Abstract
Most E2F-driven promoters are transiently activated around the G1/S transition. Although the promoter for the c-myb proto-oncogene harbors an E2F element, it is induced early in G1 following entry into the cell cycle. Furthermore, this promoter remains active throughout subsequent cell cycles. Since E2F sites function as repressor elements during G1 (due to the association of pRb with E2F factors), we investigated whether the E2F element in the c-myb promoter is regulated differently than E2F elements in promoters that are repressed during G1. By gel shift analysis, the E2F element from the c-myb promoter was found to form a unique complex, referred to as E2Fmyb-sp, which was not observed with E2F elements from several other promoters. Antibodies to DP-1, E2F1 to -5, p107, or pRb failed to either supershift or block E2Fmyb-sp complex formation. Methylation interference experiments indicate that the DNA contact residues for the E2Fmyb-sp complex are distinct from but overlapping with residues required for the binding of E2F proteins. In addition to the identification of E2Fmyb-sp, we have found that SP-1 binds to the c-myb E2F element. Functional studies revealed that E2Fmyb-sp and/or SP-1 are required to achieve full activation of the c-myb promoter in different cell types and to maintain elevated expression of the c-myb promoter during G1 in NIH 3T3 cells. These studies demonstrate that E2F elements can be regulated differently through the binding of unique sets of proteins.
The E2F family of transcription factors plays a pivotal role in the regulation of cell cycle entry and progression by restricting the expression of genes involved in cell cycle control (cyclins, cyclin-dependent kinases, and cyclin-dependent kinase inhibitors, the retinoblastoma tumor suppressor [pRb], and p107), initiation of replication (Orc1, Cdc6, and Mcms), and DNA synthesis (DNA polymerase I, thymidylate synthase (TS), thymidine kinase [TK], and dihydrofolate reductase [DHFR]) to the point of the cell cycle at which their protein products function (1, 11, 24, 28, 36, 59, 64, 72, 74). In addition, several proto-oncogenes, including c-myb, B-myb, c-myc, and N-myc, have been shown to be regulated by E2F proteins (28, 64).
E2F transcription factors are composed of two structurally related subunits, termed E2F and DP, which form heterodimeric complexes with a high affinity for specific DNA sequences (E2F elements) (5, 9, 23, 30). To date, six E2F (E2F1 through E2F6) and two DP (DP-1 and DP-2) genes have been identified in mammalian cells (12, 28). Alternate forms of the DP-2 protein (also referred to as DP-3) can be produced as the result of alternative splicing and internal translational initiation (49, 52), adding further complexity to the E2F family. E2F and DP proteins contain highly conserved DNA-binding and dimerization domains (64). The carboxyl-terminal regions of E2F1 to -5 contain potent transactivation domains, while no equivalent activity has been identified in E2F6 or in the DP proteins (12, 28, 64).
The activity of E2F factors is regulated in part through differential association with pRb family members, including pRb, p107, and p130 (28, 44, 46). pRb, p107, and p130 bind tightly to the carboxyl-terminal transactivation domains of the E2F partner, and this interaction likely blocks the association between the activation domains and transcriptional coactivators, thus inhibiting transactivation by E2F proteins (18, 22, 51, 58, 69, 70). pRb, p107, and p130, through their association with E2Fs, also exerts dominant negative effects on promoter activity, in part through their concomitant interaction with histone deacetylase molecules (6, 39, 42). The repression function of E2F-pRb complexes is important for cell cycle control in vivo since inactivation of at least one E2F family member, E2F1, leads to increased cell proliferation and tumor formation in mice (17, 71).
The various E2F family members differ in their association with pRb, p107, or p130. E2F1- to -3 associate exclusively with pRb in vivo, while E2F4 and E2F5 associate with pRb, p107, and p130 but in a cell cycle-regulated manner (46). The growth-inhibitory properties of pRb family members are regulated by phosphorylation (8, 28, 55). In quiescent or differentiated cells, pRb family members are hypophosphorylated and the majority of nuclear E2F proteins are bound to pRb family members. When cells are stimulated to proliferate, pRb family members become phosphorylated and release free, presumably active E2F (21, 46). Consistent with this model, the induction of pRb phosphorylation at the G1/S transition correlates closely with the timing of transcriptional activation of many E2F-regulated genes (35, 43).
The proto-oncogene c-myb is involved in the control of normal cell proliferation and the induction of neoplasia (40). Induced expression of c-myb has been found during proliferation of normal cells and tissue of the hemolymphopoietic system, and overexpression is observed in tumors of both hematopoietic and nonhematopoietic origin, including neuroblastoma, neuroepithelioma, and neoplasias of the lung, colon, breast, and melanoma (2, 20, 50, 62, 63, 66, 67, 73).
In normal cells, transcription of the c-myb gene is tightly regulated at transcriptional and posttranscriptional levels. To date, the majority of genes with known E2F promoter elements are activated at or near the G1/S boundary. An exception to this, however, is the proto-oncogene c-myb, which contains an E2F promoter element but is induced early during the G1 phase of the cell cycle in cells that are coming out of quiescence (65, 68). Moreover, expression of this gene remains constitutive in subsequent cycles (68). Therefore, although previous studies have shown that the c-myb gene can be induced by ectopically expressed E2F1 (53), it is able to largely escape the dominant repressive effect of pRb-E2F complexes during specific times of G1. As a first step in understanding the mechanisms governing the unique regulation of this promoter in the context of its E2F element, we investigated factor binding to the previously described E2F site within this promoter. While the E2F element from the c-myb promoter binds free, pRb-associated, and p107-associated E2F factors with affinities similar to those of other E2F elements, the c-myb E2F element also binds an apparent non-E2F-related factor which influences the regulation of its promoter. Therefore, the E2F element in the c-myb gene is subject to control by additional protein components which may contribute to the deregulated expression of c-myb in certain tumors.
MATERIALS AND METHODS
Cell culture and preparation of nuclear extracts.
X50-7 and Jurkat cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, and glutamine. U2OS and NIH 3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, penicillin, streptomycin, and glutamine. All cells were maintained at 37°C in a humidified 5% CO2-containing atmosphere.
Nuclear extracts were prepared by using a modified version of the protocol described by Dignam et al. (10). Cells were isolated and washed with phosphate-buffered saline (PBS), and the pellet was resuspended in 5 volumes of buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 5 mM dithiothreitol [DTT], 0.5 mM NaF, 0.5 mM Na3VO4, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of antipain per ml). The cell suspension was then incubated for 1 h at 4°C, and cells were lysed in a Dounce homogenizer (25 strokes). Nuclei were pelleted for 10 s in an Eppendorf Microfuge (14,000 × g), washed once in buffer A, and resuspended in 3 volumes of buffer B (20 mM HEPES [pH 7.9], 20% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 5 mM DTT, 0.5 mM NaF, 0.5 mM Na3VO4, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of antipain per ml). After incubation on ice for 30 min, cellular debris was removed by centrifugation at 4°C (14,000 × g) for 15 min, and supernatants were aliquoted and stored at −70°C.
Construction of vectors.
To generate myb(wt)-Luc, a BamHI DNA insert (−687 to +204 from the 5′ flanking region of the human c-myb gene described in reference 47) was subcloned from B1-CAT (kindly provided by Bruno Calabretta, Jefferson Cancer Institute, Philadelphia, Pa.) into the BglII site within the pGL2-Basic luciferase reporter vector, creating the myb-Luc fragment. The myb-Luc mutant derivatives were generated by site-directed mutagenesis as previously described (19), using a Bio-Rad Muta-Gene kit. The mutations introduced are shown in Fig. 5C. Sequence analysis was used to confirm the presence of the appropriate mutations.
