Abstract
Aim:
Recessive genetic variation is thought to play a role in non-Hodgkin lymphoma (NHL) etiology. Runs of homozygosity (ROH), defined based on long, continuous segments of homozygous SNPs, can be used to estimate both measured and unmeasured recessive genetic variation. We sought to examine genome-wide homozygosity and NHL risk.
Methods:
We used data from eight genome-wide association studies of four common NHL subtypes: 3061 chronic lymphocytic leukemia (CLL), 3814 diffuse large B-cell lymphoma (DLBCL), 2784 follicular lymphoma (FL), and 808 marginal zone lymphoma (MZL) cases, as well as 9374 controls. We examined the effect of homozygous variation on risk by: (1) estimating the fraction of the autosome containing runs of homozygosity (FROH); (2) calculating an inbreeding coefficient derived from the correlation among uniting gametes (F3); and (3) examining specific autosomal regions containing ROH. For each, we calculated beta coefficients and standard errors using logistic regression and combined estimates across studies using random-effects meta-analysis.
Results:
We discovered positive associations between FROH and CLL (β = 21.1, SE = 4.41, P = 1.6 × 10−6) and FL (β = 11.4, SE = 5.82, P = 0.02) but not DLBCL (P = 1.0) or MZL (P = 0.91). For F3, we observed an association with CLL (β = 27.5, SE = 6.51, P = 2.4 × 10−5). We did not find evidence of associations with specific ROH, suggesting that the associations observed with FROH and F3 for CLL and FL risk were not driven by a single region of homozygosity.
Conclusion:
Our findings support the role of recessive genetic variation in the etiology of CLL and FL; additional research is needed to identify the specific loci associated with NHL risk.
Keywords: Non-Hodgkin lymphoma, homozygosity, chronic lymphocytic leukemia, follicular lymphoma, diffuse large B-cell lymphoma, marginal zone lymphoma
INTRODUCTION
Disentangling the heritable component of non-Hodgkin lymphoma (NHL) and its subtypes is an active area of research. An early study of familial aggregation in NHL reported an increased risk of NHL among siblings, but not parents or offspring, of an index NHL case[1]. Several subsequent studies found an elevated risk of NHL associated with a first-degree family history of NHL with the highest risks for siblings[2,3]. Studies have also reported higher NHL subtype-specific risks for first-degree relatives of cases affected with a given NHL subtype[4,5], suggesting a degree of subtype specificity. In general, these findings suggest genetic factors are important in NHL etiology and, in particular, the potential role of recessively acting genetic risk alleles, but they also underscore the potential for genetic heterogeneity in susceptibility to different NHL subtypes. Part of the difficulty in characterizing risk and inheritance patterns of NHL subtypes is the limited study sample size, especially when examining specific subtypes. Genome-wide association studies (GWAS) have identified multiple susceptibility loci associated with four major subtypes of NHL[6-11], but a substantial fraction of the disease heritability remains unexplained. Most GWAS performed assume an additive model of genetic risk, which has statistical power to detect allelic associations acting through a variety of mechanisms but may not efficiently detect recessive effects, particularly as minor allele frequency and imputation quality decrease[12]. Therefore, recessively acting loci, particularly those with low minor allele frequency, could be missed by current genome-wide scans and represent potential novel disease-associated loci.
The widespread use of dense genotyping arrays has led to the identification of sizeable genomic regions consisting of consecutive homozygous SNPs in non-consanguineous populations[13]. These runs of homozygosity (ROH) vary in length, with short ROH persisting from ancient relatedness and long ROH of several megabases arising from recent parental relatedness[14]. The use of ROH as a measure of the burden of homozygosity has been demonstrated to perform better at identifying rare, recessive mutations than a conventional SNP-by-SNP analysis[15]. Furthermore, studies incorporating whole-exome sequencing have uncovered an enrichment of deleterious variants in ROH[16]. In recent years, studies have examined the association between ROH and various cancers[17-21], among other complex common diseases and traits[22]. Although no association was observed with the cumulative distribution of ROH, individual ROH were associated with the risk of childhood acute lymphoblastic leukemia[19]. Hodgkin lymphoma has been inconsistently associated with specific ROH and overall homozygosity[20,21]. To our knowledge, no studies have examined ROH in association with adult NHL.
The goal of the present study was to investigate the association of homozygosity with the risk of four major NHL subtypes: chronic lymphocytic leukemia/small chronic lymphocytic leukemia (CLL), diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), and marginal zone lymphoma (MZL). Several measures of homozygosity were tested against NHL risk using data from eight GWAS.
METHODS
We used data from eight previous GWAS of NHL[6,8,10,11] composed of cases and controls of European ancestry [Supplementary Table 1 and 2]. The National Cancer Institute (NCI) NHL GWAS included cases with one of four common NHL subtypes and controls from 22 studies of NHL: 9 prospective cohort studies, 8 population-based case-control studies, and 5 hospital- or clinic-based case-control studies or case series. These 22 studies comprising the NCI NHL GWAS were genotyped using the Illumina OmniExpress or Omni2.5 arrays and analyzed as a single study. The other seven GWAS were the University of California at San Francisco Molecular Epidemiology of Non-Hodgkin Lymphoma study (UCSF2)[23], the University of California at San Francisco Molecular Epidemiology of Non-Hodgkin Lymphoma study (UCSF1) combined with controls from the Nurses’ Health Study (NHS)[24], the Scandinavian Lymphoma Etiology Study (SCALE)[25], the Groupe d’Etude des Lymphomes de l’Adulte (GELA) study combined with controls from the European Prospective Investigation into Cancer, Chronic Diseases, Nutrition, and Lifestyles (EPIC), the Mayo Clinic Case-Control Study of Diffuse Large B-cell Lymphoma (Mayo), the Genetic Epidemiology of CLL Consortium (GEC), and the Utah Chronic Lymphocytic Leukemia Study (Utah). Genotyping was performed on commercially available Illumina and Affymetrix platforms [Supplementary Table 1]. Details, including information on quality control and data cleaning, have been previously reported[6,8,10,11]. All studies obtained informed consent from participants and were approved by their appropriate Institutional Review Boards.
Prior to analysis, additional quality control and filtering were applied to each GWAS separately, including removal of SNPs with a minor allele frequency < 0.05, > 3% missing, or Hardy-Weinberg P-value < 1 × 10−6 among controls, and removal of subjects with call rates < 97%. After quality control metrics, genotype data were available for 10,467 NHL cases, including 3061 CLL, 3814 DLBCL, 2784 FL, and 808 MZL cases, as well as 9374 controls [Supplementary Table 2].
