Abstract
Determination of the MIC, based on the activities of antibiotics against planktonic bacteria, is the standard assay for antibiotic susceptibility testing. Adherent bacterial populations (biofilms) present with an innate lack of antibiotic susceptibility not seen in the same bacteria grown as planktonic populations. The Calgary Biofilm Device (CBD) is described as a new technology for the rapid and reproducible assay of biofilm susceptibilities to antibiotics. The CBD produces 96 equivalent biofilms for the assay of antibiotic susceptibilities by the standard 96-well technology. Biofilm formation was followed by quantitative microbiology and scanning electron microscopy. Susceptibility to a standard group of antibiotics was determined for National Committee for Clinical Laboratory Standards (NCCLS) reference strains: Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 29213. Growth curves demonstrated that biofilms of a predetermined size could be formed on the CBD at specific time points and, furthermore, that no significant difference (P > 0.1) was seen between biofilms formed on each of the 96 pegs. The antibiotic susceptibilities for planktonic populations obtained by the NCCLS method or from the CBD were similar. Minimal biofilm eradication concentrations, derived by using the CBD, demonstrated that for biofilms of the same organisms, 100 to 1,000 times the concentration of a certain antibiotic were often required for the antibiotic to be effective, while other antibiotics were found to be effective at the MICs. The CBD offers a new technology for the rational selection of antibiotics effective against microbial biofilms and for the screening of new effective antibiotic compounds.
The MIC has long been the standard for antibiotic susceptibility testing. The MIC measures the actions of antibiotics against planktonic organisms and serves as an important reference in the treatment of many acute infections. Application of MICs in the treatment of chronic or device-related infections involving bacterial biofilms is often ineffective (4). Bacterial biofilms, which are microcolonies encased in extracellular polysaccharide material (slime), result from the adherence of bacteria to surfaces both in vitro (2–4, 20) and in vivo (1a, 9, 11, 13). Numerous studies have now demonstrated that biofilm-grown microorganisms have an inherent lack of susceptibility to antibiotics, whereas planktonic cultures of this same organism do not (1a, 4, 8, 10, 12, 14–17, 19–22). This resistance is lost once the biofilm is reverted to conditions that permit planktonic growth (4). The innate tolerance of microbial biofilms to antibiotic therapy has led to problems in their eradication (19) and in the management of patients with device-related infections (18). Biofilms may also interfere with the immune clearance of infectious agents (7). This difference in antibiotic susceptibility between planktonic and biofilm populations of the same organism may result from differences in the diffusion of antibiotics (17) or much more complex changes in the microbial physiology of the biofilm (4, 5, 6, 20). Several different techniques have been used to study biofilm populations (2, 3, 16). The modified Robbin’s device (MRD) has provided important information regarding biofilm physiology and antibiotic susceptibility (8, 12, 14). Morck et al. (12) demonstrated an important correlation between the antibiotic susceptibilities of biofilms in vitro using the MRD and the efficacy of antibiotic treatment in vivo. While the MRD has proven to be an effective model of biofilm formation, it is not suited for rapid antibiotic susceptibility testing in a clinical laboratory setting. In this paper we describe a new technology, the Calgary Biofilm Device (CBD), for the rapid and reproducible screening of the antibiotic susceptibilities of biofilms.
MATERIALS AND METHODS
Organisms.
Standard reference strains Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 29213, proposed for quality control use by the National Committee for Clinical Laboratory Standards (NCCLS), were used in this study. The isolates were grown in Trypticase soy broth (TSB; BBL from Fisher Canada), which was also used in the reaction vessel to initiate biofilm formation. Bacterial counts were done on Trypticase-soy agar (TSA; BDH). Antibiotic susceptibility screening and recovery of viable biofilm organisms were carried out in cation-adjusted Mueller-Hinton broth (CAMHB; BDH).
CBD.