FIG. 5.
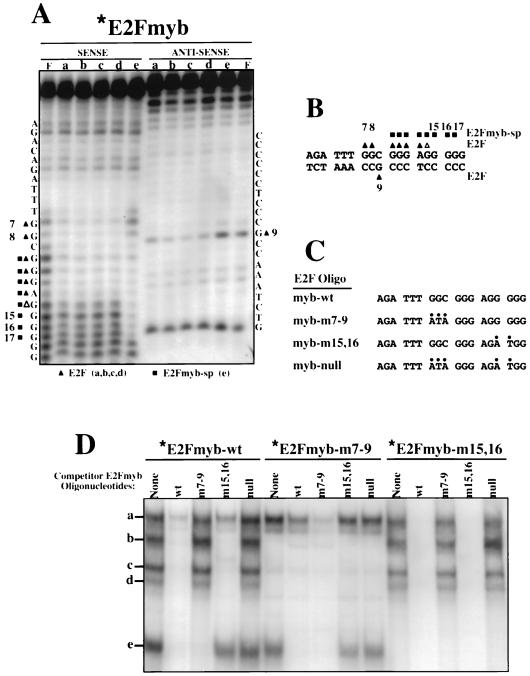
E2Fmyb-sp and E2F complexes interact with overlapping DNA sequences. (A) Methylation interference analysis of complexes a to e was performed with X50-7 nuclear extracts and the E2Fmyb probe. Sequences of the sense and antisense strands are indicated adjacent to the gels. Triangles and squares indicate bases whose methylation completely (filled) or partially (empty) blocked formation of complexes a to d (triangles) or e (squares). The cleavage pattern observed with an unbound probe (F) is also shown. (B) Summary of methylation interference analysis shown in panel A. (C) Sequence of wild-type and mutant E2Fmyb probes. For each mutant, alterations relative to the wild-type sequences are indicated by dots. (D) EMSA analysis of complexes formed between X50-7 nuclear factors and radiolabeled E2Fmyb-wt, E2Fmyb-m7-9, and E2Fmyb-m15,16 probes. Reaction mixtures were incubated in the absence (None) or in the presence of a 100-fold excess of the indicated unlabeled competitor oligonucleotides. Positions of complexes a to e are marked.
Transfections, reporter gene assays, and cell cycle analysis.
U2OS cells were transfected at 70 to 80% confluence, while NIH 3T3 cells were transfected at 10 to 15% confluence; transfections were carried out in 10-cm-diameter dishes for 16 h by the calcium phosphate method (7). Transfected cells were washed twice with PBS, refed, and incubated at 37°C and 5% CO2 for 24 h prior to analysis. Transfections included 5 μg of appropriate luciferase reporter plasmid plus 0.5 μg of pCMV-βgal (41) and 24.5 μg of carrier plasmid (pBluescript; Stratagene). Jurkat cells (7 × 106) were transfected with Superfect (Qiagen) as recommended by the manufacturer. Briefly, cells were resuspended at 2 × 106/ml in growth medium and incubated for 8 h at 37°C and 5% CO2 with a mixture of 5 μg of appropriate luciferase reporter plasmids plus 1 μg of pCMV-βgal and 20 μl of Superfect. Cells were then washed once with PBS, resuspended in 10 ml of growth medium, and incubated at 37°C and 5% CO2 for 36 h prior to analysis.
For synchronization experiments, transfected NIH 3T3 cells were placed in DMEM–10% serum for 24 h, after which they were washed twice with DMEM and starved for 48 h in DMEM–0.5% serum. Following starvation, cells were either harvested or refed with medium containing 10% serum and incubated for the indicated time periods.
Cell cycle analysis was carried out by flow cytometry. Cells were harvested, fixed in 70% ethanol, stained with propidium iodide solution (69 μM propidium iodide, 38 mM sodium citrate, 100 μg of RNase per ml), and analyzed on a Becton Dickinson flow cytometer. Luciferase assays and β-galactosidase assays (for normalization of luciferase values) were performed essentially as described by Krek et al. (30).
Electrophoretic mobility shift assays (EMSAs).
Gel shifts were performed with the following double-stranded oligonucleotides: E2Fdhfr-wt (5′-CTAGAGCAATTTCGCGCCAAACTTG-3′ and 5′-GATCCAAGTTTGGCGCGAAATTCGT-3′), E2Fdhfr-mut (5′-CTAGAGCAATTGCTCGACCAACTTG-3′ and 5′-GATCCAAGTTGGTCGAGCAATTGCT-3′), E2FE2F1-A (5′-CTAGAGCTCTTTCGCGGCAAAAAGGAG-3′ and 5′-GATCCTCCTTTTTGCCGCGAAAGAGCT-3′), E2FE2F1-B (5′-CTAGAGGATTTGGCGCGTAAAAGTGG-3′ and 5′-GATCCCACTTTTACGCGCCAAATCCT-3′), E2Fcdc2 (5′-CTAGATTTCTTTCGCGCTCTAGCCG-3′ and 5′-GATCCGGCTAGAGCGCGAAAGAAAT-3′), E2Fc-myc-wt (5′-CTAGAGAGGCTTGGCGGGAAAAAG-3′ and 5′-GATCCTTTTTCCCGCCAAGCCTCT-3′), E2Fc-myb-wt (5′-CTAGACAGATTTGGCGGGAGGGGGG-3′ and 5′-GATCCCCCCCTCCCGCCAATCTGT-3′), and TK-SP1 (5′-GATCCCGCGCCGCCCCGACT-3′ and 5′-CTAGAGTCGGGGCGGCGCGG-3′). For the E2F-c-myb mutants, the point mutations indicated in Fig. 5C were used. Oligonucleotides were synthesized by AnaGen Inc. and purified by denaturing polyacrylamide electrophoresis (54). Eluted oligonucleotides were then purified by passage through SepPak C18 chromatography columns (Waters). Complementary oligonucleotides were mixed at an equimolar ratio in 10 mM Tris (pH 7.5)–50 mM NaCl, heated to 65°C, and annealed by slow cooling to room temperature. Double-stranded oligonucleotides (100 ng) were labeled by a Klenow fill-in reaction.
For binding reactions, the following components were mixed and preincubated at room temperature for 20 min: 3 μl (5 to 10 μg) of nuclear extract, 7 μl of BFD (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM DTT), 2 μl (20 μg) of purified bovine serum albumin (New England Biolabs), 0.4 μl (2 μg) of sheared salmon sperm DNA, and 11 μl of high-pressure liquid chromatography-grade water. Unlabeled oligonucleotide competitors (100 ng) and antibodies (1 μg of purified antibodies or 1 μl of crude polyclonal antibodies) were also added to the initial mix prior to the preincubation step. Following preincubation, the labeled oligonucleotides were added (1 ng), and the mixtures were incubated for another 20 min at room temperature. Samples were then loaded directly onto a running nondenaturing 4% acrylamide–0.1% bisacrylamide gel (in 0.5× TBE [1× TBE is 90 mM Tris, 64.6 mM boric acid, and 2.5 mM EDTA, pH 8.3]) at 4°C. Retarded complexes were visualized by autoradiography (1 to 16 h at room temperature).
