SEM (n = 5). *P < 0.05 by Student’s t test. Note that the six6a KO was used as the control because it shows normal levels of cone opsin gene expression as compared to the WT [see (20)]. The expression levels of sws2 and rh2 genes are reproduced from our previous paper (20). (E) Relative expression levels of tbx2b and foxq2 in the adult eyes of the six6a/six6b/six7 TKO. All of the fish used here are in the transgenic background, Tg(-3.5opn1sw2:EGFP)kj11Tg, where EGFP is expressed in the SWS2 cone subtype. Data are represented by means ± SD (n = 3, WT; n = 2, six6a/six6b/six7 TKO). The expression levels of sws2 and rh2 genes are reproduced from our previous paper (20). (F) Relative expression levels of cone opsins and transcription factors responsible for photoreceptor gene expression in the eyes of the tbx2b (ja20) and foxq2 (ja74) mutants. Means ± SEM. *P < 0.05 by Student’s t test. The number of fish used was as follows: n = 5 (tbx2b WT), n = 4 (tbx2b mut); n = 5 (foxq2 WT), n = 5 (foxq2 mut). The expression levels of sws2 and rh2 genes in the foxq2 mutant and its control WT are reproduced from Fig. 1D.
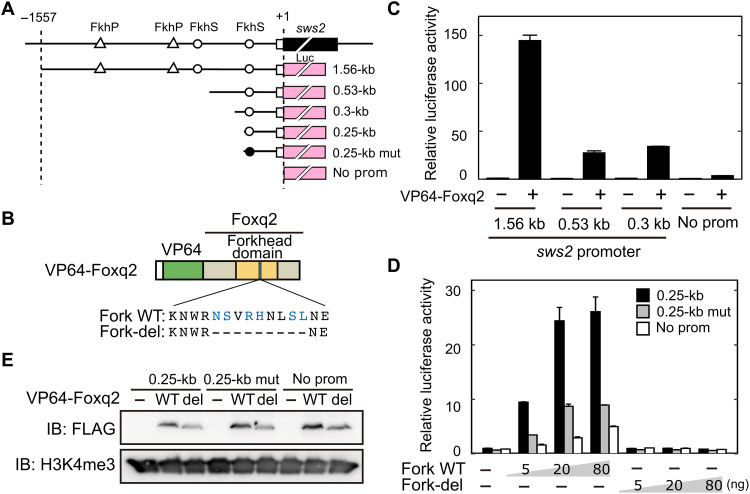
Functional interaction of Foxq2 with sws2 promoter
We then investigated functional interaction of Foxq2 with sws2 promoter. Foxq2 protein (Fig. 1C) has a conserved DNA binding domain, termed forkhead domain composed of about 100 amino acid residues (28). In Fox transcription factor family, Foxq2 is categorized into clade I forkhead proteins (29), which recognize two types of common forkhead-target DNA motifs termed FkhP (RYAAAYA) and FkhS (AHAACA) (30). Our motif scanning analysis (see Materials and Methods) revealed two FkhP motifs and two FkhS motifs present within 1.56-kb sws2 promoter region (Fig. 5A and fig. S6), which drives selective gene expression in SWS2 cones (31). In a cell-based reporter assay, the 1.56-kb sws2 promoter was transactivated by VP64-Foxq2 (Fig. 5, B and C), in which Foxq2 is N-terminally fused with four repeats of the VP16 transcriptional activator domain (Fig. 5B) (32). The transcriptional activation was attenuated by deletion of the upstream region of 1.03 or 1.26 kilo–base pair (kbp) (0.53- or 0.3-kb sws2 promoter, respectively), leaving single FkhS motif (Fig. 5, A and C). Still, we observed more than 30-fold transactivation of the 0.3-kb sws2 promoter by VP64-Foxq2, while the activation was largely reduced by the complete deletion of the promoter sequence (Fig. 5C). The VP64-Foxq2–dependent transactivation of a shorter promoter (0.25-kb sws2 promoter) was markedly decreased by introducing a 6-bp mutation in the FkhS motif (Fig. 5D). We then generated a mutant protein, termed Fork-del, lacking nine amino acid residues that are required for specific DNA binding of the forkhead domain (Fig. 5B) (30). The protein mutation abolished the ability of the transcriptional activation of the sws2 promoter (Fig. 5, B, D, and E). These results demonstrated that Foxq2 functionally interacted with the forkhead-target DNA motif in the sws2 promoter. Together with the SWS2 cone–specific expression of foxq2 (Fig. 2), Foxq2-mediated transcriptional regulation of sws2 gene would reasonably account for the selective expression of sws2 opsin in SWS2 cone subtype.
Fig. 5. DNA binding of Foxq2 to sws2 promoter.
(A) Schematic structure of the promoter-luciferase reporter constructs for zebrafish sws2. The coding (solid box) and the untranslated (open box) regions of the first exon are indicated. The translational start site is marked as +1. The forkhead binding motif (ACAACA) and its mutant (GTGGTG) are marked as open and closed circles, respectively. See also fig. S6. (B) Schematic representation of VP64-Foxq2 protein. The Foxq2 mutant protein (Fork-del) lacks the canonical Fox base–contacting residues in the forkhead domain. These critical amino acid residues are highlighted in blue according to the previous paper (30). (C and D) Transcriptional assay in human embryonic kidney 293T17 cells using the luciferase reporter containing the promoter region of zebrafish sws2 (means ± SD, n = 2). The luciferase activity derived from the firefly luciferase reporter was normalized to that from the Renilla luciferase reporter. (C) The luciferase activity in each condition was normalized to the mean value obtained from the cells transfected with both empty vector and 1.56-kb sws2 reporter vector (leftmost bar). VP64-Foxq2 expression plasmid (100 ng) was used for the transfection. (D) The amount of VP64-Foxq2 expression plasmid used is indicated. The luciferase activity in each condition was normalized to the mean value obtained from the cells transfected with both the empty expression vector and the 0.25-kb sws2 (0.25 kb) reporter vector (leftmost bar). The slight up-regulation of the luciferase expression in a reporter with no sws2 promoter might be due to an experimental artifact, such as VP64-Foxq2–dependent regulation through the reporter vector-backbone sequence(s) and/or activation of gene(s) regulating the reporter gene expression. (E) Protein expression of VP64-Foxq2 (WT) and its mutant protein (Fork-del) in the cells transfected with 80 ng of the plasmid. The antibody H3K4me3 is served as a loading control. IB, immunoblot.
