Abstract
Effective non-covalent molecular imprinting on a polymer depends on the extent of non-bonded interactions between the template and other molecules before polymerization. Here, we first determine functional monomers that can yield a doxycycline-imprinted hydrogel based on the hydrogen bond interactions at the prepolymerization step, revealed by molecular dynamics (MD) simulations, molecular docking, and simulated annealing methods. Then, acrylic acid (AA)-based doxycycline (DOX) imprinted (MIP) and non-imprinted (NIP) hydrogels are synthesized in cross-linker ethylene glycol dimethacrylate (EGDMA) ratios of 1.0, 1.5, 2.0, and 3.0 mol%. Here, molecularly imprinted polymer with 3.0 mol% EGDMA has the highest imprinting factor (1.58) and best controlled drug release performance. At this point, full-atom MD simulations of DOX–AA solutions at different EGDMA concentrations reveal that AA and EGDMA compete to interact with DOX. However, at 3.0 mol% EGDMA, AA attains numerous stable hydrogen bond interactions with the drug. This study demonstrates that the concentration of the cross-linker and functional monomer can be adjusted to increase the success of imprinting, where the interplay between these two parameters can be successfully revealed by MD simulations.
Supplementary information
The online version contains supplementary material available at 10.1007/s10965-021-02740-6.
Keywords: Imprinted hydrogel, Molecular simulations, Doxycycline
Introduction
Molecular imprinting is an effective method to recognize a molecule (a template) from structures even with similar enantio-selectivity [1]. It generates template-shaped cavities furnished with chemical moieties that can recognize and bind the molecule of interest. This technique is preferred in many applications such as bio-separation processes [2, 3], biosensors [4], diagnosis, and drug delivery systems for controlled drug release [5, 6].
The power of the molecular imprinting technique stems from covalent and/or non-covalent interactions between the template and functional monomers at the prepolymerization process [7, 8]. After polymerization, the template molecule, either a toxic pollutant [9], protein [10], or a drug [11], is removed from the polymer network, with specific recognition cavities left behind [1]. The stability of the interactions affects the formation of cavities during polymerization and their number and homogeneous distribution throughout the polymer. Computational approaches such as molecular docking [12], molecular dynamics (MD) [13, 14], virtual screening protocols [15] and quantum mechanics calculations of monomers [16], drugs [9, 17], and proteins [10], were shown to predict critical molecular interactions before the polymerization [11]. MD calculations can be very helpful not only to investigate the impact of the molecular interactions on the prepolymerization step but also to predict the template recognition abilities of the polymer [18]. Computational and experimental techniques have been popularly employed together to determine molecular recognition [19], sorption performances [20, 21], drug loading, and release performances [22, 23] of the imprinted polymers. This combinatory approach was also shown to be useful to understand the role of cross-linker density [24], and to reveal differences in molecular-level interactions of two different cross-linkers EGDMA and trimethylolpropane trimethacrylate [14]. In a recent study, computational approaches such as docking studies and MD offered the most promising functional monomer to design the accurate and rapid detector for the spike protein of SARS-CoV-2 [25].
Performance of imprinted polymer matrices was shown to depend on monomer-template interactions [26], as well as the structure of template molecules, the presence of the solvent, stoichiometry [27], reaction temperature [28], and pressure [29]. Similarly, the effect of backbone polymer chains, type of functional monomer, and monomer:template ratio [1, 30] have been reported. Even though the effect of the cross-linker in creating binding sites during the pre-polymerization step may be negligible for some systems [31], many studies underlined the critical role of the cross-linker for molecular recognition and release performance of the imprinted polymer [11, 32–37]. The cross-linker plays a role in monomer-template complexation [11], polymer morphology, and template recognition [24]. Lower frequencies of binding events between the cross-linker and template enhance imprinting factor (IF) and consequently specificity [35]. Choosing a suitable cross-linker, and a template: functional monomer or template:cross-linker ratio directly affect IF value and ameliorate selectivity of molecularly imprinted polymers [36]. Moreover, computational studies suggested that cross-linkers influence monomer-template interactions, even though they make weak hydrogen bond interactions with the template molecule compared with the functional monomer [34].
Incorporating a therapeutic into a hydrogel is a great challenge: mechanical properties, swelling behavior and hydrophilicity, drug loading, and release performances can be optimized by changing many parameters. Our motivation is to create doxycycline (DOX) imprinted contact lenses for cornea neovascularization treatment. Therapeutic soft contact lenses have been considered for the treatment of several ophthalmologic diseases, such as xerophthalmia and ocular hypertension [38, 39]. Their major advantages are their ability to increase the bioavailability of drugs, eliminating patient misusage, reducing the frequency of drug administration, and side effects of eye drops [40, 41]. Molecular imprinting [42] and drug absorption [43, 44] are the techniques to produce contact lenses with controlled drug delivery properties. Molecular imprinted therapeutic soft lenses have been developed for various drugs, such as timolol maleate [45, 46], hyaluronic acid [47], ketotifen fumarate [5], norfloxacin [48], and diclofenac [49] showing their great promise in ophthalmologic treatments [50].
DOX Hyclate (briefly DOX), a member of the tetracyclines family, is an antibiotic frequently prescribed to treat neo-vascularization, which is an inflammatory ophthalmic disease [44, 51–53]. A sustained release of the drug to the cornea is desired to surmount the low bioavailability problem as well as to maintain the drug concentration at an optimum level to prevent toxicity. At this point, molecularly imprinted contact lenses appear as an ideal solution to treat the disease with the correct dosage. For this purpose, hydrogels with imprinted DOX were developed by our group for the first time [54], and we tested these as therapeutic contact lenses in vivo [55]. First, the effects of initiator reactivity, conversion, and reaction rate on photopolymerization kinetics of DOX imprinted on AA-based hydrogels were elaborated by our group [54]. In the other study [55], DOX was imprinted on itaconic acid (ITA)-based hydrogels using a free-radical polymerization technique. Recently, we reported drug delivery performances of another tetracycline derivative—minocycline—imprinted hydrogels using computational and experimental methods [56].
This study complements our previous molecular imprinting studies on similar hydrogel matrices. Accordingly, ITA was found as the most promising functional monomer to imprint DOX on the HEMA and TEGDMA based hydrogels and was assessed in in vivo studies [55]. Then, minocycline was imprinted on AA-HEMA-EGDMA based hydrogels reaching an imprinting factor of almost 3 [56]. DOX is a member of the tetracycline family including minocycline. Both molecules share a common core of linearly fused four rings, decorated with functional groups from different positions. These groups may result in strikingly different imprinting factors, i.e. a lower or a higher IF for the same hydrogel, resulting in different drug loading and release performances. Therefore, we focus on AA-based DOX imprinting hydrogels for the following reasons; (i) results from the simulating annealing studies composed of HEMA, EGDMA, and ITA or AA, (ii) easier processing of AA, and (iii) wide commercial usage of AA in contact lens synthesis.
In this study, we aim to show the success of computational techniques in predicting the extent of interactions between the functional monomer and template molecule, in the presence of other components forming the pre-polymerization mixture. Another objective is to show the ability of molecular simulations to bring an explanation for the drug release behavior of the hydrogels based on molecular mechanics. First, different in silico methods, namely MD simulations and molecular docking are used to select a suitable functional monomer. Two main groups of functional monomers suitable for imprinted hydrogel are investigated; (i) acids including AA, ITA, and methacrylic acid (MAA), and (ii) acrylamides including n-hydroxyethyl acrylamide (HEAAm), n-hydroxymethyl acrylamide (HMAAm), and n-ethyl acrylamide (NEAAm). The effect of HEMA on the interactions between the functional monomer and DOX is further studied with the simulated annealing technique on most and least promising monomers. Then, imprinted hydrogels are synthesized using AA and 2-hydroxyethyl methacrylate (HEMA) with different amounts of cross-linker EGDMA (1.0, 1.5, 2.0, and 3.0 mol%) in presence of DOX by free radical polymerization method. NIP hydrogels are also prepared to assess the performance of imprinted hydrogels under the same conditions. The mechanism behind drug loading and release performances of the molecularly imprinted hydrogels is investigated using full-atom MD simulations of the pre-polymerization mixtures at experimental concentrations. A critical cross-linker concentration is determined for the DOX-AA-EGDMA system, revealed with the computational and experimental approaches. Finally, zero-order, first-order, Higuchi, Kopcha, and Korsmeyer-Peppas one- and two- step kinetic models are investigated to explain the DOX release behavior of MIPs with imprinting factors below or above 1. The approach taken here is the first for a solvent-free system for molecular imprinting, to the best of our knowledge.
Materials and methods
Computational studies
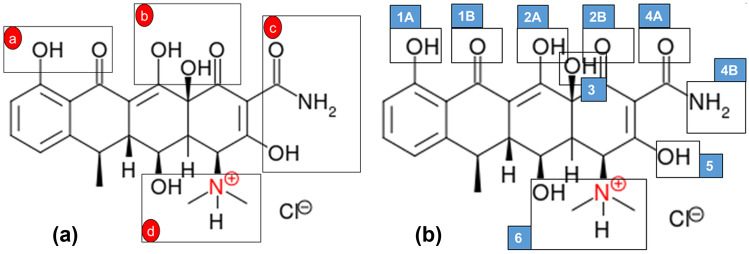
3-dimensional structures of monomers and DOX are downloaded from PubChem [57]. The functional monomers that can be used in contact lens synthesis are selected as ITA, AA, MAA, HEAAm, HMAAm, and NEAAm. Hyclate form of doxycycline (called DOX henceforth) is used in the imprinting experiments. Depending on the pKa value of DOX [58] in the polymerization media, the protonated structure of the drug is prepared using the sketch module of SYBYL-X 2.2.1 (Certara Inc., USA). DOX has many functional groups that can simultaneously establish non-covalent interactions with numerous molecules (Fig. 1).