Antibodies against E2F1 (sc-193x), E2F2 (sc-632x), E2F3 (sc-879x), E2F4 (sc-512x), E2F5 (sc-1699x), Ets-1/Ets-2 (sc-112x), and SP-1 (sc-420x) were purchased from Santa Cruz Biotechnology. The anti-c-Myc monoclonal antibody (MAb) 9E10 was kindly provided by Alberto Gandarillas (Imperial Cancer Research Fund, United Kingdom). The anti-p107 SD15 and anti-Rb N9 antibodies have been previously described (13, 60) and were kindly provided by Edward Harlow (Massachusetts General Hospital, Boston, Mass.) and William Kaelin (Dana-Farber Cancer Institute, Boston, Mass.), respectively. The anti-DP-1 polyclonal rabbit antiserum A33 was raised against a glutathione S-transferase–DP-1 fusion protein and will be described elsewhere. The preimmune serum was obtained from the rabbit prior to immunization.
Methylation interference analysis.
Methylation interference analysis was carried out essentially as previously described (4). Double-stranded DNA probes used for EMSAs were 32P labeled at either the 5′ or 3′ end as previously described (4, 54) except that 200 ng of the probe was used. One-fourth of each sample was then partially methylated with dimethyl sulfate for 3 min. For the binding reaction, the EMSA protocol was scaled up sixfold and 5 × 106 to 10 × 106 cpm of probe was used. After electrophoresis, the samples were transferred to a DEAE membrane with a semidry electrophoretic transfer unit, the membrane was exposed for autoradiography, and the relevant bands were cut out and eluted (61). The DNA was cleaved at the methylated bases by using 1 M piperidine at 90°C for 30 min. The samples were loaded onto a 10% polyacrylamide-urea sequencing gel in 1× TBE running buffer.
RESULTS
The E2F element from the c-myb promoter binds a unique factor(s) which does not interact with E2F elements from several other promoters.
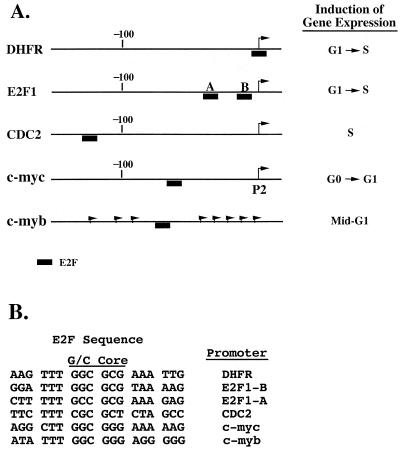
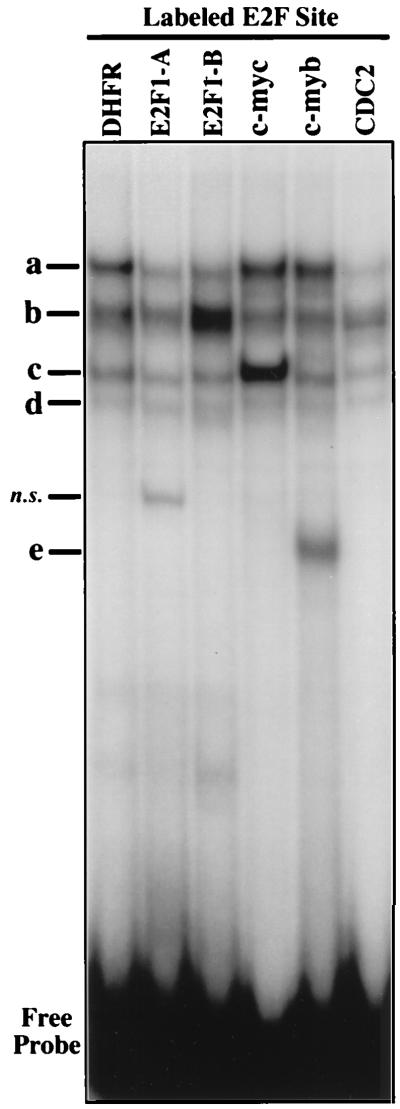
EMSA was used to compare the binding of nuclear factors to the E2F element in the c-myb promoter relative to the E2F elements from several other genes (Fig. 1). In nuclear extracts from the nontransformed human lymphoblastoid cell line X50-7, we observed four major protein complexes (a to d) which are common to each of the probes tested (Fig. 2). Interestingly, different E2F sites show different ratios of these complexes. In addition to complexes a to d, the E2F element from the c-myb promoter forms a complex (e) which is not observed with any of the other E2F elements (Fig. 2). A fifth major band was also detected with the E2FE2F1-A probe, but further analysis revealed that this band is nonspecific (data not shown).
FIG. 1.
(A) Schematic representation of several E2F-regulated cellular promoters. Arrows indicate transcription initiation sites. (B) Alignment of E2F elements from the DHFR, E2F1, cdc2, c-myc, and c-myb promoters.
FIG. 2.
EMSA of E2F elements from different promoters. Complex formation in nuclear extracts from the lymphoblastoid cell line X50-7 and the indicated radiolabeled E2F elements (Fig. 1) was analyzed by EMSA as described in Materials and Methods. Protein-DNA complexes are indicated as a to e. DHFR, c-myc, c-myb, and CDC2, oligonucleotides corresponding to the E2F sites within the DHFR, c-myc, c-myb, and cdc2 promoters, respectively; E2F1-A and E2F1-B, oligonucleotides corresponding to the two E2F sites found in the E2F1 promoter; n.s., a nonspecific complex.
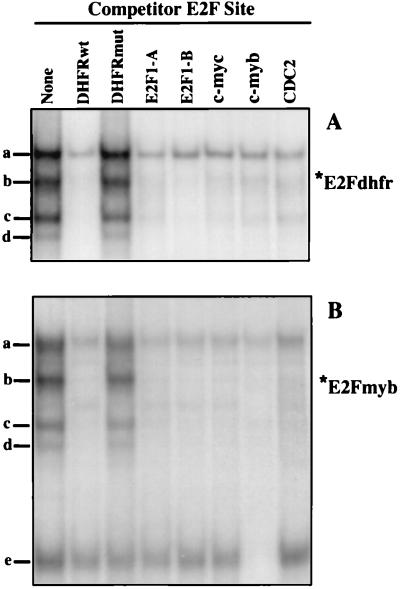
Competition experiments were performed to assess the specificity of the protein-DNA interactions observed with the E2F sites from the DHFR and c-myb promoters. As expected, the formation of complexes a to d on the DHFR E2F site are competed efficiently by each of the other E2F elements but not by a mutant DHFR E2F element, indicating the presence of E2F or E2F-related factors in these complexes (Fig. 3A). The binding of proteins in complexes a to d in assays using the c-myb E2F element are competed efficiently by each of the other E2F elements (Fig. 3B). In contrast, formation of complex e on the c-myb E2F site is competed only by a self-oligonucleotide, consistent with the apparent absence of a band with similar mobility in assays using any other labeled E2F element (Fig. 2). Therefore, the c-myb E2F element forms a complex (referred to here as E2Fmyb-sp) which does not interact with any other E2F element tested.
FIG. 3.
Identification of a DNA-binding species which interact solely with the E2F elements within the c-myb promoter. Complexes formed with X50-7 nuclear extracts and radiolabeled E2F elements from the DHFR (A) and c-myb (B) promoters were analyzed by EMSA. Reaction mixtures were preincubated in the absence (None) or in the presence of a 100-fold excess of the indicated unlabeled competitor oligonucleotides. Positions of complexes a to e are indicated. DHFRwt, c-myc, c-myb, and CDC2, oligonucleotides corresponding to the E2F sites within the DHFR, c-myc, c-myb, and cdc2 promoters, respectively; E2F1-A and E2F1-B, oligonucleotides corresponding to the two E2F sites found in the E2F1 promoter; DHFRmut, mutant DHFR probe.
