Abstract
Ferroptosis is a recently identified non-apoptotic form of cell death characterized by iron-dependent lipid peroxidation. However, the underlying exact mechanisms remain poorly understood. Here, we report that the total levels of N6-methyladenosine (m6A) modification are evidently increased upon exposure to ferroptosis-inducing compounds due to the upregulation of methylase METTL4 and the downregulation of demethylase FTO. Interestingly, RNA-seq shows that m6A modification appears to trigger autophagy activation by stabilizing BECN1 mRNA, which may be the potential mechanism for m6A modification-enhanced HSC ferroptosis. Importantly, YTHDF1 is identified as a key m6A reader protein for BECN1 mRNA stability, and knockdown of YTHDF1 could prevent BECN1 plasmid-induced HSC ferroptosis. Noteworthy, YTHDF1 promotes BECN1 mRNA stability and autophagy activation via recognizing the m6A binding site within BECN1 coding regions. In mice, erastin treatment alleviates liver fibrosis by inducing HSC ferroptosis. HSC-specific inhibition of m6A modification could impair erastin-induced HSC ferroptosis in murine liver fibrosis. Moreover, we retrospectively analyzed the effect of sorafenib on HSC ferroptosis and m6A modification in advanced fibrotic patients with hepatocellular carcinoma (HCC) receiving sorafenib monotherapy. Attractively, the m6A modification upregulation, autophagy activation, and ferroptosis induction occur in human HSCs. Overall, these findings reveal novel signaling pathways and molecular mechanisms of ferroptosis, and also identify m6A modification-dependent ferroptosis as a potential target for the treatment of liver fibrosis.
Keywords: Ferroptosis, Autophagy, Hepatic stellate cell, Liver fibrosis, m6A modification, YTHDF1
1. Introduction
Liver fibrosis is a complex physiological and pathophysiological condition associated with the mechanisms of cicatrization [1]. The deposition of extracellular matrix is a central event in liver fibrosis, and the myofibroblasts matrix is mainly produced by activated HSCs [2]. Consequently, elimination of HSCs is a main therapeutic strategy in the development of anti-fibrotic therapy [3]. We previously reported that the pathological and pathophysiological condition of liver fibrosis could alleviated by triggering the activation of apoptosis [4], senescence [5], necroptosis [6], lipocyte phenotype [7], and prevention of the proliferation [8], contractile [9], glycolytic [10] and pericyte [11] in HSCs. Interestingly, ferroptosis is thought to be a new and effective approach for scavenging HSCs in our recent research. We found that RNA binding proteins ELAVL1 and ZFP36 could trigger HSC ferroptosis to alleviate liver fibrosis by regulating autophagy pathway [12,13]. Moreover, we also revealed that mitochondrial iron metabolism pathway may regulate HSC ferroptosis via BRD7/P53/SLC25A28 axis [14]. The aim of the present study was to evaluate the potential mechanisms of ferroptosis and its role in the inhibition of liver fibrosis.
Distinct from classic morphological features of apoptosis, necrosis, necroptosis, pyroptosis, and senescence, ferroptosis is a recently identified form of programmed cell death [15]. Iron-depended lipid peroxidation, glutathione peroxidase 4 (GPX4) deletion and condensed mitochondrial membrane densities are the main characteristics of ferroptosis [16]. Attractively, system Xc- inhibition (such as sulfasalazine, sorafenib, erastin), GPX4 inhibition (such as FIN56, altretamine, RSL3, FINO2), and physiological conditions (such as cystine deprivation, amino acid starvation, high extracellular glutamate) are reported to induce ferroptosis [17]. Importantly, the crosstalk between ferroptosis and autophagy has received increasing concern, and autophagy is a targeted pathway that regulates the cellular sensitivity to ferroptosis [18]. Although transcriptional regulation of autophagy signaling pathways in ferroptosis has been elucidated well, the function of post-transcriptional regulation in ferroptosis remains poorly known.
The m6A RNA modification serves as the most abundant post-transcriptional mechanism in eukaryotic messenger RNAs (mRNAs), which is a reversible process controlled by methyltransferase, demethylases and m6A binding proteins. The methyltransferase complex catalyzes m6A mRNA methylation that contains WT1 associated protein (WTAP), methyltransferase like 3 (METTL3), METTL4, and METTL14. In contrast, alkB homolog 5 (ALKBH5) and obesity-associated protein (FTO) function as demethylases to remove m6A modifications from RNA and thus keep m6A modification in a dynamic balance. Additionally, m6A binding proteins including heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1), insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs), YTH-domain- containing protein 1/2 (YTHDC1/2), and YTH domain family 1/2/3 (YTHDF1/2/3) bind to the m6A motif to affect RNA stability or function [19]. Interestingly, exploring the post-transcriptional regulation of m6A-mediated HSC ferroptosis may provide therapeutic targets and effective diagnostic signs for liver fibrosis.
In the present study, we investigated a novel signaling pathways and molecular mechanisms of ferroptosis in liver fibrosis. Our study revealed that m6A reader YTHDF1 promoted the stability of BECN1 mRNA via recognizing the m6A binding site, thus triggering autophagy activation, and eventually leading to HSC ferroptosis. We indicated that m6A modification may be novel and critical post-transcriptional regulators of ferroptosis in liver fibrosis.
2. Materials and methods
2.1. Human liver specimens
From September 2015 and December 2020, we obtained liver resection tissues of ten patients with liver cirrhosis complicated with HCC treated with sorafenib monotherapy and liver biopsy specimens of ten patients with liver cirrhosis without any treatment were obtained in the Affiliated Hospital of Nanjing University of Chinese Medicine. The initial dose of sorafenib was 400 mg, taken orally twice daily after dinner and breakfast [21]. Subsequently, sorafinib was temporarily interrupted or reduced based on the toxicity and tolerance. Obtained written informed consent from patients. According to the electronic medical records, the data on the characteristics of patients were collected retrospectively. The diagnostic criteria for HCC and liver cirrhosis according to the American Association for the study of liver disease (AASLD) [20]. This research protocol complied with the ethical guidelines of the 1975 Declaration of Helsinki Principles and was approved by the ethics committee of Nanjing University of Chinese Medicine.
2.2. Isolation and characterization of primary human HSC
Laser capture microdissection was used to isolate primary HSCs from human liver tissue according to our reports previously [22]. Briefly, human liver tissue was stained with desmin (ab15200, Abcam) by immunofluorescence. Then, CapSure® LCM Cap (LCM0211, Thermo Fisher Scientific) was placed on the section. The cap was passed through by laser pulses, forming a thin protuberance of the thermoplastic film that connects the gap between the tissue and the cap and attaching it to the positive cells stained by the desmin. Target cells attached to the cap can be removed by lifting the cap. α-SMA (ab5694, Abcam) and platelet derived growth factor receptor beta (PDGFRB, ab32570, Abcam) were detected to confirm the characterization and purification of the obtained human HSCs.
2.3. Animal experiments
ICR mice (8-week-old, 18–22 g) were obtained from Yangzhou University (Yangzhou, China). There were 8 mice in each group and they were randomly divided into 6 groups. Mice were treated with Vehicle, CCl4, VA-Lip-control-vector + CCl4+Erastin, VA-Lip-Mettl4-shRNA + CCl4+Erastin, VA-Lip-Fto-plasmid + CCl4+Erastin, VA-Lip- Ythdf1-shRNA + CCl4+Erastin, respectively. A mixture of olive oil and carbon tetrachloride (CCl4) (9:1 (v/v)) was used to trigger liver fibrosis in mouse model by intraperitoneal injection (0.1 ml/20 g body weight), according to our previous reports [23]. Erastin was dissolved according to the instructions and given once every other day for 2 weeks by intraperitoneal injection (30 mg/kg) after the CCl4 treatment. Within 2 weeks after CCl4 treatment, VA-Lip-Mettl4-shRNA, VA-Lip-Fto-Plasmid and VA-Lip-Ythdf1-shRNA (0.75 mg/kg) were injected intravenously 3 times a week. The liver samples and blood of each group were collected at the end of the experiment. Fixation of a small portion of the liver with 4% Paraformaldehyde Fix Solution (PFS, P6148, Sigma-Aldrich) for histopathological studies. All in vivo experimental procedures were performed according to the institutional and local animal care and use committee of Nanjing University of Chinese Medicine (Nanjing, China).
2.4. Isolation and characterization of primary HSC
Primary mouse HSCs were isolated according to our previous reports [12,13]. In brief, DMEM-free solution containing pronase (2 mg/ml, PRON-RO, 10165921001, Roche) and collagenase IV (1 mg/ml, V900893, Vetec) were used to perfuse the liver in situ and following HBSS including EDTA (0.5 mM, E6758, Sigma-Aldrich). Upon completing the perfusion, the digested hepatocytes from the liver were dispersed in DMEM-free. Then, the filamentous gelatinous material was inhibited by using DNase enzyme (D4263, Sigma-Aldrich). The undigested debris was then removed by filter and centrifuged in 4 °C for 5 min at 50×g. 25% Histodenz (D2158, Sigma-Aldrich) gradient centrifugation, and the supernatant was collected to separate primary HSCs. Cells were inoculated on a culture dishes (CLS430599, Sigma-Aldrich) with a diameter of 60 mm. By detecting PDGFRB and α-SMA to identify and purity of the obtained HSCs.
2.5. Histological analyses
Liver tissue samples were paraffin embedded and 4 μm tissue sections stained with standard methods and subjected to histopathological analysis according to our reports previously [12]. Briefly, the samples were fixed in 4% PFS and then were transferred to different concentration of ethanol for dehydration. Hepatic morphology and liver fibrosis were examined by H&E, Masson, and Sirius red staining.
2.6. Reagents and antibodies
Ferrostatin-1 (S7243), necrostatin-1 (S8037), Z-VAD-FMK (S7023), liproxstatin-1 (S7699), sorafenib (S7397), erastin (S7242), RSL3 (S8155), MG-132 (S2619) were bought from Selleck Chemicals. CHX (C7698) and Act-D (129935) were purchased from Sigma-Aldrich. Anti-N6-methyladenosine antibody (ab208577), anti-METTL3 antibody (ab195352), anti-METTL4 (ab107540), anti-METTL14 antibody (ab220030), anti-FTO antibody (ab92821), anti-ALKBH5 antibody (ab195377), anti-YTHDF1 antibody (ab220162), anti-YTHDF2 antibody (ab220163), anti-YTHDF3 antibody (ab220161), anti-YTHDC2 antibody (ab220160), anti-HNRNPA2B1 antibody (ab31645), anti-ATG3 antibody (ab108282), anti-ATG4A antibody (ab223374), anti-ATG5 antibody (ab108327), anti-BECN1 antibody (ab207612), anti-ATG7 antibody (ab133528), anti-ATG9A antibody (ab108338), anti-ATG12 antibody (ab155589), anti-ATG16L1 antibody (ab187671), anti-LC3-I/II antibody (ab128025), anti-P62 antibody (ab109012), anti-NCOA4 antibody (ab86707), anti-FTH1 antibody (ab65080), and anti-beta actin (ab6276) antibody were purchased from Abcam Technology. Anti-WTAP antibody (sc-374280) was bought from Santa Cruz Biotechnology. Anti-Mouse IgG (G-21040) and anti-Rabbit IgG (G-21234) were bought from Thermo Fisher Scientific.
