Abstract
Microplastics (MPs) have been reported in the outdoor/indoor air of urban centres, raising health concerns due to the potential for human exposure. Since aerosols are considered one of the routes of Coronavirus disease 2019 (COVID-19) transmission and may bind to the surface of airborne MPs, we hypothesize that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could be associated with the levels of MPs in the air. Our goal was to quantify the SARS-CoV-2 RNA and MPs present in the total suspended particles (TSP) collected in the area surrounding the largest medical centre in Latin America and to elucidate a possible association among weather variables, MPs, and SARS-CoV-2 in the air. TSP were sampled from three outdoor locations in the areas surrounding a medical centre. MPs were quantified and measured under a fluorescence microscope, and their polymeric composition was characterized by Fourier transform infrared (FT-IR) microspectroscopy coupled with attenuated total reflectance (ATR). The viral load of SARS-CoV-2 was quantified by an in-house real-time PCR assay. A generalized linear model (GzLM) was employed to evaluate the effect of the SARS-CoV-2 quantification on MPs and weather variables. TSP samples tested positive for SARS-CoV-2 in 22 out of 38 samples at the three sites. Polyester was the most frequent polymer (80%) found in the samples. The total amount of MPs was positively associated with the quantification of SARS-CoV-2 envelope genes and negatively associated with weather variables (temperature and relative humidity). Our findings show that SARS-CoV-2 aerosols may bind to TSP, such as MPs, and facilitate virus entry into the human body.
Keywords: Airborne microplastic, SARS-CoV-2 RNA, Public health, Aerosols
1. Background
A novel human coronavirus disease known as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) emerged in December 2019 and rapidly spread worldwide, causing serious global damage to public health, and leading to severe social/economic disruptions (Akhbarizadeh et al., 2021a; Wu et al., 2020; Gralinski and Menachery, 2020). By May 2021, the pandemic was far from being controlled in Brazil, especially in large cities such as São Paulo (FIOCRUZ, 2021). With the emergence of the novel gamma and delta variants with higher transmissibility, Brazil is currently one of the world epicentres of Coronavirus disease 2019 (COVID-19). As of the date of this publication, Brazil has more than 581,150 deaths and 20,804,215 accumulated cases (FIOCRUZ, 2021).
Droplets or aerosols in the air are considered major routes of COVID-19 transmission (WHO, 2020), which seems to be affected by higher temperature and relative humidity (Biryukov et al., 2020; Sajadi et al., 2020). Aerosols are airborne particles smaller than 5 μm. There is evidence that virus particles bind to aerosols and travel over longer distances, remaining floating in the air for much longer than respiratory droplets (Zuo et al., 2020).
Whether SARS-CoV-2 aerosols can “hitchhike” in atmospheric particulate matter (PM), thereby triggering the spread of the virus, remains in question. Several methodologies have been used to sample SARS-CoV-2 in the air (Abdeldayem et al., 2022). Setti et al. (2020) showed that SARS-CoV-2 RNA is present in particulate matter (PM10). Their study was conducted at an industrial site in the Bergamo Province of northern Italy, known for high levels of air pollution and some of the most severe cases of COVID-19 in that country. Zhu et al. (2020) found significant positive associations of particulate matter 2.5 (PM2.5), particulate matter 10 (PM10), carbon monoxide (CO), nitrogen dioxide (NO2), and ozone (O3) with COVID-19 cases in China.
Airborne microplastics (MPs) are a constituent part of atmospheric PM (Levermore et al. (2020). MPs are the result of plastic degradation and are ubiquitously found in the environment due to inadequate disposal (Amato-Lourenço et al., 2020). Due to their small size and low density, MPs are also present in the air (Allen et al., 2019) and may be inhaled by humans (Amato-Lourenço et al., 2020). Airborne MPs have been found in urban centres, as shown in studies performed in London, Shanghai, and Paris (Wright et al., 2020; Liu et al., 2019; Dris et al., 2015). Atmospheric deposition of MPs in cities may range from 575 to 1008 particles/m2/day (largely composed of fibres), as shown by Wright et al. (2020). MPs are vectors of several pollutants (Hartmann et al., 2017; Akhbarizadeh et al., 2021b) as well as pathological microorganisms present in the environment (Foulon et al., 2016). Its surface associated with environmental conditions allows the adherence of microorganisms and the colonization of bacteria and viruses (Foulon et al., 2016; Mammo et al., 2020).
SARS-CoV-2 is stable on plastic surfaces for up to 72 h at room temperature (van Doremalen et al., 2020). Furthermore, the virus can remain viable and infectious in aerosols for hours (van Doremalen et al., 2020).
We hypothesize that SARS-CoV-2, in contrast to the inhalation mode of viral transmission through airborne respirable droplets, is potentially associated with airborne MPs present on total suspended particles (TSP). Viral particles have been found in aerosols and on surfaces in hospital wards (Passos et al., 2021; Liu et al., 2020). Many people circulate around large medical centres, but no studies have analysed the presence of SARS-CoV-2 and MPs in this setting.
Therefore, in this study, we quantified the SARS-CoV-2 RNA and MPs in the TSP samples collected in the area surrounding the largest medical centre in Latin America and elucidated a possible association among weather variables, MPs, and SARS-CoV-2 in the air.
2. Methods
2.1. Ethics
This study was approved by the Ethical Board of the São Paulo University Medical School (approval #: 08304519.4.0000.0065).
2.2. Study site description
As of the date of this publication, Sao Paulo megacity is the epicentre of the disease in Brazil, and the number of cases continues to increase exponentially (CVE, 2021).
The city has a population that exceeds 11.8 million people living in an area of 1521 km2 (SEADE, 2019). Additionally, Sao Paulo is affected by high concentrations of air pollutants from vehicular traffic (such as particulate matter) (CETESB, Sao Paulo State Environmental Agency, 2020) that normally exceed the World Health Organization (WHO) recommendations (WHO, 2005).
On the first day of monitoring (September 24th, 2020), the city of São Paulo counted 286,862 confirmed cases and 12,504 deaths attributed to SARS-CoV-2 (CVE, 2021).