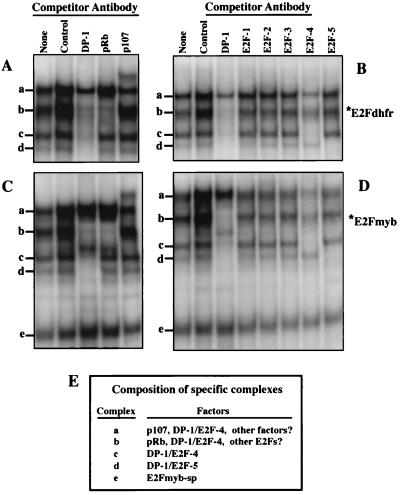
Identification of E2F and pRb family members in complexes a to d, the DHFR and c-myb E2F elements: E2Fmyb-sp is not recognized by antibodies against E2F or pRb family members.
Other investigators have found that DP-1 is a major component of the vast majority of E2F-DNA complexes (28). With X50-7 nuclear extracts and the E2F element from the DHFR promoter, a DP-1 antibody effectively inhibits the formation of complexes b to d and partially inhibits the formation of complex(es) migrating at position a (Fig. 4). (Note that crude rabbit polyclonal antiserum invariably results in a nonspecific stabilization of protein-DNA interactions in our EMSA studies, and therefore the level of binding observed with the DP-1 and pRb antisera should be compared to that of the DP-1 preimmune sera [Fig. 4, Control].) With the DHFR and c-myb probes, formation of complex b is specifically inhibited by an anti-pRb antiserum, indicating that this complex is likely composed primarily of pRb-associated E2Fs. A p107 antibody partially inhibits the formation of complex a and presents a supershifted band with each probe, indicating that p107 is a component of at least some complexes migrating at this position (Fig. 4). Importantly, the E2Fmyb-sp complex is unaffected by either the DP-1, pRb, or p107 antibody (Fig. 4). In addition, E2Fmyb-sp is not affected by the anti-E2F1, -2, -3, -4, or -5 antibody (Fig. 4). In contrast, the anti-E2F4 antibody effectively inhibits formation of complex c on the DHFR and c-myb probes. In addition, the E2F4 antibody partially decreases the level of complex a and b, indicating the presence of E2F4 in some pRb- and p107-containing complexes. The anti-E2F5 antibody specifically inhibits complex d formation with either the DHFR or the c-myb probe. Although the E2F1, -2, and -3 antibodies did not significantly affect any complex (perhaps due to low relative abundance), these antibodies were able to specifically supershift corresponding E2F complexes in extracts from cells transfected with E2F1, -2, or -3 expression vectors (data not shown). Moreover, when these antibodies were added together in combination with the E2F4 antibody, formation of complex b was completely inhibited whereas binding of complex e remained unaffected (data not shown). In conclusion, the inability of any of these antibodies to affect E2Fmyb-sp is consistent with the inability of various non-self E2F oligonucleotides to compete for binding of these factors to the c-myb E2F element. In addition, the inability of DP-1, E2F, and p107 antibodies to completely block binding of complex suggests the possibility that other factors are present in this complex.
FIG. 4.
Identification of E2F or pRb family members in E2Fdhfr and E2Fmyb protein-DNA complexes. Binding reactions were performed with X50-7 nuclear extracts and either DHFR (A and B) or c-myb (C and D) E2F site probes. Extracts were preincubated in the absence (None) or in the presence of antibodies against the indicated proteins for 20 min prior to addition of the labeled probes. Control denotes a DP-1 preimmune rabbit polyclonal antibody. A nonspecific increase in overall binding was noticed in the presence of crude rabbit polyclonal antibodies (Control, DP-1, and pRb). Positions of complexes a to e are indicated. (E) Summary of the results shown in panels A to D detailing the composition of each of the specific complexes.
Distinct nucleotide sequence recognition by E2Fmyb-sp- and E2F-containing complexes.
Methylation interference analysis revealed no differences in sequence specificity for the binding of complexes a to d to the c-myb E2F element (Fig. 5A and B). Methylation at positions 7, 8, 10, 11, 12, 13, and 14 of the sense strand or position 9 on the antisense strand each prevented binding of any of the E2F-containing complexes (a to d). In contrast, methylation of position 7 or 8 (sense strand) or 9 (antisense strand) did not affect the formation of complex e, but formation of this complex was inhibited by methylation of positions 10 to 17 of the sense strand (Fig. 5A and B). Therefore, the sequence-binding specificity of E2Fmyb-sp is distinct from but overlapping that of E2F-containing complexes.
Based on these methylation interference data, c-myb E2F site mutants were designed so as to interact specifically with either E2F-containing complexes (E2Fmyb-m15,16), E2Fmyb-sp factor(s) (E2Fmyb-m7-9), or neither (E2Fmyb-null) (Fig. 5C). As shown in Fig. 5D, addition of competitor E2Fmyb-m7-9 oligonucleotide inhibited the formation of E2Fmyb-sp- but not E2F-containing complexes (a to d) on the wild-type c-myb E2F element (E2Fmyb-wt) (although a loss of the upper portion of complex a was observed due to the loss of an SP-1-containing complex which also binds E2Fmyb-m7-9 [see Fig. 6]). In contrast, E2Fmyb-m15,16 competed for binding of complexes a (the faster-migrating portion), b, c, and d but not complex e, indicating specificity for E2F proteins. As expected, E2Fmyb-wt inhibited formation of all five complexes whereas E2Fmyb-null did not affect the formation of any complex.
FIG. 6.
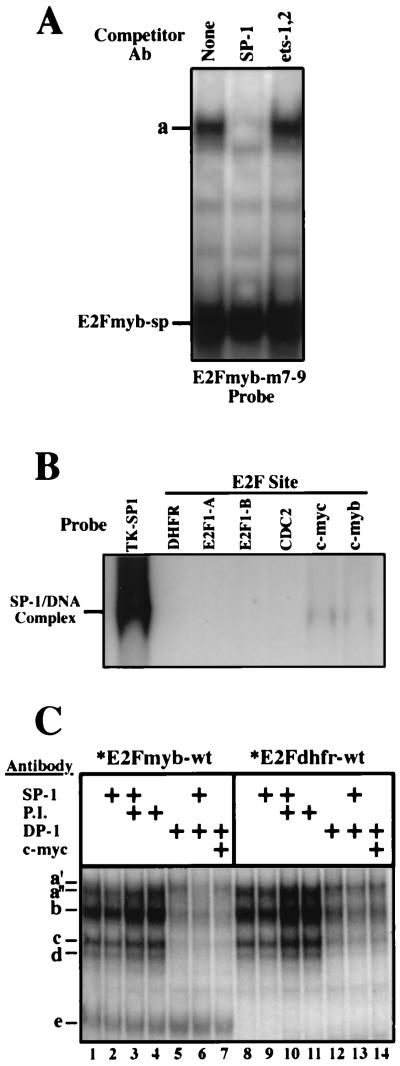
SP-1 binds to the c-myc and c-myb E2F elements. (A) Complex formation in assays employing X50-7 nuclear extracts and a radiolabeled E2Fmyb-m7-9 probe. Reaction mixtures were incubated in the absence (None) or in the presence of the indicated antibodies (Ab). (B) Binding of recombinant SP-1 to a TK-SP1 or to the indicated E2F elements. Binding reactions were performed with 0.5 footprint units of baculovirus-generated SP-1 (Promega). (C) Competition analysis of complexes formation in assays using X50-7 nuclear extracts and either an E2Fmyb-wt (lanes 1 to 7) or E2Fdhfr (lanes 8 to 14) probe. Reaction mixtures were incubated in the absence or in the presence of the indicated antibodies: anti-SP-1 (lanes 2, 3, 5, 9, 10, and 13), preimmune anti-DP-1 (P.I.; lanes 3, 4, 10, and 11), anti-DP-1 (lanes 5 to 7 and 12 to 14), and anti-c-Myc (lanes 7 and 14).
