Targeting neurons in breast cancer with anesthetic nanoparticles inhibits nerve-cancer stimulation and tumor progression.
Abstract
Neurons within the tumor microenvironment promote cancer progression; thus, their local targeting has potential clinical benefits. We designed PEGylated lipid nanoparticles loaded with a non-opioid analgesic, bupivacaine, to target neurons within breast cancer tumors and suppress nerve-to-cancer cross-talk. In vitro, 100-nm nanoparticles were taken up readily by primary neurons, trafficking from the neuronal body and along the axons. We demonstrate that signaling between triple-negative breast cancer cells (4T1) and neurons involves secretion of cytokines stimulating neurite outgrowth. Reciprocally, neurons stimulated 4T1 proliferation, migration, and survival through secretion of neurotransmitters. Bupivacaine curbs neurite growth and signaling with cancer cells, inhibiting cancer cell viability. In vivo, bupivacaine-loaded nanoparticles intravenously administered suppressed neurons in orthotopic triple-negative breast cancer tumors, inhibiting tumor growth and metastatic dissemination. Overall, our findings suggest that reducing nerve involvement in tumors is important for treating cancer.
INTRODUCTION
Tumor-infiltrating nerves from the peripheral nervous system occur in various types of cancer. Nerve/cancer interactions have been shown to support cancer development and progression (1). Cancer cells secrete neurotrophic factors that promote nerve innervation into the tumor microenvironment in a process termed neurogenesis (2). In addition, the nervous system has an essential impact on stimulating cancer cell growth, proliferation, angiogenesis, and invasion through the secretion of chemokines and neurotransmitters (3). Perineural invasion (4) of peripheral nerves into tumors and the reciprocal interactions between cancer cells and nerves suggest that targeting nerves in the tumor tissue will be beneficial for treating various cancer types, such as prostate, breast, lung, ovarian, and pancreatic cancer (1). Chemical and surgical depletion of sympathetic nerves was shown to suppress the early stage of prostate cancer development (5). Moreover, β-adrenergic blocker treatment reduced progression of breast, prostate, lung, ovarian, and melanoma cancers (6). Therefore, various therapeutic approaches that manipulate the local nerves in the tumor tissue have inhibitory effect on cancer (1, 7).
Nanotechnology is gaining attention in targeted cancer therapies and diagnostics (8–12), as well as therapeutic applications in neuronal regeneration (13). Specifically, liposomes, self-assembled lipid vesicles, are commonly used as nanoscale drug delivery systems (14, 15). Bupivacaine is a non-opioid selective sodium channel blocker that interrupts the transmission of the nerve impulse and pain signals. Encapsulation of bupivacaine into nanoparticles has been shown to reduce occurrence of systemic adverse effects after subcutaneous injection of bupivacaine (16). Micrometer-scale liposomes containing bupivacaine have been developed for prolonged local analgesia in the management of postsurgical pain (16). We hypothesized that bupivacaine-loaded nanoparticles will suppress neuronal activity in the tumor microenvironment to improve cancer management. We examined the nerve/cancer interactions and tested the ability of liposomal bupivacaine (L-BUP) to curb nerve signaling in breast cancer in vitro and in vivo models.
RESULTS
The collaborative interactions between cancer and the nervous system promote cancer progression (1). In this study, lipid nanoparticles encapsulating the non-opioid analgesic, bupivacaine, were used to suppress these interactions, thus inhibiting tumor growth and metastasis in orthotopic triple-negative breast cancer (4T1) murine models (Fig. 1A). In vitro models included cortical primary neurons and rat adrenal pheochromocytoma cells (PC12) (17).
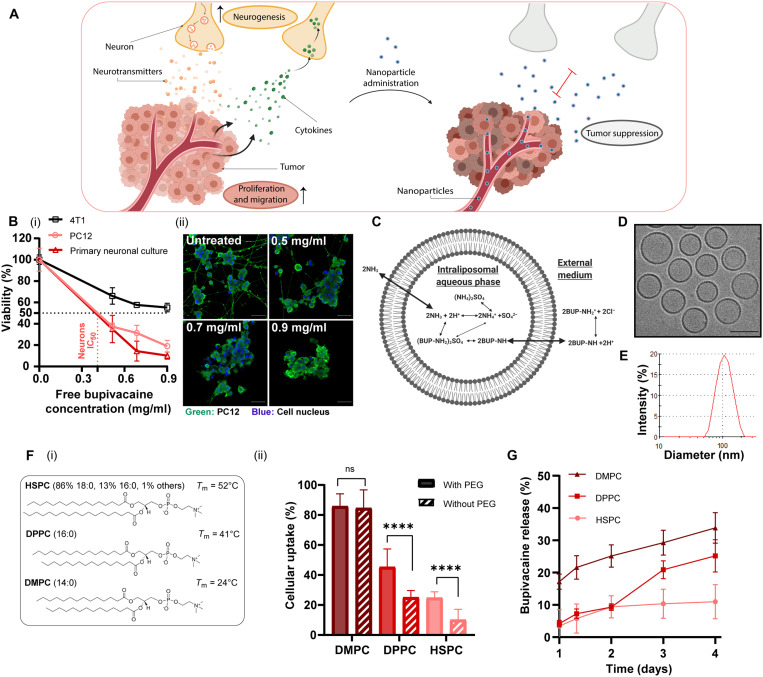
Fig. 1. Analgesic nanoparticles as a tool for treating neurons within breast cancer tumors.
Cross-talk between cancer cells and nerves supports cancer cell proliferation and migration; reciprocally, neurite growth is promoted through secretion of cytokines by cancer cells. We used lipid nanoparticles, liposomes, loaded with bupivacaine (L-BUP) to curb nerve/cancer cross-talk and inhibit tumor growth (A). Viability of primary neurons, PC12, and triple-negative breast cancer cells (4T1) after treatment with bupivacaine [(B), i] (normalized to the untreated group) and confocal imaging [(B), ii] (scale bars, 50 μm). Liposomes (100 nm) composed of HSPC (hydrogenated soybean phosphatidylcholine), cholesterol, and DSPE–PEG-2000 (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-methoxy–polyethylene glycol 2000; 55:40:5 molar ratio) were loaded with bupivacaine using ammonium sulfate gradient (C) and characterized by cryogenic transmission electron microscopy (cryo-TEM) (D) (scale bar, 100 nm) and dynamic light scattering (E) (PDI < 0.1). Various lipid formulations were compared for their neuronal uptake: HSPC, DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), and DMPC (2-dimyristoyl-sn-glycero-3-phosphocholine), with cholesterol and with/without DSPE–PEG-2000 [(F), i]. PC12 cells were incubated with rhodamine-labeled liposomes and analyzed for their cellular uptake [(F), ii]. L-BUP release profile was conducted at 37°C, comparing the stability of the different formulations (G). Results are presented as means ± SD (between two and three independent repetitions performed in at least three replicates). Two-way analysis of variance (ANOVA) was used for statistical analysis of [(B), i] and one-way ANOVA for [(F), ii, and (G)] with multiple comparisons test adjusted P value; ****P < 0.0001. IC50, median inhibitory concentration; ns, not significant.
Neurotoxicity of bupivacaine
Bupivacaine, a selective sodium channel blocker, has a neurotoxic effect that is both concentration and duration dependent (16). The median lethal concentration (LC50) of bupivacaine was evaluated in primary neurons, PC12 cells, and 4T1 triple-negative breast cancer cells. The LC50 was similar for primary neurons and PC12 cells, ~0.4 mg/ml, compared to ~0.9 mg/ml for 4T1, indicating a higher potency of bupivacaine toward neurons (Fig. 1B, i).
Bupivacaine lipid nanoparticles
Encapsulating bupivacaine in nanoparticles reduces systemic side effects and improves tumor targeting (10, 16). Bupivacaine was remotely loaded, using a transmembrane ammonium sulfate gradient (16), into 100 ± 20–nm liposomes composed of hydrogenated soybean phosphatidylcholine (HSPC), cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-methoxy–polyethylene glycol 2000 (DSPE–PEG-2000), in molar ratios of 55:40:5, respectively (Fig. 1, C to E). The encapsulation efficiency reached ~60% (fig. S2), being stable at 4°C for 12 days, with a maximal release of 4 ± 1% (fig. S1).
Nanoparticle’s lipid composition affects neuronal uptake
To study the effect the nanoparticle’ lipid composition has on neuronal uptake, we compared six different compositions of phosphatidylcholine (PC) liposomes, varying in their lipid tail length and PEG moiety (Fig. 1F). PC and cholesterol-based liposomes were chosen as candidates for neuronal delivery, as these are the main lipid components in the membranes of neurons (18). Phospholipids with shorter fatty acid chains (and a resultant lower phase transition temperature, Tm) achieved greater uptake by PC12 cells. Specifically, the uptake of 2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC; 14:0C lipid chain) liposomes was about twofold higher than 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC; 16:0C) and about threefold higher than HSPC (18:0C) (P < 0.0001). PEG, a common additive to nanoparticles used for stabilizing formulations and increasing circulation time (19), improved neuronal uptake of HSPC and DPPC liposomes, yielding about twofold higher than liposomes without PEG (P < 0.0001). Previous studies reported that PEG can improve tissue penetration (20–22); however, at the cell level, these observations are dependent on the cell type. The lipid chain length (14, 16, and 18C-long) and the PEG-2000 moiety did not affect neuronal viability after 4 and 24 hours (fig. S3). DMPC and DPPC liposomes displayed higher bupivacaine release rates at 37°C compared to HSPC liposomes (P < 0.05, from the third day; Fig. 1G). Therefore, despite their superior cellular uptake, HSPC was used as the main membrane composition of liposomes to assure a stable formulation with a greater drug retention at physiological conditions (37°C) (23). Note that adding DSPE–PEG-2000 to the membrane did not affect the bupivacaine loading efficiency into the liposomes (fig. S4).
