Abstract
Myelodysplastic syndromes (MDS) are clonal hematopoietic disorders with heterogeneous presentation, ranging from indolent disease courses to aggressive diseases similar to acute myeloid leukemia (AML). Approximately 90% of MDS patients harbor recurrent mutations , which – with the exception of mutated SF3B1 –have not (yet) been included into the diagnostic criteria or risk stratification for MDS. Accumulating evidence suggests their utility for diagnostic workup, treatment indication and prognosis. Subsequently, in patients with unexplained cytopenia or dysplasia identification of these mutations may lead to earlier diagnosis. The acquisition and expansion of additional driver mutations usually antecedes further disease progression to higher risk MDS or secondary AML and thus, can be clinically helpful to detect individuals that may benefit from aggressive treatment approaches. Here, we review our current understanding of somatic gene mutations, gene expression patterns and flow cytometry regarding their relevance for disease evolution from pre-neoplastic states to MDS and potentially AML.
Keywords: Myelodysplastic syndrome, Clonal architecture, Clonal evolution, Residual disease, Molecular targets, Measurable residual disease
SEARCH STRATEGY AND SELECTION CRITERIA.
References for this Review were identified through searches of PubMed with the search terms “measurable/minimal residual disease”, “myelodysplastic syndrome”, “clonal evolution”, “disease burden”, and “molecular” from 2005 until December 2020. Articles were also identified through searches of the authors’ own files. Only papers published in English were reviewed. The final reference list was generated on the basis of originality and relevance to the broad scope of this Review.
Alt-text: Unlabelled box
Introduction
Myelodysplastic syndromes (MDS) are clonal disorders characterized by dysplasia of at least one myeloid cell line, peripheral blood cytopenia, and a propensity to eventually progress into acute myeloid leukemia (AML) [1]. Clinically, MDS presents highly heterogeneous and may range from indolent disease courses with nearly normal life expectancies to aggressive neoplasm similar to that of AML. This clinical diversity underlines the demand for appropriate risk stratification at diagnosis and during disease course as well as personalized treatment approaches to improve outcomes and avoid evitable adverse effects of toxic treatment regimens. A growing understanding of the complex biology and sequence of biological changes in MDS yield not only the hope to improve prognostic and predictive tools for MDS patients but also to enlarge therapeutic options with novel molecular targets [2,3]. For example high teleomerase activity and telomere length have been associated with MDS, and consequently, Imetelstat - an inhibitor of telomerase activity - entered clinical studies to evaluate its potential to reduce transfusion dependence in lower risk MDS [4,5]. Several epigenetic changes have also been shown to play a role in MDS pathogenesis and to impact prognosis, such as microRNA expression patterns [6], or distinct DNA methylation profiles which may identify clusters containing outcome information independent of current MDS prognosis systems [7].
However, today not at last due to practical need in the clinical routine, methods of gene mutation assessment have developed a higher impact on clinical decision making and personalized therapy approaches. Especially the introduction of next generation sequencing (NGS) methods impacts the clinical routine and increasingly allows the identification of recurrent molecular aberrations with several related consequences [8]. Despite the fact that – with the exception of mutated SF3B1 - these mutations have not (yet) been included into the diagnostic criteria or risk stratification for MDS [1], accumulating evidence suggests their utility for diagnostic workup, treatment indication and prognostic considerations. Studies adapting high throughput sequencing of myeloid genes showed that approximately 90% of MDS patients carry at least one oncogenic mutation [3,8,9], that initiate the disease but may also change with cytotoxic treatments or during natural disease course. Hence, in patients with unexplained cytopenia, the absence of a molecular or cytogenetic alterations constitutes a negative predictive factor for the development of myeloid neoplasm [10]. However, when identifying a molecular alteration, the distinct affected gene impact the probability of developing a myeloid neoplasm. While mutations in genes encoding splicing factors, as well as JAK2 or RUNX1 show a high predictive value for the diagnosis of a myeloid neoplasm, mutations in TET2, DNMT3A, ASXL1 also frequently occur in otherwise healthy individuals [11,12] and are predictive mainly when occurring in combination with other genetic lesions. Subsequently, in patients with unexplained cytopenia and/or dysplasia identification of these mutations may lead to earlier diagnosis and – if clinically necessary – treatment initiation of MDS [10]. During time, the acquisition and expansion of additional driver mutations usually antecedes further disease progression to higher risk MDS or secondary AML and thus, can be clinically helpful to detect individuals that may benefit from aggressive treatment approaches, including allogeneic hematopoietic stem cell transplantation (HSCT) [8].
In MDS patients achieving a morphologic complete remission during treatment the longitudinal evaluation of genetic aberrations may help to estimate the depth of remission and planning of further treatment interventions. Hence, known molecular responses could aid in clinical decision-making, such as dose reductions in older patients under life-long palliative therapy or the optimal timing of an allogeneic HSCT in individuals with curative treatment approaches. Finally, after allogeneic HSCT, repetitive analyses of known molecular alteration may even allow measurable residual disease (MRD) evaluation in MDS patients, quite similar to that already established in AML [13]. However, such longitudinal studies have not been systematically performed so far. Furthermore, thresholds of evolving molecular patterns identifying “danger signals” with regard to disease evolution are not well defined. Lastly, therapeutic consequences in case of clonal evolution or increase of MRD are scarce given the paucity of approved treatment options in MDS [14].
Here, we review our current understanding of somatic gene mutations, and gene expressions in MDS and their relevance for disease evolution from pre-neoplastic states to MDS and potentially secondary AML as well as their applicability for MRD detection when applying therapy to MDS patients, and the respective treatment associations. While this approach by nature cannot be a comprehensive review of every biological aspect, we believe it represents an overview of the data with a stringent up to date clinical value.
