Abstract
Objective:
To determine whether prophylactic azithromycin is associated with the vaginal bacterial microbiome and clinical outcomes in subfertile women undergoing in vitro fertilization (IVF).
Design:
Prospective exploratory cohort study.
Setting:
Single academic fertility center.
Patients:
Subfertile women aged 18–43 years undergoing their first IVF cycle and fresh embryo transfer.
Intervention:
Primary exposure to prophylactic azithromycin (1 g orally) once at baseline.
Main Outcome Measures:
The primary outcome was the effect of azithromycin on the vaginal microbiome compared with a no-azithromycin group at 3 time points throughout the IVF cycle (baseline, retrieval, and embryo transfer). The secondary outcomes were associations of vaginal bacterial communities with clinical outcomes.
Results:
A planned a priori exploratory cohort of 27 subjects (12 in the azithromycin treatment group and 15 in the no-azithromycin group) contributed 79 vaginal swabs for the analysis as part of an ongoing randomized, controlled noninferiority trial. No specific taxa were associated with azithromycin or pregnancy at any time point. Azithromycin did not affect alpha diversity or community stability. Although there were trends of a lower bacterial load and higher percentage of Lactobacillus species in the azithromycin group at the time of transfer, these were not statistically significant. In women who did not become pregnant, the percentage of Lactobacillus species was lower (P = .048; Hodges-Lehmann estimate of difference, 0.41; 95% confidence interval, 0.08–0.65) and the change in community composition over time was higher. The percentage of Lactobacillus species at baseline was not predictive of the percentage of Lactobacillus species at the time of embryo transfer.
Conclusions:
Prophylactic azithromycin at baseline is not associated with changes in vaginal bacterial communities. Bacterial community features at the time of embryo transfer are associated with pregnancy. Bacterial community structures at baseline are not predictive of those at the time of embryo transfer.
Clinical Trial Registration Number:
Keywords: Vaginal microbiome, prophylactic antibiotics, reproductive microbiome, IVF outcomes
Approximately 1 in 8 couples receive infertility services in their lifetime. However, despite the increasing usage of these technologies, with over 270,000 in vitro fertilization (IVF) cycles initiated in 2018, the success rate, as measured using live birth rates, is just <50% in women <35 years of age (1). Studies of the female reproductive tract microbiome (defined as the collective genomes of microbes, including bacteria, viruses, and fungi) in subfertile women undergoing IVF are limited despite the fact that the characteristics of vaginal bacterial communities have been linked to miscarriage, preterm delivery, chorioamnionitis, and pre-eclampsia and women undergoing IVF are at an increased risk of adverse pregnancy outcomes (2–4).
A low-diversity, Lactobacillus-dominated microbiome in the female reproductive tract has been thought to be a sign of optimal reproductive health, whereas an increased microbial diversity has been shown to be associated with poorer reproductive outcomes (4–7). A recent prospective study of women undergoing IVF has demonstrated that a Lactobacillus-dominated endometrial environment is associated with higher rates of implantation, clinical pregnancy, and live birth (8). Additionally, infertile women undergoing IVF have been shown to be less likely to have Lactobacillus-dominated endometrial and vaginal microbiomes than healthy, asymptomatic controls (9). Taken together, these findings suggest that the female reproductive tract microbiome during the immediate preconception period plays an important role in the ability to achieve and maintain a healthy pregnancy. Although studies are being conducted to evaluate the vaginal microbiome and its relationship with reproductive outcomes, the optimal timing of predictive sampling is yet to be determined (10–13). Furthermore, the consideration of iatrogenic factors that may alter vaginal bacterial communities during an IVF cycle is lacking.
A common step in IVF protocols is to prophylactically administer antibiotics before embryo transfer to reduce the theoretical risk of an ascending bacterial infection at the time of embryo transfer; however, little evidence supports the utility of this practice (14–19). In fact, such a practice may influence IVF success rates by altering bacterial communities of the reproductive tract. Thus, it is important to assess the potential effect of prophylactic antibiotics on vaginal bacterial communities.
To address this issue, we conducted an a priori prospective exploratory cohort study based on 2 goals: to explore the effect of prophylactic azithromycin treatment on the vaginal bacterial microbiome longitudinally throughout an IVF cycle and determine whether the characteristics of the vaginal bacterial communities are associated with clinical outcomes. We hypothesized that vaginal bacterial community structures are influenced by prophylactic antibiotic exposure and that their characteristics are associated with clinical outcomes following IVF.
MATERIALS AND METHODS
This was a planned, a priori, exploratory, longitudinal cohort study conducted as a part of an ongoing randomized, controlled noninferiority trial at a single academic institution. Subjects in the parent trial (Washington University in St. Louis Institutional Review Board no. 201709018) who were registered at clinicaltrials.gov (NCT03386227) were randomly assigned to an azithromycin group or no-azithromycin group to assess the primary outcome of clinical pregnancy. The randomization in the parent trial was performed in 1:1 ratio of prophylactic azithromycin to no azithromycin in a block-randomized fashion by a computer random-number generator. Couples randomized to the antibiotic group received the current standard regimen at our clinic: 1 g of azithromycin orally once for both partners on the day of initiation of injectable gonadotropins for controlled ovarian stimulation. Couples randomized to the no-azithromycin treatment group did not receive prophylactic azithromycin. The remainder of their IVF protocol remained at the discretion of the treating reproductive endocrinologist. All the subjects received 2 g of intravenous cefazolin for surgical prophylaxis at the time of egg retrieval, which is the standard regimen at our center. The treating embryologists were blinded to the patient treatment group allocation.
The subjects were eligible for the exploratory longitudinal cohort presented here if the female partner was aged 18–43 years and undergoing her first IVF cycle with a fresh embryo transfer. The exclusion criteria included any contraindication to the antibiotic treatment, not intending to undergo embryo transfer (patients undergoing fertility preservation and oocyte donors), requiring the use of extended antibiotic coverage at the time of egg retrieval, already taking antibiotics for any reason (e.g., upper respiratory tract infection), a history of pelvic infection, using donor or frozen sperm, or planning on undergoing limited insemination (inseminating a limited number of the eggs retrieved).