Cancer cells stimulate neurite growth through cytokine secretion
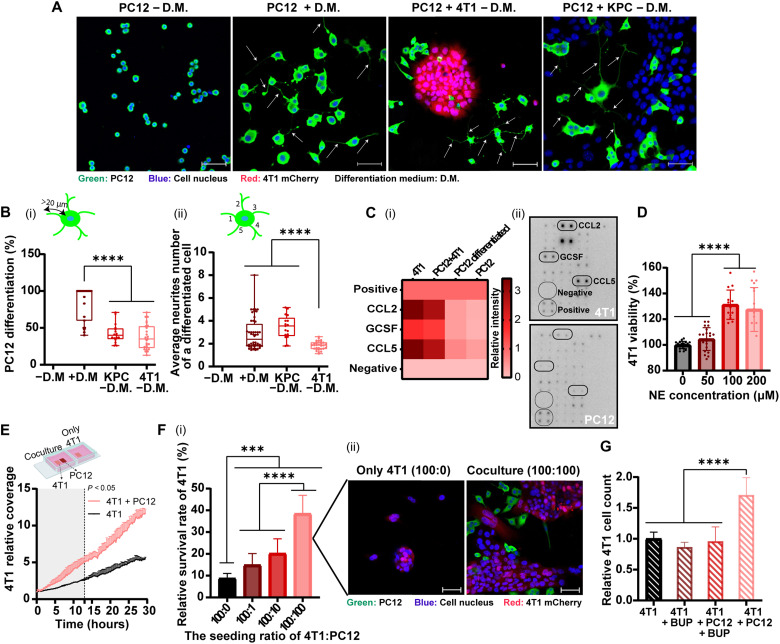
To examine the effect that different cancer cell lines have on nerve cells, PC12 cells were cocultured with either 4T1 or pancreatic ductal adenocarcinoma (KPC cell line). Cancer cells stimulated neuronal PC12 differentiation and neurite growth, compared to the untreated control (P < 0.0001), having a similar neurite stimulating effect as in neurons treated with nerve growth factor (NGF; Fig. 2, A and B). Morphometric analysis showed that 37.1 ± 16.7% and 42.6 ± 12.9% of the cells were differentiated when PC12 cells were cocultured with 4T1 or KPC cells, respectively. The average neurite number per cell was 1.8 ± 0.4 or 3.5 ± 1.0, respectively, with 4T1 or KPC (Fig. 2B). Thereby, neuronal differentiation was stimulated by the presence of both cancer cell lines, 4T1 and KPC cells. The promotion of neurite growth by cancer cells was attributed to the secretion of neurotrophic factors (24, 25). Here, we found that 4T1 cells secrete chemokines C-C motif ligand 5 (CCL5), granulocyte colony-stimulating factor (GCSF), and CCL2 (Fig. 2C and fig. S5), which increase neurite growth and neural invasion (24, 26–28). Cancer cells promoted neurogenesis (2) in a multifactorial signaling system, which included the secretion of neurotrophic factors (24, 25) and cytokines.
Fig. 2. Nerve cancer cross-talk promotes neurite growth and cancer cell proliferation, migration, and survival.
PC12 cells were cocultured with 4T1 breast cancer or pancreatic ductal adenocarcinoma (KPC) cells over 72 hours. Cell differentiation medium (D.M.) was supplemented with NGF only in the control group. PC12 neurites are highlighted with arrows (A) (scale bars, 50 μm). Morphometric analysis of the images indicate the percentage of differentiated cells [(B), i] and average number of the extending neurites [(B), ii]. Cancer cells secrete cytokines that promote neurite outgrowth, identified by a cytokine antibody array [(C), i], and was presented as a heatmap [(C), ii]. Effect of norepinephrine (NE) on 4T1 proliferation after 24 hours, normalized to the untreated group (D). The effect of PC12 on 4T1 migration, measured by time lapse then analyzed using IMARIS and normalized to the beginning coverage (E). 4T1 cell survival under starvation conditions after 96 hours, with/without PC12 cells, measured using an InCell analyzer [(F), i] and confocal microscopy [(F), ii] (scale bars, 50 μm). 4T1 cells were seeded with/without PC12 cells for 72 hours in starvation media; then, bupivacaine (0.5 mg/ml; BUP) was added for 6 hours. According to 4T1 cell count (normalized to 4T1 alone) BUP did not affect cancer cells directly but induced neuronal toxicity that affected cancer cell growth (G). Results of (B, D, and F) (three independent repetitions performed in five replicates each) and (E and G) (two independent repetitions performed in two to five replicates each) are presented as means ± SD. One-way ANOVA with adjusted P value in multiple comparisons tests was used for statistical analysis; ***P < 0.001 and ****P < 0.0001.
Neuronal signaling promotes cancer cell proliferation, migration, and survival
Nerve signaling is conducted through neurotransmitter secretion (3), such as norepinephrine (NE). Increasing concentrations of NE were used to evaluate its influence on cancer cells. 4T1 cell proliferation increased by ~30% (P < 0.0001) when treated for 24 hours with 100 and 200 μM NE, compared to the untreated group (Fig. 2D). The effect of the presence of neurons on cancer cell migration and proliferation was monitored. For this, triple-negative 4T1 breast cancer cells were seeded alone or together with neuronal PC12. Once 4T1 cells were cocultured with PC12 cells, the migration and proliferation of 4T1 cells increased compared to 4T1 alone (Fig. 2E). In addition, the coculture of 4T1 with PC12 cells contributed to cancer cell survival under starvation conditions, in a concentration-dependent manner (Fig. 2F). When 4T1 were cocultured with PC12 cell at a 1:1 or 10:1 ratio, the survival rate increased by 4.4- and 2.3-fold, respectively, compared to the survival of 4T1 that were seeded alone (P < 0.0001). Accordingly, nerve cells promote breast cancer cell proliferation, spread, and survival (1, 25).
Targeting neurons with bupivacaine reduces cancer cell viability
To suppress nerve/cancer interactions, bupivacaine was used as a neurotoxic agent. In PC12/4T1 coculture, 4T1 cell proliferation was about twofold higher compared to 4T1 cells seeded alone (P < 0.0001; Fig. 2G). However, addition of bupivacaine in neurotoxic concentration to the coculture medium annulled this effect, and cancer cell count decreased about twofold (P < 0.0001), similarly to 4T1 seeded alone treated with bupivacaine or untreated 4T1. Bupivacaine caused neuron-specific toxicity, which inhibited nerve/cancer cell interaction, followed by a decrease in cancer cell growth rates. These in vitro findings underscore the role of nerve cells in cancer development and the potential of targeting the neurons within the tumor tissue with bupivacaine to inhibit cancer progression.
The uptake of 100-nm liposomes by neurons and cancer cells
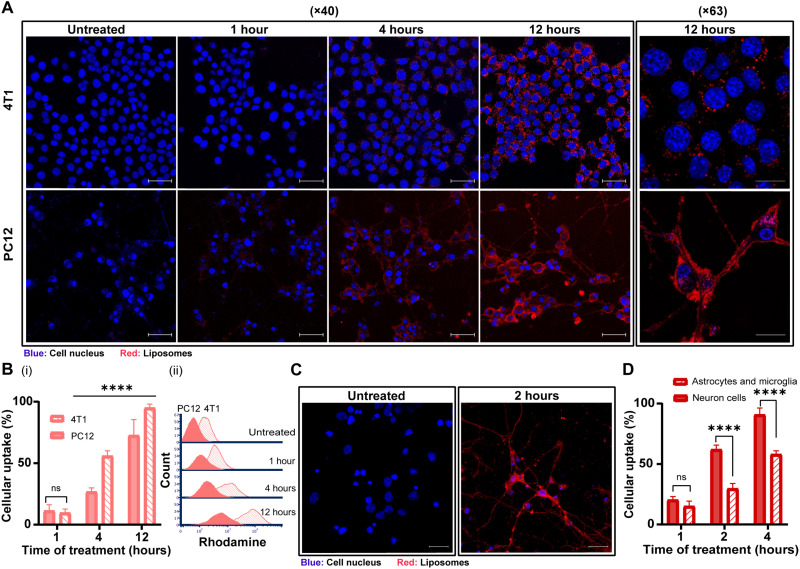
We compared the liposomal uptake in PC12 and 4T1 cells (Fig. 3). Flow cytometry analysis demonstrated an increased liposomal uptake over time by 4T1 and PC12 cells (Fig. 3B). After 1 and 12 hours, liposomal uptake by 4T1 cells increased from 9.5 ± 4.3 to 94.9 ± 3.2%, respectively, while the liposomal uptake by PC12 cells increased from 11.2 ± 5.0 to 72.6 ± 12.9%. Confocal microscopy analysis demonstrated that liposomes accumulated after 1 hour in the neuronal bodies, and after 4 hours, they were detected also along the axons (Fig. 3A). The liposomes appeared to be fused with the PC12 cell membrane, while, in 4T1 cells, liposomes were clustered in the cytoplasm (29, 30). We further examined the liposomal uptake in cortical primary neurons cells, where liposomes accumulated in both neuronal cell body and fibers after 2 hours (Fig. 3C). The liposomes were also taken up by primary astrocytes and microglia but to a lower extent than the primary neurons (Fig. 3D). Our findings emphasize the potential of nanotechnology in neurobiology research, such as neuronal targeting or recovery.
Fig. 3. The uptake of 100-nm liposomes by neuron cancer cells.
The cellular uptake of 100-nm rhodamine-labeled liposomes by PC12 and 4T1 cells over time was observed using confocal microscopy (A) (scale bars, 50 μm for ×40 magnification and 20 μm for ×63 magnification) and flow cytometry analysis [(B), i] and histogram [(B), ii]. Cellular uptake of liposomes by primary cortical neurons and astrocytes and microglia were imaged using confocal microscopy (C) (scale bars, 20 μm) and compared flow cytometry (D) analysis. Results of (B) (three independent repetitions performed in at least three replicates) and (D) (two independent repetitions performed in at least three replicates) are presented as means ± SD. One-way ANOVA with adjusted P value in multiple comparisons tests was used for statistical analysis; ****P < 0.0001.