sws2 regulation by Foxq2 downstream of Six7
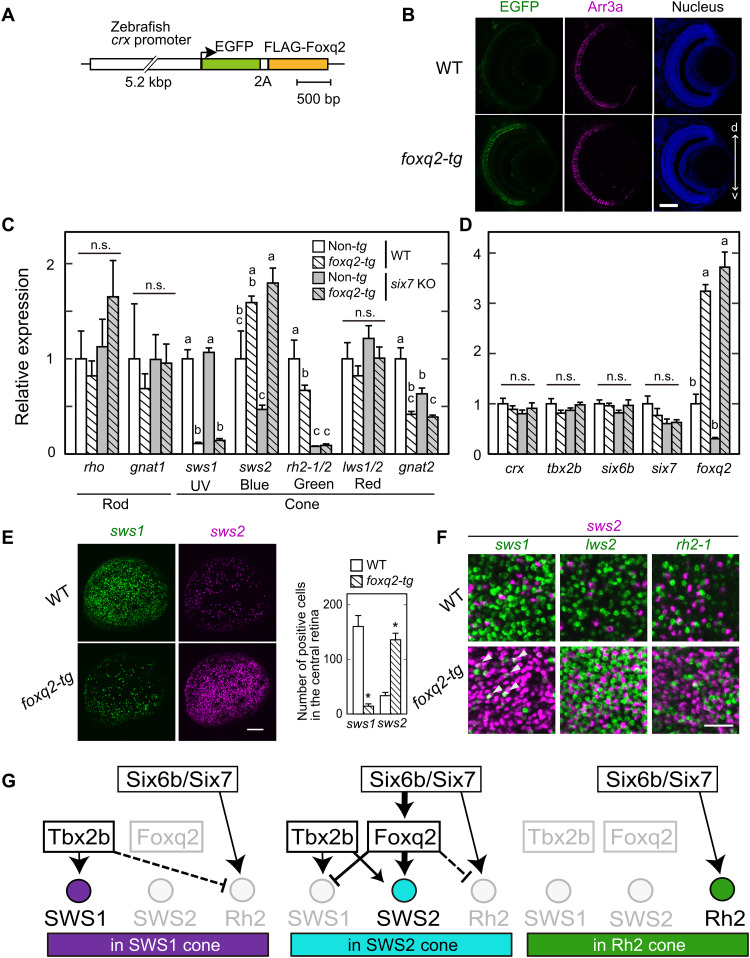
To gain deeper insight into the mechanism underlying the determination of the SWS2 subtype identity, we generated foxq2 transgenic (foxq2-tg) zebrafish, Tg(-5.2crx:EGFP-2A-FLAG-foxq2), in which both EGFP and Foxq2 expressions are driven by 5.2-kb crx promoter in all the developing and matured photoreceptor cells (Fig. 6, A and B) (14). RT-qPCR analysis of larval ocular mRNAs in three lines of foxq2-tg (ja78Tg, ja79Tg, and ja91Tg) revealed that the forced expression of foxq2 in WT background severely reduced the sws1 mRNA level (Fig. 6C and fig. S7, A and B). This phenotype was not accompanied by any detectable change in the expression levels of tbx2b and crx (Fig. 6D and fig. S7, C and D), which are regulators of sws1 expression (Fig. 1K) (16). It is most probable that Foxq2 suppresses sws1 expression in cones. In in situ HCR analysis, the forced foxq2 expression largely decreased the number of SWS1 cones and increased that of SWS2 cones (Fig. 6E and fig. S7E), while the numbers of rh2-1– and rh2-2–expressing cells were similar between the foxq2-tg (ja78Tg) and WT (fig. S7F). It should be noted that the foxq2-tg retina contained a considerable number of SWS1/SWS2 hybrid cones (sws1/sws2 double-positive cells) (Fig. 6F and fig. S7E), whereas neither LWS/SWS2 hybrid nor RH2/SWS2 hybrid cones were observed in the foxq2-tg retina (Fig. 6F and fig. S7G). These results indicate that Foxq2 controls SWS2 subtype identity by promoting sws2 expression and suppressing sws1 expression (Fig. 6G).
Fig. 6. Foxq2-mediated transcriptional regulation of sws2 downstream of Six7.
(A) Schematic drawing of a transgene construct used for generating foxq2-tg. (B) Immunofluorescent image in the foxq2-tg (ja78Tg) larvae at 5 dpf. Scale bar, 50 μm. d-v, dorsal-ventral retina. (C and D) Relative mRNA levels of opsin genes and transcription factors in the 5-dpf larval eyes of the foxq2-tg (ja78Tg) and/or six7 KO (means ± SEM). Distinct letters indicate statistically significant differences (P < 0.05 by Tukey’s multiple comparisons test). The number of fish used was as follows: n = 3 (WT), n = 5 (six7 KO), n = 4 (foxq2-tg), and n = 5 (foxq2-tg/six7 KO). (E) Expression pattern of sws1 and sws2 examined by in situ HCR using 5-dpf larval eyes of the foxq2-tg. The number of opsin gene–positive cells in the central region of the retina is indicated in a bar graph. Data are represented by means ± SEM (n = 6). Statistical significance between two genotype was determined by Student’s t test (*P < 0.05). See also fig. S7E. Scale bar, 50 μm. (F) Comparison of expression pattern of sws2 with that of sws1, rh2-2, or lws2 examined by in situ HCR using 5-dpf larval eyes of the foxq2-tg (ja78Tg). Some sws1 expression signals are colocalized with those of sws2 in the foxq2-tg eye (arrowheads). See also fig. S7 (E and G). Scale bar, 20 μm. (G) Hypothetical model of transcriptional network accounting for sws1, sws2, and rh2 opsin expression in the SWS1, SWS2, and RH2 cones. The mechanism(s) underlying the repression of rh2 expression in SWS1 and SWS2 cones remains elusive, and hence, these potential regulations are shown by broken lines.
The selective expression of foxq2 in SWS2 cones (Fig. 3, C and D), together with binding of Six6b and Six7 to foxq2 gene locus (fig. S1 and 20), suggests that sws2 expression is regulated by Foxq2 downstream of Six6b and Six7 (Fig. 6G). This possibility was explored by overexpressing foxq2 in the homozygous six7 KO (17), in which expression levels of sws2 and foxq2 are markedly reduced (Fig. 6, C and D). The forced expression of foxq2 in the six7 KO background (foxq2-tg;six7 KO) recovered the reduced expression of sws2 up to a level higher than that in the WT control (Fig. 6C). In contrast, the overexpression of foxq2 had no significant effect on the severely reduced expression levels of rh2 (rh2-1 plus rh2-2) (Fig. 6C) or on mRNA levels of crx, tbx2b, six6b, and six7, transcriptional regulators of cone opsin expression (Fig. 6D). These results demonstrated that Foxq2 regulates sws2 expression downstream of Six7, suggesting that Foxq2 is a terminal selector for SWS2 subtype identity (Fig. 6G).
Conservation of FOXQ2 gene among vertebrate species
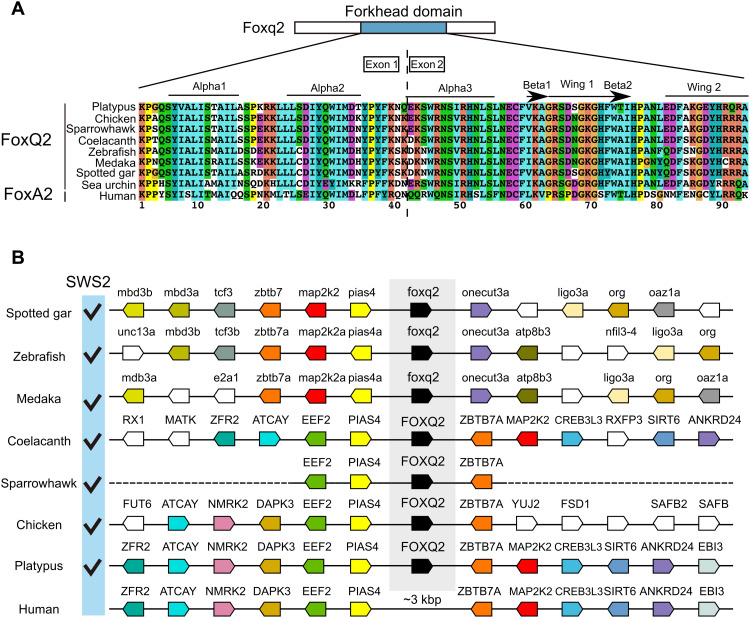
FOXQ2 gene is annotated in the genomes of a wide range of vertebrate species (Fig. 7), such as ray-finned fish (spotted gar, zebrafish, and medaka), a lobe-finned fish (coelacanth), and an avian (sparrowhawk), all of which retain SWS2 gene. We found highly conserved gene synteny around FOXQ2 locus among the vertebrates, and the synteny analysis of the human genome indicated the absence of any gene homologous to FOXQ2 between PIAS4 and ZBTB7A gene loci (Fig. 7B). BLAST search and subsequent manual annotation revealed that a mammalian species, platypus, retains a gene orthologous to FOXQ2 harboring the forkhead domain highly conserved among FOXQ2 subfamily members (Fig. 7 and fig. S8; see also Supplementary Text). The oviparous mammals including platypus diverged from marsupial and placental mammals at the earlier stage of mammalian evolution (around 200 million years ago). Notably, platypus retains SWS2 gene in its genome (33), whereas many other mammals have lost it (7, 34) most likely due to a long evolutionary history of nocturnality in mammalian ancestors (35). These lines of genomic evidence suggest that Foxq2-dependent SWS2 expression is a highly conserved regulatory mechanism that was acquired at the early stage of vertebrate evolution.