Fig. 1.
Interaction regions in a molecular docking, b molecular dynamics calculations
We employ molecular docking and molecular dynamics (MD) simulations to evaluate the candidate functional monomers commonly used in molecular imprinting methods. Molecular docking calculations are performed to calculate the interaction energies of the monomers and drug molecules, which are static. On the other hand, the MD simulations method is beneficial to understand the extent of interactions between the molecules based on their time-dependent behaviors.
Interaction energy calculations
The interaction energies between the functional monomers and DOX are calculated with molecular docking. Here, the structures of the functional monomers and DOX are geometrically optimized, and their energies are calculated using Tripos [59] and Compass [60] force fields for comparative analysis. Both force fields are appropriate for drug-monomer systems and consider detailed expressions for the non-bonded interactions. A conjugated gradient is performed for energy minimization of each structure where the energy tolerance is set to 0.0001 kcal/mol. The procedure is repeated for the monomer-DOX complexes.
DOX is expected to interact with functional monomers at four major regions (Fig. 1a) These regions contain functional groups, such as –NH, –OH, and C=O, which can build hydrogen bond interactions with the monomers. DOX-monomer complexes are then carefully positioned such that a hydrogen bond interaction can occur between these groups, maintaining a distance of ~ 1.8 Å between the hydrogen and the electronegative atom keeping an interaction angle of 180º. The interaction energy (ΔE) between the functional monomer and DOX is then calculated by,
| 1 |
Here, EDOX, Emonomer, Ecomplex are the energies of DOX, the functional monomer, and the complex, respectively. The least negative interaction energy ΔE points to the most stable complex.
Molecular dynamics simulations
Functional monomer-DOX interactions are also studied with full-atom MD simulations considering the flexibility and dynamics of all molecules in the prepolymerization step. MD calculations are performed in Materials Studio 5.0. The simulation box of size ~ 34 × 34 × 34 Å3 under periodic boundary conditions includes functional monomer and DOX molecules with a stoichiometric ratio of 16:1 mimicking the experimental conditions. Since we use the hyclate form of DOX, we add ethanol, water, and chlorine atoms to the simulation box, while keeping the total charge of the system equal to zero. The density of the box is set as 1.2 g/cm3 based on the experimental data.
Energy minimization of the periodic boxes is done with the Adopted Basis Newton–Raphson algorithm, where the convergence tolerance of force is set to 0.001 kcal/mol/Å. Compass [57] is employed as the force field with NVT ensemble (constant number of particles, volume, temperature) at 298 K. Cutoff distance for non-bonded interactions is taken as 12.0 Å. Ewald summation method is used for electrostatic interactions. The total simulation time for each MD run is 5 ns with a time step of 1 fs, which is sufficiently long to understand the extent of interactions between these small molecules. All simulations are repeated twice with different initial velocities to obtain independent trajectories.
Hydrogen bond interactions between functional monomers and DOX are monitored using the radial distribution function (RDF) g(r). From RDF plots, we report the intensity of the peaks occurring within a distance range of 1.8–2.1 Å, indicating hydrogen bond interaction between a hydrogen atom and oxygen or nitrogen atoms. The intensity of a g(r) peak is based on the frequency of a pairwise interaction during the simulation; therefore, it can be considered as a measure of probability. As we perform two independent MD runs for better sampling, we plot the intensity of the same hydrogen bond interaction g(r) peaks obtained from two MD simulations as a cumulative sum on the same plot.
The selection of a functional monomer that promises effective imprinting of DOX is based on the consensus of three different calculations; (1) interaction energy between the monomer and DOX obtained from the molecular docking in SYBYL-X (with Tripos force field) and (2) Materials Studio (with Compass force field), and (3) MD simulations in Materials Studio.
HEMA comprises the majority of the prepolymerization system and can affect the extent of functional monomer-drug interactions. To assess this, the simulated annealing technique using MD simulations is employed. In the simulated annealing, the prepolymerization system is gradually heated from a low to a high temperature, and this procedure is repeated in cycles. In this way, this method effectively samples the configurations of the drug and other monomers at different energy states and captures the monomer-drug configurations with high and low probabilities. Here, HEMA, EGDMA, DOX, and ITA or AA, compose the simulation box of which density is 1.8 g/cm3. The systems are heated and cooled gradually between 298 and 498 K in 25 cycles to reach the minimum energy configuration. The time step is 1 fs reaching a total simulation time of 1 ns. The total and potential energies are controlled if they converge at the end of the simulations.
MD simulations also are performed to understand the effect of the concentration of the cross-linker EGDMA on drug loading and release performances of the imprinted hydrogels. Four different systems with different amounts of cross-linker, 1.0, 1.5, 2.0, and 3.0 mol% are studied. Accordingly, the stoichiometric ratio of AA to DOX is set at 16:1. EGDMA to DOX ratios are 8:1, 12:1, 16:1, 24:1 following the concentration increase. These ratios correspond to pre-polymerization conditions in the experiments.
The density of the simulation box is set as 1.5 g/cm3 according to experimental data (density of pre-polymer mixture including the cross-linker is 1.0–2.0 g/cm3). All conditions and parametrization are the same with MD simulations of functional monomer—DOX systems. Non-bonded interactions between EGDMA—DOX and AA—DOX are assessed using RDF calculations.
Experimental studies
Materials
2-Hydroxyethyl methacrylate (HEMA, 97%), acrylic acid (AA, 99%), and ethylene glycol dimethacrylate (EGDMA, 98%) are purchased from Sigma-Aldrich and used in polymer synthesis directly without any purification. The thermal initiator, Vazo 52 (2,2′-azobis(2,4-dimethyl-pentanenitrile)) is supplied from Dupont. The antibiotic DOX is provided by Deva Holding. The cleansing solution is prepared with acetonitrile (gradient grade, Merck Millipore), oxalic acid dihydrate (extra pure, Merck Millipore), and methanol (technical grade) for removing DOX from cavities.
Polymer synthesis
DOX is dissolved in HEMA (97–95 mol%) for 30 min in an ultrasonic bath below 40 °C. AA (2.0 mol%) and EGDMA (1.0, 1.5, 2.0, 3.0 mol%) are added to the solution. AA: DOX molar ratio of 16:1 is used for the synthesis. The initiator Vazo 52 (0.062–0.064 mol%) is added to the reaction mixture. To dissolve the initiator, the reaction mixture is put into an ultrasonic bath for 15 min. Due to Vazo 52 is a low-temperature initiator, the temperature of the ultrasonic bath is kept under 20 °C. The oxygen in the solution is removed by bubbling nitrogen for 30 min. Then, the reaction mixture is injected into a glass mold coated with a hydrophobic solution and separated by a silicone frame 0.5 mm wide. The mold is placed into an oven for 24 h at 45 °C. NIP hydrogels are synthesized under the same conditions in the absence of DOX. Figure 2 represents DOX imprinted hydrogel synthesis. After synthesis, all samples are placed into 0.02 M oxalic acid: acetonitrile: methanol (65:15:20 v/v) solution for 24 h. Then, they are transposed to methanol and washed with water for removing DOX. All samples are dried for 48 h at 40 °C.
Fig. 2.
Schematic representation of molecular imprinting of DOX
Polymer characterization
-
Fourier transform infrared spectroscopy (FT-IR)
FT-IR spectra are measured on a Perkin Elmer spectrometer spectrum one model between 650 and 4000 cm−1 by using ATR mode.
-
Differential scanning calorimetry (DSC)
DSC experiments are carried in a cycle with Perkin Elmer DSC 4000 instrument. Nitrogen gas is used for purging at a flow rate of 20 ml/min. To determine the glass transition temperature (Tg) of polymers, the temperature scanning program starts to heat from 30 °C to 250 °C, then cools from 250 °C to 0 °C. Samples are kept at 0 °C for 15 min. After the isothermal step, they are heated from 0 °C to 250 °C again. The scanning rate is set at 20 °C/min.
-
Swelling of hydrogels
After being dried at 37 °C for 48 h, each hydrogel is first weighed and then separately kept in water for 72 h. Swelling tests of hydrogels are carried both at 4 °C and 37 °C. The equilibrium water content of hydrogels (EWC) or the swelling ratio is calculated as,2 where Ws and Wd are the weights of the swelled and dry hydrogel, respectively.
-
DOX loading and release
Dried hydrogel discs (10 mm diameter, 450 µm thickness) are immersed in 80 μM or 104 μM DOX aqueous solutions (10 ml) and kept at 4 °C for 30 h. All samples are shaken with an orbital shaker (90 rpm) while protecting from light. The initial and final concentrations of solutions are measured by Perkin Elmer Lambda 35 UV/VIS Spectrophotometer. The wavelength of maximum absorption is 274 nm for DOX.
To observe the release kinetics of DOX, MIP and NIP hydrogels are first placed in 1 mM DOX solution for 72 h at 4 °C. Then, loaded hydrogels are immersed in an artificial tear of 10 ml 0.9% NaCl solution at 37 °C. UV/VIS Spectrophotometer is used for the determination of release kinetics. All loading and release performances of hydrogels are carried out in triplicate.
-
Kinetics of drug release
There exist several models to explain drug release kinetics. In this study, we use zero-order, first-order, Higuchi, and Korsmeyer-Peppas and Kopcha models to enlighten DOX release kinetics.
Zero-order kinetic model is employed for constant drug releases, such as for osmotic systems, matrix tablets with low soluble drugs, and transdermal systems [61]. The equation for zero-order release is given by,3 Q is the released or dissolved amount of drug, Q0 is the initial amount of drug in solution, K0 is the zero-order release constant and t is time.