The molecular landscape in MDS
There are no molecular changes that are specific for MDS and many recurrent mutations are shared by other myeloid neoplasm and can sometimes even be detected in healthy individuals. Mutations involved in DNA methylation such as TET2, DNMT3A, as well as in histone modifiers ASXL1 and EZH2 are common in MDS [8], but also frequently occur in aging healthy individuals [11,12]. However, the observed composition and frequencies of mutations differ, as e.g. somatic mutations in spliceosome genes (especially SF3B1, SRSF2, U2AF1, and ZRSR2), TP53, NF1, EZH2, and BCOR have been observed more commonly in MDS, while mutations in FLT3, NPM1, DNMT3A, IDH1, and IDH2 are more common in AML [9]. Chronic myelomonocytic leukemia (CMML) is enriched with mutations in SRSF2 and TET2, atypical chronic myeloid leukemia (aCML) with mutations in ASXL1 and SETBP1, and the coexistence of JAK2 with SF3B1 is typical for MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN RS-T) [15]. Additional to the composition of mutated genes, also the genomic burden of each mutation differs and allows conclusions about the chronology in which aberrations occurred during disease evolution. Typically early events in MDS are usually found with high variant allele frequencies (VAFs) and include mutations in spliceosome genes, DNMT3A mutations, ASXL1, or TET2 [3,8,9,16,17]. Frequent subclonal events that are also known to drive disease progression to higher risk MDS or secondary AML involve mutations in transcription regulators such as RUNX1 or CUX1, as well as signal transducers (i.e. NRAS, KRAS, or CBL) or cohesin complex components (i.e. STAG2, or RAD21) [8,9,17]. Table 1 gives a broad overview of recurrently affected genes in MDS and their clinical and prognostic relevance.
Table 1.
Relevant mutations and noteworthy clinical consequences / targets in MDS.
| Mutated Gene | Clinical Associations | Prognostic and Therapeutic Relevance |
|---|---|---|
| DNMT3A | ||
| TET2 | ||
| ASXL1 | ||
| Spliceosome gene mutations | ||
| SF3B1 | ||
| IDH1 and IDH2 |
|
|
| RUNX1 | ||
| NRAS, KRAS | ||
| TP53 |
|
|
| PPM1D |
|
|
| NPM1 |
|
Besides gene mutations, also aberrant microRNA [6] and gene expression in peripheral blood or bone marrow, including BAALC (brain and acute leukemia, cytoplasmic), MN1 (meningioma-1), and WT1 (Wilm's tumor gene) have been shown to impact disease evolution and may refine prognostic information provided by the IPSS-R [18,19]. Following the description of the prognostic significance of high expressed genes correlated with a leukemic stem cell (LCS) signature in AML, Wang et al identified a LCS associated scoring system based on the expression of the 4 genes LAPTM4B, NGFRAP1, EMP1, and CPXM1 [20]. While higher LSC4 scores associated with disease risk as higher IPSS-R scores, complex cytogenetics, and mutations in RUNX1, ASXL1, and TP53, the LSC4 score also independently predicted prognosis in MDS patients irrespective of IPSS-R risks [20]. However, despite their undoubtful relevance in disease evolution and risk stratification, methodological obstacles in absolute gene quantification and generating comparable results between analyses, PCR quantification methods and laboratories so far prevented their introduction into the clinical practice.
Clonal evolution – en route to MDS
Over the past years the introduction of NGS approaches increased the possibility to identify molecular aberrations also in patients in the absence of cytogenetic aberrations and in those with only mild or absent cytopenia. In fact, age-related clonal hematopoiesis (ARCH) is driven by mutations also commonly found in MDS and AML (e.g. especially in the genes DNMT3A, TET2, ASXL1, JAK2, or TP53) [11,12,21, 22, 23]. The condition is associated with an increased risk to develop a myeloid malignancy including MDS, as well as with an increased all-cause mortality, especially by cardio-vascular events [11,12]. This observation in combination with increased detection (intentional or unintentional) of such mutations led to an increasing need to counsel and monitor individuals with detected mutations prospectively, and has prompted first centers to develop specialized clinics [24]. These findings also led to a still continuing debate on minimal diagnostic criteria for MDS and, especially in individuals with an otherwise unexplained cytopenia, the boundaries to MDS may be fluent [10].
However, to bring some clarification into the growing complexity of how to interpret somatic mutations and their prognostic relevance robust definitions were developed. All lack dysplasia in more or equal than 10% of cells, aberrant cytogenetics, or elevated blast counts and, subsequently, do not fulfill the WHO MDS disease criteria [1]. Clonal hematopoiesis of indeterminate potential (CHIP) or ARCH denotes the presence of at least one somatic mutation that is frequently found in myeloid neoplasia at a VAF ≥ 2% but without persistent cytopenia or history of an underlying hematologic disorder [25]. CHIP has been shown to associate with a higher risk to develop hematological cancers at a frequency of approximately 0.5% to 1% per year [26] as well a higher incidence of cardio-vascular events [11,12,27]. A somatic mutation at a VAF > 2% together with a cytopenia but not (yet) fulfilling diagnostic criteria for MDS constitutes a clonal cytopenia of undetermined significance (CCUS) [25]. CCUS associates with a much higher risk of progression to MDS of up to 50% to 90% in 5 years [10], especially when spliceosome gene mutations or co-mutated epigenetic regulators are affected, and with higher number and VAFs of the affected genes [10]. In fact, SF3B1-mutated CCUS patients almost invariably develop overt MDS with ring sideroblasts [28]. Still, so far close monitoring to early detect a myeloid neoplasm – if it occurs – should be the only clinical consequence, as not every precursor state will progress into a malignant disease [26].
Already of clinical importance is the presence of mutations in TP53 or PPM1D which associates with or pre-dispose to therapy-related MDS following chemo- or radiotherapy. Likely, their presence will have consequences regarding a more stringent indication for adjuvant chemotherapy in solid tumor patients with low relapse risk in the future [24,29]. However, the consequences of the presence of such mutations, while lacking signs of dysplasia, cytogenetic changes, or blood count abnormalities yet widely remain an area of very active research and great uncertainties regarding boundaries to MDS that overlap in some areas.