2.7. Plasmid construction
The pcDNA3.1-FTO plasmid, pcDNA3.1-BECN1 plasmid, METTL4 shRNA (sc-75777-SH, sc-149388-SH), YTHDF1 shRNA (sc-76945-SH, sc-155423-SH) and control vector were obtained from KeyGEN BioTECH (KG20200903-10) and Santa Cruz Biotechnology, respectively. CMV-TurboRFP-EGFP-LC3-PGK-Puro plasmid (GM-1314L204H) and pGM-CMV-GFP-hLC3 (GM-1314P101H) were obtained from Genomeditech (Shanghai, China). According to our previous reports, VA-Lip-Mettl4 -shRNA, VA-Lip-Fto-plasmid, VA-Lip-Ythdf1-shRNA, and VA-Lip-Control-vector were prepared [12]. Briefly, VA solution was formed by 50 μl DMSO add 5 mg VA (95144, Sigma). 0.14 μmol lipotrust solution (LEO-01, Hokkaido System Science) and 280 nmol VA solution were mixed in tube (AM12450, Thermo Fisher Scientific) at 25 °C. Under the condition of stirring, 12.24 nmol Mettl4-shRNA, Fto-plasmid, Ythdf1-shRNA, and Control-vector were added into VA-Lip solution. Material trapped in the filter (596–3320, Thermo Fisher Scientific) and the fractions were collected to achieve the indicated dose for in vivo use.
2.8. Construction of BECN1 mutant plasmid
Mutations of BECN1 were constructed by PCR-based methods according to previous reports [24]. The 5′-UTR, 3′UTR, and CDS of BECN1 mRNA was synthesized and subcloned into the vector pCDNA3.1. Moreover, Adenine (A) 437 and Adenine (A) 1276 were mutated to Guanine (G). Briefly, the mutant of BECN1 was conducted by the following primer pairs: (A437G mutant) BECN1, forward 5′-CAGATACTCTTT TAGGCCAGCTGGACACTCA-3′ and reverse 5′-TGAGTGTCCAGCTGGCCT AAAAGAGTATCTG-3′, (A1276G mutant) BECN1, 5′-TCTGAGGAGCAGTGGGC AAAAGCTCTCAAGT-3′ and reverse 5′-ACTTG AGAGCTTTTGCCCACTGCTC CTCAGA-3’. BECN1 WT and mutants were confirmed by DNA sequencing.
2.9. Construction of stable cell lines
According to a reported protocol, stable cell lines were constructed [25]. Briefly, the HSCs were seeded in 6-well plates (CLS3516, Sigma-Aldrich). When cells reached 70% confluence, serum-free DMEM medium was replaced. Then, the plasmid and Lipofectamine 3000 transfection reagent mixture (L3000015, Invitrogen) were added into each well to initiate transfection. After 8 h, the medium containing 5% FBS was replaced. After the beginning of the transfection approximately 48 h, the cells were digested and the cell suspensions were plated onto 25 ml culture flasks (156367, Thermo Fisher Scientific). 5 μg/ml puromycin (A1113803, Thermo Fisher Scientific) were added in the medium for 7 days to select a stable cell line, and the efficacy was measured by western blot.
2.10. Drug treatment and cell culture conditions
HSC-T6 and HSC-LX2 lines were purchased from BeNa Culture Collections (Beijing, China), the primary HSCs were isolated from mouse liver. These cell lines were cultured in a humidified atmosphere containing 5% CO2 at 37 °C and maintained in 10% FBS (A4766801, Gibco), 1% penicillin/streptomycin (C125C5, NCM Biotech), and 90% Dulbecco's Modified Eagle Medium (DMEM, 11965092, Gibco). HSC ferrotosis was induced by treating with RSL3 (2.5 μm), erastin (10 μm) and sorafenib (10 μm) for 24 h. Meanwhile, equivalent amount of DMSO without drugs were added to the cells served as a control.
2.11. Cell viability assay
2 × 103 cells were plated in 96-well plates and treated with the compounds erastin, sorafenib and RSL3. After incubation for the indicated time, the cell viability was assessed by Cell Counting Kit-8 (CCK8, C0042, Beyotime Institute of Biotechnology). Briefly, CCK8 reagent was added to each well, and the well without cells was used as the control. The cells were incubated for another 4 h at 37 °C, and the absorbance was measured at 450 nm.
2.12. Lipid ROS detection
HSCs were exposed to the compounds for the indicated time, and then stained with C11-BODIPY (D3861, Thermo Fisher Scientific) for 30 min according to the manufacturer's instructions. The well with no C11-BODIPY was used as control, then cells were washed and measured by flow cytometer (Beckman, CytoFLEX).
2.13. Lipid peroxidation assay
Cells of different treatment groups were lysed by freeze-thaw first and MDA was detected by lipid Peroxidation Assay Kit (ab118970, Abcam) according to our previous reports [12]. Briefly, the cell lysate supernatants were collected and 600 μl thiobarbituric acid (TBA) solution was added. The samples were incubated at 95 °C for 60 min. Each reaction mixture were transferred 200 μl into a 96-well plate, and the levels of MDA was detected by EnSpire Multimode Plate Reader (PerkinElmer, Waltham, MA, USA).
2.14. Iron assay
According to the manufacturer's instruction [26], Iron Assay Kit (ab83366, Abcam) was used to measure cellular iron concentration. In brief, 5 × 106 cells were seeded on to the plate and then pretreated with drugs for 24 h. Before centrifuging (13,000×g, 10 min) at 4 °C, cells were homogenized by 5 × volumes of iron assay buffer. Next, iron reducer and supernatant mixture were incubated at room temperature for 30 min. Then, each sample was added iron probe and incubated without light on the horizontal shaker for 60 min. The absorbance was detected at 593 nm by EnSpire Multimode Plate Reader (PerkinElmer, Waltham, MA, USA).
2.15. Glutathione assay
1 × 108 cells were seeded onto the plate and the relative GSH concentration were assayed according to our previous reports [12]. In brief, cells were washed with cold PBS and assayed according to the instructions using Glutathione Assay Kit (CS0260, Sigma-Aldrich). Then, 3 vol of the 5% SSA Solution were added and the samples were collected at 10000×g for 10 min. The supernatant was used to determine the amount of GSH. 150 μL GSH working mixture was added into each sample and incubated at room temperature for 5 min and then were added 50 μl of the diluted NADPH Solution. The yellow product (5-thio-2-nitrobenzoic acid) was measured at 412 nm by EnSpire Multimode Plate Reader (PerkinElmer, USA).
2.16. Quantification of RNA m6A
The total RNA was isolated by TRIzol reagent (15596018, Invitrogen) according to the instructions of the manufacturer. The total m6A RNA levels were detected by EpiQuik m6A RNA Methylation Quantitative kit (Epigentek, Farmingdale, NY, USA). Firstly, 200 ng purified PolyA + mRNA was coated on assay wells. Then, RNA high binding solution was used to bind total RNA to the strip wells. The capture and detection antibodies were used to detect the m6A levels. Detection of the enhanced signal by reading the absorbance in the microplate spectrophotometer, followed by colorimetric quantification.
2.17. Dolt blot
According to previous reports, dot blots were performed with m6A immunoblotting [27]. Briefly, RNA is mixed with the same amount of 20 × SSC buffer (Sigma-Aldrich) and incubated at 95 °C for 5 min. Then, 100 ng, 200 ng or 400 ng poly (A) + RNAs were added on Hybond N+ membranes (GE Healthcare). After UV crosslinking for 30 min. After blocking in 5% milk the membranes were then incubated with m6A antibody overnight at 4 °C. Then, they were incubated with secondary antibody and applied with ECL for development. The same amount of RNA was also added on the membrane and stained with 0.02% methylene blue for 2 h.
2.18. Western blot
For western blot analysis, HSCs were lysed in RIPA buffer containing 50 mM Tris pH 7.4, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 150 mM NaCl and protease inhibitors (P8340, Sigma-Aldrich). Protein concentration from supernatant was quantified by the BCA Protein Assay Kit (23250, Thermo Fisher Scientific). Proteins were separated and transferred to PVDF membranes, and blocked for 30 min in 5% milk and blotted with related antibodies in 4 °C overnight. The membranes were incubated with secondary antibody for 1 h. Protein bands were exposed using the chemiluminescence system (Bio-Rad, Hercules, CA).
2.19. RNA extraction and real-time PCR
TRIzol reagent was used to isolated the total RNA from liver tissues or cells according to the instructions (15596018, Invitrogen). The purity and concentration of RNA were measured using Nanodrop. Then, Hifair® II 1st Strand CDNA Synthesis Super Mix (11123ES60, YEASEN) was used to synthesize the template complementary DNA from total RNA. The QuantiTect SYBR Green PCR Kit Hieff qPCR SYBR Green Master Mix (11202ES08, YEASEN) was used to analyze the cDNA by qRT-PCR on ABI 7500 system (Applied Biosystems). Fold change was calculated using 2−ddCt and GAPDH levels were taken for normalization. The primers were showed in Table 1.
Table 1.
Primer Sequences used for qRT-PCR.
| Genes | Forward primer | Reverse primer |
|---|---|---|
| Human | ||
| METTL3 | 5′-CAAGCTGCACTTCAGACGAA-3′ | 5′-GCTTGGCGTGTGGTCTTT-3′ |
| METTL14 | 5′-AGAAACTTGCAGGGCTTCCT-3′ | 5′-TCTTCTTCATATGGCAAATTTTCTT-3′ |
| METTL4 | 5′-TATCCCTCTTGGTCTGTGGAG-3′ | 5′-ACCTTCGTAGGGCTTTTTGTG-3′ |
| WTAP | 5′-GGCGAAGTGTCGAATGCT-3′ | 5′-CCAACTGCTGGCGTGTCT-3′ |
| ALKBH5 | 5′-CGGCGAAGGCTACACTTACG-3′ | 5′-CCACCAGCTTTTGGATCACCA-3′ |
| FTO | 5′-ACTTGGCTCCCTTATCTGACC-3′ | 5′-TGTGCAGTGTGAGAAAGGCTT-3′ |
| YTHDF1 | 5′-CAAGCACACAACCTCCATCTTCG-3′ | 5′-GTAAGAAACTGGTTCGCCCTCAT-3′ |
| YTHDC1 | 5′-AGATGGGTCTGTCAGATCTGGT-3′ | 5′-TCTGAACCTGCATATGACTCTGAT-3′ |
| ATG3 | 5′-CAGGCATGCTGAGGTGATGA-3′ | 5′-CGTTAACAGCCATTTTGCCACT-3′ |
| ATG4A | 5′-AAACCCCTGCTGCTCATTGT-3′ | 5′-ATGCCCCTAAAGACTGTGGC-3′ |
| ATG5 | 5′-AAGACCTTCTGCACTGTCCATC-3′ | 5′-TGCAATCCCATCCAGAGTTGC-3′ |
| ATG7 | 5′-TTGAGCGGCGGATCCAATTC-3′ | 5′-ATCTGGTGTCCATCAGCTTCAG-3′ |
| BECN1 | 5′-AGGTACCGACTTGTTCCCTA-3′ | 5′-TCCATCCTGTACGGAAGACA-3′ |
| ATG9A | 5′-AGGGCTGGAGAGGAGCACATAC-3′ | 5′-GGCGACGTGCACCAACAG-3′ |
| ATG12 | 5′-TGGAGGGGAAGGACTTACGG-3′ | 5′-TGGATGGTTCGTGTTCGCTC-3′ |
| ATG14 | 5′-CTGTACGTGGCTGTGGAGC-3′ | 5′-TCTTGCTTGCTCTTAAGTCGGC-3′ |
| ATG16 | 5′-CATCTCGGAGCAACTGAGGC-3′ | 5′-CCAGTTGAGCTAACTCCCCAC-3′ |
| GAPDH | 5′-AAATCCCATCACCATCTTCCAG-3′ | 5′-AGGGGCCATCCACAGTCTTCT-3′ |
| Rat | ||
| Mettl3 | 5′-CTTTAGCATCTGGTCTGGGCT-3′ | 5′-CCTTCTTGCTCTGCTGTTCCT-3′ |
| Mettl14 | 5′-CTGGACCCACATTTGCGAGC-3′ | 5′-GGAAGCCCTGCAAGTTTCTCT-3′ |
| Mettl4 | 5′-CTCGCCGAAGTTCTGAAAGACTA-3′ | 5′-AAACGAGCTGGGGTAAAGGC-3′ |
| Wtap | 5′-TAAGCTTGCAGAGCCCCCAC-3′ | 5′-CATCTTGTACCCCGAGACGC-3′ |
| Alkbh5 | 5′-TGAGATCCCGGAATGGGTGC-3′ | 5′-CCGAAGCAAAGTGCCGAGTC-3′ |
| Fto | 5′-TCATGGGACATCGAGACGCC-3′ | 5′-TCCAAGGTGCCTGTTGAGCA-3′ |
| Ythdf1 | 5′-GGACAGTCCAATCCGAGTAACA-3′ | 5′-GTGAGATACGGGATGGGAGG-3′ |
| Ythdc1 | 5′-TGCCAAAGCAAAGGGTGTATG-3′ | 5′-TGATGACAATCTGGCAAAACCTT-3′ |
| Atg3 | 5′-GAGGCTACCCTAGACACCAGA-3′ | 5′-TGTTAAAGGCTGCCGTTGCT-3′ |
| Atg4a | 5′-TTCGCCTGGGCATAAACCAA-3′ | 5′-GAAGGTCTGGGTTGTGTGAGG-3′ |
| Atg5 | 5′-TGATCCCGGTAGACCCAACC-3′ | 5′-CCACACGTCTCGAAGCACAT-3′ |
| Atg7 | 5′-GCGGATGTATGGACCCCAAA-3′ | 5′-GGAGATCTTGGCGTTGTCCA-3′ |
| Becn1 | 5′-TTGGCCAATAAGATGGGTCTGAA-3′ | 5′-TGTCAGGGACTCCAGATACGAGTG-3′ |
| Atg9a | 5′-GCGAGGCTGGTAACTGGAATC-3′ | 5′-TACGCATCGCGAGCTCACTT-3′ |
| Atg12 | 5′-CTGTCCAAGCACTCATCGACT-3′ | 5′-ATCCCCATGCCTGTGATTTGC-3′ |
| Atg14 | 5′-GCCATGGAAGGAAAACGGCT-3′ | 5′-AAACACGCCAACCGATCGTA-3′ |
| Atg16 | 5′-AATATCTTTGGGAGACGCTCTGTC-3′ | 5′-CTGCATTGACCTCTCCGTCAT-3′ |
| Actb | 5′-TTCTACAATGAGCTGCGTGTG-3′ | 5′-GGGGTGTTGAAGGTCTCAAA-3′ |
2.20. Trypan blue staining
According to our previous reports [12], trypan blue staining was assayed. In brief, HSC-LX2 cells were cultured in 24-well plates and petreated with vehicle, erastin, sorafenib and RSL3 for 24 h. After treated indicated time, the cells were stained with trypan blue solution (KGY015, KeyGEN BioTECH). ImageJ software was used to quantify trypan blue-positive ratio in 10 random regions.