In the first-order kinetic model release rate is concentration dependent. This model is described by,4
where K is the first-order release constant.5
The Higuchi model explains drug release based on Fick’s law. According to this model; the initial concentration of the drug in the matrix is much higher than its solubility; drug diffuses only in one dimension; particle size of the drug molecules is much smaller than matrix thickness; swelling and dissolution of the matrix are negligible and drug diffusivity is constant [61].
Dissolution in a homogenous matrix can be modeled by,
| 6 |
where Qs is the drug solubility in the matrix, ft is the amount of drug released in time t per unit area, and D is the diffusivity of drug in the matrix [61, 62]. Equation 6 can be simplified as,
| 7 |
where KH is the Higuchi rate constant.
To determine drug release kinetics, the first 60% portion of the drug release data is used in the Korsmeyer-Peppas model. The equation for the Korsmeyer-Peppas model is expressed as,
| 8 |
where Qt is the drug concentration in the release solution at time t, Q∞ is the equilibrium concentration of the drug in the release solution, K is the rate constant and n is the release exponent showing release mechanisms for cylindrical matrices as determined in Table 1.
Table 1.
Release exponent values for Korsmeyer-Peppas model
| Release exponent (n) | Drug transport mechanism | Rate as a function of time |
|---|---|---|
| 0.5 | Fickian diffusion | t−0.5 |
| 0.45 < n < 0.89 | Non-Fickian transport | tn − 1 |
| 0.89 | Case II transport | Zero-order release |
| > 0.89 | Super case II transport | tn − 1 |
The Kopcha model reveals the amount of diffusion and erosion of the systems and presents their effects on drug release with
| 9 |
where Mt is the amount of drug dissolved at time t, A is the diffusional constant and B is the erosion constant. The A/B ratio explains the mechanism of drug release. A/B > 1 suggests that drug release is controlled by diffusion, whereas A/B < 1 suggests that erosion is dominant. If A/B = 1, both diffusion and erosion control the release mechanism [63].
Results and discussion
Selection of a suitable functional monomer for imprinting
A good imprint on a polymer is a template-shaped cavity accommodating the chemical moieties that can recognize and select the template. Stable non-covalent interactions between the template and functional monomers before polymerization, shape the chemical and physical properties of the imprinted cavity on the polymer. This in turn affects the loading/release mechanism of the template molecules in the polymer. Therefore, the functional monomer having the highest interaction energy with the template molecule is usually suggested as the most suitable monomer for polymer synthesis [64]. However, the presence of other components, such as the cross-linker can also affect the extent of interactions between the functional monomer and the template [56], which cannot be predicted from the calculations considering only the functional monomer and template. For a reliable decision on the most suitable functional monomer for imprinting, all components in the system should be considered.
Here, an approach of three major steps is employed: (1) molecular docking to determine interaction energies for evaluating the functional monomers for imprinting, (2) full-atom MD simulations of the functional monomer-DOX systems to assess the findings of step 1, and (3) simulated annealing calculations to consider the effect of main chain monomer HEMA and cross-linker EGDMA on the monomer-DOX interactions.
Interaction energies between the functional monomers and DOX are calculated with molecular docking using two different force fields (Compass and Tripos) for the regions indicated in Fig. 1a. Both force fields are popularly used in the selection of functional monomers for molecular imprinting [65]. Therefore, their agreement on the results can improve the reliability of the calculations. Results displayed in Table 2 indicate that both force fields mostly agree on the interaction energies with similar order of magnitudes. Here, there exist some exceptions particularly for the regions (a) and (b). DOX accommodates an –OH group at position (b), neighboring two =O groups at positions (a) and (b) (Fig. 1a). This –OH group has the capability of making an intermolecular hydrogen bond with the functional monomer, as well as intramolecular hydrogen bonds with two =O groups. According to the orientation of the –OH, the interaction energy of the complex may differ [66]. The interaction energies are calculated after an energy minimization that can force the –OH group to make a hydrogen bond with one of the three above-mentioned alternatives. Consequently, the results are evaluated considering all four positions (i.e., a summation of interaction energies at positions a, b, c, and d). Tripos and Compass both suggest the order from best interacting monomer to least interacting with the drug as ITA > HEAAm > HMAAm > NEAAm > MAA > AA. Nonetheless, when the magnitudes of interaction energies are considered, all investigated functional monomer—DOX complexes with different configurations suggest that the investigated functional monomers can imprint DOX.
Table 2.
Interaction energies between functional monomers and DOX
| Functional monomer | Regions | Tripos (kcal/mol) | Compass (kcal/mol) |
|---|---|---|---|
| ITA | a | −16.29 | −13.48 |
| b | −7.78 | −18.63 | |
| c | −12.35 | −12.55 | |
| d | −13.01 | −13.00 | |
| AA | a | −12.47 | −8.91 |
| b | −5.63 | −4.62 | |
| c | −6.51 | −8.86 | |
| d | −7.06 | −8.78 | |
| MAA | a | −8.96 | −11.15 |
| b | −5.18 | −4.85 | |
| c | −8.02 | −8.53 | |
| d | −7.77 | −7.53 | |
| HEAAm | a | −7.39 | −8.82 |
| b | −5.02 | −16.94 | |
| c | −15.16 | −10.19 | |
| d | −15.71 | −16.18 | |
| HMAAm | a | −12.08 | −13.16 |
| b | −9.52 | −10.72 | |
| c | −9.29 | −11.49 | |
| d | −8.46 | −9.91 | |
| NEAAm | a | −8.59 | −10.95 |
| b | −9.28 | −7.37 | |
| c | −10.51 | −7.44 | |
| d | −11.45 | −11.02 |
Molecular docking calculations do not consider the flexibility and dynamics of the molecules, which can affect both the extent and magnitude of non-bonded interactions. To address this, full-atom MD simulations are conducted for the functional monomers—DOX systems. Figure 3 represents a summary of all detected hydrogen bond interactions between functional monomers (interacting groups are circled on the figure) and DOX from six different positions detailed in Fig. 1b. Here, the intensity of the g(r) peaks at a hydrogen bond interaction distance ~ 1.8–2.1 Å are shown as bar plots.
Fig. 3.
RDF values for hydrogen bond interactions between DOX and a ITA, b AA, c MAA, d HEAAm, e HMAAm, f NEAAm
Among the functional monomers, ITA makes the most stable hydrogen bond interactions with DOX from all six positions, owing to two hydroxyl groups (Fig. 3a). This result agrees with the molecular docking calculations.
AA interacts with the drug from all available positions as well (Fig. 3b), but with less stability compared to ITA. Particularly, the carbonyl (C=O) group at position 4A and hydroxyl (–OH) group at position 6 of DOX make stable hydrogen bond interactions with AA. Due to intramolecular hydrogen bond interactions on DOX between positions 1A and 1B, AA could not establish stable interactions from this region. Nonetheless, AA has a considerable amount of hydrogen bond interactions with the drug from all positions.
MAA is one of the most preferred functional monomers for MIPs [4, 29, 45]. The interaction of MAA and DOX is investigated with RDF analysis, presented in Fig. 3c. MAA shows a similar trend as AA, in terms of the extent of hydrogen bond interactions with DOX. Especially positions 4A and 6 are highlighted for non-covalent interactions with the drug.
HEAAm monomer accommodates three functional groups (–NH, C=O, –OH) that can make hydrogen bond interactions with DOX. Figure 3d shows that positions 5 and 6 come forward with their more stable hydrogen bond interactions when compared to other regions on the drug. Flexible –CH2OH group of HEAAm increases the mobility of the monomer, hence –OH moiety can interact with functional groups of DOX.
In general, HMAAm (Fig. 3e) shows less interest in the DOX molecule when compared to HEAAm. On the other hand, position 2 on DOX is an attractive site for HMAAm, whereas HEAAm has weaker interactions. The final acrylamide monomer, NEAAm (Fig. 3f) generally has weak hydrogen bond interactions with DOX, when compared to other acrylamide derivatives.
Figure 4 summarizes all results for functional monomer selection using computational approaches. Here, interaction energies (kcal/mol) from molecular dockings and cumulative g(r) intensities from MD simulations (detailed in Fig. 3) are shown. Most importantly, the general trend from these calculations is the same. Accordingly, among the functional monomers, ITA promises to yield effective imprinting on the hydrogel, while it is followed by HEAAm. AA and MAA are expected to lead to lower imprinting of the drug on the hydrogel.
Fig. 4.
Comparison of in silico methods for the functional monomer selection
At the end of the interaction energy calculations and MD simulations, both investigating only functional monomers and DOX, ITA is determined as the most promising functional monomer to imprint DOX. Indeed, in our previous study, ITA was shown to successfully imprint DOX on hydrogels using TEGDMA as cross-linker releasing DOX in a controlled manner [55]. On the other hand, AA is determined to have the least interaction with DOX (Fig. 4) among the candidate monomers. While these calculations show the capacity of the investigated functional monomers to imprint DOX, the cross-linker EGDMA and the main chain monomer HEMA should be also considered since they constitute the majority of the prepolymerization solution. EGDMA and HEMA accommodate electronegative groups suitable for hydrogen bond interaction with DOX. The competition between the functional monomer, EGDMA, and HEMA can lead to a surprising result, which may not be predicted from the docking and MD simulations of the functional monomer-DOX systems.