Clonal evolution – transforming from MDS to secondary AML
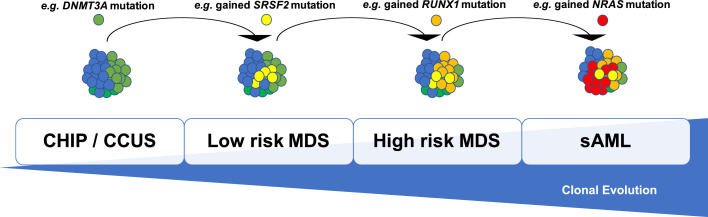
Frequency differences in the mutated genes in low risk MDS, compared to high risk MDS and also to secondary AML indicate that the order of mutation acquisition is not random during progression (Figure 1). Several studies with paired MDS and secondary AML samples have shown that signaling gene mutations (NRAS, KRAS, FLT3, PTPN11) and myeloid transcription factors (i.e. CEBPA, RUNX1), as well as TP53, and cohesin complex components (STAG2, RAD21), along with new cytogenetic abnormalities, expand or emerge at the time of disease progression [9,16,17,26,30,31]. In many cases, disease progression is associated with clonal evolution, typically defined by the expansion of a subclone with a unique set of mutation patterns that drive disease progression to high risk MDS or secondary AML. Especially the combination of STAG2 mutations and NRAS mutations may play an important role in progress to secondary AML in a subgroup of patients [32]. Furthermore, mutations in the genes FLT3, PTPN11, NRAS, and NPM1 have been associated with a fast progression from MDS to AML [30,33], and mutations in ASXL1 with progression from CMML to secondary AML [34]. Thus, monitoring tumor burden and clonal evolution using serial sequencing may provide advantages over using e.g. the blast count - which often underestimates the tumor burden - and may allow for early detection of disease progression prior to clinical deterioration [35].
Fig. 1.
Schematic display of a hierarchical, branching model of the natural MDS disease course. Acquisition of additional genetic abnormalities over time which gradually lead to a more aggressive disease phenotype.
Molecular markers predicting treatment response
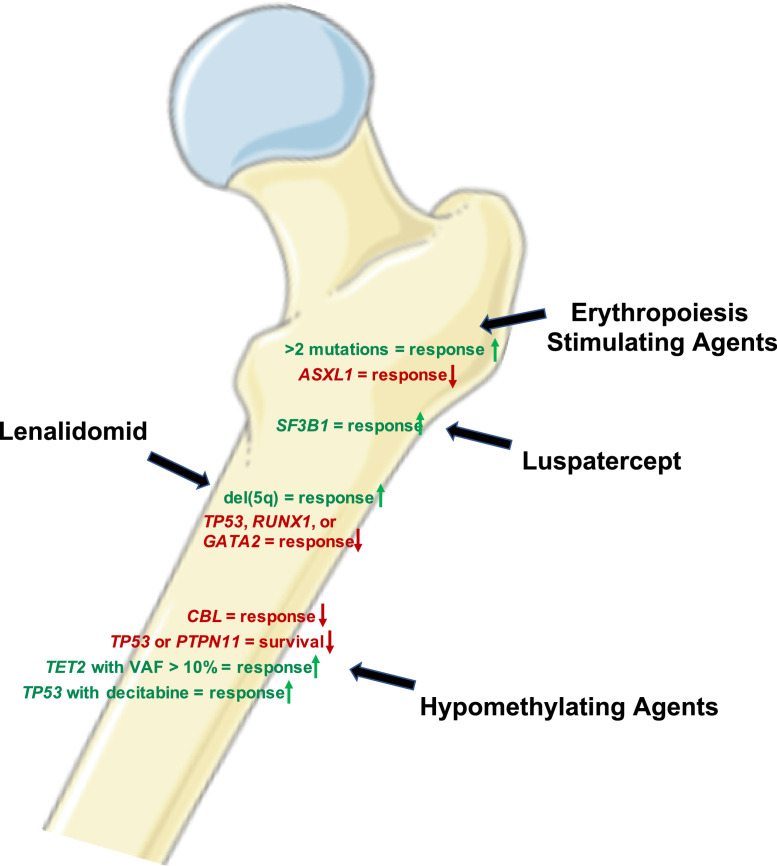
Molecular aberrations underlie the pathogenic mechanism driving MDS, and subsequently, can be utilized to estimate treatment responses (Figure 2). A long-known example in MDS is the 5q-syndrome with its particular phenotype through haploinsufficiency of a variety of genes, subsequent high response rates to lenalidomide [36], and resistance mechanism through development of TP53 mutations with their underlying pathogenesis [37]. Also the presence of RUNX1 and GATA2 mutations may render del(5q) MDS unresponsive to lenalidomide induced megakaryocytic differentiation and apoptosis [38]. Accordingly, SF3B1-mutated MDS not only shows a relatively favorable prognosis in general, but also constitutes a condition with a high likelihood to respond to luspatercept with abolishment of transfusion requirement, a recently approved treatment option for low risk MDS patients [28,39].
Fig. 2.
Genetic factors able to predict responses for different MDS treatment options (Graphic was constructed using Servier Medical Art).
In low risk MDS treatment with erythropoiesis stimulating agents (ESA) or luspatercept aims at improving anemia. However, the molecular changes that impact responses is only poorly investigated. While Kosmider et al did not identify a specific typical MDS mutation, they showed that the presence of more than 2 mutations associated with a lower likelihood of responses [40], while a recent American society of Hematology (ASH) abstract with darbepoetin suggested that in this context, ASXL1 mutations might be linked to lower response rates [41].
Also in patients with high risk MDS, in whom hypomethylating agents (HMA) remain the current standard of care with clinical responses of approximately 40% to 50% and complete remission rates of 10% to 15%, molecular alterations may allow response prediction. HMA inhibit DNA methyltransferases which leads to a decreased methylation of DNA cytosine residues. Of the genes frequently mutated in MDS that encode proteins involved in epigenetic regulations, TET2 mutations at VAFs >10% have been repeatedly shown to predict response to HMA [42,43], especially in the context of unmutated ASXL1 [42], but did not result in longer survival in 2 analyses. In contrast, a third study suggested similar response rates but significantly longer survival in patients with either TET2 or EZH2 mutations under treatment with azacitidine in two independent cohorts [44]. Additionally, in a murine model Tet2 loss sensitized cells to treatment with azacitidine in vivo which supports these clinical observations [42]. On the other hand, CBL mutations were linked to a lower response rate to HMA while TP53 and PTPN11 mutations resulted in shorter survival, but did not affect initially response to treatment [42,44]. In fact, Bejar et al especially pointed out that they were not able to identify a mutation profile predicting treatment failure of HMA [42]. Another study suggested that the use of decitabine may improve response rates in TP53 mutated MDS patients compared to standard chemotherapy and may provide a bridge to allogeneic HSCT as curative treatment option for some patients [45]. For the rare but aggressive MDS subgroup with mutated NPM1 a potential benefit of intensive chemotherapy and HSCT has been described, indicating that these patients should be treated similar to patients diagnosed with AML [46]. However, combination therapies of venetoclax and HMA have been shown to have high activity in NPM1-mutated AML as well as in higher risk MDS [47]. Subsequently, it will be interesting to investigate this combination also in this rare MDS subgroup harboring NPM1 mutations. Certainly, many of the described clinical observations would benefit from prospective trials further confirming the observed results.