When the E2Fmyb-m7-9 was used as a probe, complex e was detected (Fig. 5D). However, complex a was also observed with E2Fmyb-m7-9, and this activity corresponds to the binding of SP-1, which is shown below to bind both E2Fmyb-m7-9 and E2Fmyb-wt (see Fig. 6). The binding of proteins in complexes a and e to the E2Fmyb-m7-9 probe is inhibited by an excess of cold E2Fmyb-wt and E2Fmyb-m7-9 but not E2Fmyb-m15,16 or E2Fmyb-null oligonucleotide (Fig. 5D).
With E2Fmyb-m15,16 as a probe, only complexes a to d are observed; these complexes are competed by the E2Fmyb-wt and E2Fmyb-m15,16 but not the E2Fmyb-m7-9 or E2Fmyb-null oligonucleotide (Fig. 5D). Therefore, E2Fmyb-m15,16 specifically binds E2F-containing protein complexes.
SP-1 binds to the E2F element in the c-myb promoter.
As shown in Fig. 5D, binding of a factor(s) with the mobility of complex a is evident with the E2Fmyb-m7-9 probe. To date, we have tested a panel of different mutant oligonucleotides and have been unable to genetically dissociate the binding of E2Fmyb-sp-containing factor(s) and a factor(s) which binds with the mobility of complex a (data not shown). This suggests that the factor(s) that gives rise to complex a with E2Fmyb-m7-9 as a probe interacts with sequences similar to those required for the formation of the E2Fmyb-sp complex. We have tested a variety of antibodies which recognize transcription factors that are known to bind GC-rich sequences in EMSAs in an attempt to identify this factor (data not shown and Fig. 6). As shown in Fig. 6A, an antibody that recognizes SP-1 abrogates the binding of proteins in complex a to E2Fmyb-m7-9 but does not affect the binding of E2Fmyb-sp. In contrast, an antibody which recognizes the transcription factors Ets-1 and Ets-2 does not affect the binding of either complex a or complex e. Therefore, SP-1 is a major component of complex a with E2Fmyb-m7-9 as a probe. As shown in Fig. 6B, purified recombinant human SP-1 binds to both E2Fmyb and E2Fmyc but not to E2Fdhfr, E2Fcdc2, E2FE2F1-A, or E2FE2F1-B. However, the apparent affinity is significantly lower than the observed affinity for a previously described SP-1 site (TK-SP1).
We next tested whether SP-1 present in X50-7 nuclear extracts interacts with the E2Fmyb-wt probe. As shown in Fig. 6C, complex a consists of two closely migrating complexes, a′ and a", which are specifically inhibited in the presence of anti-SP-1 (lane 2) and DP-1 (lane 5) antibodies, respectively. Moreover, the combination of SP-1 and DP-1 antibodies (lane 6) completely eliminates formation of both complexes, while the combination of either SP-1 plus a preimmune DP-1 serum (lane 3) or DP-1 plus c-Myc (lane 7) blocked the formation of only complex a′ or a", respectively. On the other hand, addition of the SP-1 antibody did not affect any complex observed with the E2Fdhfr and E2FE2F1-A probes (which do not bind SP-1 [Fig. 6B]), while addition of the anti-DP-1 antibody partially decreases the amount of complex a in assays using these probes (Fig. 6C and data not shown). We conclude that cellular SP-1 binds to the E2F site in the c-myb promoter and makes up a fraction of the complexes that migrate at position a but does not interact with the E2F sites in the DHFR, cdc2, and E2F1 promoters.
Analysis of E2Fmyb-sp complex in c-myb promoter function.
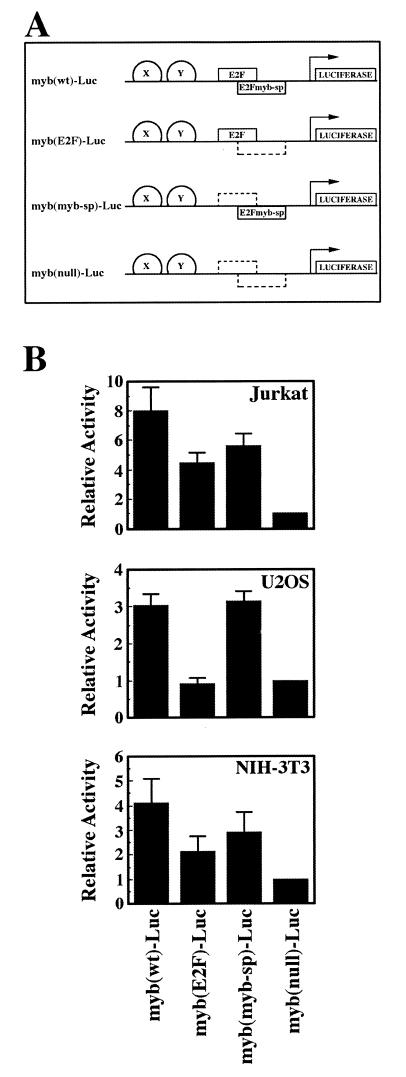
We have shown that a factor with biochemical properties distinct from those of known E2F proteins interacts with the DNA-binding site that overlaps the E2F element in the c-myb promoter. Using mutant or wild-type oligonucleotide probes, we have determined that E2Fmyb-sp is significantly expressed in every mammalian cell type tested to date, including 14 human cell lines from different tissues, primary T lymphocytes, and NIH 3T3 cells (data not shown). We therefore assessed the relative influence of E2F factors versus E2Fmyb-sp and/or SP-1 on c-myb promoter activity in several cell lines. The c-myb promoter was linked to a luciferase reporter gene, and mutations were introduced into the E2F element of each promoter which were shown in Fig. 5C to result in selective factor binding (Fig. 7A).
FIG. 7.
Functional regulation of the c-myb promoter through the E2F element in asynchronous cells. (A) Schematic representation of the luciferase reporter plasmids used in panel B and Fig. 8. Wild-type (solid box) and mutant (dashed box) E2F and E2Fmyb-sp binding sites are shown as boxes; the presence of other elements in the c-myb promoter which were left unmodified is represented as semicircles labeled X and Y. (B) The indicated myb-Luc reporter plasmids containing wild-type or mutant E2F elements were cotransfected with pCMV-βgal into asynchronously growing Jurkat, U2OS, and NIH 3T3 cells; 40 h later, cell extracts were prepared and luciferase and β-galactosidase assays were performed. Luciferase values were normalized for β-galactosidase activity and represent the means of at least four different experiments. Luciferase activities of the indicated promoter reporter plasmids relative to cells transfected with myb(null)-Luc ± standard deviation (error bars) are shown.