2.21. Transmission electron microscopy
According to our previous reports [12], morphological characteristics of HSC ferroptosis was performed by transmission electron microscopy assay. In brief, HSC-LX2 cells (20,000 cells/well) were cultured and inoculated onto 4-well Chambered Coverglass (155382, Thermo Fisher Scientific). Olympus EM208S transmission electron microscope was used to acquire the images.
2.22. RNA sequencing
Total RNA was isolated from HSC-LX2 cells by TRIzol reagent (15596018, Invitrogen). For each sample, TruSeq Stranded Total RNA with 1 μg RNA was used for library preparation with Ribo-Zero Gold kit (MRZG12324, Illumina). TopHat v1.4.1 was used to align the reads of each sample with the human reference genome (GRCh38.p10). EdgeR version 3.08 was performed for differential gene expression. The Benjamini-Hochburg method was used to calculate the adjusted P values. We used fold change <0.65 for downregulationn and fold change >1.4 for upregulatio as cutoffs.
2.23. MeRIP qPCR
According to a reported protocol, m6A immunoprecipitation (MeRIP) qPCR was performed [27]. In brief, normal rabbit IgG and 5 μg m6A antibody were conjugated to 50 μl Protein A/G Plus Agarose overnight at 4 °C, respectively. The antibody was incubated with 100 μg fragmented total RNA in immunoprecipitation buffer (0.5% NP40, 50 mM Tris-HCl and 750 mM NaCl) added 40U RNase inhibitor (2313Q, Takara) at 4 °C overnight. After incubation with 300 μl elution buffer (0.05% SDS, 1 mM EDTA and 5 mM Tris-HCl) with 8.4 μg proteinase K (9034, Takara) at 50 °C for 1.5 h, RNAs were eluted from the beads. Following TRIzol and ethanol precipitation, the enrichment was measured by real-time PCR after the input and m6A-enriched RNAs were reversely transcribed.
2.24. Immunofluorescence
For immunofluorescence staining [12], HSC-LX2 were cultured in 24-well plates and treated with the compounds for the indicated time. Then, they were incubated with antibody against LC3 (ab48394, Abcam) overnight. Thereafter, FITC-labeled secondary antibodies (ab6717, Abcam Technology) were added. Nuclear counterstaining was assayed with DAPI (KGA215-50, KeyGEN BioTECH) and the images were taken with the fluorescence microscopy or confocal microscopy.
2.25. Long-lived protein degradation analysis
According to our previous reports [13], LC-MS/MS was used to determine the rate of long-lived protein degradation. In brief, the HSC-LX2 cells transfected with METTL4 shRNA or FTO plasmid, and labeled with L-[2H4] lysine, or L-[U–13C6, 15N4] arginine and l-arginine and l-lysine, L-[U–13C6, 14N4] arginine and L-[U–13C6, 15N2] lysine (74-79-3, Cambridge Isotope Laboratories; 23128, Sigma-Aldrich). Then, the cell scraping buffer containing 1 mM sodium orthovanadate (S6508, Sigma-Aldrich), 0.25 M sucrose (BP818, Sigma-Aldrich), 5 mM β-glycerophosphate (G5422, Sigma-Aldrich), 5 mM NaF (S7920, Sigma-Aldrich), and protease inhibitor mixture (04693132001, Roche) were added. Before samples were concentrated on spin tubes (CLS8160, Sigma-Aldrich), the mixed cells were centrifuged and lysed in 2% benzonase (E1014, Sigma-Aldrich), 2 M thiourea (PHR1758, Sigma-Aldrich), and 6 M urea (U5378, Sigma-Aldrich). The mixed proteins were separated by SDS-PAGE. Gel lanes were cut into 15 pieces, and the samples were digested in the gel. The peptide mixtures obtained were STAGE-tipped. LC-MS/MS was used to detect the identification and relative quantification of peptides.
2.26. Cycloheximide chase assay
To measure BECN1 protein stability, HSC-LX2 cells were treated with 100 μg/ml cycloheximide (C7698, Sigma-Aldrich) during indicated times after treatment with 10 μM erastin for 24 h. The stability of BECN1 was detected through western blot analysis as described previously [28].
2.27. RIP-RT-PCR
According to a reported protocol [28], RNA immunoprecipitation (RIP) was performed. In brief, cells were irradiated twice at 254 nm with 400 mJ/cm2 by Stratalinker. After washing with cold PBS, cells were lysed in high salt lysis buffer (0.5 mM DTT, 0.2% NP-40, 300 mM NaCl, 20 mM Tris-HCl PH 7.6, 200U/ml RNase inhibitor, and protease inhibitor) for 30 min at 4 °C. After treatment with 1 U RNase T1(EN0541, Thermo Fisher Scientific) at 24 °C for 15 min, the supernatant was incubated with Protein A/G Plus Agarose (sc-2003, Santa Cruz Biotechnology), or anti-YTHDF1 antibody in 500 μl 1 × IP buffer supplemented with RNase inhibitors overnight at 4 °C. Then, 100 μL of elution buffer (20 mg/ml Proteinase K, 0.05% SDS, 1 mM EDTA pH 8.0, 5 mM Tris-HCL pH 7.5) was added for 2 h at 50 °C. RNAs associated with YTHDF1 were recovered by TRIzol and ethanol precipitation and detected by real-time PCR.
2.28. Biotinylated RNA pull-down assay
According to our previous reports [13], the proteins bound to the RNA and synthesis of biotinylated transcripts were detected. Briefly, template for PCR amplification of the 5ʹ-UTR, CDS, and 3ʹ-UTR of BECN1 mRNA were obtained from cDNA of HSC-LX2 cells. Cells were washed twice with cold PBS, harvested and lysed in cell lysis. Then, whole cell lysates (120 μg per sample) were incubated with biotinylated transcripts (9 μg) for 30 min at 30 °C. Paramagnetic streptavidin-conjugated Dynabeads (11206D, Dynal) was used to isolate the complexes and analyzed by western blot.
2.29. Luciferase reporter system construction
As described previously [12], 3′-UTR, 5′-UTR and CDS of human BECN1 were cloned into pGL3-Luc vector (E1751, Promega). Then, pGL3-Luc-BECN1-3′UTR, pGL3-Luc-BECN1-CDS and pGL3-Luc-BECN1-5′UTR plasmids were transfected into cells. To monitor transfection efficiencies, the luciferase reporter vectors were transfected in cells together with phRL-null (E2231, Promega).
2.30. Calculations and statistics
The matched controls was used for individual animal experiments and cell experiments, performed in duplicate or triplicate and repeated 3 times, and the data were pooled. Data were expressed as mean ± standard error of the mean (SEM). Statistical analysis was performed using either one-way or Student's t-test (two-group comparison) analyses of variance followed by Student-Newman-Keuls test (more than two groups). In all analysis, a probability of less than 0.05 was considered to indicate statistical significance. GraphPad Prism 8 (GraphPad software version 8.0) was used for all statistical analyses.
