Abstract
CCAAT/enhancer binding protein α (C/EBPα) is a transcription factor involved in creating and maintaining the adipocyte phenotype. We have shown previously that insulin stimulates dephosphorylation of C/EBPα in 3T3-L1 adipocytes. Studies to identify the insulin-sensitive sites of phosphorylation reveal that a C/EBPα peptide (amino acids H215 to K250) is phosphorylated on T222, T226, and S230 in vivo. The context of these phosphoamino acids implicates glycogen synthase kinase 3 (GSK3), whose activity is known to be repressed in response to insulin, as a potential kinase for phosphorylation of T222 and T226. Accordingly, GSK3 phosphorylates the predicted region of C/EBPα on threonine in vitro, and GSK3 uses C/EBPα as a substrate in vivo. In addition, the effect of pharmacological agents on GSK3 activity correlates with regulation of C/EBPα phosphorylation. Treatment of 3T3-L1 adipocytes with the phosphatidylinositol 3-kinase inhibitor wortmannin results in phosphorylation of C/EBPα, whereas treatment with the GSK3 inhibitor lithium results in dephosphorylation of C/EBPα. Collectively, these data indicate that insulin stimulates dephosphorylation of C/EBPα on T222 and T226 through inactivation of GSK3. Since dephosphorylation of C/EBPα in response to lithium is blocked by okadaic acid, strong candidates for the T222 and T226 phosphatase are protein phosphatases 1 and 2a. Treatment of adipocytes with insulin alters the protease accessibility of widespread sites within the N terminus of C/EBPα, consistent with phosphorylation causing profound conformational changes. Finally, phosphorylation of C/EBPα and other substrates by GSK3 may be required for adipogenesis, since treatment of differentiating preadipocytes with lithium inhibits their conversion to adipocytes.
In view of the prevalence of obesity and obesity-related diseases, such as type II diabetes, it is important to understand the molecular basis for adipose tissue development and metabolism. Traditionally, adipocytes were known to play an important role in lipid homeostasis through their ability to store triacyglycerol and release free fatty acids and glycerol in response to changing energy needs. It is now recognized that adipocytes play a more central role in metabolism through their secretion of factors that regulate food intake and metabolic efficiency (46). The negative health consequences of excess white adipose tissue have been well documented (49), but the effects of an absence of fat have only been studied recently. Mice without white adipose tissue were created by directing expression to adipocytes of a dominant-negative protein that forms dimers with members of the C/EBP and AP-1 families, but which does not bind DNA (37). These mice have abnormal growth rates and enlarged internal organs and die prematurely. In addition, they exhibit a number of metabolic defects, including diabetes, with reduced leptin and increased serum glucose, insulin, free fatty acids, and triacylglycerols. The widespread effects of blocking C/EBP and AP-1 activities in adipose tissue highlight the importance of studying these transcription factors in adipocyte differentiation and metabolism.
The molecular events associated with preadipocyte differentiation have been most thoroughly studied in 3T3-L1 cells (reviewed in references 9, 13, 32, 34, and 46). Treatment of preadipocytes with inducers of differentiation stimulates a rapid and transient increase in C/EBPβ and C/EBPδ, which in turn mediates the transcriptional activation of peroxisomal proliferator-activated receptor γ (PPARγ) during the 2 days following stimulation of differentiation. Expression of PPARγ, probably in conjunction with C/EBPβ, induces expression of C/EBPα to its maximal level 4 to 5 days after initiation of differentiation. Together, C/EBPα and PPARγ activate the transcription of genes involved in creating and maintaining the adipocyte phenotype (e.g., adipocyte lipid binding protein [422/aP2], stearoyl coenzyme A desaturase I, insulin-responsive glucose transporter [GLUT4], and leptin) (6, 24, 25, 31). The expression of PPARγ and C/EBPα remains elevated in adipocytes through positive self- and cross-regulation.
C/EBPα not only plays an important role in preadipocyte differentiation, but also regulates gene expression in fully differentiated adipocytes. Although C/EBPα-deficient fibroblasts acquire the morphological characteristics of adipocytes upon ectopic expression and activation of PPARγ, these C/EBPα−/− adipocytes are resistant to insulin. Further analyses revealed that these cells fail to increase glucose uptake in response to insulin because of impaired expression of insulin receptor and insulin receptor substrate 1 (50). Likewise, NIH-3T3 cells, which differentiate into adipocytes without expression of C/EBPα, are insulin resistant. In this case, it was found that the cells do not acquire the ability to transport glucose in response to insulin because of impaired expression of GLUT4 (17). Thus, C/EBPα is essential for the acquisition of insulin sensitivity by adipocytes. Given the requirement of C/EBPα for insulin responsivity, it is not surprising that insulin feeds back to inhibit this transcription factor. In 3T3-L1 adipocytes, insulin suppresses transcription of the C/EBPα gene and stimulates dephosphorylation of C/EBPα protein (22, 31). These events closely correlate with suppression of GLUT4 gene expression, perhaps as part of the mechanism of desensitization to insulin. In this study, we investigate further the dephosphorylation of C/EBPα by insulin.
C/EBPα is a member of the basic region-leucine zipper (bZIP) family of transcription factors (reviewed in reference 35). The C-terminal zipper domain mediates the formation of homodimers as well as heterodimers with family members and perhaps a subset of other bZIP transcription factors (e.g., ATF2) (44). The basic region is adjacent to the zipper domain and binds specific DNA sequences. The N terminus of C/EBPα contains multiple transactivation domains that work synergistically to transactivate C/EBPα-dependent promoters. The region N terminal to bZIP contains four regions that are highly conserved throughout evolution and that are separated by linker regions enriched in glycines and prolines (18). Conserved regions 1, 2, and 3 loosely correspond to transactivation domains identified previously (19, 39–41). Each conserved region is capable of transactivating the leptin promoter when fused to the C/EBPα bZIP domain (18). Conserved region 4 is the most highly conserved region outside the bZIP and probably plays a regulatory role in C/EBPα function. In the present study, three sites of phosphorylation were identified within this region, T222, T226, and S230, two of which are regulated by insulin. We present evidence that glycogen synthase kinase 3 (GSK3) phosphorylates T222 and T226, causing a conformational change in C/EBPα. Upon treatment with insulin, T222 and T226 are dephosphorylated through inactivation of GSK3 and activation of protein phosphatase 1 (PP1) or PP2A. Finally, we present evidence that GSK3 is required for adipocyte development, since inhibition of GSK3 activity with lithium blocks, not only C/EBPα phosphorylation, but also preadipocyte differentiation.
MATERIALS AND METHODS
Cell culture and transfection.
Mouse 3T3-L1 preadipocytes (20) and human embryonic kidney 293T cells (a kind gift from Mitchell Lazar) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL) supplemented with 10% calf serum, as described previously (22). Cells were transfected by calcium phosphate coprecipitation, as described previously (24). Expression plasmids, at the amounts indicated, were supplemented with pcDNA3.1(−) (Invitrogen), such that the total DNA was 20 μg/10-cm-diameter plate. Cells were lysed 24 to 48 h following transfection. Constitutively active GSK3 (S9A) was obtained from Peter J. Roach (Indiana University School of Medicine) (16).
C/EBPα expression plasmids.
To increase the ease of manipulating the mouse C/EBPα gene (5), four unique restriction sites were created through introduction of silent mutations. C/EBPα from the NruI site (+5 nucleotide [nt]) to the EcoRV site (+2111 nt) was subcloned into pBluescript (KS+) [pBS(KS+)] (Stratagene), and the following silent mutations were made by site-directed mutagenesis (QuickChange; Stratagene): a KpnI site was generated by mutation of C531 to G, a PstI site was generated by mutation of A774 to G, an SphI site was generated by mutation of C822 to T, and an XhoI site was generated by mutation of G1074 to C. In addition, the translational start site for C/EBPα was optimized, as described previously (29), to maximize expression of full-length p42C/EBPα, while minimizing translation from internal methionines. A p30C/EBPα expression vector was constructed by excising the first 180 nt of the gene, which includes the start sites of translation for p42C/EBPα and p40C/EBPα, but not the start site for p30C/EBPα.