Consequently, before the hydrogel synthesis, we conduct a final simulation, namely simulated annealing using MD simulations. This approach is employed to conduct an effective sampling and to reveal the favorable low- and unfavorable high-energy configurations of functional monomer-drug in the presence of other major components of the prepolymerization solution, i.e. EGDMA and HEMA. Here, we investigate ITA and AA, the most and the least promising functional monomers to imprint DOX according to Fig. 4, respectively. The prepolymerization solution consisting of DOX, ITA/AA, HEMA, and EGDMA is subjected to 25 cycles of heating from 298 to 498 K. Here, 298 K is the experimental imprinting temperature, and 498 K is a high temperature to sample high-energy states. The simulations converge as shown in Supplementary Fig. S1. Figure 5 indicates hydrogen bond interactions of DOX in two investigated systems for the whole temperature range. The backbone of HEMA mostly interacts from its carbonyl group (C=O) with DOX. In the system with ITA (Fig. 5a), DOX makes hydrogen bond interactions with the functional monomer (ITA), cross-linker (EGDMA), and backbone monomer (HEMA) to similar extents (Fig. 5a inset). Most importantly, in the other system with AA (Fig. 5b), the extent of hydrogen bond interactions between DOX and the functional monomer (AA) is similar to those from the system with ITA. However, the extents of hydrogen bond interactions are altered with both EGDMA and HEMA. Here, DOX seems to interact with HEMA more. These results suggest that even though ITA seems highly promising to obtain better imprints of DOX based on functional monomer-drug interactions (Fig. 4), other components in the solution may as well interact with DOX to ultimately affect the imprinting.
Fig. 5.
Summary of RDF analysis for the simulated annealing simulations of prepolymerization systems including EGDMA, HEMA and, a ITA, or b AA. Sums of g(r) values calculated for each group are given in the insets
Simulated annealing simulations thus suggest that ITA and AA will result in imprinted hydrogels with similar imprinting factors for DOX. In our previous study, ITA was shown to successfully imprint DOX on hydrogels using TEGDMA as a cross-linker releasing DOX in a controlled manner [55]. Besides ITA, AA is also considered as a suitable functional monomer to imprint DOX on hydrogels since it provides a three-dimensional contour around the drug in the prepolymerization step that can further serve to obtain templates on the hydrogel. AA is widely used in commercial contact lens production with cross-linker EGDMA due to its hydrophilicity, its processibility in hydrogel synthesis, and its popularity in numerous imprinting studies [67–69]. In this line, we previously synthesized minocycline imprinted hydrogels with AA and EGDMA and obtained hydrogels with high imprinting factors reaching 3 [56]. While minocycline is a tetracycline derivative as DOX, they have different functional groups on the same core of four infused rings, which can result in strikingly different imprinted hydrogels with different drug release performances. Here, we decide to investigate DOX imprinting on AA-based hydrogels with different cross-linker concentrations that allow a direct comparison with minocycline imprinted hydrogels [56] synthesized under the same conditions used in this study. Accordingly, this study aims to reveal molecular details to explain the similarities/differences in imprinting small molecules from the same family.
Experimental step
DOX MIP and NIP hydrogels are synthesized with different amounts of cross-linker EGDMA. Then, these are characterized and their drug delivery performances are tested. DOX release kinetics of hydrogels are evaluated by zero-order, first-order, Higuchi, Korsmeyer-Peppas and Kopcha kinetic models. We finally conduct MD simulations to reveal the effect of cross-linker concentration on DOX imprinting.
Hydrogel synthesis and characterization
FT-IR results show that hydrogels are successfully synthesized (Fig. 6). In all samples, the characteristic stretching peak of C=C at 1637 cm−1 belonging to the vinyl group is disappeared after the completion of polymerization. –OH stretching of AA seems to be between 3000 and 2500 cm−1 wavelength. The presence of 2960 cm−1 (stretching) and 1720 cm−1 (stretching) peaks belonging to HEMA, AA and EGDMA confirm the presence of –CH and C=O groups in the copolymer structure, respectively. Groups corresponding to the FT-IR peaks are shown in Fig. 2.
Fig. 6.
FT-IR spectrum of the hydrogel (before and after synthesis)
DSC analysis shows that the thermal properties of MIP and NIP hydrogels with the same cross-linker content are not significantly different (Table 3). As expected, increasing the cross-linker amount in hydrogel matrices increases Tg values of all hydrogel samples. Also, the effects of the cleansing procedure on hydrogels are explored by DSC and swelling test (Table 4). Hydrogel samples, which are treated with cleansing solution and methanol, have slightly lower Tg values compared to others. The cleansing procedure does not apparently disrupt the cross-linked structure of the hydrogel. However, lower Tg values imply a low amount of solvent molecules entrapped in the hydrogel.
Table 3.
Tg values of hydrogels
| Code | Tg (ºC) | |
|---|---|---|
| Washed | Unwashed | |
| NIP 1 | 113.6 | 115.1 |
| NIP 1.5 | 113.7 | 116.9 |
| NIP 2 | 114.2 | 117.9 |
| NIP 3 | 114.9 | 119.8 |
| MIP 1 | 114.9 | 115.7 |
| MIP 1.5 | 114.4 | 116.2 |
| MIP 2 | 116.0 | 116.6 |
| MIP 3 | 117.9 | 120.1 |
Table 4.
Water content of hydrogels with their mean values ± standard deviation (n = 3)
| Code | EWC (%) at 4 °C | EWC (%) at 37 °C | |
|---|---|---|---|
| Cleansed | Cleansed | Not cleansed | |
| NIP 1 | 41.5±0.7 | 42.6±1.1 | 35.5±0.5 |
| NIP 1.5 | 40.4±0.5 | 40.7±0.4 | 33.6±1.0 |
| NIP 2 | 37.0±0.6 | 40.0±0.8 | 33.3±0.3 |
| NIP 3 | 32.8±0.9 | 33.7±1.0 | 30.8±0.1 |
| MIP 1 | 43.1±0.5 | 42.9±0.8 | 33.5±1.0 |
| MIP 1.5 | 40.8±0.2 | 40.3±0.7 | 34.2±0.2 |
| MIP 2 | 37.3±1.0 | 36.4±0.5 | 32.3±1.0 |
| MIP 3 | 34.9±0.9 | 33.4±1.0 | 30.9±0.1 |
The swelling behavior of hydrogels is determined at both drug loading temperature (4 °C) and human body temperature (37 °C). The equilibrium water contents (EWC) are given in Table 4. The water content of the hydrogels does not change significantly with temperature, except NIP 2. As expected, increasing the cross-linker amount decreases the EWC of the hydrogels. Also, water adsorption capacity after the cleansing procedure increases due to the cavities formed after removing the drug molecules. The slight increase in the EWC % value for NIP2 at 37 °C implies a structural difference when compared to the other hydrogels. At the prepolymerization step, the molecules can adopt an orientation to create intermolecular spaces. The intermolecular space in the NIP2 structure may be high when compared to other NIPs and MIPs. Therefore, more water can fill these spaces, i.e. more swelling than expected. Another plausible explanation may be the positioning of the monomers in the hydrogel structure. In MIPs, functional monomers would be next to each other around the template. After the removal of DOX, these neighboring functional monomers would have a high potential to make hydrogen bond interactions with each other. However, in NIP2, the monomers may be randomly distributed and oriented in the hydrogel, where they are more prone to hydrogen-bond interaction with water molecules. A more detailed discussion on the effect of EGDMA % on the molecular interactions governing the hydrogel performance will be given in the following section.
DOX loading
The drug loading capacity of MIP and NIP hydrogels is explored by calculating the imprinting factor (IF). IF expresses the recognition ability of MIPs according to the interactions between the template molecule and template-shaped cavities on the polymer. Equations (10) and (11) are used for calculating IF [38].
| 10 |
| 11 |
Here, Q is adsorption capacity, C0 is initial concentration, Ce is equilibrium concentration, V is adsorption volume, and m is the weight of the hydrogel. As experiments are performed in triplicate, average amounts of the loadings are used in the calculations.
Drug loading capacity and IF values are studied for two different drug concentrations, 80 μM and 104 μM loading solutions. At high concentrations, the loading capacity of all samples significantly enhances when compared to loading at low concentrations (Fig. 7a). IF values of the MIPs are also affected (Fig. 7b).
Fig. 7.
a DOX loading capacity of samples at different concentrations, b IF values of MIPs at different concentrations
For the NIPs hydrogels, drug loading capacity decreases with increasing the cross-linker ratio (Fig. 7a). This observation is more apparent for the low concentrated solution. Here, the absence of DOX molecules in the prepolymerization step may lead to a random distribution of functional monomer AA in the polymer structure. Also, increasing the EGDMA ratio may obstruct the interaction between AA and DOX molecules leading to a smaller extent of interactions. The same tendency is reported by Hiratani and Alvarez-Lorenzo [64] that discuss hindered interactions between functional monomer and timolol at NIPs due to cross-linker. On the other hand, at high concentrated drug loading solutions (104 μM), the effect of the cross-linker concentration on drug loading is not clear. Here, the NIP samples may be forced to intake the maximum amount of drug reaching full capacity. The same effect of cross-linker is also observed for MIP hydrogels, except for MIP3 (Fig. 7a). Intriguingly, for both low and high drug loading concentrations, MIP hydrogel containing 3.0% EGDMA (MIP3), uptakes more drug when compared to its NIP counterpart (NIP3). Also, the IF value is higher than 1.0 only for MIP3 (Fig. 7b), which indicates that this cross-linker percentage is critical for the drug loading performance of the imprinted hydrogel.
EGDMA contains functional groups that can interact with DOX. To shed light on the effect of cross-linker concentration on the drug loading of MIPs, the competition between AA and EGDMA for binding DOX is investigated using MD simulations. The experimental stoichiometric ratios of AA:DOX: EGDMA are used in the simulations.