3. Results
3.1. m6A modification is up-regulated during HSC ferroptosis
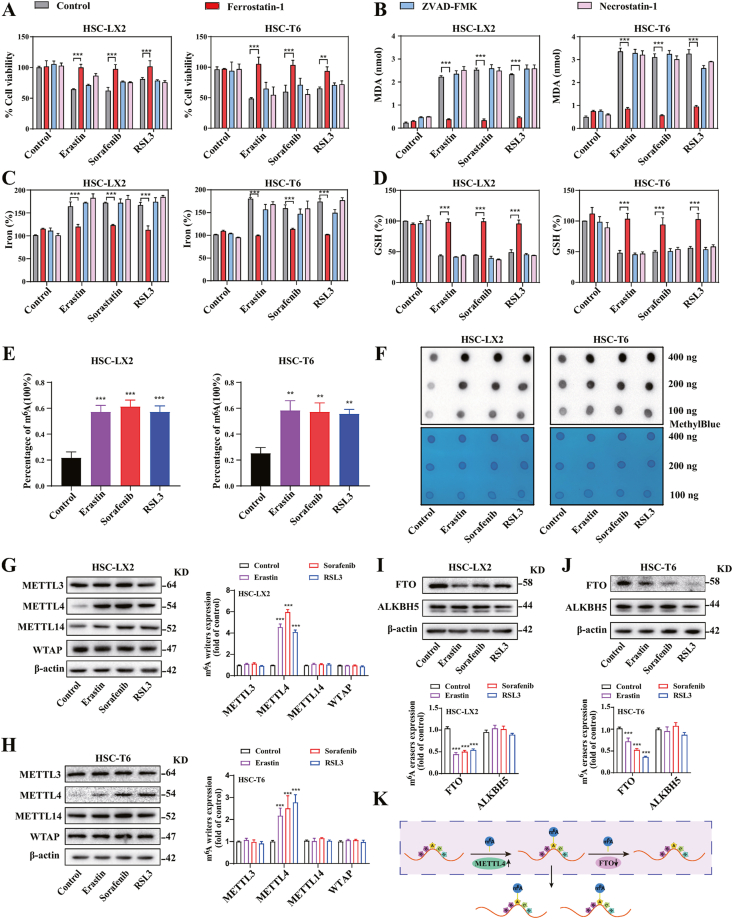
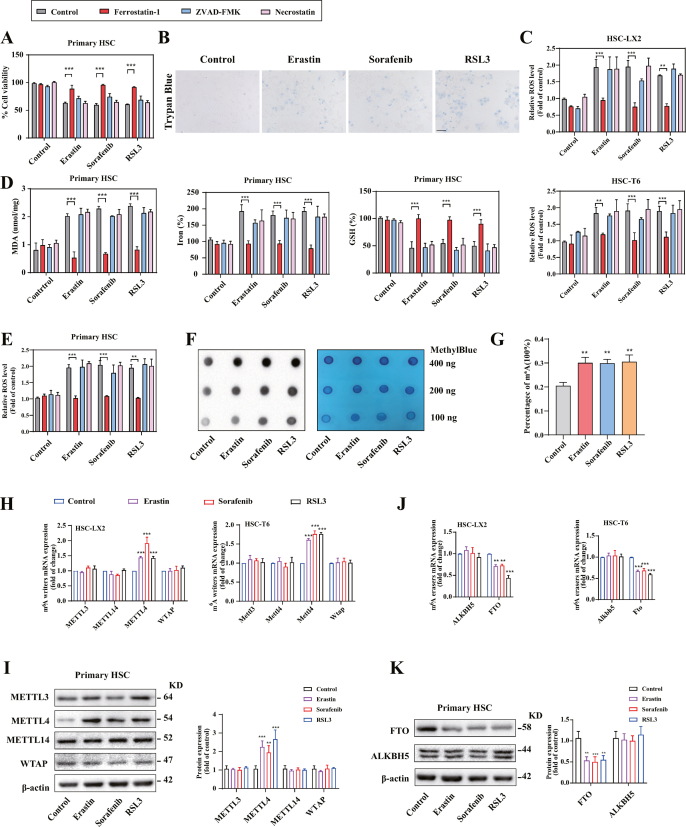
Our previous studies showed that preclinical and clinical drugs (e.g., sorafenib and erastin) can induce HSC ferroptosis [12]. Consistent with our previous findings [16,29], treatment with erastin, sorafenib, and RSL3 evidently inhibited cell viability in both rat (HSC-T6), human (HSC-LX2) HSC cells and primary HSCs, but ferroptosis inhibitor (ferrostatin-1) rather than necroptosis inhibitor (necrostatin-1) and apoptosis inhibitor (ZVAD-FMK) completely rescued the cell growth inhibition (Fig. 1A) (Fig. S1A). Moreover, trypan blue staining indicated that erastin, sorafenib, and RSL3 treatment markedly resulted in an increase in the dead cells (Fig. S1B). Lipid peroxidation, iron overload, lipid ROS accumulation, and glutathione (GSH) depletion are hallmarks of ferroptotic cells [29]. As expected, treatment with erastin, sorafenib, and RSL3 dramatically increased the end products of lipid peroxidation MDA (Fig. 1B), intracellular redox-active iron overload (Fig. 1C), GSH depletion (Fig. 1D), and lipid ROS accumulation (Fig. S1C). Importantly, ferrostain-1, but not necrostatin-1 and ZVAD-FMK, significantly impaired these classical ferroptotic events in HSC ferroptosis (Fig. 1A–D) (Figs. S1C–E). Overall, these results fully confirmed that erastin, sorafenib, and RSL3 can trigger HSC ferroptosis in vitro.
Fig. 1.
m6A modification is up-regulated during HSC ferroptosis. HSC-T6 and HSC-LX2 cells were treated with RSL3 (2.5 μM), sorafenib (10 μM), and erastin (10 μM), with or without the addition of Ferrostatin-1 (1 μM), Necrostatin-1 (10 μM), ZVAD-FMK (10 μM) for 24 h. (A) Cell Counting Kit-8 kit was used to determine the Cell viability (**, p < 0.01, ***, p < 0.001, n = 3 in every group). (B-D) MDA production, iron accumulation, and GSH levels were measured by commercial kits (***, p < 0.001, n = 3 in every group). (E, F) m6A RNA Methylation Quantitative kit and dot blot were used to detect the m6A modification (**, p < 0.01, ***, p < 0.001, n = 3 in every group). (G, H) Western blot showed the protein levels of m6A writers. (***, p < 0.001, n = 3 in every group). (I, J) The protein levels of m6A erasers were determined by western blot (***, p < 0.001, n = 3 in every group). (K) The mechanism of the increased m6A modification in RNA was showed.
m6A modification can regulate a variety of cell fates including apoptosis, necroptosis, senescence, and endothelium-mesenchymal transition (EMT). Whether m6A modification is involved in the regulation of HSC ferroptosis? To test this potential possibility, we first examined the levels of m6A modification during HSC ferroptosis. Attractively, m6A RNA methylation quantification assay and dot blot showed a significant increase in m6A modification levels after treatment with sorafenib, erastin, and RSL3 (Fig. 1E and F) (Figs. S1F and 1G). Of note, the protein and mRNA expression of METTL4 rather than other methyltransferase was significantly upregulated (Fig. 1G and H) (Figs. S1H and 1I), whereas the level of FTO rather than other demethylase was markedly downregulated in HSC ferroptosis (Fig. 1I and J) (Figs. S1J and S1K). Collectively, these findings suggested that m6A modification was increased due to the dysregulation of writer METTL4 and eraser FTO during HSC ferroptosis (Fig. 1K).
3.2. Inhibition of m6A modification confers resistance to HSC ferroptosis
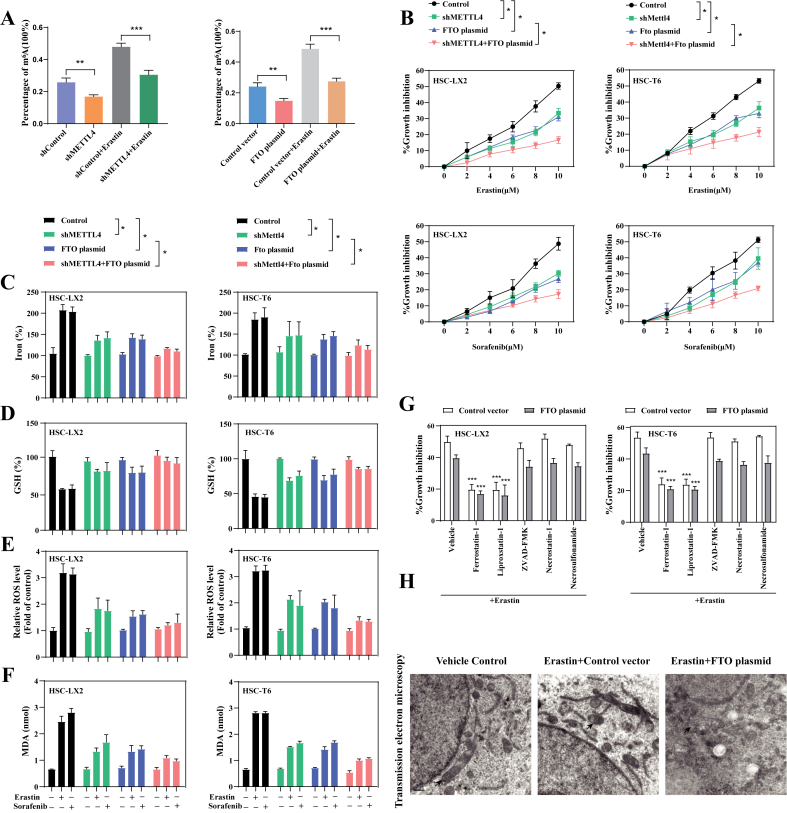
HSC-LX2 cells stably transfected with METTL4 shRNA or FTO plasmid were used to investigate whether upregulated m6A modification was directly involved in the induction of ferroptosis (Figs. S2A and 2B). m6A RNA methylation quantification assay confirmed that both METTL4 knockdown and FTO overexpression could completely impair erastin-induced up-regulation of m6A modification in HSC-LX2 (Fig. 2A). Moreover, METTL4 shRNA or FTO plasmid significantly abolished the growth inhibition in HSC-LX2 and HSC-T6 cells by erastin and sorafenib treatment (Fig. 2B). Furthermore, the results showed that m6A modification inhibition by METTL4 shRNA and FTO plasmid evidently damaged erastin- and sorafenib-induced redox-active iron accumulation (Fig. 2C), GSH depletion (Fig. 2D), lipid ROS generation (Fig. 2E), and MDA production (Fig. 2F). Interestingly, in the presence of FTO plasmid, ferroptosis inhibitors including liproxstatin-1 and ferrostain-1 markedly reversed growth inhibition induced by erastin(Fig. 2G), whereas ZVAD-FMK, necrostatin-1, and necrosulfonamide (a potent necroptosis inhibitor that targets mixed lineage kinase domain-like protein), did not effectively reverse this process (Fig. 2G). More importantly, transmission electron microscopy assays confirmed that erastin treatment substantially shanked mitochondria and decreased the number of ridges, whereas FTO plasmid completely abolished the typical changes of ferroptotic cells (Fig. 2H). Overall, these data showed that up-regulation of m6A modification by ferriptosis inducer treatment may contribute to HSC ferroptosis, but not necroptosis and apoptosis.
Fig. 2.
Inhibition of m6A modification confers resistance to HSC ferroptosis. METTL4 shRNA or FTO plasmid were stably transfected into HSC-LX2 cells followed by erastin (10 μM) treatment for 24 h. (A) The m6A levels were detected by m6A RNA Methylation Quantitative kit (**, p < 0.01, ***, p < 0.001, n = 3 in every group). (B) METTL4 shRNA or FTO plasmid were transfected into HSC-LX2 and HSC-T6 cells followed by sorafenib (10 μM) or erastin (10 μM) treatment for 24 h. Cell Counting Kit-8 kit was used to determine the Cell viability (*, p < 0.05, n = 3 in every group). (C–F) Iron accumulation, GSH depletion, lipid ROS level and MDA production were assayed (*, p < 0.05, n = 3 in every group). (G) FTO plasmid transfected into HSC-LX2 and HSC-T6 cells were treated with erastin (10 μM) with or without the indicated inhibitors (Liproxstatin-1, 100 nM; Ferrostatin-1, 1 μM; Necrostatin-1, 10 μM; ZVAD-FMK, 10 μM; Necrosulfonamide, 0.5 μM) for 24 h. Cell viability was assayed by Cell Counting Kit-8. (***, p < 0.001, n = 3 in every group). (H) Control vector or FTO plasmid were transfected into HSC-LX2 cells and treated with erastin (10 μM) for 24 h. Transmission electron microscopy was used to examin the typical changes of ferroptotic cells. Scale bars: 0.2 μm. Representative photographs were showed.