We used PLINK1.9[26,27] to identify ROH; specifically, we used the two-step command --homozyg. In the first step, PLINK1.9 identifies directly genotyped SNPs that are possibly within an ROH by looking at 50-SNP sliding windows across the genome and flagging all SNPs that are encompassed by at least 5% of fully homozygous windows. For this step, we allowed one heterozygous SNP and up to five SNPs with no calls within each window to account for a small amount of possible genotyping error and loss. In the second step, ROH are identified from these sliding windows by requiring a minimum number of consecutive homozygous SNPs. We required at least 100 consecutive homozygous SNPs for each ROH and that these SNPs span at least 1500 kilobases (kb), with at least one SNP every 50 kb and the maximum gap between SNPs of 5000 kb. These parameters were selected with reference to the “ROH_1.5Mb” ROH calling parameters used by Gazal et al.[28] We restricted analyses to the autosomal chromosomes.
To estimate the extent of homozygosity across the genome, we calculated the fraction of the autosome covered by ROH (FROH) by summing the lengths of ROH and dividing by 3 × 109 base pairs as the approximate size of the autosome for all GWAS. As another measure to assess homozygosity, we also quantified and tested differences in relatedness across the genome in our study using a variant of the inbreeding coefficient, F3[29]. F3, which estimates the correlation between uniting gametes, is an alternative to ROH-based estimates with potentially reduced bias and standard errors[30]. We estimated F3 using the -ibc command in PLINK1.9. To estimate the association of FROH and F3 with NHL, we then estimated beta coefficients and standard errors for each GWAS using logistic regression, adjusting for age, sex (except in the UCSF1/NHS study, where all controls were female), fraction of missing SNPs, and the ten principal components of ancestry to account for population stratification. The fraction of missing SNPs was calculated for each participant as the number of SNPs without calls divided by the total number of SNPs genotyped on the array that passed quality control metrics. Associations were combined across GWAS for each subtype of NHL using random-effects meta-analysis implemented with the command “metan” in STATA v15.
After determining ROH as described above, we also tested whether specific genomic regions encompassed by ROH were associated with risk of each of the four NHL subtypes. We divided each autosomal chromosome into “bins” of 500 kb in length. We then calculated the midpoint of each identified ROH and assigned it to the corresponding bin. Each study participant in the analysis was therefore categorized as either homozygous (exposed) or heterozygous (unexposed) at each bin across the autosome. We calculated beta coefficients and standard errors for the association between presence of an ROH in each bin and risk of NHL subtype within each GWAS using logistic regression, adjusting for age, sex (except in the UCSF1/NHS study), fraction of missing SNPs, and ten principal components of ancestry. Results were combined across GWAS using METAL[31], and multiple-testing adjustment was performed using a Bonferroni correction.
RESULTS
Table 1 presents summary statistics for the ROH, FROH, and F3 by GWAS. Among participants, the median total length of ROH ranged from 11,535 to 23,014 kb depending on the GWAS. The median number of ROH per individual ranged from 4 in the UCSF2 GWAS, which used an older and less dense GWAS chip, to 8 in the GEC GWAS, which included familial CLL cases. Median FROH ranged from 0.38% to 0.77% of the autosome. Median F3 ranged from −6.99 × 10−4 to 3.02 × 10−3 [Table 1].
Table 1.
Summary of genome-wide homozygosity measures by GWAS and case-control status*
| Subtype | Study | No. cases |
No. controls |
Median ROH total length (kb) |
IQR ROH total length (kb) |
Median number of ROH |
IQR number of ROH |
Median FROH |
IQR FROH | Median F3 | IQR F3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CLL | NCI | Cases | 2140 | 18593 | 13057-25486 | 7 | 5-9 | 0.62% | 0.44%-0.79% | 2.57 × 10−3 | −1.34 × 10−3−7.49 × 10−3 |
| Controls | 6105 | 18045 | 12674-24396 | 7 | 5-9 | 0.60% | 0.42%-0.81% | 1.48 × 10−3 | −2.07 × 10−3−5.36 × 10−3 | ||
| GEC | Cases | 387 | 23014 | 16496-29054 | 8 | 6-10 | 0.77% | 0.55%-0.97% | 6.41 × 10−4 | −3.66 × 10−3−4.85 × 10−3 | |
| Controls | 294 | 21920 | 14852-28391 | 8 | 6-10 | 0.73% | 0.50%-0.95% | −6.99 × 10−4 | −4.90 × 10−3−3.06 × 10−3 | ||
| UCSF2 | Cases | 213 | 12993 | 7893-19530 | 4 | 3-6 | 0.43% | 0.26%-0.65% | 2.73 × 10−3 | −0.63 × 10−3−8.83 × 10−3 | |
| Controls | 746 | 12173 | 7523-17111 | 4 | 3-6 | 0.41% | 0.25%-0.57% | 1.51 × 10−3 | −2.25 × 10−3−5.72 × 10−3 | ||
| Utah | Cases | 321 | 18806 | 13535-24431 | 7 | 5-9 | 0.63% | 0.45%-0.81% | 3.69 × 10−4 | −2.79 × 10−3−4.71 × 10−3 | |