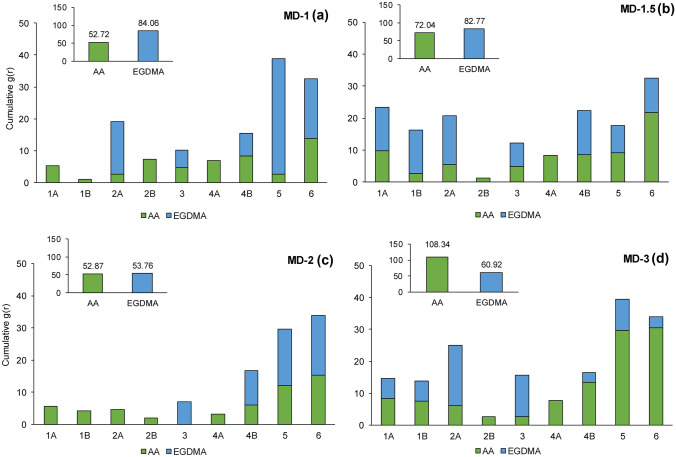
During the MD simulations mimicking the prepolymerization medium, DOX makes hydrogen bond interaction both with AA and EGDMA, competing for the same regions on the drug. The stability of these molecular interactions is the basis for drug release performances of the hydrogels. The extent of hydrogen bond interactions is quantified by RDF analysis for DOX-AA, and DOX-EGDMA pairs. Figure 8a shows the intensity of g(r) peaks at ~ 2.0 Å indicating hydrogen bond interactions, for 1.0% EGDMA concentration simulations (MD-1). Functional monomer AA interacts with the drug molecules from all available positions, especially dominating positions 1, 2, and 4. EGDMA competes with AA, especially at positions 3 and 6, but occupies position 5 of DOX for hydrogen bond interactions. Overall, as seen from the inset of Fig. 8a, hydrogen bond interactions between the carbonyl group of EGDMA and the functional groups of DOX are more stable than those between AA and DOX.
Fig. 8.
Cumulative intensity (g(r)) of radial distribution function. Sums of g(r) values calculated for each group are given in the insets. Sums of g(r) values calculated for each group are given in the insets
When the amount of EGDMA is increased to 1.5% (MD-1.5 in Fig. 8b), EGDMA starts to interact with the DOX molecule at position 1, but the stability of the interaction between EGDMA and DOX decreases at both positions 5 and 6. When all positions are considered (Fig. 8b inset), the frequency of interactions between AA and drug molecules seems to increase and get closer to that of drug-EGDMA pairs, when compared to MD-1 (Fig. 8a).
In 2.0% EGDMA concentration (MD-2), AA visits the drug molecules from all positions, except 3, while competing with EGDMA particularly at positions 5 and 6. Interestingly, cumulative hydrogen bond interaction g(r) intensities for AA and EGDMA are similar as shown in Fig. 8c inset. Moreover, the frequency of all pair-wise interactions seems to decrease at 2.0% cross-linker concentration when compared to 1.0% and 1.5% (insets in Fig. 8a–c).
In MD-3 (3.0% EGDMA), AA strongly interacts with DOX for hydrogen bonds, except from positions 2 and 3. At this cross-linker ratio, AA wins its competition with EGDMA and effectively surrounds DOX molecules, which may serve to obtain a good and nearly complete shape of templates on the polymer (Fig. 8d). At this cross-linker concentration, smaller molecular weight AA can easily diffuse in the prepolymerization mixture, where its interactions with DOX are facilitated by larger EGDMA molecules creating a crowding environment [70].
As revealed by MD simulations, with the dominance of the cross-linker at the molecular level, DOX molecules can hardly leave the cavities of imprinted hydrogel structure during drug release. This explains the ineffectiveness of AA leading to a low IF value of imprinted samples under the threshold value of 1. AA becomes dominant only when the cross-linker ratio is 3.0%, and the IF value of MIP3 is higher than 1.
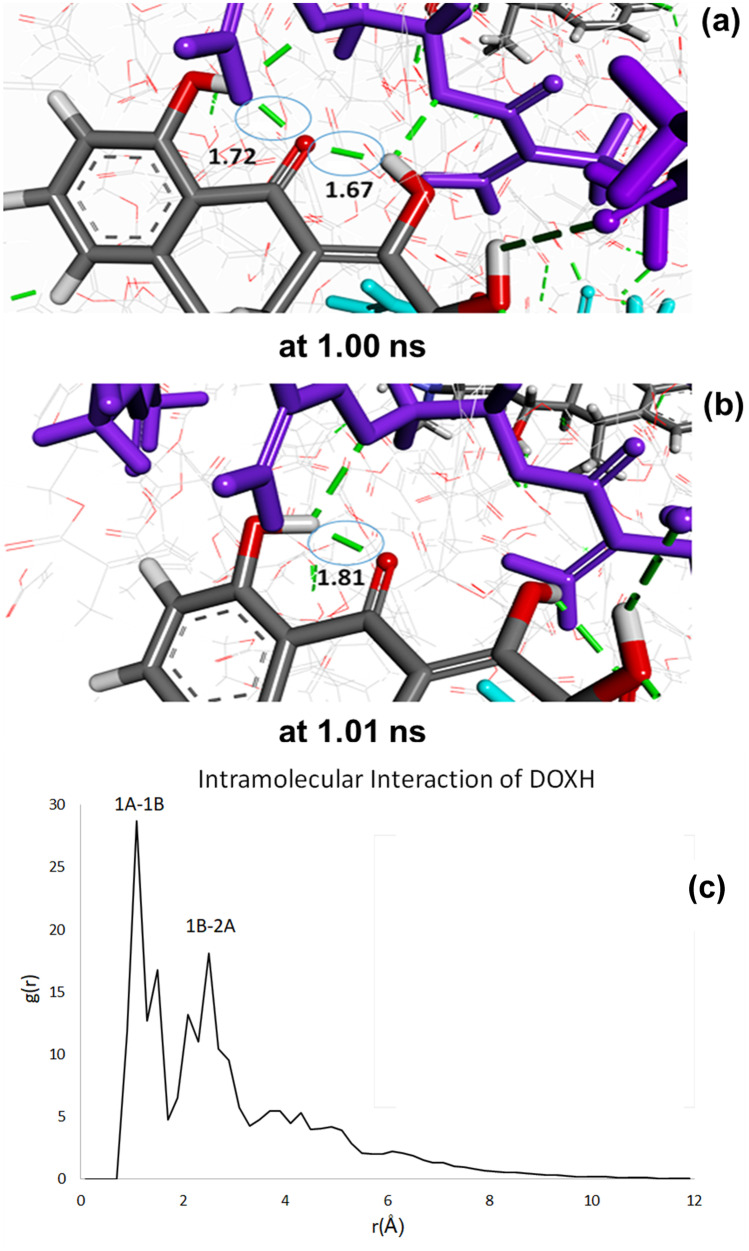
MD simulations also reveal molecular mechanisms behind low IF values of the hydrogels. Adjacent functional groups of DOX make strong intra-molecular interactions and occasionally interact with AA molecules, which is illustrated by successive time steps during MD simulations (Fig. 9). As a result, the stability of inter-molecular interactions between DOX and functional monomers, especially from positions 1 and 2 seems to be not maintained. When compared to our published study, the members of the same drug family, DOX and minocycline, differentiate non-negligibly since their functional groups, and these can affect the interactions with functional monomers and thereby the imprinting performance. IF values of DOX imprinted hydrogels are lower than minocycline imprinted hydrogels of which IF is over 3, lacking intra-molecular hydrogen bond interactions [56].
Fig. 9.
a Intra-molecular interaction at 1.00 ns, b at 1.01 ns, c Radial distribution function of DOX. CPK coloring is used for DOX, while EGDMA and AA are shown in purple and cyan, respectively
DOX release
After loading drug molecules at 1 mM for 72 h, the release kinetics of hydrogel samples are observed for 48 h in 0.9% NaCl solution at 37 ºC. To mimic the human eye physiological conditions, kinetic studies are carried directly on wet hydrogels in the solution. The amount of DOX release from different hydrogel samples is given in Fig. 10. The highest amount of DOX is released from NIP 1 among other hydrogels. There are two main reasons for this behavior; DOX molecules can easily diffuse from low cross-linker ratio hydrogel, and there is no molecular cavity specific for DOX structure to control release kinetics. As expected, the amount of released DOX (AR) decreases for both MIP and NIP hydrogels when we increase the cross-linker ratio in polymer matrices. This also leads to the rigidity of the structure as indicated by increasing values of Tg. At this point, DOX release and loading performances support each other.
Fig. 10.
DOX release performances of NIPs and MIPs. Data expressed as mean ± SD (n = 3)
When we focus on the release kinetics of NIP 1 and MIP 1 (Fig. 10a), both hydrogels display similar drug release kinetics at the beginning. After 5 h, drug release amounts diverge, where NIP 1 releases slightly more DOX at the end of 48 h. For both NIP 1 and MIP 1, DOX release reaches a steady-state after 24 h. When the cross-linker ratio is increased, differences in AR for MIP and NIP hydrogels become smaller (Fig. 10a–c). Amounts of DOX release are almost the same for NIP 2 and MIP 2. When the cross-linker EGDMA concentration is 3.0% in polymer matrices, drug release for MIP 3 is slightly higher than that of NIP 3 (Fig. 10d).
MD studies for 1, 1.5 and 2% EDGMA ratios explain why the difference in maximum release amounts between NIP 1–MIP 1, NIP 1.5–MIP 1.5, and NIP 2–MIP 2 diminishes (Fig. 8). At a lower EGDMA concentration, AA molecules cannot effectively surround the DOX due to the dominance of EGDMA—drug interactions, which results in poor release performance of the MIP when compared to NIP. With increasing cross-linker concentration, the stability of hydrogen bond interactions between AA and drug molecules increases, whereas the stability of interactions between EGDMA and DOX decreases. When the ratio of EGDMA is set to 3.0 mol%, functional monomer AA makes more stable hydrogen bond interactions with DOX in particular at positions 4B, 5, and 6 (Fig. 8d). At this concentration, AA is assumed to be more effective to build DOX-shaped cavities. Both loading (Fig. 7) and release results (Fig. 10) support this inference.