The subjects were enrolled from February 2018 to September 2018. The primary exposure for this analysis was to azithromycin or no azithromycin, and the primary outcome was the vaginal microbiome’s (viral and bacterial) characteristics (20). The secondary outcomes were IVF cycle characteristics, clinical pregnancy rate, and live birth rate. The analysis of this cohort study was performed according to The Strengthening the Reporting of Observational Studies in Epidemiology guidelines (20, 21). Written informed consent was obtained from each subject before enrollment.
A convenience sample size targeting 50 patients for enrollment was initially estimated. Because of low recruitment in the parent trial, 27 subjects were enrolled in the exploratory study. The STATA 12.0 (College Station, TX) statistical program was used to compare baseline and demographic characteristics between those who received azithromycin and those who did not using the Student’s t-test, chi-square test, or Fisher’s exact test, as appropriate.
Serial vaginal swabs were obtained at 3 time points across the IVF cycle in a standardized fashion: immediately before baseline ultrasound (before azithromycin administration to the treatment group); immediately before egg retrieval (before cefazolin administration to all the patients); and immediately before embryo transfer. A sterile speculum was inserted into the vaginal canal, and a dry, sterile, dual-tipped rayon swab (Starplex Scientific, Etobicoke, Ontario, Canada) was applied 3–5 times to both the lateral sidewalls of the vaginal canal. The swab was immediately placed into a sterile collection tube and stored at −80°C until batch analysis.
The QIAmp BiOStic Bacteremia DNA Kit (Qiagen, Germantown, MA) was used to extract DNA. To survey the bacterial communities of each sample, amplicons were generated from the V4 region of 16S rRNA gene, and Illumina sequencing adapters were added in a single-step polymerase chain reaction (PCR) to create sequencing libraries (22). Nucleotide sequence reads were generated on the Illumina MiSeq instrument (Illumina, Inc,. San Diego, CA) (2 × 250 base pair sequences). The sequences were processed and classified using mothur software, as described (22). Chimeric sequences were screened using UCHIME (23). The sequences were classified using a naïve Bayesian classifier (24) with the Ribosomal Database Project training set, version 9 (25). The Lactobacillus species was identified based on the National Center for Biotechnology Information’s Basic Local Alignment Search Tool nucleotide sequence alignment with reference genomes (26). The sequences were subsampled to 4,627 sequences for each sample. One sample collected just before embryo transfer could not be amplified and sequenced and was excluded. The bacterial community structures were analyzed with the R statistical programming language using Vegan and Labdsv libraries for ecological analyses (27, 28). Alpha diversity (including richness, Shannon’s and Simpson’s diversity indices, and Pielou’s evenness) was calculated. Alpha diversity assesses the characteristics of the community within each sample based on criteria such as the number of different taxa present and their relative abundance. Beta diversity (Bray-Curtis dissimilarity), which is a measure of dissimilarity in the community structure of 2 samples, was also calculated. Nonmetric multidimensional scaling plots were generated using the Labdsv library to visually cluster the samples based on dissimilarity in the community structures. Adonis (Vegan) was used to perform permutational multivariate analysis of variants associated with Bray-Curtis dissimilarities. Bacterial load quantification was performed by quantitative PCR assay using the V4 region’s amplification primers (F-551-GTGYCAGCMGCCGCGGTAA, R-806-GGACTACNVGGGTWTCTAAT) and TB Green qPCR Advantage Premix (Takara Bio USA, Inc., Mountainview, CA) with the following cycle conditions: 95°C, 30 seconds; 40 cycles at 95°C, 5 seconds; 55°C, 20 seconds; and 72°C, 90 seconds (29). An estimate of the Lactobacillus bacterial load was calculated by multiplying the percentage of Lactobacillus determined a relative abundance assay by the total bacterial load determined using the quantitative PCR assay, as described (30).
The Wilcoxon’s rank sum test was used to compare diversity metrics, bacterial loads, and the relative abundance of Lactobacillus species between the azithromycin and no-azithromycin groups and between subjects with clinical pregnancy and those with a negative cycle outcome. For the Wilcoxon’s rank sum test, Hodges-Lehmann (HL) estimate of difference was used to determine 95% confidence interval (CI). A generalized linear model was used to explore the features of the bacterial communities (random effects tested: percentage of Lactobacillus and antibiotic treatment arm) associated with IVF outcomes (response variables tested: clinical pregnancy and live birth). All analyses were performed using the R (www.r-project.org) and STATA 12.0 (College Station, TX) statistical programs.
RESULTS
Twenty-seven subjects contributed 79 vaginal swabs for the analysis, including 27 at the baseline, 26 before egg retrieval, and 26 before embryo transfer. The mean age of our group was 34 ± 4.4 years, and the mean body mass index was 30.74 ± 7.47 kg/m2. There were no significant differences between the treatment and no-azithromycin groups in the baseline or clinical characteristics (Table 1), cycle-level variables (Table 2), IVF outcomes (Table 3), and clinical outcomes in terms of the rates of clinical pregnancy, implantation, miscarriage, or live birth (Table 4). No adverse side effects were reported for either group.
TABLE 1.
Demographic and clinical characteristics of study participants.