The C/EBPα gene was also engineered to encode proteins in which T222, T226, and S230 have been mutated to either alanines or serines. To create these mutations, the TTS (wild type), AAA, SSS, STS, ATS, and AAS oligonucleotides were synthesized and then subcloned into the newly created PstI and SphI sites of C/EBPα (Table 1). These C/EBPα mutants were subcloned from pBS(KS+) into pcDNA 3.1(+) (Invitrogen) by using BamHI and HindIII sites, and the resultant expression vectors were used in transient transfections.
TABLE 1.
Oligonucleotides synthesized and then subcloned into newly created PstI and SphI sites of C/EBPα
| Oligonucleotide | Sequence |
|---|---|
| Wild type; TTS | GCCTGGCCACCCCACACCGCCGCCCACGCCCGTGCCCAGCCCGCATG |
| Mutant | |
| AAA | GCCTGGCCACCCCGCACCGCCGCCCGCGCCCGTGCCCGCCCCACATG |
| SSS | GCCTGGCCACCCCAGCCCGCCGCCCAGCCCCGTGCCCAGCCCACATG |
| STS | GCCTGGTCACCCCAGCCCGCCGCCCACGCCCGTGCCCAGCCCACATG |
| ATS | GCCTGGTCACCCCGCCCCGCCGCCCACGCCCGTGCCCAGCCCGCATG |
| AAS | GCCTGGGCACCCCGCACCGCCGCCCGCGCCCGTGCCCAGCCCACATG |
To make His-tagged, full-length C/EBPα (His-p42C/EBPα), the NruI-HindIII fragment (containing C/EBPα +5 nt to +2079 nt) was excised from C/EBPα, and the HindIII site was filled in with Klenow fragment prior to subcloning into the PvuII site of pEBVHisA (Invitrogen). Similarly, His-tagged p18C/EBPα (His-p18C/EBPα; also referred to as His-p18TTS), His-tagged p12C/EBPα (His-p12C/EBPα), and His-tagged p10C/EBPα (His-p10C/EBPα) were constructed by inserting the MluI-EcoRV fragment (+698 nt to +2111 nt) into the PvuII site of pEBVHisA, the SmaI-SmaI fragment (+861 nt to +1293 nt) into the XhoI site of pEBVHisC, and the BanI-SmaI fragment (+931 nt to +1293 nt) into the PvuII site of pEBVHisB, respectively. His-p18SSS and His-p18AAA were constructed with the p42C/EBPαSSS and p42C/EBPαAAA mutants, respectively. Briefly, after the MluI site was filled in, the MluI-HindIII fragment was subcloned into the EcoRV and HindIII sites of pcDNA3.1(−). This fragment was then excised by using PmeI and HindIII and subcloned into the PvuII and HindIII sites of pEBVHisA. His-p18STS, His-p18ATS, and His-p18AAS were created as described above for His-p18TTS with the primers indicated.
In vivo phosphorylation.
293T cell monolayers were preincubated in phosphate-free DMEM (Gibco-BRL) for 30 min. These cells were then incubated in phosphate-free DMEM that was supplemented with 32Pi (Amersham; 0.5 mCi/ml; 4 ml/10-cm-diameter plate) for 3 h. After being rinsed twice with phosphate-buffered saline, the cells were lysed in urea lysis buffer (8 M urea, 0.5 M NaCl, 45 mM Na2HPO4, 5 mM NaH2PO4, 10 mM imidazole [pH 8.0]) for protein purification.
Purification of His-tagged proteins.
To identify sites of phosphorylation in C/EBPα, 100 10-cm-diameter plates of 293T cells were transiently transfected with expression plasmids encoding either His-p42C/EBPα or His-p18C/EBPα. These plates were rinsed with phosphate-buffered saline, and then each was lysed in 0.5 ml of urea lysis buffer and sonicated. Pooled lysates were incubated with 2 ml of ProBond nickel resin (Invitrogen) for 2 h at room temperature. Nickel resin was washed three times with 10 bed volumes of wash buffer (8 M urea, 0.4 M NaCl, 17.6 mM Na2HPO4, 32.4 mM NaH2PO4, 10 mM imidazole [pH 6.75]), and bound proteins were eluted by being washed two times in 5 bed volumes of elution buffer (8 M urea, 0.4 M NaCl, 6 mM Na2HPO4, 44 mM NaH2HPO4, 10 mM imidazole [pH 5.30]). The solution containing these eluted proteins was adjusted to pH 8.0, and the proteins were incubated a second time with nickel resin. This purification process was repeated twice with the following changes: for each of the subsequent rounds, the bed volume was decreased by half, and for the final wash, the stringency was increased by raising the concentration of imidazole from 10 to 50 mM. This protocol was also used in a scaled-down version to purify His-C/EBPα from 12 plates of transfected 293T cells labeled with 32Pi. Unlabeled and labeled products from this purification procedure were pooled and concentrated by centrifugation (Centricon-10; Amicon). Purification products were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). To optimize protein purification and monitor protein purity, proteins were visualized by silver staining (2), and C/EBPα was detected by immunoblot analysis. For protein recovery, SDS-PAGE gels were zinc stained according to the manufacturer’s instructions (Pierce). His-C/EBPα was identified by its mobility and excised from the gel. Samples were then sent to the Protein Structure Core Facility at the University of Nebraska Medical Center for analyses. In-gel cleavage was performed by using vapor cyanogen bromide followed by trypsin. Peptides were separated on a Vydac C18 column and sequenced with an ABI 477 protein sequencer, as described previously (48).
Immunoblot analysis.
Cell lysis and immunoblotting for C/EBPα were performed as described previously (22) with a polyclonal C/EBPα antibody that was raised against a synthetic peptide (amino acids 253 to 265) (29).
Phosphoamino acid analysis.
Purified His-tagged forms of truncated or full-length C/EBPα that had been phosphorylated either in vitro or in vivo were separated by SDS-PAGE. After transfer onto PVDF (polyvinylidene difluoride)-Plus membrane (Micron Separations, Inc.) and visualization by autoradiography, His-tagged C/EBPα was excised and hydrolyzed in 6 M HCl for 60 min at 110°C. Acid was removed by evaporation in a Speedvac and with two 0.5-ml washes of water. Samples were resuspended in 10 μl of water containing 1 ng of phosphoserine, phosphotyrosine, and phosphothreonine (Sigma) and were spotted onto cellulose plates (Kodak). Amino acids were separated by thin-layer electrophoresis for 45 min at 20 mA in pH 2.5 buffer (5.9% [vol/vol] glacial acetic acid, 0.8% [vol/vol] formic acid [88%], 0.3% [vol/vol] pyridine, 0.3 mM EDTA) (21). Phosphoamino acid standards were visualized by ninhydrin staining, and 32P-labeled phosphoamino acids were detected by autoradiography.
In vitro kinase reactions.
Protein purification for in vitro kinase assays of His-p42C/EBPα, His-p18C/EBPα, His-p12C/EBPα, and His-p10C/EBPα was performed as described above, except that His-tagged forms of C/EBPα were not eluted from nickel resin following the last purification. Rather, purified C/EBPα proteins were washed with kinase buffer and then incubated in kinase buffer with or without 5 U of GSK3 (New England Biolabs) for 60 min at 37°C. The nickel resin was washed extensively with wash buffer, and proteins were eluted in elution buffer and separated by SDS-PAGE as described above. Proteins were transferred to PVDF-Plus membrane (22), and the blot was subjected to autoradiography and immunoblot analyses.
Preparation of nuclei.