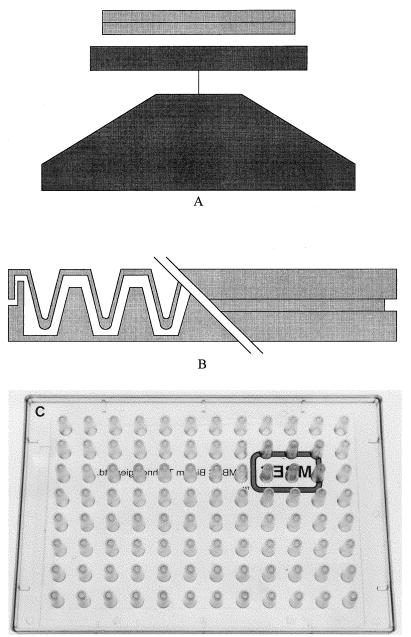
The CBD (Fig. 1) consists of a two-part reaction vessel. The top component forms a lid that has 96 pegs and that is sealed on the top so that the pegs can be removed without opening the vessel and allowing possible contamination. The pegs are designed to sit in the channels of the bottom component of the reaction vessel and to fit into the wells of a standard 96-well plate. The bottom of the vessel serves to channel the flow of medium across the pegs to create consistent shear force across all pegs, resulting in the formation of equivalent biofilms at each peg site. The CBD is commercially available as the MBEC Assay System, with both the device and method of use the property of and available through MBEC Biofilms Technology Ltd., Calgary Alberta, Canada.
FIG. 1.
CBD for biofilm antibiotic susceptibility testing. (A) Tilt table used to create the shear force required for biofilm formation. The table can be placed in incubators to control temperature or oxygen tension. (B) Cutaway view of the device showing the pins sitting in the channels of the incubation tray. (C) Top plate of the device.
Biofilm formation.
The inoculum was established by the direct colony suspension method from 18- to 24-h TSA plates, standardized with McFarland standards, and validated by determination of viable counts on TSA plates. Biofilm formation was carried out at 35°C and 95% relative humidity on a rocking table (Red Rocker model; Hoefer Instrument Co.) such that fluid flowed along the channels of the CBD, generating the required shear force across all pegs. Biofilm formation was determined by obtaining viable counts on TSA plates following disruption of the biofilm by sonication (12). The bacteria could be removed either from individual pegs, broken from the lid, or from all pegs at one time, by sonication for 5 min on high with an Aquasonic (model 250HT; VWR Scientific) sonicator.
Biofilm growth curves.
Biofilms of each isolate were initiated as described above. At specific time points two pegs from different points on the lid were removed, placed in microcentrifuge tubes containing 200 μl of TSB, and sonicated as described above. Viable counts were determined on TSA plates. The same procedure was used to control for the numbers of CFU per peg in all antibiotic susceptibility tests prior to exposure to antibiotic.
Antibiotic susceptibility tests. (i) Antibiotic preparation.
Amikacin (ICN), ampicillin (Sigma), aztreonam (ICN), cefazolin (Sigma), cefotaxime (Sigma), ceftazidime (Eli Lilly), ciprofloxacin (Bayer), clindamycin-HCl (Pharmacia & Upjohn), gentamicin sulfate (Sigma), imipenem (Merck-Frost), oxacillin (Sigma), penicillin G (Sigma), piperacillin (Sigma), tobramycin (Sigma), and vancomycin (Sigma) were prepared as stock solutions of 6,200 μg/ml, and the stock solutions were stored at −80°C. Working solutions were prepared in CAMHB at a concentration of 1,024 μg/ml, and from these working solutions serial twofold dilutions were made in CAMHB in the wells of the 96-well plate.
(ii) Biofilm susceptibility testing.