Neurons are integral in breast cancer tumors
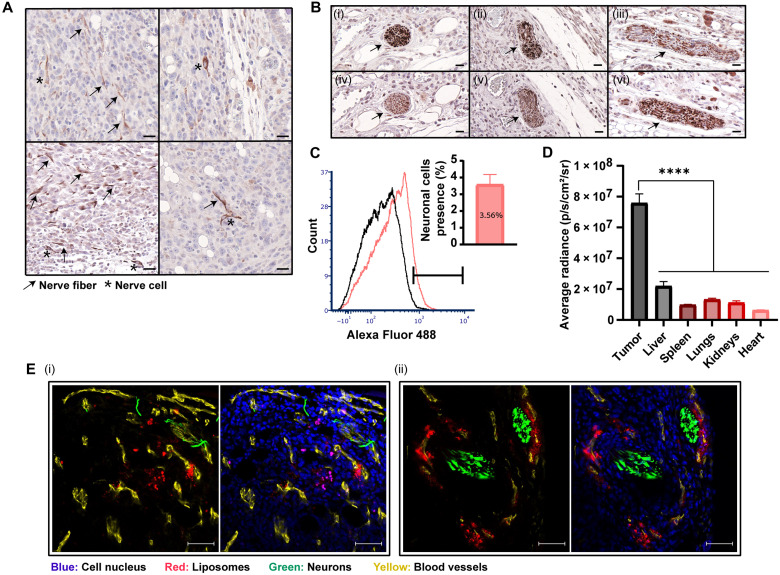
Identifying neurons in 4T1 breast cancer in vivo model was conducted using two different immunohistochemistry-specific neuronal markers, anti–βIII-tubulin and anti-PGP9.5 (Fig. 4, A and B). The nerve trunk, as well as neuronal cell bodies and axons, was detected and distributed throughout the non-necrotic areas of the tumor tissue. Quantitative flow cytometry analysis of tumors dissociated into single-cell suspensions after staining for adrenergic neurons (using anti–tyrosine hydroxylase) demonstrated that 3.6 ± 0.6% of the total tumor cell population were neurons (Fig. 4C). Nerves inside 4T1 tumor tissue demonstrate their active innervation into the tumor tissue and their potential involvement in tumor development (2, 25).
Fig. 4. Biodistribution of 100-nm liposomes to orthotopic breast cancer tumors and their delivery to tumor neurons.
Detection of nerve fibers and cells in 4T1 tumor tissue sections was performed using immunohistochemistry staining of anti–βIII-tubulin. Nerve fibers and cells are represented by arrows and stars, respectively (A) (scale bars, 20 μm). Coimmunostaining of serial sections (i and iv, ii and v, and iii and vi) of the 4T1 tumor tissue with anti-PGP9.5 and anti–βIII-tubulin demonstrated nerve trunk presence in breast cancer (B) (scale bars, 20 μm). Detection of adrenergic neurons with anti–tyrosine hydroxylase in 4T1 tumor tissue was performed using flow cytometry (C). Rhodamine-labeled liposomes were intravenously injected to mice bearing orthotopic 4T1 tumors. Twenty-four hours after administration, nanoparticle biodistribution to different tissues was quantified using IVIS (In Vivo Imaging System) analysis (D). Liposomal accumulation within the tumor tissue was also demonstrated by fluorescent histology (E) (scale bars, 50 μm). Nerve infiltration (βIII-tubulin+) and liposome accumulation are associated with blood vessels presence (CD31+). Results of (C) (n = 10) and (D) (n = 3) are presented as means ± SEM. One-way ANOVA with adjusted P value in multiple comparisons tests was used for statistical analysis; ****P < 0.0001.
Nanoparticle delivery to tumor neurons
We explored the capacity of liposomes to target neurons within the tumor tissue. Rhodamine-labeled liposomes were intravenously injected to mice bearing orthotopic 4T1 tumors. Twenty-four hours after administration, the tumors were excised and the liposomal biodistribution was assessed using ex vivo IVIS (In Vivo Imaging System) imaging (Fig. 4D and fig. S6A) and fluorescent histology (Fig. 4F). Increased liposomal accumulation at the tumor tissue was recorded compared to healthy tissues (Fig. 4D). Histological examination of the tumor tissue demonstrated nerve infiltration into the tumor and liposome distribution with the tumor neurons (Fig. 4F and fig. S6B). These findings are attributed to the infiltration of sympathetic nerve fibers into the tumor with its vasculature (2, 6) and to the preferential delivery of nanomedicines to the tumor microenvironment through its blood supply (10). Therefore, on the basis of our data, the intravenous treatment of bupivacaine-loaded liposomes enables the specific targeting of neurons within the tumor tissue. Previous clinical studies have demonstrated the biodistribution of nanoparticles to human tumors (31–35). However, the enhanced permeation and retention (EPR) effect in humans can vary between patients and among tumor types (34, 36). Therefore, the heterogeneity of the EPR should be evaluated in patients before the usage of the nanomedicines in the clinic. Next-generation nanomedicine will be tailored to the specific biomarkers of the patient (37). In addition, the liposome membrane was labeled with a Cy-5 lipid fluorophore and loaded with a magnetic resonance imaging contrast agent gadolinium (Gd) to track the biodistribution of the carrier and of the payload. The liposomes were intravenously injected to mice bearing orthotopic 4T1 tumors, and the biodistribution to the tumor, brain, and blood was quantified 24 and 48 hours after administration (fig. S7A). Quantitative elemental analysis Inductively coupled plasma (ICP) of Gd in the samples indicated that 11.3 ± 1.2% and 3.8 ± 2.0% of the injected dose accumulated in the tumor, 24 and 48 hours after the injection, respectively, while 12.2 ± 3.9% and 1.7 ± 0.6% were detected in plasma. Gd levels in the brain were below the limit of detection (<0.01 parts per million), suggesting negligible blood-brain barrier penetration in this model. Similar findings were noticed in the whole-animal IVIS imaging (fig. S7B).
Nanoparticle bupivacaine inhibits tumor growth and metastases
While the median lethal dose for intravenous administration of free bupivacaine in mice is 6 to 8 mg/kg body weight (38), the liposomal form of the drug allowed administering higher doses in a safe manner. L-BUP was administered at two different concentrations, 10 mg/kg body weight and 3.5 mg/kg. To compare the efficacy of L-BUP with other common treatment, liposomal doxorubicin (L-DOX) was administered at 4 mg/kg. No signs of toxicity were observed during the treatment with L-BUP, and mice’s weight increased gradually similarly to the untreated group (fig. S8). The higher dose of L-BUP at 10 mg/kg inhibited tumor growth, similarly to L-DOX (Fig. 5, B and C), while L-BUP at 3.5 mg/kg did not affect tumor progression (P < 0.05). Therefore, a higher dose of bupivacaine is needed to achieve a therapeutic effect. This is attributed to the fact that only a fraction of the injected nanoparticles reach the tumor bed, where the lower L-BUP dose was below the neuronal therapeutic threshold (fig. S7) (39).
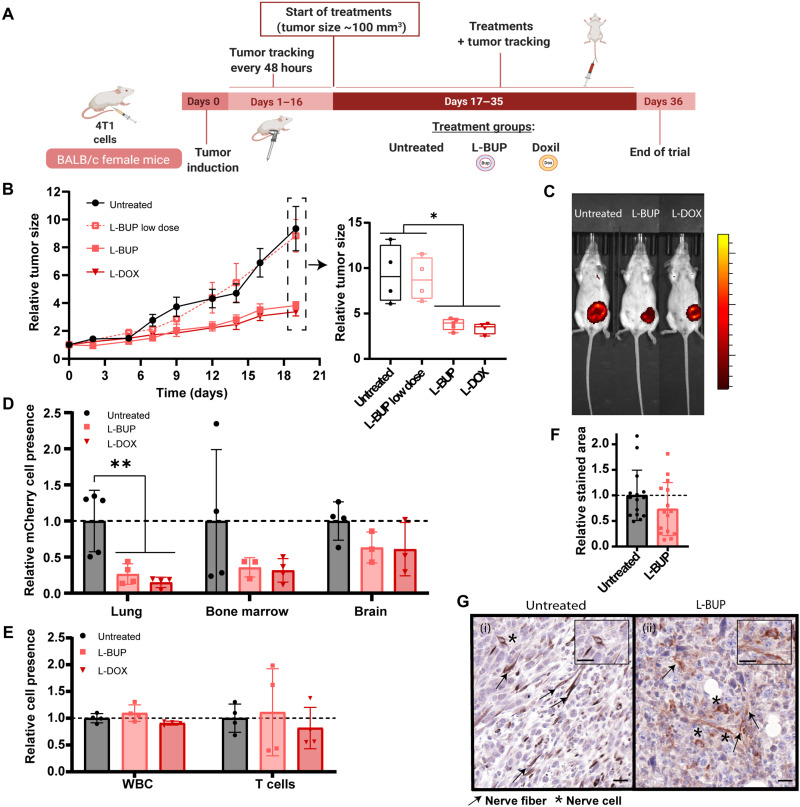
Fig. 5. L-BUP inhibits tumor growth and metastases.