Nuclei were purified from 3T3-L1 preadipocytes or adipocytes by a procedure modified from that of Dignam et al. (15). Briefly, 3T3-L1 cells were washed with 5 ml of phosphate-buffered saline, and then 2 ml of hypotonic lysis buffer (20 mM Tris [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 1 mM dithiothreitol [DTT]) with 2-μl/ml protease inhibitors (PIC 1, which is 1-mg/ml leupeptin, 1-mg/ml antipain, and 10-mg/ml benzamidine in aprotinin; and PIC 2, which is 1-mg/ml chymostatin and 1-mg/ml pepstatin A in dimethyl sulfoxide [DMSO]), and phosphatase inhibitors (30 mM β-glycerophosphate, 1 mM sodium orthovanadate, and 1-mg/ml p-nitrophenylphosphate). Cells were scraped in 1 ml of hypotonic lysis buffer, and Igepal CA-630 was added (1:100 [vol/vol] for preadipocytes, 1:67 [vol/vol] for adipocytes) prior to Dounce homogenization (25 strokes). Disruption of plasma membranes was verified by trypan blue staining and light microscopy. Nuclei were pelleted in a microcentrifuge at 5,000 × g for 1 min. The supernatant was removed, the nuclei-containing pellet was resuspended in 1 ml of hypotonic lysis buffer, and the sample was centrifuged as described above. The process of resuspension and centrifugation was repeated. For quantification of nuclei, 1 5-μl aliquot was lysed in 0.5% SDS and vortexed, and A260 was measured. Nuclear pellets were lysed in isoelectric focusing buffer or immunoblot lysis buffer or used for the protease accessibility assay.
Protease accessibility assay.
To assess the sensitivity of nuclear C/EBPα to protease, nuclei from 3T3-L1 adipocytes were resuspended in hypotonic lysis buffer at 60 A260/ml. Increasing amounts of trypsin in 20 μl of phosphate-buffered saline were added to 30-μl aliquots of nuclei on ice for 30 min. The final concentrations of trypsin were 0, 8, 24, 80, and 240 μg/ml. Reactions were terminated with 50 μl of 2× immunoblot lysis buffer (2% SDS and 120 mM Tris [pH 6.8]) and heated to 100°C for 10 min. Samples were subjected to SDS-PAGE and immunoblot analysis for C/EBPα.
Alkaline phosphatase treatment of samples.
Nuclei were lysed in an SDS-containing buffer (1% SDS, 60 mM Tris [pH 6.8]) at a concentration of approximately 20 mg of protein/ml. Nuclear proteins were incubated with alkaline phosphatase (Boehringer Mannheim) at a concentration of approximately 1 U/mg of protein in the SDS-containing buffer for 1 h at 37°C. Samples were mixed with an equal volume of isoelectric focusing buffer prior to electrophoretic separation.
Isoelectric focusing.
Nuclei were lysed in isoelectric focusing buffer (9 M urea, 1% Igepal CA-630, 1% DTT) at a concentration of approximately 10 mg of protein/ml. This nuclear lysate was sonicated and then centrifuged at 4°C for 15 min, and the supernatant was collected for further analysis. Polyacrylamide-urea mini gels were made according to the manufacturer’s instructions (Bio-Rad) with a 1:1 mixture of pH 3 to 10 and pH 5 to 8 ampholytes (Bio-Rad). Protein samples (approximately 20 μg) were loaded and focused for 15 min at 100 V, 15 min at 200 V, and then 1 h at 450 V. Proteins were then transferred onto PVDF-plus membrane for immunoblot analysis.
Preadipocyte differentiation and Oil Red-O staining.
3T3-L1 preadipocytes were induced to differentiate as described previously (47), except that the medium was not supplemented with insulin on day 4 or thereafter (30). Oil Red-O staining was performed essentially as described previously (42). Briefly, cell monolayers were washed with phosphate-buffered saline, fixed in 3.7% formaldehyde for 2 min, washed with H2O, incubated with Oil Red-O solution for 1 h at room temperature, and then washed with H2O.
RESULTS
Identification of T222, T226, and S230 as phosphoamino acids.
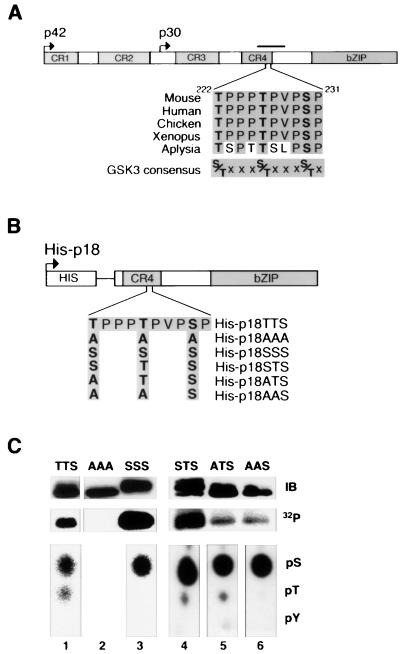
We have shown previously that C/EBPα contains amino acids that become dephosphorylated in response to insulin (22, 31). To determine which amino acids in C/EBPα are phosphorylated, we labeled the cellular phosphate pool and purified 32P-labeled C/EBPα for analyses. In these experiments, 293T cells were transiently transfected with expression vectors encoding His-tagged C/EBPα constructs. Transfected cells were incubated with 32Pi, and His-tagged forms of C/EBPα were purified on a nickel resin. Eluted proteins were separated by SDS-PAGE, and C/EBPα was excised from the gel. C/EBPα was subjected to in-gel cleavage by treatment with cyanogen bromide and trypsin. The C/EBPα peptides were separated by high-performance liquid chromatography. Following this separation, labeled peptides were sequenced by Edman degradation to identify the phosphoamino acids. Using this approach, in three independent experiments, the peptide spanning amino acids H215 through K250 was identified as a phosphopeptide (solid bar in Fig. 1A). Amino acid analysis of this phosphopeptide revealed that it contains both phosphoserine and phosphothreonine. Edman degradation demonstrated that this peptide is phosphorylated on T222 and S230.
FIG. 1.
Identification of phosphorylation sites in C/EBPα. (A) A schematic diagram of C/EBPα. Regions that are highly conserved across species (conserved regions 1 to 4 and bZIP domain) are shaded. Bent arrows denote translational start sites for two predominant C/EBPα species (p42C/EBPα and p30C/EBPα). A phosphopeptide identified in three independent labeling experiments is indicated by the solid bar. Amino acids within a region of this phosphopeptide are shown for mouse, human, chicken, Xenopus, and Aplysia proteins. The consensus sequence for GSK3 is given. (B) Schematic representation of the His-tagged C/EBPα fusion proteins used in subsequent labeling experiments. The C/EBPα amino acid sequence from T222 to P231 is given, and the C/EBPα mutants in which T222, T226, and S230 were mutated to either alanines or serines are illustrated. (C) His-p18TTS (lane 1), His-p18AAA (lane 2), His-p18SSS (lane 3), His-p18STS (lane 4), His-p18ATS (lane 5), and His-p18AAS (lane 6) were expressed in 293T cells, labeled with 32P in vivo, purified, and separated by SDS-PAGE. These samples were subjected to immunoblot analysis for C/EBPα (top panel; IB) and autoradiography (middle panel; 32P). Phosphoamino acid analysis was performed with the remainder of the His-tagged C/EBPα mutants (bottom panels). The positions of phosphoserine (pS), phosphothreonine (pT), and phosphotyrosine (pY) are indicated. His-p18AAA contained no detectable 32P.
The identified amino acids are within a consensus sequence of phosphorylation by GSK3 (Fig. 1A) (26). In addition, since insulin inhibits GSK3 activity in adipocytes (11), GSK3 is a particularly good candidate for modulating sites that are dephosphorylated upon insulin treatment. We therefore considered the possibility that GSK3 is a C/EBPα kinase. For many GSK3 substrates, phosphorylation of a priming site at the n position is required for subsequent phosphorylation by GSK3 at the n − 4 position (43). Applying this hierarchical model to C/EBPα, we predicted that phosphorylation of S230 by an unspecified kinase would be necessary for phosphorylation at T226 by GSK3. Thereafter, phosphorylation of T226 would serve as the priming site for GSK3-mediated phosphorylation of T222. We identified phosphate on both T222 and S230, but we were unable to conclusively identify T226 as a phosphoamino acid by using Edman degradation. 32P at this site may be difficult to detect because slower phosphate turnover at T226 relative to T222 and S230 could result in lower specific activity upon metabolic labeling, or because more rapid dephosphorylation of T226 than of the other sites could result in lower relative stoichiometry of phosphorylation.