Biofilms were formed on the lid of the CBD as described above and were then transferred to a standard 96-well plate in which dilutions of the specified antibiotics were prepared in CAMHB. Antibiotic plates were incubated overnight at 35°C, after which the lid was removed, rinsed in phosphate-buffered saline, and placed in a second 96-well plate containing CAMHB. The biofilm was removed from the CBD lid by sonication as described above, a new plate cover was added, and the viability of the biofilm was determined after 24 h of incubation at 35°C either by obtaining plate counts or by reading the turbidity at 650 nm in a 96-well plate reader (Molecular Devices from Fisher Canada). The minimal biofilm eradication concentration (MBEC) was defined as the minimal concentration of antibiotic required to eradicate the biofilm.
(iii) MIC assays.
MICs were determined according to the 1997 guidelines of NCCLS. The concentration of antibiotic required to prevent the growth of a planktonic population was also derived from the CBD by measuring the turbidity at 650 nm after incubation of the isolate in antibiotics for 24 h. In this case the MIC (CBD) was defined as the lowest concentration of antibiotic in which a planktonic bacterial population could not be established by shedding of bacteria from the biofilm.
SEM.
Pins were broken from the lid and allowed to air dry overnight. Samples were then fixed with 2.5% glutaraldehyde in phosphate-buffered saline (0.2 M; pH 7.4) and were prepared for scanning electron microscopy (SEM) on a Hitachi model 450 scanning electron microscope as reported previously (12).
Statistics.
The biofilms that formed on each of the pegs were compared by one-way analysis of variance by applying the Bartlett’s test for homogeneity of variances and the Tukey-Kramer multiple-comparisons test.
RESULTS
Biofilm formation on the CBD.
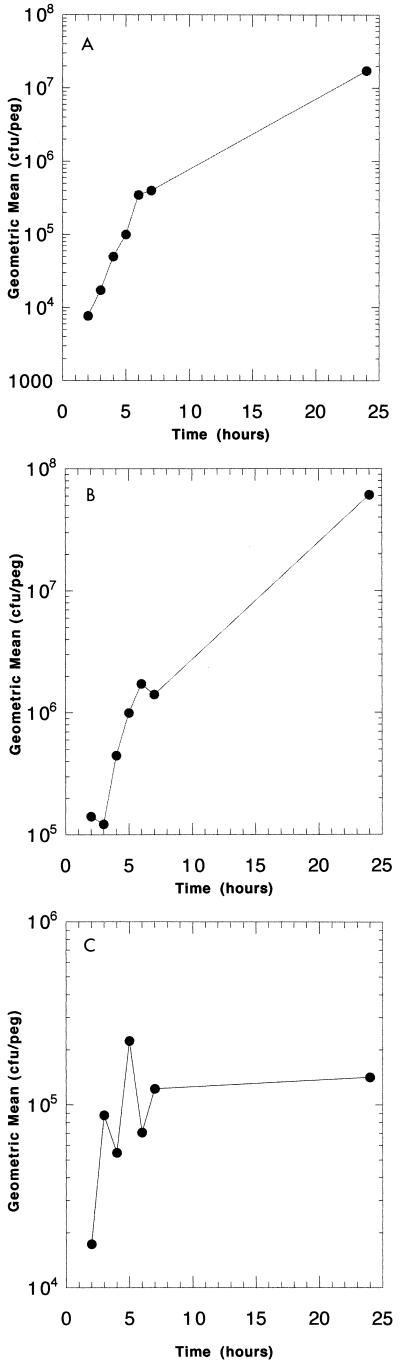
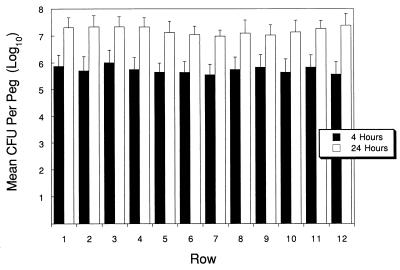
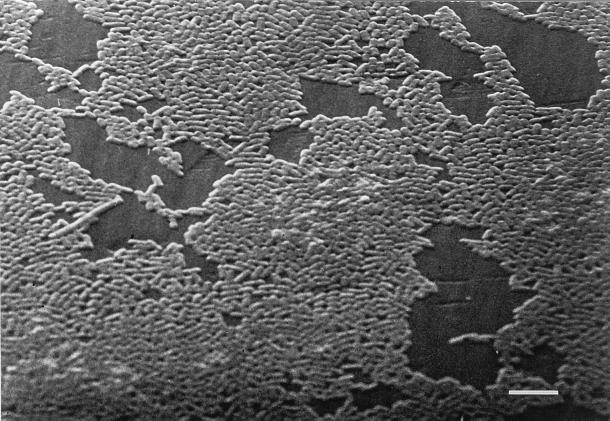
Biofilm growth curves for E. coli, P. aeruginosa, and S. aureus are shown in Fig. 2. Densities of 4 × 105 organisms per peg were reached by E. coli in 6 h, P. aeruginosa in 4 h, and S. aureus in 7 h. E. coli and P. aeruginosa reached final biofilm concentrations of 3 × 107 to 5 × 107 after 24 h, and S. aureus reached a maximal density of 1 × 105 to 2 × 105 CFU/peg. To test the variability