| Controls | 403 | 17452 | 12288-22866 | 6 | 5-8 | 0.58% | 0.41%-0.76% | 9.47 × 10−4 | −2.09 × 10−3−3.89 × 10−3 | ||
| DLBCL | NCI | Cases | 2621 | 18046 | 12671-24990 | 7 | 5-9 | 0.60% | 0.42%-0.83% | 2.11 × 10−3 | −1.48 × 10−3−6.54 × 10−3 |
| Controls | 6105 | 18199 | 12753-24519 | 7 | 5-9 | 0.61% | 0.43%-0.82% | 1.46 × 10−3 | −2.05 × 10−3−5.39 × 10−3 | ||
| Mayo | Cases | 393 | 16586 | 12167-21774 | 6 | 5-8 | 0.55% | 0.41%-0.73% | 1.67 × 10−4 | −3.03 × 10−3−3.07 × 10−3 | |
| Controls | 172 | 16451 | 11368-23319 | 6 | 4-8 | 0.55% | 0.38%-0.78% | 2.19 × 10−4 | −2.55 × 10−3−2.94 × 10−3 | ||
| UCSF2 | Cases | 253 | 11535 | 5957-19153 | 4 | 3-6 | 0.38% | 0.20%-0.64% | 1.69 × 10−3 | −1.62 × 10−3−6.60 × 10−3 | |
| Controls | 745 | 12182 | 7554-17158 | 4 | 3-6 | 0.41% | 0.25%-0.57% | 1.48 × 10−3 | −2.20 × 10−3−5.82 × 10−3 | ||
| GELA/EPIC | Cases | 547 | 15526 | 10391-21466 | 6 | 4-7 | 0.52% | 0.35%-0.72% | 1.27 × 10−3 | −2.27 × 10−3−5.00 × 10−3 | |
| Controls | 525 | 17327 | 11782-23437 | 6 | 4-8 | 0.58% | 0.39%-0.78% | 2.56 × 10−3 | −0.87 × 10−3−6.14 × 10−3 | ||
| FL | NCI | Cases | 2085 | 17838 | 12387-24319 | 7 | 5-9 | 0.59% | 0.41%-0.81% | 1.41 × 10−3 | −2.14 × 10−3−6.15 × 10−3 |
| Controls | 6105 | 18110 | 12753-24512 | 7 | 5-9 | 0.60% | 0.43%-0.82% | 1.49 × 10−3 | −2.06 × 10−3−5.41 × 10−3 | ||
| UCSF1/NHS | Cases | 119 | 17058 | 12645-22150 | 7 | 5-8 | 0.57% | 0.42%-0.74% | 8.51 × 10−5 | −3.57 × 10−3−3.05 × 10−3 | |
| Controls | 340 | 17144 | 12100-24513 | 6.5 | 5-9 | 0.57% | 0.40%-0.82% | 6.35 × 10−4 | −3.05 × 10−3−4.29 × 10−3 | ||
| UCSF2 | Cases | 209 | 13621 | 8911-19655 | 5 | 3-7 | 0.45% | 0.30%-0.66% | 3.02 × 10−3 | −1.34 × 10−3−7.58 × 10−3 | |
| Controls | 745 | 12071 | 7517-17115 | 4 | 3-6 | 0.40% | 0.25%-0.57% | 1.50 × 10−3 | −2.16 × 10−3−5.76 × 10−3 | ||
| SCALE | Cases | 371 | 12582 | 7643-19776 | 5 | 3-7 | 0.42% | 0.25%-0.66% | 3.02 × 10−3 | −0.50 × 10−3−7.52 × 10−3 | |
| Controls | 790 | 12132 | 7506-17654 | 5 | 3-6 | 0.40% | 0.25%-0.59% | 1.22 × 10−3 | −1.60 × 10−3−5.00 × 10−3 | ||
| MZL | NCI | Cases | 808 | 18097 | 12831-24555 | 7 | 5-9 | 0.60% | 0.43%-0.82% | 1.83 × 10−3 | −1.54 × 10−3−7.30 × 10−3 |
| Controls | 6102 | 18268 | 12811-24623 | 7 | 5-9 | 0.61% | 0.43%-0.82% | 1.46 × 10−3 | −2.01 × 10−3−5.38 × 10−3 |
Median and interquartile range are provided for runs of homozygosity, fraction of runs of homozygosity, and correlation between uniting gametes (F3). GWAS: Genome-wide association studies; IQR: interquartile range; ROH: runs of homozygosity; FROH: fraction of runs of homozygosity; CLL: chronic lymphocytic leukemia; DLBCL: diffuse large B-cell lymphoma; FL: follicular lymphoma; MZL: marginal zone lymphoma.
We discovered a positive association between the risk of CLL and increased homozygosity as measured by FROH (β = 21.1, 95% SE = 4.41, P = 1.6 × 10−6) and F3 (β = 27.5, SE = 6.51, P = 2.4 × 10−5) [Table 2] with limited evidence of between-study heterogeneity (Phet = 0.42 and 0.11, respectively) [Supplementary Table 3]. As CLL is an indolent lymphoma and there is a potential for tumor DNA contamination in the blood drawn for genotyping, we performed a sensitivity analysis using only CLL cases and controls from prospective nested case-control studies (ATBC, CPS-II, EPIC, HPFS, MCCS, NHS, NYU-WHS, PLCO, and WHI) from the NCI NHL GWAS, where the DNA was often collected many years prior to diagnosis. Despite the reduction in the number of CLL cases (n = 2140 to 889), the estimated association parameters for FROH and F3 were similar (FROH: β = 21.3, SE = 7.76, P = 6.04 × 10−3; F3: β = 22.5, SE = 5.45, P = 3.57 × 10−5).
Table 2.
Risk of NHL subtypes associated with measures of genome-wide homozygosity, FROH and F3*
| FROH | F3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subtype | β | SE | P-value | I 2 | P het | β | SE | P-value | I 2 | P het |
| CLL | 21.14 | 4.41 | 1.59 × 10−6 | 0.0% | 0.42 | 27.46 | 6.51 | 2.44 × 10−5 | 49.7% | 0.11 |
| DLBCL | 0.04 | 10.89 | 1.0 | 72.5% | 0.01 | 1.96 | 9.57 | 0.84 | 82.4% | 0.001 |
| FL | 11.39 | 5.82 | 0.02 | 5.3% | 0.37 | 13.19 | 8.01 | 0.10 | 64.2% | 0.04 |
| MZL | −0.87 | 7.88 | 0.91 | 6.40 | 5.20 | 0.22 | ||||
Estimates of the log odds (β), standard error (SE), and P-value are provided for the association between FROH and F3 and each subtype, adjusted for age, sex (except UCSF1/NHS), percentage of missing SNPs, and principal components and combined using random effects meta-analysis. The I2 statistic provides an estimate of heterogeneity in association estimates across GWAS, and Phet is the P-value for heterogeneity among studies. FROH: Fraction of runs of homozygosity; NHL: non-Hodgkin lymphoma; CLL: chronic lymphocytic leukemia; DLBCL: diffuse large B-cell lymphoma; FL: follicular lymphoma; MZL: marginal zone lymphoma.