Once the tumors reached 100-mm3, L-BUP at a high dose (10 mg/kg) or low dose (3.5 mg/kg), or L-DOX (4 mg/kg body weight, positive control), was intravenously administered (A). Tumors were sized every 2 or 3 days (B) (normalized to day 0) and were imaged by IVIS at the end of the trial (C). L-BUP treatment reduced metastases formation in lung (P < 0.01), bone marrow (P < 0.6), and brain (P < 0.8), compared to the untreated group. Metastases were detected by dissociating the tissue into a single-cell suspension and then detecting 4T1 mCherry using flow cytometry (D) (normalized to the untreated group). Moreover, the flow cytometry analysis indicated that there was no change in the level of total white blood cell (CD31+) and T cell (CD45+/CD3+) count in the tumor tissue in the different treatments (E) (normalized to the untreated group). Nerve density in 4T1 tumor tissue sections, with and without L-BUP treatment, was evaluated by immunohistochemistry staining and analysis of anti–βIII-tubulin (F) (normalized to the untreated group). Nerve structure was damaged because of L-BUP treatment [(G), ii] compared to the untreated group [(G), i]. Nerve fibers are represented by arrows, and nerve cells are represented by stars (G) (scale bars, 20 μm). Results of (B) (4 ≤ n ≤ 6) are presented as means ± SEM. Results of (D to F) (3 ≤ n ≤ 5) are presented as means ± SD. Two-tailed unpaired Student’s t test was used for statistical analysis of (B and F) and one-way ANOVA for (D and E) with adjusted P value in multiple comparisons tests; *P < 0.05 and **P < 0.01.
Five weeks after tumor induction mice were euthanized, and the lungs, brain, and bone marrow were resected and dissociated into single cells for quantitative detection of metastasis by flow cytometry (Fig. 5D). L-BUP inhibited metastases in the lung (P < 0.01), bone marrow (P < 0.6), and the brain (P < 0.8) in comparison to the untreated group. The L-BUP treatment was as effective as the L-DOX treatment in preventing metastasis in the lung, bone marrow, and brain (non-significant statistical difference between L-BUP and L-DOX).
The effect of the different treatments on the infiltration of immune cells (CD45+) and T cells (CD45+/CD3+) was examined using flow cytometry (Fig. 5E). All the treatment groups had a similar immune cell profile, suggesting that the observed therapeutic effect of L-BUP is related to nonimmune mechanisms, such as the involvement of the nervous system, and not as a result of immune response activation.
The difference in nerve density within the tumor tissue between L-BUP and the untreated group was examined by immunohistochemistry staining of anti–βIII-tubulin in different sections. Somewhat reduced tumor nerve density was recorded in the L-BUP treatment group (P ≈ 0.2; Fig. 5F). However, the integrity of all the observed neurons in the L-BUP treatment group were compromised (Fig. 5G), similarly to neurons exposed to the bupivacaine in vitro (Fig. 1B, ii). Immunohistochemistry staining of anti–lactic dehydrogenase (LDH) and hematoxylin and eosin (H&E) staining indicated that the L-BUP did not induce direct toxicity to the cancer cells, contrarily to the L-DOX–treated tumors that were highly necrotic (fig. S9) (40). The non-necrotic areas of the tumor accounted for 43.5 ± 15.4% and 18.5 ± 3.0% in the L-BUP and L-DOX treatment groups, respectively (P < 0.05), and 43.5 ± 13.5% of the untreated control group. The molecular indication for glycolytic metabolism in cancer cells of the L-BUP–treated tumors (positive for anti-LDH staining), suggests that tumor size reduction following the L-BUP treatments was mainly independent of direct toxicity of bupivacaine to the cancer cells. Overall, we used bupivacaine-loaded liposomes to suppress nerves within the tumor microenvironment and demonstrated that L-BUP inhibited tumor growth and metastatic dissemination without causing any toxic effect nor sever immunogenic response.
DISCUSSION
Nerves actively infiltrate the tumor microenvironment and promote tumor development and progression through neurotransmitter secretion. Their recruitment is driven by the cancer cells, through the secretion of neurotrophic factors and cytokines (1, 24). Here, we present the use of L-BUP as a mean to reduce tumor-associated neurons and total tumor mass. In coculture of PC12 and 4T1, free bupivacaine reduced the viability of the neuronal cells. While previous studies focused on local administration of bupivacaine to surgical and cancer sites (16, 38, 41), here, we developed a nanoscale PEGylated L-BUP formulation to improve tumor uptake through intravenous administration. Liposomes (100 nm) intravenously administered to mice bearing orthotopic 4T1 tumors were distributed throughout the tumor and specifically with the tumor neurons, curbing tumor growth. Hence, L-BUP may be leveraged as an alternative tool for treating breast cancer. In addition, non-opioid L-BUP (38) may be a new strategy for dealing with cancer pain owing to sensory nerves in the tumor microenvironment (1).
Overall, we demonstrate the collaborative interactions between nerves and cancer and the potential of analgesic nanotechnology to suppress these interactions. This study suggests that targeting nerves in the tumor tissue using non-opioid anesthetic nanoparticles is a potential new clinical approach that can improve breast cancer therapy.
MATERIALS AND METHODS
Bupivacaine-loaded liposomes preparation
Lipid mixture of HSPC or DPPC or DMPC (Lipoid, Ludwigshafen, Germany), cholesterol (Sigma-Aldrich, Rehovot, Israel), and DSPE–PEG-2000 (Lipoid, Ludwigshafen, Germany) in molar percentages of 55:40:5 was dissolved in warm absolute ethanol. Bupivacaine was actively loaded into the liposomes using the ammonium sulfate gradient method (16). Shortly, once all the lipids were completely dissolved in ethanol, the lipid suspension was added into 250 mM ammonium sulfate solution to a final concentration of 50 mM total lipids, which corresponds to 1.5 × 1013 liposomes/ml. To obtain homogenous 100-nm liposomes, the liposome mixture was extruded through 400-, 200-, 100-, and 80-nm pore-size polycarbonate membranes (Whatman, Newton, MA, USA) using a LIPEX extruder (Northern Lipids, Vancouver, Canada) at a temperature that is above the Tm (70°, 60°, or 50°C for HSPC, DPPC, and DMPC, respectively). The liposome solution was dialyzed against saline (150 mM NaCl at pH 5.5) (1:1000 volume ratio) using a 12- to 14-kDa dialysis membrane (Spectrum Laboratories Inc., USA) at 4°C and exchanged three times (after 1 hour, 4 hours, and overnight). For active loading, bupivacaine hydrochloride (Sigma-Aldrich, Rehovot, Israel) was dissolved in saline at pH 5.5 and added to the ammonium sulfate liposomes to reach a final concentration of 5 mg/ml. The mixture was incubated at 800 rpm for 1 hour at a temperature that is above the Tm. The non-encapsulated bupivacaine was removed by dialysis against saline in the same manner as described above.
Liposomes size analysis included mean diameter (in nm) and particle size distribution polydispersity index (PDI) measurements that were carried out by dynamic light scattering using a Zetasizer Nano ZSP (Malvern, UK). Particles concentration (liposomes/ml) measurements were carried out using a Zetasizer Ultra (Malvern, UK).
Rhodamine-labeled liposomes preparation
Rhodamine-labeled liposomes were prepared using ethanol injection method, as described above. Shortly, 16:0 Liss Rhod PE (Avanti Polar Lipids, Alabaster, AL, USA) was added to the lipid mixture at 0.1% molar ratio. As for the buffers, the lipid mixture was injected into phosphate-buffered saline (PBS; Sigma-Aldrich, St. Louis, USA). To compare between different liposomal compositions, liposomes composed of HSPC, DMPC, and DPPC were prepared in the same molar ratios as mentioned previously. As for the liposomes without DSPE–PEG-2000, the composition was HSPC:cholesterol in molar percentages of 60:40. In addition, when preparing the DMPC-liposomes, 14:0 Liss Rhod PE was used instead of 16:0.
DOX-loaded liposomes preparation
DOX (TEVA, Israel) was actively loaded into 100-nm liposomes using the ammonium sulfate gradient method (39). The dissolved lipid mixture of 55:40:5 of HSPC, cholesterol, and DSPE–PEG-2000 was injected into 120 mM ammonium sulfate solution to reach a final concentration of 50 mM total lipids, which corresponds to 1.5 × 1013 liposomes/ml. The liposome mixture was extruded as described above and then dialyzed against 10% w/w sucrose (1:1000 volume ratio) with three exchanges during 24 hours. For active loading, DOX was dissolved in 10% (w/w) sucrose and added to the ammonium sulfate liposomes to reach a final concentration of 2 mg/ml. The mixture was incubated in 65°C at 800 rpm for 1 hour. The non-encapsulated DOX was removed by dialysis against 10% (w/w) sucrose in the same manner as previously described.
Cryogenic transmission electron microscopy
Cryogenic transmission electron microscopy (cryo-TEM) imaging was performed using a FEI (Thermo Fisher Scientific) Talos 200C high-resolution TEM (Technion Center for Electron Microscopy of Soft Matter, at the Wolfson Department of Chemical Engineering). Specimen preparation was carried out at controlled conditions of 25°C and 100% relative humidity in a Controlled environment vitrification system. A drop of diluted L-BUP (0.5 mM) was placed on a carbon-coated perforated polymer film, supported on a 200-mesh TEM grid, and mounted on a tweezer. The drop was thinned into a film with a filter paper–covered metal strip and then was quickly plunged into liquid ethane at its freezing point (−183°C). The grid was transferred under controlled conditions into a Gatan 626 (Gatan, Pleasanton, CA) cryo-holder and imaged at −175°C. Images were recorded digitally using an FEI Falcon III highly sensitive direct-imaging camera. A Volta “phase plate” was used to enhance image contrast.