of biofilms formed at each peg site, the biofilms were grown to reach a specific density, as predicted by the growth curve. The lid was then sonicated and the contents of each well were plated. No difference was seen between the biofilms that formed on the pegs of each row, as shown for P. aeruginosa in Fig. 3. Table 1 compares the mean, median, standard deviation, lower and upper 95% confidence intervals, and maximum and minimum counts found across the device at 4 and 24 h of growth of P. aeruginosa, and the results validate the equivalence of each of the 96 biofilms produced. The data in Fig. 3 represent data from four separate experiments. SEM of the biofilm formed by E. coli on the CBD (Fig. 4) demonstrates the attached bacteria and the formation of a biofilm.
FIG. 2.
Growth curves of E. coli ATCC 25922 (A), P. aeruginosa ATCC 27853 (B), and S. aureus ATCC 29213 (C) demonstrating that a biofilm of a specific size can be produced over a specific period of growth on the CBD. The number of bacteria per peg was determined by breaking pegs from the lid at appropriate times and determining the bacterial number as described in Materials and Methods.
FIG. 3.
Mean number of CFU of P. aeruginosa per peg on the MBEC device determined by plating bacteria sonicated from each peg position for biofilms grown for 4 or 24 h, as described in Materials and Methods. The values obtained were compared by analysis of variance and Bartlett’s test for homogeneity of variance. No significant difference was found between rows (P = 0.9982).
TABLE 1.
Colonization at each peg site of MBEC assay system for P. aeruginosa grown for 4 or 24 ha
| Parameter | Log10 count/peg
|
|
|---|---|---|
| 4 h | 24 h | |
| Mean | 5.725 | 7.202 |
| Median | 5.778 | 7.204 |
| SD | 0.448 | 0.383 |
| Lower 95% CIb | 5.634 | 7.124 |
| Upper 95% CI | 5.816 | 7.280 |
| Minimum | 5.000 | 6.477 |
| Maximum | 6.602 | 7.954 |
Biofilms of P. aeruginosa grown for 4 or 24 h as described in Materials and Methods were sonicated and plated. The variability between pegs was determined by statistical procedures as described in Materials and Methods.
CI, confidence interval.
FIG. 4.
Scanning electron micrograph of an E. coli biofilm formed on the MBEC device. Pins from the device were broken off and fixed as described in Materials and Methods. Bar, 50 μm.
Antibiotic susceptibility of E. coli ATCC 25922.