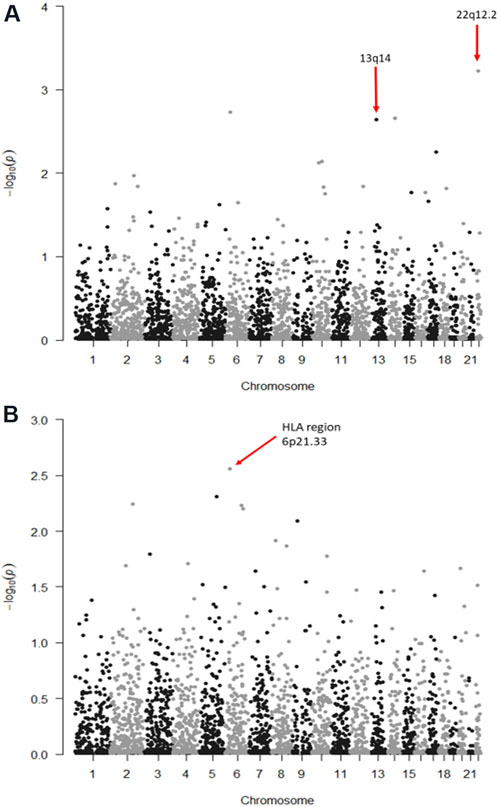
Figure 1A shows the P-values (−log10) from the meta-analysis assessing the associations of CLL with ROH centered in each 500 kb bin in the autosome. No bins reached statistical significance after correction for multiple testing (CLL Bonferroni alpha level = 0.05/45,590 bins = 1.1 × 10−5); the most significantly associated bin was located at chromosome 22q12.2 (P = 5.92 × 10−4). However, one of the top ten associated bins overlapped with the chromosome 13q14 region, a region where a somatic deletion is often seen in CLL[32]. To test whether the association between FROH and CLL was due to mosaicism at 13q14, we excluded the 45 CLL cases identified to have this deletion from the NCI NHL GWAS. After removal of these 45 cases, the association between FROH and CLL was slightly attenuated but remained statistically significant (β = 16.34, SE = 4.74, P = 5.71 × 10−4), similar to the results from the entire NCI NHL GWAS for CLL (β = 18.9, SE = 4.76, P = 6.73 × 10−5).
Figure 1.
Manhattan plots of log10(P) values for the association between runs of homozygosity (ROH) and the risk of chronic lymphocytic leukemia (CLL) (A) and follicular lymphoma (FL) (B). Here, ROH were divided into 500-kb bins across each chromosome, and each bin was tested for its association with CLL or FL. No bins reached statistical significance after correction for multiple testing.
As structural alterations in chromosomes, such as trisomy 12, are a hallmark of CLL and could potentially be mistakenly called as ROH using this method, we conducted a sensitivity analysis excluding chromosomes 12 and 13, which are frequently altered in CLL, from the meta-analysis[33]. After excluding these two chromosomes, the results for the association between FROH and CLL were found to be similar (β = 19.3, SE = 4.4, P = 9.0 × 10−6) to previous results for all 22 autosomal chromosomes. Similarly, for F3, exclusion of chromosomes 12 and 13 led to results that are similar in magnitude and remain statistically significant (β = 17.6, SE = 7.4, P = 0.02). Between-study heterogeneity (I2 = 62.2%, Phet = 0.05) was detected in this sensitivity analysis, largely attributable to the GEC GWAS.
We also identified positive associations between FL risk and increased FROH (β = 11.4, SE = 5.82, P = 0.02) but not F3 (β = 13.2, SE = 8.01, P = 0.10) [Table 2]. There was evidence of between-study heterogeneity in the meta-analysis of F3 and FL (I2 = 64.2%, P = 0.04) due to the USCF1/NHS GWAS, but not in the meta-analysis of FROH and FL (I2 = 5.3%, P = 0.37) [Supplementary Table 3]. We performed a sensitivity analysis using only cases and controls collected from prospective nested case-control studies (ATBC, CPS-II, EPIC, HPFS, MCCS, NHS, NYU-WHS, PLCO, and WHI) as part of the NCI NHL GWAS. This resulted in a reduction of the number of cases (n = 2085 to 521) and statistical power, but the association between FROH and risk of FL was qualitatively similar (β = 14.1, SE = 11.4, P = 0.21). Given the importance of the HLA region in FL risk, we conducted a sensitivity analysis removing chromosome 6 from estimations of FROH and F3 and repeating the meta-analysis of the four GWAS. The results for both FROH (β = 12.5, SE = 6.29, P = 0.047) and F3 (β = 11.1, SE = 9.26, P = 0.23) were found to be similar to the between-study heterogeneity observed for F3 (I2 = 73.1%, P = 0.01) due to the UCSF1/NHS GWAS.
Figure 1B shows the P-values (−log10) from the meta-analysis of logistic regression models estimating the associations of FL with ROH centered in each 500-kb bin in the autosome. No bins reached statistical significance assuming a Bonferroni-corrected alpha level. The most significant association was for a bin overlapping the HLA region 6p21.33 (P = 2.78 × 10−3).
No associations were observed with either FROH or F3 and the risk of DLBCL or MZL [Table 2]; however, between-study heterogeneity in DLBCL risk was present for both FROH (I2 = 72.5%, Phet = 0.01) and F3 (I2 = 82.4%, Phet = 0.001) [Supplementary Table 3]. In a leave-one-out sensitivity analysis [Supplementary Table 4], removal of either the NCI NHL GWAS or the GELA/EPIC GWAS, which had association results in opposite directions, reduced the I2 among the remaining three GWAS to 0%. Without the GELA/EPIC GWAS, there were significant associations between FROH and DLBCL (β = 13.1, SE = 4.62, P = 0.004) and between F3 and DLBCL (β = 13.4, SE = 3.18, P = 2.4 × 10−5).
DISCUSSION
To our knowledge, the present study represents the first analysis of the association between homozygosity and risk of four major NHL subtypes. We assessed genome-wide homozygosity using F3 and FROH, where the beta coefficient represents the log odds for a one-unit change in the fraction of ROH. We discovered that increased genome-wide homozygosity was associated with an increased risk of CLL and FL. We estimated that a 0.1% increase in the fraction of ROH was associated with a 2% increase in CLL risk and 1% increase in FL risk compared to those without any ROH. While we did not identify specific regions containing novel associations with these four subtypes of NHL, our analyses suggest that recessive genetic variation is likely to play a role in the risk of these two NHL subtypes and may account for a fraction of the unexplained heritability of these diseases. Our study did not find convincing evidence that recessive variation is a significant contributor to the risk of DLBCL and MZL, further supporting the hypothesis that each NHL subtype is likely to have its own genetic architecture of disease susceptibility.
Evidence suggests that CLL has high heritability with first-degree relatives having a 6-8.5-fold increase in risk of CLL[5,34]. Approximately 17%-25% of the familial relative risk of CLL is explained by currently known risk loci[6,9], leaving a sizable fraction that remains undiscovered. We discovered strong associations between CLL and genome-wide homozygosity as measured by FROH and F3. These findings suggest that recessive genetic variants are likely to contribute to risk.