This study was carried out in the outdoor areas of the largest medical centre in Latin America (including a medical school). This area is characterized by a high volume of vehicular and pedestrian traffic (CET, 2020), in addition to having bus stops and access to the subway. During the SARS-CoV-2 pandemic, the Central Institute of Clinical Hospital of the Faculty of Medicine of the University of São Paulo and the Institute of Infectiology – Emilio Ribas were appointed as reference centres for COVID-19 in the state of Sao Paulo. In these hospitals, 1100 hospital beds (360 intensive care beds) were allocated exclusively to COVID-19. In addition, the emergency room of the hospitals attends around 90 patients per day suspect of COVID-19.
2.3. Total suspended particles (TSP) sampling
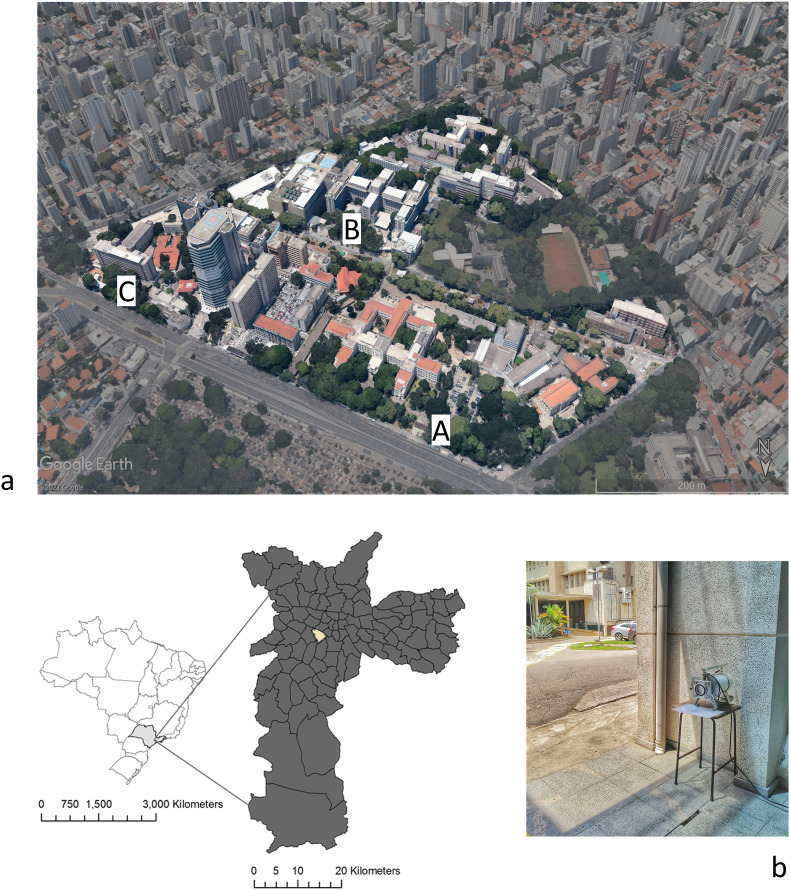
The samplings occurred from September 24th to November 1st, 2020, from three outdoor locations in the areas surrounding the hospital complex, A (−23°33′15.8″S 46°40′13.8″W), B (−23°33′24.0″S 46°40′12.4″W) and C (−23°33′20.0″S 46°40′05.4″W) (Fig. 1 a). Site “C" was located at an infectious disease hospital that shared the main entrance for patients and staff. During the sampling period, an average circulation of 2623 people in location A, 4104 people in location B, and 882 people in location C were recorded. TSP samples were collected for 24 h using a Handi-vol sampler (Energética, Brazil) operating at a flow rate of 3 L/min on a fibreglass filter (porosity <1 μm, diameter: 110 mm) (Whatman®, Whatman Labware Products, Maidstone, Kent, UK)) (Fig. 1b). The samplings were carried out at a height of 1.25 m because it corresponds to the standard breathing height of an adult (Sharma and Kumar, 2020). Filters were weighed before and after particle collection. After sampling, the filters were immediately stored at −20 °C before the analysis. All the materials used in handling the filters (tweezers, Petri dishes, among others) were autoclaved before sampling and opened at the time of collection.
Fig. 1.
(a) Overview of the hospital complex and the sampling site locations in the Sao Paulo municipality, (b) TSP sampling at site 3.
Weather data, including the temperature (°C) and relative humidity (%), were obtained from the station closest to the sampling points provided by the Emergency Management Centre of Sao Paulo Municipality (CGE).
2.4. Quality control and evaluation of microplastic sample processing
A plastic-free approach was applied to guarantee the quality of the results, allowing us to evaluate the sources of variability and error and to increase the reliability of the collected data. All procedures were performed in accordance with the protocols recommended by Rochman et al. (2019). The analyses were performed at the Experimental Air Pollution Laboratory (LIM05) of the Faculty of Medicine of the University of São Paulo and at the Molecular Spectroscopy Laboratory - Department of Fundamental Chemistry of the Institute of Chemistry - University of São Paulo. Briefly, all materials were washed thoroughly with Milli-Q® (Merck KGaA, Darmstadt, Germany) water three times before use. The glass materials were cleaned with a 1:1 nitric acid solution. All materials and samples were covered with aluminium foil (before and after processing) to avoid airborne sample contamination. Cotton lab coats (100%) and clean nitrile gloves were used during all procedures. The samples were processed in a clean laminar flow cabinet. A procedural blank was used for every ten samples during the whole procedure to account for contamination during sampling and laboratory protocols. The amounts in the procedural blanks were subtracted from TSP samples according to their chemical identifier (Rochman et al., 2019).
2.5. Quantification of microplastics in TSP samples
Half of the filters were stained with the lipophilic dye Nile Red (Sigma-Aldrich®, Saint Louis, Missouri, USA). This dye has been used in studies with microplastics being adsorbed on plastic surfaces and making them fluorescent when irradiated with UVA light (Erni-Cassola et al., 2017). Preparation of staining solution was carried out using 1 mg Nile Red (C26H18N2O2 ultrapure) for each 1 mL of chloroform and then mixed in a volumetric flask (Tamminga et al., 2017). The solution was transferred to brown glass bottles for storage. The filters were stained with 1 mL of Nile Red solution in two sequences of 0.5 mL each and covered with watch glass for at least 24 h at room temperature (Tamminga et al., 2017). The coloured filters were analysed under a fluorescence microscope (Olympus BX51) under U.V. and an attached Olympus camera (Olympus Co, St Laurent, Quebec, Canada) (Fig. S4 – supplemental material). The quantified MPs were classified as fibres or particles due to their distinct structural characteristics (e.g., total surface area) and possible deleterious effects on human health (Amato-Lourenço et al., 2020). To identify and quantify man-made fluorescent particles/fibres, we adopted the recommendations described by Hidalgo-Ruz et al. (2012). Particle/fibre lengths were recorded using Image Pro-Plus 6.0 software. The lower observation limit was 50 μm.