| Variable | No antibiotic (n = 15) | Antibiotic (n = 12) | P value |
|---|---|---|---|
| Age (y), mean (SD) | 32.27 (3.8) | 35.25 (4.67) | .07 |
| BMI (kg/m2), mean (SD) | 30.94 (8.1) | 30.48 (7) | .88 |
| Race, no. (%) | 1 | ||
| White | 14 (93.3) | 12 (100) | |
| Asian | 1 (6.7) | 0 (0) | |
| Ethnicity, no. (%) | |||
| Non-Hispanic | 15 (100) | 12 (100) | |
| Duration of infertility (mo), median (IQR) | 24 (18, 48) | 18 (8, 32) | .20 |
| Diagnosis, no. (%) | .35 | ||
| Male factor | 5 (33.3) | 3 (25) | |
| Unexplained | 1 (6.7) | 4 (33.3) | |
| Tubal factor | 2 (13.3) | 1 (8.3) | |
| Endometriosis | 1 (6.7) | 1 (8.3) | |
| Ovulatory dysfunction | 1 (6.7) | 1 (8.3) | |
| PCOS | 4 (26.7) | 0 | |
| DOR | 1 (6.7) | 2 (16.7) | |
| Protocol, no. (%) | .05 | ||
| Long agonist | 9 (60) | 7 (58.3) | |
| Antagonist | 5 (33.3) | 1 (8.3) | |
| Flare | 0 | 4 (33.3) | |
| Luteal E2 flare | 1 (6.7) | 0 | |
| Prior live birth, no. (%) | 1a | ||
| Yes | 3 (20) | 2 (16.7) | |
| No | 12 (80) | 10 (83.3) |
Note: BMI = body mass index; DOR = diminished ovarian reserve; E2 = estradiol; IQR = interquartile range; SD = standard deviation; PCOS polycystic ovary syndrome; SD standard deviation.
Fisher’s exact test.
TABLE 2.
Cycle-level variables.
| Variable | No antibiotic (n = 15) | Antibiotic (n = 12) | P value |
|---|---|---|---|
| AMH (ng/mL), mean (SD) | 2.16 (1.15) | 1.47 (0.62) | .07 |
| Total gonadotropin (U), mean (SD) | 2,525 (1,315) | 2,893 (1,005) | .43 |
| Peak estradiol (pg/mL), mean (SD) | 2,255 (1,068) | 2,005 (825) | .51 |
| Day of embryo transfer, no. (%)a | .24b | ||
| 3 | 7 (50) | 9 (75) | |
| 5 | 7 (50) | 3 (25) | |
| No. of embryos transferred, median (IQR) | 1 (1, 2) | 2 (1, 2) | .06 |
Note: AMH = antimüllerian hormone; IQR = interquartile range; SD = standard deviation.
Out of 14 in the no-antibiotic group.
Fisher’s exact test.
TABLE 3.
IVF outcomes.
| Variable | No antibiotic (n = 15) | Antibiotic (n = 12) | P value |
|---|---|---|---|
| No. of eggs retrieved | 14 (5, 20) | 10 (7, 14) | .25 |
| No. of mature eggs | 11 (4, 14) | 9 (7, 11) | .52 |
| Fertilization rate, mean (SD) | 6.3 (4.5) | 6.7 (3.3) | .83 |
| No. of blastocysts | 1 (0, 4) | 0 (0, 3) | .32 |
| No. cryopreserved | 1 (0, 3) | 0 (0, 3) | .27 |
| Blastocyst utilization ratea | 0.25 (0, 0.5) | 0 (0, 0.5) | .26 |
Note: IQR = interquartile range; IVF = in vitro fertilization; SD = standard deviation.
All values are expressed as median (IQR) unless designated otherwise. Mann-Whitney U test (nonparametric) was performed for discrete numbers.
Out of 14 in the no-antibiotic group.
TABLE 4.
Clinical outcomes.
| Variable | No antibiotic (n = 15) | Antibiotic (n = 12) | P value |
|---|---|---|---|
| Clinical pregnancy, no. (%) | 9 (60) | 7 (58.3) | .93 |
| Implantation rate, mean (SD) | 0.57 (0.47) | 0.4 (0.42) | .3 |
| Miscarriage, no. (%) | 0 | 1 (3.7) | .44a |
| Live births, no. (%) | 9 (60) | 5 (41.7) | .45 |
Note: SD = standard deviation.
Fisher’s exact test.