3.3. Reduced ferroptosis by m6A modification inhibition is associated with autophagy inactivation
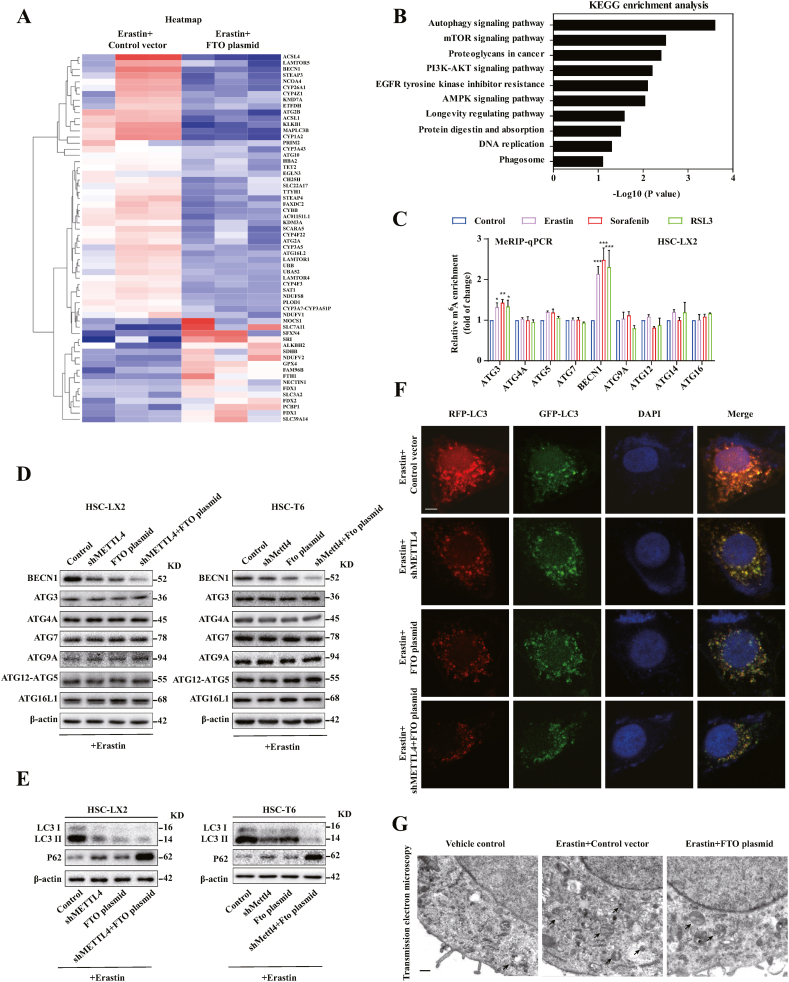
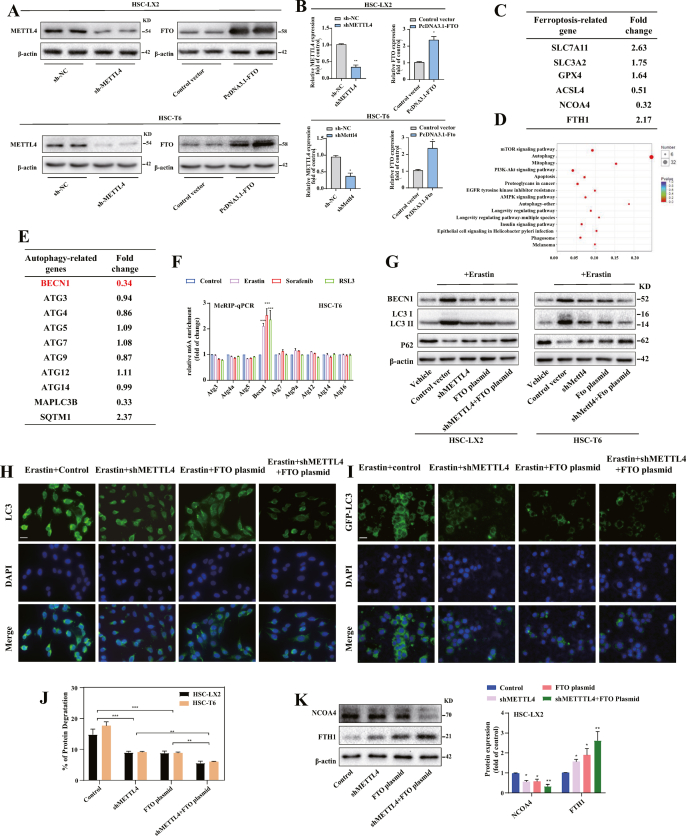
We further performed RNA sequencing (RNA-seq) to further explore the molecular mechanism by which m6A modification promotes HSC ferroptosis and to identify the target genes involved in the glutamine metabolic pathway, non-classical autophagy pathway and classical iron metabolic pathway. The results showed that 424 mRNAs were significantly increased (fold change >1.4), whereas 317 mRNAs were apparently decreased (fold change <0.65) in FTO overexpressed HSC-LX2 cells as shown in the heat map (Fig. 3A). As expected, some ferroptosis-related genes, such as SLC7A11 (2.63 fold), SLC3A2 (1.75 fold), GPX4 (1.64 fold), were identified among these genes (Fig. S2C). These positive outcomes validated our screen approach. Importantly, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and GO analysis fully indicated that autophagy signaling was potentially regulated by m6A modification during HSC ferroptosis (Fig. 3B) (Fig. S2D). Remarkably, we found that the expression of BECN1, a crucial target gene in autophagy signaling, decreased significantly upon FTO overexpression (Fig. S2E). Moreover, m6A RNA-immunoprecipitation (MeRIP) qPCR revealed that the levels of m6A modification in BECN1, but not in ATG3, ATG4A, ATG5, ATG7, ATG9A, ATG12 and ATG16L1, were increased in HSC ferroptosis (Fig. 3C) (Fig. S2F). Consistently, the reduction of m6A modification by FTO plasmid or METTL4 shRNA markedly reduced the protein expression of BECN1 without large differences in other autophagy-related genes (Fig. 3D). It is generally accepted that LC3-II conversion and autophagosome formation require the activation of the conserved BECN1 complex. As expected, the expression of BECN1 and the conversion of LC3-II were increased by erastin treatment (Fig. S2G). While the conversion of LC3-I to LC3-II in erastin induced HSCs was eliminated following reduced m6A modification (Fig. 3E). Meanwhile, immunofluorescence staining and GFP-LC3 puncta formation assay suggested that the down-regulation of m6A modification significantly reduced the expression of endogenous and exogenous LC3 in HSC-LX2 cells (Figs. S2H and S1I). Indeed, HSC-LX2 cells were transfected with CMV-TurboRFP-EGFP- LC3-PGK-Puro plasmid to detect autophagy flux, and the results showed that FTO plasmid and METTL4 shRNA substantially blocked autophagy flux during HSC ferroptosis (Fig. 3F). Besides, transmission electron microscopy assays further confirmed that the down-regulation of m6A modification by FTO plasmid reduced autophagic vesicles in HSC ferroptosis (Fig. 3G). Interestingly, the reduction of m6A modification significantly inhibited the autophagic degradation of long-lived protein in the long-lived protein degradation assay (Fig. S2J). Notably, NCOA4-dependent FTH1 degradation plays a central role in facilitating ferritinophagy [30]. As expected, FTO plasmid and METTL4 shRNA markedly impaired erastin-induced upregulation of NCOA4 and downregulation of FTH1 in HSC-LX2 cells (Fig. S2K). Altogether, these data suggested that the induction of ferroptosis by m6A modification was associated with autophagy activation.
Fig. 3.
Reduced ferroptosis by m6A modification inhibition is associated with autophagy inactivation. (A) FTO plasmid was transfected into HSC-LX2 cells and treated with erastin (10 μM) for 24 h. Total RNA was isolated for RNA-Seq. Clustering of HSC-LX2 cells were demonstrated by microarray heat map. The significantly differentially expressed mRNAs were analysied by hierarchical cluster: gray, no change; bright blue, underexpression; bright red, overexpression (FTO plasmid, n = 3; Control vector, n = 3). (B) Differentially expressed mRNAs were enriched by KEGG enrichment analysis in FTO plasmid group (Control vector, n = 3; FTO plasmid, n = 3). (C) The levels of m6A modification in autophagy-related gene were determined by MeRIP qPCR (*, p < 0.05, **, p < 0.01, ***, p < 0.001, n = 3 in every group). (D) METTL4 shRNA or FTO plasmid transfected into HSC-T6 and HSC-LX2 cells were treated with erastin (10 μM) for 24 h. Western blot showed the protein expression of BECN1, ATG3, ATG4A, ATG7, ATG9A, ATG5-ATG12 and ATG16L1 (n = 3 in every group). (E) Western blot was used to determine the expression of LC3-I/II and p62 (n = 3 in every group). (F) METTL4 shRNA or FTO plasmid with CMV-TurboRFP- EGFP-LC3-PGK-Puro plasmid were transferred into HSC-LX2 cells by erastin (10 μM) treatment for 24 h. The fluorescence spots were detected. Representative photographs were showed. Scale bars: 50 μm. (G) HSC-LX2 cells transfected with FTO plasmid or control vector by erastin (10 μM) treatment for 24 h. Transmission electron microscopy was used to examine the autolysosomes or autophagosomes. Representative photographs were showed. Scale bars: 0.2 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Induction of autophagy by BECN1 plasmid impairs m6A modification inhibition-induced resistance to HSC ferroptosis
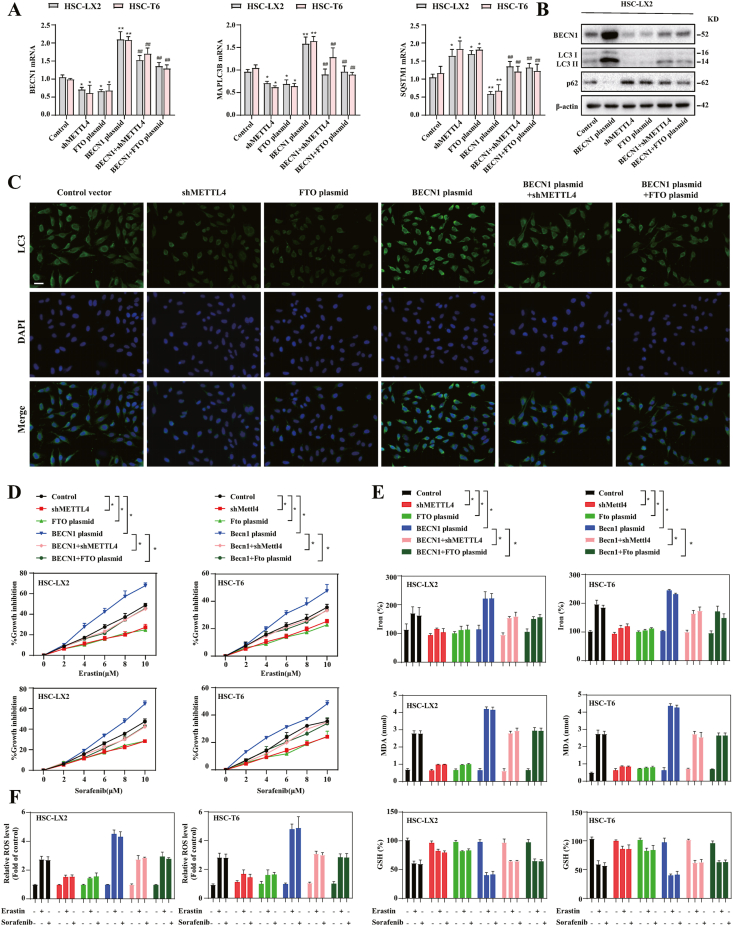
We further to estimate whether autophagy inactivation mediated m6A modification inhibition-induced ferroptosis resistance, BECN1 plasmid was employed to activate autophagy. As expected, the results confirmed that BECN1 plasmid completely impaired m6A modification inhibition-induced BECN1 downregulation during HSC ferroptosis (Fig. 4A and B). Moreover, BECN1 overexpression restored the generation of autophagosome (Fig. 4A–C), and autophagic flux (Fig. 4A–C) in the presence of m6A modification reduction. Indeed, erastin and sorafenib-induced growth inhibition was damaged by FTO plasmid and METTL4 shRNA, whereas the BECN1 plasmid dramatically promoted growth inhibition during HSC ferroptosis (Fig. 4D). Similarly, pretreatment with METTL4 shRNA or FTO plasmid completely abolished iron accumulation (Fig. 4E), MDA production (Fig. 4E), GSH depletion (Fig. 4E) and lipid ROS accumulation (Fig. 4F) in HSCs by erastin- and sorafenib treatment. However, m6A modification inhibition did not markedly impair the ferroptotic events in the presence of BECN1 plasmid (Fig. 4E and F). Taken together, these findings showed that activation of autophagy by BECN1 plasmid impaired the resistance of m6A modification inhibition to HSC ferroptosis.