Then, the filter samples were analysed using Bruker's Vertex 80v model FT-IR infrared spectrometer, and Ominic software, version 8.1.0.10. The chemical composition was verified through the spectral match with the “HR Hummel Polymer and Additives” and “Aldrich condensed phase” libraries. Minor corrections such as baseline adjustment and removal of the carbon dioxide (CO2) peak were performed after obtaining the spectra to improve the visualization of the spectra. Samples whose match was over 60% were considered. Due to the high volume and time constraints, 5% of fibres/particles in each sample were randomly analysed as conducted by Wright et al. (2020).
2.6. SARS-CoV-2 quantification
2.6.1. DNA∕RNA extraction
Nucleic acids were extracted from half of the TSP fibreglass filters. Briefly, the filters were incubated for 3 h at 56 °C in AL buffer (Qiagen, Hilden, Germany) and 100 mg/mL proteinase K (Qiagen, Hilden, Germany). Then, DNA and RNA were extracted using a Magna Pure Compact Nucleic Acid Isolation kit (Magna Pure Compact, Roche Diagnostics GmbH, Germany) according to the manufacturer's instructions.
2.6.2. Real-time reverse-transcription PCR
SARS-CoV-2 RNA was quantified by an in-house real-time PCR assay that amplified part of the envelope protein (E) (Corman Victor et al., 2020) and nucleocapsid protein (N1) (CDC, 2021) genes. Positive and negative controls were included in all amplification reactions. As a positive control, synthetic RNA from SARS-CoV-2 Standard (Exact Diagnostics SARS-CoV-2 Standard, Cat Number #COV019) and RNA extracted from inactivated SARS-CoV-2. Negative controls consisted of the reaction with all reagents and eluents without a sample.
Real-time PCR was performed using StepOne System equipment (Applied Biosystems, Foster City, CA, USA) using conditions described previously in Corman Victor et al., 2020). Primer and probe sequences are presented in Supplemental Material - Table S1). Standard curves were generated from serial dilutions (1:10) of SARS-CoV-2 RNA (Supplemental Material Figs. S2 and S3) and converted to genomic units per TSP. Samples were considered positive if amplification of target regions had a cycle threshold value (Ct) less than 40.
2.7. Statistical analyses
Descriptive data are presented as medians, means, minimum and maximum values. The nonparametric data (SARS-CoV-2 envelope and nucleocapsid) were log transformed to comply with the assumptions of variance of homogeneity and residual normality. Analysis of variance (one-way analysis of variance (ANOVA)) followed by a Tukey post hoc test was used to test for differences among MPs, TSP, SARS-CoV-2 envelope, and nucleocapsid quantification variables considering each sampling site.
Exponential regression was performed considering SARS-CoV-2 and total MPs quantification. A generalized linear model (GzLM) (Mccullagh, 1984) was employed to evaluate the obtained SARS-CoV-2 quantification on MPs and weather variables. The GzLM was fitted using the log-link function and Poisson distribution. Among the GzLM families, the most commonly used in studies on the impact of air pollution on health is Poisson regression (Conceição et al., 2001). The models were evaluated for normal distribution of the residuals, and the random effects and respective confidence intervals of the estimated variables were calculated. Akaike's information criterion (AIC) was applied to indicate a better fitting model. Statistical analyses were performed using IBM® SPSS® Statistics software (version 26 IBM Corp., Chicago, IL, USA). A value of p < 0.05 was considered statistically significant.
3. Results
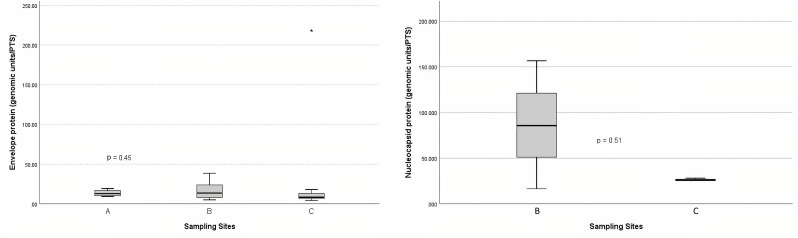
Descriptive results for the quantification of SARS-CoV-2 envelope and nucleocapsid genes, MPs (fibres and particles), TSP, and weather data at sites A, B, and C are summarized in Table 1 . The SARS-CoV-2 envelope was detected in 22 out of 38 samples (Ct < 40) at the three sites (site A; N = 5, site B; N = 10 and site C; N = 7), while the SARS-CoV-2 nucleocapsid was detected in just 6 samples at sites B (N = 3) and C (N = 3).
Table 1.
Descriptive data for each sampling site (N = number of samples; S.D. = standard deviation).