Molecular markers in transplanted MDS patients
As of today, allogeneic HSCT remains the only treatment option capable to fully eradicate and cure MDS. Generally, allogeneic HSCT is mainly applied in high risk MDS, where it has demonstrated improved long term survival compared to other treatment options, especially HMA [48]. In contrast, in low risk MDS patients the expected longer survival and a substantial transplant-related toxicity has led to refrain from allogeneic HSCT for the majority of patients in this group. However, clinical factors, as well as the presence of TP53, RUNX1, or ASXL1 mutations may modify the approach and the center specific non-relapse mortality (NRM) is a key parameter when considering allogeneic HSCT in low risk MDS [49]. Higher intensity conditioning may be preferable for disease control, but the preferred regimen especially in an older and comorbid patient population is still under debate [50, 51, 52]. Another important aspect is the tumor burden of the patients going into transplantation. Many centers use HMA or AML-induction-like therapies, depending on the pre-transplant tumor burden and patient age to reduce blast levels prior to allogeneic HSCT. However, here the best approach is also up to debate and the existing data is not uncontroversial [53,54]. Given all these uncertainties the growing knowledge of MDS biology may aid in refining and individualizing the therapeutic approaches.
Due to the descripted therapeutic approach in general, the mutational landscape in transplanted MDS patients is skewed towards aberrations more frequently found in high risk MDS [55]. In a pivotal study by Lindsley et al TP53 mutations – present in 19% of the patients in the analyzed cohort - associated with shorter overall survival and higher relapse rates following allogeneic HSCT, independently of the applied conditioning regimen [55]. RAS-pathway mutations (defined as NRAS, KRAS, PTPN11, CBL, NF1, RIT1, FLT3, and KIT) on the other hand associated with shorter overall survival following allogeneic HSCT in younger patients (<40 yr) only after reduced intensity conditioning (RIC). Thus, patients with RAS-pathway mutations may benefit from myeloablative conditioning (MAC) in this group. The presence of JAK2 mutations were associated with higher rates of death without relapse, regardless of the conditioning regimen – although the reason remains unknown. In an Italian study including 274 MDS patients somatic mutation in ASXL1, RUNX1, or TP53 were independently associated with unfavorable outcomes and shorter survival after allogeneic HSCT [2]. Furthermore, in another smaller study of 87 MDS patients who underwent allogeneic HSCT for MDS, mutations in the genes TP53, TET2, and DNMT3A associated with shorter overall survival. Noteworthy, in this study all 18 TP53-mutant patients died within five years after HSCT [56]. Intriguingly, the microenvironment of TP53 mutant MDS shows an immune privileged, evasive phenotype with a PDL1 overexpression which may contribute also to an inferior graft-versus-disease response after allogeneic HSCT [57]. These findings provide a possible target for an immune-checkpoint-based approach to improve outcomes in TP53-mutated MDS patients destined for allogeneic HSCT.
MRD detection in MDS
Until today, only a few published studies fill the relative void of MRD assessment in MDS patients. Most of them have been conducted in the context of an allogeneic HSCT, which currently remains the only treatment capable to fully eradicate the disease. Still, post-transplant relapse or progression remains a major cause of transplant failure, and longitudinal MRD testing following allogeneic HSCT will help to identify patients at risk of disease progression. So far, known potential MRD markers in MDS include (CD34-lineage specific) chimerism analysis [58,59], flow cytometry [60], and WT1 expression levels [60, 61, 62], but all only included small patient numbers. In terms of traceable driver mutations, a recent study of 53 patients including 14 with MDS analyzed circulating tumor DNA by sensitive digital droplet PCR assays for individual driver mutations after allogeneic HSCT. Increasing levels of the individual mutational burden between one month and three months was a sensitive predictor of relapse following allogeneic HSCT [63]. In another study that searched bone marrow for known gene mutations adapting NGS 30 days after performing HSCT, the risk of disease progression was higher among patients in whom mutations were detected compared to those in whom these mutations were not detected [64]. In a recent paper that analyzed the impact of NGS-based mutation negativity during disease course in a heterogenous patient population that included 95 MDS patients with different treatments, including allogeneic HSCT, it was shown that achieving NGS-based mutation negativity associated with improved outcomes [65], which was especially true for the loss of TP53 mutations [65]. First prospective analyses already utilized MRD to pre-emptively treat impeding relapse and showed feasibility to potentially prevent or delay hematological relapse. The RELAZA2 trial tested an azacitidine-based MRD-guided therapy in 53 AML and MDS patients that achieved complete remission following either intensive chemotherapy or allogeneic HSCT with a traceable molecular marker (i.e. mainly NPM1 mutations) or CD34+ peripheral blood chimerism [66]. Rautenberg et al monitored 35 MDS and AML patients for peripheral blood WT1 expression following allogeneic HSCT [62], also initiating treatment with azacitidine in patients with elevated levels. After 6 cycles of azacitidine treatment, 37% of patients achieved a WT1 level normalization which correlated with improved outcomes. Noteworthy, these studies mostly used peripheral blood for MRD monitoring avoiding unpleasant bone marrow biopsies for the patients. However, so far large studies analyzing MRD in MDS following remission achievement including in the context of an allogeneic HSCT are missing and, thus, it remains an open question, which methods, targets, material (peripheral blood vs bone marrow) or time-point would be most informative.
For the detection of MDS-associated mutations for MRD analysis in the context of cytoreductive therapy without HSCT, data remains limited – likely also due to the difficulty to discriminate between CHIP and malignant populations. However, often VAF levels of MDS-associated mutations clearly exceed the bone marrow blast count and also lower-risk MDS patients with a normal blast count can have an molecular or cytogenetic aberrations in nearly all bone marrow cells [9]. This indicates that the blast count in MDS patients may frequently underestimate the actual disease burden. Subsequently, close monitoring of known mutations may provide additional prognostic information, albeit not comparable to the extent known from acute leukemias.
Conclusion and future perspectives
Our knowledge on MDS biology is getting increasingly complex and continuously growing at an unprecedented pace. Zealously running NGS panels for MDS-related mutations leading to CHIP or CCUS diagnosis in (still) apparently healthy individuals result in challenging discussions and decisions regarding further actions [24]. Especially the detection of recurrent mutations, gene expression patterns, and flow cytometric analyses have resulted in continuously improved risk stratification and the development of personalized therapies. With the growing knowledge of the complex MDS and pre-MDS biology also comes the opportunity for therapeutic trials which will help to clinically address actionable genetic changes in MDS.