E. coli biofilms that were grown for 6 h and that reached counts of 1.5 × 106 CFU/peg were used for susceptibility testing. Two pegs from all plates used in these studies were sonicated, and plate counts were determined to ensure that the appropriate biofilm had been developed during the incubation period. The MICs and MBECs of penicillins (ampicillin and piperacillin), cephalosporins (cefazolin and cefotaxime), aminoglycosides (gentamicin and tobramycin), quinolones (ciprofloxacin), and trimethoprim-sulfamethoxazole for E. coli ATCC 25922 are presented in Table 2. Similar results were obtained in at least two independent studies with each organism. The MICs of ampicillin, piperacillin, cefazolin, ciprofloxacin, and trimethoprim-sulfamethoxazole either obtained by the NCCLS standard method or derived from the CBD were similar and within the NCCLS quality control (QC) range for this organism. The NCCLS method-derived MICs of cefotaxime and tobramycin fell within the NCCLS QC range; however, the MICs derived from the CBD were approximately 10-fold higher for cefotaxime and 4-fold higher for tobramycin. The MIC of gentamicin was 1 dilution above the maximum QC value when the MIC was derived by NCCLS methods and 1 further dilution higher when it was derived from the CBD. The biofilm formed by this E. coli strain proved to be significantly more resistant to antibiotics than a planktonic culture of the same organism. The MBECs were 1,000-fold or more greater than the MICs for ampicillin, ciprofloxacin, cefazolin, cefotaxime, and trimethoprim-sulfamethoxazole. The MBEC of piperacillin fell in the intermediate range of susceptibility for MIC assays. The gentamicin concentration required for biofilm eradication was high, i.e., within resistant levels in MIC assays, but was within ranges achievable in humans. The most active antibiotic against the E. coli biofilm was tobramycin, with an MBEC of 2 μg/ml; the organism would be considered sensitive under NCCLS standards for an MIC assay.
TABLE 2.
Antibiotic susceptibility of E. coli ATCC 25922 as a planktonic population (MIC) and as a biofilm population (MBEC) as derived by the NCCLS assay and an assay with the CBD
| Antibiotic | MIC (μg/ml)a
|
MBEC (μg/ml)
|
||
|---|---|---|---|---|
| NCCLS assaya | Assay with CBDa | A650a | 0 CFU/pegb | |
| Ampicillin | 2 | 4 | >1,024 | >1,024 |
| Piperacillin | 1 | 4 | 32 | 32 |
| Cefazolin | 1 | 4 | >1,024 | >1,024 |
| Cefotaxime | 0.06 | 1 | 256 | 256 |
| Ciprofloxacin | 0.004 | 0.008 | 8 | 8 |
| Gentamicin | 2 | 4 | 16 | 16 |
| Tobramycin | 1 | 4 | 2 | 2 |
| Trimethoprim-sulfa-methoxazole | 0.06/1.14 | 0.12/2.28 | 64/1,216 | 32/608 |
The values were obtained by measuring the turbidity at 650 nm (A650) on a 96-well plate reader.
The values were obtained by determination of plate counts.
Antibiotic susceptibility of P. aeruginosa ATCC 27853.
P. aeruginosa biofilms of 3 × 106 CFU/peg were formed in 4 h of incubation. The MICs and MBECs of a penicillin (piperacillin), a cephalosporin (ceftazidime), a quinalone (ciprofloxacin), aminoglycosides (amikacin, gentamicin, and tobramycin), monobactams (aztreonam), and carbapenems (imipenem), which are typically used against P. aeruginosa, were obtained. The MICs obtained by NCCLS methods fell within the prescribed QC range, as did the MICs derived from the CBD, for all antibiotics except gentamicin, which required 1 higher dilution (Table 3). Again, a marked difference between the MICs and the MBECs of most antibiotics was seen. Aztreonam, ceftazidime, imipenem, and piperacillin showed no activity at concentrations 1,000-fold or more greater than the MIC. The concentration of gentamicin required to eliminate the biofilm was 60-fold greater than the MIC. The other aminoglycosides (tobramycin and amikacin) were effective against the biofilm at concentrations within the susceptible range for the MIC assay. Ciprofloxacin required a concentration of 4 μg/ml to eradicate the biofilm, but this concentration falls outside the susceptible range by MIC standards but still represents obtainable drug levels.
TABLE 3.