| Site | N | Variables | Mean | S.D. | Median | Minimum | Maximum |
|---|---|---|---|---|---|---|---|
| A | 12 | Temperature (°C) | 27.7 | 1.4 | 30.8 | 21.8 | 34.7 |
| Relative humidity (%) | 37.5 | 5.1 | 31.0 | 16.0 | 68.0 | ||
| Envelope protein gene quantification - (genomic units/TSP) | 13.66 | 1.93 | 12.86 | 9.24 | 19.28 | ||
| Nucleocapsid protein gene quantification - (genomic units/TSP) | – | – | – | – | – | ||
| Total MPs (n/m³) | 14.02 | 1.53 | 12.96 | 9.03 | 24.95 | ||
| MPs fibres (n/m³) | 13.85 | 1.36 | 13.08 | 9.03 | 24.02 | ||
| MPs particles (n/m³) | 0.23 | 0.08 | 0.23 | 0.00 | 0.93 | ||
| TSP (mg/m³) | 5.75 | 2.91 | 520 | 2.08 | 11.11 | ||
| B | 16 | Temperature (°C) | 31.4 | 1.8 | 32.9 | 17.1 | 38.0 |
| Relative humidity (%) | 35.5 | 5.3 | 27.0 | 16.0 | 76.0 | ||
| Envelope protein gene quantification - (genomic units/TSP) | 17.21 | 3.32 | 13.50 | 4.99 | 38.52 | ||
| Nucleocapsid protein gene quantification - (genomic units/TSP) | 86.26 | 40.45 | 85.66 | 16.50 | 156.64 | ||
| Total MPs (n/m³) | 15.90 | 1.18 | 15.28 | 8.80 | 23.38 | ||
| MPs fibres (n/m³) | 15.30 | 1.13 | 14.58 | 8.80 | 22.68 | ||
| MPs particles (n/m³) | 0.10 | 0.05 | 0.00 | 0.00 | 0.70 | ||
| TSP (mg/m³) | 5.44 | 2.45 | 6.25 | 1.39 | 9.03 | ||
| C | 10 | Temperature (°C) | 27.8 | 1.0 | 27.7 | 22.8 | 33.1 |
| Relative humidity (%) | 42.5 | 4.0 | 44.0 | 13.0 | 54.0 | ||
| Envelope protein gene quantification - (genomic units/TSP) | 38.70 | 29.95 | 8.25 | 4.30 | 218.15 | ||
| Nucleocapsid protein gene quantification - (genomic units/TSP) | 26.43 | 0.83 | 25.84 | 25.38 | 28.09 | ||
| Total MPs (n/m³) | 14.03 | 1.57 | 15.05 | 6.71 | 20.37 | ||
| MPs fibres (n/m³) | 13.89 | 1.50 | 14.81 | 7.18 | 19.91 | ||
| MPs particles (n/m³) | 0.09 | 0.05 | 0.00 | 0.00 | 0.46 | ||
| TSP (mg/m³) | 6.55 | 2.93 | 6.02 | 4.40 | 10.65 | ||
The envelope concentration (genomic units/TSP) ranged from 9.24 to 19.28 at site A, 4.99 to 38.52 at site B, and 4.30 to 218.15 at site C. The nucleocapsid concentration (genomic units/TSP) ranged from 16.50 to 156.64 at site B and 25.38 to 28.09 at site C. The envelope and nucleocapsid concentrations were not significantly different according to the sites (p > 0.05) (Fig. 2 ).
Fig. 2.
Envelope and nucleocapsid protein concentrations according to the sites.
The number of MPs fibres compared to the MP particles found was significantly higher at the three sites (p < 0.001). On site A, fibres ranged from 9.03 to 24.02 units/m³, while particles ranged from 0 to 0.93 units/m³. On Site B, the fibres ranged from 8.80 to 22.68 units/m³, while the particles ranged from 0 to 0.70 units/m³. On Site C, the fibres ranged from 7.18 to 19.91 units/m³, while the particles ranged from 0 to 0.46 units/m³. Regarding the size of the obtained MPs, fibres length ranged from 50.01 to 1579.43 μm (mean value = 162.01 (±1.57)) and particles diameter ranged from 50.12 to 877.09 μm (mean value = 110.23 (±4.20)).
The amounts of MPs (particles and fibres) and TSP were not significantly different among the three sampling sites (p > 0.05).
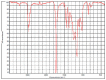
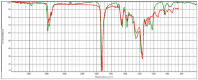
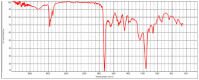
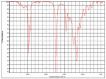
Polyester was the most frequent polymer (80.4%), followed by poly(ethyl acrylate) (2.8%), poly(butyl acrylate:acrylic acid) (2.8%), poly(amide-6,6:amide-6) (2.8%), poly(acrylonitrile:vinylidene chloride) (2.8%), polyamide resin (2.8%), polyether urethane (2.8%), poly(1,4-butylene terephthalate) (2.8%). There was no statistical difference between the types of polymers found in the three sites. When compared to the reference spectral library of plastic particles, the detected MPs particles/fibres had signs of weathering. Microphotographs and the FTIR spectra of the main types of microplastics found in the TSP filters are presented in Table 2 . Figure data on all the analysed filters are presented in the supplementary information, Table S4.
Table 2.
Microphotographs and FTIR spectra of the main types of microplastics.
| Sample | Image | Spectrum | Reference spectrum | Spectrum overlay | Chemical characterization |
|---|---|---|---|---|---|
| 02covid_part1 |  |
 |
 |
 |
Poly(ethyl acrylate) HR Hummel Polymer and Additives (match: 83%) |
| hv04_part1_ATR |  |
 |
 |
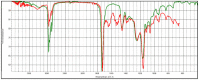 |
Poly(butyl acrylate:acrylic acid), HR Hummel Polymer and Additives (match: 83%) |
| hv13covid_1 |  |
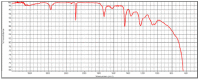 |
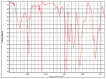 |
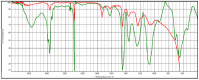 |
Poly(acrylonitrile:vinylidene chloride) HR Hummel Polymer and Additives (71%) |
| hvcovid26_1 |  |
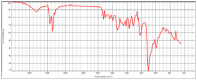 |
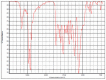 |
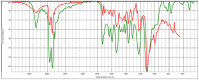 |
Polyether urethane HR Hummel Polymer and Additives (71%) |
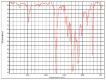
| hv02cov_07 |  |
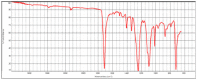 |
 |
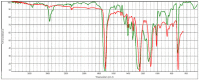 |
Polyester, terephthalic HR Hummel Polymer and Additives (match: 72%) |
MPs particles were detected in the three procedural blanks (n = 3), while fibres were observed in all of them (n = 11). Their respective sizes are shown in Supplemental Material Table S5. It was not possible to identify the chemical composition of the fibres found in the procedural blanks due to the low spectral match with the reference values.
The daily maximum air temperature oscillated from 17.1 °C to 39.0 °C in this period, whereas the minimum relative humidity ranged from 13% to 76%.