CLL is a hematologic malignancy characterized by large-scale chromosomal alterations[35] and is often preceded by a long-lasting pre-malignant stage of monoclonal B-cell lymphocytosis (MBL)[36]. It is possible that cases with diagnosed CLL at the time of enrollment or those with unidentified MBL had tumor DNA in their blood samples, and that some mosaic events may have been erroneously picked up as ROH or homozygous genotypes due to the calling method utilized. This could have inflated our association results, but our sensitivity analyses suggested that any influence of mosaicism on our findings was likely to be small. We excluded CLL cases with DNA collected after diagnosis in the NCI NHL GWAS and saw similar results to our primary analysis, suggesting that tumor contamination is unlikely to be responsible for the observed association. Further sensitivity analysis excluding individuals with known 13q14 deletion in NCI NHL GWAS showed that the associations between FROH and F3 and CLL were only slightly attenuated. We also removed chromosomes 12 and 13 from the calculation of FROH and F3, as these two chromosomes are frequently altered in newly diagnosed CLL[33], and again saw results similar to the primary analysis. While we cannot exclude the possibility that some of the association could be due to erroneous homozygosity calling of structural variation, the robustness of the associations indicates that potential bias from these sources probably does not explain the observed association.
We did not identify any specific homozygous regions that were associated with CLL risk after adjustment for multiple comparisons, but we observed nominal associations with ROH at chromosomes 22q12.2 and 13q14.2. The chromosome 22q12.2 region includes POZ/BTB and AT hook containing zinc finger 1 (PATZ1), which is a transcription factor that interacts with p53 and may modulate p53-mediated apoptosis[37]. The region also contains phosphoinositide-3-kinase interacting protein 1 (PIK3IP1), which inhibits antitumor immunity[38]. The chromosome 13q14.2 region contains several genes of potential interest, including RB transcriptional corepressor 1 (RB1) and cysteinyl-leukotriene receptor 2 (CYSLTR2). RB1 is a known tumor suppressor, which is frequently somatically deleted in CLL[32]. CYSLTR2 is part of the leukotriene pathway and thought to play a role in lymphocyte proliferation and maturation[39].
For FL, we discovered a positive association between FROH and risk of FL, suggesting recessive genetic variation is a contributor to risk. Previous epidemiologic work has shown that family history of NHL is a risk factor for FL[40], and first-degree relatives have a 4-6-fold increase risk of FL[4,5]. The largest GWAS of FL to date detected multiple genetic loci associated with risk, with the strongest associations seen in the HLA region[10]. Consistent with our findings, a previous study of first-degree relatives suggested that FL may follow an autosomal recessive mode of inheritance[5]. In sensitivity analyses, we showed that results for FROH are similar after excluding the entirety of chromosome 6, suggesting that our findings were not due to the HLA region. This finding supports the role of non-HLA variation in the etiology of FL and provides evidence that there may be additional rare, recessive loci that are associated with the risk of FL. We observed similar results after limiting to cases with prospectively collected DNA, suggesting that tumor contamination is unlikely. Although we did not find any individual regions (based on 500-kb bins of ROH) that were significantly associated with the risk of FL after adjustment for multiple testing, we observed nominal evidence for homozygosity overlapping the HLA region at chromosome 6p21.33. We previously reported that homozygosity for classical HLA class II alleles, such as HLA-DRB1, was associated with an increased risk of FL compared to individuals with heterozygosity[41]. These findings are consistent with the hypothesis that HLA homozygosity may increase risk by reducing an individual’s ability to recognize a diverse set of foreign antigens.
DLBCL shows a strong association with family history of NHL in epidemiologic studies[42], and it has been previously estimated that 16% of the variation in DLBCL is due to common genetic variants[8]. Overall, we did not observe an association between DLBCL and homozygosity, suggesting the recessive variation may not have a strong role in DLBCL susceptibility, a finding consistent with a previous study of DLBCL among first-degree relatives[5]. The meta-analysis for DLBCL was affected by substantial between-study heterogeneity (Phet = 0.01 for FROH and Phet = 0.001 for F3). In the leave-one-out meta-analysis, removing either the NCI NHL GWAS or the GELA/EPIC GWAS reduced the I2 among the remaining three GWAS to 0%, and significant associations were observed with FROH and F3 after removing GELA/EPIC. The GELA/EPIC GWAS was different from the other studies in that it included patients from clinical trials, who were slightly younger and less likely to be female than the other studies [Supplementary Table 2]. In addition, this GWAS combined cases from French clinical trials with controls from various European countries in the EPIC cohort. It is possible that the degree of population matching, while adequate for a GWAS, is not sufficient for analyses of F3 and FROH, which are known to be especially sensitive to population stratification[43]. Even after adjustment using principal components, some population substructure may have affected the GELA/EPIC study, leading to results that differ from the other DLBCL studies. Our analysis of FL similarly included one GWAS that combined the cases and controls from 2 different United States studies. Although we did observe some heterogeneity among the FL studies for F3, we did not observe such heterogeneity for FROH and FL, suggesting the F3 may be more sensitive to population substructure than FROH. The patients in the UCSF1/NHS study were slightly younger and less likely to be female than the cases in the NCI GWAS, which could have also contributed to the heterogeneity in results.
DLBCL is known to have substantial disease heterogeneity[44,45]. It is possible that a greater understanding of the heterogeneity of DLBCL and a further molecular subtype-specific analysis, which was not possible for the present study, may allow a better elucidation of any association between ROH and disease risk. While the analysis of F3 and DLBCL was similarly affected by between-study heterogeneity [Supplementary Table 3], a potential advantage of F3 over FROH as a measure of homozygosity is its expected smaller variance and bias[30]. In addition, the method of calculating FROH used in this study does not count homozygosity below a length threshold of 1500 kb[28]. It is possible that a portion of the recessive genetic variation in DLBCL resides in short, common ROH of ancient ancestry[14] that was unmeasured by our approach.
Although family history of NHL or other hematologic cancers has been associated with increased risk of MZL[46], we did not find evidence linking FROH or F3 to MZL. Our published GWAS of MZL found two independent loci conferring risk, both in the HLA region of the genome[11], and a follow-up study reported that homozygosity at class I HLA-B and -C and class II HLA-DRB1 loci was associated with increased risk of MZL[41]. Although our study did not detect an association with homozygosity more broadly across the genome, local homozygosity, at least in select regions, is still likely to play a role in risk. Of the four subtypes of NHL examined in the present study, MZL is the least common. Our power to detect an association with MZL was limited due to the small sample size, which was possibly compounded by the known heterogeneity within MZL, with multiple recognized subtypes[47]. Given that we expect ROH to capture the effects of small, scattered, recessive genotypes, a larger sample size is likely needed to elucidate the genetic etiology of MZL. As other highly heritable traits, such as certain autoimmune diseases and atopy, are associated with the risk of MZL[46], identifying shared genetic architecture with these more common phenotypes may be a complementary strategy to further elucidate the underlying genetic architecture.