Author contributions
MJ and SSch wrote the first draft of the manuscript. ASK, KHM, and UP contributed to the writing and editing and all authors finalized the manuscript.
Footnotes
Funding: None
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia [Internet] Blood. American Society of Hematology. 2016:2391–2405. doi: 10.1182/blood-2016-03-643544. [cited 2020 Jul 22] [DOI] [PubMed] [Google Scholar]
- 2.Della Porta M.G., Gallì A., Bacigalupo A., Zibellini S., Bernardi M., Rizzo E., Allione B., Van Lint M.T., Pioltelli P., Marenco P. Clinical effects of driver somatic mutations on the outcomes of patients with myelodysplastic syndromes treated with allogeneic hematopoietic stem-cell transplantation. J Clin Oncol. 2016;34:3627–3637. doi: 10.1200/JCO.2016.67.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haferlach T., Nagata Y., Grossmann V., Okuno Y., Bacher U., Nagae G., Schnittger S., Sanada M., Kon A., Alpermann T. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. Nature Publishing Group; 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steensma D.P., Fenaux P., Van Eygen K, Raza A., Santini V. Imetelstat Achieves Meaningful and Durable Transfusion Independence in High Transfusion – Burden Patients With Lower-Risk Myelodysplastic Syndromes in a Phase II Study. J Clin Oncol Clin Oncol. 2020 doi: 10.1200/JCO.20.01895. [DOI] [PubMed] [Google Scholar]
- 5.Williams J., Heppel N.H., Britt-Compton B., Grimstead J.W., Jones R.E., Tauro S., Bowen D.T., Knapper S., Groves M., Hills R.K. Telomere length is an independent prognostic marker in MDS but not in de novo AML. Br J Haematol. 2017;178:240–249. doi: 10.1111/bjh.14666. [DOI] [PubMed] [Google Scholar]
- 6.Sokol L., Caceres G., Volinia S., Alder H., Nuovo G.J., Liu C.G., Mcgraw K., Clark J.A., Sigua C.A., Chen D.T. Identification of a risk dependent microRNA expression signature in myelodysplastic syndromes. Br J Haematol. 2011;153:24–32. doi: 10.1111/j.1365-2141.2011.08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reilly B., Tanaka T.N., Diep D., Yeerna H., Tamayo P., Zhang K., Bejar R. DNA methylation identifies genetically and prognostically distinct subtypes of myelodysplastic syndromes. Blood Adv. 2019;3:2845–2858. doi: 10.1182/bloodadvances.2019000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E., Gerstung M., Malcovati L., Tauro S., Gundem G., Van Loo P., Yoon C.J., Ellis P., Wedge D.C., Pellagatti A. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter M.J., Shen D., Shao J., Ding L., White B., Kandoth C., Miller C.A., Niu B., McLellan M.D., Dees N.D. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia [Internet] Nature Publishing Group. 2013;27:1275–1282. doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malcovati L., Gallì A., Travaglino E., Ambaglio I., Rizzo E., Molteni E., Elena C., Ferretti V.V., Catricalà S., Bono E. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood. 2017;129:3371–3378. doi: 10.1182/blood-2017-01-763425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/nejmoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., Purcell S.M. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/nejmoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuurhuis G.J., Heuser M., Freeman S., Béne M.C., Buccisano F., Cloos J., Grimwade D., Haferlach T., Hills R.K., Hourigan C.S. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 2018;131:1275–1291. doi: 10.1182/blood-2017-09-801498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platzbecker U. Treatment of MDS. Blood. 2019;133:1096–1107. doi: 10.1182/blood-2018-10-844696. [DOI] [PubMed] [Google Scholar]
- 15.Palomo L., Meggendorfer M., Hutter S., Twardziok S., Adema V., Fuhrmann I., Fuster-Tormo F., Xicoy B., Zamora L., Acha P. Molecular landscape and clonal architecture of adult myelodysplastic /myeloproliferative neoplasms. Blood. 2020 doi: 10.1182/blood.2019004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bejar R., Stevenson K., Abdel-Wahab O., Galili N., Nilsson B., Garcia-Manero G., Kantarjian H., Raza A., Levine R.L., Neuberg D. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsley R.C., Mar B.G., Mazzola E., Grauman P.V., Shareef S., Allen S.L., Pigneux A., Wetzler M., Stuart R.K., Erba H.P. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thol F., Yun H., Sonntag A.K., Damm F., Weissinger E.M., Krauter J., Wagner K., Morgan M., Wichmann M., Göhring G. Prognostic significance of combined MN1, ERG, BAALC, and EVI1 (MEBE) expression in patients with myelodysplastic syndromes. Ann Hematol. 2012;91:1221–1233. doi: 10.1007/s00277-012-1457-7. [DOI] [PubMed] [Google Scholar]
- 19.Pellagatti A., Benner A., Mills K.I., Cazzola M., Giagounidis A., Perry J., Malcovati L., Della Porta M.G., Jädersten M., Verma A. Identification of gene expression-based prognostic markers in the hematopoietic stem cells of patients with myelodysplastic syndromes. J Clin Oncol. 2013;31:3557–3564. doi: 10.1200/JCO.2012.45.5626. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y.H., Lin C.C., Yao C.Y., Hsu C.L., Hou H.A., Tsai C.H., Chou W.C., Tien H.F. A 4-gene leukemic stem cell score can independently predict the prognosis of myelodysplastic syndrome patients. Blood Adv. 2020;4:644–654. doi: 10.1182/bloodadvances.2019001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young A.L., Challen G.A., Birmann B.M., Druley T.E. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. Nature Publishing Group; 2016;7 doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimm J., Bill M., Jentzsch M., Beinicke S., Häntschel J., Goldmann K., Schulz J., Cross M., Franke G.–N., Behre G. Clinical impact of clonal hematopoiesis in acute myeloid leukemia patients receiving allogeneic transplantation. Bone Marrow Transplant. 2019;54 doi: 10.1038/s41409-018-0413-0. [DOI] [PubMed] [Google Scholar]