To determine whether C/EBPα is phosphorylated on T226, we designed a series of truncated C/EBPα mutants for use in phosphoamino acid analysis experiments. Specifically, we created a His-tagged expression vector for an N-terminally truncated form of C/EBPα, His-p18TTS, and ones in which T222, T226, and S230 were converted to combinations of alanines and serines (His-p18SSS, His-p18AAA, His-p18STS, His-p18ATS, and His-p18AAS), as shown schematically in Fig. 1B. 293T cells were transiently transfected with expression vectors for His-p18C/EBPα proteins. Two days later, cells were labeled with 32Pi, and His-tagged C/EBPα proteins were purified on a nickel resin. Immunoblot analysis of the purified products revealed that the C/EBPα proteins were expressed and purified (Fig. 1C, top panel). Autoradiography of these immunoblots revealed that each of these C/EBPα proteins, except His-p18AAA, is phosphorylated in vivo (Fig. 1C, middle panel). His-p18AAA (lane 2) contains no detectable phosphate, indicating that p18TTS is not phosphorylated on any sites other than T222, T226, and S230. Upon phosphoamino acid analysis, we found that His-p18TTS (lane 1), His-p18STS (lane 4), and His-p18ATS (lane 5) are phosphorylated on serine and threonine, whereas His-p18SSS (lane 3) and His-p18AAS (lane 6) are phosphorylated on serine alone (Fig. 1C, bottom panel). Loss of phosphothreonine upon conversion of either His-p18STS to His-p18SSS or His-18ATS to His-18AAS indicates that T226 is phosphorylated in vivo. This finding, together with results from our initial mapping studies (above), demonstrates that C/EBPα is phosphorylated on T222, T226, and S230.
GSK3 phosphorylates C/EBPα in vitro.
To investigate further whether GSK3 phosphorylates C/EBPα, we used purified C/EBPα as a substrate for GSK3 in vitro. For these assays, His-tagged versions of full-length or truncated forms of C/EBPα (Fig. 2A) were purified from 293T cells. Wild-type p42C/EBPα and p18C/EBPα contain the GSK3 consensus sites of phosphorylation, whereas p12C/EBPα and p10C/EBPα do not. Purified His-tagged forms of C/EBPα were combined, and duplicate kinase assays were performed in either the absence or presence of GSK3 prior to separation by SDS-PAGE. Immunoblot analysis revealed that the amounts of C/EBPα proteins in each reaction varied by less than a factor of 2 (Fig. 2B). Autoradiography revealed that p42C/EBPα and p18C/EBPα, but not p12C/EBPα and p10C/EBPα, are substrates for phosphorylation by GSK3 (Fig. 2C). Phosphoamino acid analysis demonstrated that phosphorylation of p42C/EBPα by GSK3 in vitro occurs only on threonine (Fig. 2D). These findings indicate that GSK3 phosphorylates threonine specifically in a region between amino acids R192 and G246 in vitro and are consistent with GSK3 phosphorylating T222 and T226.
FIG. 2.
GSK3 phosphorylates C/EBPα in vitro. (A) Schematic representation of His-tagged C/EBPα fusion proteins used as substrates in kinase assays in vitro. A putative region of GSK3 phosphorylation is denoted by the solid bar. (B) His-tagged p42, p18, p12, and p10 C/EBPα were expressed in 293T cells and purified. These proteins were mixed together, and kinase reactions were performed in the absence (−) or presence (+) of recombinant GSK3. Proteins were separated by SDS-PAGE (15% polyacrylamide gel) and subjected to immunoblot analysis with antibody specific for C/EBPα. The migration of His-tagged p42, p18, p12, and p10 C/EBPα is indicated. (C) The immunoblot in panel B was subjected to autoradiography. (D) 32P-labeled His-p42C/EBPα was excised, and phosphoamino acid analysis was performed. Phosphoamino acid standards (pS, pT, and pY) are indicated. Similar results were obtained in three independent experiments.
GSK3 phosphorylates C/EBPα in vivo.
Since the steady-state stoichiometry of protein phosphorylation depends upon the balance of kinase and phosphatase activities, we reasoned that overexpression of a C/EBPα kinase would increase the proportion of C/EBPα that is phosphorylated. To assess whether GSK3 is a C/EBPα kinase in a cellular context, 293T cells were transfected with expression vectors encoding either p30C/EBPα alone (Fig. 3, lanes 1 to 4) or p30C/EBPα and constitutively active GSK3 (Fig. 3, lanes 5 to 8). p30C/EBPα is an alternate translation product which arises from initiation at the third start codon (29). This form was used because phosphorylation-induced shifts in p30C/EBPα can be visualized by SDS-PAGE (22). Two days after transfection, cells were not treated or treated with the GSK3 inhibitor lithium for 1 h prior to lysis. Samples were separated by SDS-PAGE, and C/EBPα was detected by immunoblot analysis. When expressed alone, p30C/EBPα exists predominantly as a single high-mobility form (Fig. 3, lanes 1 and 3). Inhibition of GSK3 activity by treatment of these cells with lithium results in the loss of the weak band that crowns the p30C/EBPα species (Fig. 3, lanes 2 and 4). This observation suggests that, under these conditions, a small proportion of p30C/EBPα is phosphorylated due to endogenous GSK3 activity in 293T cells. Overexpression of constitutively active GSK3 dramatically increases the proportion of the upper species (Fig. 3, lanes 5 and 7). Inhibition of GSK3 activity by 1 h of lithium treatment results in the loss of this upper species. These data suggest that GSK3 phosphorylates C/EBPα in vivo.
FIG. 3.
p30C/EBPα is a substrate for GSK3 in vivo. 293T cells were transfected with p30C/EBPα alone or both p30C/EBPα and constitutively active GSK3. Forty-eight hours later, these cells were not treated (−) or treated with 25 mM LiCl (+) for 1 h prior to lysis. Samples were separated by SDS-PAGE (11.5% polyacrylamide gel) and subjected to immunoblot analyses for C/EBPα. Similar results were obtained in three independent experiments.
Phosphorylation of C/EBPα correlates with the regulation of GSK3 activity by insulin, wortmannin, and lithium.
We have demonstrated that GSK3 phosphorylates C/EBPα in vitro and in vivo. We next tested whether the ability of insulin to inhibit GSK3 (38) is responsible for insulin-dependent dephosphorylation of C/EBPα in 3T3-L1 adipocytes. In many cell types, including adipocytes, activation of the insulin receptor is known to stimulate phosphatidylinositol (PI) 3-kinase activity, which, through the actions of other signaling molecules (possibly Akt [10] or PDK1 [14]), causes phosphorylation and inhibition of GSK3 (11). If GSK3 is the insulin-sensitive C/EBPα kinase, then we would expect inhibition of PI 3-kinase with wortmannin to stimulate GSK3 and therefore cause phosphorylation of C/EBPα. Conversely, we would expect inhibition of GSK3 activity with lithium to result in dephosphorylation of C/EBPα.