Fig. 4.
Induction of autophagy by BECN1 plasmid impairs m6A modification inhibition-induced resistance to HSC ferroptosis. (A) BECN plasmid and METTL4 shRNA or FTO plasmid were transfected into HSC-T6 and HSC-LX2 cells and trested with erastin (10 μM) for 24 h. Real-time PCR were used to measure the mRNA levels of BECN1, MAPILC3B and SQSTM1 (*, p < 0.05, **, p < 0.01, compared with Control group, ##, p < 0.01, compared with BECN1 plasmid group, n = 3 in every group). (B) Western blot showed the protein expression of p62 and LC3-I/II (n = 3 in every group). (C) The endogenous LC3 levels were measured by immunofluorescence (n = 3 in every group). (D) HSC-LX2 and HSC-T6 cells transfected with BECN plasmid and METTL4 shRNA or FTO plasmid were treated with erastin (10 μM) or sorafenib (10 μM) for 24 h. Cell viability was assayed by Cell Counting Kit-8 (n = 3 in every group, *, p < 0.05). (E, F) MDA production, Iron accumulation, GSH depletion and lipid ROS level were assayed by commercial kits (*, p < 0.05, n = 3 in every group).
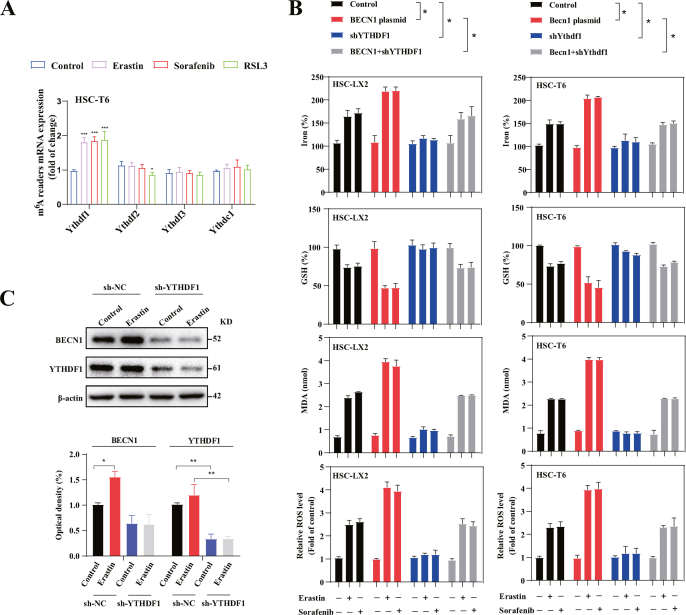
3.5. m6A reader protein YTHDF1 promotes autophagy activation and BECN1 mRNA stability via recognizing the m6A binding site
It has been reported that m6A modification functions in condition of its recognition by m6A reader proteins [31]. Which m6A reader protein mediates the regulation of autophagy-dependent ferroptosis by m6A modification? To address this problem, we performed an unbiased screen among m6A reader proteins. Interestingly, the results showed that YTHDF1 protein and mRNA rather than other m6A readers were significantly upregulated during HSC ferroptosis (Fig. 5A and B and S3A). Moreover, the results showed that pretreatmet with YTHDF1 shRNA dramatically impaired the induction of BECN1 plasmid on HSC ferroptosis (Fig. S3B). YTHDF1 can recognize m6A methylated mRNA and promotes the post-transcriptional regulation of its targets [32]. Indeed, western blot showed that YTHDF1 shRNA attenuated erastin-induced expression of BECN1 (Fig. S3C). Growing evidence suggest that YTHDF1 binds to the m6A sites of short-lived human transcripts, promoting mRNA stability and translation. We observed that the mRNA half-life of BECN1 in HSC-LX2 cells transfected with YTHDF1 plasmid was prolonged compared with control vector group (Fig. 5C). Consistently, HSC-LX2 cells treated with translation inhibitor cycloheximide showed no significant difference in the protein half-life of BECN1 between YTHDF1 plasmid and control vector group (Fig. 5D). In order to verify whether YTHDF1 bound to the BECN1 mRNA in HSC LX2 cells, the interaction between YTHDF1 and BECN1 mRNA was detected by RNP IP. As shown in Fig. 5E, compared with GAPDH PCR product the BECN1 PCR products were highly enriched in YTHDF1 samples, demonstrating a direct bind of YTHDF1 to BECN1 mRNA. Furthermore, the results of mRNA-protein precipitation showed that YTHDF1 could readily interact with the BECN1 CDS rather than 3′-UTR and 5′-UTR transcripts of BECN1 (Fig. 5F). MeRIP qPCR also showed that m6A modification was significantly enriched in BECN1 CDS (Fig. 5G). Importantly, the luciferase activity was significantly increased in HSC-LX2 cells transfected with the BECN1 CDS (Fig. 5H). Attractively, detailed primary sequence analysis revealed two 5-nucleotide consensus sequence (AGACC, GGACA) in CDS of the BECN1 mRNA sequence (Fig. 5I). To confirm the roles of m6A binding sites on the BECN1 CDS region, BECN1-CDS-WT, BECN1-CDS-Mut1 (A437G) and BECN1-CDS-Mut2 (A1276G) were generated as shown in Fig. 5J. RNP IP assays showed that BECN1-CDS-Mut2, but not BECN1-CDS-WT and BECN1-CDS-Mut1, completely impaired the binding of YTHDF1 and BECN1 mRNA (Fig. 5K). Consistently, the RNA stability assay revealed that YTHDF1 plasmid prolonged the half-life of BECN1-CDS-WT mRNA and BECN1-CDS-Mut1 mRNA, but not BECN1-CDS-Mut2 mRNA (Fig. 5L). Besides, western blot results also indicated that BECN1-CDS-Mut2 evidently damaged erastin-induced upregulation of BECN1 protein (Fig. 5M). Collectively, these data indicated that YTHDF1 plasmid promoted BECN1 mRNA stability via binding to the m6A binding site CDS regions at A1276.
Fig. 5.
m6A reader protein YTHDF1 promotes autophagy activation and BECN1 mRNA stability via recognizing the m6A binding site. HSC-T6 and HSC-LX2 cells were added sorafenib (10 μM), erastin (10 μM), and RSL3 (2.5 μM) and treated for 24 h. (A) Western blot was used to assay the protein levels of m6A readers (n = 3 in every group). (B) The mRNA levels of m6A readers were determined by real-time PCR (***, p < 0.001, n = 3 in every group). (C) YTHDF1 plasmid or control vector were transfected into HSC-LX2 cells and treated with erastin (10 μM) for 24 h, and then were pretreated with Act-D (5 μg/ml) for indicated times. Real-time PCR was used to measure the remaining BECN1 mRNA levels (*, p < 0.05, n = 3 in every group). (D) YTHDF1 plasmid or control vector were transfected into HSC-LX2 and treated with erastin (10 μM) for 24 h. Western blot was used to determine the protein levels of BECN1 at different times under the CHX (100 μg/ml) treatment (n = 3 in every group). (E) The binding of YTHDF1 and BECN1 mRNA was measured by ribonucleoprotein immunoprecipitation (RNP IP) (**, p < 0.01, n = 3 in every group). (F) biotinylated transcripts of the BECN1 mRNA 3′-UTR, CDS, 5′-UTR were used for mRNA affinity isolation assay (n = 3 in every group). (G) HSC-LX2 cells were transfected with control vector or YTHDF1 plasmid and by erastin (10 μM) treatment for 24 h. The levels of m6A modification in BECN1 mRNA 3′-UTR, CDS and 5′-UTR were determined by MeRIP qPCR (**, p < 0.01, n = 3 in every group). (H) HSC-LX2 cells were stably transfected with empty vector (pGL3)or luciferase constructs carrying the genes encoding 5′-UTR-BECN1, CDS-BECN1, 3′-UTR-BECN1. Then cells were added erastin (10 μM) and treated for 24 h, and luciferase activities were measured (**, p < 0.01, n = 3 in every group). (I) BECN1 mRNA m6A binding site was showed. (J) Schematic representation of positions of m6A motifs and mutation in CDS within BECN1 mRNA were showed. (K) Binding of YTHDF1 with the BECN1-CDS-WT, BECN1-CDS-Mut1 (A437G) and BECN1-CDS-Mut2 (A1276G) in HSC-LX2 were analyzed by YTHDF1 RNP IP (**, p < 0.01, n = 3 in every group). (L) HSC-LX2 cells were transfected with control vector or YTHDF1 plasmid and BECN1-CDS-WT, BECN1-CDS-Mut1 and BECN1-CDS-Mut2 plasmid, and then were pretreated with Act-D (5 μg/ml) for indicated times. Real-time PCR was used to measure the remaining BECN1 mRNA (*, p < 0.05, n = 3 in every group). (M) After transfected with BECN1-CDS-WT, BECN1-CDS-Mut1 and BECN1-CDS-Mut2 plasmid, HSC-LX2 cells were treated with erastin (10 μM) for 48 h. The protein expression of BECN1 was measured by western blot (*, p < 0.05, ***, p < 0.001, n = 3 in every group).