- 23.Rothenberg-Thurley M., Amler S., Goerlich D., Köhnke T., Konstandin N.P., Schneider S., Sauerland M.C., Herold T., Hubmann M., Ksienzyk B. Persistence of pre-leukemic clones during first remission and risk of relapse in acute myeloid leukemia. Leukemia [Internet]. Springer US; 2018;32:1598–1608. doi: 10.1038/s41375-018-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steensma D.P., Bolton K.L. What To Tell Your Patient With Clonal Hematopoiesis And Why: Insights From Two Specialized Clinics. Blood. 2020;136:1623–1631. doi: 10.1182/blood.2019004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steensma D.P., Bejar R., Jaiswal S., Lindsley R.C., Sekeres M.A., Hasserjian R.P., Ebert B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeZern A.E., Malcovati L., Ebert B.L. CHIP, CCUS, and Other Acronyms: Definition, Implications, and Impact on Practice. Am Soc Clin Oncol Educ B. 2019:400–410. doi: 10.1200/edbk_239083. [DOI] [PubMed] [Google Scholar]
- 27.Jaiswal S., Natarajan P., Silver A.J., Gibson C.J., Bick A.G., Shvartz E., McConkey M., Gupta N., Gabriel S., Ardissino D. Clonal Hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malcovati L., Stevenson K., Papaemmanuil E., Neuberg D., Bejar R., Boultwood J., Bowen D.T., Campbell P.J., Ebert B.L., Fenaux P. SF3B1-mutant myelodysplastic syndrome as a distinct disease subtype - A Proposal of the International Working Group for the Prognosis of Myelodysplastic Syndromes (IWG-PM) Blood [Internet] 2020 doi: 10.1182/blood.2020004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coombs C.C., Zehir A., Devlin S.M., Kishtagari A., Syed A., Jonsson P., Hyman D.M., Solit D.B., Robson M.E., Baselga J. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell. 2017;21:374–382.e4. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T., Tyndel M.S., Kim H.J., Ahn J.S., Choi S.H., Park H.J., Kim Y.K., Yang D.H., Lee J.J., Jung S.H. The clonal origins of leukemic progression of myelodysplasia. Leukemia [Internet] Nature Publishing Group. 2017;31:1928–1935. doi: 10.1038/leu.2017.17. [DOI] [PubMed] [Google Scholar]
- 31.Murphy D.M., Bejar R., Stevenson K., Neuberg D., Shi Y., Cubrich C., Richardson K., Eastlake P., Garcia-Manero G., Kantarjian H. NRAS mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia. 2013;27:2077–2081. doi: 10.1038/leu.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martín-Izquierdo M., Abáigar M., Hernández-Sánchez J.M., Tamborero D., López-Cadenas F., Ramos F., Lumbreras E., Madinaveitia-Ochoa A., Megido M., Labrador J., Sánchez-Real J. Co-occurrence of cohesin complex and Ras signaling mutations during progression from myelodysplastic syndromes to secondary acute myeloid leukemia. Haematologica. 2020 doi: 10.3324/haematol.2020.248807. haematol.2020.248807 p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K., Jabbour E., Wang X., Luthra R., Bueso-Ramos C., Patel K., Pierce S., Yang H., Wei Y., Daver N. Dynamic acquisition of FLT3 or RAS alterations drive a subset of patients with lower risk MDS to secondary AML. Leukemia. 2013;27:2081–2083. doi: 10.1038/leu.2013.165. [DOI] [PubMed] [Google Scholar]
- 34.Gelsi-Boyer V., Trouplin V., Roquain J., Adélaïde J., Carbuccia N., Esterni B., Finetti P., Murati A., Arnoulet C., Zerazhi H. ASXL1 mutation is associated with poor prognosis and acute transformation in chronic myelomonocytic leukaemia. Br J Haematol. 2010;151:365–375. doi: 10.1111/j.1365-2141.2010.08381.x. [DOI] [PubMed] [Google Scholar]
- 35.Menssen A.J., Walter M.J. Genetics of progression from MDS to secondary leukemia. Blood. 2020;136:50–60. doi: 10.1182/blood.2019000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenaux P., Giagounidis A., Selleslag D., Beyne-Rauzy O., Mufti G., Mittelman M., Muus P., Te Boekhorst P., Sanz G., Del Cañizo C. A randomized phase 3 study of lenalidomide versus placebo in RBC transfusion-dependent patients with Low-/Intermediate-1-risk myelodysplastic syndromes with del5q. Blood. 2011;118:3765–3776. doi: 10.1182/blood-2011-01-330126. [DOI] [PubMed] [Google Scholar]
- 37.Ebert B.L. Molecular dissection of the 5q deletion in myelodysplastic syndrome. Semin Oncol. 2011;38:621–626. doi: 10.1053/j.seminoncol.2011.04.010.Molecular. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Høyer S., Deng Y., Parker J., Jiang J., Mo A., Docking T.R., Gharaee N., Li J., Umlandt P., Fuller M., Jädersten M. Loss of lenalidomide-induced megakaryocytic differentiation leads to therapy resistance in del(5q) myelodysplastic syndrome. Nat Cell Biol [Internet]. Springer US; 2020;22:526–533. doi: 10.1038/s41556-020-0497-9. [DOI] [PubMed] [Google Scholar]
- 39.Fenaux P., Platzbecker U., Mufti G.J., Garcia-Manero G., Buckstein R., Santini V., Díez-Campelo M., Finelli C., Cazzola M., Ilhan O. Luspatercept in patients with lower-risk myelodysplastic syndromes. N Engl J Med. 2020;382:140–151. doi: 10.1056/NEJMoa1908892. [DOI] [PubMed] [Google Scholar]
- 40.Kosmider O., Passet M., Santini V., Platzbecker U., Andrieu V., Zini G., Beyne-Rauzy O., Guerci A., Masala E., Balleari E., Bulycheva E., Dreyfus F., Fenaux P. Are somatic mutations predictive of response to erythropoiesis stimulating agents in lower risk myelodysplastic syndromes? Haematologica. 2016;101:e280–e283. doi: 10.3324/haematol.2016.142695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanamoto H., Morita Y., Ichikawa M., Nannya Y., Shibayama H., Maeda Y., Hata T., Miyamoto T., Kawabata H., Takeuchi K., Tanaka H., Kishimoto J., Miyano S. ASXL1 Mutations Predict a Poor Response to Darbepoetin Alfa in Anemic Patients with Low-Risk MDS: A Multicenter. Phase II Study. Blood. 2020;136(S1):28–29. [Google Scholar]