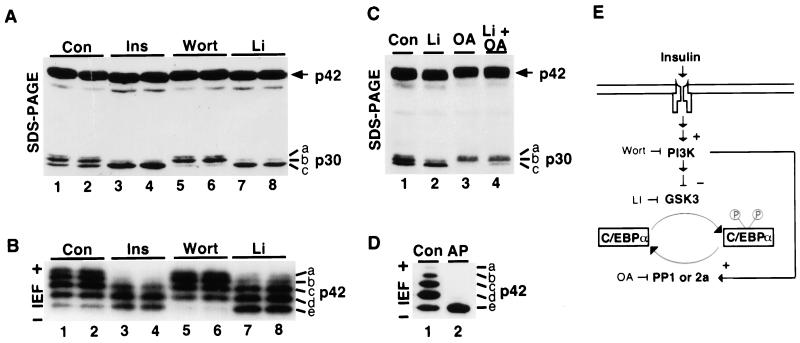
In these experiments, we treated 3T3-L1 adipocytes with wortmannin or lithium and then assessed the phosphorylation of the two predominant C/EBPα translation products, p30C/EBPα and p42C/EBPα, by immunoblot analysis. In the absence of treatment (Fig. 4A, lanes 1 and 2), p30C/EBPα has three distinct mobilities upon SDS-PAGE (bands a to c). We have shown previously that these differences in mobility are due to phosphorylation. Treatment of samples with alkaline phosphatase in vitro causes the top two bands to disappear with a proportionate increase in the bottom band (22). Treatment with either insulin (Fig. 4A, lanes 3 and 4) or lithium (Fig. 4A, lanes 7 and 8) causes dephosphorylation of p30C/EBPα (loss of bands a and b with an increase in band c), whereas treatment with wortmannin (Fig. 4A, lanes 5 and 6) results in net phosphorylation of C/EBPα (loss of band c). Therefore, insulin appears to stimulate dephosphorylation of two sites within p30C/EBPα through inhibition of GSK3.
FIG. 4.
Regulation of C/EBPα phosphorylation by pharmacological agents. (A) 3T3-L1 adipocytes were not treated (Con, lanes 1 and 2) or were treated with 167 nM insulin for 60 min (Ins; lanes 3 and 4), 200 nM wortmannin for 60 min (Wort; lanes 5 and 6), or 25 mM LiCl for 30 min (Li; lanes 7 and 8). Whole-cell lysates were separated by SDS-PAGE (11.5% polyacrylamide gel) and subjected to immunoblot analysis for C/EBPα. The migration of p42C/EBPα and p30C/EBPα is indicated. p30C/EBPα migrates as three bands (a to c). (B) 3T3-L1 adipocytes were treated as in panel A, except that nuclei were prepared. Nuclear proteins were separated by isoelectric focusing and subjected to immunoblot analysis for C/EBPα. Acidic (+) and basic (−) ends of the blot are indicated. Similar results were obtained in three independent experiments. Since p30C/EBPα is more basic (predicted pI of 9.7) than p42C/EBPα (predicted pI of 7.6), p30C/EBPα is not resolved into discreet bands when focused to equilibrium with standard ampholytes (pH range of 3 to 10). p42C/EBPα can be resolved into five bands (a to 3) by this technique. (C) 3T3-L1 adipocytes were not treated (Con; lane 1) or were treated with 25 mM LiCl (Li; lane 2), 1.5 μM okadaic acid (OA; lane 3), or both 25 mM LiCl and 1.5 μM okadaic acid (lane 4) for 1 h. Whole-cell lysates were separated by SDS-PAGE (11.5% polyacrylamide gel) and subjected to immunoblot analysis for C/EBPα. (D) Nuclear lysates were prepared from 3T3-L1 adipocytes and either not treated (Con; lane 1) or treated with alkaline phosphatase (AP; lane 2) prior to isoelectric focusing and immunoblot analysis for C/EBPα. (E) Model of signaling pathway through which insulin, wortmannin, lithium, and okadaic acid regulate the phosphorylation of C/EBPα.
Treatment of adipocytes with insulin, wortmannin, or lithium appears to cause analogous changes in the phosphorylation of p42C/EBPα. However, phosphorylation-induced changes in the mobility of this larger protein are difficult to resolve by SDS-PAGE. Thus, to determine whether p42C/EBPα is regulated similarly to p30C/EBPα, a parallel experiment was performed in which 3T3-L1 adipocyte lysates were analyzed by isoelectric focusing (Fig. 4B). p42C/EBPα from unstimulated 3T3-L1 adipocytes is resolved into at least five bands by this technique (Fig. 4B, lanes 1 and 2 [forms a to e]). Treatment of adipocytes with either insulin (Fig. 4B, lanes 3 and 4) or lithium (Fig. 4B, lanes 7 and 8) results in a decrease in the two most acidic forms of p42C/EBPα (bands a and b). In contrast, treatment of cells with wortmannin (Fig. 4B, lanes 5 and 6) causes loss of the most basic form of p42C/EBPα (band e). To confirm that the presence of multiple p42C/EBPα bands arises from the phosphorylation of p42C/EBPα, adipocyte lysates were treated with alkaline phosphatase in vitro prior to isoelectric focusing. Upon this treatment, the acidic forms of C/EBPα were lost and there was accumulation of its most basic form (Fig. 4D, lane 2). This observation indicates that the most basic species is C/EBPα without phosphate and suggests that the acidic bands represent C/EBPα with increasing amounts of phosphate. The simplest interpretation of these data is that p42C/EBPα, like p30C/EBPα, is dephosphorylated at two sites following insulin treatment. In addition, the regulation of p30C/EBPα and p42C/EBPα phosphorylation by wortmannin and lithium is consistent with the hypothesis that GSK3 is the insulin-sensitive C/EBPα kinase.
Phosphorylation of C/EBPα correlates with the regulation of PP1 and PP2A by okadaic acid.
Dephosphorylation of C/EBPα requires not only inhibition of GSK3, but also activity of a C/EBPα phosphatase. PP1 is a good candidate, since the activity of this phosphatase is induced by insulin in 3T3-L1 adipocytes, and since treatment of adipocytes with okadaic acid, which inhibits PP1 and PP2A, results in accumulation of hyperphosphorylated C/EBPα (31). To determine whether PP1 or PP2A is directly responsible for dephosphorylation of the sites within p30C/EBPα, 3T3-L1 adipocytes were not treated or treated with lithium, okadaic acid, or lithium and okadaic acid (Fig. 4C, lanes 1 to 4). Since lithium stimulates only minimal dephosphorylation in the presence of okadaic acid, PP1 or PP2A is likely responsible for dephosphorylation of C/EBPα at these sites. Moreover, since induction of PP1 activity by insulin is inhibited by wortmannin (3), our finding that wortmannin stimulates phosphorylation of C/EBPα in the presence of lithium is consistent with wortmannin also inhibiting C/EBPα phosphatase activity (not shown). Taken together, these data are consistent with the model proposed in Fig. 4E, in which stimulation of PI 3-kinase by insulin results in simultaneous inhibition of the C/EBPα kinase (GSK3) and activation of the C/EBPα phosphatase (PP1 or PP2A) to cause dephosphorylation of C/EBPα.
Phosphorylation alters C/EBPα conformation.
To determine whether phosphorylation alters the three-dimensional structure of C/EBPα, we compared the accessibilities to protease of specific C/EBPα residues after treatment of adipocytes with insulin or wortmannin. The sensitivity to proteolysis depends on the tertiary structure of the protein and the extent to which the protease-sensitive sites are protected or exposed through protein-protein or protein-DNA interactions. For instance, interaction of C/EBPα with DNA protects the bZIP domain from tryptic digestion (data not shown and reference 45). Trypsin cleaves R-X and K-X and is predicted to cut C/EBPα at 36 sites, of which 24 are in the bZIP domain and are therefore resistant to proteolysis. Although partial digestion of the 10 sites outside the bZIP domain by trypsin likely gives a large number of different C/EBPα fragments, the anti-peptide C/EBPα antibody used in our experiments only recognizes those that contain amino acids 253 to 265, a region just N terminal to the bZIP domain. Thus, only C-terminal proteolytic fragments of C/EBPα are observed in this protease accessibility assay.