Fig. 7. FOXQ2 gene conservation among animal species.
(A) Sequence alignment of forkhead domain of FOXQ2 and FOXA2 proteins. Secondary structure elements are indicated at the top according to the FOXA2-DBD/DBE2 complex in the previous study (63). (B) Genomic environment of FOXQ2 genes in vertebrate species. Orthologous genes contributing to the conserved order of FOXQ2 genes among vertebrates are similarly color-coded. The presence of SWS2 gene in each species is indicated as checkmarks according to the previous study (64). Accession numbers for the genomic location of FOXQ2 in each species are indicated in table S6.
DISCUSSION
Under the solar light irradiation, both terrestrial and aquatic environments on the earth’s surface are enriched with blue-to-green region of visible spectrum (36), which is detected by cone photoreceptor cells expressing middle-wavelength–sensitive opsin genes, SWS2 and RH2. Their expression requires transcription factors, Six6 and Six7, in zebrafish (17, 20), and the six6/six7 mutant fish lacking both SWS2 and RH2 cones show severely reduced survival rate due to impairment of visually driven foraging behavior (20). A recent study of in vivo calcium imaging of zebrafish cone photoreceptors reported that SWS2 and RH2 cones, but not SWS1 or LWS cones, display strong spectral opponency and efficiently extract chromatic information from the natural light spectrum (37). In this way, a combination of spectrally distinctive SWS2 and RH2 cones particularly plays an important role in the tetrachromatic visual system. The present study explored the transcriptional regulatory logic that defines SWS2 and RH2 cone identities. For this purpose, cone-enriched transcription factors were comprehensively identified by the transcriptome analysis with purified cone cells (versus purified rod cells; data file S1). Subsequent functional analyses of these transcription factors demonstrated that Foxq2 is indispensable for sws2 expression and probably for formation and/or maturation of SWS2 cones (Fig. 1). We pursued expression profiling of the cone-enriched transcription factors among the four isolated cone subtypes (Fig. 3, A to C). foxq2 is selectively expressed in SWS2 cone, whereas all cone subtypes express six6b and six7 (Fig. 3C), which are required for both sws2 and rh2 gene expression (20). A transcriptional network is deciphered in which Foxq2 acts as a downstream regulator of Six7 (Fig. 6, A to D) and regulates sws2 expression (Fig. 5). A wide range of vertebrate species retain both FOXQ2 and SWS2 gene (Fig. 7). These lines of evidence demonstrate that Foxq2 determines SWS2 cone identity in zebrafish (Fig. 6G) and suggest that Foxq2-dependent sws2 expression is a highly conserved regulatory mechanism that was acquired at the early stage of vertebrate evolution.
Quantitative comparison of transcription factor expression among the four isolated cone subtypes (Fig. 3C) provides valuable information about terminal differentiation of each cone subtype in zebrafish. foxq2, a crucial regulator of sws2 expression, is selectively expressed in SWS2 cones, indicating that Foxq2 is a bona fide terminal selector of SWS2 cone. On the other hand, tbx2b, known as a master regulator of SWS1 cone (16), is expressed in not only SWS1 cones but also SWS2 cones (Fig. 3C), in which Tbx2b acts as a supportive regulator of sws2 expression (Fig. 1F). The expression of the SWS1 master regulator in SWS2 cones implies that some mechanism should suppress sws1 misexpression in SWS2 cones. The sws1 suppression should be mediated by Foxq2 because the forced expression of foxq2 markedly reduced the number of SWS1 cone cells and the sws1 mRNA level (Fig. 6, C and E). It is likely that Foxq2 also suppresses rh2 misexpression in SWS2 cones (Figs. 1D and 6C and fig. S2). Thus, Foxq2 has dual functions acting as an activator of sws2 transcription and as a suppressor of sws1 and rh2 genes in SWS2 cones (Fig. 6G) and in developing cones. The dual functions of Foxq2 would enhance robustness of SWS2 cone identity. Meanwhile, thrb is predominantly expressed in LWS cone subtype (Fig. 3C), being consistent with the widely accepted idea that Thrb is a master regulator for the LWS cone (13–15). Collectively, Foxq2, Tbx2b, and Thrb can be defined as the terminal selectors (38) that govern cell fate determination of SWS2, SWS1, and LWS cone subtypes, respectively.
In contrast to these three terminal transcriptional selectors, six7 is widely expressed among the four cone subtypes (Fig. 3C) and thus six7 is unlikely to be a terminal selector of RH2. Although Six7 is indispensable for expression of rh2 [rh2-1, rh2-2, rh2-3, and rh2-4; Fig. 6C and (17)], terminal differentiation of RH2 cone might be mediated by an unidentified transcription factor. An alternative idea is that Rh2 cone identity is established by both (i) the presence of six7 and (ii) the absence of the terminal selectors such as tbx2b (for SWS1), foxq2 (for SWS2), and thrb (for LWS). In this scenario, differentiation toward Rh2 cones should be a default pathway in the cone development because Rh2 cone differentiation is governed by six7 (17) and six7 expression begins as early as that of crx, a master regulator of cone and rod development (17, 39). It is worth noting that the “Rh2-default” hypothesis is consistent with molecular phylogeny of cone opsin genes, in which RH2 subfamily diverged from SWS2 at the latest step of the molecular evolution of the cone opsins (4, 5, 40). Building on these assumptions, we propose that the tetrachromatic color vision system in ancestral vertebrates used the RH2-default mechanism of cone differentiation as is observed in zebrafish. In mammals, on the other hand, RH2 and SWS2 genes have been lost, and hence, it is likely that the RH2-default mechanism has been modified to a “SWS1-default” mechanism. In the latter mechanism, the presence or absence of only one terminal selector, THRB, directs differentiation between LWS and SWS1 cones as shown in mice (8, 13).
FOXQ2 gene is evolutionarily conserved not only among vertebrates but also in many invertebrates (29, 30, 41). In embryos of the invertebrates, FOXQ2 is responsible for specification and positioning of anterior neuroectoderm (42, 43). The anterior neuroectoderm develops into central nervous system, in which a subset of sensory cells express a photoreceptive protein, c-opsin, that is orthologous to vertebrate cone and rod opsins (44, 45). FOXQ2 might regulate differentiation of the c-opsin–expressing cells from progenitor cells in the anterior neuroectoderm. A transcriptional network in the anterior neuroectoderm of invertebrates also includes six3, a gene orthologous to vertebrate six3, six6, and six7 (42, 43). The presence of the Six3/6/7-Foxq2 transcriptional network in both vertebrate and invertebrate species suggests that their common ancestor used this transcriptional network for development of the light-sensitive cells. In the vertebrate lineage, the Six3/6/7-Foxq2 transcriptional network might have been co-opted for establishment of SWS2 cone identity during or after the appearance of a full set of cone opsins, thereby conferring high-acuity discrimination of blue-to-green region of visible light.
MATERIALS AND METHODS
Zebrafish
The Ekkwill strain was used as the WT zebrafish. All appropriate ethical approval and licenses were obtained from Institutional Animal Care and Use Committees of The University of Tokyo. All the procedures were conducted according to the local guidelines of The University of Tokyo. Zebrafish were raised in a 14-hour light/10-hour dark cycle and fed twice per day with live baby brine shrimps. Embryos were raised at 28.5°C in egg water (artificial seawater diluted 1.5:1000 in water). Mutant strains and transgenic lines used in the present study are listed in table S1.
