Abstract
Objective
To characterize the Staphylococcus aureus strains colonizing healthy Spanish children.
Methods
Between March and July 2018, 1876 Spanish children younger than 14 years attending primary healthcare centers were recruited from rural and urban areas. Staphylococcus aureus colonization of the anterior nostrils was analyzed. MecA and mecC genes, antibiotic susceptibility, and genotyping according to the spa were determined in all strains, and the following toxins were examined: Panton‐Valentine leucocidin (pvl), toxic shock syndrome toxin (tst), and exfoliative toxins (eta, etb, etd). Multilocus sequence typing (MLST) and staphylococcal cassette chromosome (SCCmec) typing were performed on methicillin‐resistant Staphylococcus aureus (MRSA) strains, as well as pulsed‐field gel electrophoresis (PFGE).
Results
619 strains were isolated in 1876 children (33%), and 92% of them were sent for characterization to the Spanish National Centre of Microbiology (n = 572). Twenty (3.5%) of these strains were mecA‐positive. Several spa types were detected among MRSA, being t002 the most frequently observed (30%), associating with SCCmec IVc. Among MSSA, 33% were positive for tst, while only 0.73% were positive for pvl. The 20 MRSA strains were negative for pvl, and 6 (30%) harbored the tst gene.
Conclusions
methicillin‐resistant Staphylococcus aureus nasal colonization in Spanish children is rare, with t002 being the most observed spa type, associated with SCCmec IVc. None of the MRSA strains produced pvl, but up to 30% of S. aureus strains were positive for tst.
Keywords: methicillin‐resistant, molecular characterization, nasal colonization, Staphylococcus aureus
This is one of the largest epidemiological studies in which we have screened up to 1876 healthy Spanish children for methicillin‐resistant Staphylococcus aureus (MRSA) colonization. All the isolated strains have been characterized by analyzing their genotype, the presence of mecA and mecC genes, and their antibiotic susceptibility. In addition, Panton‐Valentine leucocidin, toxic shock syndrome toxin, and exfoliative toxins production were analyzed. This epidemiological surveillance study leads to a better understanding of the strain flow and the rate of MRSA nasal carriage among Spanish children, and the prevalence of virulence genes and antimicrobial sensitivities of colonizing strains throughout the country.

1. INTRODUCTION
Staphylococcus aureus causes most of the skin and soft tissue infections (SSTIs) in children, such as boils, carbuncles, abscesses (Barrios Lopez et al., 2013), and other more complicated infections, including septic arthritis, osteomyelitis, necrotizing fasciitis, pneumonia, endocarditis, and sepsis (Lowy, 1998; Tong et al., 2015). The nasal cavity is considered the primary anatomical site for S. aureus colonization (Kluytmans et al., 1997; Verhoeven et al., 2014), although it can also be found in the axilla, perineum, rectum, and throat. Strains of methicillin‐resistant Staphylococcus aureus (MRSA) present affinity to human skin and mucous thanks to the phenol‐soluble modulin (PSM) peptides PSMα1 and PSMα2 (Joo et al., 2011). In the last decades, an increase in community‐associated MRSA infections (CA‐MRSA) has been observed, causing SSTIs in healthy individuals, often children (Watkins et al., 2012). In Spain, the first cases were reported in 2006 (Broseta et al., 2006), although very few data regarding these infections in the pediatric population are yet available in many European countries (Del Rosal et al., 2020). Healthy children could constitute an MRSA reservoir in the community, and an elevated rate of colonization increases the risk of future infections (Turner et al., 2019). Methicillin‐sensitive S. aureus (MSSA) strains producing Panton‐Valentine leucocidin (PVL) represent a public health threat, given virulence factors frequently transfer between strains (Rasigade et al., 2010). Data regarding MSSA and MRSA colonization in children are scarce in Spain. We recently studied the prevalence of nasal colonization of MSSA and MRSA in community‐dwelling Spanish children, analyzing risk factors for this colonization, in a community nationwide surveillance study: Colonization by S. aureus in the Community (COSACO) (Del Rosal et al., 2020). The observed prevalence of S. aureus colonization was 33% and of MRSA 1.44%. In this manuscript, we report the characterization of the S. aureus strains isolated in the COSACO study.
2. METHODS
2.1. Study participants and samples
We conducted a nationwide, observational, cross‐sectional multicenter study on children from community settings throughout Spain (the COSACO study; Del Rosal et al., 2020). Between March and July 2018, we recruited patients younger than 14 years attending primary healthcare centers without active infections. Seventy primary healthcare centers from all Spanish regions and in both rural and urban settings took part in the study. According to region features and population density, at least two healthcare centers (from rural, urban, and mixed areas) were selected to take part in the study. Nasal swabs from every patient were collected following the study protocol (Del Rosal et al., 2020). Cultures of nasal swabs, strain isolation, and antimicrobial susceptibility testing of isolates were performed by 27 hospital‐based microbiology laboratories.
2.2. Methicillin resistance detection and staphylococcal cassette chromosome mec typing
For detecting methicillin resistance, a disk diffusion method with cefoxitin (30 µg) was employed, as well as susceptibility testing or disk diffusion (Del Rosal et al., 2020). Also, minimum inhibitory concentrations (MICs) for MRSA strains were studied with E‐test® strips (Biomerieux). The methods and antimicrobial susceptibility interpretations were following the European Committee on Antimicrobial Susceptibility Testing guidelines (EUCAST, 2018). When an isolate showed an inhibition zone smaller than 20 mm surrounding the cefoxitin disk (Kateete et al., 2019), we screened the presence of the mecA and mecC genes using primers by polymerase chain reaction (PCR). S. aureus strains were considered MRSA if the mecA or mecC gene was detected. Cefoxitin‐resistant strains without mecA/mecC detection were excluded from the analysis. MRSA isolates were characterized by pulsed‐field gel electrophoresis (PFGE), spa typing, multilocus sequence typing (MLST), and staphylococcal cassette chromosome mec (SCCmec) typing. We screened for the presence of genes that code for the following virulence factors by multiplex PCR reactions: pvl, tst, eta, etb, and etd. The mecA gene was detected by PCR, as described by Geha et al. (1994). The mecC gene was studied according to Garcia‐Alvarez et al. (2011). SCCmec types were determined using a multiplex PCR strategy, which generates an amplification pattern for each SCCmec structural type (Oliveira & de Lencastre, 2002). Additional typing of the isolates was performed by 2 different PCR methods to detect the SCCmec IV subtypes IVa, IVb, IVc, IVd, and IVh (Milheirico et al., 2007) and SCCmec type V (Zhang et al., 2005). PCR‐positive controls were used as previously described (Vindel et al., 2009).
2.3. Pulsed‐field gel electrophoresis, spa typing, BURP analysis, multilocus sequence typing, and toxin study
All the strains of MSSA and MRSA were characterized for spa typing, which is the established method based on sequencing of a single polymorphic variable‐number tandem repeat, namely the repeat region of the S. aureus protein A gene. This spa typing method is highly discriminatory, and it has been used for studying the nature of different outbreaks and for assigning strains to phylogenetic lineages in epidemiological studies. The spa type was assigned by employing the Ridom StaphType software, and, besides this, clustering of the isolates was performed by the BURP (Based Upon Repeat Pattern) algorithm implemented in StaphType. Isolates with spa types with more than five repeats were clustered into different groups (CCs), with the calculated cost between members of a group being ≤6 (Mellmann et al., 2007).
We determined the bacterial MLST profiles as described in Enright et al., 2000. Allelic profiles and sequence types were assigned using the MLST database (http://www.mlst.net). The presence of genes coding for the virulence factors pvl, tst, eta, etb, and etd was screened by multiplex PCR reactions following the method described by Vandenesch et al., 2003.
All MRSA isolates and those with frequently occurring spa types, like t002‐CC002 strains, were genotyped by PFGE after SmaI digestion of chromosomal DNA, prepared using a modification of the protocol described by Cookson et al. (2007). Analysis of the gels was performed according to the criteria of Tenover et al. (1995), and a dendrogram was constructed with Molecular Analyst software (Bio‐Rad) using the Dice correlation coefficient and the unweighted pair‐group method, with averages having a tolerance position of 0.8%. Positive controls of various PFGE profiles were used as previously described by our group (Vindel et al., 2009). In addition, a prototype USA300 strain kindly provided by Professor Herminia de Lencastre was also used as a PFGE control.
3. RESULTS
3.1. Bacterial isolates and patient population
A total of 1876 patients (aged 7.01 ± 4.38 years) were enrolled. The prevalence of S. aureus colonization was 33% (95% CI: 30.8–35.1) and of MRSA 1.44% (95% CI: 0.78–2.1) (Del Rosal et al., 2020). A total of 572 S. aureus strains (92% of the isolated strains were from the COSACO study) were ultimately sent for molecular characterization to the National Centre of Microbiology (Majadahonda, Spain) (1 sample/child); 20 (3.5%) of them were mecA‐positive (MRSA) and were isolated from throughout the country (Figure 1). Colonization by S. aureus was more frequently observed in older children, males, urban settings, and in those with chronic diseases. In addition, there was a higher MRSA colonization rate among children living in rural environments (Del Rosal et al., 2020).
FIGURE 1.
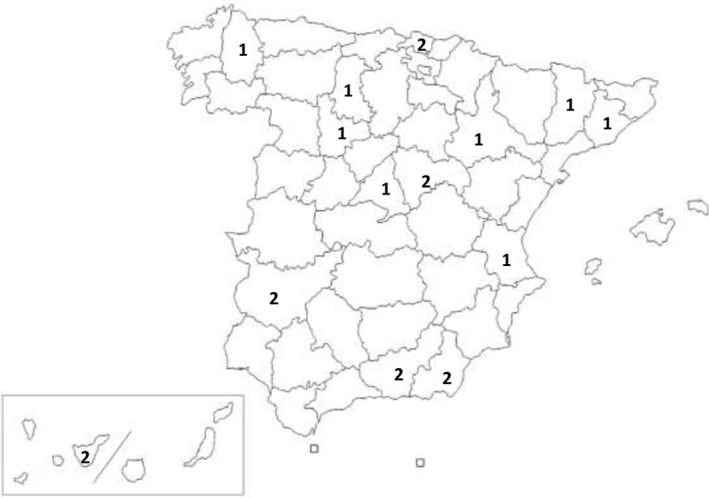
Map of Spain indicating the location of the twenty Staphylococcus aureus mecA‐positive strains. The homogeneous distribution of the strains in different areas of Spain is shown: 3.50% (20/572)
3.2. Population structure of the community‐associated methicillin‐resistant Staphylococcus aureus
Twenty oxacillin‐resistant strains were confirmed to carry the mecA gene. In general, these strains presented a low level of oxacillin resistance, with only 4 strains having a MIC ≥32 mg/L. The MRSA strains were isolated from 8 girls and 12 boys (Table 1).
TABLE 1.
Characterization of MRSA strains
| Cosaco sample number | MEC A | SCC MEC | Subtype IV | Toxins | PFGE |
SPA‐type clonal clusters |
MLST |
Antimicrobials MIC (mg/l) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVL | TSST | ETA | ETB | ETD | O | V | T | L | D | E | CM | SXT | |||||||
| 62 | + | IV | IVc | − | − | − | − | − | E1 | t002‐CC002 | Undefined | 8 | 1 | 1 | 1.50 | 0.12 | 0.12 | 0.023 | 0.023 |
| 87 | + | IV | IVc | − | − | − | − | − | E1 | t002‐CC002 | ST5115 | 4 | 1 | 1.50 | 2 | 0.12 | 0.12 | 0,094 | 0.032 |
| 109 | + | IV | IVc | − | − | − | − | − | G1 | t4100‐Singleton | ST72 | 32 | 1.50 | 1.50 | 1.50 | 0.25 | 0.12 | 0.094 | 0.032 |
| 133 | + | IV | IVc | − | − | − | − | − | F1 | t012‐CC012 | ST30 | 6 | 1 | 0.75 | 1.5 | 0.19 | 0.12 | 0.064 | 0.047 |
| 134 | + | IV | IVc | − | − | − | − | − | F1 | t012‐CC012 | ST30 | 4 | 1 | 0.75 | 1.5 | 0.12 | 0.12 | 0.094 | 0.047 |
| 147 | + | IV | IVa | − | + | − | − | − | E6 | t002‐CC002 | ST5 | 6 | 1 | 1 | 2 | 0.19 | 0.12 | 0.094 | 0.032 |
| 150 | + | IV | IVc | − | − | − | − | − | E7 | t002‐CC002 | ST5 | 12 | 1 | 1 | 2 | 0.12 | 0.12 | 0.094 | 0.032 |
| 168 | + | IV | IVc | − | − | − | − | − | E5 | t067‐CC002 | ST125 | >256 | 1.50 | 1.50 | 2 | 0.19 | 0.12 | 0.094 | 0.047 |
| 171 | + | IV | IVc | − | − | − | − | − | G2 | t20101‐Singleton | ST72 | 16 | 1.50 | 1.50 | 2 | 0.19 | 0.12 | 0.125 | 0.047 |
| 181 | + | IV | IVa | − | − | − | − | − | K | t4407‐ Singleton | ST6 | 24 | 1 | 1.50 | 2 | 0.19 | 24 | 0.094 | 0.032 |
| 232 | + | IV | IVa | − | − | − | − | − | F2 | t1507‐Singleton | ST30 | 16 | 1 | 1 | 0.75 | 0.12 | 0.25 | 0.125 | 0.032 |
| 233 | + | IV | IVc | − | − | − | − | − | E3 | t450‐Singleton | ST5115 | 3 | 1 | 1 | 2 | 0.19 | 0.12 | 0.064 | 0.032 |
| 258 | + | IV | IVc | − | + | − | − | − | L | t2532‐CC002 | ST125 | 1 | 1 | 0.50 | 1.50 | 0.12 | 12 | 0.094 | 0.032 |
| 320 | + | IV | IVc | − | − | − | − | − | E4 | t067‐CC002 | ST125 | 64 | 1 | 1 | 1.5 | 0.19 | 12 | 0.094 | 0.032 |
| 385 | + | II | ND | − | + | − | − | − | N | t223‐CC223 | ST22 | 1.50 | 1 | 0.75 | 1 | 0.12 | 0.12 | 0.094 | 0.032 |
| 443 | + | IV | IVc | − | + | − | − | − | E4 | t002‐CC002 | ST5 | 4 | 1 | 1.50 | 2 | 0.19 | 0.12 | 0.094 | 0.023 |
| 453 | + | IV | IVc | − | + | + | + | − | E2 | t002‐CC002 | ST5 | 12 | 1 | 1 | 2 | 0.19 | 12 | 0.190 | 0.032 |
| 464 | + | IV | IVa | − | − | − | − | − | O | t790‐CC223 | ST22 | 1.50 | 1 | 0.75 | 1.50 | 0.19 | 0.09 | 0.094 | 0.047 |
| 472 | + | ND | ND | − | − | − | − | − | F3 | t4867‐CC002 | ST34 | 1.50 | 1 | 0.75 | 0.75 | 0.12 | 0.25 | 0.094 | 0.094 |
| 537 | + | ND | ND | − | + | − | − | − | Q | t021‐CC012 | Undefined | 48 | 1 | 1 | 1.50 | 0.25 | 0.19 | 0.094 | 0.047 |
Antimicrobials: CM, clindamycin; D, daptomycin; E, erythromycin; L, linezolid; O, oxacillin; SXT, cotrimoxazole; T, teicoplanin; V, vancomycin. Mec A: gene mecA. MLST: multilocus sequence typing, an unambiguous procedure for characterizing isolates of bacterial species using the sequences of internal fragments of 7 housekeeping genes. ND: not determined. PFGE: pulsed‐field gel electrophoresis (E1, E2, E3, E4, different among them in 1, 2, 3 bounds; closely related); (F1 and F2, closely related among themselves); (G1 and G2, 2 different bands, closely related strains); the remainder of the strains have different profiles. SCC MEC: staphylococcal cassette chromosome mec. Spa‐type: the composition of the variable number of tandem repeats in the 3´ end of the staphylococcal protein A gene (spa). Clonal clusters (CCs), according to BURP analysis. Toxins: PVL, Panton‐Valentine leucocidin; TSST, toxic shock syndrome toxin; ETA, exfoliative toxin A; ETB, exfoliative toxin B.
3.2.1. Molecular characterization of methicillin‐resistant Staphylococcus aureus
Several PFGE profiles and their respective dendrogram were obtained (Table 1; Figure 2). According to Tenover (≥80% similitude) [21], they belonged to 8 types, with the following subtypes: E (7), F (2), G (2), K (1), L (1), N (1), Q (1), and Z (1). The 2 G subtypes were similar to the USA300 profile, with a similitude grade of 84.6%. Thirteen different spa types were observed in the 20 MRSA strains, belonging to 3 clonal complexes or clonal clusters: CC002 (t002, t067, t2532, t4867), CC012 (t012, t021), CC223 (t223, t790) and 5 singleton (t4407, t20101, t1507, t450) (Table 1). Among the MSSA strains (Table 2), the most prevalent was t012‐CC012, with 41 strains (27 harbored the tst gene); t021‐CC012 had 25 (17 with the tst gene); t375‐CC375 had 23 (with no tst gene); t166‐CC166 had 22 (with 18 tst genes); t223 had 21 (with 17 tst genes); and unknown had 26 (with 10 tst genes). Only 1 strain (t355‐Singleton) was positive for the pvl gene (Table 2). To study a possible relationship among the strains with the same spa type, we performed a PFGE of all t002‐CC002 strains (6 MRSA and 14 MSSA), given this was the most frequently observed spa type in MRSA strains and, besides this, t002 occupies the third place in the list of Ridom SpaServer (MSSA and MRSA) database frequencies. They proved to be different clones (according to Tenover criteria) (Figure 3).
FIGURE 2.
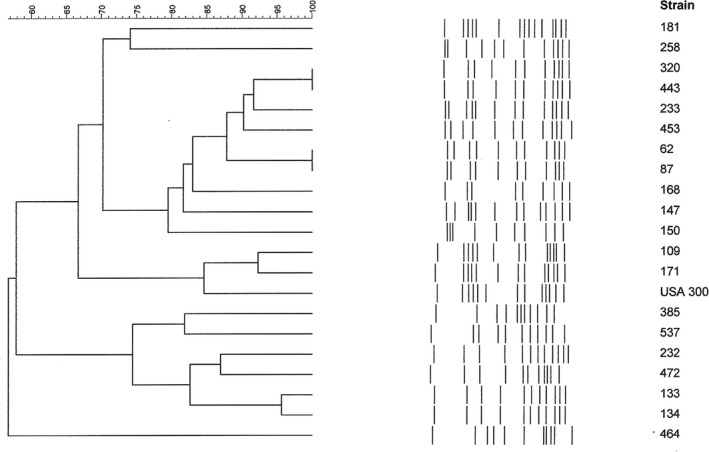
PFGE pattern of MRSA isolates. Dendrogram resulting from the comparison of PFGE band restriction patterns of the 20 MRSAs (mecA +) with the USA300 clone band pattern
TABLE 2.
Frequency of spa‐types/clonal complexes among isolated Staphylococcus aureus
| Clonal complexes | spa‐type | MRSA | MSSA |
|---|---|---|---|
| F | F | ||
| CC015 | t069, t073, t1170, t1268, t1523, t1618, t1985, t2536, t2869, t3085, t3537, t620, t630, t6606, t6704, t7669, t798 | 0 | 1 |
| t031, t302, t728 | 0 | 2 | |
| t050 | 0 | 3 | |
| t230 | 0 | 4 | |
| t1451 | 0 | 8 | |
| t571 | 0 | 12 | |
| t015 | 0 | 15 | |
| CC012 | t093, t1626, t1654, t2147, t253, t2557, t2579, t297, t318, t3382, t363, t6511, t7085, t822, t840, t932 | 0 | 1 |
| t037, t1070, t7785 | 0 | 2 | |
| t018, t122 | 0 | 4 | |
| t019 | 0 | 10 | |
| t021** | 1 | 25 | |
| t012* | 2 | 41 | |
| CC002 | t1107, t1277, t151, t2066, t2168, t2264, t242, t509, t653, t8348 | 0 | 1 |
| t045, t1094 | 0 | 2 | |
| t2167 | 0 | 3 | |
| t2532, t4867 | 1 | 0 | |
| t067 | 2 | 1 | |
| t002 | 6 | 12 | |
| CC166 | t3905, t3909, t6390, t864, t884 | 0 | 1 |
| t136, t352, t862 | 0 | 2 | |
| t369, t3906 | 0 | 3 | |
| t1057 | 0 | 5 | |
| t240 | 0 | 11 | |
| t166*** | 0 | 21 | |
| CC084 | t1875, t1957, t228, t2325, t279, t368, t393, t803 | 0 | 1 |
| t094, t1877 | 0 | 2 | |
| t085 | 0 | 3 | |
| t084 | 0 | 15 | |
| CC159 | t1425, t1688, t1994, t269, t3204, t3454 | 0 | 1 |
| t272, t645 | 0 | 3 | |
| t159 | 0 | 5 | |
| CC223 | t1433, t309, t4565 | 0 | 1 |
| t712, t233 | 0 | 2 | |
| t9606 | 0 | 3 | |
| t005 | 0 | 4 | |
| t790 | 1 | 0 | |
| t223**** | 1 | 22 | |
| CC330/065 | t130, t330, t560, t706 | 0 | 1 |
| t4545, t880 | 0 | 2 | |
| t065 | 0 | 12 | |
| CC081 | t056, t081, t1315, t472, t7475 | 0 | 1 |
| t078 ψ | 0 | 4 | |
| CC616 | t1722, t616, t825 | 0 | 1 |
| t1406 | 0 | 2 | |
| t364 | 0 | 5 | |
| t493 | 0 | 19 | |
| CC008 | t068, t8163 | 0 | 1 |
| t024 | 0 | 2 | |
| t701 | 0 | 4 | |
| t008 | 0 | 8 | |
| CC375 | t1691, t525 | 0 | 1 |
| t375 | 0 | 23 | |
| Singleton | t1194, t1328, t1639, t1721, t20074 #, t20077, t20078 #, t20079, t20080 Ω, t20081, t20082 Φ, t20083 #, t20088 #, t20094, t20095, t20096 #, t3072, t355 ƿ, t4463, t4600, t818, t937 | 0 | 1 |
| t1507, t20101, t4100, t4407, t450 | 1 | 0 | |
| t267, t20076 | 0 | 2 | |
| t127, t189 | 0 | 3 | |
| t20073 | 0 | 4 | |
| t209 Φ Φ | 0 | 6 | |
| No founder | t091, t10234, t10389, t1152, t11791, t1192, t12198, t12228, t12238, t12818, t12831, t13748, t1456, t1537, t1617, t1642, t1784, t18809, t1977, t2088, t2435, t248, t2637, t275, t287, t3272, t333, t339, t3638, t399, t4523, t515, t529, t5891, t638, t693, t777, t8025, t809, t929, t949, t9931 | 0 | 1 |
| t026, t20076, t4446, t458, t528, t 5738, t748, t779 | 0 | 2 | |
| t1445 ƿ ƿ ƿ | 0 | 3 | |
| t148 | 0 | 4 |
F, Frequency, the number of times that the spa lineage occurred in 545 MSSA and 20 MRSA. # 1 toxic shock syndrome toxin (TSST) positive; *27 TSST positives; **17 TSST positives; *** 18 TSST positives; **** 17 TSST positives; Φ 1 exfoliative toxin A (ETA) positive; Φ Φ 5 ETA positives; Ω 1 exfoliative toxin D (ETD) positive; ψ 3 ETD positives; ƿ 1 PVL positive; ƿ ƿ ƿ 3 PVL positives
FIGURE 3.
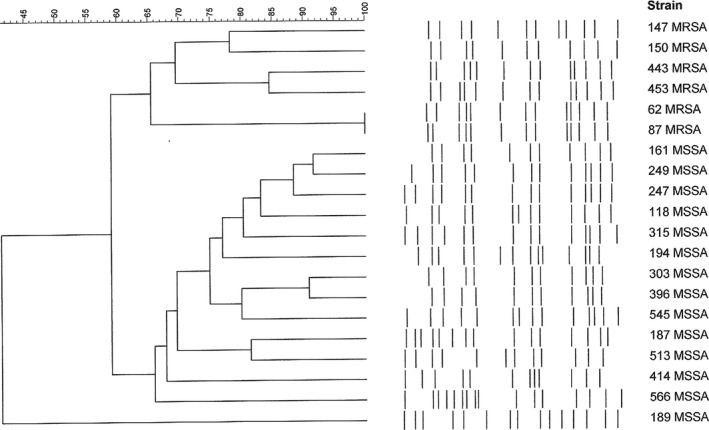
PFGE pattern of t002 S. aureus strains. Dendrogram resulting from the comparison of PFGE band restriction patterns of the 6 MRSAs with 14 MSSA isolates; both groups harbor the t002 spa‐type. PFGE clusters were defined based on a similarity of 80% or higher
We compared the MRSA strains according to their MLST profiles. The most abundant MLST types were ST5 (4 strains) and ST125 (3 strains). Two of the former 3 strains harbored the spa type t067‐CC002 and presented a high oxacillin MIC. Seventeen of the 20 MRSA strains harbored SCCmec IV (85%); among them, 13 were IVc subtypes.
We analyzed the distribution of the S. aureus strains according to toxin production. None of the MRSA strains was pvl‐positive, 6 were tst positive and only 1 harbored eta and etb genes. On the contrary, 4 of the MSSA strains were positive for pvl (1 harbored spa type t355‐Singleton and 3 the t1445‐Singleton type), 180 were positive for the tst gene (28 harbored spa type t012‐CC012; 18 type t166‐CC166; 17 type t021‐CC012 and t223‐CC223; and 2 the t002‐CC002 type), among the more representative spa types (Table 3).
TABLE 3.
Distribution of toxins among all the Staphylococcus aureus strains a
| mecA strains (−) (n = 545) | mecA strains (+) (n = 20) | |||
|---|---|---|---|---|
| (+) | (‐) | (+) | (−) | |
| PVL | 4 (0.73%) | 541 (99.27%) | 0 | 20 (100%) |
| TSST | 180 (33.03%) | 365 (66.97%) | 6 (30%) | 14 (70%) |
| ETA | 23 (4.22%) | 522 (95.78%) | 1 (5%) | 19 (95%) |
| ETB | 8 (1.47%) | 537 (98.53%) | 1 (5%) | 19 (95%) |
| ETA+ETB | 4 (0.72%) | 541 (99.27%) | 0 | 20(100%) |
| ETD | 10 (1.83%) | 535 (98.17%) | 0 | 20 (100%) |
ETA, exfoliative toxin A; ETB, exfoliative toxin B; ETD, exfoliative toxin D; PVL, Panton‐Valentine leucocidin; TSST, toxic shock syndrome toxin.
Seven more mecA (−) and mecC (−) strains are not included because they have decreased sensitivity to oxacillin that should be more widely studied. Two of these strains were TSST (+) and 1 ETD (+).
3.2.2. Resistance profile (antimicrobial susceptibility)
All the MRSA strains were sensitive to vancomycin, teicoplanin, linezolid, daptomycin, clindamycin, and trimethoprim‐sulfamethoxazole, although four were resistant to erythromycin, according to the European Committee on Antimicrobial Susceptibility Testing [15]. Two of these strains were ST125 and 1 ST6, and the other was ST5.
4. DISCUSSION
In our COSACO study, the MRSA prevalence in Spanish children was 1.4%. This prevalence was higher among younger children and in those living in rural areas (Del Rosal et al., 2020). In this manuscript, we have focused on molecular characterization and toxin production from 572 strains, 92% of the S. aureus strains isolated in the COSACO study. Regarding MRSA strains, t002‐CC002 was the most frequently observed spa type, associated with the SCCmec IVc type. In MSSA, the most prevalent spa types were t012‐CC012, t021‐CC012, and t375‐CC375. Up to 30% of the MSSA and MRSA strains were positive for tst. The detection of pvl was exceptionally in MSSA strains; none of the MRSA strains were positive for the pvl gene. In our study, we did not detect mecA or mecC in 7 of 27 cefoxitin‐resistant strains. Some cefoxitin‐resistant S. aureus strains are not linked to the presence of mec genes, being the resistance caused by hyperproduction of β‐lactamase or mutations in the penicillin‐binding protein 4 promoter or other genes (Argudín et al., 2018). Given we have focused on the characterization of mecA/mecC‐positive MRSA, these 7 strains were excluded from the analysis. A strong evolutionary relationship has previously been revealed between clinical and nasal colonization isolates (Lamers et al., 2011). In addition, nasal decolonization of MRSA has been reported to reduce the progression to infection (van Rijen et al., 2008). For this reason, the performance of surveillance studies like ours is relevant.
Children present a higher risk of infection from a certain type of SCCmec, such as SCCmec type IV, compared to adults (David & Daum, 2010). In Israel, Rokney et al. found that the clone ST5‐t002‐CC002‐IV‐pvl+ was the main cause of community‐associated staphylococcal infections in the pediatric population (Rokney et al., 2019). Some 17.8% of these strains were non‐susceptible to erythromycin and clindamycin. Similarly, we observed that 85% of the MRSA strains in our study were SCCmec type IV; among these, 35% were t002‐CC002. However, only 1 of our strains was erythromycin‐resistant, and none of them were pvl‐positive or clindamycin‐resistant. In a study performed in Colombian children, the authors observed that MSSA strains presented more diverse and frequent virulence genes than MRSA strains. In addition, SCCmec type IVc was the most commonly observed one among MRSA strains (Jiménez et al., 2011). In our study, we observed a similar distribution.
As for toxins, we must pay attention to the tst gene, which is present in up to 30% of the mecA‐negative and mecA‐positive strains that colonized the children in our study. Eight PFGE profiles were obtained among the 20 MRSA mecA‐positive strains. It is necessary to point out that 2 G subtypes were similar to the USA300 profile; although this is a low number, it is necessary to keep in mind.
We also used PFGE to compare strains with the same spa type. For this reason, we chose t002‐CC002 strains (6 MRSA and 14 MSSA), given this was the most abundant spa‐type observed among the MRSA strains and the third in relative global frequencies of spa‐type occurrences in the Ridom SpaServer (MSSA and MRSA). We compared the spa types of the COSACO MRSA strains with the MRSA strains received from various geographic areas around the country in the National Reference Laboratory of Staphylococci, from the National Centre of Microbiology (Majadahonda, Spain). As in the COSACO study, we also found t002‐CC002 and t067‐CC002 spa types among the strains received in the center during the same study period (data not shown); spa types t012‐CC012, t021‐CC012, and t223‐CC223 were represented in this group of strains, as we had observed in the COSACO MRSA strains. Most of these strains were isolated in adults in both in‐hospital and community‐based populations. Other authors have also reported that t067‐CC002 and t002‐CC002 are dominant among MRSA strains isolated in Spain, in contrast to the relatively low frequency of these spa types in other European countries (Vindel et al., 2009).
One of our limitations is that we have only sampled the anterior nares, not studying other sites of colonization. Therefore, we could have underestimated the prevalence of S. aureus colonization in our population. Also, we have studied only one isolate from each patient. Le et al. studied whether one isolate is enough for identifying individuals who are S. aureus colonized in the community (Le et al., 2018). Among their participants, 89% intranasally carried a single S. aureus spa type, 81.4% when they analyzed mecA‐positive strains. According to this, our results could have slightly underestimated the prevalence.
It has been suggested that older children are more prone to colonization with S. aureus (Lamaro‐Cardoso et al., 2009). Other authors have studied younger children, including only children younger than 5 years of age (Kateete et al., 2019; Tavares et al., 2010). In our case, we included patients up to 14 years of age recruited during a short period (4 months who lived in both rural and urban settings throughout the country. This approach has lead to a better understanding of the strain flow and allowed us to demonstrate that the prevalence of MRSA nasal carriage among children was low in the country. Our study suggests the need for permanent active surveillance of the mentioned carrier strains.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Federico Roman: Formal analysis (lead); Investigation (equal); Supervision (equal); Writing‐original draft (equal). Ana Mendez‐Echevarria: Conceptualization (lead); Funding acquisition (lead); Investigation (equal); Methodology (equal); Supervision (equal); Writing‐original draft (supporting); Writing‐review & editing (lead). Teresa Del Rosal: Conceptualization (supporting); Funding acquisition (supporting); Investigation (equal); Supervision (equal); Writing‐review & editing (equal). Cesar Garcia‐Vera: Conceptualization (equal); Funding acquisition (supporting); Investigation (supporting); Methodology (equal); Validation (supporting); Writing‐review & editing (supporting). Luis Escosa‐Garcia: Conceptualization (supporting); Data curation (supporting); Funding acquisition (supporting); Methodology (supporting); Writing‐review & editing (equal). Martin Agud: Data curation (equal); Investigation (equal); Writing‐review & editing (equal). Fernando Chaves: Conceptualization (equal); Methodology (equal); Writing‐original draft (supporting); Writing‐review & editing (equal). Jose Gutierrez‐Fernandez: Data curation (equal); Investigation (equal); Writing‐review & editing (equal). Enrique Ruiz de Gopegui: Data curation (equal); Investigation (equal); Validation (equal); Writing‐review & editing (equal). Guillermo Ruiz‐Carrascoso: Data curation (equal); Investigation (equal); Validation (equal); Writing‐review & editing (equal). Maria del Carmen Ruiz‐Gallego: Data curation (equal); Validation (equal); Writing‐review & editing (equal). Albert Bernet: Data curation (equal); Investigation (equal); Validation (equal); Writing‐review & editing (equal). Sara Maria Quevedo: Data curation (equal); Investigation (equal); Validation (equal); Writing‐review & editing (equal). Ana Maria Fernandez‐Verdugo: Data curation (equal); Investigation (equal); Writing‐review & editing (equal). Talia Sainz: Conceptualization (equal); Methodology (equal); Writing‐review & editing (equal). Cristina Calvo: Conceptualization (equal); Data curation (equal); Investigation (equal); Methodology (equal); Writing‐review & editing (equal).
ETHICS STATEMENT
Informed consent was obtained from the parents or guardians of all the children before their inclusion, as well as from those patients aged 12 years or older. The study was approved by the Clinical Research Ethics Committee at La Paz University Hospital, Madrid (Ref.: PI18/00372).
ACKNOWLEDGEMENTS
To all members of the COSACO STUDY GROUP, who have collaborated in the recruitment of patients and collected samples and clinical data: M.R. Albañil‐ Ballesteros, M. Alcaraz‐Quiñonero, E. Álvarez‐Bueno, J. Anillo‐Lago, J.M. Azcona‐Gutiérrez, F. Baquero‐Artigao, J. Blanco‐ Gonzalez, A. Bercedo‐Sanz, J.V. Bernad‐Usoz, M. Biosca‐ Pamies, A. Bonet‐Garrosa, C. Borja‐Pérez, B. Botella‐Serrano, M.A. Caballero‐Morales, M. Carballal‐Mariño, M.T. Calvo‐Lorenzo, J.E. Callejas‐Pozo, A. Caro‐Gómez, I. Casares‐ Alonso, G. Castillo‐Aguas, C. Cayuela, J.L. Díaz de Tuesta, J. Díez‐Sebastián, B. Domínguez, M. Duelo‐Marcos, Y. Escanciano, C. Esteban‐Redondo, F.J. Fernández‐ López, M. Fernández‐Rodríguez,M.I. Galán Navarro, A. Gallego‐Iborra, F. García‐García, A. García‐Merino, R. García‐ Perez, M. Gasca‐Santiyán, I. Gil‐Alexandres, S. Giner‐Almaraz, M. Giribet‐ Folch, M.C. González‐Velasco, R. Gómez Casares, A. Gómez‐Cobiá, D. Gros‐ Esteban, B. Guillot‐Roselló, J.A. Heras‐Galindo, A.B. Hidalgo‐ Calero, A. Iofrio De Arece, R. Jiménez Ales, J Jové‐Naval, M.N Larrosa‐Escartín, A. Leal‐Negredo, C. Lobelle, I. López‐ Hernández, A. Lorente García‐Mauriño, M. Lozano‐Vergara, S. Marfil‐Olink, L. Martínez, M. Martínez, R.M. Masvidal, N. Menéndez‐González, J. Mengual‐Gil, R. Mínguez‐Verdejo, I. Miranda, C. Mourelo, M.E. Muñoz‐Hiraldo, B. Nogueira‐González, V. Ortiz‐Revuelta, M.A. Ordoñez‐ Alonso, P. Oscola, M.L. Padilla‐Esteban, I. Perez‐García, J. Pita‐Carretero, D. Quílez ‐Agreda, R.M. Ramirez‐ Gavira, MJ. Ramos, E. Rico‐Mari, S. Rodríguez‐ Moldes, E. Rodríguez‐Salinas, E. Ruiz‐Chercoles, C.A Salas‐Venero, F. Sanatorio, E. Sanchez‐Almeida, N. Sánchez‐Cordero, M.D. Sánchez‐Díaz, M. Sanchez‐Echenique, B. Sancho‐Madrid, C. Seral, C.I. Servera‐ Ginard, S. Solaz‐Barrios, E. Suarez‐Vicente, M.L. Torres‐Alvarez de, C. Torres‐Lloret, I. Ubeda‐Sansano, M.J. Vaquerizo‐ Pollino, M.D. Villar‐ Fernández, O. Rubio‐Remiro, B. Viñado, A. Viudes de Velasco, A. Yagüe. This study has been supported by the Spanish Ministry of Science and Innovation – Instituto de Salud Carlos III, and Fondos FEDER of the EU [Grant Nº PI18CIII/00372; Fondo de Investigaciones Sanitarias‐Spanish Health Research Fund (ISCIII)]; Grant Award “Jose María Corretger” from the Spanish Society for Pediatric Infectious Diseases; Grant Research Award from the Spanish Association of Pediatric Primary Care; and a Small Grant Award from the European Society for Pediatric Infectious Diseases.
Román, F. , Mendez‐Echevarria, A. , Del Rosal, T. , Garcia‐Vera, C. , Escosa‐Garcia, L. , Agud, M. , Chaves, F. , Gutiérrez‐Fernández, J. , Ruiz de Gopegui, E. , Ruiz‐Carrascoso, G. , Ruiz‐Gallego, M. D. C. , Bernet, A. , Quevedo, S. M. , Fernández‐Verdugo, A. M. , Sainz, T. , & Calvo, C. (2021). Characterization of methicillin‐resistant Staphylococcus aureus strains colonizing the nostrils of Spanish children. MicrobiologyOpen, 10, e1235. 10.1002/mbo3.1235
DATA AVAILABILITY STATEMENT
All data are provided in full in this paper with the exception, of the study protocol and the main demographic data of the screened population, which are available at: https://doi.org/10.2147/IDR.S282880.
REFERENCES
- Argudín, M. A. , Roisin, S. , Nienhaus, L. , Dodémont, M. , de Mendonça, R. , Nonhoff, C. , Deplano, A. , & Denis, O. (2018). Genetic diversity among Staphylococcus aureus isolates showing oxacillin and/or cefoxitin resistance not linked to the presence of mec Genes. Antimicrobial Agents and Chemotherapy, 62(7), e00091–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios Lopez, M. , Gomez Gonzalez, C. , Orellana, M. A. , Chaves, F. , & Rojo, P. (2013). Staphylococcus aureus abscesses: methicillin‐resistance or Panton‐Valentine leukocidin presence? Archives of Disease in Childhood, 98, 608–610. 10.1136/archdischild-2012-302695 [DOI] [PubMed] [Google Scholar]
- Broseta, A. , Chaves, F. , Rojo, P. , & Otero, J. R. (2006). Emergence of a single clone of community‐associated methicillin‐resistant Staphylococcus aureus in southern Madrid children. Enfermedades Infecciosas Y Microbiología Clínica, 24, 31–65. [DOI] [PubMed] [Google Scholar]
- Cookson, B. D. , Robinson, D. A. , Monk, A. B. , Murchan S., Deplano A., de Ryck R., Struelens M. J., Scheel C.,Fussing V., Salmenlinna S., Vuopio‐Varkila J., Cuny C., Witte W., Tassios P. T., Legakis N. J., van Leeuwen W., van Belkum A., Vindel A., Garaizar J., … Enright M. C. (2007). Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin resistant Staphylococcus aureus strains: the HARMONY collection. Journal of Clinical Microbiology, 45, 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, M. Z. , & Daum, R. S. (2010). Community‐associated methicillin‐resistant Staphylococcus aureus epidemiology and clinical consequences of an emerging epidemic. Clinical Microbiology Reviews, 23, 616–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rosal, T. , Méndez‐Echevarría, A. , Garcia‐Vera, C. , Escosa‐Garcia, L. , Agud, M. , Chaves, F. , Román, F. , Gutierrez‐Fernandez, J. , Ruiz de Gopegui, E. , Ruiz‐Carrascoso, G. , Ruiz‐Gallego, M. D. C. , Bernet, A. , Quevedo, S. M. , Fernández‐Verdugo, A. M. , Díez‐Sebastian, J. , & Calvo, C. , The COSACO Study Group (2020). Staphylococcus aureus nasal colonization in Spanish children. THE COSACO nationwide surveillance study. Infect Drug Resist, 13, 4643–4651. 10.2147/IDR.S282880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, M. C. , Day, N. P. , Davies, C. E. et al (2000). Multilocus sequence typing for characterization of methicillin‐resistant and methicillin‐susceptible clones of Staphylococcus aureus . J Clin Microb, 38, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2018). European committee on antimicrobial susceptibility testing breakpoint tables for interpretation of MICs and zone diameters. European Committee Antimicrobia Susceptibility Testing. [Google Scholar]
- García‐Alvarez, L. , Holden, M. T. , Lindsay, H. et al (2011). Methicillin‐resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. The Lancet Infectious Diseases, 11, 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geha, D. J. , Uhl, J. R. , Gustaferro, C. A. , & Persing, D. H. (1994). Multiplex PCR for identification of methicillin‐resistant staphylococci in the clinical laboratory. Journal of Clinical Microbiology, 32, 1768–1772. 10.1128/jcm.32.7.1768-1772.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez, J. N. , Ocampo, A. M. , Venegas, J. M. , Rodríguez, E. A. , Garcés, C. G. , Patiño, L. A. , Ospina, S. , & Correa, M. M. (2011). Characterisation of virulence genes in methicillin susceptible and resistant Staphylococcus aureus isolates from a paedriatic population in a university hospital of Medellín, Colombia. Memórias do Instituto Oswaldo Cruz, 106, 980–985. [DOI] [PubMed] [Google Scholar]
- Joo, H. S. , Cheung, G. Y. , & Otto, M. (2011). Antimicrobial activity of community associated methicillin‐resistant Staphylococcus aureus is caused by phenol‐soluble modulin derivatives. Journal of Biological Chemistry, 286, 8933–8940. 10.1074/jbc.M111.221382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kateete, D. P. , Bwanga, F. , Seni, J. , Mayanja, R. , Kigozi, E. , Mujuni, B. , Ashaba, F. K. , Baluku, H. , Najjuka, C. F. , Källander, K. , Rutebemberwa, E. , Asiimwe, B. B. , & Joloba, M. L. (2019). CA‐MRSA and HA‐MRSA coexist in community and hospital settings in Uganda. Antimicrobial Resistance & Infection Control, 8, 94. 10.1186/s13756-019-0551-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluytmans, J. , van Belkum, A. , & Verbrugh, H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clinical Microbiology Reviews, 10, 505–520. 10.1128/CMR.10.3.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaro‐Cardoso, J. , de Lencastre, H. , Kipnis, A. , Pimenta, F. C. , Oliveira, L. S. C. , Oliveira, R. M. , Nouer, S. S. , Aires‐de‐Sousa, M. , Milheiriço, C. , & Sgambatti‐Andrade, A. L. (2009). Molecular epidemiology and risk factors for nasal carriage of Staphylococcus aureus and methicillin‐resistant S. aureus in infants attending day care centers in Brazil. Journal of Clinical Microbiology, 47, 3991–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers, R. P. , Stinnett, J. W. , Muthukrishan, G. , Parkinson, C. L. , & Cole, A. M. (2011). Evolutionary analysis of Staphylococcus aureus identity genetic relationships between nasal carriage and clinical isolates. PLoS One 6, e16426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, T. T. , Nadimpalli, M. , Wu, J. , Heaney, C. D. , & Stewart, J. R. (2018). Challenges in estimating characteristics of Staphylococcus aureus nasal carriage among humans enrolled in surveillance studies. Frontiers in Public Health, 6, 163. 10.3389/fpubh.2018.00163.eCollection [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy, F. D. (1998). Staphylococcus aureus infection. N Engl J Med., 339, 520–32. [DOI] [PubMed] [Google Scholar]
- Mellmann, A. , Weniger, T. , Berssenbrugge, C. , Rothganger, J. , Sammeth, M. , Stoye, J. , & Harmsen, D. (2007). Based upon repeat pattern (BURP): an algorithm to characterize the long‐term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiology, 7, 98. 10.1186/1471-2180-7-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milheirico, C. , Oliveira, D. C. , & de Lencastre, H. (2007). Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin‐resistant Staphylococcus aureus: ´SCCmec IV multiplex´. Journal of Antimicrobial Chemotherapy, 60, 42–48. 10.1093/jac/dkm112 [DOI] [PubMed] [Google Scholar]
- Oliveira, D. C. , & de Lencastre, H. (2002). Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin‐resistant Staphylococcus aureus . Antimicrobial Agents and Chemotherapy, 46, 2155–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasigade, J. P. , Laurent, F. , Lina, G. , Meugnier, H. , Bes, M. , Vandenesch, F. , Etienne, J. , & Tristan, A. (2010). Global distribution and evolution of Panton‐Valentine leukocidin‐positive methicillin‐susceptible Staphylococcus aureus, 1981–2007. Journal of Infectious Diseases, 201, 1589–1597. [DOI] [PubMed] [Google Scholar]
- Rokney, A. , Baum, M. , Ben‐Shimol, S. , Sagi, O. , Anuka, E. , Agmon, V. , Greenberg, D. , Valinsky, L. , & Danino, D. (2019). Dissemination of the Methicillin‐resistant Staphylococcus aureus Pediatric Clone (ST5‐t002‐IV‐PVL+) as a Major Cause of Community‐associated Staphylococcal Infections in Bedouin Children, Southern Israel. The Pediatric Infectious Disease Journal, 38, 230–235. 10.1097/INF.0000000000002126 [DOI] [PubMed] [Google Scholar]
- Tavares, D. A. , Sá‐Leão, R. , Miragaia, M. , & de Lencastre, H. (2010). Large screening of CA‐MRSA among Staphylococcus aureus colonizing healthy young children living in two areas (urban and rural) of Portugal. BMC Infectious Diseases, 10, 110. 10.1186/1471-2334-10-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover, F. C. , Arbeit, R. D. , Goering, R. V. et al (1995). Review interpreting chromosomal DNA restriction patterns produced by pulsed‐field gel electrophoresis: criteria for bacterial strains typing. Journal of Clinical Microbiology, 33, 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, S. Y. , Davis, J. S. , Eichenberger, E. , Holland, T. L. , & Fowler, V. G. Jr (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clinical Microbiology Reviews, 28, 603–661. 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, N. A. , Sharma‐Kuinkel, B. K. , Maskarinec, S. A. , Eichenberger, E. M. , Shah, P. P. , Carugati, M. , Holland, T. L. , & Fowler, V. G. Jr (2019). Methicillin‐resistant Staphylococcus aureus: an overview of basic and clinical research. Nature Reviews Microbiology, 17, 203–218. 10.1038/s41579-018-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijen, M. , Bonten, M. , Wenzel, R. , & Kluytmans, J. (2008). Mupirocin ointment for preventing Staphylococcus aureus infections in nasal carriers. Cochrane Database Systematic Review, Cd006216. 10.1002/14651858.CD006216.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch, F. , Naimi, T. , Enright, M. C. , Lina, G. , Nimmo, G. R. , Heffernan, H. , Liassine, N. , Bes, M. , Greenland, T. , Reverdy, M. E. et al (2003). Community‐acquired methicillin‐resistant Staphylococcus aureus carrying Panton‐Valentine leukocidin genes, worldwide emergence. Emerging Infectious Diseases, 9, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven, P. O. , Gagnaire, J. , Botelho‐Nevers, E. , Grattard, F. , Carricajo, A. , Lucht, F. et al (2014). Detection and clinical relevance of Staphylococcus aureus nasal carriage: an update. Expert Review of Anti‐Infective Therapy, 12, 75–89. [DOI] [PubMed] [Google Scholar]
- Vindel, A. , Cuevas, O. , Cercenado, E. et al (2009). Methicillin resistant Staphylococcus aureus in Spain: molecular epidemiology and utility of different typing methods. Journal of Clinical Microbiology, 47, 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, R. R. , David, M. Z. , & Salata, R. A. (2012). Current concepts on the virulence mechanisms of methicillin‐resistant Staphylococcus aureus . Journal of Medical Microbiology, 61, 1179–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , McClure, J. A. , Elsayed, S. et al (2005). Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin‐resistant Staphylococcus aureus . Journal of Clinical Microbiology, 43, 5026–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are provided in full in this paper with the exception, of the study protocol and the main demographic data of the screened population, which are available at: https://doi.org/10.2147/IDR.S282880.