We generated 16S rRNA gene sequencing data for 26 samples at each time point; 1 sample collected before embryo transfer could not be amplified for sequencing. Most of the communities were Lactobacillus-dominant. L. crispatus and L. iners were the 2 most prevalent species (Fig. 1), although we also observed L. gasseri, L. johnsonii, and L. jensenii. Unexpectedly, L. rhamnosis, which is sometimes considered a probiotic, was prevalent in a sample from a woman who did not report taking probiotics. Some samples also contained more diverse communities, which included Gardnerella, Prevotella, Ureaplasma, and others (Fig. 1).
FIGURE 1.
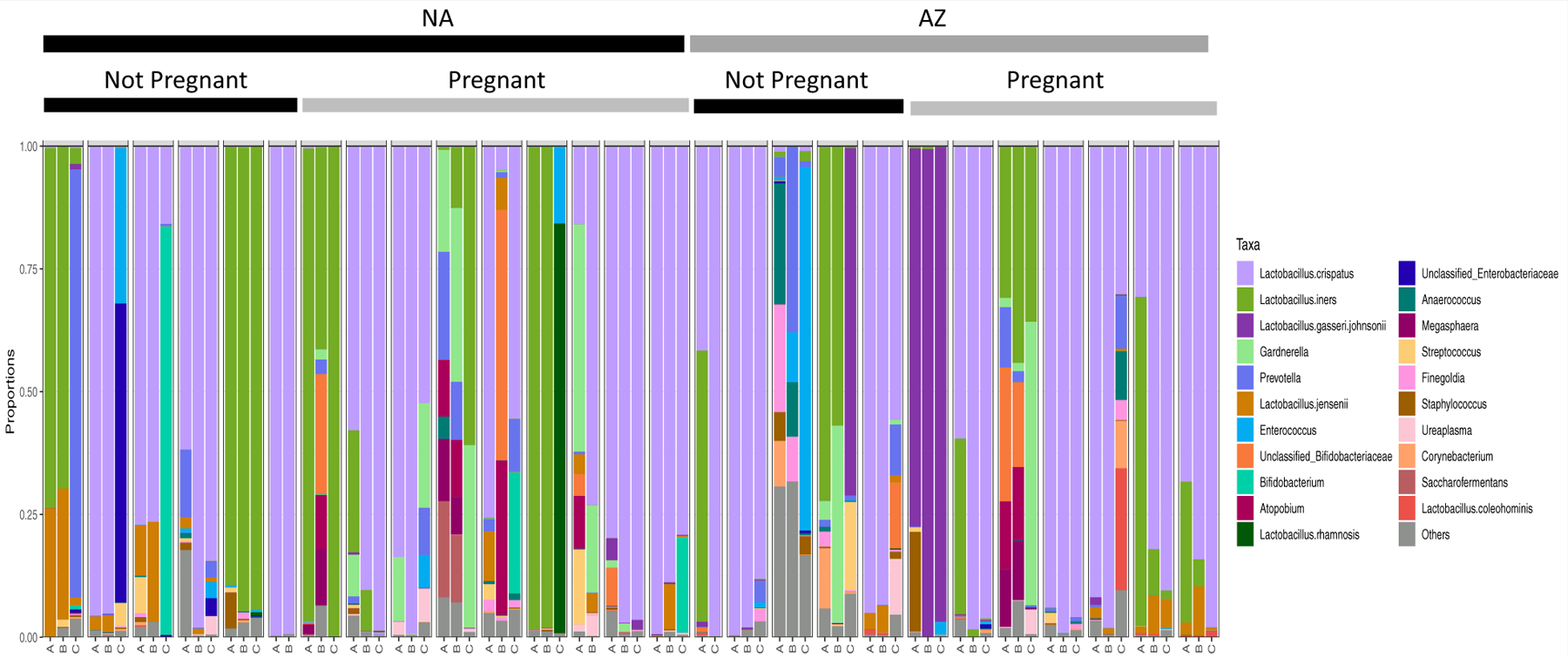
Vaginal bacterial community structures in patients undergoing IVF over 3 sampling intervals. The relative abundance of each taxon is represented by a different color. The sequential samples obtained from each subject are represented in a group, with sample A collected at baseline, B just before egg retrieval, and C just before embryo transfer. Subjects who did not undergo prophylactic azithromycin treatment are marked with a black bar labeled “NA,” and those receiving azithromycin are marked with a gray bar labeled “AZ.” Within each of these groups, subjects who did or did not become pregnant are annotated. According to Linear discriminant analysis Effect Size analysis, no specific taxa were associated with azithromycin treatment or pregnancy. IVF = in vitro fertilization.
Community composition was typically dynamic over the 3 sampling intervals (Fig. 1 and Supplemental Fig. 1, available online). No specific taxa were associated with azithromycin treatment or pregnancy at any of the 3 time points. The nonmetric multidimensional scaling analysis did not reveal associations between community composition and the time point at which the samples were collected (baseline, egg retrieval, or embryo transfer) (Supplemental Fig. 1A), azithromycin treatment (Supplemental Fig. 1B), or clinical pregnancy, as determined by the cycle’s outcome (Supplemental Fig. 1C).
Azithromycin treatment did not significantly affect the bacterial load. The baseline bacterial loads of the communities were similar for the azithromycin and no-azithromycin groups (Supplemental Fig. 2A, available online), and they remained similar at the time of egg retrieval (Supplemental Fig. 2B). There was a trend of lower bacterial loads in the azithromycin group compared with those in the no-azithromycin group before embryo transfer, but it was not statistically significant (P = .09; HL, 0.46; 95% CI, −0.04 to 1.14) (Supplemental Fig. 2C).
Azithromycin treatment also did not affect the Shannon diversity of the communities. The baseline Shannon diversities of the communities were similar for the azithromycin and no-azithromycin groups (Fig. 2A), and they remained similar at the time of egg retrieval and before embryo transfer (Fig. 2B and 2C). The same was true for other measures of alpha diversity as well (Supplemental Fig. 3, available online).
FIGURE 2.
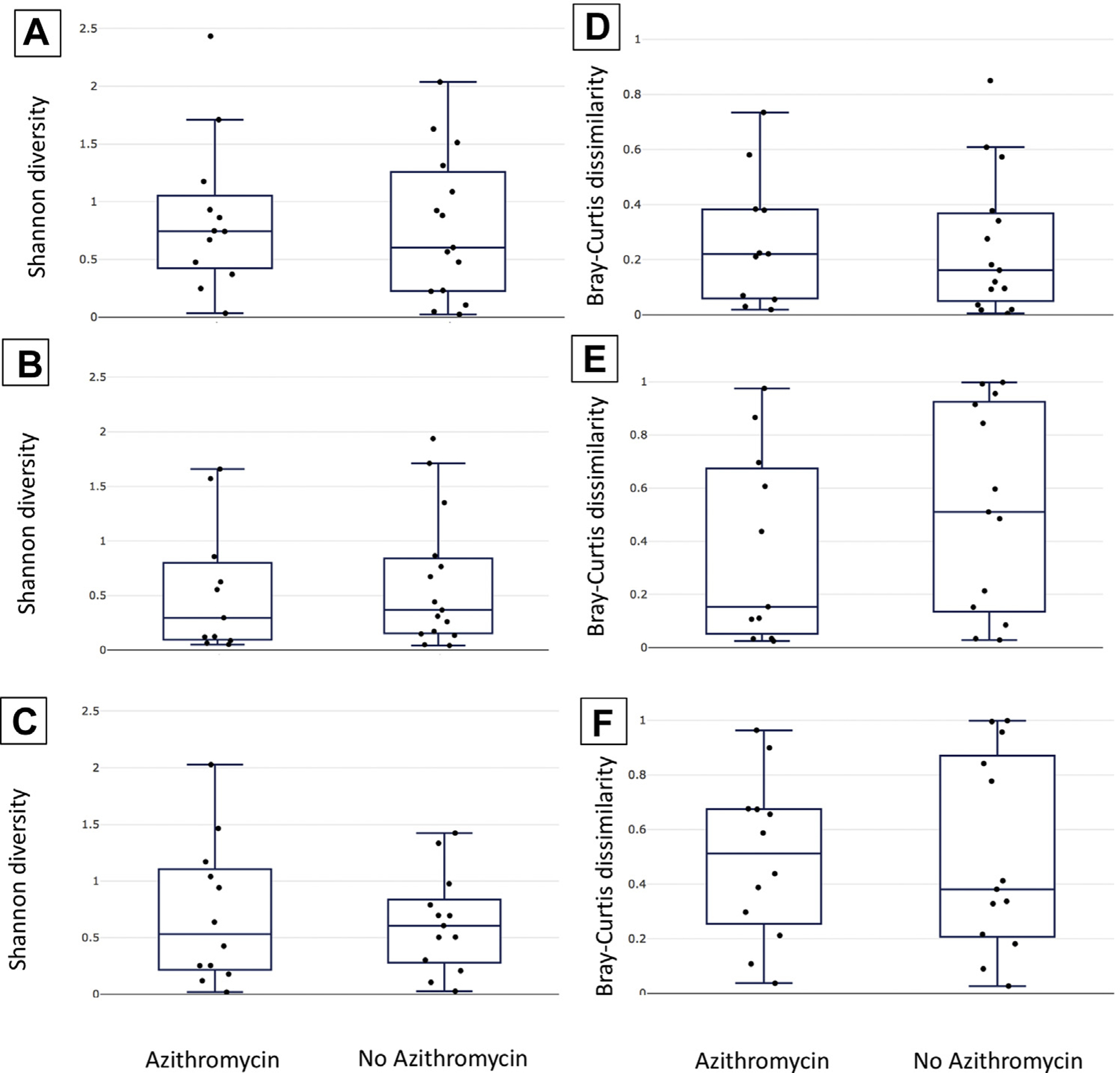
Alpha diversity and community stability were not associated with azithromycin treatment. Shannon diversity (A–C) and Bray-Curtis dissimilarity (D–F) are plotted using box and whisker plots to compare the azithromycin and no-azithromycin groups. The median is indicated by a horizontal line, quartiles by boxes, and minimum and maximum by whiskers. Shannon diversities are shown for samples collected at the time of enrollment (panel A), egg retrieval (panel B), and embryo transfer (panel C). Bray-Curtis dissimilarities (representing the degree of change in community structure) are shown for the time period between enrollment and egg retrieval (D), egg retrieval and embryo transfer (E), and enrollment and embryo transfer (F).