Lipids and bupivacaine concentration measurements
High-performance liquid chromatography (HPLC; 1260 Infinity, Agilent Technologies, Santa Clara, CA) equipped with an evaporative light scattering detector (ELSD) and ultraviolet detectors was used to quantify the lipid composition of the liposomes and the loaded bupivacaine concentration. Absorption was measured at a wavelength of 230 nm. The separation was achieved using Luna C18 column (5 mm, 100 Å; Phenomenex LTP, Aschaffenburg, Germany). The mobile phase consisted of three solutions: A, 0.2% trifluoroacetic acid (TFA) in water; B, 0.2% TFA in methanol; and 0.2% TFA in isopropanol. Bupivacaine separation was achieved by starting conditions of 80% A and 20% B, followed by a linear gradient up to 10% A and 90% B for 15 min at 40°C with constant flow rate of 1.5 ml/min. Then, lipid separation was completed by using the last solvent composition for five more minutes and then by gradually changing the composition to 80% B and 20% C within 22 min at 40°C with constant flow rate of 2.2 ml/min. The solvent composition gradually returned to the opening conditions within 18 min. ELSD settings were adjusted to 1.6 SLM of the inert gas flow, 60°C as the nebulizer temperature, and 80°C as the evaporator temperature. Sample was prepared by dilution of the liposomes 1:20 in methanol, and injection volume was 10 μl. Linear calibration curves were obtained according to the peak areas in the chromatograms and used for quantification of analyzed samples concentration.
To verify the loaded bupivacaine concentration in another method, the liposomes were diluted 50-fold with ethanol (to release the encapsulated bupivacaine) and measured at a wavelength of 230 nm by plate reader (Tecan, Mannedorf, Switzerland). Calibration curve was prepared by diluting bupivacaine hydrochloride together with the ammonium sulfate liposomes in ethanol.
The encapsulation efficiency (60%) was determined as the percentage of the final loaded bupivacaine concentration according to HPLC (3 mg/ml) to the initial bupivacaine concentration that was used for the remote loading (5 mg/ml). The drug-to-lipid mole ratio in the nanoparticles (0.3) was calculated by dividing the loaded bupivacaine concentration (9.2 mM) by the final lipid concentration (30.5 mM), as was determined by HPLC (fig. S2).
Bupivacaine release profile
L-BUP was dialyzed against saline (1:10 volume ratio) at 4° or 37°C at 200 rpm. The level of free bupivacaine in the extraliposomal buffer was quantified at the desired time intervals by measuring 100 μl of sample at the plate reader in 230-nm wavelength. The sample was then returned back to the incubation until the next measurement. The percentage of bupivacaine released was determined by the ratio of free bupivacaine released to total encapsulated bupivacaine concentration.
Gadolinium liposomes biodistribution
Cy-5–labeled liposomes loaded with gadolinium (Gd) were also prepared at the ethanol injection method as described before. Briefly, 0.2% molar percentage of DSPE–PEG-2000–Cy-5 (Biochempeg, Watertown, USA) was added to the lipid mixture, which was then injected into PBS solution containing diethylenetriaminepentaacetic acid gadolinium(III) (167 mg/ml) dihydrogen salt hydrate (Sigma-Aldrich, Rehovot, Israel) to reach a final concentration of 50 mM total lipids, which corresponds to 1.5 × 1013 liposomes/ml. The liposome mixture was extruded as described above and then dialyzed against PBS (1:1000 volume ratio) with three exchanges during 24 hours. Once the 4T1 mCherry tumor reached 100 mm3, 200 μl of the liposomes was intravenously injected to mice bearing orthotopic 4T1 tumors, and 24 and 48 hours after injection, the mice were euthanized and the tumor, brain, and blood were collected. The blood was collected using syringe rinsed with heparin and then centrifuged for 2 min at 1000g, and only the plasma was collected. The brain extraction was conducted after PBS perfusion. All the tissues were weighed and burned at 550°C for 5 hours, and their ashes were dissolved in 1% nitric acid. Then, the mixtures were filtered through 0.45-μm syringe filters, and the samples were analyzed using inductively coupled plasma optical emission spectroscopy (5110 ICP-OES; Agilent, CA, USA). Calibration curve was prepared from a dilution of calibration standard (Sigma-Aldrich, Rehovot, Israel) in 1% nitric acid. Gd emission was measured at 342.246 nm, and on the basis of the calibration curve, concentrations were calculated using ICP-OES software. The values were divided by the injected dose concentration to determine the percentage out of the injected dose. The final presented values were normalized by dividing by the organ’s weight.
Cell culture
All cells were cultured in 37°C in a humidified atmosphere and 5% CO2 in air, and every 2 days, a fresh medium was added to the cells. Triple-negative breast cancer cell line (4T1) [American Type Culture Collection (ATCC)] were grown in RPMI 1640 (Sigma-Aldrich, Rehovot, Israel) supplemented with 10% (v/v) of fetal bovine serum (FBS; Biological Industries, Beit Haemek, Israel), 1% (v/v) penicillin-streptomycin solution [100 IU/ml of Penicillin G Sodium Salt and streptomycin sulfate (100 μg/ml)] (Pen-Strep), and 1% (v/v) l-glutamine (Biological Industries, Beit Haemek, Israel). 4T1 cells expressing mCherry were developed (42) and provided by R. Satchi-Fainaro (Cancer Research and Nanomedicine Laboratory, Tel Aviv University). Puromycin was added to cell medium in a final concentration of 10 μg/ml.
KPC cells were established in the laboratory of S. K. Batra. KPC cells were LSL KrasG12D/+;LSLTrp53R172H/+ of pancreatic carcinomas, along with inactivation of the ;Pdx-1-Cre (KPC) transgenic mice (43). Cells were cultured in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) supplemented with 10% (v/v) of FBS, 1% (v/v) Pen-Strep, and 1% (v/v) l-glutamine.
PC12 cells (ATCC) were provided by O. Shefi (Neuroengineering and Regeneration laboratory, Faculty of Engineering, Bar-Ilan). Cells were cultured in suspension in the RPMI medium supplemented with 10% (v/v) horse serum (HS; Biological Industries, Beit Haemek, Israel), 5% (v/v) FBS, 1% (v/v) Pen-Strep, 1% (v/v) l-glutamine, and 0.2% (v/v) amphotericin B (Biological Industries, Beit Haemek, Israel). To induce their differentiation, PC12 cells were seeded on collagen type l (Corning Inc., NY, USA)–coated plates in serum-reduced media (1% HS) supplemented with murine β-NGF (50 ng/ml; PeproTech, Israel).
Primary cultures of neonate rat pups (P0 to P4) cortical neurons were established and provided by S. Berlin (Department of Neuroscience, The Ruth and Bruce Rappaport Faculty of Medicine, Technion—Israel Institute of Technology) and were produced as was described previously (44). These cells were used for confocal analysis of liposomal uptake. Primary cultures of embryonic day 13.5 (E13.5) or E16.5 mouse embryo cortical neurons were provided by Y. Buganim (Department of Developmental Biology and Cancer Research, Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School) and were produced as was previously described (45). These cells were used for flow cytometry analysis of liposomal uptake. Cells were plated into six-well plate coated with poly-l-lysine (0.1 mg/ml). Neurons were grown in Neurobasal medium (Gibco, Ireland) plus B27 supplement (Gibco, Ireland) and GlutaMAX (Gibco, Ireland) until day 5; then, medium was replaced with Neurobasal medium plus B27 with Cytosine β-d-arabinofuranoside (Ara-C) at 10 μM, to avoid the proliferation of glial cells, and without GlutaMAX for another 11 days, up to 14 days in vitro, where neuronal maturity is considered to be reached.
Breast cancer model
Eight- to 10-week-old BALB/c female mice (Envigo, Jerusalem, Israel) were used as breast cancer animal models. Fifty microliters of 4 × 105 4T1 cells was subcutaneously injected into the fourth mammary fat pad to obtain primary tumor model. All animal experiments were approved by, and in compliance with, the Inspection Committee on the Constitution of the Animal Experimentation at the Technion, Israel Institute of Technology.
Confocal imaging
Confocal microscopy (LSM 700, Zeiss, Germany) was used to examine several experiments such as coculture studies, liposomal uptake, etc. For tissue culture imaging, PC12 cells were stained overnight with rabbit polyclonal anti–tyrosine hydroxylase (Abcam, Cambridge, UK), following a staining with donkey polyclonal anti-rabbit IgG H&L conjugated Alexa Fluor 488 (Abcam, Cambridge, UK). Antibodies were diluted by 1:1000 in a blocking serum. All cells were stained with Hoechst (1 μg/ml) for nuclei labeling. Acquisition was performed using the ZEN software and applying the 405-, 488-, 555-, and 639-nm lasers.
Imaging and morphometric analysis of PC12
To study the effect of cancer cells on nerve cells, PC12 cells were cocultured with 4T1 mCherry and KPC. Cells were seeded in an optical μ-slide eight-well collagen IV–coated plate (ibidi, Madison, WI) in serum-reduced media for 72 hours, at a density of 3 × 104 cells per well for PC12 cells and at a density of 1.5 × 104 cells per well for 4T1 and KPC. As a positive control to neuronal differentiation, 24 hours from seeding, NGF was added to the medium of some of the PC12 cells that were seeded alone (17). After 72 hours from seeding, PC12 cells were stained and imaged using a confocal microscope. All the confocal images were taken to quantitative morphometric analysis by the IMARIS software, which enables semiautomatic tracing of neurites and length measurements. The analysis included two accepted morphological parameters at the single-cell level: (i) the percentage of differentiated cells out of the total cell population and (ii) the average number of neurites, which can be either axons or dendrites, extending from the cell body of a differentiated cell. A cell was regarded as differentiated when its neurite length exceeded 20 μm from the nucleus. Morphological parameters and statistics were measured for at least 20 fields of three independent repetitions.