Although there exist slight differences between some DOX release profiles, such as for the NIP3–MIP3 pair, the drug release profiles between NIP–MIP pairs are expected to be more significant to assess the performance of MIPs. While NIPs are generally used as control systems to determine the imprinting efficiency in MIPs, the presence of non-template background sites formed by the functional monomer with high binding capacity should be considered [71]. Here, a functional monomer that can dimerize such as AA, can reduce the number of background sites both in NIP and MIP. This provides rather a good basis for estimating the imprinting efficiency. However, the dimerization of AA can also result in a lower amount of template-shaped cavities in MIP, which would be reflected in the drug release profiles that are used to understand the success of imprinting in this study. In this respect, the release profiles of MIP-NIP pairs seem to be similar plausibly due to a low number of template-shaped sites; but DOX is successfully imprinted using 3 mol% EGDMA with IF > 1. To support the dimerization tendency of AA, we investigate the hydrogen bond interactions between AA molecules in the MD simulations. Cumulative g(r) values at a distance of 1.7 Å indicate the presence of AA-AA dimerization in the prepolymerization mixtures of MIP1, MIP1.5, MIP2, MIP3 systems (Supplementary Fig. S2). Here, the extent of hydrogen bond interactions increases with the increasing amount of EGDMA.
To reveal the similarities and differences between the different drug release mechanisms of NIPs and MIPs, the release data are evaluated by several kinetics models and discussed in the next section.
Release kinetics
Zero-order, first-order, Higuchi, Korsmeyer-Peppas, and Kopcha kinetic models are used to analyze the in vitro release data and evaluate the release kinetics. Model parameters K, n, A, B, and regression coefficient (R2) values are summarized in Table 5. Based on kinetic models, zero-order and first-order kinetics are not sufficient to explain the drug release kinetic due to low R2 values. The Higuchi model provides a better description of DOX release from hydrogels, when compared to zero- and first-order models. To clarify the release kinetics, the Korsmeyer-Peppas model is applied both in one-step and two-steps approaches. At the one-step approach, rate exponents of DOX release are calculated for 215 min (i.e. cumulative 60% DOX release). R2 is not high enough for one step Korsmeyer-Peppas model. The best correlation coefficient values are obtained for two-step Korsmeyer-Peppas and Kopcha release models. At the two-steps approach, the rate exponent n1 (first 55 min) and n2 (55–215 min) are calculated to determine the drug transport mechanism.
Table 5.
DOX release rate constants in 0.9% NaCl and lachrymal fluid obtained by the fitting of the release profiles to the zero-order, first-order, Higuchi, Korsmeyer–Peppas and Kopcha Kinetics for imprinted and non-imprinted hydrogels
| Code | Zero-Order | First-Order | Higuchi | Korsmeyer-Peppasa one step |
Korsmeyer-Peppasb two steps |
Kopcha | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K | R2 | K | R2 | K | R2 | n | R2 | n1 n2 |
|
A | B | A/B | R2 | ANIP/AMIP | |
| NIP 1 | 0.3541 | 0.9145 | -0.0011 | 0.9327 | 3.4964 | 0.9488 | 0.9562 | 0.8709 |
1.6869 0.4171 |
0.9957 0.9833 |
0.1769 | -0.0018 | -98.28 | 0.9767 | 1.03 |
| MIP 1 | 0.2985 | 0.8874 | -0.0012 | 0.9385 | 3.6540 | 0.9560 | 0.8356 | 0.8941 |
1.4023 0.4151 |
0.9983 0.9908 |
0.1719 | -0.0018 | -95.50 | 0.9806 | |
| NIP 1.5 | 0.2827 | 0.8732 | -0.0012 | 0.9389 | 3.5019 | 0.9411 | 1.1048 | 0.8407 |
2.1762 0.5702 |
0.9973 0.8220 |
0.1498 | -0.0015 | -99.87 | 0.9658 | 1.00 |
| MIP 1.5 | 0.3072 | 0.8929 | -0.0011 | 0.9134 | 3.6130 | 0.9426 | 0.8120 | 0.9034 |
1.2736 0.3986 |
0.9936 0.9563 |
0.1496 | -0.0016 | -93.50 | 0.9766 | |
| NIP 2 | 0.3174 | 0.8835 | -0.0013 | 0.9352 | 3.7343 | 0.9468 | 1.2072 | 0.8381 |
2.2162 0.4328 |
0.9958 0.9919 |
0.1337 | -0.0015 | -89.13 | 0.9694 | 1.04 |
| MIP 2 | 0.3054 | 0.8731 | -0.0012 | 0.9230 | 3.6631 | 0.9418 | 0.9921 | 0.8636 |
1.7836 0.4321 |
0.9928 0.9661 |
0.1287 | -0.0014 | -91.93 | 0.9698 | |
| NIP 3 | 0.3435 | 0.8746 | -0.0014 | 0.9072 | 3.9445 | 0.9248 | 1.3552 | 0.7964 |
2.6891 0.4252 |
0.9916 0.9792 |
0.0965 | -0.0011 | -87.73 | 0.9573 | 0.90 |
| MIP 3 | 0.3448 | 0.8782 | -0.0014 | 0.9160 | 4.1146 | 0.9384 | 1.0354 | 0.8708 |
1.7507 0.4220 |
0.9964 0.9471 |
0.1077 | -0.0013 | -82.85 | 0.9627 | |
aKorsmeyer-Peppas was fitted to first 60% drug release
bKorsmeyer-Peppas was fitted to first 60% drug release into two steps
According to the Higuchi model, increasing the cross-linker ratio increases the rate constant (K) for NIPs. Excluding MIP 1.5, MIP samples show a similar tendency, increasing cross-linker increases K proportionally. In addition, MIP3 has the highest K value among hydrogel samples considering the Higuchi model.
According to the two-step Korsmeyer-Peppas model, DOX release takes place in two different drug transport mechanisms. For both MIP and NIP hydrogels, the super case transport II profile controls DOX transport during the first 55 min. After 55 min, drug release occurs following Fickian diffusion. When we evaluate the n values by considering the Korsmayer-Peppas (two-step) drug release model, we see that the n value increases with an increase in cross-linker ratio for NIPs at the first 55 min. But after 55 min, at the Fick diffusion zone, the trend changes for NIPs. In the case of MIPs, n values of samples are as follows at both the Super case II transport and Fick diffusion zones; MIP2 > MIP3 > MIP1 > MIP1.5.
Model constants A and B in the Kopcha model are obtained for each sample with relatively high correlation. As shown in Table 5, the ratio of A/B are calculated to be higher than 1, which shows that the release mechanism is predominately diffusion-based. In this study, the ratio of diffusion rate constants for each NIP and MIP pair (ANIP/AMIP) prepared in the same amount of cross-linker was also calculated to compare the diffusion rates of the pairs. The calculated ratios are given in Table 5. Except for the NIP3-MIP3 pair, the ratio of the diffusion rate constants is almost 1, indicating the equal diffusion rate for each pair. On the other hand, the ratio for the NIP3-MIP3 pair is about 0.9, which is an indication of the relatively rapid diffusion of MIP3 when compared to NIP3.
Conclusions
In this study, molecular docking with two different force fields and MD simulations are used to understand DOX—monomer interactions at the atomic level. With the sight of these studies, a suitable functional monomer and critical cross-linker ratio can be detected. Here, we show that both interaction energy calculations for possible configurations and full-atom MD simulations, agree on the extent of molecular interactions between functional monomers and DOX. While this approach is popularly employed to determine the functional monomer for imprinting, we show that the presence of other components in the prepolymerization step, i.e. HEMA and cross-linker EGDMA significantly affects the extent of interactions between the drug and the functional monomers. To this end, we suggest that the “most promising” monomer to imprint DOX, ITA, and the “least promising” monomer AA will lead to similar imprinting factors and drug release performances.
In the experimental part, DOX MIP and NIP hydrogel synthesis is carried out with HEMA, EGDMA, and AA, which are commercial monomers for contact lens production. With a systematic approach, we observe a critical cross-linker concentration, namely 2.0%, up to which NIP hydrogels have similar performances as MIPs. Among all imprinted hydrogels, samples synthesized with 3.0% cross-linker (MIP 3) adsorb more DOX with respect to NIP3. In addition, an IF value > 1.0 indicates that DOX imprinting is achieved in MIP 3, which also displays the best-controlled DOX release. The molecular mechanism behind this observation is revealed by MD studies, which suggest that at 3.0% EGDMA concentration, AA monomers effectively surround drug molecules and maintain their interaction stabilities, leading to better molecular cavities on the polymer. We also show that DOX accommodates stable intra-molecular hydrogen-bonding interactions, which may lead to relatively low IF values. Indeed, minocycline that is from the same tetracycline family, lacks this structural detail and can be imprinted with higher IF values using the same functional monomer and cross-linker [53].
This complementary computational and experimental study also shows that a critical cross-linker concentration exists for hydrogels as was shown in a similar study [53], and MD simulations provide valuable insights on the molecular mechanisms governing molecularly imprinted polymers so that they can save an important amount of time and money before experiments.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge Dr. Ozlem Gürses for her comments and valuable suggestions about the treatment of corneal neovascularization. The authors would also like to especially thank Deva (Istanbul, Turkey) and Assis. Prof. Pelin Suzgun (Marmara University, Faculty of Pharmacy) for providing DOX.