We hypothesized that there is a greater change in the community structures of the samples collected at the baseline and at the time of egg retrieval between women who received azithromycin and those who did not. To test this, we calculated the Bray-Curtis dissimilarities between samples collected from the same woman over time, and then, we compared the values of the group that received azithromycin with those of the group that did not. We found that there was no difference in the community stability between the groups associated with azithromycin treatment from the baseline to egg retrieval (Fig. 2D), egg retrieval to embryo transfer (Fig. 2E), or the baseline to embryo transfer (Fig. 2F).
When we stratified each group of women based on whether they became pregnant or had a live birth, we observed that there were trends in several features that were associated with outcomes independent of azithromycin treatment. In the samples taken just before embryo transfer, the Shannon diversity was slightly higher and bacterial loads were lower in the group of women who did not become pregnant compared with the group of women who did become pregnant (Fig. 3A and 3B, respectively). Additionally, just before embryo transfer, the percentage of Lactobacillus species in the community was lower in women who did not become pregnant (P = .048; HL, 0.41; 95% CI, 0.08–0.65) in the azithromycin group (Fig. 3C), and this trend held true when we considered the percentage of all Lactobacillus species, excluding L. iners, a species that has been shown to be associated with adverse pregnancy outcomes (Supplemental Fig. 4, available online) (4–6, 8, 9). The estimated total Lactobacillus load followed the same trends as the total bacterial loads and percentage of Lactobacillus species, consistent with our findings of a positive correlation between bacterial load and the percentage of Lactobacillus species (Supplemental Fig. 5, available online). Finally, the change in the community composition from the baseline to just before embryo transfer was higher in those who did not become pregnant (Fig. 3D). The same trends were observed when women with live births were compared with those without live births (Supplemental Figs. 5 and 6, available online).
FIGURE 3.
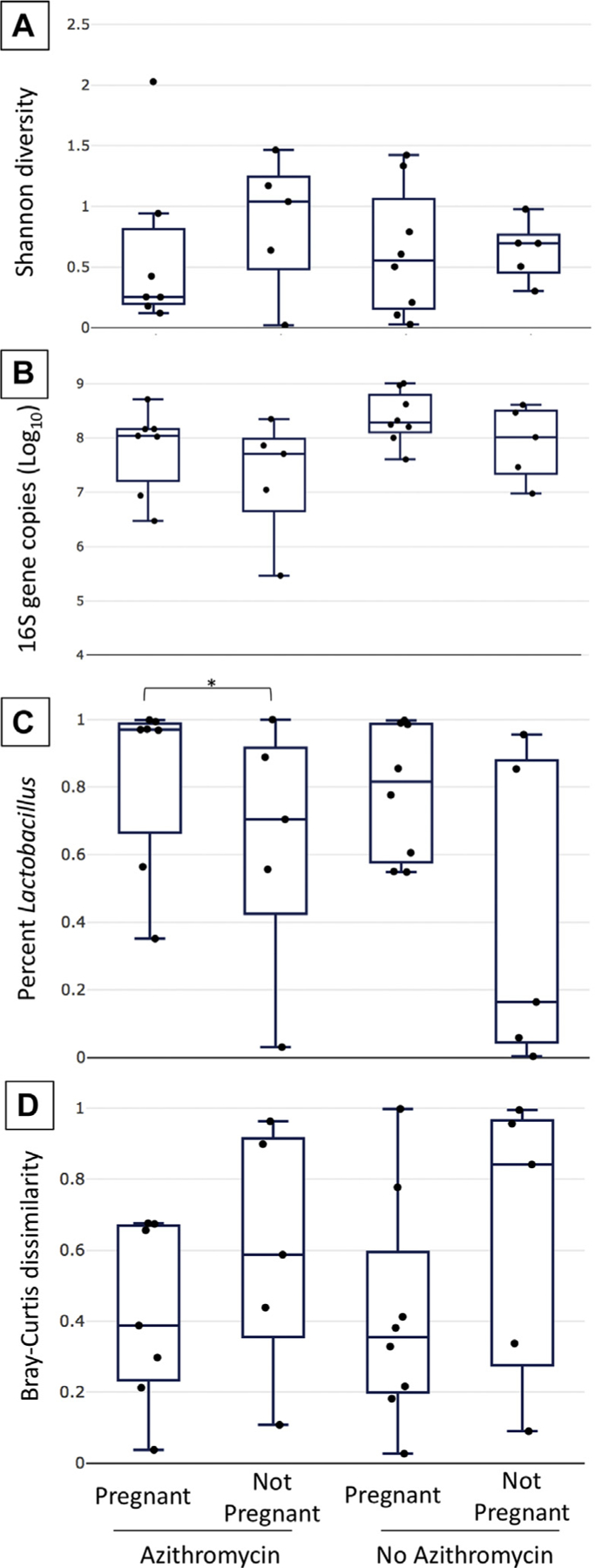
Bacterial community characteristics associated with pregnancy. Shannon diversity (A), 16S rRNA gene copy number (B), the percentage of Lactobacillus species in the samples (C), and the Bray-Curtis dissimilarity measure (D) are represented by box and whisker plots to compare groups based on azithromycin treatment and pregnancy outcome. The median is indicated by a horizontal line, quartiles by boxes, and minimum and maximum by whiskers. In the azithromycin group, the percentage of Lactobacillus species was statistically significantly different between the pregnant and nonpregnant groups (P = .048; HL, 0.41; 95% CI, 0.08–0.65). CI = confidence interval; HL = Hodges-Lehmann estimate of difference.