Antibiotic susceptibility of P. aeruginosa ATCC 27853 as a planktonic population (MIC) and as a biofilm population (MBEC) as derived by the NCCLS assay and an assay with the CBD
| Antibiotic | MIC (μg/ml)
|
MBEC (μg/ml)
|
||
|---|---|---|---|---|
| NCCLS assaya | Assay with CBDa | A650a | 0 CFU/pegb | |
| Amikacin | 2 | 4 | 16 | 16 |
| Aztreonam | 2 | 4 | >1,024 | >1,024 |
| Ceftazidime | 1 | 2 | >1,024 | >1,024 |
| Ciprofloxacin | 0.25 | 0.25 | 4 | 4 |
| Gentamicin | 2 | 4 | 128 | 128 |
| Imipenem | 1 | 4 | >1,024 | >1,024 |
| Piperacillin | 2 | 16 | >1,024 | >1,024 |
| Tobramycin | 0.5 | 1 | 2 | 2 |
The values were obtained by measuring the turbidity at 650 nm (A650) on a 96-well plate reader.
The values were obtained by determination of plate counts.
Antibiotic susceptibility of S. aureus ATCC 29213.
S. aureus biofilms of 2 × 105 CFU/peg were formed in 6 h of incubation. The susceptibility of this QC isolate to penicillins (penicillin and oxacillin), a cephalosporin (cefazolin), a quinolone (ciprofloxacin), an aminoglycoside (gentamicin), a glycopeptide (vancomycin), and clindamycin was determined. The MICs of all the antibiotics obtained by the NCCLS protocol fell within the QC range, as did the MICs of all antibiotics as derived from the CBD (Table 4). Again, the MICs were not always predictive of the MBECs. The MBECs of penicillin, oxacillin, cefazolin, ciprofloxacin, clindamycin, and vancomycin were 100- to 1,000-fold higher than the MICs for planktonic populations of this organism. Only the aminoglycoside gentamicin had activity against the S. aureus biofilm.
TABLE 4.
Antibiotic susceptibility of S. aureus ATCC 29213 as a planktonic population (MIC) and as a biofilm population (MBEC) derived by the NCCLS assay and an assay with the CBD
| Antibiotic | MIC (μg/ml)
|
MBEC (μg/ml)
|
||
|---|---|---|---|---|
| NCCLS assaya | Assay with CBDa | A650a | 0 CFU/pegb | |
| Cefazolin | 0.5 | 0.5 | >1,024 | >1,024 |
| Ciprofloxacin | 0.25 | 0.5 | 512 | 512 |
| Clindamycin | 0.12 | 0.25 | 128 | 256 |
| Gentamicin | 0.5 | 0.5 | 2 | 2 |
| Oxacillin | 0.12 | 0.25 | >1,024 | >1,024 |
| Penicillin | 1 | 4 | 128 | 128 |
| Vancomycin | 1 | 1 | >1,024 | >1,024 |
The values were obtained by measuring the turbidity at 650 nm (A650) on a 96-well plate reader.
The values were obtained by determination of plate counts.
DISCUSSION
The CBD produced 96 equivalent biofilms, making it the first assay system truly amenable to antibiotic susceptibility testing for adherent bacterial populations. The reproducibilities of the results for biofilms formed on each of the pegs of the CBD demonstrate the equivalence of the biofilms formed at each of the sites for susceptibility testing. In addition, the reproducible growth curves obtained for each isolate demonstrate that biofilms of a predicted size can be formed on each peg of the CBD lid (Fig. 2). It is therefore possible to select a target biofilm size for antibiotic susceptibility testing and to expose the biofilm to multiple antibiotics in a single assay. SEM of the biofilms that formed (Fig. 3) on the CBD demonstrates the typical biofilm appearance, as seen on the modified Robbin’s device (MRD) or on catheter surfaces (12, 13). Therefore, the major advantages offered by the CBD are its multiple equivalent biofilms that can be used for testing and its ease of use. The CBD requires no pumps or tubing, making the process much simpler to set up than the MRD, and eliminates a major source of possible contamination. The availability of multiple testing sites greatly reduced the time required to determine the antibiotic susceptibilities of biofilms from weeks with the MRD to 3 days with the CBD. The CBD is also amenable to automation because it is built on the typical platform for 96-well plates. There were no differences in the MBECs for all three organisms obtained either by reading of the turbidity at 650 nm or by quantitative bacteriology. This eliminates the need in most cases to do quantitative microbiology to obtain MBECs, again contributing to the ease of use of the assay and contributing to its automation potential.