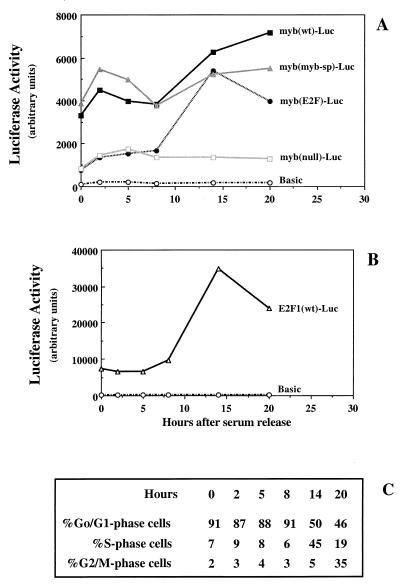
The results shown in Fig. 7B indicate that E2Fmyb-sp and/or SP-1 contributes to the activity of the c-myb promoter in several cell lines, although the relative contribution of E2F and either E2Fmyb-sp/SP-1 may be cell line dependent. E2F factors play a lesser role than E2Fmyb-sp/SP-1 in the overall level of promoter activity observed in asynchronous U2OS cells, since myb(E2F)-Luc and myb(null)-Luc have similar activities which are lower than those of myb(wt)-Luc or myb(myb-sp)-Luc (Fig. 7B). On the other hand, in asynchronously growing Jurkat and NIH 3T3 cells, the activity of myb(wt)-Luc is higher than those of myb(E2F)-Luc and myb(myb-sp)-Luc, which are higher (three- to sixfold) than that of myb(null)-Luc but similar to each other. This suggests that E2F and either E2Fmyb-sp or SP-1 may each contribute to the activation of the c-myb promoter (perhaps at different phases of the cell cycle). To test this possibility, we performed cell synchronization experiments in NIH 3T3 cells. Although these cells do not enter a G0 state identical to that of primary fibroblasts (8), they are a useful system to address events that occur after the G0/G1 transition. Accordingly, the wild-type c-myb promoter is active in serum-starved NIH 3T3 cells and does not dramatically increase following serum stimulation. Significantly, the activity of myb(wt)-Luc closely parallels the activity of myb(myb-sp)-Luc throughout the cell cycle (Fig. 8A). Moreover, like the activity of myb(wt)-Luc, the activity of myb(myb-sp)-Luc is significantly higher in G0/G1 (0, 2, 4, and 8 h) than that of a mutant which selectively binds E2F proteins [myb(E2F)-Luc], indicating that E2Fmyb-sp and/or SP-1 are likely responsible for the observed activity of the c-myb promoter during G1. During S and G2/M phases (14 and 20 h after readdition of serum), E2F and/or either E2Fmyb-sp or SP-1 may contribute to the elevated activity of this promoter, since myb(E2F)-Luc and myb(myb-sp)-Luc are more active than myb(null)-Luc but less active than myb(wt)-Luc (Fig. 8A).
FIG. 8.
Activation of the c-myb promoter by E2Fmyb-sp and/or SP-1 through the E2F element during the G1 phase of the cell cycle. (A) The indicated myb-Luc reporter plasmids containing wild-type or mutant E2F elements were cotransfected with pCMV-βgal into NIH 3T3 cells. The cells were placed in low (0.5%) serum 4 h after the removal of the calcium phosphate precipitates. The cells remained in low serum for >48 h to induce quiescence, at which point serum was added (time zero). At the indicated time points, cells were removed for cell cycle analysis by flow cytometry and for determination of luciferase and β-galactosidase activities. (B) An E2F1 luciferase reporter plasmid containing wild-type E2F elements was analyzed in parallel as a control. Luciferase values from a representative experiment (normalized for β-galactosidase activity) (A and B) and percentages of cells in each phase of the cell cycle at the indicated time points (C) are shown.
DISCUSSION
A large number of promoters contain potential E2F-binding sites which, in many cases, have been shown to be key controlling elements governing the regulated expression of their corresponding genes. E2F sites in the cyclin A and the mouse TK promoters function as repressor elements in growth-arrested cells and as activators following the G1/S transition (29, 56). On the other hand, although mutation of the E2F binding site in several other promoters (B-myb, E2F1, E2F2, cdc6 and orc-1) was shown to result in a dramatic increase in activity in G0 and/or G1, the activity of such mutants during S phase was not elevated relative to wild-type promoters (26, 27, 33, 34, 45, 48, 57, 72). Therefore, while the activation function of E2F proteins bound to these promoters was not always clearly manifested under the experimental conditions used, the influence of the dominant repressor function of pRb family members was consistently apparent.
While most genes studied to date which contain E2F elements are induced near the G1/S phase transition, some of the genes which contain E2F elements are regulated either earlier (e.g., c-myb and c-myc) or later (e.g., the cyclin A gene and cdc2) in the cell cycle. The timing of cyclin A promoter activation is regulated through the coordinate action of E2F proteins and another factor, CDF-1, which binds to an element which is contiguous to the E2F element (37, 38). We have found that the E2F element in the c-myb promoter binds distinct factors relative to the E2F elements from several other promoters. Specifically, we have identified a novel binding activity, E2Fmyb-sp, which interacts with the c-myb E2F element but not several other E2F elements. In addition, we have shown that SP-1 binds to the c-myb E2F element. With recombinant protein, however, the affinity of SP-1 for the c-myb E2F element is significantly lower than its affinity for a prototypical SP-1-binding site. Despite a low affinity for the c-myb E2F element, the high abundance of SP-1 compared to E2F factors may still allow it to contribute to c-myb promoter activation under some circumstances. In this regard, SP-1 has been shown to mediate serum responsiveness for other promoters such as the ornithine decarboxylase gene promoter (32) and could possibly contribute such a role in c-myb regulation.
We have not determined the identifies of the factor(s) in the E2Fmyb-sp complex. CDF-1, a factor known to participate in the timing of activation of other E2F element-containing genes, is likely not E2Fmyb-sp since (i) CDF-1 interacts with DNA through a bipartite binding site whose sequence is distinct from the myb-sp interaction sequence; and (ii) CDF-1 is a repressor factor (37), while E2Fmyb-sp appears to be an activator in every cell type tested to date. A transcription factor database search suggested that MZF-1 (myeloid zinc finger 1) may bind to the DNA sequence recognized by E2Fmyb-sp. However, MZF-1 and E2Fmyb-sp do not appear to be the same factor since UV cross-linking experiments indicated that the molecular mass of E2Fmyb-sp is ca. 30 kDa (7a) whereas that of MZF-1 is ca. 56 kDa (3). In addition, MZF-1 has been shown to be a transcriptional repressor in nonhematopoietic cells (16), while we have shown here that E2Fmyb-sp is a transcriptional activator in different cellular types, including the nonhematopoietic U2OS and NIH 3T3 cell lines. Although the inability to genetically dissociate the binding of E2Fmyb-sp and SP-1 suggests that E2Fmyb-sp could be a member of the SP-1 family, the low molecular weight of E2Fmyb-sp suggests that it is different from the known prototypical SP-1 family members. In addition, gel shift competition experiments showed that an excess of cold oligonucleotide that binds SP-1 family members (TK-SP1) did not compete for binding of E2Fmyb-sp but efficiently inhibited the binding of SP-1 family members to itself (7a).
The DNA recognition sequence of E2Fmyb-sp clearly overlaps the core E2F element sequence. Methylation interference analysis indicates that binding of E2Fmyb-sp factor to DNA requires direct contact with nucleotides that are required for the binding of E2F molecules. This indicates that the binding of E2Fmyb-sp and that of E2F are likely mutually exclusive, suggesting that competition for binding to the c-myb E2F element dictates the mode of regulation governing the expression of the c-myb gene.