When we evaluated the data using a generalized linear model, we found that a higher percentage of Lactobacillus species just before embryo transfer was associated with higher rates of clinical pregnancy (P = .045; HL, 3.01; 95% CI, 0.33–6.44) and live birth (P = .019; HL, 5.36; 95% CI, 1.74–11.18). No association was found at the baseline or at the time of egg retrieval. Additionally, the percentage of Lactobacillus species was not stable in the individuals throughout the IVF cycle (Fig. 1 and Supplemental Fig. 7, available online).
DISCUSSION
To our knowledge, this is the first study to examine the effect of antibiotics on vaginal bacterial communities in subfertile women undergoing IVF with a fresh embryo transfer. The results showed that in vaginal microbiome samples taken at the time of egg retrieval and embryo transfer, changes in the taxonomic composition, alpha diversity, and beta diversity are not associated with azithromycin exposure at the time of gonadotropin initiation. This result could be due to the lack of a marked effect of azithromycin on the vaginal bacterial communities; an effect that is masked by rapid reconstitution of the vaginal microbiome after treatment; or an effect that is masked by the dynamic changes occurring due to the prescribed treatments.
There were trends of lower bacterial loads and higher percentages of Lactobacillus species in the azithromycin group at the time of embryo transfer compared with those in the no-azithromycin group, although these were not statistically significant. The potential effect of prophylactic antibiotic administration in all the subjects at the time of oocyte retrieval must be taken into consideration as well because its individual and potentiated effects could not be determined and may have influenced the bacterial load. Taken together, these data suggest that azithromycin administered at the start of the cycle has at the most a minor impact on bacterial community features overall.
Routine antibiotic prophylaxis during an IVF cycle to improve embryo transfer success rates is commonly performed, but little evidence exists to support its use. A Cochrane review conducted in 2012 analyzed randomized, controlled trials from the literature, and although 4 studies were identified for potential inclusion, 3 were excluded (18). Only 1 randomized, controlled trial that was powered to determine a difference in catheter contamination rate has addressed this issue. Although antibiotics significantly reduced the catheter contamination rate (49.4% in the medicated group vs. 62.3% in the nonmedicated group, P = .03), there was no significant difference in the clinical pregnancy rate (56.1% vs. 61.5%, P = .10) (16). Our data and this study suggest that prophylactic antibiotics may not be necessary or useful in IVF treatment.
The effect of broad-spectrum antibiotic exposure in the immediate preconception period has not been well described, but it might result in an unintended outcome of altering the follicular and endometrial microbiomes, which has received increasing attention for playing a significant role in reproductive competence and assisted reproductive technology outcomes. Approximately 40% of pregnant women in the United States receive antibiotics before delivery to prevent maternal and neonatal infectious complications (31). Although the benefit of antibiotics during pregnancy cannot be denied, prolonged or inappropriate exposure can have real and adverse consequences, including increased prevalence of antibiotic resistance, and, through influence on the microbiome, can result in longstanding neonatal immunologic and metabolic consequences (32). One possible mechanism for these consequences is that broad-spectrum antibiotics do not discriminate between protective and pathogenic bacteria, and the absence of protective bacteria may have an equally deleterious effect as that of pathogenic bacteria. The minimal association we observed between antibiotic treatment and bacterial community might be due to the antibiotic type or the duration or timing of exposure.
Few studies have examined longitudinal changes in the bacterial microbiome throughout an IVF cycle (13, 33). Here, we demonstrated that bacterial community structures at baseline are not predictive of those at the time of embryo transfer, suggesting that the timing of sampling is important to improve clinical outcomes. Prior work has demonstrated that vaginal bacterial communities undergo dynamic changes throughout the menstrual cycle, which are thought to be influenced by corresponding hormonal changes (11, 34). Given the substantial hormonal shifts experienced throughout an IVF cycle, a limited number of studies, including the study we have presented here, have had similar findings (13, 33). Jakobsson et al. (33) have demonstrated that vaginal Lactobacillus became dominated 1 of 3 species of normal vaginal flora (L. crispatus, L. gasseri, and/or L. jensenii) with rising serum estradiol levels. Hyman et al. (13) assessed 4 time points throughout an IVF cycle and 1 during the first trimester of pregnancy in 30 infertile women undergoing IVF and a fresh embryo transfer. Their study demonstrated that species diversity varies with changes in hormonal milieu, as measured by serum progesterone and estradiol levels. Consistent with these observations, we found a decreasing trend of alpha diversity at the time of egg retrieval compared with that at the other 2 time points, with Lactobacillus dominance in most samples. This occurred in both the azithromycin and no-azithromycin groups and was likely driven by peak hormonal changes at that time point. Hyman et al. (13) concluded that vaginal bacterial diversity at the time of transfer, as opposed to other time points, is most associated with clinical outcomes, with higher diversity in women who do not become pregnant. However, all the subjects in both these studies received prophylactic antibiotics and/or glucocorticoids as a routine part of treatment; therefore, their potential effects could not be determined (13, 33). Our data support these conclusions and suggest that prophylactic antibiotics do not affect the associations between microbial community structures and pregnancy outcomes after IVF.