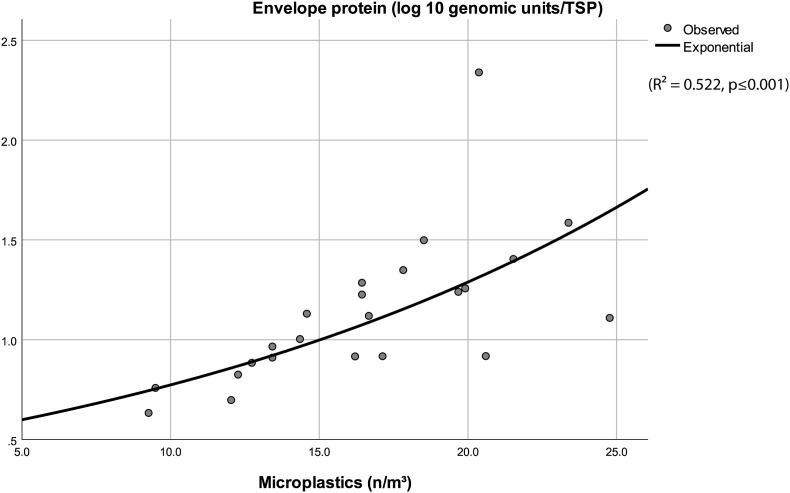
The exponential regression results demonstrated that the number of MPs (fibres + particles) can statistically significantly predict the SARS-CoV-2 envelope concentration (genomic units per TSP) (R2 = 0.522, p ≤ 0.001) (Fig. 3 ). The regression equation is presented in the supplementary material (Equation S(6)). However, the same result was not observed with the nucleocapsid concentration or considering the TSP and genomic particles.
Fig. 3.
Correlation curve of the envelope protein and total microplastics (R2 = 0.522, ***p < 0.001).
The GzLM analysis showed that the total amount of MPs was positively associated with the amount of the SARS-CoV-2 envelope and negatively associated with weather variables (temperature and relative humidity) (Table 3 ).
Table 3.
Coefficients estimated with the generalized linear model (GzLM). β = beta coefficient, S.E. = standard error.
| Predictor | Β | S.E. | p-value |
|---|---|---|---|
| (Intercept) | 8.21 | 1003 | <0.001 |
| Total MPs | 0.067 | 0.015 | <0.001 |
| Temperature | −0.172 | 0.021 | <0.001 |
| Humidity | −0.037 | 0.006 | <0.001 |
Dependent variable: envelope protein (genomic units per TSP).
4. Discussion
This is the first evidence showing that the amount of SARS-CoV-2 is significantly correlated with the number of MPs fibres in the air. Previous evidence indicates that plastic surfaces are carriers of the virus and promote prolonged survival when compared to other materials (van Doremalen et al., 2020). Our data suggest that MPs may also be carriers of SARS-CoV-2 in the air.
The issue of the airborne particulate matter-associated microbiome, particularly viruses, in urban environments remains largely underinvestigated (Jiang, 2015). The capability of SARS-CoV-2 to bind particulate matter, including MPs, could occur through the mechanism of adsorption (Qu et al., 2020). The large surface area of fibres would facilitate their role as carriers. In addition, Corpet (2021) proposed the idea that plastics, by being waterproof substances, prolong virus survival by avoiding more rapid drying. Consistent with this idea, the protein coating that MPs acquire during environmental exposure, curiously known as eco-corona, could indeed prolong SARS-CoV-2 survival and facilitate MPs internalization to cells. (Ramsperger et al., 2020; Pathan et al., 2020).
Due to the FTIR limit of detection, we found and characterized more fibres than particles in air samples. We cannot, however, exclude that MP particles are present in the urban air, as previously suggested by Wright et al. (2020). The most frequently identified polymer in our samples was polyester, which is the main fabric for medical aprons. The high flux of the hospital staff wearing medical aprons on the sampling sites could therefore explain our results. The inhalation of long MP fibres would result in their entrapment in the upper respiratory tract, which is also one of the entry sites of the coronavirus. Further worrying is the fact that the weathering of MPs facilitates their internalization into cells (Ramsperger et al., 2020).
We found that lower temperature and humidity were associated with an increase in the presence of SARS-CoV-2 envelope RNA. These data are in accordance with previous epidemiological data. Coccia (2021) reported that the number of infected people was higher in Italian cities with 100 days exceeding the limits set for PM10 or ozone, having a low wind speed and lower temperature. In addition, Ma et al. (2020) demonstrated a negative association between COVID-19 mortality and temperature.
This study has some limitations that should not be ignored. It is possible that the apparent high ratio of positive samples was due to the fact that samples were collected surrounding large medical facilities treating COVID-19 patients. Thus, our results cannot be extrapolated to other sites. We did not assess viral viability or infectivity since our filter material inactivated the virus rapidly (Pan et al., 2019). Other studies have shown that the virus is rapidly inactivated under environmental conditions such as exposure to sunlight and high temperature (Carraturo et al., 2020; Biryukov et al., 2020) however, in plastic particles, the virus can remain viable for 72 h. We found that the concentration of the nucleocapsid was lower than that of the envelope. Passos et al. (2021) also found few positives when testing the samples by only the nucleocapsid. Setti et al. (2020) obtained many more positives for SARS-CoV-2 envelope RNA than for the other regions tested. The ranges in envelope and nucleocapsid concentrations in the different sampling sites may be related to site characteristics and environmental factors affecting degradation in the target genetic material.
The detection of SARS-CoV-2 genetic material in the air seems to vary according to several conditions. Whereas Setti et al. (2020) found viral particles in an outdoor industrial area in Bergamo, Linillos-Pradillo et al. (2021) and Chirizzi et al. (2021) could not find genomic particles in Madrid and parts of Italy. On the other hand, we and others (Passos et al., 2021) found viral particles in hospital surroundings, whereas Piana et al. (2021) only found genomic particles surrounding infected patients but not in other parts of the hospital. Influencing factors include weather, levels of air pollution, social interaction, restriction of economic activities, and indoor ventilation systems. It must be stressed that in Brazil, due to governmental denialism, strict social isolation and severe restrictions in economic activities never occurred, which may have influenced our and Passos et al.’s (2021) results, both in Brazilian large cities. Whether in situations of a high number of infected individuals with social circulation present, airborne transmission is more likely to occur remains to be determined.
5. Conclusion
In summary, we showed that the presence of SARS-CoV-2 RNA surrounding a large medical centre in Brazil was associated with the number of MP fibres in the air, high temperature, and humidity. We, therefore, suggest that MP's may be a carrier and increase virus survival in the air, thereby facilitating the entry of the virus into the human body. We should still expect substantial increases in environmental MPs due to the widespread use of disposables (including protection devices) during the pandemic. We believe our findings open venues for further research, also in other environments than medical facilities. It seems that the COVID-19 pandemic and MPs are complementary mirrors of the Anthropocene period (O'Callaghan-Gordo and Antó, 2020; Enrique De-la-Torre et al., 2021).