Using reporter plasmids bearing defined mutations within the E2F element of the c-myb promoter, we have demonstrated that E2Fmyb-sp and/or SP-1 proteins contribute more significantly to c-myb promoter activity than E2F factors in cycling U2OS. The same was also observed in X50-7 cells (our unpublished observations). In contrast, both E2F and E2Fmyb-sp/SP-1 could potentially contribute similarly to c-myb promoter activation in cycling Jurkat and NIH 3T3 cells, suggesting that each may play a role at different phases of the cell cycle. In this regard, our results indicate that E2Fmyb-sp and/or SP-1 are required for full activity of the c-myb promoter during the G1 phase of the cell cycle, while both E2F and E2Fmyb-sp/SP-1 could potentially contribute similarly to the overall activity of the c-myb promoter during the S phase. These data suggest that binding of each factor might prevail over binding of the others at different phases of the cell cycle. Although by EMSA analysis we did not observe significant differences in the E2F/E2Fmyb-sp/SP-1 DNA-binding ratio throughout the cell cycle (7a), it is possible that factors governing chromatin structure determine site occupancy by either E2F or E2Fmyb-sp/SP-1 in vivo. Alternatively, protein modifications which control DNA binding affinities of these factors may not be maintained during preparation of nuclear extracts. The differences observed in the contribution of E2F factors in the overall activity of the c-myb promoter among different cell lines might be attributed to differences in cell growth, which might be more vigorous in NIH 3T3 and Jurkat cells than in U2OS or X50-7 cells. In addition, Hofferer et al. (25) have shown that UV or gamma irradiation induces the level of E2F DNA-binding activity (likely by increasing E2F protein levels), suggesting that the cell environment may also affect the relative contribution of E2F to the activity of the c-myb promoter. Therefore, a number of factors regulating the DNA-binding activity of the various proteins that interact with the c-myb E2F element will dictate whether they are subject to control by either E2Fmyb-sp, SP-1, or E2F protein.
Results from previous experiments suggest that a cyclin A-associated kinase negatively regulates the DNA-binding activity of E2F1, -2, and -3 through its interaction with a cyclin-binding domain that is present in the amino termini of these factors, while the absence of this domain in E2F4 and -5 allows these members to escape this regulatory pathway (28, 31). In this report, we have provided evidence suggesting the existence of a more general mechanism that negatively regulates the interaction of all E2F family members with certain DNA-binding sites through the occupation of these sites by non-E2F factors.
Finally, our findings may be relevant to the possible role of c-myb overexpression in oncogenesis. Since E2Fmyb-sp or SP-1 is required for full activation of the c-myb gene, it is possible that deregulated expression of these factors in some cases contributes to the overexpression of c-myb which has been observed in multiple tumors (2, 14, 15, 20, 50, 62, 63, 66, 67, 73).
ACKNOWLEDGMENTS
We thank Bruno Calabretta and Teresa Bellón for kindly providing the B1-CAT reporter plasmid, and we thank Alberto Gandarillas, Ed Harlow, and William Kaelin for gifts of the anti-c-Myc MAb 9E10, anti-p107 MAb SD15, and anti-Rb antibody N9, respectively. We also thank Irene Garcia-Higuera, Daniel Dwyer, Maria A. Escudero, Eun-Joo Jung, and Antonio Rodriguez for invaluable advice during this project, and we thank James de Caprio, Irene Garcia-Higuera, and Antonio Rodriguez for critically reading the manuscript.
This work was supported by postdoctoral fellowships from the Human Frontier Science Program Organization and the Spanish Ministry of Education and Science to M.R.C. and research grants from the American Cancer Society (RPG-97-065-01-VM) and the National Institutes of Health (R01 GM48045) to E.F.
REFERENCES
- 1.Adams P D, Kaelin W K. The cellular effects of E2F overexpression. Curr Top Microbiol Immunol. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- 2.Alitalo K, Winqvist R, Lin C C, De La Chapelle A, Schwab M, Bishop J M. Aberrant expression of an amplified c-myb oncogene in two cell lines from a colon carcinoma. Proc Natl Acad Sci USA. 1984;81:4534–4538. doi: 10.1073/pnas.81.14.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen K E, de la Luna S, Kerkhoven R M, Bernards R, La Thangue N B. Distinct mechanisms of nuclear accumulation regulate the functional consequence of E2F transcription factors. J Cell Sci. 1997;110:2819–2831. doi: 10.1242/jcs.110.22.2819. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 5.Bandara L R, Buck V M, Zamanian M, Johnston L H, La Thangue N B. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 1993;12:4317–4324. doi: 10.1002/j.1460-2075.1993.tb06116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Campanero M R, Flemington E K. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Campanero, M. R., and E. K. Flemington. Unpublished data.
- 8.Cobrinik D. Regulatory interactions among E2Fs and cell cycle control proteins. Curr Top Microbiol Immunol. 1996;208:32–61. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 9.Cress W D, Nevins J R. Interacting domains of E2F1, DP1, and the adenovirus E4 protein. J Virol. 1994;68:4213–4219. doi: 10.1128/jvi.68.7.4213-4219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duronio R J, Bonnette P C, O’Farrell P H. Mutations of the Drosophila dDP, dE2F, and cyclin E genes reveal distinct roles for the E2F-DP transcription factor and cyclin E during the S-phase transition. Mol Cell Biol. 1998;18:141–151. doi: 10.1128/mcb.18.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 13.Dyson N, Dembski M, Fattaey A, Ngwu C, Ewen M, Helin K. Analysis of p107-associated proteins: p107 associates with a form of E2F that differs from pRB-associated E2F-1. J Virol. 1993;67:7641–7647. doi: 10.1128/jvi.67.12.7641-7647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erisman M D, Rothberg P G, Diehl R E, Morse C C, Spandorfer J M, Astrin S M. Deregulation of the c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol Cell Biol. 1985;5:1969–1976. doi: 10.1128/mcb.5.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escot C, Theillet C, Lidereau R, Spyratos F, Champeme M H, Gest J, Callahan R. Genetic alterations of the c-myc protooncogene (MYC) in human primary breast carcinomas. Proc Natl Acad Sci USA. 1986;83:4834–4838. doi: 10.1073/pnas.83.13.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagan R, Flint K J, Jones N. Phosphorylation of E2F-1 modulates its interaction with the retinoblastoma gene product and the adenoviral E4 19 KDa protein. Cell. 1994;78:799–811. doi: 10.1016/s0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 17.Field S J, Tsai F-Y, Kuo F, Zubiaga A M, Kaelin W G, Livingston D M, Orkin S H, Greenberg M E. E2F1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 18.Flemington E K, Speck S H, Kaelin W G. E2F-1 mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foss K, McClain W H. Rapid site-directed mutagenesis in plasmids. Gene. 1987;59:285–290. doi: 10.1016/0378-1119(87)90336-2. [DOI] [PubMed] [Google Scholar]
- 20.Griffin C A, Baylin S B. Expression of the c-myb oncogene in human small cell lung carcinoma. Cancer Res. 1985;45:272–275. [PubMed] [Google Scholar]
- 21.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 22.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helin K, Wu C-L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative transactivation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 24.Hiyama H, Iavarone A, Reeves S A. Regulation of the cdk inhibitor p21 gene during cell cycle progression is under the control of the transcription factor E2F. Oncogene. 1998;16:1513–1523. doi: 10.1038/sj.onc.1201667. [DOI] [PubMed] [Google Scholar]