3.6. HSC-specific inhibition of m6A modification impairs erastin-induced HSC ferroptosis in murine liver fibrosis
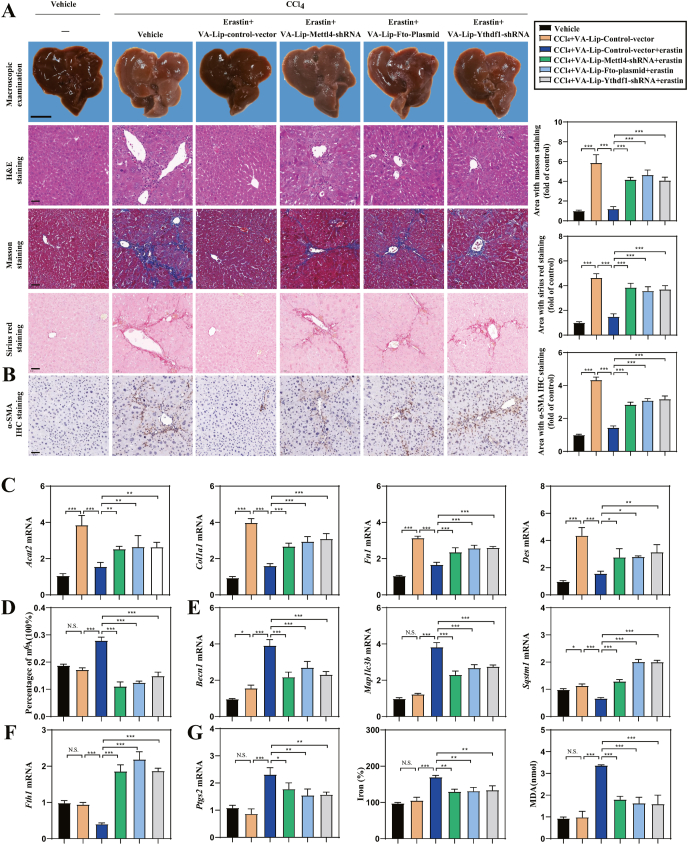
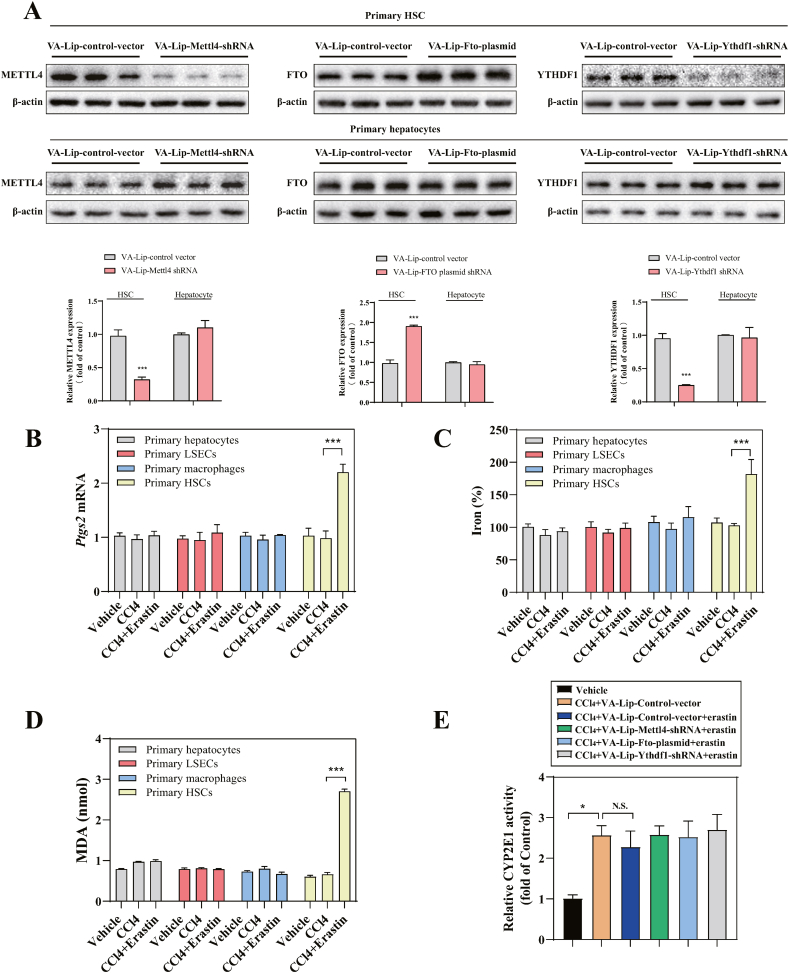
To evaluate whether the inhibition of m6A modification impaired ferroptosis in vivo, vitamin A-coupled liposomes carrying Mettl4 shRNA (VA-Lip-Mettl4-shRNA), Fto plasmid (VA-Lip-Fto-plasmid) and Ythdf1 shRNA (VA-Lip-Ythdf1-shRNA) were generated to inhibit HSC-specific m6A modification in CCl4-induced mouse model of liver fibrosis (Fig. S4A). Macroscopic examination showed that fibrotic pathological was changed in the model group of liver fibrosis compared with the control group, whereas ferroptosis inducer erastin treatment alleviated CCl4-induced liver fibrosis (Fig. 6A). Interestingly, pretreatment with VA-Lip-Mettl4-shRNA, VA-Lip-Fto-plasmid and VA-Lip-Ythdf1-shRNA completely damaged the improvement of erastin on liver fibrosis (Fig. 6A). Moreover, hematoxylin and eosin (H&E) staining, masson staining, and sirius red staining indicated that erastin treatment markedly reduced collagen deposition in the central vein (Fig. 6A). Attractively, VA-Lip-Mettl4-shRNA, VA-Lip-Fto-plasmid and VA-Lip-Ythdf1-shRNA evidently reversed the effect of erastin on liver fibrosis (Fig. 6A). Immunohistochemistry staining revealed that VA-Lip-Mettl4-shRNA, VA-Lip-Fto-plasmid and VA-Lip-Ythdf1-shRNA completely abolished the inhibition of erastin on α-SMA (actin, alpha 2, smooth muscle, aorta) expression (Fig. 6B). Furthermore, the results demonstrated that VA-Lip-Mettl4-shRNA, VA-Lip-Fto-plasmid and VA-Lip- Ythdf1-shRNA completely eliminated the mRNA expression of Acta2, Col1a1, Fn1, and Des,which were inhibited by erastin (Fig. 6C).
Fig. 6.
HSC-specific inhibition of m6A modification impairs erastin-induced HSC ferroptosis in murine liver fibrosis. Mice of 6 groups were treated with Vehicle, CCl4, CCl4+VA-Lip-control-vector + Erastin, CCl4+VA-Lip-Mettl4-shRNA + Erastin, CCl4+VA-Lip-Fto-Plasmid + Erastin, CCl4+VA-Lip-Ythdf1-shRNA + Erastin. (A) Macroscopic examination was used to observe the pathological changes of the livers. Scale bars: 1 cm. Histopathological study was performed by H&E, Masson, and Sirius Red staining. Representative photographs were showed. Scale bars: 50 μm. (***, p < 0.001, n = 6 in every group). (B) Immunohistochemical staining of α-SMA was determined. Representative photographs were showed. Scale bars: 50 μm.(***, p < 0.001, n = 6 in every group). (C) Real-time PCR was measured to determine the mRNA expression of liver fibrosis markers (Acta2, Col1a1, Fn1, and Des) (*, p < 0.05, **, p < 0.01, ***, p < 0.001, n = 6 in every group). (D) The m6A levels were determined by m6A RNA Methylation Quantitative kit (n = 6 in every group, ***, p < 0.001, N.S., not significant). (E, F) Real-time PCR was used to determine the mRNA expression of autophagy markers (Becn1, Map1lc3b, Sqstm1, and Fth1) (*, p < 0.05, ***, p < 0.001, N.S., not significant, n = 6 in every group). (G) The mRNA expression of Ptgs2, iron accumulation and MDA production were determined (*, p < 0.05, **, p < 0.01, ***, p < 0.001, N.S., not significant, n = 6 in every group). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We previously reported that ferroptosis inducer erastin could alleviate liver fibrosis by inducing ferroptosis in HSCs, but not in liver sinusoidal endothelial cells, hepatocytes, and macrophages [12]. Then, primary HSCs, hepatocytes, macrophages, and liver sinusoidal endothelial cells (LSECs) were isolated from the livers of fibrotic mice to further elucidate the role of m6A modification in erastin-induced ferroptosis in HSC. Importantly, VA-Lip-Mettl4-shRNA, VA-Lip-Fto-plasmid and VA-Lip- Ythdf1-shRNA significantly prevented the upregulation of erastin on m6A modification (Fig. 6D). Besides, VA-Lip-Mettl4-shRNA, VA-Lip-Fto-plasmid and VA-Lip-Ythdf1-shRNA blocked the activation of ferritinophagy by erastin treatment, characterized by decreased Becn1 and Map1lc3b expression and increased Sqstm1 and Fth1 expression (Fig. 6E and F). More importantly, inhibition of m6A modification by VA-Lip-Mettl4-shRNA, VA-Lip-Fto-plasmid and VA-Lip-Ythdf1-shRNA completely impaired erastin-induced HSC ferroptosis characterized by Ptgs2 (prostaglandin-endoperoxide synthase 2) expression, redox-active iron reduction and MDA elimination (Fig. 6G) in primary HSCs. Interestingly, erastin failed to increase the expression of the ferroptosis marker PTGS2 in primary hepatocytes, macrophages, and LSECs (Fig. S4B). Moreover, treatment with erastin could not trigger redox-active iron reduction and MDA elimination in primary hepatocytes, macrophages, and LSECs (Figs. S4C and 4D). We also examined whether Erastin administration suppressed fibrosis by reducing hepatic CYP2E1 activity. The results showed that Erastin administration did not reduce CYP2E1 activity (Fig. S4E). In conclusion, these results supported the hypothesis that HSC-specific m6A modification inhibition in murine liver fibrosis impaired erastin-induced HSC ferroptosis.
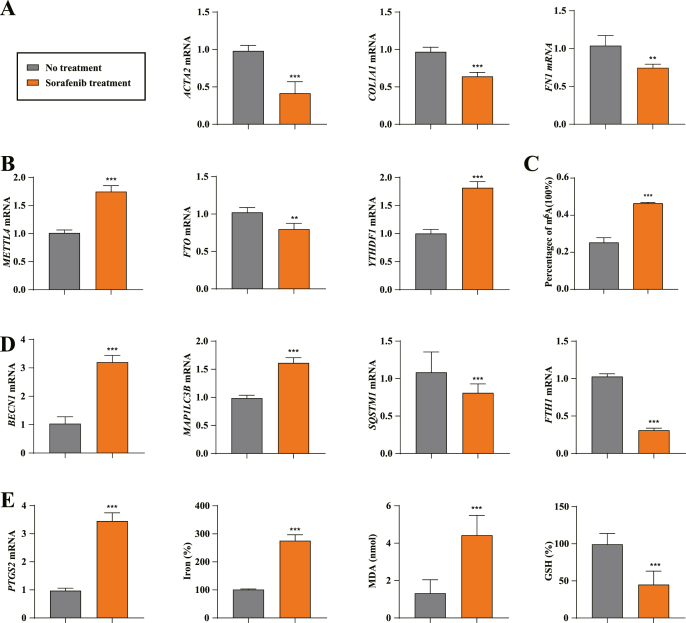
3.7. m6A modification upregulation, autophagy activation, and ferroptosis induction occur in human HSCs receiving sorafenib monotherapy
To evaluate the potential mechanism of m6A modification clinically, the partial hepatectomy specimens from 10 patients with cirrhotic complicated with HCC treated with sorafenib monotherapy and liver biopsy samples from 10 patients with cirrhotic who did not receive any treatment were analyzed. Then, laser capture microdissection was used to isolated the primary HSCs from the collected liver tissue according to previous reports. Consistent with previous findings, the mRNA levels of fibrosis markers ACTA2, COL1A1 and FN1 were significantly decreased in hepatectomy samples treated with sorafenib compared with untreated liver biopsy samples (Fig. 7A). As expected, real-time PCR analysis showed that the expression of METTL4 and YTHDF1 were upregulated, but the expression of FTO was downregulated in sorafenib monotherapy samples (Fig. 7B). m6A quantified assay demonstrated a significant increase in m6A modification levels in primary HSCs treated with sorafenib compared to untreated controls (Fig. 7C). Furthermore, sorafenib treatment may upregulate the levels of ferritinophagy markers BECN1 and MAP1LC3B, and downregulate the levels of the ferritinophagy substrates SQSTM1 and FTH1 in human HSCs (Fig. 7D). Importantly, sorafenib treatment increased the expression of ferroptosis marker PTGS2 in primary HSCs, as well as ferroptotic events including redox-active iron overload, lipid peroxidation and GSH deletion (Fig. 7E). Taken together, these findings suggested that sorafenib monotherapy contributed to the upregulation of m6A modification, activation of autophagy, and induction of ferroptosis in human HSCs from patients with fibrotic.
Fig. 7.
m6A modification upregulation, autophagy activation, and ferroptosis induction occur in human HSCs receiving sorafenib monotherapy. (A, B) laser capture microdissection (LCM) was used to isolated the primary human HSCs from the collected liver tissue. ACTA2, FN1, COL1A1, METTL4, FTO, and YTHDF1 mRNA expression were determined by real-time PCR (No treatment, n = 10; Sorafenib treatment, n = 10, **, p < 0.01, ***, p < 0.001). (C) The m6A levels were determined by m6A RNA Methylation Quantitative kit (No treatment, n = 10; Sorafenib treatment, n = 10, ***, p < 0.001). (D) Real-time PCR was used to determine the mRNA expression of autophagy markers BECN1, MAP1LC3B, SQSTM1, and FTH1 (No treatment, n = 10; Sorafenib treatment, n = 10, ***, p < 0.001). (E) The PTGS2 mRNA expression, iron accumulation, MDA production and GSH depletion were determined (No treatment, n = 10; Sorafenib treatment, n = 10, ***, p < 0.001).