- 42.Bejar R., Lord A., Stevenson K., Bar-Natan M., Pérez-Ladaga A., Zaneveld J., Wang H., Caughey B., Stojanov P., Getz G. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124:2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itzykson R., Kosmider O., Cluzeau T., Mansat-De Mas V., Dreyfus F., Beyne-Rauzy O., Quesnel B., Vey N., Gelsi-Boyer V., Raynaud S. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25:1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 44.Tobiasson M., McLornan D.P., Karimi M., Dimitriou M., Jansson M., Ben Azenkoud A, Jädersten M., Lindberg G., Abdulkadir H., Kulasekararaj A. Mutations in histone modulators are associated with prolonged survival during azacitidine therapy. Oncotarget. 2016;7:22103–22115. doi: 10.18632/oncotarget.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welch J.S., Petti A.A., Miller C.A., Fronick C.C., O'Laughlin M., Fulton R.S., Wilson R.K., Baty J.D., Duncavage E.J., Tandon B. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med. 2016;375:2023–2036. doi: 10.1056/nejmoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montalban-Bravo G., Kanagal-Shamanna R., Sasaki K., Patel K., Ganan-Gomez I., Jabbour E., Kadia T., Ravandi F., DiNardo C., Borthakur G. NPM1 mutations define a specific subgroup of MDS and MDS/MPN patients with favorable outcomes with intensive chemotherapy. Blood Adv. 2019;3:922–933. doi: 10.1182/bloodadvances.2018026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ball B.J., Famulare C.A., Stein E.M., Tallman M.S., Derkach A., Roshal M., Gill S.I., Manning B.M., Koprivnikar J., McCloskey J. Venetoclax and hypomethylating agents (HMAs) induce high response rates in MDS, including patients after HMA therapy failure. Blood Adv. 2020;4:2866–2870. doi: 10.1182/bloodadvances.2020001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kroeger N., Sockel K., Wolschke C., Bethge W., Schlenk R.F., Wolf D., Stadler M., Kobbe G., Wulf G., Bug G. Prospective Multicenter Phase 3 Study Comparing 5-Azacytidine (5-Aza) Induction Followed By Allogeneic Stem Cell Transplantation Versus Continuous 5-Aza According to Donor Availability in Elderly MDS Patients (55-70 years) (VidazaAllo Study) Blood. 2018;132(Suppl: 208) [Google Scholar]
- 49.Novak P., Zabelina T., Wolschke C., Ayuk F., Christopeit M., Kröger N. Allogeneic Stem Cell Transplantation for Patients with Lower-Risk Myelodysplastic Syndrome. Biol Blood Marrow Transplant. 2020;26:2047–2052. doi: 10.1016/j.bbmt.2020.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Jentzsch M., Döhring C., Linke R., Hille A., Grimm J., Pönisch W., Vucinic V., Franke G.-N., Behre G., Niederwieser D. Comparison of non-myeloablative and reduced-intensity allogeneic stem cell transplantation in older patients with myelodysplastic syndromes. Am J Hematol. 2019;94:1344–1352. doi: 10.1002/ajh.25636. [DOI] [PubMed] [Google Scholar]
- 51.Kröger N., Iacobelli S., Franke G.N., Platzbecker U., Uddin R., Hübel K., Scheid C., Weber T., Robin M., Stelljes M. Dose-reduced versus standard conditioning followed by allogeneic stem-cell transplantation for patients with myelodysplastic syndrome: A prospective randomized phase III study of the EBMT (RICMAC Trial) J Clin Oncol. 2017;35:2157–2164. doi: 10.1200/JCO.2016.70.7349. [DOI] [PubMed] [Google Scholar]
- 52.Scott B.L., Pasquini M.C., Logan B.R., Wu J., Devine S.M., Porter D.L., Maziarz R.T., Warlick E.D., Fernandez H.F., Alyea E.P. Myeloablative versus reduced-intensity hematopoietic cell transplantation for acute myeloid leukemia and myelodysplastic syndromes. J Clin Oncol. 2017;35:1154–1161. doi: 10.1200/JCO.2016.70.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warlick E.D., Cioc A., DeFor T., Dolan M., Weisdorf D. Allogeneic Stem Cell Transplantation for Adults with Myelodysplastic Syndromes: Importance of Pretransplant Disease Burden. Biol Blood Marrow Transplant [Internet]. American Society for Blood and Marrow Transplantation; 2009;15:30–38. doi: 10.1016/j.bbmt.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 54.De Witte T., Bowen D., Robin M., Malcovati L., Niederwieser D., Yakoub-Agha I., Mufti G.J., Fenaux P., Sanz G., Martino R. Allogeneic hematopoietic stem cell transplantation for MDS and CMML: Recommendations from an international expert panel. Blood. 2017;129:1753–1762. doi: 10.1182/blood-2016-06-724500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindsley R.C., Saber W., Mar B.G., Redd R., Wang T., Haagenson M.D., Grauman P.V., Hu Z.-H., Spellman S.R., Lee S.J. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med. 2017;376:536–547. doi: 10.1056/nejmoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bejar R., Stevenson K.E., Caughey B., Lindsley R.C., Mar B.G., Stojanov P., Getz G., Steensma D.P., Ritz J., Soiffer R. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32:2691–2698. doi: 10.1200/JCO.2013.52.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sallman D.A., McLemore A.F., Aldrich A.L., Komrokji R.S., McGraw K.L., Dhawan A., Geyer S., Hou H.-A., Eksioglu E.A., Sullivan A. TP53 mutations in myelodysplastic syndromes and secondary AML confer an immunosuppressive phenotype. Blood. 2020 doi: 10.1182/blood.2020006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenow F., Berkemeier A., Krug U., Müller-Tidow C., Gerss J., Silling G., Groth C., Wieacker P., Bogdanova N., Mesters R. CD34 + lineage specific donor cell chimerism for the diagnosis and treatment of impending relapse of AML or myelodysplastic syndrome after allo-SCT. Bone Marrow Transplant. 2013;48:1070–1076. doi: 10.1038/bmt.2013.2. [DOI] [PubMed] [Google Scholar]