Consistent with other studies, the percentage of Lactobacillus species at the time of embryo transfer was predictive of clinical pregnancy in our cohort. In our study, this association was observed in the sample taken at the time of embryo transfer but not in the samples taken at the baseline or at the time of egg retrieval. Similarly, a prospective study of 35 patients undergoing IVF has found that a Lactobacillus-dominated endometrial environment at the time of embryo transfer is associated with higher rates of implantation (60.7% vs. 23.1%, P = .02), pregnancy (70.6% vs. 33.3%, P = .03), ongoing pregnancy (58.8% vs. 13.3%, P = .02), and live birth (58.8% vs. 6.7%, P = .002) (8). This is in contrast to a recent prospective study by Koedooder et al. (35), which concluded that vaginal microbiome profiling before the start of an IVF cycle can be used to predict the cycle’s outcome. The vaginal microbiome of 192 women was analyzed within 3 months before IVF cycle initiation. Women with a low percentage of Lactobacillus species in their vaginal sample had a lower likelihood of successful implantation. In their cohort, a higher relative abundance of L. jensenii (>35%) was associated with unfavorable outcomes. A high relative abundance of L. jensenii was uncommon in the samples obtained from our cohort; therefore, we were unable to confirm this association in our baseline samples or extrapolate it to the other time points we evaluated. In the study by Koedooder et al. (35), the investigators also found that a higher relative abundance of L. crispatus was associated with poorer outcomes in women with Lactobacillus-dominant communities. We did not observe an association between outcomes and any Lactobacillus species at any of our time points. It is possible that the baseline bacterial community characteristics of their study population were different from those of ours or that our small sample size limited our ability to generalize their results to our population. Approximately 9% of the population in the study by Koedooder et al. (35) was exposed to antibiotics at some point during the 3 months leading up to IVF (medication, dose, and timing were not reported), and the samples were self-collected, which may also have limited the accuracy of the results. Prior studies have not commented on or examined the potential effect of antibiotics on Lactobacillus, which is an important consideration.
The strengths of our study include its novel findings regarding the effect of prophylactic antibiotics on the vaginal bacterial microbiome in subjects with diverse infertility diagnoses who were undergoing a fresh embryo transfer. This is one of the few studies that have examined bacterial trends longitudinally throughout an IVF cycle, and it suggests that the timing of sampling is important for accurate characterization of clinical outcomes and determining their association with the timing of sampling. Further, we included live birth outcomes, which is arguably the most important clinical outcome.
There are several limitations of this study that must be considered. Given the exploratory nature and small number of women included, this study was not powered to detect significant differences in the clinical endpoints. This exploratory study revealed that the effect sizes were small, and, therefore, future studies will require larger groups of subjects to be appropriately powered. Antibiotic prophylaxis protocols vary widely by institution, and our clinic’s administration of azithromycin at the baseline may not be generalizable to other practices. Although the groups were randomized, there was no placebo control. Additionally, we routinely administered cefazolin at the time of egg retrieval, which may have acted as a potential confounder, and we could not determine the exact impact this had on antibiotic exposure at the baseline or in isolation. This study included a predominantly young, non-Hispanic cohort undergoing a fresh embryo transfer, which may have further limited its generalizability to other populations of women undergoing a frozen embryo transfer; racial and ethnic differences in the microbiome have been noted (36). Although the study was limited to patients undergoing their first fresh IVF cycle, different stimulation protocols were used. Finally, the mean age was higher and antimüllerian hormone levels were lower in the azithromycin group, which may have contributed to a potential bias.
In conclusion, this small, prospective, exploratory, pilot study shows that in vaginal microbiome samples taken at the time of egg retrieval and fresh embryo transfer, changes in taxonomic composition, alpha diversity, and beta diversity are not associated with azithromycin exposure at the time of gonadotropin initiation. Although there were trends of lower bacterial loads and higher percentages of Lactobacillus species in the azithromycin group at the time of embryo transfer compared with those in the no-azithromycin group, these were not statistically significant. Further, we demonstrated that bacterial community structures at baseline are not predictive of those at the time of embryo transfer, which highlights the importance of timing in the assessment of vaginal microbiome to determine its associations with reproductive outcomes. Future studies should include a larger, more racially and ethnically diverse sample cohort stratified by age and stimulation protocol and should further evaluate the timing of sampling in relation to antibiotic exposure and embryo transfer to optimize IVF outcomes.
Supplementary Material
Acknowledgments
Supported by the National Institute of Child Health and Human Development of the National Institutes of Health (NIH) (grant no. T32HD055172) as a part of the Reproductive Epidemiology Training Program at Washington University in St. Louis. (A.M.E.)
Supported by the Washington University Institute of Clinical and Translational Sciences, which is, in part, supported by the NIH/National Center for Advancing Translational Sciences (NCATS) (CTSA grant no. UL1TR002345).
Supported by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital.
Footnotes
A.M.E. has nothing to disclose. M.J.S. has nothing to disclose. B.S.B. has nothing to disclose. J.K.R. has nothing to disclose. B.N.H. has nothing to disclose. H.G. has nothing to disclose. E.S.J. has nothing to disclose. K.M.W. has nothing to disclose.