Quantification of 4T1 cell confluence in PC12/4T1 coculture
To study the effect of nerve cells on cancer cells, PC12 cells were cocultured with 4T1 in serum-reduced media for 96 hours. 4T1 mCherry cells were seeded in 24-well plates at a density of 7 × 104 cells per well, and PC12 cells were seeded together with 4T1 in increasing density of 0, 7 × 102, 7 × 103, and 7 × 104 cells per well. Each plate was scanned using GE InCell Analyzer2000 to obtain random images (13 fields per well and six wells per each treatment) using 4′,6-diamidino-2-phenylindole (DAPI) and Texas Red channels. The plates were scanned 24 hours from seeding (was set as start point) and again after 96 hours (was set as end point). Before each scanning, cells were stained with Hoechst (1 μg/ml ) for nuclei labeling. The images were analyzed using the InCell software to quantify the amount of nuclei of 4T1 mCherry cells. To determine the percentage of the survival rate of 4T1 mCherry cells, the obtained value for each well at the end point of the experiment was divided by the value of the same well at the start point.
A similar experiment was conducted with the presence of bupivacaine. PC12 cells were cocultured with 4T1 mCherry in serum-reduced media in 24-well plates at a density of 10 × 104 cells per well for each type cell. After 72 hours from seeding, the medium was replaced to a fresh reduced medium without or with bupivacaine (0.5 mg/ml), and the cells were incubated for another 6 hours. Then, cell nuclei were stained with Hoechst (1 μg/ml), and each plate was scanned by GE InCell Analyzer2000 using DAPI and Texas Red channels. The obtained images were analyzed using the InCell software to quantify the amount of 4T1 mCherry cells according to their intensity. To determine the relative cell count of 4T1 mCherry cells, all of the values were normalized to the value of the untreated group (4T1 mCherry cells that were seeded alone).
Cell viability assay
Cell viability was determined using a commercial 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) viability assay (Sigma-Aldrich, Rehovot, Israel). That assay was used to determine how highly 4T1 proliferation was stimulated by NE bitartrate (Sigma-Aldrich, Rehovot, Israel). 4T1 cells were seeded in 96-well plates at a density of 1 × 104 cells per well. After 24 hours, the medium was replaced with serum-reduced medium for another 24 hours. Then, the medium was replaced with fresh serum-reduced medium supplemented with an increasing concentration of NE (0, 50, 100, and 200 μM), and the cells were incubated for 24 hours. Then, the medium was removed, and the MTT assay was conducted.
MTT assay was also used to determine the toxicity of bupivacaine on 4T1 and PC12. 4T1 cells were seeded in 96-well plates at a density of 2.5 × 104 cells per well 2 days before the experiment. PC12 cells were seeded separately in 96-well plates at a density of 3 × 104 cells per well 6 days before the experiment and treated two times with NGF to be fully differentiated. Both cell types were treated with an increasing concertation of bupivacaine (0, 0.5, 0.7, and 0.9 mg/ml) for 6 hours. Then, the medium was removed, and the MTT assay was conducted. This experiment was also imaged by confocal microscopy, as described above. The toxicity of bupivacaine was also evaluated on cortical primary neuronal culture. The cells were seeded in 96-well plate at a density of 5 × 104, and at day 10 in vitro, bupivacaine (B Braun Medical Ltd.) was added to the media with an increasing concentrations of 0, 0.5, 0.7, and 0.9 mg/ml for 6 hours. Then, the medium was removed, and the MTT assay was conducted.
In addition, MTT assay was also used to examine the toxicity effect of PEG on PC12 cells. The cells were seeded, treated with NGF as described previously, and incubated for 4 and 24 hours with liposomes in a final concentration of 0.5 mM total lipids, which corresponds to 1.5 × 1011 liposomes/ml. Then, the medium was removed, and the MTT assay was conducted.
Migration assay
PC12 and 4T1 mCherry cells were seeded separately in two-well silicone insert (ibidi, Madison, WI) on an optical μ-slide eight-well collagen IV–coated plate (ibidi, Madison, WI). As a control, 4T1 mCherry cells were seeded alone. PC12 cells were seeded at a density of 8.5 × 104 cells per well in serum-reduced media supplemented with NGF, and 24 hours later, mCherry cells were seeded at a density of 3.5 × 104 cells per well. Twenty-four hours later, the media and the silicone insert were removed, the wells were filled with medium, and the plate was imaged using ZEISS Cell Observer SD for live imaging using bright-field and CY3 channel. The cells were monitored every 15 min for 30 hours, and the images were analyzed by IMARIS software to obtain the area cover of 4T1 mCherry cells. All the values were normalized to the area cover measured at the beginning of the experiment.
Cytokine kit array
PC12 cells were cocultured with 4T1 for 72 hours. Cells were seeded in six-well plate in serum-reduced media at a density of 4 × 105 cells per well for PC12 cells and at a density of 2.5 × 105 cells per well for 4T1. In addition, 4T1 cells were seeded alone at a density of 3 × 105 cells per well. Cytokine profile of the conditioned medium was determined using Mouse Cytokine Antibody Array (Abcam, Cambridge, UK). The membranes were imaged using FUSION FX 6 (VILBER, France). Semiquantitative data were assessed using the ROI tool in Fiji software, and the obtained values were normalized to the positive control of each membrane.
Evaluation of liposomal cellular uptake
PC12 cells were seeded 6 days before the experiment and treated twice with NGF to be fully differentiated. PC12 and 4T1 were incubated for different time durations with rhodamine-labeled liposomes in a final concentration of 0.5 mM total lipids, which corresponds to 1.5 × 1011 liposomes/ml. Then, the culture medium was removed, and the cells were rinsed with PBS three times, incubated with 0.25% trypsin-EDTA for 5 min, and centrifuged at 400g for 5 min. The cell pellet was resuspended in fresh medium. Cellular uptake was detected in the mCherry channel after acquisition of 10,000 single cells per sample using a flow-activated cell sorter (FACS; FACSAria III, BD Biosciences). Analyses were conducted using FCS Express (De Novo software).
Primary neurons were incubated for different time durations with rhodamine-labeled liposomes in a final concentration of 5 mM total lipids, which corresponds to 1.5 × 1012 liposomes/ml. After detaching and rinsing the cells as described above, cells were incubated in 10% FBS for 30 min at room temperature (RT). Subsequently, the cells were incubated with specific neuronal marker CD24-FITC (fluorescein isothiocyanate) clone M1/69 antibody (STEMCELL Technologies, Canada) (46) diluted 1:100 for 30 min on ice. Then, the cells were rinsed with PBS three times and resuspended in PBS with 5% (v/v) FBS. Cellular uptake was detected in the mCherry (liposomes) and FITC (neurons) channels using a Beckman Coulter Gallios cytometer.
In vivo biodistribution
4T1 cells were injected to the mammary fat pad of BALB/c female mice as was described above. Once the tumor reached 200 mm3, 200 μl of rhodamine-labeled liposomes were intravenously injected. Twenty-four hours after injection, mice were euthanized and tumors, livers, spleens, lungs, kidneys, and hearts were extracted and imaged by IVIS. Moreover, liposome accumulation was visualized using fluorescence histology imaging.
IVIS imaging
As part of the biodistribution experiment, mice were euthanized and the extracted tissues were imaged ex vivo using the IVIS SpectrumCT Pre-Clinical In Vivo Imaging System (PerkinElmer, MA, USA) at an excitation of 570 nm and emission of 620 nm, binning of 2, f-stop of 2, and 8-s exposure (fig. S6A) for rhodamine-labeled liposomes and at an excitation of 675 nm and emission of 720 nm, binning of 2, f-stop of 2, and 1-s exposure for Cy-5–labeled liposomes loaded with Gd. Quantitative data from all images were obtained using ROI tool in Living Image software. A control (noninjected) mouse was used for analysis, and its average radiance of each tissue was subtracted from the average radiance of the injected mice.
In addition, at the end of the efficacy experiment 4T1 mCherry tumors were imaged by a whole-animal IVIS imaging at an excitation of 570 nm and emission of 620 nm, binning of 8, f-stop of 2, and 0.5-s exposure.
Immunohistochemistry analysis
4T1 tumors were fixated using formalin solution, neutral-buffered, 10% histological tissue fixative (Sigma-Aldrich, Rehovot, Israel) for at least 24 hours before embedding in paraffin and sectioning. Tissue sections were deparaffinized in a xylene ethanol gradient [xylene, xylene/ethanol (1:1 v/v), absolute ethanol, 95% ethanol, 70% ethanol, and 50% ethanol] for 3 min each and lastly placed in distilled water (DW). Antigen retrieval was done in 10 mM tri-sodium citrate solution at pH 6 tittered with HCl. Ready-to-use normal goat serum (2.5%; Vector Laboratories) was used for blocking. Incubation with primary antibody was at 4°C overnight with either rabbit anti–βIII-tubulin, rabbit anti-PGP9.5, or rabbit anti-LDH (1:2000; Abcam, Cambridge, UK). Tissue sections were rinsed three times with DW and then were incubated for 30 min in 0.3% hydrogen peroxide solution for blocking of endogenous peroxidase activity. Tissue sections were then washed in DW and incubated with ready-to-use secondary goat anti-rabbit antibody conjugated to horseradish peroxidase (MP7451 kit, Vector Laboratories) for 40 min at RT. Tissue sections were rinsed three times with DW, then, for color development, were incubated with DAB solution (SK4105 kit, Vector Laboratories) for 3 min, washed with DW, and counterstained with hematoxylin. Slides were scanned using 3DHistech Panoramic 250 Flash III automated slide scanner (3DHistech, Budapest, Hungary). Quantitative data from the slides were assessed using color deconvolution tool with H DAB vector in Fiji software. The images were adjusted to the same threshold, and the stained area was measured. Regarding the measurement of the nerve density within the tissue, the obtained values were normalized to the untreated group. Regarding the measurement of the LDH expression, which evaluates the glycolytic metabolism, the obtained values are presented as the percentage of the stained area out of the tissue section. H&E staining was conducted according to the standard protocol, and the area of the viable regions was measured using CaseViewer software.