Nuclei were isolated from 3T3-L1 adipocytes that had been treated for 1 h with insulin to induce dephosphorylation or wortmannin to induce phosphorylation (Fig. 3) (22). Nuclei were incubated with increasing concentrations of trypsin on ice for 30 min, and then nuclear proteins were separated by SDS-PAGE for immunoblot analysis (Fig. 5). Endogenous C/EBPα proteins, p42C/EBPα and p30C/EBPα, were observed in undigested samples from insulin-treated (Fig. 5, lane 1) or wortmannin-treated (Fig. 5, lane 2) adipocytes. The effects of phosphorylation on mobility of C/EBPα are not as well resolved as in Fig. 3, because the proportion of acrylamide in the gel was higher. Treatment of nuclei with increasing concentrations of trypsin resulted in C/EBPα digestion products of decreasing size. Of interest, the trypsin fingerprints from insulin- and wortmannin-treated cells, at any given trypsin concentration, are substantially different. For example, dephosphorylated C/EBPα from insulin-treated adipocytes shows cleavage at three distinct sites in the N terminus (Fig. 5, bands a to c) that are not observed with phosphorylated C/EBPα from wortmannin-treated adipocytes. In addition, more widespread changes in sensitivity to protease were observed in two other regions (Fig. 5, complex d and e). Although some differences in band pattern of complex d and e can be explained through differential phosphorylation altering mobility of C/EBPα upon SDS-PAGE, the loss of bands a to c after wortmannin treatment suggests that phosphorylation of C/EBPα alters the sensitivity of these sites to trypsin. It is conceivable that altered cleavage of some sites is due to phosphorylation per se (i.e., by inhibiting the interactions between trypsin and C/EBPα). However, the magnitude and widespread nature of the differential sensitivity suggest that phosphorylation causes a change in the three-dimensional structure of C/EBPα, thereby altering the accessibility of protease to many sites of cleavage. Since the accessibility of C/EBPα to trypsin was assessed in nuclei, changes in protease accessibility could also reflect differential interactions with transcriptional coactivators or other nuclear proteins.
FIG. 5.
Insulin and wortmannin alter the trypsin sensitivity of C/EBPα. 3T3-L1 adipocytes were treated for 1 h with 167 nM insulin or 250 nM wortmannin, as indicated. Nuclei were purified from cells and incubated for 30 min on ice in the absence (lanes 1 and 2) or presence of various concentrations of trypsin (lanes 3 and 4, 8 μg/ml; lanes 5 and 6, 24 μg/ml; lanes 7 and 8; 80 μg/ml; lanes 9 and 10, 240 μg/ml). Samples were separated by SDS-PAGE (15% polyacrylamide gel) and subjected to immunoblot analysis for C/EBPα. Because the C/EBPα antibody used in these experiments was raised against an epitope located near the bZIP domain, which is protected from trypsinization, these partially digested C/EBPα fragments are N-terminal truncation products.
Lithium inhibits preadipocyte differentiation.
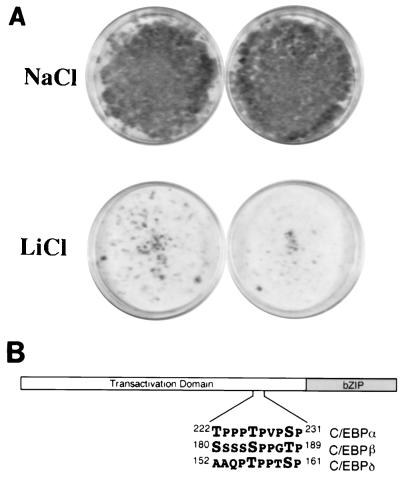
Given the importance of C/EBPα in preadipocyte differentiation (13) and the established role of GSK3 in the development of tissues in other species (4), we considered the possibility that GSK3 activity is required for adipogenesis. To assess this putative role for GSK3, we investigated whether the GSK3 inhibitor lithium could block preadipocyte differentiation (Fig. 6A). 3T3-L1 preadipocytes were induced to differentiate under standard conditions, with isobutylmethylxanthine, dexamethasone, insulin, and fetal calf serum, in the presence of either 25 mM LiCl or 25 mM NaCl as a control. Thereafter, cells were continuously incubated in the presence of LiCl or NaCl over the course of differentiation. Eight days after the induction of differentiation, the degree of adipogenesis was assessed qualitatively by staining cellular lipid droplets with Oil Red-O. Upon induction, 3T3-L1 preadipocytes differentiated almost completely, irrespective of the presence of NaCl (Fig. 6A, top). In contrast, lithium treatment almost completely inhibited adipocyte differentiation (Fig. 6A, bottom). Expression of adipocyte markers C/EBPα and 422/aP2 was also inhibited by lithium (data not shown). In addition to GSK3, lithium is known to inhibit inositol monophosphatase (27) and may inhibit other enzymatic activities as well. Nevertheless, the observed inhibition of adipogenesis by lithium is consistent with the hypothesis that GSK3 activity is required for adipocyte differentiation.
FIG. 6.
(A) Lithium inhibits preadipocyte differentiation. 3T3-L1 preadipocytes were induced to differentiate as described in Methods and Materials, except that on days 1, 2, 4, and 6, 25 mM NaCl or LiCl was added to the medium. On day 8, cells were fixed and stained with Oil Red-O to visually assess the accumulation of lipid droplets. (B) Schematic representation of a generalized C/EBP transcription factor. The region of C/EBPα that is phosphorylated by GSK3 and putative GSK3 recognition sequences in C/EBPβ and C/EBPδ are given.
To address potential targets of GSK3 activity in preadipocyte differentiation, we considered known regulators of adipogenesis. The current model of 3T3-L1 differentiation involves coordinated expression of four transcription factors, C/EBPβ, C/EBPδ, PPARγ, and C/EBPα (13). Our work shows that at least one of these, C/EBPα, is phosphorylated by GSK3. As a first step to determine whether the others are likewise regulated by GSK3, we inspected their sequences for putative GSK3 sites. C/EBPβ and C/EBPδ, but not PPARγ, contain sequence elements which resemble GSK3 recognition sequences (Fig. 6B). Next, we determined whether the phosphorylation of C/EBPβ is sensitive to lithium. Consistent with the hypothesis that C/EBP family members are regulated by GSK3, lithium treatment of cells caused dephosphorylation of C/EBPβ as assessed by immunoblot analysis (41a). For each of these C/EBPs, the putative sites of GSK3 phosphorylation are located just N terminal to the bZIP. In addition to this conservation among C/EBP family members, within C/EBPα, C/EBPβ, and C/EBPδ, the region of putative GSK3 phosphorylation is highly conserved across species. Collectively, these data raise the possibility that GSK phosphorylation of C/EBPs may be required for preadipocyte differentiation.
DISCUSSION
Our analyses have led to the discovery that several sites within conserved region 4 of C/EBPα are phosphorylated in vivo (Fig. 1). Two sites, T222 and T226, are phosphorylated by GSK3, whereas S230 is phosphorylated by an unknown kinase. In addition, this work delineates a part of the signaling mechanism through which C/EBPα phosphorylation is regulated. Previous work (38) has established that insulin, acting through signaling intermediates, including PI 3-kinase, causes inhibition of GSK3 in adipocytes. We report here that inactivation of GSK3 leads to dephosphorylation of two sites, T222 and T226 (Fig. 4). Furthermore, since okadaic acid treatment of adipocytes blocks the lithium-induced dephosphorylation of C/EBPα, it is likely that PP1 or PP2A is responsible for dephosphorylation of the insulin-sensitive sites (Fig. 4D). Our work also shows that insulin and wortmannin alter the protease accessibility of C/EBPα, thereby implying that phosphorylation at T222 and T226 causes a conformational change in the structure of C/EBPα (Fig. 5). Finally, we report here that lithium inhibits preadipocyte differentiation (Fig. 6), suggesting that GSK3-mediated phosphorylation of C/EBPα and other transcription factors, such as C/EBPβ, is required for adipogenesis.