The MICs obtained by NCCLS standard protocols and those obtained with the CBD were similar for almost all antibiotics tested. This is an interesting observation in that the planktonic population tested for antibiotic susceptibility by the assay with the CBD is one that was continually shed from the biofilm, whereas a prescribed inoculum was tested by the NCCLS assay. The data could be interpreted to indicate that the MIC is predictive of antibiotic efficacy against bacteria being seeded from a nidus of infection, such as a biofilm on a catheter or a line. This is consistent with what is often seen in recurrent infections. Antibiotics that are seen to be effective in MIC assays are able to produce symptomatic relief by eliminating the planktonic population; however, because the biofilm is not eliminated by antibiotic treatment, reinfection occurs once the antibiotic is removed.
A clear difference in antibiotic susceptibility was seen between planktonic populations of each of the bacteria tested and the biofilm populations of the same organism. These results obtained with the CBD are in keeping with previously reported results obtained with the MRD and by other methods of biofilm production (8, 12, 14, 22). The data produced by Morck et al. (12), who used the MRD, have recently been compared to data derived from the CBD, with the only difference being that the current technology allows a more detailed analysis of susceptibility because the number of samples that could be handled was greatly increased (1). As a biofilm, each of the isolates had an unique susceptibility to the group of antibiotics tested. E. coli ATCC 25922 was the most sensitive to tobramycin, with the other aminoglycoside tested (gentamicin) and the β-lactam piperacillin showing some activity against E. coli. The P. aeruginosa ATCC 27853 biofilm was the most susceptible to the aminoglycosides tobramycin and amikacin but was not nearly as susceptible to gentamicin. Eradication of the Pseudomonas biofilm could also be obtained with ciprofloxacin at obtainable concentrations. The biofilm of S. aureus ATCC 29213 proved to be very difficult to eradicate, with only gentamicin proving to be effective at achievable drug concentrations.
It is clear that the antibiotic susceptibilities of planktonic populations, as determined by MIC methodologies, are not necessarily applicable to effective treatment of the same organism once a biofilm has been established. One problem faced in selecting alternative antibiotic treatment has been the lack of an easy, reproducible assay which could provide a measure of antibiotic activity against a biofilm. The CBD provides a new technology that can be applied to recalcitrant, recurrent, or device-related infections caused by organisms for which MICs have not provided clinically relevant information. The CBD should also prove to be important in the development of new antibiotics selected for their efficacies against biofilms.
ACKNOWLEDGMENTS
The studies were funded by Natural Science and Engineering Research Council of Canada grants to H. Ceri and M. E. Olson.
REFERENCES
- 1.Ceri, H., et al. Unpublished data.