- 25.Hofferer M, Wirbelauer C, Humar B, Krek W. Increased levels of E2F-1-dependent DNA binding activity after UV- or γ-irradiation. Nucleic Acids Res. 1999;27:491–495. doi: 10.1093/nar/27.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsiao K-M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 27.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 28.Johnson D G, Schneider-Broussard R. Role of E2F in cell cycle control and cancer. Front Biosci. 1998;3:447–458. doi: 10.2741/a291. [DOI] [PubMed] [Google Scholar]
- 29.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krek W, Livingston D M, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 31.Krek W, Xu G, Livingston D M. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 32.Kumar A P, Butler A P. Serum responsive gene expression mediated by Sp1. Biochem Biophys Res Commun. 1998;252:517–523. doi: 10.1006/bbrc.1998.9676. [DOI] [PubMed] [Google Scholar]
- 33.Lam E W-F, Bennett J D, Watson R J. Cell-cycle regulation of human B-myb transcription. Gene. 1995;160:277–281. doi: 10.1016/0378-1119(95)00184-8. [DOI] [PubMed] [Google Scholar]
- 34.Lam E W-F, Watson R J. An E2F-binding sites mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Thangue N B. E2F and the molecular mechanisms of early cell-cycle control. Biochem Soc Trans. 1996;24:54–59. doi: 10.1042/bst0240054. [DOI] [PubMed] [Google Scholar]
- 36.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu N, Lucibello F C, Engeland K, Muller R. A new model of cell cycle-regulated transcription: repression of the cyclin A promoter by CDF-1 and anti-repression by E2F. Oncogene. 1998;16:2957–2963. doi: 10.1038/sj.onc.1201838. [DOI] [PubMed] [Google Scholar]
- 38.Liu N, Lucibello F C, Korner K, Wolfraim L A, Zwicker J, Muller R. CDF-1, a novel E2F-unrelated factor, interacts with cell cycle-regulated repressor elements in multiple promoters. Nucleic Acids Res. 1997;25:4915–4920. doi: 10.1093/nar/25.24.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 40.Luscher B, Eisenman R N. New light on Myc and Myb. Part II. Myb. Genes Dev. 1990;4:2235–2241. doi: 10.1101/gad.4.12b.2235. [DOI] [PubMed] [Google Scholar]
- 41.MacGregor G R, Caskey C T. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 1989;17:2365. doi: 10.1093/nar/17.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 43.Muller R. Transcriptional regulation during the mammalian cell cycle. Trends Genet. 1995;11:173–178. doi: 10.1016/S0168-9525(00)89039-3. [DOI] [PubMed] [Google Scholar]
- 44.Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 45.Neuman E, Flemington E K, Sellers W R, Kaelin W G. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 47.Nicolaides N C, Gualdi R, Casadevall C, Manzella L, Calabretta B. Positive autoregulation of c-myb expression via Myb binding sites in the 5′ flanking region of the human c-myb gene. Mol Cell Biol. 1991;11:6166–6176. doi: 10.1128/mcb.11.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ormondroyd E, de la Luna S, La Thangue N B. A new member of the DP family, DP3, with distinct protein products suggests a regulatory role for alternative splicing in the cell cycle transcription factor DRTF1/E2F. Oncogene. 1995;11:1437–1446. [PubMed] [Google Scholar]
- 50.Pelicci P G, Lanfrancone L, Brathwaite M D, Wolman S R, Dalla-Favera R. Amplification of the c-myb oncogene in a case of human acute myelomonocytic leukemia. Science. 1984;224:1117–1121. doi: 10.1126/science.6585957. [DOI] [PubMed] [Google Scholar]
- 51.Qin X, Chittenden T, Livingston D M, Kaelin W G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 52.Rogers K T, Higgins P D R, Milla M M, Phillips R S, Horowitz J M. DP-2, a heterodimeric partner of E2F: identification and characterization of DP-2 proteins expressed in vivo. Proc Natl Acad Sci USA. 1995;92:7594–7599. doi: 10.1073/pnas.93.15.7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sala A, Nicolaides N C, Engelhard A, Bellon T, Lawe D C, Arnold A, Graña X, Giordano A, Calabretta B. Correlation between E2F-1 requirement in the S phase and E2F-1 transactivation of cell cycle-related genes in human cells. Cancer Res. 1994;54:1402–1406. [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Sardet C, LeCam L, Fabbrizio E, Vidal M. E2Fs and the retinoblastoma protein family. In: Ghysdael J, Yaniv M, editors. Oncogenes as transcriptional regulators. Berlin, Germany: Birkhauser Verlag; 1997. pp. 1–63. [Google Scholar]
- 56.Schulze A, Zerfass K, Spitkovsky S, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sears R, Ohtani K, Nevins J R. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sellers W R, Rodgers J W, Kaelin W G. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shan B, Zhu X, Chen P-L, Durfee T, Yang Y, Sharp D, Lee W-H. Molecular cloning of cellular genes encoding retinoblastoma-associated proteins: identification of a gene with properties of the transcription factors E2F. Mol Cell Biol. 1992;12:5620–5631. doi: 10.1128/mcb.12.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. The transcription factor E2f interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 61.Singh H, Sen R, Baltimore D, Sharp P A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 62.Slamon D J, Boone T C, Murdock D C, Keith D E, Press M F, Larson R A, Souza L M. Studies of the human c-myb gene and its product in human acute leukemias. Science. 1986;233:347–351. doi: 10.1126/science.3014652. [DOI] [PubMed] [Google Scholar]
- 63.Slamon D J, DeKernion J B, Verma I M, Cline M J. Expression of cellular oncogenes in human malignancies. Science. 1984;224:256–262. doi: 10.1126/science.6538699. [DOI] [PubMed] [Google Scholar]
- 64.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 65.Stern J B, Smith K A. Interleukin-2 induction of T-cell G1 progression and c-myb expression. Science. 1986;233:203–206. doi: 10.1126/science.3523754. [DOI] [PubMed] [Google Scholar]
- 66.Thiele C J, Cohen P S, Israel M A. Regulation of c-myb expression in human neuroblastoma cells during retinoic acid-induced differentiation. Mol Cell Biol. 1988;8:1677–1683. doi: 10.1128/mcb.8.4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thiele C J, McKeon C, Triche T J, Ross R A, Reynolds C P, Israel M A. Differential protooncogene expression characterizes histopathologically indistinguishable tumors of the peripheral nervous system. J Clin Investig. 1987;80:804–811. doi: 10.1172/JCI113137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson C B, Challoner P B, Neiman P E, Groudine M. Expression of the c-myb proto-oncogene during cellular proliferation. Nature. 1986;319:374–380. doi: 10.1038/319374a0. [DOI] [PubMed] [Google Scholar]
- 69.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 70.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 71.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Tumor induction and tissue atrophy in mice lacking E2F1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 72.Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins J R, Williams R S. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yokota J, Tsunetsugu-Yokota Y, Bultifora H, Fevre C L, Cline M J. Alterations of the myc, myb and Ha-ras proto-oncogenes in cancers are frequent and show clinical correlations. Science. 1986;231:261–265. doi: 10.1126/science.3941898. [DOI] [PubMed] [Google Scholar]
- 74.Zhu L, Zhu L, Xie E, Chang L-S. Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol Cell Biol. 1995;15:3552–3562. doi: 10.1128/mcb.15.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]