Purification of rod and cone photoreceptor cells
For isolation of rod and cone photoreceptor cells, we used the transgenic zebrafish lines, Tg(rho:egfp)ja2 (46) and Tg(gnat2:egfp)ja23 (17), which express EGFP in rods and all cone subtypes, respectively. The rod and cone cells were isolated as described previously (17, 20, 47). Briefly, retinas were dissected from dark-adapted adult fish under dim red light. The isolated retinas were digested with 0.25% trypsin, deoxyribonuclease I (10 U/ml), 2 mM MgCl2, and 2 mM EGTA in Ca2+-free Ringer’s solution for 30 min at 37°C. The reaction was terminated by adding soybean trypsin inhibitor (final 0.5%) and fetal bovine serum (final 10%). The dissociated cells were filtrated through a 35-μm nylon mesh (Falcon). EGFP-positive cells were isolated with a fluorescence activating cell sorter (FACSAria, BD Biosciences) by the following three parameters: forward scatter, side scatter, and green fluorescence. The isolated cells were directly collected into 800 μl of TRIzol reagent (Thermo Fisher Scientific) in 1.5-ml microtubes.
For isolation of cone subtypes, we used four lines of transgenic zebrafish: Tg(-5.5opn1sw1:EGFP)kj9 (48), Tg(-3.5opn1sw2:EGFP)kj11 (31), Tg(RH2-2/GFP-PAC)kj4 (49), and Tg(-0.6opn1lw1-lws2:GFP)kj19 (50), which express EGFP in SWS1, SWS2, RH2, and LWS cone subtypes, respectively. The cone photoreceptor isolation was carried out as described above with some modifications as follows. The isolated cells were directly collected into 450 μl of the lysis buffer (RNeasy Lysis Buffer, Qiagen) in 1.5-ml microtubes.
Microarray
Total RNA was isolated from the sorted cells using an RNeasy Extraction kit (Qiagen). Quality and quantity of the resulting RNA were assessed using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific) and an Agilent 2100 Bioanalyzer (Agilent Technologies). Microarray analysis was performed using the Agilent 4 × 44 k Zebrafish microarray according to the manufacturer’s protocol for the two-color method. Cy3- or Cy5-labeled complementary RNA (cRNA) probe was synthesized from 150 ng of total RNA using the Quick-Amp Labeling Kit (Agilent Technologies). Quantity of the resulting labeled cRNA was assessed using a NanoDrop ND-2000 spectrophotometer. Equal amounts of Cy3- and Cy5-labeled cRNA (825 ng) from two different samples were hybridized to zebrafish microarrays (Agilent Zebrafish Oligo Microarrays version 2, G2519F-019161) for 17 hours at 60°C. To compare gene expression profiles between cone and rod samples, we analyzed two independent biological replicates as follows: (i) Cy3-rod#1 cRNA and Cy5-cone#1 cRNA and (ii) Cy5-rod#2 cRNA and Cy3-cone#2 cRNA. The hybridized arrays were then washed and scanned using an Agilent microarray scanner (G2505 C; Agilent Technologies). Data were extracted from the scanned image using Feature Extraction version 10.5.1.1 (Agilent Technologies). We then listed differentially expressed genes with Microsoft Excel as follows: (i) We excluded any probes whose signals in two arrays were all determined as negative, meaning that probes were not flagged as “WellAboveBG” in any of two arrays. (ii) We selected all the probes whose signal intensities vary largely between the photoreceptor cell types according to the following thresholds of the averaged ratios: 10-fold increase for cone compared with rod and 4-fold increase for rod compared with cone. (iii) We checked whether both of the two independent biological replicates showed similar expression profiles and selected probes whose absolute values of the ratios in the two biological replicates were both >2.0 for rod/cone or both >3.0 for cone/rod. The lower threshold was given for the rod/cone ratio because a small but noticeable level of rod contamination was detected in cone samples such as gnat1 (Fig. 1A). We set these threshold values on the basis of the ratios observed for known cone-specific genes. The microarray datasets are available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (GEO no. GSE168749).
Generation of mutant zebrafish
The foxq2, thrb, and nr2f6b mutants were generated with a CRISPR-Cas9 system. The single-guide RNA (sgRNA) sequences (table S2) were designed to target exon 1 or exon 2 using CRISPRdirect (https://crispr.dbcls.jp/), CHOPCHOP (https://chopchop.cbu.uib.no), or CRISPRscan (www.crisprscan.org). The sgRNA was synthesized by a cloning-free method as previously described (51). For generating Cas9 mRNA, the template plasmid DNA, pCS2−nCas9n (Addgene, no. 47929) (52) or pCS2+hSpCas9 (Addgene, no. 51815) (53), was used for in vitro transcription. Cas9 mRNA was synthesized with the SP6 mMESSAGE mMACHINE Kit (Ambion) and purified with the RNeasy Mini kit (Qiagen). The solution containing 200-pg Cas9 mRNA and 25-pg sgRNA was injected into the cytoplasm of the one cell-stage embryos.
The tbx2b, e2f7, and nfia mutants were generated by transcription activator–like effector nucleases (TALENs) as previously described (17, 20). To target each of the three genes, a pair of the TAL effector repeats (table S2) were designed to target exon 2 or exon 3 with the Golden Gate assembly methods. TALEN mRNA was synthesized using the SP6 mMESSAGE mMACHINE Kit (Ambion) and purified with the RNeasy Mini kit (Qiagen). The solution containing 200 pg each of the two TALEN mRNAs was injected into the cytoplasm of the one-cell stage embryos.
The injected fish (F0) were crossed with the WT zebrafish. The resultant F1 offspring were screened for the presence of CRISPR-Cas9– or TALEN-induced mutations by a combination of PCR and subsequent enzyme digestion for thrb, nr2f6b, tbx2b, e2f7, and nfia mutants or by heteroduplex mobility assay for foxq2 mutants as described previously (54). To sequence-verify mutations, genomic sequences surrounding the mutations were amplified by nested PCR, and the resultant PCR products were sequenced directly. After isolation of mutant zebrafish and verification of the mutations, the mutant genotypes were confirmed by a combination of PCR and subsequent enzyme digestion except for the foxq2 (ja74) gene locus. The foxq2ja74 mutant genotype was determined by PCR with two pairs of primers: (i) foxq2_Fw1A (5′-TGGCT AAACG AACAA ACACG-3′) and foxq2_Rv1WT (5′-ATGGA TTGAC ATTGT CCTCT G-3′) and (ii) foxq2_Fw1mutA (5′-GCTGG AAGAG CAGAA CAATG-3′) and foxq2_Rv1A (5′-GGAAA TGAGG GCAAT GTAGG-3′). PCR primers used for amplification are listed in table S3.
The mutant larvae and its siblings were dissected into anterior and posterior segments; the posterior parts were used for genotyping, while the anterior segments were soaked in RNAlater (Sigma-Aldrich) for RT-qPCR analysis or fixed with 4% paraformaldehyde (PFA) in Ca2+- and Mg2+-free Dulbecco’s phosphate-buffered saline (D-PBS) for cryosectioning.
RT-qPCR analysis
Zebrafish were anesthetized by chilling on ice, and their tissues were collected during the light phase of the light-dark cycle and soaked in RNAlater. After genotyping described in the previous section, the larval eyes were isolated with a needle. A pair of two larval eyes or one adult eye was considered a biological replicate for each genotype. RNA extraction and reverse transcription were conducted as described previously (17, 20). In short, RNA was extracted and purified with the RNeasy Micro Kit or RNeasy Mini Kit (Qiagen). In all the experiments except for Fig. 1A, the extracted RNA was reverse-transcribed into cDNA with the oligo(dT)15 primer with GoScript Reverse Transcriptase (Promega). In Fig. 1A, the reverse transcription was conducted with SuperScript II (Thermo Fisher Scientific) using anchored (dT)16 primers. The reverse-transcribed cDNA was subjected to qPCR using GoTaq qPCR Master Mix (Promega) and the StepOnePlus Real-Time PCR system (Applied Biosystems) following the manufacturers’ protocols. Expression levels were calculated by the relative standard curve method. The standard curve was prepared with serial dilutions of cDNA samples reverse-transcribed from total RNA of zebrafish eye. The transcript levels were normalized to beta-actin 2 (actb2) transcript levels in all the figures. Primers used for qPCR are listed in table S4 and in our previous studies (17, 20). Total transcript level of RH2 (rh2-1 and rh2-2) or LWS (lws1 and lws2) opsin genes at the larval stage was measured with a set of PCR primer, which amplifies both rh2-1 and rh2-2 opsin genes (referred to here as rh2-1/2) or both lws1 and lws2 opsin genes (referred to here as lws1/2). Expression levels of all the transcript isoforms of thrb were measured in Fig. 1A, while, in the rest of the experiments, we measured expression levels of a transcript isoform of thrb, thrb2, which is essential for lws expression in mouse and zebrafish (13, 14).