This study had both limitations and strengths. The use of FROH as a measure of homozygosity is known to require large sample sizes[30]. We were able to combine eight GWAS across four subtypes of NHL to increase our sample size and provide a fairly comprehensive analysis of recessive inheritance. However, as discussed, we may have had insufficient sample size to detect associations for MZL. Further, we could not separately examine clinically relevant molecular subtypes of DLBCL. ROH patterns are exquisitely sensitive to population history[48]. Thus, we restricted our study population to individuals of European ancestry and adjusted for principal components of ancestry. This limited the potential for problematic population stratification in our study sample, but it also reduced the generalizability of the findings to individuals of other ancestries. To date, the optimal approach to analyze specific ROH to identify genomic regions with novel, recessively acting risk alleles has not been established, but both the ROH calling procedure and the use of F3 are accepted as means of identifying recent relatedness in outbred European populations[28,30] and can provide evidence for the presence of recessive loci. Although it is possible that the associations between FROH and F3 and CLL are due in part to DNA contamination by tumor cells, our sensitivity analyses suggest that germline homozygosity is likely an independent risk factor for CLL. Finally, although our study provides clues as to the genetic etiology of NHL, the clinical value of these findings is uncertain. Additional studies are needed to further elucidate the role of recessive variation in NHL risk.
In conclusion, we provide new evidence for the role of recessive genetic variation in the risk of CLL and FL in outbred European-ancestry populations. The knowledge that recessive variation in disease susceptibility to NHL is likely to be present suggests that further studies should be undertaken to identify the specific loci responsible for the associations reported here. As GWAS increase in sample size, they will have greater statistical power to identify recessive genetic variants-associated risk and further characterize the underlying genetic architecture of specific NHL subtypes.
Supplementary Material
Acknowledgements
ATBC - The ATBC Study is supported by the Intramural Research Program of the United States National Cancer Institute, National Institutes of Health, Department of Health and Human Services.
BCCA - Canadian Institutes for Health Research (CIHR); Canadian Cancer Society; Michael Smith Foundation for Health Research.
CPS-II - The Cancer Prevention Study-II (CPS-II) Nutrition Cohort is supported by the American Cancer Society. Genotyping for all CPS-II samples were supported by the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program.
ELCCS - Blood Cancer UK, United Kingdom.
ENGELA - Association pour la Recherche contre le Cancer (ARC), Institut National du Cancer (INCa), Fondation de France, Fondation contre la Leucémie, Agence nationale de sécurité sanitaire de l’alimentation, de l’environnement et du travail (ANSES).
EPIC - Coordinated Action (Contract #006438, SP23-CT-2005-006438); HuGeF (Human Genetics Foundation), Torino, Italy; Cancer Research UK. The coordination of EPIC is financially supported by the European Commission (DG-SANCO) and the International Angency for Research on Cancer. The national cohorts are supported by the Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l'Education National, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Centre (DKFZ), Federal Ministry of Education and Research (BMBF), Deutsche Krebshilfe, Deutsches Krebsforschungszentrum and Federal Ministry of Education and Research (Germany); the Hellenic Health Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), PI131/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia (no. 6236) and Navarra, ISCIII RETIC ((RD06/0020) (Spain); Swedish Cancer Society, Swedish Research Council and Country Councils of Skåne and Västerbotten (Sweden); Cancer Research UK [14136 to K.T. Khaw, N.J. Wareham; C570/A16491 to RCT and C8221/A19170 to T. Key (EPIC-Oxford)], Medical Research Council [10001433 to K.T. Khaw, N.J. Wareham, MR/M012190/1 to T. Key (EPIC-Oxford)] (United Kingdom).
EpiLymph - European Commission (grant references QLK4-CT-2000-00422 and FOOD-CT-2006-023103); We thank CERCA programme/Generalitat de Catalunya for institutional support. This work was supported by Spanish Ministry of Economy and Competitiveness - Carlos III Institute of Health cofunded by FEDER funds/European Regional Develpment Fund (ERDF) - a way to build Europe (grant references CIBERESP, PI17/01280, PI20/00288) with the support of the Secretariat for Universities and Research of the Ministry of Business and Knowledge of the Government of Catalonia (2017SGR1085) who had no role in the data collection, analysis or interpretation of the results; the NIH (contract NO1-CO-12400); the Compagnia di San Paolo-Programma Oncologia; the Federal Office for Radiation Protection grants StSch4261 and StSch4420, the José Carreras Leukemia Foundation grant DJCLS-R12/23, the German Federal Ministry for Education and Research (BMBF-01-EO-1303); the Health Research Board, Ireland and Cancer Research Ireland; Czech Republic supported by MH CZ - DRO (MMCI, 00209805) and MEYS - NPSI - LO1413; Fondation de France and Association de Recherche Contre le Cancer.
GEC/Mayo GWAS - National Institutes of Health (CA118444, CA148690, CA92153). Intramural Research Program of the NIH, National Cancer Institute. Veterans Affairs Research Service. Data collection for Duke University was supported by a Leukemia & Lymphoma Society Career Development Award, the Bernstein Family Fund for Leukemia and Lymphoma Research, and the National Institutes of Health (K08CA134919), National Center for Advancing Translational Science (UL1 TR000135).
HPFS (Walter C. Willet) - The HPFS was supported in part by National Institutes of Health grants U01 CA167552, R01 CA149445, and R01 CA098122. We would like to thank the participants and staff of the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required.
Iowa-Mayo SPORE - NCI Specialized Programs of Research Excellence (SPORE) in Human Cancer (P50 CA97274); National Cancer Institute (P30 CA086862, P30 CA15083); Henry J. Predolin Foundation.
Italian GxE - Italian Association for Cancer Research (AIRC, Investigator Grant 11855) (PC); Fondazione Banco di Sardegna 2010-2012, and Regione Autonoma della Sardegna (LR7 CRP-59812/2012) (MGE).
Mayo Clinic Case-Control - National Institutes of Health (R01 CA92153 and CA200703); National Cancer Institute (P30 CA015083).
MCCS - The Melbourne Collaborative Cohort Study recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 396414 and 1074383 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database.
MSKCC - Geoffrey Beene Cancer Research Grant, Lymphoma Foundation (LF5541); Barbara K. Lipman Lymphoma Research Fund (74419); Robert and Kate Niehaus Clinical Cancer Genetics Research Initiative (57470); U01 HG007033; ENCODE; U01 HG007033.
NCI-SEER - Intramural Research Program of the National Cancer Institute, National Institutes of Health, and Public Health Service (N01-PC-65064, N01-PC-67008, N01-PC-67009, N01-PC-67010, N02-PC-71105).