Funding
This work was supported by the Scientific and Technological Research Council of Turkey (TÜBİTAK) [Grant Number 114M459].
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ozge Kurkcuoglu, Email: olevitas@itu.edu.tr.
F. Seniha Güner, Email: guners@itu.edu.tr.
References
- 1.Byrne ME, Park K, Peppas NA. Molecular imprinting within hydrogels. Adv Drug Deliv Rev. 2002;54:149–161. doi: 10.1016/S0169-409X(01)00246-0. [DOI] [PubMed] [Google Scholar]
- 2.Trotta F, Biasizzo M, Caldera F. Molecularly imprinted membranes. Membranes. 2012;2:440–477. doi: 10.3390/membranes2030440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshikawa M, Tharpa K, Dima Ş-O. Molecularly imprinted membranes: past, present, and future. Chem Rev. 2016;116:11500–11528. doi: 10.1021/acs.chemrev.6b00098. [DOI] [PubMed] [Google Scholar]
- 4.Ertürk G, Mattiasson B. Molecular imprinting techniques used for the preparation of biosensors. Sensors. 2017;17:1–17. doi: 10.3390/s17020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tieppo A, White CJ, Paine AC, et al. Sustained in vivo release from imprinted therapeutic contact lenses. J Control Release. 2012;157:391–397. doi: 10.1016/j.jconrel.2011.09.087. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Wang X, Lu W, et al. Molecular imprinting: perspectives and applications. Chem Soc Rev. 2016;45:2137–2211. doi: 10.1039/C6CS00061D. [DOI] [PubMed] [Google Scholar]
- 7.Hashim SNNS, Boysen RI, Schwarz LJ, et al. A comparison of covalent and non-covalent imprinting strategies for the synthesis of stigmasterol imprinted polymers. J Chromatogr A. 2014;1359:35–43. doi: 10.1016/j.chroma.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 8.Cowen T, Stefanucci E, Piletska E, et al. Synthetic mechanism of molecular imprinting at the solid phase. Macromolecules. 2020;53:1435–1442. doi: 10.1021/acs.macromol.9b01913. [DOI] [Google Scholar]
- 9.Mojica E-RE, Autschbach J, Bright FV, Aga DS. Tetracycline speciation during molecular imprinting in xerogels results in class-selective binding. Analyst. 2011;136:749–755. doi: 10.1039/C0an00707b. [DOI] [PubMed] [Google Scholar]
- 10.Boroznjak R, Reut J, Tretjakov A, et al. A computational approach to study functional monomer-protein molecular interactions to optimize protein molecular imprinting. J Mol Recognit. 2017;30:1–9. doi: 10.1002/jmr.2635. [DOI] [PubMed] [Google Scholar]
- 11.Nicholls IA, Andersson HS, Charlton C, et al. Theoretical and computational strategies for rational molecularly imprinted polymer design. Biosens Bioelectron. 2009;25:543–552. doi: 10.1016/j.bios.2009.03.038. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Zhang W, Wu Z, et al. Theoretical design, preparation, and evaluation of Ginkgolide B molecularly imprinted polymers. J Sep Sci. 2020;43:514–523. doi: 10.1002/jssc.201900675. [DOI] [PubMed] [Google Scholar]
- 13.Nicholls IA, Andersson HS, Golker K, et al. Rational design of biomimetic molecularly imprinted materials: theoretical and computational strategies for guiding nanoscale structured polymer development. Anal Bioanal Chem. 2011;400:1771–1786. doi: 10.1007/s00216-011-4935-1. [DOI] [PubMed] [Google Scholar]
- 14.Shoravi S, Olsson GD, Karlsson BCG, Nicholls IA. On the influence of crosslinker on template complexation in molecularly imprinted polymers: a computational study of prepolymerization mixture events with correlations to template-polymer recognition behavior and nmr spectroscopic studies. Int J Mol Sci. 2010;15:10622–10634. doi: 10.3390/ijms150610622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karim K, Breton F, Rouillon R, et al. How to find effective functional monomers for effective molecularly imprinted polymers? Adv Drug Deliv Rev. 2005;57:1795–1808. doi: 10.1016/j.addr.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Yu X, Zeng H, Wan J, Cao X. Computational design of a molecularly imprinted polymer compatible with an aqueous environment for solid phase extraction of chenodeoxycholic acid. J Chromatogr A. 2020;1609:460490. doi: 10.1016/j.chroma.2019.460490. [DOI] [PubMed] [Google Scholar]
- 17.Rostamizadeh K, Vahedpour M, Bozorgi S. Synthesis, characterization and evaluation of computationally designed nanoparticles of molecular imprinted polymers as drug delivery systems. Int J Pharm. 2012;424:67–75. doi: 10.1016/j.ijpharm.2011.12.054. [DOI] [PubMed] [Google Scholar]
- 18.Janczura M, Sobiech M, Giebułtowicz J, Luliński P. Computational and experimental designing of imprinted sorbent for the determination of nitroxidative stress products: an analysis of 4-hydroxyphenylacetic acid conversion. J Mater Sci. 2021;56:8439–8460. doi: 10.1007/s10853-021-05847-9. [DOI] [Google Scholar]
- 19.Xu W, Wang Y, Huang W, et al. Computer-aided design and synthesis of CdTe@SiO2 core-shell molecularly imprinted polymers as a fluorescent sensor for the selective determination of sulfamethoxazole in milk and lake water. J Sep Sci. 2017;40:1091–1098. doi: 10.1002/jssc.201601180. [DOI] [PubMed] [Google Scholar]
- 20.Khurana L, Balasubramanian K. Adsorption potency of imprinted Starch/PVA polymers confined ionic liquid with molecular simulation framework. J Environ Chem Eng. 2016;4:2147–2154. doi: 10.1016/j.jece.2016.03.032. [DOI] [Google Scholar]
- 21.Silva CF, Menezes LF, Pereira AC, Nascimento CS. Molecularly imprinted polymer (MIP) for thiamethoxam: a theoretical and experimental study. J Mol Struct. 2021;1231:129980. doi: 10.1016/j.molstruc.2021.129980. [DOI] [Google Scholar]
- 22.Kong Y, Wang N, Ni X, et al. Molecular dynamics simulations of molecularly imprinted polymer approaches to the preparation of selective materials to remove norfloxacin. J Appl Polym Sci. 2016;133:1–11. doi: 10.1002/app.42817. [DOI] [Google Scholar]
- 23.Suksuwan A, Lomlim L, Rungrotmongkol T, et al. The composite nanomaterials containing (R)-thalidomide-molecularly imprinted polymers as a recognition system for enantioselective-controlled release and targeted drug delivery. J Appl Polym Sci. 2015;132:1–21. doi: 10.1002/app.41930. [DOI] [Google Scholar]
- 24.Golker K, Nicholls IA. The effect of crosslinking density on molecularly imprinted polymer morphology and recognition. Eur Polym J. 2016;75:423–430. doi: 10.1016/j.eurpolymj.2016.01.008. [DOI] [Google Scholar]
- 25.Cubuk H, Ozbil M, Cakir Hatir P. Computational analysis of functional monomers used in molecular imprinting for promising COVID-19 detection. Comput Theor Chem. 2021;1199:113215. doi: 10.1016/j.comptc.2021.113215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Sun B, Li Y, Chang W. Study properties of molecular imprinting polymer using a computational approach. Analyst. 2003;128:944–949. doi: 10.1039/B212731H. [DOI] [Google Scholar]
- 27.Andersson LI, Müller R, Vlatakis G, Mosbach K (2016) Mimics of the binding sites of opioid receptors obtained by molecular imprinting of enkephalin and morphine. Proc Natl Acad Sci USA 92:4788–4792. 10.1073/pnas.92.11.4788 [DOI] [PMC free article] [PubMed]
- 28.Sellergren B, Shea KJ. Influence of polymer morphology on the ability of imprinted network polymers to resolve enantiomers. J Chromatogr A. 1993;635:31–49. doi: 10.1016/0021-9673(93)83112-6. [DOI] [Google Scholar]
- 29.Sellergren B, Dauwe C, Schneider T. Pressure-induced binding sites in molecularly imprinted network polymers. Macromolecules. 1997;30:2454–2459. doi: 10.1021/ma960745i. [DOI] [Google Scholar]
- 30.Hu X, Hao L, Wang H, et al. Hydrogel contact lens for extended delivery of ophthalmic drugs. Int J Polym Sci. 2011 doi: 10.1155/2011/814163. [DOI] [Google Scholar]
- 31.O'Mahony J, Wei S, Molinelli A, Mizaikoff B. Imprinted polymeric materials. Insight into the nature of prepolymerization complexes of quercetin imprinted polymers. Anal Chem. 2006;78:6187–6190. doi: 10.1021/ac060446j. [DOI] [PubMed] [Google Scholar]
- 32.Shoravi S, Olsson GD, Karlsson BCG, et al. In silico screening of molecular imprinting prepolymerization systems: oseltamivir selective polymers through full-system molecular dynamics-based studies. Org Biomol Chem. 2016;14:4210–4219. doi: 10.1039/C6OB00305B. [DOI] [PubMed] [Google Scholar]
- 33.Iturralde I, Paulis M, Leiza JR. The effect of the crosslinking agent on the performance of propranolol imprinted polymers. Eur Polym J. 2014;53:282–291. doi: 10.1016/j.eurpolymj.2014.02.003. [DOI] [Google Scholar]
- 34.Olsson GD, Karlsson BCG, Shoravi S, et al. Mechanisms underlying molecularly imprinted polymer molecular memory and the role of crosslinker: Resolving debate on the nature of template recognition in phenylalanine anilide imprinted polymers. J Mol Recognit. 2012;25:69–73. doi: 10.1002/jmr.2147. [DOI] [PubMed] [Google Scholar]