A hierarchical model of phosphorylation has been described for several GSK3 substrates (7, 43). If this model holds true for C/EBPα, then phosphorylation of S230 is required for subsequent phosphorylation of T226 and then T222. Results from two experiments indicate that phosphorylation by GSK3 need not occur in this requisite order. First, a glutathione S-transferase–C/EBPα fusion protein purified from bacteria (and therefore not phosphorylated) is phosphorylated by GSK3 in vitro on threonine alone (not shown). Second, based on the hierarchical model, we would expect neither the p30TTA mutant nor the p30AAA mutant to be phosphorylated by GSK3. However, we observed that while p30AAA exists as a single dephosphorylated band upon SDS-PAGE, p30TTA migrates as a doublet whose low-mobility band is rapidly lost after lithium treatment (not shown). Together, these experiments suggest that T222 and T226 are substrates for GSK3 even when S230 is mutated to alanine. Despite this negative evidence, it remains possible that phosphorylation of S230 changes the affinity of C/EBPα for recognition by GSK3 and may therefore serve a modulatory role.
Phylogenetic analysis of C/EBPα has allowed us to define four highly conserved regions in the N-terminal transactivation domain, and we believe that the regions of greatest functional importance are likely to be found therein (18). Each of conserved regions 1, 2, and 3 has intrinsic transactivation ability, suggesting that these regions may directly interact with coactivators or with the basal transcriptional machinery. In contrast, the fourth conserved region, although the most highly conserved, has no known function. Our finding that GSK3 phosphorylates three sites within conserved region 4 suggests that C/EBPα activity may be regulated through this mechanism. However, the precise role of this modification has proved difficult to uncover. Mutation of T222 and T226 to alanines does not change the ability of C/EBPα to transactivate leptin or C/EBPα promoters in reporter gene assays (data not shown). Furthermore, altering the phosphorylation status of C/EBPα by cotransfection of GSK3 also does not influence the ability of C/EBPα to transactivate in these experiments (data not shown). Finally, ectopic expression of the T222A, T226A mutant, like its wild-type counterpart, is sufficient to induce spontaneous differentiation of preadipocytes (data not shown), demonstrating that phosphorylation of these sites is not required for activation of chromatin-embedded genes. Thus, GSK3-mediated phosphorylation does not, in itself, dramatically alter the activity of C/EBPα in our assays. Since the protease accessibility of C/EBPα was determined in nuclei, the effects of phosphorylation on sensitivity of C/EBPα to trypsin (Fig. 5) may be the result of conformational changes or differential interactions with nuclear proteins.
It has been reported that protein kinase C can phosphorylate specific sites within the basic region of C/EBPα in vitro (33). This modification is an attractive mechanism for the regulation of C/EBPα activity, since addition of a negative charge to the basic region profoundly reduces its affinity for DNA. However, we have no evidence from labeling experiments that this phosphorylation occurs in vivo. Phosphate incorporation into His-p18AAA was undetectable in our studies, suggesting that T222, T226, and S230 are the only phosphorylated residues in the C-terminal 18 kDa of C/EBPα (Fig. 1C).
We have shown that phosphorylation of C/EBPα by GSK3 is regulated by insulin. In addition to its role in insulin signaling, GSK3 is a mediator in the Wnt signaling pathway, where it has been shown to be important in the specification of cell fate (reviewed in references 4 and 12). Wnts are a family of secreted glycoproteins which, probably through activation of frizzled receptors, stimulate a signaling cascade resulting in the inactivation of GSK3. Improper expression of Wnts has severe developmental consequences. For instance, overexpression of Wnt-1 results in the formation of two-headed tadpoles (36). Wnts are also important for the formation of mesodermal derivatives such as Xenopus myoblasts, in which dominant-negative expression of Wnt blocks MyoD expression and impairs skeletal muscle formation (23). It is tempting to speculate that Wnt signaling may also be important in the differentiation of mesodermal derivatives into adipocytes by regulating the activity of transcription factors such as the C/EBP family. As shown in Fig. 6B, C/EBPβ and C/EBPδ contain GSK3 consensus sequences and may, like C/EBPα, be phosphorylated by GSK3. Our finding that lithium prevents adipogenesis supports the hypothesis that GSK3 activity is required for the differentiation of 3T3-L1 preadipocytes. Although the presence of receptors for Wnt on 3T3-L1 preadipocytes has not been reported, other cell models with adipogenic potential, NIH 3T3 cells and CH310T1/2, respond to Wnts (1, 8). We propose that regulation by GSK3 of C/EBPα phosphorylation and possibly preadipocyte differentiation is controlled not only by insulin, but also by Wnts and other ligands.
Insulin has rapid effects on the flow of carbon through metabolic pathways by regulating the activity of metabolic enzymes through stimulating their phosphorylation or dephosphorylation. Insulin also has longer-term effects on metabolism by altering gene expression to regulate the amount of enzyme or regulatory proteins available for metabolism. GSK3 acts as a node through which insulin signals to regulate both carbohydrate metabolism and adipocyte gene expression. The best-characterized role of GSK3 is in mediating the effects of insulin on glycogen metabolism through its phosphorylation and inhibition of glycogen synthase (28). It is now apparent that GSK3 also mediates the effects of insulin on adipocyte gene expression, since C/EBPα, a transcription factor required for acquisition of insulin sensitivity, is phosphorylated by GSK3. Dephosphorylation of the GSK3 sites in both glycogen synthase and C/EBPα appears to be mediated by PP1, since both activities are sensitive to okadaic acid and wortmannin. Thus, insulin appears to use reciprocal regulation of GSK3 and PP1 activities to coordinately regulate short- and long-term effects on adipocyte metabolism.
ACKNOWLEDGMENTS
This work is supported by a research grant to O.A.M. from the NIDDK, National Institutes of Health (RO1-DK51563). S.E.R. and R.L.E. are supported by predoctoral fellowships from the Natural Sciences and Engineering Research Council of Canada.
We thank Lawrence Argetsinger, Christin Carter-Su, Susanne Mandrup, Barbara Nicke, Jessica Schwartz, and John Williams for critical review of the manuscript. In addition, we thank Robert Lewis for phosphoamino acid analysis of peptide H215-K250, Mitchell Lazar for suggesting that we treat differentiating preadipocytes with lithium, and M. Daniel Lane for providing antiserum to C/EBPα.
REFERENCES
- 1.Bafico A, Gazit A, Wu-Morgan S S, Yaniv A, Aaronson S A. Characterization of Wnt-1 and Wnt-2 induced growth alterations and signaling pathways in NIH 3T3 fibroblasts. Oncogene. 1998;16:2819–2825. doi: 10.1038/sj.onc.1201797. [DOI] [PubMed] [Google Scholar]
- 2.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Virology. 1987;8:93–99. [Google Scholar]
- 3.Brady M J, Bourbonais F J, Saltiel A R. The activation of glycogen synthase by insulin switches from kinase inhibition to phosphatase activation during adipogenesis in 3T3-L1 cells. J Biol Chem. 1998;273:14063–14066. doi: 10.1074/jbc.273.23.14063. [DOI] [PubMed] [Google Scholar]
- 4.Cadigan K M, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 5.Christy R J, Kaestner K H, Geiman D E, Lane M D. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA. 1991;88:2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W H, Friedman A D, Nakabeppu Y, Kelly T J, Lane M D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 7.Chu B, Soncin F, Price B D, Stevenson M A, Calderwood S K. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem. 1996;271:30847–30857. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- 8.Cook D, Fry M J, Hughes K, Sumathipala R, Woodgett J R, Dale T C. Wingless inactivates glycogen synthase kinase-3 via an intracellular signalling pathway which involves a protein kinase C. EMBO J. 1996;15:4526–4536. [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelius P, MacDougald O A, Lane M D. Regulation of adipocyte development. Annu Rev Nutr. 1994;14:99–129. doi: 10.1146/annurev.nu.14.070194.000531. [DOI] [PubMed] [Google Scholar]
- 10.Cross D A E, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 11.Cross D A E, Watt P W, Shaw M, van der Kaay J, Downes C P, Holder J C, Cohen P. Insulin activates protein kinase B, inhibits glycogen synthase kinase-3 and activates glycogen synthase by rapamycin-insensitive pathways in skeletal muscle and adipose tissue. FEBS Lett. 1997;406:211–215. doi: 10.1016/s0014-5793(97)00240-8. [DOI] [PubMed] [Google Scholar]
- 12.Dale T C. Signal transduction by the Wnt family of ligands. Biochem J. 1998;329:209–223. doi: 10.1042/bj3290209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darlington G J, Ross S E, MacDougald O A. The role of C/EBP genes in adipocyte differentiation. J Biol Chem. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 14.Delcommenne M, Tan C, Gray V, Rue L, Woodgett J, Dedhar S. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc Natl Acad Sci USA. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eldar-Finkelman H, Argast G M, Foord O, Fischer E H, Krebs E G. Expression and characterization of glycogen synthase kinase-3 mutants and their effect on glycogen synthase activity in intact cells. Proc Natl Acad Sci USA. 1996;93:10228–10233. doi: 10.1073/pnas.93.19.10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Jack A K, Hamm J K, Pilch P F, Farmer S R. Reconstitution of insulin-sensitive glucose transport in fibroblasts requires expression of both PPARγ and C/EBPα. J Biol Chem. 1999;274:7946–7951. doi: 10.1074/jbc.274.12.7946. [DOI] [PubMed] [Google Scholar]
- 18.Erickson, R. L., K. A. Longo, S. E. Ross, N. Hemati, and O. A. MacDougald. Structure and function of C/EBPα. In Proceedings of the Steenbock Symposium. IOS Press, Amsterdam, The Netherlands, in press.