Discuss: You can discuss this article with its authors and other readers at https://www.fertstertdialog.com/posts/xfss-d-20-00075
REFERENCES
- 1.Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, Kim YM. Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am J Obstet Gynecol 2015;213:S29–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stout MJ, Zhou Y, Wylie KM, Tarr PI, Macones GA, Tuuli MG. Early pregnancy vaginal microbiome trends and preterm birth. Am J Obstet Gynecol 2017;217:356.e1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox C, Eichelberger K. Maternal microbiome and pregnancy outcomes. Fertil Steril 2015;104:1358–63. [DOI] [PubMed] [Google Scholar]
- 4.Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The vaginal microbiome and preterm birth. Nat Med 2019;25:1012–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green KA, Zarek SM, Catherino WH. Gynecologic health and disease in relation to the microbiome of the female reproductive tract. Fertil Steril 2015; 104:1351–7. [DOI] [PubMed] [Google Scholar]
- 7.Al-Nasiry S, Ambrosino E, Schlaepfer M, Morre SA, Wieten L, Voncken JW, et al. The interplay between reproductive tract microbiota and immunological system in human reproduction. Front Immunol 2020;11:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno I, Codoner FM, Vilella F, Valbuena D, Martinez-Blanch JF, et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol 2016;215:684–703. [DOI] [PubMed] [Google Scholar]
- 9.Kyono K, Hashimoto T, Nagai Y, Sakuraba Y. Analysis of endometrial microbiota by 16S ribosomal RNA gene sequencing among infertile patients: a single-center pilot study. Reprod Med Biol 2018;17:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koedooder R, Singer M, Schoenmakers S, Savelkoul PHM, Morre SA, de Jonge JD, et al. The ReceptIVFity cohort study protocol to validate the urogenital microbiome as predictor for IVF or IVF/ICSI outcome. Reprod Health 2018;15:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 2012;4: 132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno I, Franasiak JM. Endometrial microbiota-new player in town. Fertil Steril 2017;108:32–9. [DOI] [PubMed] [Google Scholar]
- 13.Hyman RW, Herndon CN, Jiang H, Palm C, Fukushima M, Bernstein D, et al. The dynamics of the vaginal microbiome during infertility therapy with in vitro fertilization-embryo transfer. J Assist Reprod Genet 2012;29:105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanchin R, Harmas A, Benaoudia F, Lundkvist U, Olivennes F, Frydman R. Microbial flora of the cervix assessed at the time of embryo transfer adversely affects in vitro fertilisation outcome. Fertil Steril 1990;5:866–70. [DOI] [PubMed] [Google Scholar]
- 15.Egbase PE, al-Sharhan M, al-Othman S, al-Mutawa M, Udo EE, Grudzinskas JG. Incidence of microbial growth from the tip of the embryo transfer catheter after embryo transfer in relation to clinical pregnancy rate following in-vitro fertilization and embryo transfer. Hum Reprod 1996;11: 1687–9. [DOI] [PubMed] [Google Scholar]
- 16.Brook N, Khalaf Y, Coomarasamy A, Edgeworth J, Braude P. A randomized controlled trial of prophylactic antibiotics (co-amoxiclav) prior to embryo transfer. Hum Reprod 2006;21:2911–5. [DOI] [PubMed] [Google Scholar]
- 17.Kaye L, Bartels C, Bartolucci A, Engmann L, Nulsen J, Benadiva C. Old habits die hard: retrospective analysis of outcomes with use of corticosteroids and antibiotics before embryo transfer. Fertil Steril 2017;107:1336–40. [DOI] [PubMed] [Google Scholar]
- 18.Kroon B, Hart RJ, Wong BM, Ford E, Yazdani A. Antibiotics prior to embryo transfer in ART. Cochrane Database Syst Rev 2012:CD008995. [DOI] [PubMed] [Google Scholar]
- 19.Tachedjian G, Aldunate M, Bradshaw CS, Cone RA. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res Microbiol 2017;168:782–92. [DOI] [PubMed] [Google Scholar]
- 20.Eskew AM, Stout MJ, Bedrick BS, Riley JK, Omurtag KR, Jimenez PT, et al. Association of the eukaryotic vaginal virome with prophylactic antibiotic exposure and reproductive outcomes in a subfertile population undergoing in vitro fertilisation: a prospective exploratory study. BJOG 2020;127:208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elm EV, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Int J Surg 2014;12:1495–9. [DOI] [PubMed] [Google Scholar]
- 22.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013;79:5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 2014;42:D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 27.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, Minchin PR, et al. Vegan: community ecology package. R package version 2.5-6. 2015;2–3. [Google Scholar]
- 28.Roberts DW. Labdsv: ordination and multivariate analysis for ecology. R package version 2.0-1. 2007;1. [Google Scholar]
- 29.Liu CM, Aziz M, Kachur S, Hsueh PR, Huang YT, Keim P, et al. BactQuant: an enhanced broad-coverage bacterial quantitative real-time PCR assay. BMC Microbiol 2012;12:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tettamanti Boshier FA, Srinivasan S, Lopez A, Hoffman NG, Proll S, Fredricks DN, et al. Complementing 16S rRNA gene amplicon sequencing with total bacterial load to infer absolute species concentrations in the vaginal microbiome. mSystems 2020;5:e00777–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ledger WJ, Blaser MJ. Are we using too many antibiotics during pregnancy? BJOG 2013;120:1450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramasethu J, Kawakita T. Antibiotic stewardship in perinatal and neonatal care. Semin Fetal Neonatal Med 2017;22:278–83. [DOI] [PubMed] [Google Scholar]
- 33.Jakobsson T, Forsum U. Changes in the predominant human Lactobacillus flora during in vitro fertilisation. Ann Clin Microbiol Antimicrob 2008;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickey RJ, Abdo Z, Zhou X, Nemeth K, Hansmann M, Osborn TW 3rd, et al. Effects of tampons and menses on the composition and diversity of vaginal microbial communities over time. BJOG 2013;120:695–704. [DOI] [PubMed] [Google Scholar]
- 35.Koedooder R, Singer M, Schoenmakers S, Savelkoul PHM, Morre SA, de Jonge JD, et al. The vaginal microbiome as a predictor for outcome of in vitro fertilization with or without intracytoplasmic sperm injection: a prospective study. Hum Reprod 2019;34:1042–54. [DOI] [PubMed] [Google Scholar]
- 36.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, et al. The compositionand stability of the vaginalmicrobiotaofnormal pregnant women is different from that of nonpregnant women. Microbiome 2014;2:1–9.24468033 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