Data availability
For primer sequences and standard curve data, see the Supplemental Material. All other data or materials can be obtained from the corresponding author upon request.
Credit author statement
Luís Fernando Amato-Lourenço – Conceptualization, Data curation, Writing- Reviewing and Editing, Statistics. Natália de Souza Xavier Costa- Methodology, Data curation, Writing – original draft preparation. Kátia Cristina Dantas - Methodology, Data curation, Writing – original draft preparation. Luciana dos Santos Galvão – Methodology, Investigation. Fernando Negri Moralles – Methodology, Investigation. Suzette Cleuza Ferreira Spina Lombardi - Methodology. Alfredo Mendroni Júnior - Methodology. José Angelo Lauletta Lindoso – Methodology, Writing – original draft preparation. Rômulo Augusto Ando – Methodology, Investigation. Felipe Gallego Lima - Methodology. Regiani Carvalho-Oliveira - Methodology, Data curation, Writing – original draft preparation. Thais Mauad – Supervision, Data curation, Writing- Reviewing and Editing
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was financially supported by the São Paulo Research Foundation (FAPESP- project number: 19/03397-5 and FMUSP-HC: Propostas nº 01/2020 - Pesquisas em Covid-19 Sistema FMUSP-HC). We would like to thank the Emergency Management Centre of Sao Paulo Municipality (CGE) for making weather data available, Dr. E. Durigon and Dr. D. Durigon (Institute of Biomedical Sciences - University of Sao Paulo), Brazil for kindly providing inactivated SARS-CoV-2 to this study and Dr. Walcy Rosolia Teodoro (Rheumatology Division of the Hospital das Clínicas HCFMUSP) for making the fluorescence microscope available.
Footnotes
This paper has been recommended for acceptance by Da Chen.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envpol.2021.118299.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abdeldayem O.M., Dabbish A.M., Habashy M.M., Mostafa M.K., Elhefnawy M., Amin L., Al-Sakkari E.G., Ragab A., Rene E.R. Viral outbreaks detection and surveillance using wastewater-based epidemiology, viral air sampling, and machine learning techniques: a comprehensive review and outlook. 2022 Jan;803:149834. doi: 10.1016/j.scitotenv.2021.149834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhbarizadeh R., Dobaradaran S., Nabipour I., Tangestani M., Abedi D., Javanfekr F., Jeddi F., Zendehboodi A. Abandoned Covid-19 personal protective equipment along the Bushehr shores, the Persian Gulf: an emerging source of secondary microplastics in coastlines. Mar. Pollut. Bull. 2021 Jul;168:112386. doi: 10.1016/j.marpolbul.2021.112386. Epub 2021 Apr 19. PMID: 33901902; PMCID: PMC8055201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhbarizadeh R., Dobaradaran S., Amouei Torkmahalleh M., Saeedi R., Aibaghi R., Faraji Ghasemi F. Suspended fine particulate matter (PM2.5), microplastics (MPs), and polycyclic aromatic hydrocarbons (PAHs) in air: their possible relationships and health implications. Environ. Res. 2021 Jan;192:110339. doi: 10.1016/j.envres.2020.110339. Epub 2020 Oct 14. PMID: 33068583. [DOI] [PubMed] [Google Scholar]
- Allen S., Allen D., Phoenix V.R., et al. Atmospheric transport and deposition of microplastics in a remote mountain catchment. Nat. Geosci. 2019;12:339–344. doi: 10.1038/s41561-019-0335-5. [DOI] [Google Scholar]
- Amato-Lourenço L.F., Dos Santos Galvão L., de Weger L.A., Hiemstra P.S., Vijver M.G., Mauad T. An emerging class of air pollutants: potential effects of microplastics to respiratory human health? Sci. Total Environ. 2020 Dec 20;749:141676. doi: 10.1016/j.scitotenv.2020.141676. Epub 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biryukov J., Boydston J.A., Dunning R.A., Yeager J.J., Wood S., Reese A.L., Ferris A., Miller D., Weaver W., Zeitouni N.E., Phillips A., Freeburger D., Hooper I., Ratnesar-Shumate S., Yolitz J., Krause M., Williams G., Dawson D.G., Herzog A., Dabisch P., Wahl V., Hevey M.C., Altamura L.A. Increasing temperature and relative humidity accelerates inactivation of SARS-CoV-2 on surfaces. mSphere. 2020 Jul 1;5(4) doi: 10.1128/mSphere.00441-20. e00441-20. PMID: 32611701; PMCID: PMC7333574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraturo F., Del Giudice C., Morelli M., et al. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020;265(Pt B):115010. doi: 10.1016/j.envpol.2020.115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC, Centers for Disease Control and Prevention . 2021. Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Primers and Probes.https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html Available from: Accessed on March 2021. [Google Scholar]
- CET, Companhia de Engenharia de Tráfego . 2020. Mobilidade no Sistema Viário Principal Volumes e Velocidades 2019: Relatórios Corporativos, Desempenho do Sistema Viário.http://www.cetsp.com.br/sobre-a-cet/relatorios-corporativos.aspx Available from: Accessed on March 2021. [Google Scholar]
- CETESB, Sao Paulo State Environmental Agency . 2020. Qualidade do ar no Estado de São Paulo 2019.https://cetesb.sp.gov.br/ar/publicacoes-relatorios/ Available from: [Google Scholar]
- Chirizzi D., Conte M., Feltracco M., Dinoi A., Gregoris E., Barbaro E., La Bella G., Ciccarese G., La Salandra G., Gambaro A., Contini D. SARS-CoV-2 concentrations and virus-laden aerosol size distributions in outdoor air in north and south of Italy. Environ. Int. 2021 Jan;146:106255. doi: 10.1016/j.envint.2020.106255. Epub 2020 Nov 12. PMID: 33221596; PMCID: PMC7659514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição MSC, Saldiva PHN, Singer JM. GLM and GAM model for analyzing the association between atmospheric pollution and morbidity-mortality markers: an introduction based on data from the city of São Paulo. Rev. bras. epidemiol. 2001;4(3) doi: 10.1590/S1415-790X2001000300007. [DOI] [Google Scholar]