In situ hybridization
In situ hybridization using ocular sections was carried out as described previously (17, 20). In short, zebrafish were anesthetized by chilling on ice and subjected to dissection the light phase of the light-dark cycle. The larval anterior segments or the adult eyes were fixed in 4% PFA in D-PBS overnight at 4°C. Before the fixation, the adult eyes were enucleated and poked with tweezers to make a tiny hole in the cornea. After sucrose infiltration and optimal cutting temperature compound embedding, the 10-μm frozen ocular sections were prepared with a cryostat. The cryosections were pretreated with proteinase K and hybridized with digoxigenin-labeled cRNA probes, and the hybridization signals were visualized by nitro blue tetrazolium–bromochloroindolyl phosphate staining. The images were acquired with an upright microscope (Axioplan2, Carl Zeiss). The cRNA probes were generated as described in our previous study (17, 20).
Immunofluorescent labeling of flat-mounted retina of adult zebrafish
Immunofluorescent labeling of flat-mounted retina was carried out as described (20) with some modifications. Briefly, retinas were isolated from dark-adapted adult zebrafish, and each of the retinas was processed to have four small radial cuts with equal spacing. Each retina was placed onto a piece of Parafilm sheet in a ~50-μl drop of fixative (4% PFA in D-PBS with 5% sucrose) and incubated at room temperature for 15 min. The fixed retina was then covered with another piece of Parafilm sheet over which a 1.5-g weight (1.5-ml tube containing water) was placed. After 30-min incubation at room temperature, the flattened retina was placed in 1 ml of fixative for another 45 min at room temperature. After the fixation, the retina was washed with 5% sucrose in D-PBS three times for 20 min each and then placed in 300 μl of antibody diluent (1% Triton X-100, 1% Tween 20, 1% dimethyl sulfoxide in D-PBS) containing the zpr1 antibody (diluted 1:100, Zebrafish International Resource Center, Eugene) for ~18 hours at room temperature. The retina was then washed with the antibody diluent three times for 10 min each and treated with goat anti-mouse immunoglobulin G (IgG) antibody conjugated to Alexa Fluor 568 (diluted to 10 μg/ml; Molecular Probes, A-11004) in the antibody diluent for ~18 hours at room temperature. This second antibody reaction was performed in the presence of 10 μM DRAQ5 (DR5050, BioStatus) for staining of the cell nucleus. The retina was then washed with the antibody diluent three times for 10 min each, mounted onto an agarose-coated slide glass with the photoreceptor layer facing down, and cover-slipped with VECTASHIELD Mounting Medium. Fluorescent images of the immuno-stained retina were captured with a confocal laser scanning microscope (TCS SP8, Leica).
Hybridization chain reaction
FISH (fluorescence in situ hybridization) was conducted by using HCR v3.0 technology (55) according to the manufacturer’s instruction (Molecular Instruments). Whole-mount larvae [3 days postfertilization (dpf) or 5 dpf] were fixed in 4% PFA in D-PBS with 0.1% Tween 20 overnight at 4°C. Flat-mounted retinas were prepared as described above. Probe sets were listed in data file S2. Two probe sets, opn1mw1UTR and opn1mw2UTR, were designed against 3′ untranslated region (3′UTR) to distinguish expressions of these two genes, while other probe sets were designed against coding sequences. Combinations of Alexa Fluor–conjugated hairpins and probe sets used for each experiment were described in table S5. Fluorescent images of the whole-mount larvae were imaged with a light sheet fluorescence microscopy (Z.1, Zeiss). For cone mosaic analysis in the larval stage, eyes were dissected from the animal after HCR. Whole larval eyes and flat-mounted retinas were incubated with 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml) for nuclear staining. Whole larval eyes were embedded in 1% low melting point agarose and imaged with a confocal laser scanning microscope (FV3000, Olympus). Flat-mounted retinas were cover-slipped with PermaFluor Mountant (Thermo Fisher Scientific) and observed with a confocal laser scanning microscope (FV3000, Olympus).
The number of cone opsin positive cells in each fluorescent image was counted using Analyzed Particle function in ImageJ (Fiji) version 2.1.0. First, the central region of the retina was cropped from the image (87.63 μm by 87.63 μm). The trimmed images were converted to 8-bit black and white images (“Convert to Mask”). The binarized objects were filled (“Fill Holes”), and connected components were cut apart into separate ones (“Watershed”). After a threshold was selected, Analyze Particles settings were set to size 5 to 100 (μm2) and circularity 0.05 to 1.00. Six images from three individuals (two eyes each) for each genotype were processed for the quantification in Fig. 6E and fig. S7, while three images from three individuals were processed in Fig. 2E.
TUNEL staining
One head of an adult fish was subjected to cryosectioning for each genotype in a single experiment. Frozen ocular sections (10 μm thick) were prepared as described in the previous section for in situ hybridization. The cryosections were subjected to TUNEL staining by using the Click-iT TUNEL Alexa Fluor 594 Kit (Invitrogen) according to the manufacturer’s protocol. The stained sections were coverslipped with PermaFluor Mountant (Thermo Fisher Scientific) after incubation with DAPI (1 μg/ml) for nuclear staining, and observed with a confocal laser scanning microscope (FV3000, Olympus).
Immunohistochemistry
Immunohistochemistry with ocular sections was carried out as described previously (17, 20). Briefly, ocular cryosections were prepared as described in the previous section. The cryosections were pretreated with a blocking solution and then incubated with a primary antibody diluted in the blocking solution overnight at 4°C. After washed with PBS [10 mM Na-phosphate buffer, 140 mM NaCl, and 1 mM MgCl2 (pH 7.4)], the treated sections were immersed again with the blocking solution and then incubated for 4 hours at room temperature with a secondary antibody and with DAPI (3 μg/ml) for staining of the cell nuclei. The stained sections were coverslipped with VECTASHIELD Mounting Medium (Vector Laboratories) and imaged with a confocal laser scanning microscope (TCS SP8, Leica). The primary antibody used is mouse monoclonal antibody Zpr1 (diluted 1:400; Zebrafish International Resource Center, Eugene) against arrestin 3a. The secondary antibody used is goat anti-mouse IgG antibody conjugated with Alexa Fluor 568 (diluted to 2 μg/ml; Molecular Probes, A-11004).
Luciferase assay
For constructing the firefly luciferase reporter plasmids, the 1.56-kb sws2 upstream region with its 5′UTR was amplified by PCR. The amplified fragment was ligated using the In-Fusion cloning kit (Takara) into the pGL4.13[luc2/SV40] (Promega, E6681) digested with Hind III and Bgl II. PCR primers used were as follows: IF-sws2-1.5kFwBgl (5′-CGAGG ATATC AGATC TAACG ATGTT TGCTG TTTGT TC-3′) and IF-sws2-RvHind (5′-CCGGA TTGCC AAGCT TCTTG CTTGT AATTG GTGCC C-3′). The 0.53-kb sws2 reporter was constructed in a manner similar to the 1.56-kb sws2 reporter with following primers: IF_532_sw2_FwXho (5′-GCTCG CTAGC CTCGA GCAAC TCTCA AGTAT TTAAG G-3′) and IF-sws2-RvHind. The 1.56-kb sws2 reporter was truncated by PCR to generate the 0.3- and 0.25-kb sws2 reporter by using the following PCR primers: sws2_300_Fw (5′-TCTTG TACTG CGCAG ATGTA G-3′), 250sw2_Fw (5′-GAAAC TTTGT GTGTA GCTGA TG-3′), and pGL4.13_Rv (5′-AGATC TGATA TCCTC GAGGC TAG-3′). For generating the pGL4 vector having no basal promoter, the SV40 promoter in the pGL4.13 vector was removed by a combination of enzyme digestion and self-ligation; the pGL4.13 vector was double digested with Hind III and Xho I, treated T4 DNA polymerase to make a blunt end, and self-ligated. The nucleotide mutations on the potential Foxq2-binding motif were introduced into the 0.25-kb SWS2 reporter by PCR. The promoter sequence in each of these resultant constructs was sequenced to confirm that no unintended mutation was incorporated into the promoter region.