Fluorescence immunohistochemistry analysis
4T1 tissue sections (10 μm thick) were fixed in 4% paraformaldehyde for 10 min, followed by 10 min of permeabilization with 0.1% Triton X-100. Normal donkey serum (10%; Abcam, Cambridge, UK) in PBST (PBS with 0.025% Triton X-100) was used for blocking for 2 hours. Tissue sections were incubated overnight at 4°C in blocking buffer containing rabbit anti–βIII-tubulin (1:1000; Abcam, Cambridge, UK) for neurons detection and goat anti-CD31 (1:200; R&D Systems, Minneapolis) for blood vessels detection. Following overnight incubation, sections were rinsed three times with PBST and then incubated with blocking buffer containing species-appropriate secondary antibodies donkey anti-rabbit Alexa Fluor 488 and donkey anti-goat Alexa Fluor 647 (1:500; Abcam, Cambridge, UK) for 2 hours in RT. Tissue sections were rinsed three times with PBST, then mounted with DAPI-containing Fluoromount (BioLegend, California), coverslipped, and stored in the dark at 4°C until confocal imaging.
In vivo therapeutic efficacy
4T1 mCherry cells were subcutaneously injected, as described above. Once the tumor reached 100 to 200 mm3 (about 17 days after 4T1 injection), mice were divided to four different groups (five mice each) as follows: untreated, L-DOX (4 mg/kg body weight), and L-BUP in high (10 mg/kg) and low (3.5 mg/kg) dose. All of the treatments were administered by intravenous injection for 18 days. The high dose of L-BUP was injected twice a week, and the rest of the treatments were injected once a week. Mice weight and the tumor dimension were measured every 2 to 3 days. The tumor volume was measured using a caliper and calculated as (width2 × length)/2. To compare mice weight and tumor dimension between the various treatments group, each mouse’s tumor size and the body weight were normalized to the initial size and weight that were measured at day 0 (the day before the first treatment).
Metastases detection
Mice were euthanized, and the brain, lungs, bone marrow, and tumor were extracted and held in RPMI at 4°C until initiating the organ’s dissociation. Lungs and tumors were enzymatically and physically dissociated using a gentleMACS machine (Miltenyi Biotec, Bergisch Gladbach, Germany) and dissociation kits (Miltenyi Biotec) following the machine dissociation protocols.
Brain tissue (47, 48) was cut to small pieces and enzymatically digested by incubation in RPMI supplemented with collagenase D (0.4 mg/ml; Sigma-Aldrich) and deoxyribonuclease I (0.2 mg/ml; Sigma-Aldrich) for 30 min in 37°C at 100 rpm. Then, the tissue was physically dissociated using gentleMACS. The suspension was passed through a 70-μm cell strainer (BD Biosciences, CA, USA) and centrifuged at 400g for 5 min. The pellet was resuspended with 7 ml of RPMI; then, 3 ml of 10:90 v/v of PBSX10:Percoll (GE Healthcare Bio-Sciences; Sigma-Aldrich) was added, and the tube was gently mixed. The mixture was centrifuged at 400g for 25 min in 4°C to form Percoll gradient; the supernatant was discarded, and the pellet of the cells was resuspended in PBS.
Bone marrow cells were harvested from the intact tibia and femur. The bone marrow was flushed out with PBS through a 25-gauge needle. Then, the PBS with the stromal cells were filtered in a 70-nm strainer and centrifuged at 500g for 5 min.
The single-cell suspension of each tissue was obtained by passing the cell suspension through a 35-μm cell strainer (BD Biosciences, CA, USA). After dissociating of the organs into a single-cell suspension, the 4T1 mCherry cells were detected in the mCherry channel using FACSAria and analyzed with FCS Express. Following acquisition of 10,000 single cells per sample, the average percentage of positive mCherry cells for each tissue was normalized to the average percentage measured at the control group.
Quantification of immune cells in the tumor tissue
After dissociation of the tumor tissue into single-cell suspension as was described above, cells suspension was incubated with CD45-FITC and CD3-APC (BioLegend, California) diluted 1:200 for 1 hour in the dark on ice. Cells were washed three times with PBS and analyzed using FACSAria. For each test sample, 10,000 single cells were acquired, and data analysis was performed using FCS Express. The average percentage of the positive population was normalized to the average percentage measured at the control group.
Nerve detection
4T1 cells were injected to the mammary fat pad of BALB/c female mice as was described above. Once the tumor reached 300 to 500 mm3 (about 3 weeks from 4T1 injection), or the treatments of the efficacy experiment were completed (about 36 days from 4T1 injection), mice were euthanized. Single-cell suspension in addition to tissue sections were produced from the tumors, and nerve detection was conducted by immunohistochemistry and flow cytometry analyses. As for the flow cytometry analysis, cells suspension was incubated with rabbit anti–tyrosine hydroxylase (Abcam, Cambridge, UK), following a staining with donkey anti-rabbit Alexa Fluor 488 (Abcam, Cambridge, UK). Antibodies were diluted by 1:1000 in a blocking serum. The analysis was performed using BD LSR-II Analyzer (Biosciences, San Jose, CA, USA), and data analysis was obtained using FCS Express.
Statistical analysis
All statistical analyses—Student’s t test, one-way ANOVA, two-way analysis of variance (ANOVA), and three-way ANOVA—were performed using Prism GraphPad software. Difference below P = 0.05 was considered statistically significant.
Acknowledgments
We thank A. Rolls from the Faculty of Medicine, Technion and T. Ben-Shaanan from University of California, San Francisco, USA for professional input and helpful discussions that greatly contributed to the paper. We also thank O. Adir and G. Kaneti for assistance in Fiji software analyses and I. Davidovich from the Technion Center for Electron Microscopy of Soft Matter for the cryo-TEM measurements. Images in this paper were created with BioRender.com and Adobe Illustrator. Funding: This project received funding from the European Union’s Horizon 2020 research and innovation program under the grant agreement no. 680242-ERC (Next-Generation Personalized Diagnostic Nanotechnologies for Predicting Response to Cancer Medicine). The authors also acknowledge the support of Israel Innovation Authority for the Nofar Grant (67967), the Israel Science Foundation (1778/13, 1421/17); The Israel Ministry of Economy for a Kamin Grant (52752, 69230); the Israel Ministry of Science Technology and Space—Office of the Chief Scientist (3-11878); the Israel Ministry of Science, Technology and Space (3-16963; 3-17418); the Israel Cancer Association (2015-0116); Leventhal 2020 COVID19 Research Fund (ATS #11947), the German-Israeli Foundation for Scientific Research and Development for a GIF Young grant (I-2328-1139.10/2012); the European Union FP-7 IRG Program for a Career Integration Grant (908049); the Phospholipid Research Center Grant (ASC-2018-062/1-1); the Ministry of Agriculture and Rural Development—Office of the Chief Scientist (323/19); the Louis family Cancer Research Fund; a Mallat Family Foundation grant; The Unger Family Fund; a Carrie Rosenblatt Cancer Research Fund; the Technion Integrated Cancer Center (TICC); the Russell Berrie Nanotechnology Institute; and the Lorry I. Lokey Interdisciplinary Center for Life Sciences and Engineering. A.S. acknowledges the Alon and Taub Fellowships. M.K. and M.S. wish to thank TEVA Pharmaceuticals The National Forum for Bio-Innovators (NFBI) for the Doctoral Fellowship. M.K. also wishes to thank to the Technion Integrated Cancer Center (TICC) Rubinstein scholarship and Robert B. Kalmansohn Fellowship Fund in the Emerson Life Sciences Building. M.P. wishes to thank the Israeli Ministry of Science and Technology for the Shulamit Aloni Doctoral Fellowship. H.A.-M. wishes to thank the Baroness Ariane de Rothschild Women Doctoral Program. A.O. wishes to thank the Basque Country Government for the Post-Doctoral Fellowship. Author contributions: A.S. supervised and directed the research. M.K. wrote the manuscript in addition to designing, performing, and analyzing all the experiments. M.S. and S.K. helped perform and analyze the in vitro and in vivo experiments. M.P., H.A.-M., and P.M.-R. helped perform the in vivo experiments. Y.B. and A.O. helped design and conduct the experiments on primary neurons. N.D. assisted in performing and analyzing the imaging experiments. D.H. helped with the histology staining and analyzing. J.S. and J.S.-R. assisted in data analysis. All authors helped improve the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Correspondence and requests for materials should be addressed to A.S. (avischroeder@gmail.com).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
REFERENCES AND NOTES
- 1.Zahalka A. H., Frenette P. S., Nerves in cancer. Nat. Rev. Cancer 20, 143–157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Q., Yang Y., Liang X., Du G., Liu L., Lu L., Dong J., Han H., Zhang G., The clinicopathological significance of neurogenesis in breast cancer. BMC Cancer 14, 484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mancino M., Ametller E., Gascon P., Almendro V., The neuronal influence on tumor progression. Biochim. Biophys. Acta 1816, 105–118 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Liebig C., Ayala G., Wilks J. A., Berger D. H., Albo D., Perineural invasion in cancer: A review of the literature. Cancer 115, 3379–3391 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Magnon C., Hall S. J., Lin J., Xue X., Gerber L., Freedland S. J., Frenette P. S., Autonomic nerve development contributes to prostate cancer progression. Science 341, 1236361 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Cole S. W., Nagaraja A. S., Lutgendorf S. K., Green P. A., Sood A. K., Sympathetic nervous system regulation of the tumour microenvironment. Nat. Rev. Cancer 15, 563–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amit M., Takahashi H., Dragomir M. P., Lindemann A., Gleber-Netto F. O., Pickering C. R., Anfossi S., Osman A. A., Cai Y., Wang R., Knutsen E., Shimizu M., Ivan C., Rao X., Wang J., Silverman D. A., Tam S., Zhao M., Caulin C., Zinger A., Tasciotti E., Dougherty P. M., El-Naggar A., Calin G. A., Myers J. N., Loss of p53 drives neuron reprogramming in head and neck cancer. Nature 578, 449–454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anselmo A. C., Mitragotri S., Nanoparticles in the clinic: An update. Bioeng. Transl. Med. 4, e10143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroeder A., Heller D. A., Winslow M. M., Dahlman J. E., Pratt G. W., Langer R., Jacks T., Anderson D. G., Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer 12, 39–50 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Brannon-Peppas L., Blanchette J. O., Nanoparticle and targeted systems for cancer therapy. Adv. Drug Deliv. Rev. 64, 206–212 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Mo R., Gu Z., Tumor microenvironment and intracellular signal-activated nanomaterials for anticancer drug delivery. Mater. Today 19, 274–283 (2016). [Google Scholar]