NHS (Meir J. Stampfer) - The NHS was supported in part by National Institutes of Health grants CA186107, CA87969, CA49449, CA149445, CA098122 and CA134958. We would like to thank the participants and staff of the Nurses' Health Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
NSW - NSW was supported by grants from the Australian National Health and Medical Research Council (ID990920), the Cancer Council NSW, and the University of Sydney Faculty of Medicine.
NYU-WHS - National Cancer Institute (R01 CA098661, P30 CA016087); National Institute of Environmental Health Sciences (ES000260).
PLCO - This research was supported by the Intramural Research Program of the National Cancer Institute and by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS.
SCALE - Swedish Cancer Society (2009/659). Stockholm County Council (20110209) and the Strategic Research Program in Epidemiology at Karolinska Institutet. National Institutes of Health (5R01 CA6966902); Plan Denmark.
UCSF2 - The UCSF studies were supported by the NCI, National Institutes of Health, CA1046282 and CA154643. The collection of cancer incidence data used in this study was supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology, and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement #1U58 DP000807-01 awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the authors, and endorsement by the State of California, the California Department of Health Services, the National Cancer Institute, or the Centers for Disease Control and Prevention or their contractors and subcontractors is not intended nor should be inferred.
UTAH/Sheffield - National Institutes of Health CA134674. Partial support for data collection at the Utah site was made possible by the Utah Population Database (UPDB) and the Utah Cancer Registry (UCR). Partial support for all datasets within the UPDB is provided by the Huntsman Cancer Institute (HCI) and the HCI Cancer Center Support grant, P30 CA42014. The UCR is supported in part by NIH contract HHSN261201000026C from the National Cancer Institute SEER Program with additional support from the Utah State Department of Health and the University of Utah. Partial support for data collection in Sheffield, UK was made possible by funds from Yorkshire Cancer Research and the Sheffield Experimental Cancer Medicine Centre. We thank the NCRI Haemato-oncology Clinical Studies Group, colleagues in the North Trent Cancer Network the North Trent Haemato-oncology Database.
UTAH-MM - National Institutes of Health CA134674, the Leukemia Lymphoma Society 6067-09, and pilot funds from the Huntsman Cancer Institute (HCI). Partial support for data collection at the Utah site was made possible by the Utah Population Database (UPDB) and the Utah Cancer Registry (UCR). Partial support for all datasets within the UPDB is provided by the HCI Comprehensive Cancer Center Support grant, P30 CA42014 (M Beckerle). The UCR is supported in part by NIH contract HHSN261201000026C from the National Cancer Institute SEER Program with additional support from the Utah State Department of Health and the University of Utah.
WHI - WHI investigators are: Program Office - (National Heart, Lung, and Blood Institute, Bethesda, Maryland) Jacques Rossouw, Shari Ludlam, Dale Burwen, Joan McGowan, Leslie Ford, and Nancy Geller; Clinical Coordinating Center - (Fred Hutchinson Cancer Research Center, Seattle, WA) Garnet Anderson, Ross Prentice, Andrea LaCroix, and Charles Kooperberg; Investigators and Academic Centers - (Brigham and Women's Hospital, Harvard Medical School, Boston, MA) JoAnn E. Manson; (MedStar Health Research Institute/Howard University, Washington, DC) Barbara V. Howard; (Stanford Prevention Research Center, Stanford, CA) Marcia L. Stefanick; (The Ohio State University, Columbus, OH) Rebecca Jackson; (University of Arizona, Tucson/Phoenix, AZ) Cynthia A. Thomson; (University at Buffalo, Buffalo, NY) Jean Wactawski-Wende; (University of Florida, Gainesville/Jacksonville, FL) Marian Limacher; (University of Iowa, Iowa City/Davenport, IA) Robert Wallace; (University of Pittsburgh, Pittsburgh, PA) Lewis Kuller; (Wake Forest University School of Medicine, Winston-Salem, NC) Sally Shumaker; Women’s Health Initiative Memory Study - (Wake Forest University School of Medicine, Winston-Salem, NC) Sally Shumaker. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, United States Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C.
YALE - National Cancer Institute (CA62006); National Cancer Institute (CA165923).
Disclaimer:
This project was funded by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH and in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Footnotes
Availability of data and materials
NCI NHL GWAS data used in this manuscript has been deposited in the dbGaP repository with accession code phs000801.v2.p1. Other relevant data used in this study are available upon request.
Conflicts of interest
All authors declare that there are no conflicts of interest.
Ethical approval and consent to participate
All studies obtained informed consent from participants and were approved by their respective Institutional Review Boards as listed. These are ATBC: (NCI Special Studies Institutional Review Board); BCCA: UBC BC Cancer Agency Research Ethics Board; CPS-II: American Cancer Society; ELCCS: Northern and Yorkshire Research Ethics Committee; ENGELA: IRB00003888 - Comite d’ Evaluation Ethique de l’Inserm IRB # 1; EPIC: Imperial College London; EpiLymph: International Agency for Research on Cancer, HPFS: Harvard School of Public Health (HSPH) Institutional Review Board; Iowa-Mayo SPORE: University of Iowa Institutional Review Board; Italian GxE: Comitato Etico Azienda Ospedaliero Universitaria di Cagliari; Mayo Clinic Case-Control: Mayo Clinic Institutional Review Board; Mayo GEC: Mayo Clinic Institutional Review Board, #07-005788-03; MCCS: Cancer Council Victoria’s Human Research Ethics Committee; MD Anderson: University of Texas MD Anderson Cancer Center Institutional Review Board; MSKCC: Memorial Sloan-Kettering Cancer Center Institutional Review Board; NCI-SEER (NCI Special Studies Institutional Review Board); NHS: Partners Human Research Committee, Brigham and Women’s Hospital, NSW: NSW Cancer Council Ethics Committee; NYU-WHS: New York University School of Medicine Institutional Review Board; PLCO: (NCI Special Studies Institutional Review Board); SCALE: Scientific Ethics Committee for the Capital Region of Denmark; SCALE: Regional Ethical Review Board in Stockholm (Section 4) IRB#5; UCSF1/UCSF2: University of California San Francisco Committee on Human Research; UTAH: University of Utah; WHI: Fred Hutchinson Cancer Research Center; Yale: Human Investigation Committee, Yale University School of Medicine.