- Corman Victor M., Olfert Landt, Marco Kaiser, Richard Molenkamp, Adam Meijer, Chu Daniel K.W., Tobias Bleicker, Sebastian Brünink, Schneider Julia, Schmidt Marie Luisa, Mulders Daphne G.J.C., Haagmans Bart L., van der Veer Bas, van den Brink Sharon. Lisa Wijsman, Gabriel Goderski, Jean-Louis Romette, Ellis Joanna, Maria Zambon, Malik Peiris, Herman Goossens, Chantal Reusken, Koopmans Marion P.G., Christian Drosten. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet D.E. Why does SARS-CoV-2 survive longer on plastic than on paper? Med. Hypotheses. 2021;146:110429. doi: 10.1016/j.mehy.2020.110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia M. How do low wind speeds and high levels of air pollution support the spread of COVID-19? Atmos Pollut Res. 2021 Jan;12(1):437–445. doi: 10.1016/j.apr.2020.10.002. Epub 2020 Oct 7. PMID: 33046960; PMCID: PMC7541047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CVE . 2021. Centro de Vigilância Epidemiológica "Prof. Alexandre Vranjac". Situação Epidemiológica: Boletins Diário COVID-19. São Paulo, SP.https://www.saude.sp.gov.br/cve-centro-de-vigilancia-epidemiologica-prof.-alexandre-vranjac/areas-de-vigilancia/doencas-de-transmissao-respiratoria/coronavirus-covid-19/situacao-epidemiologica Available from: Accessed on: March 2021. [Google Scholar]
- Dris R., Gasperi J., Rocher V., Saad M., Renault N., Tassin B. Microplastic contamination in an urban area: a case study in Greater Paris. Environ. Chem. 2015;12(5):592. [Google Scholar]
- De-la-Torre G.E., Dioses-Salinas D.C., Pizarro-Ortega C.I., Santillán L. New plastic formations in the Anthropocene. Sci. Total Environ. 2021 Feb 1;754:142216. doi: 10.1016/j.scitotenv.2020.142216. Epub 2020 Sep 6. PMID: 33254855. [DOI] [PubMed] [Google Scholar]
- Erni-Cassola G., Gibson M.I., Thompson R.C., Christie-Oleza J.A. Lost, but found with nile red: a novel method for detecting and quantifying small microplastics (1 mm to 20 μm) in environmental samples. Environ. Sci. Technol. 2017 Dec 5;51(23):13641–13648. doi: 10.1021/acs.est.7b04512. Epub 2017 Nov 20. PMID: 29112813. [DOI] [PubMed] [Google Scholar]
- Fiocruz Fundação Oswaldo Cruz. Rio de Janeiro; 2021. MonitoraCOVID-19. Painéis Estados.https://bigdata-covid19.icict.fiocruz.br/ Available from: [Google Scholar]
- Foulon V., Le Roux F., Lambert C., Huvet A., Soudant P., Paul-Pont I. Colonization of polystyrene microparticles by Vibrio crassostreae: light and electron microscopic investigation. Environ. Sci. Technol. 2016;50(20):10988–10996. doi: 10.1021/acs.est.6b02720. [DOI] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020 Jan 24;12(2):135. doi: 10.3390/v12020135. PMID: 31991541; PMCID: PMC7077245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann N.B., Rist S., Bodin J., Jensen L.H., Schmidt S.N., Mayer P., Meibom A., Baun A. Microplastics as vectors for environmental contaminants: exploring sorption, desorption, and transfer to biota. Integrated Environ. Assess. Manag. 2017 May;13(3):488–493. doi: 10.1002/ieam.1904. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Ruz V., Gutow L., Thompson R.C., Thiel M. Microplastics in the marine environment: a review of the methods used for identification and quantification. Environ. Sci. Technol. 2012 Mar 20;46(6):3060–3075. doi: 10.1021/es2031505. Epub 2012 Mar 2. PMID: 22321064. [DOI] [PubMed] [Google Scholar]
- Jiang W, et al. Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat. Protoc. 2015;10(5):768–779. doi: 10.1038/nprot.2015.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levermore J.M., Smith T.E.L., Kelly F.J., Wright S.L. Detection of microplastics in ambient particulate matter using Raman spectral imaging and chemometric analysis. Anal. Chem. 2020 Jul 7;92(13):8732–8740. doi: 10.1021/acs.analchem.9b05445. Epub 2020 Jun 22. PMID: 32568507. [DOI] [PubMed] [Google Scholar]
- Linillos-Pradillo B., Rancan L., Ramiro E.D., Vara E., Artíñano B., Arias J. Determination of SARS-CoV-2 RNA in different particulate matter size fractions of outdoor air samples in Madrid during the lockdown. Environ. Res. 2021 Apr;195:110863. doi: 10.1016/j.envres.2021.110863. Epub 2021 Feb 17. PMID: 33609549; PMCID: PMC7888991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Wang X., Fang T., Xu P., Zhu L., Li D. Source and potential risk assessment of suspended atmospheric microplastics in. Shanghai. Sci. Total Environ. 2019;675:462–471. doi: 10.1016/j.scitotenv.2019.04.110. Epub 2019 Apr 9), 207. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Xu K., Ho K.F., Kan H., Fu Q., Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020 Jun;582(7813):557–560. doi: 10.1038/s41586-020-2271-3. Epub 2020 Apr 27. PMID: 32340022. [DOI] [PubMed] [Google Scholar]
- Ma Y., Zhao Y., Liu J., He X., Wang B., Fu S., Yan J., Niu J., Zhou J., Luo B. Effects of temperature variation and humidity on the death of COVID-19 in Wuhan, China. Sci. Total Environ. 2020 Jul 1;724:138226. doi: 10.1016/j.scitotenv.2020.138226. Epub 2020 Mar 26. PMID: 32408453; PMCID: PMC7142681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammo F.K., Amoah I.D., Gani K.M., Pillay L., Ratha S.K., Bux F., Kumari S. Microplastics in the environment: interactions with microbes and chemical contaminants. Sci. Total Environ. 2020 Nov 15;743:140518. doi: 10.1016/j.scitotenv.2020.140518. Epub 2020 Jul 2. PMID: 32653705. [DOI] [PubMed] [Google Scholar]
- Mccullagh P. Generalized linear models. Europ. J. Oper. Res. 1984;16(3):285–292. doi: 10.1016/0377-2217(84)90282-0. [DOI] [Google Scholar]