- 1a.Chaud C, Lueet J E, Rohner P, Herrmann M, Auckenthaler R. Resistance of S. aureus recovered from infected foreign body in-vivo to killing by antimicrobials. J Infect Dis. 1991;163:1369–1373. [PubMed] [Google Scholar]
- 2.Christensen G D, Simpson W A, Bisno A, Beachy E. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37:318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen G D, Simpson W A, Younger J J, Baddour L M, Barrett F F, Melton D M, Beachey E. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 5.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of bacterial biofilms. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 6.Evans D J, Brown M R W, Allison D G, Gilbert P. Susceptibility of bacterial biofilms to tobramycin: role of specific growth rate and phase in the division cycle. J Antimicrob Chemother. 1990;25:585–591. doi: 10.1093/jac/25.4.585. [DOI] [PubMed] [Google Scholar]
- 7.Johnson G M, Lee D A, Regelman W E, Gray E D, Peter G, Quie P G. Interference with granulocyte function by Staphylococcus epidermidis slime. Infect Immun. 1986;54:13–20. doi: 10.1128/iai.54.1.13-20.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumon H, Ono N, Ilda M, Nickel J C. Combination effect of fosfomycin and ofloxacin against Pseudomonas aeruginosa growing in biofilms. Antimicrob Agents Chemother. 1995;39:1038–1044. doi: 10.1128/aac.39.5.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam J, Chan R, Lam K, Costerton J W. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen T, Fiehn N-E. Resistance of Streptococcus sanguis biofilms to antimicrobial agents. APMIS. 1996;104:280–284. [PubMed] [Google Scholar]
- 11.Marrie T J, Costerton J W. Scanning and transmission electron microscopy of in situ bacterial colonization of intravenous and intraatrial catheters. J Clin Microbiol. 1984;19:687–693. doi: 10.1128/jcm.19.5.687-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morck D W, Lam K, McKay S G, Olson M E, Prosser B, Ellis B D, Cleeland R, Costerton J W. Comparative evaluation of fleroxacin, ampicillin, trimethoprim-sulfamethoxazole, and gentamicin as treatments of catheter-associated urinary tract infections in a rabbit model. Int J Antimicrob Agents. 1994;4:S21–S27. doi: 10.1016/0924-8579(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 13.Nickel J C, Gristina A C, Costerton J W. Electron microscopy of infected Foley catheter. Can J Surg. 1985;28:50–54. [PubMed] [Google Scholar]
- 14.Raad I, Darouiche R, Hachem R, Sacilowski M, Bodey G P. Antibiotics and prevention of microbial colonization of catheters. Antimicrob Agents Chemother. 1995;39:2397–2400. doi: 10.1128/aac.39.11.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid G, Sharman S, Advikolanu K, Tieszer C, Martin R, Bruce A W. Effects of ciprofloxacin, norfloxacin, and ofloxacin on in vitro adhesion and survival of Pseudomonas aeruginosa AK1 on urinary catheters. Antomicrob Agents Chemother. 1994;38:1490–1495. doi: 10.1128/aac.38.7.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosser B T, Taylor D, Cix P A, Cluland R. Methods for evaluating of antibiotics on bacterial biofilm. Antimicrob Agents Chemother. 1987;31:1502–1506. doi: 10.1128/aac.31.10.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart P S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warren J W, Mucie H L, Berquist E J, Hoppes J M. Sequelae and management of urinary tract infection in the patient requiring chronic catheterization. J Urol. 1981;125:1–8. doi: 10.1016/s0022-5347(17)54874-0. [DOI] [PubMed] [Google Scholar]
- 19.Widmer A F, Wiestner A, Frei R, Zimmerli W. Killing of nongrowing and adherent Escherichia coli determines drug efficacy in device-related infections. Antimicrob Agents Chemother. 1991;35:741–746. doi: 10.1128/aac.35.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson M. Susceptibility of oral bacterial biofilms to antimicrobial agents. J Med Microbiol. 1996;44:79–87. doi: 10.1099/00222615-44-2-79. [DOI] [PubMed] [Google Scholar]
- 21.Wright T L, Ellen R P, Lacroix J-M, Sinnadurai S, Mittelman M W. Effects of metronidazole on Porphyromonas gingivalis biofilms. J Periodont Res. 1997;32:473–477. doi: 10.1111/j.1600-0765.1997.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 22.Yassien M, Khardori N, Ahmedy A, Toama M. Modulation of biofilms of Pseudomonas aeruginosa by quinolones. Antimicrob Agents Chemother. 1995;38:2262–2268. doi: 10.1128/aac.39.10.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]