- 35.Muhammad T, Nur Z, Piletska EV, et al. Rational design of molecularly imprinted polymer: the choice of cross-linker. Analyst. 2012;137:2623. doi: 10.1039/c2an35228a. [DOI] [PubMed] [Google Scholar]
- 36.Tom LA, Schneck NA, Walter C. Improving the imprinting effect by optimizing template:monomer:cross-linker ratios in a molecularly imprinted polymer for sulfadimethoxine. J Chromatogr B Anal Technol Biomed Life Sci. 2012;909:61–64. doi: 10.1016/j.jchromb.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Henthorn DB, Peppas NA. Molecular simulations of recognitive behavior of molecularly imprinted intelligent polymeric networks. Ind Eng Chem Res. 2007;46:6084–6091. doi: 10.1021/ie061369l. [DOI] [Google Scholar]
- 38.Alvarez-Rivera F, Concheiro A, Alvarez-Lorenzo C. Epalrestat-loaded silicone hydrogels as contact lenses to address diabetic-eye complications. Eur J Pharm Biopharm. 2018;122:126–136. doi: 10.1016/j.ejpb.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 39.Maulvi FA, Lakdawala DH, Shaikh AA, et al. In vitro and in vivo evaluation of novel implantation technology in hydrogel contact lenses for controlled drug delivery. J Control Release. 2016;226:47–56. doi: 10.1016/j.jconrel.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Farkouh A, Frigo P, Czejka M. Systemic side effects of eye drops: a pharmacokinetic perspective. Clin Ophthalmol. 2016;10:2433–2441. doi: 10.2147/OPTH.S118409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowman RJC, Cope J, Nischal KK. Ocular and systemic side effects of brimonidine 0.2% eye drops (Alphagen®) in children. Eye. 2004;18:24–26. doi: 10.1038/sj.eye.6700520. [DOI] [PubMed] [Google Scholar]
- 42.Tashakori-Sabzevar F, Mohajeri SA. Development of ocular drug delivery systems using molecularly imprinted soft contact lenses. Drug Dev Ind Pharm. 2015;41:703–713. doi: 10.1016/j.biomaterials.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 43.Rosa dos Santos JF, Alvarez-Lorenzo C, Silva M, et al. Soft contact lenses functionalized with pendant cyclodextrins for controlled drug delivery. Biomaterials. 2009;30:1348–1355. doi: 10.1089/jop.2010.0099. [DOI] [PubMed] [Google Scholar]
- 44.Gordon MK, Desantis A, Deshmukh M, et al. Doxycycline hydrogels as a potential therapy for ocular vesicant injury. J Ocul Pharmacol Ther. 2010;26:407–419. doi: 10.3109/03639045.2014.948451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiratani H, Fujiwara A, Tamiya Y, et al. Ocular release of timolol from molecularly imprinted soft contact lenses. Biomaterials. 2005;26:1293–1298. doi: 10.1016/j.biomaterials.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 46.Hiratani H, Alvarez-Lorenzo C. The nature of backbone monomers determines the performance of imprinted soft contact lenses as timolol drug delivery systems. Biomaterials. 2004;25:1105–1113. doi: 10.1016/S0142-9612(03)00622-7. [DOI] [PubMed] [Google Scholar]
- 47.Ali M, Byrne ME. Controlled release of high molecular weight hyaluronic acid from molecularly imprinted hydrogel contact lenses. Pharm Res. 2009;26:714–726. doi: 10.1007/s11095-008-9818-6. [DOI] [PubMed] [Google Scholar]
- 48.Alvarez-Lorenzo C, Yañez F, Barreiro-Iglesias R, Concheiro A. Imprinted soft contact lenses as norfloxacin delivery systems. J Control Release. 2006;113:236–244. doi: 10.1016/j.jconrel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Tieppo A, Pate KM, Byrne ME. In vitro controlled release of an anti-inflammatory from daily disposable therapeutic contact lenses under physiological ocular tear flow. Eur J Pharm Biopharm. 2012;81:170–177. doi: 10.1016/j.ejpb.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Silva D, de Sousa HC, Helena Gil M, et al. Moxifloxacin imprinted silicon based hydrogels for sustained ocular release. Ann Med. 2019;51:103–103. doi: 10.1080/07853890.2018.1562708. [DOI] [Google Scholar]
- 51.Jovanovic V, Nikolic L. The effect of topical doxycycline on corneal neovascularization. Curr Eye Res. 2014;39:142–148. doi: 10.3109/02713683.2013.833246. [DOI] [PubMed] [Google Scholar]
- 52.Kim SW, Ha BJ, Kim EK, et al. The effect of topical bevacizumab on corneal neovascularization. Ophthalmology. 2008 doi: 10.1016/j.ophtha.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Dan L, Shi-long Y, Miao-li L, et al. Inhibitory effect of oral doxycycline on neovascularization in a rat corneal alkali burn model of angiogenesis. Curr Eye Res. 2008;33:653–660. doi: 10.1080/02713680802245772. [DOI] [PubMed] [Google Scholar]
- 54.Dalgakiran D, Inan T, Güner FS. Investigation of photoinduced polymerization of doxycycline-imprinted hydrogels: effect of template on initiator reactivity, conversion, and reaction rate. Turk J Chem. 2017;41:862–873. doi: 10.3906/kim-1701-37. [DOI] [Google Scholar]
- 55.Erkal Ilhan S, Kürkçüoğlu Ö, Inan T, et al. Preparation and determination of in vivo and in vitro performance of doxycycline imprinted contact lenses for corneal neovascularization treatment. J Turk Chem Soc Sect A Chem. 2018 doi: 10.18596/jotcsa.428846. [DOI] [Google Scholar]
- 56.Eroglu B, Dalgakiran D, Inan T, et al. A computational and experimental approach to develop minocycline-imprinted hydrogels and determination of their drug delivery performances. J Polym Res. 2018;25(2018):258. doi: 10.1007/s10965-018-1647-7. [DOI] [Google Scholar]
- 57.Bolton EE, Chen J, Kim S, et al. PubChem3D: a new resource for scientists. J Cheminform. 2011;3:1–15. doi: 10.1186/1758-2946-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Şanli S, Şanli N, Alsancak G. Determination of protonation constants of some tetracycline antibiotics by potentiometry and LC methods in water and acetonitrile-water binary mixtures. J Braz Chem Soc. 2009;20:939–946. doi: 10.1590/S0103-50532009000500020. [DOI] [Google Scholar]
- 59.Clark M, Cramer RD, Van Opdenbosch N. Validation of the general purpose Tripos 5.2 force field. J Comput Chem. 1989;10:982–1012. doi: 10.1002/jcc.540100804. [DOI] [Google Scholar]
- 60.Sun H. COMPASS: an ab initio force-field optimized for condensed-phase applications overview with details on alkane and benzene compounds. J Phys Chem B. 1998;5647:7338–7364. doi: 10.1021/jp980939v. [DOI] [Google Scholar]
- 61.Dash S, Murthy PN, Nath L, Chowdhury P. Kinetic modeling on drug release from controlled drug delivery systems. Acta Pol Pharm. 2010;67:217–223. [PubMed] [Google Scholar]
- 62.Costa P, Lobo JMS. Modelling and comparison of dissolution profiles. Eur J Pharm Sci. 2001;13:123–133. doi: 10.1016/S0928-0987(01)00095-1. [DOI] [PubMed] [Google Scholar]
- 63.Chevalier E, Viana M, Artaud A, Haddouchi S, Chulia D. A novel application of the T-cell for flow-through dissolution: the case of bioceramics used as ibuprofen carrier. Talanta. 2009;77(4):1545–1548. doi: 10.1016/j.talanta.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 64.Hiratani H, Alvarez-Lorenzo C. T imolol uptake and release by imprinted soft contact lenses made of N, N-diethylacrylamide and methacrylic acid. J Control Release. 2002;83:223–230. doi: 10.1016/S0168-3659(02)00213-4. [DOI] [PubMed] [Google Scholar]
- 65.Suryana S, Mutakin, Rosandi Y, Hasanah AN. An update on molecularly imprinted polymer design through a computational approach to produce molecular recognition material with enhanced analytical performance. Molecules. 2021;26(7):1891. doi: 10.3390/molecules26071891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santos OMM, Silva DM, Martins FT, et al. Protonation pattern, tautomerism, conformerism, and physicochemical analysis in new crystal forms of the antibiotic doxycycline. Cryst Growth Des. 2014;14:3711–3726. doi: 10.1021/cg500877z. [DOI] [Google Scholar]
- 67.Nicolescu TV, Sarbu A, Ghiurea M, Donescu D. Influence of crosslinker / porogen ratio upon imprinted polymer parameters. UPB Sci Bull Ser. 2011;73:163–172. doi: 10.1002/app.37528. [DOI] [Google Scholar]
- 68.Hui A, Sheardown H, Jones L. Acetic and acrylic acid molecular imprinted model silicone hydrogel materials for ciprofloxacin-hcl delivery. Materials. 2012;5:85–107. doi: 10.3390/ma5010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feás X, Seijas JA, Vázquez-Tato MP, et al. Syntheses of molecularly imprinted polymers: molecular recognition of cyproheptadine using original print molecules and azatadine as dummy templates. Anal Chim Acta. 2009;631:237–244. doi: 10.1016/j.aca.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 70.Kuznetsova IM, Turoverov KK, Uversky VN. What macromolecular crowding can do to a protein. Int J Mol Sci. 2014;15:23090–23140. doi: 10.3390/ijms151223090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Song D, Lanni LM, Shimizu KD. Importance of functional monomer dimerization in the molecular imprinting process. Macromolecules. 2010;43(15):6284–6294. doi: 10.1021/ma101013c. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations.