Abstract
The aim of this study was to evaluate self-replicating RNA lipid nanoparticles (saRNA LNPs) to neutralize SARS-CoV-2 variants delta (B.1.617 lineage) and alpha (B.1.1.7 lineage). Before immunization of mice with saRNA LNPs, we saw high expression of S-protein at both mRNA and protein levels after transfection of HEK293T/17 cells with saRNA LNPs. After oral immunization of BALB/c mice with 0.1 – 10 µg saRNA LNPs , a high quantity of SARS-CoV-2 specific IgG and IgA antibodies were seen with a dose-dependent pattern. Importantly, the ratio of IgG2a/IgG1 in serum of vaccinated mice showed Th1/Th2 skewing response. We also found that the secreted antibodies could neutralize SARS-CoV-2 variants delta (B.1.617 lineage) and alpha (B.1.1.7 lineage). Re-stimulated splenocytes of vaccinated mice showed high secretion of IFN-γ, IL-6, and TNF- α . The authors think that although the preclinical study confirmed the efficacy of saRNA LNPs against SARS-CoV-2, the actual efficacy and safety of the oral vaccine must be evaluated in clinical trials.
Keywords: Oral vaccine, Self-replicating RNA, B.1.1.7 lineage, B.1.617 lineage
1. Introduction
Vaccines have different formulations and they are becoming more advanced every day [1]. The use of nucleic acids and nanoparticles are two examples of advanced materials, recently used in vaccine production [2]. Early studies focused on the use of DNA instead of RNA because RNA is less stable than DNA [3]. DNA vaccines produced poor results in human clinical trials [4] and this led to using RNA. Of course, this change in strategy was attributed to the success of cancer immunotherapy by mRNA molecules [5], [6]. Moreover, the success of developing mRNA-based COVID-19 vaccines opens a new window to vaccine design [7], [8]. There are currently two types of RNA vaccines, including conventional mRNA and self-replicating RNA (saRNA) [9]. These RNAs are produced in vitro and encode pathogen antigens [10].
To produce a saRNA, the sequence of an RNA-dependent RNA polymerase (RDRP) complex from viral origin to amplify saRNA, as well as the 5́ and 3́ untranslated regions (UTR), is needed for its replication in cytoplasm [11]. After expression of saRNA, activation of immune system is occurred [12]. The production of saRNA vaccines is not very complicated and can be easily integrated into a production line [13]. For example, Hekele et al. designed and produced saRNA vaccine for influenza H7N9 within eight days [14]. There are several formulations that can be used to deliver saRNA vaccines, including cationic polymers [15], lipopolyplexes [16], and lipid nanoparticles (LNPs) [17]. Interestingly, because of the self-replicating property of saRNA, a high immune response can be achieved with low doses of saRNA vaccines [18]. Importantly, the site of action for saRNA is the cytoplasm and it does not require to enter the cell nucleus. Indeed, there is no risk of integration of saRNA into the genome [5], [6], [19].
Although a variety of COVID-19 vaccines have been developled and some are being used globally, the majority of the population of developing countries still have not been vaccinated because of lack of funds, no infrastructure, and social issues [20]. They need a simple and inexpensive vaccine. Therefore, we sought to design, produce, and evaluate an oral vaccine that could be easily used. The aim of this study was to evaluate saRNA LNPs in mice to neutralize SARS-CoV-2 variants delta (B.1.617) and alpha (B.1.1.7) variants.
2. Materials and methods
2.1. Plasmid construct
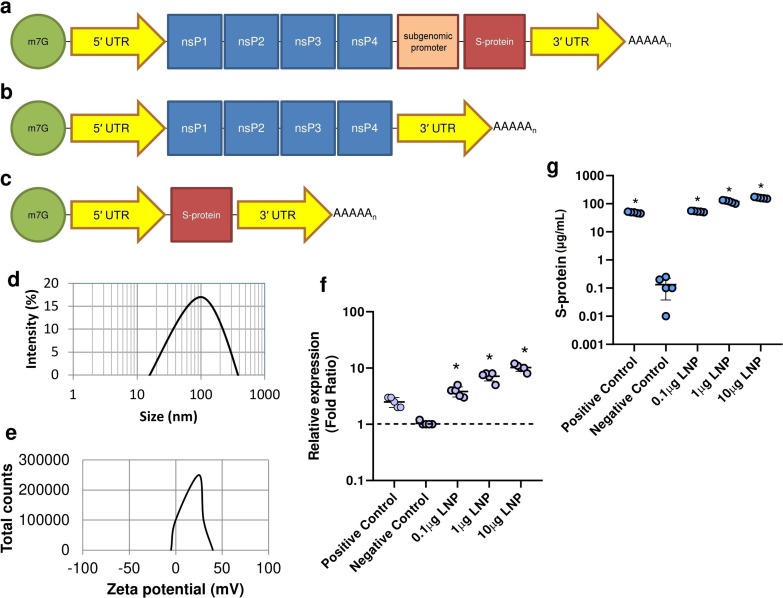
To synthesize the saRNA construct, a common bacterial plasmid vector with a T7 promoter was used. The required sequence for saRNA construct was synthesized and sub-cloned by Biomatik, Canada. Based on (Fig. 1 a), saRNA construct has 5′ UTR (GenBank accession number: NC_001449), nsP1-4 (GenBank accession number: NC_001449), sub-genomic promoter (GenBank accession number: NC_001449), S-protein (GenBank accession number: MZ571142.1), 5′ UTR (GenBank accession number: NC_001449), and polyA tail. The negative control construct (Fig. 1 b) has the same structure except for the sub-genomic promoter and the S-protein coding sequence. Positive control construct (Fig. 1 c) has 4 main parts, including 5′ UTR (GenBank accession number: NC_001449), S-protein (GenBank accession number: MZ571142.1), 3′ UTR (GenBank accession number: NC_001449), and polyA tail. Supplementary 1 shows the full length sequence of saRNA construct used in this study.
Fig. 1.
The schematic diagrams of saRNA (a), negative control (b), and positive control construct (c). Abbreviation: 7-Methylguanosine (m7G); Untranslated region (UTR); Non-structural protein (nsP), Spike protein (s-protein). The size distribution (d) and zeta potential (e) of saRNA LNPs by DLS apparatus. HEK293T/17 cells were treated with saRNA LNPs at 0.1–10 μg and the expression of S-protein was evaluated by Real-time PCR (f) and ELISA (g). All data were shown as mean ± SD. * indicates significance difference with P < 0.05 when compared with negative controls using a one-way ANOVA adjusted for multiple comparisons with n = 5.
2.2. Synthesis of linear saRNA
The plasmid encoding saRNA construct was transformed into E. coli (institute Pasteur, Iran), cultured in Luria Broth with 100 μg/mL carbenicillin (Sigma Aldrich, UK). Plasmids were purified using a Plasmid Plus MaxiPrep kit (QIAGEN, UK) and their concentration and purity were measured on a NanoDrop spectrophotometer (ThermoFisher, UK). Then, cloned plasmids were linearized using MluI for 3 h at 37 °C. Then, saRNA transcripts were produced using 1 μg of linearized DNA template in a MEGAScript™ reaction (Ambion, UK) for 1 h at 37 °C. One μg linear saRNA was mixed with 1 μM ScriptCap™ (CellScript, WI, USA) for 1 h at 37 °C. Synthesized saRNA was purified by LiCl precipitation, re-suspended in RNA storage buffer, and stored at − 80 °C. To evaluate the purity of synthesized saRNA, the A260/A280 ratio was measured using the NanoDrop spectrophotometer (ThermoFisher, UK) [21].
2.3. Encapsulation of saRNA in LNPs
To encapsulate saRNA, we used a simple chemical process [21] in which 0.1, 1, and 10 μg of purified saRNA were separately mixed with an ethanolic lipid mixture of 1,2-dilinoleyl oxy-3-dimethylaminopropane, 1,2-diastearoyl-sn-glycero-3-phosphocholine, cholesterol, and 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 at a ratio of 10:48:2:40 at pH 4.0. The mixture was vigorously stirred by a T-mixer and then placed in a dialysis bag to purify overnight. Then, the size distribution and zeta potential of the produced LNPs were determined by dynamic light scattering (DLS) (Malvern Instruments Ltd, Malvern, UK). The encapsulation of saRNA was confirmed by gel electrophoresis and the entrapment efficiency was measured by a NanoDrop spectrophotometer (ThermoFisher, UK) at 260 nm. For positive and negative controls, the same encapsulation process was also performed.
2.4. The expression of saRNA in HEK293T/17 cells
In Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco) containing 10% fetal bovine serum (FBS) (Gibco), 1% L-glutamine (Thermo Fisher Scientific), and 1% penicillin–streptomycin (Thermo Fisher Scientific), HEK293T/17 cells were cultured at 37 °C for 5 days. Then, cells were separately incubated with 0.1–10 µg saRNA, negative control, and positive control LNPs. After 24 h, the expression of S-protein was confirmed by Real-time PCR and ELISA (Supplementary 2).
2.5. Immunization
All procedures related to animal experiments used in this section were approved by the ethical committee of Sirjan School of Medical Sciences, Sirjan, Iran (Ethical code: IR.SIRUMS.REC.1400.001). We used 10 BALB/c mice aged 6–8 weeks in each study group and they were orally immunized with 0.1, 1, and 10 µg saRNA LNPs at weeks 1 and 3. For each mouse, 100 µL of vaccine mixture was orally administered using a needle-free insulin syringe. In the control groups, BALB/c mice were orally immunized with 10 µg positive control LNPs and 10 µg negative control LNPs at weeks 1 and 3 as described. Serum samples were collected at weeks 2, 4, and 6 and the spleen of vaccinated mice was removed at week 6.
2.6. Recovered COVID-19 patient samples
Here, the serum samples of recovered COVID-19 patients (n = 10) were obtained from Zahedan University of Medical Sciences, Zahedan, Iran. All of them had been infected with the delta variant. Written informed consent was given from all participants (ethical codes: IR.ZAUMS.REC.1399.317 and IR.ZAUMS.REC.1399.316). All recovered patients showed a negative PCR test at the time of sampling.
2.7. Serum levels of antibodies in mice and recovered COVID-19 patients
A semi-quantitative ELISA was used to determine the levels of IgG, IgG1, IgG2a, and IgA antibodies in the sera of vaccinated mice. Also, the levels of IgG and IgA antibodies were determined in the sera of recovered COVID-19 patients.
First, high binding ELISA plates (Biomat, Italy) were coated with SARS-CoV-2 S- protein recombinant antigen (Sigma-Aldrich) at 1 μgmL−1. After washing plates, 50 μL of diluted serum samples collected from immunized mice and recovered COVID-19 patients were separately added to wells. After incubation for 1 h at 37 °C, plates were washed with PBS and then 100 μL of the following secondary antibody was separately added: 1) anti-mouse IgG-HRP, 2) anti-mouse IgG1-HRP, 3) anti-mouse IgG2a-HRP, 4) anti-mouse IgA-HRP, 5) anti-human IgG-HRP, 6) anti-human IgA-HRP (Southern Biotech). After incubation and washing with PBS, 50 μL of 3,3′, 5,5′-tetramethylbenzidine was added and then the reactions was stopped by adding 50 μL of 10% sulfuric acid. Finally, the absorbance of each well was read by a Spectrophotometer at 450 nm (BioTek Industries) and serum levels of antibodies was measured using a standard curve.
2.8. Wild-type viral neutralization assay
To evaluate the ability of vaccinated mice or recovered COVID-19 patients to neutralize SARS-CoV-2 virus, wild-type viral neutralization assay was applied according to McKey et al [21]. SARS-CoV-2 variant B.1.1.7 and variant B.1.617 were first isolated from COVID-19 patients. Then, they were cultured on Caco2 cells in DMEM (Gibco) containing 10% FBS (Gibco), 1% L-glutamine (Thermo Fisher Scientific), and 1% penicillin–streptomycin (Thermo Fisher Scientific) for 5 days at 37 °C. Finally, propagated viruses were purified by Caesium chloride gradient centrifugation. In the next step, all sera were first incubated at 56 °C for 30 min and were serially diluted in DMEM (Gibco, Thermo Fisher Scientific) with 1% penicillin–streptomycin (Thermo Fisher Scientific), and 0.3% BSA fraction V (Thermo Fisher Scientific). Serum dilutions were separately incubated with 100 TCID50 per well of SARS-CoV-2 variant B.1.1.7 and variant B.1.617 for 1 h at room temperature. Then, they were transferred to 96-well plates pre-seeded with HEK293T/17 cells and incubated at 37 °C for 5 days. After incubation, 100 µL of crystal violet (Sigma-Aldrich) was added to each well and scored for cytopathic effect. The neutralization titer was calculated as the reciprocal of the highest serum dilution at which full virus neutralization occurred.
2.9. IFN-γ ELISpots
Based on the Mouse IFN-γ ELISpotPLUS kit (Mabtech), anti-IFN-γ -pre-coated plates were first blocked with 10% FBS (Gibco), and then 2.5 × 105 splenocytes of vaccinated mice and 1 μgmL−1 SARS-CoV-2 peptides (AGX819, Sigma-Aldrich) were added. Plates were incubated overnight at 37 °C and 5% CO2. After incubation, biotinylated cytokine-specific detection antibodies (Mabtech), streptavidin-enzyme conjugate (Mabtech), and substrate (Mabtech) were added. Finally, each well was examined under an optical microscope (Zeiss, Germany) and the number of stained cells was calculated.
2.10. Secretion of IL-6 and TNF-α
The level of IL-6 and TNF-α in serum samples of immunized mice, recovered COVID-19 patients, and the supernatant of activated splenocytes was measured by ELISA kit. Briefly high binding ELISA plates (Biomat, Italy) were separately coated with anti-mouse and anti-human IL-6 and TNF-α (Southern Biotech) and then the corresponding samples were separately added. After incubation for 1 h at 37 °C, plates were washed by PBS, and then 100 μL of secondary antibodies, including anti-mouse IL-6-HRP (Southern Biotech), anti-human IL-6-HRP (Southern Biotech), anti-mouse TNF-α-HRP (Southern Biotech), and anti-human TNF-α-HRP (Southern Biotech) were added. Then, 50 μL of 3,3′, 5,5′-tetramethylbenzidine was added and incubated for 15 min at room temperature. Finally, the reaction was stopped by adding 50 μL of 10% sulfuric acid and the absorbance of each well was read by a Spectrophotometer at 450 nm (BioTek Industries). The level of IL-6 and TNF-α was determined by a standard curve.
2.11. Statistical analysis
GraphPad Prism (version 8.4) was used to prepare graphs and statistics. One-way ANOVA was used and P values<0.05 were considered significant (with n = 10 biologically independent mice and n = 10 recovered COVID-19 patients). All data are shown as mean ± standard deviation (SD).
3. Results
3.1. LNP characterization
Synthesized linear saRNA was encapsulated in LNPs and then characterized. The average particle size and zeta potential of saRNA LNPs were 100 ± 5 nm and + 22 ± 0.6 mV, respectively (Fig. 1 d and Fig. 1 e). The highest entrapment efficiency of saRNA was 67 ± 2 %.
3.2. The expression of S-protein
Before immunization of mice with saRNA LNPs, their efficacy was verified in HEK273T/17 cells. High expression of S-protein was observed at both mRNA and protein levels after transfection of HEK293T/17 cells with saRNA LNPs (Fig. 1 f and Fig. 1 g). It was found that the relative expression of S-protein was increased with increasing LNP dose.
3.3. Serum levels of IgG, IgG1, IgG2a, and IgA antibodies
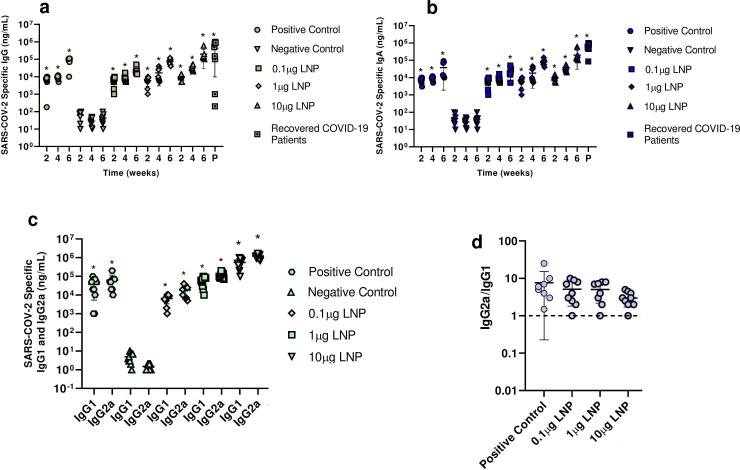
BALB/c mice were orally immunized with 0.1–10 µg saRNA LNPs at weeks 1 and 3 and their serum samples were then collected at weeks 2, 4, and 6. ELISA assay showed a high quantity of SARS-CoV-2 specific IgG, IgG1, IgG2a, and IgA antibodies in a dose-dependent manner in serum of mice (Fig. 2 a-c). Interestingly the levels of IgG and IgA antibodies in mice immunized with 10 μg saRNA LNPs was almost close to antibody levels in recovered COVID-19 patients (P > 0.05). Here, significant differences were found between the level of antibodies in vaccinated mice or recovered COVID-19 patients and negative control (P < 0.05). A Th1/Th2 skewing response (Fig. 2 d) was also observed in mice vaccinated with positive control and 0.1–10 μg of saRNA LNPs . It means that all mice vaccinated with saRNA LNPs and positive control LNPs showed a Th1-biased response.
Fig. 2.
The serum level of IgG (a), IgA (b), IgG1 and IgG2a (c) against SARS-CoV-2 in mice vaccinated with 0.1–10 μg saRNA LNPs. The ratio of IgG2a/IgG1 was used to find Th1/Th2 skewing responses in vaccinated mice (d). * indicates significance difference at p < 0.05 when compared with negative control using a one-way ANOVA adjusted for multiple comparisons with n = 10 biologically independent mice and recovered COVID-19 patients.
3.4. Wild-type viral neutralization assay
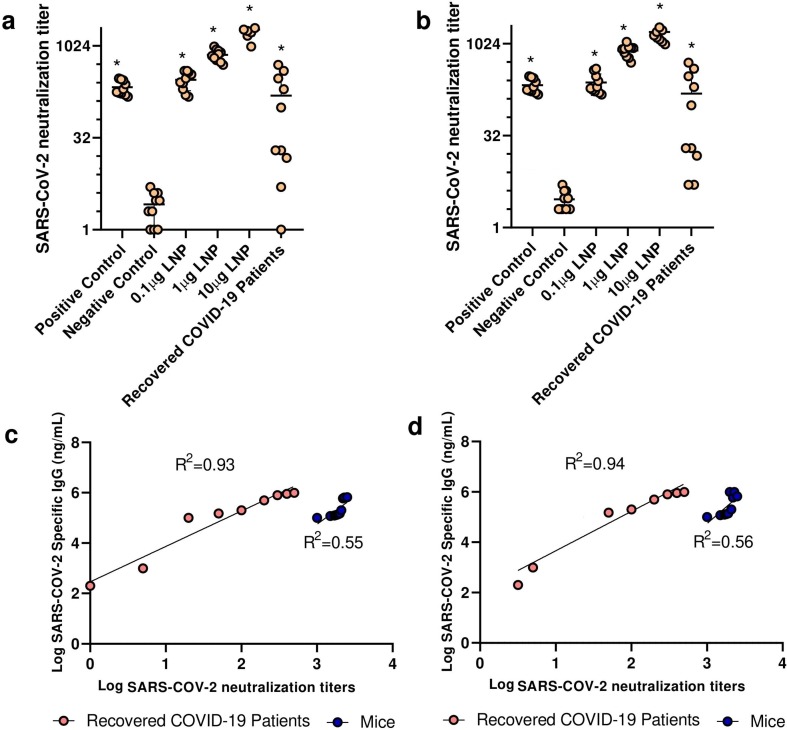
SARS-CoV-2 variants B.1.1.7 and B.1.617 were isolated and cultured on Caco2 cells. In the next step, serum dilutions were separately incubated with 100 TCID50 per well of both SARS-CoV-2 variants. Then, they were transferred to pre-seeded well with HEK293T/17 cells and incubated for 5 days. After incubation, the cytopathic effect was recorded and the neutralization titer was calculated. We observed a high viral neutralization titer in mice vaccinated with saRNA LNPs at 0.1–10 μg in a linear dose-dependent manner. We also found that the secreted antibodies induced by saRNA LNPs could neutralize both SARS-CoV-2 variants B.1.1.7 (alpha) and B.1.617 (delta) (Fig. 3 a-b). Interestingly, secreted antibodies detected in recovered COVID-19 patients could neutralize both variants. A significant positive correlation was observed between SARS-CoV-2 specific IgG and SARS-CoV-2 neutralization titer in both vaccinated mice and recovered COVID-19 patients (Fig. 3 c-d).
Fig. 3.
The viral neutralization titer against SARS-CoV-2 variants B.1.1.7 (a) and B.1.617 (b) in mice vaccinated with 0.1–10 μg saRNA LNPs and recovered COVID-19 patients. A significant positive correlation was seen between SARS-CoV-2 specific IgG and SARS-CoV-2 neutralization titer of secreted antibodies against SARS-CoV-2 variants B.1.1.7 (c) and B.1.617 (d) in both vaccinated mice and recovered COVID-19 patients. * indicates significance difference at p < 0.05 when compared with negative control using a one-way ANOVA adjusted for multiple comparisons with n = 10 biologically independent mice and recovered COVID-19 patients.
3.5. Cellular and cytokine responses
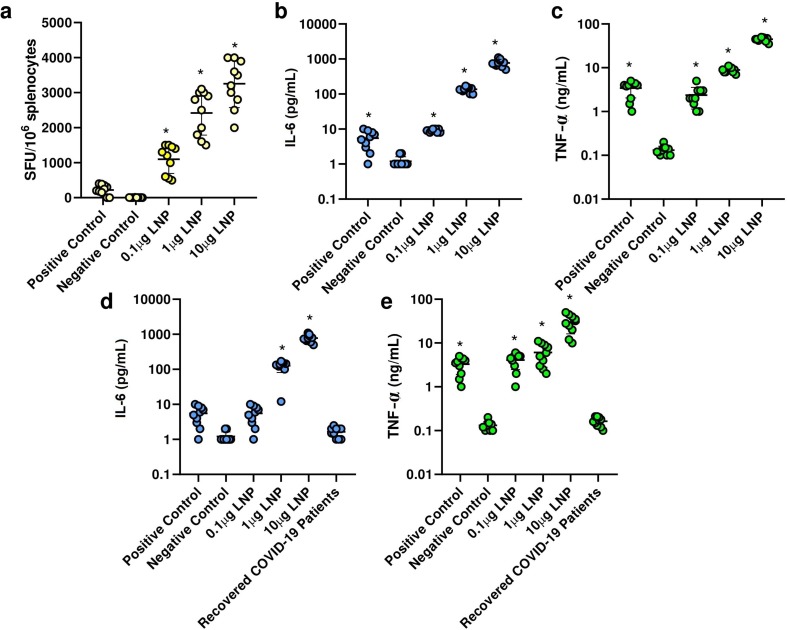
Splenocytes of vaccinated mice were first activated with SARS-CoV-2 peptides and then added to anti-IFN-γ-pre-coated plates. After incubation for overnight at 37 °C, detection antibodies, streptavidin-enzyme conjugate, and substrate were added. Finally, each well was examined under an optical microscope and the number of stained cells was calculated. We found that re-stimulated splenocytes of vaccinated mice had a high IFN-γ secretion in a linear dose-dependent manner (Fig. 4 a). There was a significant difference between mice vaccinated with 10 μg saRNA LNPs and other groups (P < 0.05).
Fig. 4.
Splenocytes of vaccinated mice re-stimulated with SARS-CoV-2 peptides had a high IFN-γ secretion in a linear dose-dependent manner (a). The secretion of IL-6 (b) and TNF-α (c) by re-stimulated splenocytes. The serum level of IL-6 (d) and TNF-α (e) in vaccinated mice (at week 6) and in recovered COVID-19 patients (one week after recovery). * indicates significance difference at p < 0.05 when compared with negative control groups using a one-way ANOVA adjusted for multiple comparisons with n = 10 biologically independent mice and recovered COVID-19 patients.
We also measured the secretion of IL-6 and TNF-α by re-stimulated splenocytes and the serum levels of IL-6 and TNF-α in vaccinated mice and recovered COVID-19 patients. It was shown that in both supernatants of re-stimulated splenocytes and vaccinated mouse sera, there was a high level of IL-6 and TNF-α in a dose-dependent manner (Fig. 4 b-e). Here, significant differences were observed between mice vaccinated with 10 μg saRNA LNPs and other vaccinated groups (P < 0.05). We found significant differences between the level of IL-6 and TNF-α in the serum of recovered COVID-19 patients compared with vaccinated mice by saRNA LNPs (P < 0.05).
4. Discussion
In this study, we sought to develop a saRNA-based oral vaccine that could simultaneously fight both the SARS-COV-2 variants B.1.1.7 (alpha) and B.1.617 (delta). Vaccines based on saRNA have already been used for some infectious diseases [14], [22]. This technology has also been studied for COVID-19, showing some advantages versus conventional mRNA [21], [23]. The strength of saRNA-based vaccines is that they can induce potent immune responses at low doses because of their self-replicating property. Of course, these vaccines, like other vaccines, may have side effects and must be evaluated. Currently, although various vaccines against SARS-COV-2 are designed with different platforms [24], most people of un-developed and developing countries still have not been vaccinated, which can be due to various reasons, such as economic and social problems [20]. Unfortunately, the genomic structure of SARS-COV-2 is not stable and it is susceptible to mutations [25]. This phenomenon leads to a significant reduction of vaccine efficacy with any platform [26].
In this study, the designed saRNA had alphavirus replicase, RDRP, as well as the gene coding for the S-protein. The replicase was required for saRNA amplification and mediates the production of this RNA [27]. This leads to the production of a large number of S-proteins that be taken up by antigen presenting cells (APCs). Of course, it should be noted that saRNAs, in order to efficiently be delivered to cells, must be protected. One of the best ways is to use LNPs [21] and they can be optimized for target cells [28]. We first tested the efficacy of saRNAs in HEK293T cells and then in mice. The high expression of S-protein at both mRNA and protein levels was observed after transfection of HEK293T/17 cells. This valuable finding gave us hope that the saRNA LNPs might stimulate the immune system and could act as an effective vaccine. In this study, the vaccine was orally administered because our goal was to stimulate the immune cells located in the mucosa. Oral administration is much easier than injectable vaccines and is especially more acceptable by children. After vaccination of mice with saRNA LNPs, we saw high levels of IgG, IgG1, IgG2a, and IgA antibodies against SARS-CoV-2 in a dose-dependent manner. Interestingly, the concentration of IgG and IgA antibodies in the serum of recovered COVID-19 patients was approximately equal to antibody concentration in the serum of mice vaccinated with saRNA LNPs at 10 μg. Another important finding was Th1/Th2 skewing or Th1-biased response. Moreover, a high viral neutralization titer was seen in mice vaccinated with saRNA LNPs at 0.1–10 μg and in recovered COVID-19 patients. Interestingly, these antibodies could neutralize both variants of SARS-CoV-2 and this shows that the designed system (saRNA LNPs) has been fully expressed. Importantly, re-stimulated splenocytes of vaccinated mice showed a high IFN-γ secretion in a linear, dose-dependent manner. Supernatants of re-stimulated splenocytes and vaccinated mouse sera contained a high level of IL-6 and TNF-α in a dose-dependent manner.
Before us, McKay et al. had shown that saRNA SARS-CoV-2 LNP vaccine induces remarkably high and dose-dependent SARS-CoV-2 specific antibody titers in mouse sera, as well as robust neutralization of both a pseudo-virus and wild-type virus [21]. Also, Spencer et al showed vaccination with saRNA and adenoviral COVID vaccines induce robust immune responses in mice. They demonstrated that two-dose heterologous vaccination was better than single-dose. Neutralizing titers after heterologous prime-boost were at least comparable or higher than the titers measured after homologous prime-boost vaccination with viral vectors [23]. It is important to know the intestinal immune responses against SARS-CoV-2 is more effective than others [29]. Some studies on COVID-19 animal models [30] and COVID-19 patients with gastrointestinal symptoms [31] revealed that intraepithelial CD8 + lymphocytes and lamina propria residing CD4 + and CD8 + effector T cells are significantly expanded compared with healthy controls [32]. Importantly, inflammatory dendritic cells are significantly reduced in the lamina propria of COVID-19 patients with gastrointestinal symptoms [32]. These data suggest that intestinal infection with SARS-CoV2 alters immune signatures and leads to a more favorable immune response. Interestingly, the serum levels of IL-6 and IL-17 are lower in COVID-19 patients with GI symptoms compared with COVID-19 patients without GI symptoms. Another interesting point is that the presence of the virus in the GI tract may trigger a long-term production of anti-viral IgA antibodies, compared with IgG and IgM antibodies [33], [34].
Taken together, the oral vaccine, based on saRNA LNPs, could induce a Th1-biased response to produce a high quantity of SARS-CoV-2 specific IgG and IgA antibodies. We also found that the produced antibodies could neutralize SARS-CoV-2 variants B.1.1.7 (alpha) and B.1.617 (delta).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank Dr. Cristian Smerdou (Cima Universidad de Navarra, Spain) and PD.Dr. Vladimir Temchura, Institute of Clinical and Molecular Virology, Friedrich-Alexander-University Erlangen-Nürnberg, 91054 Erlangen, Germany for valuable helps. We thank the Zahedan University of Medical Sciences, Zahedan, Iran for providing sera from recovered COVID-19 patients.
Funding
This work was financially supported by Sirjan School of Medical Sciences, Sirjan, Iran (grant number: 99000071).
Ethical approval
All experiments were under the guidelines of the National Institute of Health, the provisions of the Declaration of Helsinki, and the ethics committee of Sirjan School of Medical Sciences, Sirjan, Iran. (Ethical code: IR.SIRUMS.REC.1400.001).
Informed consent
Informed consent was obtained from all individuals included in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.108231.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.D’Amico C., Fontana F., Cheng R., Santos H.A. Development of vaccine formulations: past, present, and future. Drug Delivery and Translational Research. 2021;11(2):353–372. doi: 10.1007/s13346-021-00924-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen Y., Hao T., Ou S., Hu C., Chen L. Applications and perspectives of nanomaterials in novel vaccine development. MedChemComm. 2018;9(2):226–238. doi: 10.1039/c7md00158d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumaragurubaran K., Kaliaperumal K. DNA vaccine: the miniature miracle. Veterinary World. 2013;6(3):228. doi: 10.5455/vetworld.10.5455/vetworld.2013.228-232. [DOI] [Google Scholar]
- 4.Hobernik D., Bros M. DNA vaccines—how far from clinical use? Int. J. Mol. Sci. 2018;19(11):3605. doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilboa E., Vieweg J. Cancer immunotherapy with mRNA-transfected dendritic cells. Immunol. Rev. 2004;199(1):251–263. doi: 10.1111/j.0105-2896.2004.00139.x. [DOI] [PubMed] [Google Scholar]
- 6.Schlake T., Thess A., Thran M., Jordan I. mRNA as novel technology for passive immunotherapy. Cell. Mol. Life Sci. 2019;76(2):301–328. doi: 10.1007/s00018-018-2935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teo S.P. Review of COVID-19 mRNA Vaccines: BNT162b2 and mRNA-1273. Journal of Pharmacy Practice. 2021;08971900211009650 doi: 10.1177/08971900211009650. [DOI] [PubMed] [Google Scholar]
- 9.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28 [Google Scholar]
- 10.Phimister E.G., Fuller D.H., Berglund P. Amplifying RNA vaccine development. N. Engl. J. Med. 2020;382(25):2469–2471. doi: 10.1056/NEJMcibr2009737. [DOI] [PubMed] [Google Scholar]
- 11.Ferrero D., Ferrer-Orta C., Verdaguer N. Viral RNA-dependent RNA polymerases: a structural overview. Virus Protein and Nucleoprotein Complexes. 2018:39–71. doi: 10.1007/978-981-10-8456-0_3. [DOI] [PubMed] [Google Scholar]
- 12.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., Reece S.T., Sahin U., Tregoning J.S. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26(2):446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson N.A., Kester K.E., Casimiro D., Gurunathan S., DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. npj Vaccines. 2020;5(1):1–6. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hekele A., Bertholet S., Archer J., Gibson D.G., Palladino G., Brito L.A., Otten G.R., Brazzoli M., Buccato S., Bonci A., Casini D., Maione D., Qi Z.-Q., Gill J.E., Caiazza N.C., Urano J., Hubby B., Gao G.F., Shu Y., De Gregorio E., Mandl C.W., Mason P.W., Settembre E.C., Ulmer J.B., Craig Venter J., Dormitzer P.R., Rappuoli R., Geall A.J. Rapidly produced SAM® vaccine against H7N9 influenza is immunogenic in mice. Emerging Microbes Infect. 2013;2(1):1–7. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blakney A.K., Zhu Y., McKay P.F., Bouton C.R., Yeow J., Tang J., Hu K., Samnuan K., Grigsby C.L., Shattock R.J. Big is Beautiful: Enhanced saRNA delivery and immunogenicity by a higher molecular weight, bioreducible, cationic polymer. ACS Nano. 2020;14(5):5711–5727. doi: 10.1021/acsnano.0c00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perche F., Clemençon R., Schulze K., Ebensen T., Guzmán C.A., Pichon C. Neutral lipopolyplexes for in vivo delivery of conventional and replicative RNA vaccine. Molecular Therapy-Nucleic Acids. 2019;17:767–775. doi: 10.1016/j.omtn.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goswami R., Chatzikleanthous D., Lou G., Giusti F., Bonci A., Taccone M., Brazzoli M., Gallorini S., Ferlenghi I., Berti F., O’Hagan D.T., Pergola C., Baudner B.C., Adamo R. Mannosylation of LNP results in improved potency for self-amplifying RNA (SAM) vaccines. ACS Infect. Dis. 2019;5(9):1546–1558. doi: 10.1021/acsinfecdis.9b00084. [DOI] [PubMed] [Google Scholar]
- 18.Lundstrom K. The Potential of Self-amplifying RNA Vaccines for Infectious Diseases and COVID-19. Vaccine Research. 2020;7(1):25–37. [Google Scholar]
- 19.Blakney A.K., Ip S., Geall A.J. An update on self-amplifying mRNA vaccine development. Vaccines. 2021;9(2):97. doi: 10.3390/vaccines9020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jentsch P.C., Anand M., Bauch C.T. Prioritising COVID-19 vaccination in changing social and epidemiological landscapes: a mathematical modelling study. Lancet. Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luisi K., Morabito K.M., Burgomaster K.E., Sharma M., Kong W.-P., Foreman B.M., Patel S., Fisher B., Aleshnick M.A., Laliberte J., Wallace M., Ruckwardt T.J., Gordon D.N., Linton C., Ruggiero N., Cohen J.L., Johnson R., Aggarwal K., Ko S.-Y., Yang E.S., Pelc R.S., Dowd K.A., O’Hagan D., Ulmer J., Mossman S., Sambor A., Lepine E., Mascola J.R., Pierson T.C., Graham B.S., Yu D. Development of a potent Zika virus vaccine using self-amplifying messenger RNA. Sci. Adv. 2020;6(32):eaba5068. doi: 10.1126/sciadv.aba5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer A.J., McKay P.F., Belij-Rammerstorfer S., Ulaszewska M., Bissett C.D., Hu K., Samnuan K., Blakney A.K., Wright D., Sharpe H.R., Gilbride C., Truby A., Allen E.R., Gilbert S.C., Shattock R.J., Lambe T. Heterologous vaccination regimens with self-amplifying RNA and adenoviral COVID vaccines induce robust immune responses in mice. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-23173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Tenchov R., Smoot J., Liu C., Watkins S., Zhou Q. A comprehensive review of the global efforts on COVID-19 vaccine development. ACS Cent. Sci. 2021;7(4):512–533. doi: 10.1021/acscentsci.1c00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Na W., Moon H., Song D. A comprehensive review of SARS-CoV-2 genetic mutations and lessons from animal coronavirus recombination in one health perspective. J of Microbiology. 2021;59(3):332–340. doi: 10.1007/s12275-021-0660-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noh J.Y., Jeong H.W., Shin E.-C. SARS-CoV-2 mutations, vaccines, and immunity: implication of variants of concern. Signal Transduction and Targeted Therapy. 2021;6(1):1–2. doi: 10.1038/s41392-021-00623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J., Eygeris Y., Gupta M., Sahay G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021;170:83–112. doi: 10.1016/j.addr.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26(9):363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weidinger C., Hegazy A.N., Glauben R., Siegmund B. COVID-19—from mucosal immunology to IBD patients. Mucosal Immunol. 2021;14(3):566–573. doi: 10.1038/s41385-021-00384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Choy K.-T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., Peiris M., Yen H.-L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung W.K., To K.-F., Chan P.K., Chan H.L., Wu A.K., Lee N., Yuen K.Y., Sung J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125(4):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livanos A.E., Jha D., Cossarini F., Gonzalez-Reiche A.S., Tokuyama M., Aydillo T., Parigi T.L., Ramos I., Dunleavy K., Lee B. Gastrointestinal involvement attenuates COVID-19 severity and mortality. MedRxiv. 2020 [Google Scholar]
- 33.Gaebler C., Wang Z., Lorenzi J.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Z. Wang, J. Lorenzi, F. Muecksch, S. Finkin, C. Viant, C. Gaebler, M. Cipolla, H. Hoffmann, T. Oliveira, D. Oren, neutralization by dimeric IgA, Sci. Transl. Med 13. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.