To generate the expression plasmid of Foxq2, we first inserted a FLAG epitope tag into the pCAG vector (gifted from T. Matsuda). The resultant vector was named as pCAG-FLAG. We then amplified the cDNA fragments of foxq2 from retinal cDNAs of adult zebrafish and cloned them into the EcoRV-treated pCAG-FLAG vector. These plasmids were named as drFoxq2/pCAG-FLAG. For generating the VP64-Foxq2 expression plasmid, we first cloned the multiple repeats of the herpes simplex VP16 activation domain (synthesized DNA fragments) into the Xho I–treated pCAG-FLAG vector. The resultant plasmid, named as pCAG-FLAG-VP64N, was treated with the EcoRV and ligated with the PCR-amplified cDNA fragment. The nucleotide deletion in the DNA binding domain of Foxq2 was introduced by PCR. The DNA sequence for drFoxq2/pCAG-FLAG-VP64N is available in the DDBJ/EMBL/NCBI (accession no. LC633544).
Human embryonic kidney (HEK) 293T17 cells were grown in the Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). HEK293T17 cells in 24-well plates were transiently transfected with polyethyleneimine (Polysciences, no. 24765). The firefly luciferase plasmid harboring the sws2 promoter described above was used as a reporter plasmid (10 ng per well), while the Renilla luciferase plasmid, pGL4.74[hRluc/TK] (Promega, E6921), was simultaneously transfected as an internal control reporter (0.5 ng per well). The amount of the expression plasmid of Foxq2 used for the transfection is indicated in the figure legends. The total amount of DNA transfected in each well was equally adjusted by adding the empty expression plasmids. The transfected cells were collected 36 to 48 hours after the transfection, rinsed with PBS(−) (137 mM NaCl, 2.69 mM KCl, 5.5 mM Na2HPO4, and 1.47 mM KH2PO4), and lysed with 100 μl of passive lysis buffer (Promega). Of this lysate, 10 μl was used for the dual luciferase assay, and the rest was further lysed in an SDS–polyacrylamide gel electrophoresis (PAGE) sampling buffer for the immunoblot analysis described below. The dual luciferase assay was conducted with Dual-Luciferase Reporter Assay System (Promega) and a GloMax Multi Detection System (Promega) according to the manufacturer’s protocols. The luciferase activity derived from the firefly luciferase reporter was normalized to that from the Renilla luciferase reporter.
Immunoblot analysis
Immunoblot analysis was carried out as described previously (20). In short, proteins lysed in an SDS-PAGE sampling buffer were separated on a gel by SDS-PAGE, transferred to a Immobilon-P transfer membrane (Millipore), and probed with primary antibodies overnight at 4°C. The bound primary antibodies were detected by horseradish peroxidase–conjugated secondary antibodies in combination with an enhanced chemiluminescence detection system using Western Lightning Chemiluminescence Reagent (PerkinElmer Life Sciences) or ImmunoStar (Wako Pure Chemical Industries). Chemiluminescent images were acquired with ImageQuant LAS 4000 mini (GE Healthcare). The primary antibodies used were as follows: anti-FLAG antibody (diluted to 0.8 μg/ml; Sigma-Aldrich, F3165) and anti-H3K4me3 antibody (diluted 1:5000; Upstate, 07-473). The secondary antibodies used were as follows: horseradish peroxidase–conjugated anti-mouse IgG (diluted to 0.2 μg/ml; Kirkegaard & Perry Laboratories, 074-1816) and horseradish peroxidase–conjugated anti-rabbit IgG (diluted to 0.2 μg/ml; Kirkegaard & Perry Laboratories, 074-1516).
Generation of transgenic zebrafish
To construct a Foxq2 transgene plasmid, the FLAG-Foxq2 coding sequence was amplified by PCR from the plasmid, drFoxq2/pCAG-FLAG (described in the section of Luciferase assay). The amplified fragment was then cloned with an In-Fusion HD cloning kit into the pT2drCrx5.2kGP2ASix7 (20) digested by Xho I and Bam HI. The resultant plasmid, named pT2drCrx5.2kGP2AFoxq2, was used for the generation of transgenic zebrafish. The foxq2 transgenic zebrafish, ja78Tg, ja79Tg, and ja91Tg strains, were generated with the Tol2-based transgenesis system (56). The Tol2 transposase mRNA was transcribed from pCS-TP in vitro using the SP6 mMESSAGE mMACHINE Kit (Ambion) and purified with the RNeasy Mini kit (Qiagen). The purified Tol2 mRNA and the plasmid DNA were mixed and diluted to a final concentration of 25 ng/μl for each in 0.05% phenol red solution. About 1 nl of the DNA/RNA solution was injected into each of WT embryos at the one-cell stage. Fluorescence-positive embryos were isolated and raised to adulthood. The raised F0 founder fish were crossed with the WT fish, and subsequent F1 embryos were screened by the presence of fluorescence at four or 5 dpf. The transgenic lines were established from individual F0 fish. The foxq2-tg larval fish and its siblings were genotyped by ocular EGFP fluorescence just before the sampling and soaked in RNAlater for RT-qPCR analysis or fixed with 4% PFA in D-PBS for cryosectioning. The GFP-negative siblings were used as a control. To unambiguously identify the transgenic larvae according to ocular EGFP expression, synthesis of melanin pigment was inhibited by treating embryos with the egg water containing 0.003% 1-phenol-2-thiourea from 24 hours to 5 days after fertilization.
Motif scanning
The sws2 promoter (fig. S6) was sequence-verified by traditional Sanger sequencing and used for the motif scanning. Fox and Crx binding profiles, each represented as a matrix consisting of nucleotide counts per position, i.e., position frequency matrix, were retrieved from the JASPAR CORE database (http://jaspar.genereg.net). These retrieved binding profiles (total of 11 profiles) were composed of one binding profile of Crx in mouse (57) and two binding profiles (FkhP and FkhS) for each of five Fox proteins in mouse (FoxA2, FoxL1, FoxK1, FoxJ1, and FoxJ3) (58). These frequency matrices were used to construct position-dependent letter-probability matrices that describe the probability of each possible letter at each position in the pattern with the simplest background model assuming that each letter appears equally frequently in the dataset. The sws2 promoter was scanned for individual matches to each of the Crx and Fox motifs with FIMO v5.1.1 (59). Biased distribution of individual letters in the promoter sequences was normalized by a zeroth-order model of Markov background probabilities constructed with the fasta-get-Markov tool in the MEME suite v5.1.1 (59) using nucleotide sequences between 1000-bp upstream and 1000-bp downstream of a transcription start site for all protein-coding genes in zebrafish (genome assembly GRCz11, Ensembl Release 98). All motif occurrences with P < 1 × 10−4 are indicated in fig. S6. The motif scanning results and matrix identities (IDs) of Fox and Crx in the database are included in data file S3.
BLAST searches and phylogenetic analysis
For identifying FOXQ2 genes in platypus and chicken, tBLASTn search (Ensembl web tools) was conducted against genome sequences of platypus (reference genome ID: mOrnAna1.p.v1) and chicken (reference genome ID: GRCg6a) using the amino acid sequence of forkhead domain of zebrafish Foxq2 (Ensembl protein ID: ENSDARP00000119225.2) as the query. We retrieved nucleotide sequences in intergenic regions showing the higher alignment score than any other regions encoding members of Fox families. The retrieved nucleotide sequences of FOXQ2 genes were mapped onto two distinct genomic regions but were adjacently located in the region of the same chromosome, where any other gene is not annotated. We thus assumed that these mapped regions are two exons of FOXQ2 genes. Consistently, the forkhead domain of zebrafish Foxq2 is encoded in two exons. We then manually annotated exon-exon junctions of FOXQ2 genes according to the GT/AG mRNA processing rule. The annotated cDNA sequence of the forkhead domain of FOXQ2 gene was translated into a protein sequence and used for constructing a phylogenetic tree described in the next paragraph. The nucleotide sequences and annotations of FOXQ2 genes are summarized in data file S4.
For constructing a phylogenetic tree, FOXQ2 amino acid sequences in spotted gar, medaka, coelacanth, and sparrow hawk were retrieved from Ensembl database (Ensembl Release 101). FOXQ2 sequences in platypus and chicken identified by our blast searching were also used for the phylogenetic tree construction. Amino acid sequences of other members of Fox subfamilies were retrieved by performing tblastn searches (NCBI) against RefSeq RNA transcripts in purple sea urchin, zebrafish, chicken, platypus, and human using the amino acid sequence of forkhead domain of zebrafish Foxq2 as the query sequence with E <1 × 10−20. Among the retrieved cDNA sequences, members of representative Fox subfamily (FOXA, FOXB, FOXC, FOXF, FOXJ, and FOXQ) were selected and translated into amino acid sequences. The resultant sequences of Fox proteins were aligned by multiple sequence alignment programs: G-INS-i program in MAFFT v7.471 under default settings (60). The aligned sequences were trimmed, remaining the sequences of the forkhead domain. Alignment gaps were manually inspected and deleted. The maximum likelihood tree was inferred by RAxML-NG v1.0.2 (61). The best tree was selected out of 40 alternative runs on 20 random- and 20 parsimony-based starting trees (--tree pars{20}, rand{20} option). The amino acid replacement models of Le-Gascuel (LG) with gamma distribution (G4) were selected using the Akaike information criterion implemented in ModelTest-NG version x.y.z (62). The bootstrap values were obtained from sampling 500 times. The amino acid sequences used for the construction of phylogenetic tree are listed in data file S4. Accession numbers for genome assemblies and Fox genes are also provided in data file S4.
Statistical analysis
Sample sizes were determined on the basis of prior literature and best practices in the field, and no statistical methods were used to predetermine sample size. A two-tailed unpaired t test was used to determine the statistical significance between two datasets (Excel). Tukey-Kramer honestly significant difference test was used to determine the statistical significance among multiple datasets (the “stats” package in R, version 3.6.1).
Acknowledgments
We are grateful to members of the Fukada Lab for valuable discussion, especially to M. Nagata for technical assistance. We thank the National BioResource Project (NBRP) Zebrafish for providing the transgenic line, and S. Kawamura, The University of Tokyo, for the zebrafish opsin transgenic lines, Tg(-5.5opn1sw1:EGFP)kj9, Tg(-3.5opn1sw2:EGFP)kj11, Tg(RH2-2/GFP-PAC)kj4, and Tg(-0.6opn1lw1-lws2:GFP)kj19. We also thank T. Matsuda for providing the pCAG empty vector, M. Kinoshita for the pCS2+hSpCas9 vector, and W. Chen for the pCS2–nCas9n vector. We also thank members of the FACS core laboratory, The University of Tokyo, for helping sorting cells. Funding: This work was supported by JSPS KAKENHI grant number JP16J01681 (to Y.O.), JSPS KAKENHI grant number JP19K16196 (to T.S.), JSPS KAKENHI grant number JP19K06758 (to D.K.), JSPS KAKENHI grant number JP17H06096 (to Y.F.), and Research Foundation for Opto-Science and Technology (to D.K.). Author contributions: Conceptualization: Y.O., T.S., D.K., and Y.F. Microarray: T.S. Photoreceptor cell isolation: Y.O. and T.S. Plasmid generation: Y.O. and T.S. Fish generation and RT-qPCR: Y.O. and T.S. Immunohistochemistry: Y.O. and D.K. In situ hybridization chain reaction: T.S. In situ hybridization: Y.O. TUNEL staining: T.S. Luciferase assay: Y.O. Phylogenetic analysis: Y.O. Supervision: D.K. and Y.F. Writing—original draft: Y.O. and T.S. Writing—review and editing: Y.O., D.K., and Y.F. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Microarray data and nucleotide sequence of Foxq2 have been deposited in the databases indicated in Materials and Methods.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S8
Tables S1 to S6
Legends for data files S1 to S4
References
Other Supplementary Material for this manuscript includes the following:
Data files S1 to S4
REFERENCES AND NOTES
- 1.Lamb T. D., Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Goldsmith T. H., Optimization, constraint, and history in the evolution of eyes. Q. Rev. Biol. 65, 281–322 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Baden T., Osorio D., The retinal basis of vertebrate color vision. Annu. Rev. Vis. Sci. 5, 177–200 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Okano T., Kojima D., Fukada Y., Shichida Y., Yoshizawa T., Primary structures of chicken cone visual pigments: Vertebrate rhodopsins have evolved out of cone visual pigments. Proc. Natl. Acad. Sci. U.S.A. 89, 5932–5936 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama S., Evolution of dim-light and color vision pigments. Annu. Rev. Genomics Hum. Genet. 9, 259–282 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Collin S. P., Knight M. A., Davies W. L., Potter I. C., Hunt D. M., Trezise A. E. O., Ancient colour vision: Multiple opsin genes in the ancestral vertebrates. Curr. Biol. 13, R864–R865 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Davies W. I. L., Collin S. P., Hunt D. M., Molecular ecology and adaptation of visual photopigments in craniates. Mol. Ecol. 21, 3121–3158 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Swaroop A., Kim D., Forrest D., Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 11, 563–576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cepko C., Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci. 15, 615–627 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Furukawa T., Morrow E. M., Cepko C. L., Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91, 531–541 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Mears A. J., Kondo M., Swain P. K., Takada Y., Bush R. A., Saunders T. L., Sieving P. A., Swaroop A., Nrl is required for rod photoreceptor development. Nat. Genet. 29, 447–452 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Haider N. B., Naggert J. K., Nishina P. M., Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum. Mol. Genet. 10, 1619–1626 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Ng L., Hurley J. B., Dierks B., Srinivas M., Saltó C., Vennström B., Reh T. A., Forrest D., A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 27, 94–98 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Suzuki S. C., Bleckert A., Williams P. R., Takechi M., Kawamura S., Wong R. O. L., Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl. Acad. Sci. U.S.A. 110, 15109–15114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eldred K. C., Hadyniak S. E., Hussey K. A., Brenerman B., Zhang P.-W., Chamling X., Sluch V. M., Welsbie D. S., Hattar S., Taylor J., Wahlin K., Zack D. J., Johnston R. J. Jr., Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 362, eaau6348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Delfin K., Morris A. C., Snelson C. D., Gamse J. T., Gupta T., Marlow F. L., Mullins M. C., Burgess H. A., Granato M., Fadool J. M., Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc. Natl. Acad. Sci. U.S.A. 106, 2023–2028 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa Y., Shiraki T., Kojima D., Fukada Y., Homeobox transcription factor Six7 governs expression of green opsin genes in zebrafish. Proc. Biol. Sci. 282, 20150659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinen A., Hamaoka T., Yamada Y., Kawamura S., Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163, 663–675 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takechi M., Kawamura S., Temporal and spatial changes in the expression pattern of multiple red and green subtype opsin genes during zebrafish development. J. Exp. Biol. 208, 1337–1345 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Ogawa Y., Shiraki T., Asano Y., Muto A., Kawakami K., Suzuki Y., Kojima D., Fukada Y., Six6 and Six7 coordinately regulate expression of middle-wavelength opsins in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 116, 4651–4660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renninger S. L., Gesemann M., Neuhauss S. C. F., Cone arrestin confers cone vision of high temporal resolution in zebrafish larvae. Eur. J. Neurosci. 33, 658–667 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Raymond P. A., Barthel L. K., A moving wave patterns the cone photoreceptor mosaic array in the zebrafish retina. Int. J. Dev. Biol. 48, 935–945 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Müller C. W., Herrmann B. G., Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature 389, 884–888 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Snelson C. D., Santhakumar K., Halpern M. E., Gamse J. T., Tbx2b is required for the development of the parapineal organ. Development 135, 1693–1702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkov L. I., Kim-Han J. S., Saunders L. M., Poria D., Hughes A. E. O., Kefalov V. J., Parichy D. M., Corbo J. C., Thyroid hormone receptors mediate two distinct mechanisms of long-wavelength vision. Proc. Natl. Acad. Sci. U.S.A. 117, 15262–15269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raymond P. A., Barthel L. K., Bernardos R. L., Perkowski J. J., Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev. Biol. 6, 36 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross J. M., Dowling J. E., Tbx2b is essential for neuronal differentiation along the dorsal/ventral axis of the zebrafish retina. Proc. Natl. Acad. Sci. U.S.A. 102, 4371–4376 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark K. L., Halay E. D., Lai E., Burley S. K., Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420 (1993). [DOI] [PubMed] [Google Scholar]
- 29.Larroux C., Luke G. N., Koopman P., Rokhsar D. S., Shimeld S. M., Degnan B. M., Genesis and expansion of metazoan transcription factor gene classes. Mol. Biol. Evol. 25, 980–996 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa S., Gisselbrecht S. S., Rogers J. M., Hartl D. L., Bulyk M. L., DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl. Acad. Sci. U.S.A. 110, 12349–12354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takechi M., Seno S., Kawamura S., Identification of cis-acting elements repressing blue opsin expression in zebrafish UV cones and pineal cells. J. Biol. Chem. 283, 31625–31632 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Beerli R. R., Segal D. J., Dreier B., Barbas C. F., Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. U.S.A. 95, 14628–14633 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies W. L., Carvalho L. S., Cowing J. A., Beazley L. D., Hunt D. M., Arrese C. A., Visual pigments of the platypus: A novel route to mammalian colour vision. Curr. Biol. 17, R161–R163 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Bowmaker J. K., Evolution of vertebrate visual pigments. Vision Res. 48, 2022–2041 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Jacobs G. H., Evolution of colour vision in mammals. Philos. Trans. R. Soc. B Biol. Sci. 364, 2957–2967 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cronin T. W., Bok M. J., Photoreception and vision in the ultraviolet. J. Exp. Biol. 219, 2790–2801 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Yoshimatsu T., Bartel P., Schröder C., Janiak F. K., St-Pierre F., Berens P., Baden T., Ancestral circuits for vertebrate colour vision emerge at the first retinal synapse. bioRxiv , 2020.10.26.356089 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobert O., Regulatory logic of neuronal diversity: Terminal selector genes and selector motifs. Proc. Natl. Acad. Sci. U.S.A. 105, 20067–20071 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Shen Y., Rest J. S., Raymond P. A., Zack D. J., Isolation and characterization of a zebrafish homologue of the cone rod homeobox gene. Invest. Ophthalmol. Vis. Sci. 42, 481–487 (2001). [PubMed] [Google Scholar]
- 40.Ebrey T., Koutalos Y., Vertebrate photoreceptors. Prog. Retin. Eye Res. 20, 49–94 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Fritzenwanker J. H., Gerhart J., Freeman R. M. Jr., Lowe C. J., The Fox/Forkhead transcription factor family of the hemichordate Saccoglossus kowalevskii. EvoDevo 5, 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinigaglia C., Busengdal H., Leclère L., Technau U., Rentzsch F., The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol. 11, e1001488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Range R. C., Wei Z., An anterior signaling center patterns and sizes the anterior neuroectoderm of the sea urchin embryo. Development 143, 1523–1533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamb T. D., Collin S. P., Pugh E. N. Jr., Evolution of the vertebrate eye: Opsins, photoreceptors, retina and eye cup. Nat. Rev. Neurosci. 8, 960–976 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A. W., Wittbrodf J., Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869–871 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Asaoka Y., Mano H., Kojima D., Fukada Y., Pineal expression-promoting element (PIPE), a cis-acting element, directs pineal-specific gene expression in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 99, 15456–15461 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mano H., Asaoka Y., Kojima D., Fukada Y., Brain-specific homeobox Bsx specifies identity of pineal gland between serially homologous photoreceptive organs in zebrafish. Commun. Biol. 2, 364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takechi M., Hamaoka T., Kawamura S., Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish. FEBS Lett. 553, 90–94 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Tsujimura T., Chinen A., Kawamura S., Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 12813–12818 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsujimura T., Hosoya T., Kawamura S., A single enhancer regulating the differential expression of duplicated red-sensitive opsin genes in zebrafish. PLOS Genet. 6, e1001245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gagnon J. A., Valen E., Thyme S. B., Huang P., Ahkmetova L., Pauli A., Montague T. G., Zimmerman S., Richter C., Schier A. F., Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLOS ONE 9, e98186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jao L.-E., Wente S. R., Chen W., Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. 110, 13904–13909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ansai S., Kinoshita M., Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open. 3, 362–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ota S., Hisano Y., Muraki M., Hoshijima K., Dahlem T. J., Grunwald D. J., Okada Y., Kawahara A., Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells 18, 450–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi H. M. T., Schwarzkopf M., Fornace M. E., Acharya A., Artavanis G., Stegmaier J., Cunha A., Pierce N. A., Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawakami K., Takeda H., Kawakami N., Kobayashi M., Matsuda N., Mishina M., A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7, 133–144 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Corbo J. C., Lawrence K. A., Karlstetter M., Myers C. A., Abdelaziz M., Dirkes W., Weigelt K., Seifert M., Benes V., Fritsche L. G., Weber B. H. F., Langmann T., CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 20, 1512–1525 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Badis G., Berger M. F., Philippakis A. A., Talukder S., Gehrke A. R., Jaeger S. A., Chan E. T., Metzler G., Vedenko A., Chen X., Kuznetsov H., Wang C.-F., Coburn D., Newburger D. E., Morris Q., Hughes T. R., Bulyk M. L., Diversity and complexity in DNA recognition by transcription factors. Science 324, 1720–1723 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S., MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A., Wren J., RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darriba D., Posada D., Kozlov A. M., Stamatakis A., Morel B., Flouri T., ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J., Dantas Machado A. C., Guo M., Sagendorf J. M., Zhou Z., Jiang L., Chen X., Wu D., Qu L., Chen Z., Chen L., Rohs R., Chen Y., Structure of the forkhead domain of FOXA2 bound to a complete DNA consensus site. Biochemistry 56, 3745–3753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lagman D., Ocampo Daza D., Widmark J., Abalo X. M., Sundström G., Larhammar D., The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evol. Biol. 13, 238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshizawa T., Shichida Y., Fukada Y., Biochemical and photochemical analyses of retinal proteins in chicken cone cells. Pure Appl. Chem. 63, 171–176 (1991). [Google Scholar]
- 66.Gillard G. B., Grønvold L., Røsæg L. L., Holen M. M., Monsen Ø., Koop B. F., Rondeau E. B., Gundappa M. K., Mendoza J., Macqueen D. J., Rohlfs R. V., Sandve S. R., Hvidsten T. R., Comparative regulomics supports pervasive selection on gene dosage following whole genome duplication. Genome Biol. 22, 103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S8
Tables S1 to S6
Legends for data files S1 to S4
References
Data files S1 to S4