- 19.Friedman A D, McKnight S L. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 20.Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell. 1974;3:127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- 21.Groblewski G E, Wishart M J, Yoshida M, Williams J A. Purification and identification of a 28-kDa calcium-regulated heat-stable protein. J Biol Chem. 1996;271:31502–31507. doi: 10.1074/jbc.271.49.31502. [DOI] [PubMed] [Google Scholar]
- 22.Hemati N, Ross S E, Erickson R L, Grobelwski G E, MacDougald O A. Signaling pathways through which insulin regulates CCAAT/enhancer binding protein α (C/EBPα) phosphorylation and gene expression in 3T3-L1 adipocytes: correlation with GLUT4 gene expression. J Biol Chem. 1997;41:25913–25919. doi: 10.1074/jbc.272.41.25913. [DOI] [PubMed] [Google Scholar]
- 23.Hoppler S, Brown J D, Moon R T. Expression of a dominant-negative Wnt blocks induction of MyoD in Xenopus embryos. Genes Dev. 1996;10:2805–2817. doi: 10.1101/gad.10.21.2805. [DOI] [PubMed] [Google Scholar]
- 24.Hwang C-S, Mandrup S, MacDougald O A, Geiman D E, Lane M D. Transcriptional activation of the obese gene by CCAAT/enhancer binding proteinα. Proc Natl Acad Sci USA. 1996;93:873–877. doi: 10.1073/pnas.93.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaestner K H, Christy R J, Lane M D. Mouse insulin-responsive glucose transporter gene: characterization of the gene and trans-activation by the CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:251–255. doi: 10.1073/pnas.87.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennelly P J, Krebs E G. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 27.Klein P S, Melton D A. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence J C, Roach P J. New insights into the role and mechanism of glycogen synthase activation by insulin. Diabetes. 1997;46:541–547. doi: 10.2337/diab.46.4.541. [DOI] [PubMed] [Google Scholar]
- 29.Lin F-T, MacDougald O A, Diehl A M, Lane M D. A 30 kilodalton alternative translation product of the CCAAT/enhancer binding protein α message: transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci USA. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDougald O A, Cornelius P, Lin F-T, Chen S S, Lane M D. Glucocorticoids reciprocally regulate expression of the CCAAT/enhancer-binding protein α and δ genes in 3T3-L1 adipocytes and white adipose tissue. J Biol Chem. 1994;269:19041–19047. [PubMed] [Google Scholar]
- 31.MacDougald O A, Cornelius P, Liu R, Lane M D. Insulin regulates transcription of the CCAAT/enhancer binding protein (C/EBP) α, β, and δ genes in fully-differentiated 3T3-L1 adipocytes. J Biol Chem. 1995;270:647–654. doi: 10.1074/jbc.270.2.647. [DOI] [PubMed] [Google Scholar]
- 32.MacDougald O A, Lane M D. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 33.Mahoney C W, Shuman J, McKnight S L, Chen H-C, Huang K-P. Phosphorylation of CCAAT-enhancer binding protein by protein kinase C attenuates site-selective DNA binding. J Biol Chem. 1992;267:19396–19403. [PubMed] [Google Scholar]
- 34.Mandrup S, Lane M D. Regulating adipogenesis. J Biol Chem. 1997;272:5367–5370. doi: 10.1074/jbc.272.9.5367. [DOI] [PubMed] [Google Scholar]
- 35.McKnight S L. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. CCAAT/enhancer binding protein; pp. 771–795. [Google Scholar]
- 36.McMahon A P, Moon R T. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell. 1989;58:1075–1084. doi: 10.1016/0092-8674(89)90506-0. [DOI] [PubMed] [Google Scholar]
- 37.Moitra J, Mason M M, Olive M, Krylov D, Gavrilova O, Marcus-Samuels B, Feigenbaum L, Lee E, Aoyama T, Eckhaus M, Reitman M L, Vinson C. Life without white fat: a transgenic mouse. Genes Dev. 1998;12:3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moule S K, Welsh G I, Edgell N J, Foulstone E J, Proud C G, Denton R M. Regulation of protein kinase B and glycogen synthase kinase-3 by insulin and β-andrenergic agonists in rat epididymal fat cells. J Biol Chem. 1997;272:7713–7719. doi: 10.1074/jbc.272.12.7713. [DOI] [PubMed] [Google Scholar]
- 39.Nerlov C, Ziff E B. CCAAT/enhancer binding protein-α amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nerlov C, Ziff E B. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-α on the serum albumin promoter. Genes Dev. 1994;8:350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 41.Pei D, Shih C. An “attenuator domain” is sandwiched by two distinct transactivation domains in the transcription factor C/EBP. Mol Cell Biol. 1991;11:1480–1487. doi: 10.1128/mcb.11.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Piwien de Pilipuk, G., S. E. Ross, O. A. MacDougald, and J. Schwartz. Unpublished data.
- 42.Preece A. A manual for histogic technicians. Boston, Mass: Little Brown; 1972. [Google Scholar]
- 43.Roach P J. Control of glycogen synthase by hierarchical protein phosphorylation. FASEB J. 1990;4:2961–2968. [PubMed] [Google Scholar]
- 44.Shuman J D, Cheong J, Coligan J E. ATF-2 and C/EBPα can form a heterodimeric DNA binding complex in vitro. J Biol Chem. 1997;272:12793–12800. doi: 10.1074/jbc.272.19.12793. [DOI] [PubMed] [Google Scholar]
- 45.Shuman J D, Vinson C R, McKnight S L. Evidence of changes in protease sensitivity and subunit exchange rate on DNA binding by C/EBP. Science. 1990;249:771–778. doi: 10.1126/science.2202050. [DOI] [PubMed] [Google Scholar]
- 46.Spiegelman B M, Flier J S. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 47.Student A K, Hsu R Y, Lane M D. Induction of fatty acid synthetase synthesis in differentiating 3T3-L1 preadipocytes. J Biol Chem. 1980;255:4745–4750. [PubMed] [Google Scholar]
- 48.Volle D J, Fulton J A, Chaika O V, McDermott K, Luang H, Steinke L A, Lewis R E. Phosphorylation of the kinase suppressor of Ras by associated kinases. Biochemistry. 1999;38:5130–5137. doi: 10.1021/bi983050d. [DOI] [PubMed] [Google Scholar]
- 49.Wickelgren I. Obesity: how big a problem? Science. 1998;280:1364–1367. doi: 10.1126/science.280.5368.1364. [DOI] [PubMed] [Google Scholar]
- 50.Wu Z, Rosen E D, Brun R, Hauser S, Adelmant G, Troy A E, McKeon C, Darlington G J, Spiegelman B M. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]