- 59.Bornhäuser M., Oelschlaegel U., Platzbecker U., Bug G., Lutterbeck K., Kiehl M.G., Schetelig J., Kiani A., Illmer T., Schaich M. Monitoring of donor chimerism in sorted CD34+ peripheral blood cells allows the sensitive detection of imminent relapse after allogeneic stem cell transplantation. Haematologica. 2009;94:1613–1617. doi: 10.3324/haematol.2009.007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mo X.D., Qin Y.Z., Zhang X.H., Xu L.P., Wang Y., Yan C.H., Chen H., Chen Y.H., Han W., Wang F.R. Minimal residual disease monitoring and preemptive immunotherapy in myelodysplastic syndrome after allogeneic hematopoietic stem cell transplantation. Ann Hematol [Internet]. Annals of Hematology; 2016;95:1233–1240. doi: 10.1007/s00277-016-2706-y. [DOI] [PubMed] [Google Scholar]
- 61.Yoon J.H., Jeon Y.W., ah Yahng S, Shin S.H., Lee S.E., Cho B.S., Lee D.G., Eom K.S., Kim H.J., Lee S. Wilms Tumor Gene 1 Expression as a Predictive Marker for Relapse and Survival after Hematopoietic Stem Cell Transplantation for Myelodysplastic Syndromes. Biol Blood Marrow Transplant [Internet]. Elsevier Inc. 2015;21:460–467. doi: 10.1016/j.bbmt.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 62.Rautenberg C., Bergmann A., Pechtel S., Fischermanns C., Haas R., Germing U., Kobbe G., Schroeder T. Wilm's Tumor 1-guided preemptive treatment with hypomethylating agents for molecular relapse of AML and MDS after allogeneic transplantation. Bone Marrow Transplant [Internet]. Springer US; 2020 doi: 10.1038/s41409-020-01039-2. [DOI] [PubMed] [Google Scholar]
- 63.Nakamura S., Yokoyama K., Shimizu E., Yusa N., Kondoh K., Ogawa M., Takei T., Kobayashi A., Ito M., Isobe M. Prognostic impact of circulating tumor DNA status post–allogeneic hematopoietic stem cell transplantation in AML and MDS. Blood. 2019;133:2682–2695. doi: 10.1182/blood-2018-10-880690. [DOI] [PubMed] [Google Scholar]
- 64.Duncavage E.J., Jacoby M.A., Chang G.S., Miller C.A., Edwin N., Shao J., Elliott K., Robinson J., Abel H., Fulton R.S., Fronick C.C., O'Laughlin M., Heath S.E. Mutation Clearance after Transplantation for Myelodysplastic Syndrome. N Engl J Med. 2018;379:1028–1041. doi: 10.1056/nejmoa1804714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yun S., Geyer S.M., Komrokji R.S., Al Ali N.H., Song J., Hussaini M., Sweet K.L., Lancet J.E., List A.F., Padron E. Prognostic significance of serial molecular annotation in myelodysplastic syndromes (MDS) and secondary acute myeloid leukemia (sAML) Leukemia [Internet]. Springer US; 2020 doi: 10.1038/s41375-020-0997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Platzbecker U., Middeke J.M., Sockel K., Herbst R., Wolf D., Baldus C.D., Oelschlägel U., Mütherig A., Fransecky L., Noppeney R. Measurable residual disease-guided treatment with azacitidine to prevent haematological relapse in patients with myelodysplastic syndrome and acute myeloid leukaemia (RELAZA2): an open-label, multicentre, phase 2 trial. Lancet Oncol. 2018;19:1668–1679. doi: 10.1016/S1470-2045(18)30580-1. [DOI] [PubMed] [Google Scholar]
- 67.Walter M.J., Ding L., Shen D., Shao J., Grillot M., McLellan M., Fulton R., Schmidt H., Kalicki-Veizer J., O'Laughlin M. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thol F., Friesen I., Damm F., Yun H., Weissinger E.M., Krauter J., Wagner K., Chaturvedi A., Sharma A., Wichmann M. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J Clin Oncol. 2011;29:2499–2506. doi: 10.1200/JCO.2010.33.4938. [DOI] [PubMed] [Google Scholar]
- 69.Abdel-Wahab O., Pardanani A., Patel J., Wadleigh M., Lasho T., Heguy A., Beran M., Gilliland D.G., Levine R.L., Tefferi A. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia. 2011;25:1200–1202. doi: 10.1038/leu.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brian Dalton W., Helmenstine E., Pieterse L., Li B., Gocke C.D., Donaldson J., Xiao Z., Gondek L.P., Ghiaur G., Gojo I. The K666N mutation in SF3B1 is associated with increased progression of MDS and distinct RNA splicing. Blood Adv. 2020;4:1192–1196. doi: 10.1182/bloodadvances.2019001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patnaik M.M., Hanson C.A., Hodnefield J.M., Lasho T.L., Finke C.M., Knudson R.A., Ketterling R.P., Pardanani A., Tefferi A. Differential prognostic effect of IDH1 versus IDH2 mutations in myelodysplastic syndromes: A Mayo Clinic Study of 277 patients. Leukemia. Nature Publishing Group. 2012;26:101–105. doi: 10.1038/leu.2011.298. [DOI] [PubMed] [Google Scholar]
- 72.Montalban-Bravo G., Garcia-Manero G., Jabbour E. Therapeutic choices after hypomethylating agent resistance for myelodysplastic syndromes. Curr Opin Hematol. 2018;25:146–153. doi: 10.1097/MOH.0000000000000400. [DOI] [PubMed] [Google Scholar]
- 73.Gaidzik V.I., Teleanu V., Papaemmanuil E., Weber D., Paschka P., Hahn J., Wallrabenstein T., Kolbinger B., Köhne C.H., Horst H.A. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2160–2168. doi: 10.1038/leu.2016.126. [DOI] [PubMed] [Google Scholar]
- 74.Bernard E., Nannya Y., Hasserjian R.P., Devlin S.M., Tuechler H., Medina-Martinez J.S., Yoshizato T., Shiozawa Y., Saiki R., Malcovati L. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med. 2020;26:1549–1556. doi: 10.1038/s41591-020-1008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sallman D.A., DeZern A.E., Steensma D.P., Sweet K.L., Cluzeau T., Sekeres M.A., Garcia-Manero G., Roboz G.J., McLemore A.F., McGraw K.L. Phase 1b/2 Combination Study of APR-246 and Azacitidine (AZA) in Patients with TP53 mutant Myelodysplastic Syndromes (MDS) and Acute Myeloid Leukemia (AML) Blood. 2018;132(Suppl: 3091) [Google Scholar]