- O'Callaghan-Gordo C., Antó J.M. COVID-19: the disease of the anthropocene. Environ. Res. 2020 Aug;187:109683. doi: 10.1016/j.envres.2020.109683. Epub 2020 May 15. PMID: 32425223; PMCID: PMC7227607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Lednicky J.A., Wu C.Y. Collection, particle sizing and detection of airborne viruses. J. Appl. Microbiol. 2019 Dec;127(6):1596–1611. doi: 10.1111/jam.14278. Epub 2019 Jun 26. PMID: 30974505; PMCID: PMC7167052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan S.I., Arfaioli P., Bardelli T., Ceccherini M.T., Nannipieri P., Pietramellara G. Soil pollution from micro- and nanoplastic debris: a hidden and unknown biohazard. Sustainability. 2020;12(18):7255. doi: 10.3390/su12187255. [DOI] [Google Scholar]
- Passos R.G., Silveira M.B., Abrahão J.S. Exploratory assessment of the occurrence of SARS-CoV-2 in aerosols in hospital facilities and public spaces of a metropolitan center in Brazil. Environ. Res. 2021 Apr;195:110808. doi: 10.1016/j.envres.2021.110808. Epub 2021 Jan 26. PMID: 33513382; PMCID: PMC7835608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piana A., Colucci M.E., Valeriani F., Marcolongo A., Sotgiu G., Pasquarella C., Margarucci L.M., Petrucca A., Gianfranceschi G., Babudieri S., Vitali P., D'Ermo G., Bizzarro A., De Maio F., Vitali M., Azara A., Romano F., Simmaco M., Romano Spica V. Monitoring COVID-19 transmission risks by quantitative real-time PCR tracing of droplets in hospital and living environments. mSphere. 2021 Jan 6;6(1) doi: 10.1128/mSphere.01070-20. e01070-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu G., Li X., Hu L., Jiang G. An imperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID-19) Environ. Sci. Technol. 2020;54(7):3730–3732. doi: 10.1021/acs.est.0c01102. [DOI] [PubMed] [Google Scholar]
- Ramsperger A.F.R.M., Narayana V.K.B., Gross W., Mohanraj J., Thelakkat M., Greiner A., Schmalz H., Kress H., Laforsch C. Environmental exposure enhances the internalization of microplastic particles into cells. Sci. Adv. 2020 Dec 9;6(50) doi: 10.1126/sciadv.abd1211. PMID: 33298447; PMCID: PMC7725476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman C.M., Brookson C., Bikker J., Djuric N., Earn A., Bucci K., Athey S., Huntington A., McIlwraith H., Munno K., De Frond H., Kolomijeca A., Erdle L., Grbic J., Bayoumi M., Borrelle S.B., Wu T., Santoro S., Werbowski L.M., Zhu X., Giles R.K., Hamilton B.M., Thaysen C., Kaura A., Klasios N., Ead L., Kim J., Sherlock C., Ho A., Hung C. Rethinking microplastics as a diverse contaminant suite. Environ. Toxicol. Chem. 2019;38(4):703–711. doi: 10.1002/etc.4371. [DOI] [PubMed] [Google Scholar]
- Sajadi M.M., Habibzadeh P., Vintzileos A., Shokouhi S., Miralles-Wilhelm F., Amoroso A. Temperature, humidity, and latitude analysis to estimate potential spread and seasonality of coronavirus disease 2019 (COVID-19) JAMA Netw Open. 2020 Jun 1;3(6) doi: 10.1001/jamanetworkopen.2020.11834. PMID: 32525550; PMCID: PMC7290414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setti L., Passarini F., De Gennaro G., Barbieri P., Perrone M.G., Borelli M., Palmisani J., Di Gilio A., Torboli V., Fontana F., Clemente L., Pallavicini A., Ruscio M., Piscitelli P., Miani A. SARS-Cov-2RNA found on particulate matter of Bergamo in Northern Italy: first evidence. Environ. Res. 2020 Sep;188:109754. doi: 10.1016/j.envres.2020.109754. Epub 2020 May 30. PMID: 32526492; PMCID: PMC7260575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEADE . 2019. SEADE. Indicadores do Estado de São Paulo.http://www.seade.gov.br/ Available online at: (2019) [Google Scholar]
- Sharma A., Kumar P. Quantification of air pollution exposure to in-pram babies and mitigation strategies. Environ. Int. 2020 Jun;139:105671. doi: 10.1016/j.envint.2020.105671. [DOI] [PubMed] [Google Scholar]
- Tamminga M., Hengstmann E., Fischer E.K. Nile red staining as a subsidiary method for microplastic quantification: a comparison of three solvents and factors influencing application reliability. SDRP J. Earth Sci. Environ. Stud. 2017;2 doi: 10.15436/JESES.2.2.1. [DOI] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020 Apr 16;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, World Health Organization . 2020. Coronavirus Disease (COVID-19): How Is it Transmitted? World Health Organization.https://www.who.int/news-room/q-a-detail/coronavirus-disease-covid-19-how-is-it-transmitted Available from: [Google Scholar]
- WHO, World Health Organization . World Health Organization; 2005. WHO Air Quality Guidelines for Particulate Matter, Ozone, Nitrogen Dioxide and Sulfur Dioxide: Global Update 2005. [DOI] [PubMed] [Google Scholar]
- Wright S.L., Ulke J., Font A., Chan K.L.A., Kelly F.J. Atmospheric microplastic deposition in an urban environment and an evaluation of transport. Environ. Int. 2020 Mar;136:105411. doi: 10.1016/j.envint.2019.105411. Epub 2019 Dec 27. PMID: 31889555; PMCID: PMC7013824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. Epub 2020 Feb 3. Erratum in: Nature. 2020 Apr;580(7803):E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Xie J., Huang F., Cao L. Association between short-term exposure to air pollution and COVID-19 infection: evidence from China. Sci. Total Environ. 2020 Jul 20;727:138704. doi: 10.1016/j.scitotenv.2020.138704. Epub 2020 Apr 15. PMID: 32315904; PMCID: PMC7159846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y.Y., Uspal W.E., Wei T. Airborne transmission of COVID-19: aerosol dispersion, lung deposition, and virus-receptor interactions. ACS Nano. 2020 Nov 25 doi: 10.1021/acsnano.0c08484. acsnano.0c08484. Epub ahead of print. PMID: 33236896; PMCID: PMC7724984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For primer sequences and standard curve data, see the Supplemental Material. All other data or materials can be obtained from the corresponding author upon request.