REFERENCES
- 1.Chatterjee N, Hartge P, Cerhan JR, et al. Risk of non-Hodgkin's lymphoma and family history of lymphatic, hematologic, and other cancers. Cancer Epidemiol Biomarkers Prev 2004;13:1415–21. [PubMed] [Google Scholar]
- 2.Chang ET, Smedby KE, Hjalgrim H, et al. Family history of hematopoietic malignancy and risk of lymphoma. J Natl Cancer Inst 2005;97:1466–74. [DOI] [PubMed] [Google Scholar]
- 3.Wang SS, Slager SL, Brennan P, et al. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10 211 cases and 11 905 controls from the International Lymphoma Epidemiology Consortium (InterLymph). Blood 2007;109:3479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldin LR, Björkholm M, Kristinsson SY, Turesson I, Landgren O. Highly increased familial risks for specific lymphoma subtypes. Br J Haematol 2009;146:91–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altieri A, Bermejo JL, Hemminki K. Familial risk for non-Hodgkin lymphoma and other lymphoproliferative malignancies by histopathologic subtype: the Swedish Family-Cancer Database. Blood 2005;106:668–72. [DOI] [PubMed] [Google Scholar]
- 6.Berndt SI, Camp NJ, Skibola CF, et al. Meta-analysis of genome-wide association studies discovers multiple loci for chronic lymphocytic leukemia. Nat Commun 2016;7:10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berndt SI, Skibola CF, Joseph V, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet 2013;45:868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerhan JR, Berndt SI, Vijai J, et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet 2014;46:1233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Law PJ, Berndt SI, Speedy HE, et al. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat Commun 2017;8:14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skibola CF, Berndt SI, Vijai J, et al. Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am J Hum Genet 2014;95:462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijai J, Wang Z, Berndt SI, et al. A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat Commun 2015;6:5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lettre G, Lange C, Hirschhorn JN. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet Epidemiol 2007;31:358–62. [DOI] [PubMed] [Google Scholar]
- 13.Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet 2006;15:789–95. [DOI] [PubMed] [Google Scholar]
- 14.Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet 2012;91:275–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller MC, Visscher PM, Goddard ME. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics 2011;189:237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szpiech ZA, Xu J, Pemberton TJ, et al. Long runs of homozygosity are enriched for deleterious variation. Am J Hum Genet 2013;93:90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enciso-Mora V, Hosking FJ, Houlston RS. Risk of breast and prostate cancer is not associated with increased homozygosity in outbred populations. Eur J Hum Genet 2010;18:909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spain SL, Cazier JB, Houlston R, Carvajal-Carmona L, Tomlinson I; CORGI Consortium. Colorectal cancer risk is not associated with increased levels of homozygosity in a population from the United Kingdom. Cancer Res 2009;69:7422–9. [DOI] [PubMed] [Google Scholar]
- 19.Hosking FJ, Papaemmanuil E, Sheridan E, et al. Genome-wide homozygosity signatures and childhood acute lymphoblastic leukemia risk. Blood 2010;115:4472–7. [DOI] [PubMed] [Google Scholar]
- 20.Thomsen H, Inacio da Silva Filho M, Fuchs M, et al. Evidence of inbreeding in Hodgkin lymphoma. PLoS One 2016;11:e0154259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sud A, Cooke R, Swerdlow AJ, Houlston RS. Genome-wide homozygosity signature and risk of Hodgkin lymphoma. Sci Rep 2015;5:14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi PK, Esko T, Mattsson H, et al. Directional dominance on stature and cognition in diverse human populations. Nature 2015;523:459–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conde L, Halperin E, Akers NK, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet 2010;42:661–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vivo I, Prescott J, Setiawan VW, et al. ; Australian National Endometrial Cancer Study Group. Genome-wide association study of endometrial cancer in E2C2. Hum Genet 2014;133:211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smedby KE, Foo JN, Skibola CF, et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21.32 and suggests shared genetic susceptibility with diffuse large B-cell lymphoma. PLoS Genet 2011;7:e1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gazal S, Sahbatou M, Perdry H, Letort S, Génin E, Leutenegger AL. Inbreeding coefficient estimation with dense SNP data: comparison of strategies and application to HapMap III. Hum Hered 2014;77:49–62. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yengo L, Zhu Z, Wray NR, et al. Detection and quantification of inbreeding depression for complex traits from SNP data. Proc Natl Acad Sci U S A 2017;114:8602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edelmann J, Holzmann K, Miller F, et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood 2012;120:4783–94. [DOI] [PubMed] [Google Scholar]
- 33.Puiggros A, Blanco G, Espinet B. Genetic abnormalities in chronic lymphocytic leukemia: where we are and where we go. Biomed Res Int 2014;2014:435983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldin LR, Björkholm M, Kristinsson SY, Turesson I, Landgren O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin's lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica 2009;94:647–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malek SN. The biology and clinical significance of acquired genomic copy number aberrations and recurrent gene mutations in chronic lymphocytic leukemia. Oncogene 2013;32:2805–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawstron AC, Bennett FL, O'Connor SJ, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med 2008;359:575–83. [DOI] [PubMed] [Google Scholar]
- 37.Valentino T, Palmieri D, Vitiello M, Pierantoni GM, Fusco A, Fedele M. PATZ1 interacts with p53 and regulates expression of p53-target genes enhancing apoptosis or cell survival based on the cellular context. Cell Death Dis 2013;4:e963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Wang J, Wang X, et al. Pik3ip1 is a negative immune regulator that inhibits antitumor T-cell immunity. Clin Cancer Res 2019;25:6180–94. [DOI] [PubMed] [Google Scholar]
- 39.Paulucci BP, Pereira J, Picciarelli P, Levy D, di Francesco RC. Expression of CysLTR1 and 2 in maturating lymphocytes of hyperplasic tonsils compared to peripheral cells in children. Inflammation 2016;39:1216–24. [DOI] [PubMed] [Google Scholar]
- 40.Linet MS, Vajdic CM, Morton LM, et al. Medical history, lifestyle, family history, and occupational risk factors for follicular lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr 2014;2014:26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang SS, Carrington M, Berndt SI, et al. HLA Class I and II diversity contributes to the etiologic heterogeneity of non-Hodgkin lymphoma subtypes. Cancer Res 2018;78:4086–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerhan JR, Kricker A, Paltiel O, et al. Medical history, lifestyle, family history, and occupational risk factors for diffuse large B-cell lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr 2014;2014:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howrigan DP, Simonson MA, Keller MC. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics 2011;12:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–11. [DOI] [PubMed] [Google Scholar]
- 45.Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005;105:1851–61. [DOI] [PubMed] [Google Scholar]
- 46.Bracci PM, Benavente Y, Turner JJ, et al. Medical history, lifestyle, family history, and occupational risk factors for marginal zone lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr 2014;2014:52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Geneva: IARC Press; 2008. [Google Scholar]
- 48.McQuillan R, Leutenegger AL, Abdel-Rahman R, et al. Runs of homozygosity in European populations. Am J Hum Genet 2008;83:359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.