- 12.Byrne J. D., Betancourt T., Brannon-Peppas L., Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv. Drug Deliv. Rev. 60, 1615–1626 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Marcus M., Karni M., Baranes K., Levy I., Alon N., Margel S., Shefi O., Iron oxide nanoparticles for neuronal cell applications: Uptake study and magnetic manipulations. J. Nanobiotechnol. 14, 37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lammers T., Kiessling F., Hennink W. E., Storm G., Drug targeting to tumors: Principles, pitfalls and (pre-) clinical progress. J. Control. Release 161, 175–187 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Lammers T., Hennink W., Storm G., Tumour-targeted nanomedicines: Principles and practice. Br. J. Cancer 99, 392–397 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant G. J., Barenholz Y., Piskoun B., Bansinath M., Turndorf H., Bolotin E. M., DRV liposomal bupivacaine: Preparation, characterization, and in vivo evaluation in mice. Pharm. Res. 18, 336–343 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Gordon J., Amini S., White M. K., General overview of neuronal cell culture. Methods Mol. Biol. 1078, 1–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vance J. E., Campenot R. B., Vance D. E., The synthesis and transport of lipids for axonal growth and nerve regeneration. Biochim. Biophys. Acta (BBA) 1486, 84–96 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Suk J. S., Xu Q., Kim N., Hanes J., Ensign L. M., PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 99, 28–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nance E. A., Woodworth G. F., Sailor K. A., Shih T. Y., Xu Q., Swaminathan G., Xiang D., Eberhart C., Hanes J., A dense poly(ethylene glycol) coating improves penetration of large polymeric nanoparticles within brain tissue. Sci. Transl. Med. 4, 149ra119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calvo P., Gouritin B., Villarroya H., Eclancher F., Giannavola C., Klein C., Andreux J. P., Couvreur P., Quantification and localization of PEGylated polycyanoacrylate nanoparticles in brain and spinal cord during experimental allergic encephalomyelitis in the rat. Eur. J. Neurosci. 15, 1317–1326 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Calvo P., Gouritin B., Chacun H., Desmaële D., D’Angelo J., Noel J.-P., Georgin D., Fattal E., Andreux J. P., Couvreur P., Long-circulating PEGylated polycyanoacrylate nanoparticles as new drug carrier for brain delivery. Pharm. Res. 18, 1157–1166 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Beltrán-Gracia E., López-Camacho A., Higuera-Ciapara I., Velázquez-Fernández J. B., Vallejo-Cardona A. A., Nanomedicine review: Clinical developments in liposomal applications. Cancer Nanotechnol. 10, (2019). [Google Scholar]
- 24.Amit M., Na’ara S., Gil Z., Mechanisms of cancer dissemination along nerves. Nat. Rev. Cancer 16, 399–408 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Jobling P., Pundavela J., Oliveira S. M., Roselli S., Walker M. M., Hondermarck H., Nerve-cancer cell cross-talk: A novel promoter of tumor progression. Cancer Res. 75, 1777–1781 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Gao T., Shen Z., Ma C., Li Y., Kang X., Sun M., The CCL5/CCR5 chemotactic pathway promotes perineural invasion in salivary adenoid cystic carcinoma. J. Oral Maxillofac. Surg. 76, 1708–1718 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Dobrenis K., Gauthier L. R., Barroca V., Magnon C., Granulocyte colony-stimulating factor off-target effect on nerve outgrowth promotes prostate cancer development. Int. J. Cancer 136, 982–988 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Edman L. C., Mira H., Arenas E., The beta-chemokines CCL2 and CCL7 are two novel differentiation factors for midbrain dopaminergic precursors and neurons. Exp. Cell Res. 314, 2123–2130 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Sahay G., Alakhova D. Y., Kabanov A. V., Endocytosis of nanomedicines. J. Control. Release 145, 182–195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canton I., Battaglia G., Endocytosis at the nanoscale. Chem. Soc. Rev. 41, 2718–2739 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Lee H., Shields A. F., Siegel B. A., Miller K. D., Krop I., Ma C. X., Russo P. M. L., Munster P. N., Campbell K., Gaddy D. F., Leonard S. C., Geretti E., Blocker S. J., Kirpotin D. B., Moyo V., Wickham T. J., Hendriks B. S., 64Cu-MM-302 positron emission tomography quantifies variability of enhanced permeability and retention of nanoparticles in relation to treatment response in patients with metastatic breast cancer. Clin. Cancer Res. 23, 4190–4202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S.-J., Huang W.-S., Chuang C.-M., Chang C.-H., Lee T.-W., Ting G., Chen M.-H., Chang P. M.-H., Chao T.-C., Teng H.-W., Chao Y., Chen Y.-M., Lin T.-P., Chang Y.-J., Chen S.-J., Huang Y.-R., Lan K.-L., A phase 0 study of the pharmacokinetics, biodistribution, and dosimetry of 188 Re-liposome in patients with metastatic tumors. EJNMMI Res. 9, 46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrington K., Rowlinson-Busza G., Syrigos K., Uster P., Abra R., Stewart J., Biodistribution and pharmacokinetics of 111In-DTPA-labelled pegylated liposomes in a human tumour xenograft model: Implications for novel targeting strategies. Br. J. Cancer 83, 232–238 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrington K. J., Mohammadtaghi S., Uster P. S., Glass D., Peters A. M., Vile R. G., Stewart J. S., Effective targeting of solid tumors in patients with locally advanced cancers by radiolabeled pegylated liposomes. Clin. Cancer Res. 7, 243–254 (2001). [PubMed] [Google Scholar]
- 35.Puvanakrishnan P., Park J., Chatterjee D., Krishnan S., Tunnell J. W., In vivo tumor targeting of gold nanoparticles: Effect of particle type and dosing strategy. Int. J. Nanomedicine 7, 1251–1258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golombek S. K., May J. N., Theek B., Appold L., Drude N., Kiessling F., Lammers T., Tumor targeting via EPR: Strategies to enhance patient responses. Adv. Drug Deliv. Rev. 130, 17–38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward D. M., Shodeinde A. B., Peppas N. A., Innovations in biomaterial design toward successful RNA interference therapy for cancer treatment. Adv. Healthc. Mater. 10, 2100350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bupivacaine Hydrochloride Injection, USP, Hospira, Inc. (U.S. Food and Drug Administration (FDA). Center for Drug Evaluatuion and Research) (2009); www.accessdata.fda.gov/drugsatfda_docs/label/2010/071165s020lbl.pdf.
- 39.Abumanhal-Masarweh H., Koren L., Zinger A., Yaari Z., Krinsky N., Kaneti G., Dahan N., Lupu-Haber Y., Suss-Toby E., Weiss-Messer E., Schlesinger-Laufer M., Shainsky-Roitman J., Schroeder A., Sodium bicarbonate nanoparticles modulate the tumor pH and enhance the cellular uptake of doxorubicin. J. Control. Release 296, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ju C., Zhou J., Miao H., Chen X., Zhang Q., Bupivacaine suppresses the progression of gastric cancer through regulating circ_0000376/miR-145-5p axis. BMC Anesthesiol. 20, 275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Exadaktylos A. K., Buggy D. J., Moriarty D. C., Mascha E., Sessler D. I., Can anesthetic technique for primary breast cancer surgery affect recurrence or metastasis? J. Am. Soc. Anesthesiol. 105, 660–664 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller K., Clementi C., Polyak D., Eldar-Boock A., Benayoun L., Barshack I., Shaked Y., Pasut G., Satchi-Fainaro R., Poly(ethylene glycol)-paclitaxel-alendronate self-assembled micelles for the targeted treatment of breast cancer bone metastases. Biomaterials 34, 3795–3806 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Zinger A., Koren L., Adir O., Poley M., Alyan M., Yaari Z., Noor N., Krinsky N., Simon A., Gibori H., Krayem M., Mumblat Y., Kasten S., Ofir S., Fridman E., Milman N., Lubtow M. M., Liba L., Shklover J., Shainsky-Roitman J., Binenbaum Y., Hershkovitz D., Gil Z., Dvir T., Luxenhofer R., Satchi-Fainaro R., Schroeder A., Collagenase nanoparticles enhance the penetration of drugs into pancreatic tumors. ACS Nano 13, 11008–11021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. Berlin, E. Y. Isacoff, Optical control of glutamate receptors of the NMDA-kind in mammalian neurons, with the use of photoswitchable ligands, in Biochemical Approaches for Glutamatergic Neurotransmission (Neuromethods, Humana Press, 2018), chap. 10, pp. 293–325. [Google Scholar]
- 45.Kaech S., Banker G., Culturing hippocampal neurons. Nat. Protoc. 1, 2406–2415 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Pruszak J., Ludwig W., Blak A., Alavian K., Isacson O., CD15, CD24, and CD29 define a surface biomarker code for neural lineage differentiation of stem cells. Stem Cells 27, 2928–2940 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korin B., Dubovik T., Rolls A., Mass cytometry analysis of immune cells in the brain. Nat. Protoc. 13, 377–391 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Korin B., Avraham S., Azulay-Debby H., Farfara D., Hakim F., Rolls A., Short-term sleep deprivation in mice induces B cell migration to the brain compartment. Sleep 43, zsz222 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9