4. Discussion
Ferroptosis is a newly discovered form of regulated cell death caused by iron-dependent lipid peroxidation, which is essentially different from other well-characterized types of cell death [33]. Further recognition of ferroptosis in the development of liver fibrosis has brought new perspectives for diagnosis and treatment. Attractively, Li et al. recently reported that artemether-induced ferroptosis alleviates liver fibrosis via IRP2/Iron/ROS axis [34]. Furthermore, Kuo et al. found that chrysophanol attenuates liver fibrosis induced by hepatitis B virus X protein by regulating HSC ferroptosis [35]. Consistent with previous researches, we found that ferroptosis inducers significantly inhibited cell viability and alleviated CCl4-induced liver fibrosis by inducing HSC ferroptosis. Although ferroptosis brings new perspectives for the treatment and prevention of liver fibrosis, the precise role of ferroptosis in liver fibrosis has not been defined and there are still many drawbacks that need to be noticed. Of note, we previously found that ferroptosis inducers sorafenib and erastin could trigger MDA production, lipid ROS accumulation, and redox-active iron overload in HSCs, but not hepatocytes, macrophages, and liver sinusoidal endothelial cells [13]. In fact, many studies have confirmed that sorafenib treatment may exert anti-fibrotic activity in preclinical and clinical studies. Wang et al. showed that sorafenib can inhibit intrahepatic fibrosis, hydroxyproline accumulation and collagen deposition [36]. The sensitivity of liver cells to ferroptosis inducers was different, and specifically targeting HSC ferroptosis will be the research direction in the future.
m6A has been identified as the predominant internal modification of mRNAs in eukaryotic uncovering the consensus motif RRACH [37]. The m6A motifs are typically enriched in the 3′-UTR and CDS, which may play a critical role in regulating precursor mRNA maturation, translation and degradation [37]. A large number of studies have fully confirmed that m6A modification is closely related with various human diseases, and serves as an important role in regulating cell fate including apoptosis, necroptosis, senescence and EMT [27,38]. Interestingly, Wang et al. demonstrated that m6A modification promotes adipogenesis through activating autophagy [39]. Moreover, Niu et al. reported that m6A demethylases FTO is up-regulated in human breast cancer, and silencing FTO can significantly inhibit breast cancer cell proliferation and promote cell apoptosis [40]. However, the function and mechanism of m6A modification in ferroptosis remain unknown. In the present study, we reported that m6A modification was increased due to the upregulation of methylase METTL4 and the downregulation of demethylase FTO during HSC ferroptosis. Although more experiments are needed to confirm the exact role of m6A modification in HSC ferroptosis, our results showed a novel function of m6A modification in addition to regulating apoptosis, necroptosis, senescence and EMT.
Recent studies indicated that cellular autophagy plays an important role in ferroptosis [41,42]. In particular, the activation of selective autophagy such as NCOA4-mediated ferritinophagy, RAB7A-facilitated lipophagy [43], ARNTL-induced clockophagy [44], and PINK1-activated mitophagy [45], has been reported to promote ferroptosis. In the current study, we found that m6A modification triggered autophagy activation by stabilizing BECN1 mRNA. BECN1 plasmid could impair the inhibitory action of m6A modification inhibition on HSC ferroptosis. With a better understanding of autophagy-dependent ferroptosis, autophagy may have potential as a target for antifibrotic therapy.
m6A-binding proteins with YTH domain, including YTHDF1/2/3, YTHDC1/2, have been recognized to be the “readers” of m6A motif, which can regulate gene expression through recognize and bind to the m6A modified transcript [46]. Accumulating evidence has demonstrated that YTHDF1 can recognize m6A methylated mRNA and promote the post-transcriptional regulation of its targets [32]. In HCC, YTHDF1 contributes to the translation of autophagy-related genes ATG2A and ATG14 by binding to m6A modified mRNA, thus facilitating autophagy and malignancy of HCC [47]. In non-small cell lung cancer (NSCLC), YTHDF1 deficiency inhibits the proliferation of NSCLC cell and the formation of xenograft tumor by regulating the translational efficiency of CDK2, CDK4, and cyclin D1 [48]. In mouse brain, YTHDF1 facilitates translation of m6A-methylated neuronal mRNAs in response to neuronal stimulation, and this process contributes to learning and memory [49]. During EMT, YTHDF1 promotes the translation activation of m6A-increased Snail mRNA [28]. Importantly, the physiological role of YTHDF1 for the regulation of other tissue needs to be deciphered to distinguish the differential effect of specific m6A modifications during the development of disease. In the present study, we found that YTHDF1 promoted the activation of autophagy and BECN1 mRNA stability via recognizing the m6A binding site within BECN1 CDS at A1276. The mutation of BECN1 m6A sites could abrogate the YTHDF1-mediated BECN1 mRNA stability, and in turn prevented HSC ferroptosis. Our results reveal a new perspective on the potential mechanism by which YTHDF1 regulates ferroptosis.
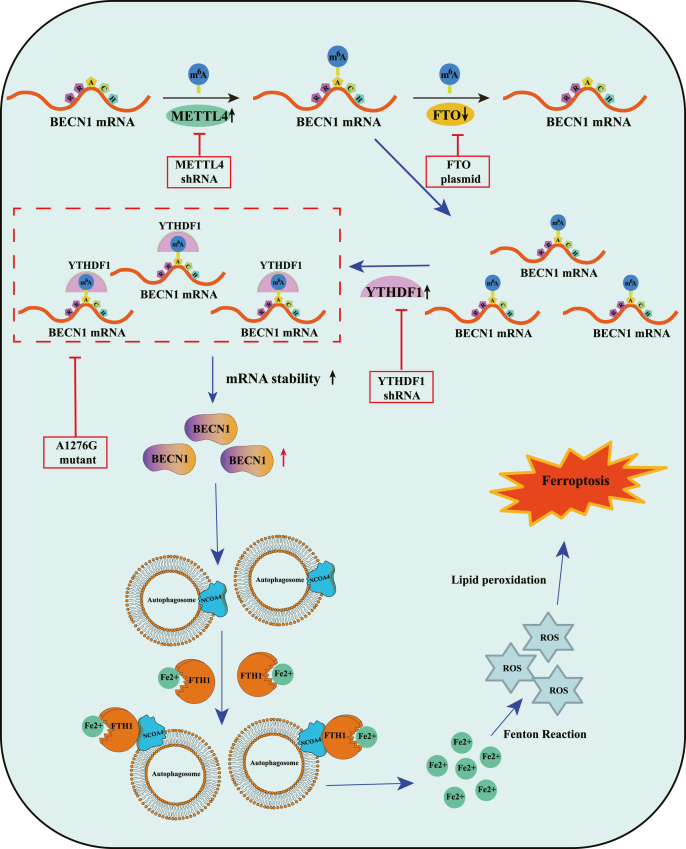
In summary, we illustrated that the interaction of BECN1 mRNA and m6A modification under ferroptosis inducer treatment enhanced autophagic ferritin degradation, and ultimately causing ferroptosis (Fig. 8). Further analyses of post-transcriptional regulation mechanisms of ferroptosis may provide valuable insights into therapeutic and diagnostic approaches to regulate HSC survival and death in liver fibrosis.
Fig. 8.
m6A modification induces HSC ferroptosis by regulating autophagy signaling pathway. The upregulation of methylase METTL4 and the downregulation of demethylase FTO increased the levels of m6A modifications in BECN1 mRNA. m6A reader YTHDF1 promoted BECN1 mRNA stability via recognizing the m6A binding site, thus triggering autophagy activation, and eventually leading to HSC ferroptosis.
Author contributions
Min Shen did the experiment and analyzed the data. Yujia Li, Yingqian Wang, Jiangjuan Shao, Feng Zhang and Guoping Yin participated in experiments. Anping Chen, Zili Zhang and Shizhong Zheng conceived the idea of study and manuscript writing. All authors had access to the study data and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (82000572, 82073914, 82173874), the Natural Science Foundation of Jiangsu Province (BK20200840), the Major Project of the Natural Science Research of Jiangsu Higher Education Institutions (19KJA310005), General Projects of the Natural Science Research of Jiangsu Higher Education Institutions (20KJB310003), the Joint Project of Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica and Yangtze River Pharmaceutical (JKLPSE202005), the Natural Science Foundation of Nanjing University of Chinese Medicine (NZY82000572), the Open Project of Chinese Materia Medica First-Class Discipline of Nanjing University of Chinese Medicine (2020YLKX023, 2020YLKX022),and Jiangsu Graduate Research and Practice Innovation Program (KYCX21_1738). We are very grateful for the help provided by the experiment center for science and technology of Nanjing University of Chinese Medicine.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102151.
Contributor Information
Zili Zhang, Email: zilizhang@njucm.edu.cn.
Shizhong Zheng, Email: nytws@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- 1.Breitkopf-Heinlein K., Syn W.K. Harnessing liver progenitors in the treatment of liver fibrosis: a step in the right direction? Gut. 2020;69(6):975–976. doi: 10.1136/gutjnl-2019-320203. [DOI] [PubMed] [Google Scholar]
- 2.Gandhi C.R. Hepatic stellate cell activation and pro-fibrogenic signals. J. Hepatol. 2017;67(5):1104–1105. doi: 10.1016/j.jhep.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang S., Friedman S.L. Hepatic fibrosis: a convergent response to liver injury that is reversible. J. Hepatol. 2020;73(1):210–211. doi: 10.1016/j.jhep.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bian M., He J., Jin H., Lian N., Shao J., Guo Q., Wang S., Zhang F., Zheng S. Oroxylin A induces apoptosis of activated hepatic stellate cells through endoplasmic reticulum stress. Apoptosis. 2019;24(11–12):905–920. doi: 10.1007/s10495-019-01568-2. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z., Yao Z., Zhao S., Shao J., Chen A., Zhang F., Zheng S. Interaction between autophagy and senescence is required for dihydroartemisinin to alleviate liver fibrosis. Cell Death Dis. 2017;8(6) doi: 10.1038/cddis.2017.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia Y., Wang F., Guo Q., Li M., Wang L., Zhang Z., Jiang S., Jin H., Chen A., Tan S., Zhang F., Shao J., Zheng S. Curcumol induces RIPK1/RIPK3 complex-dependent necroptosis via JNK1/2-ROS signaling in hepatic stellate cells. Redox Biol. 2018;19:375–387. doi: 10.1016/j.redox.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z., Zhao S., Yao Z., Wang L., Shao J., Chen A., Zhang F., Zheng S. Autophagy regulates turnover of lipid droplets via ROS-dependent Rab25 activation in hepatic stellate cell. Redox Biol. 2017;11:322–334. doi: 10.1016/j.redox.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Lu C., Xu W., Zhang F., Shao J., Zheng S. Nrf2 knockdown attenuates the ameliorative effects of ligustrazine on hepatic fibrosis by targeting hepatic stellate cell transdifferentiation. Toxicology. 2016;365:35–47. doi: 10.1016/j.tox.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Xu W., Lu C., Zhang F., Shao J., Yao S., Zheng S. Dihydroartemisinin counteracts fibrotic portal hypertension via farnesoid X receptor-dependent inhibition of hepatic stellate cell contraction. FEBS J. 2017;284(1):114–133. doi: 10.1111/febs.13956. [DOI] [PubMed] [Google Scholar]
- 10.Wang F., Jia Y., Li M., Wang L., Shao J., Guo Q., Tan S., Ding H., Chen A., Zhang F., Zheng S. Blockade of glycolysis-dependent contraction by oroxylin a via inhibition of lactate dehydrogenase-a in hepatic stellate cells. Cell Commun. Signal. 2019;17(1):11. doi: 10.1186/s12964-019-0324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F., Lu S., He J., Jin H., Wang F., Wu L., Shao J., Chen A., Zheng S. Ligand activation of PPARγ by ligustrazine suppresses pericyte functions of hepatic stellate cells via SMRT-Mediated transrepression of HIF-1α. Theranostics. 2018;8(3):610–626. doi: 10.7150/thno.22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z., Guo M., Li Y., Shen M., Kong D., Shao J., Ding H., Tan S., Chen A., Zhang F., Zheng S. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16(8):1482–1505. doi: 10.1080/15548627.2019.1687985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Yao Z., Wang L., Ding H., Shao J., Chen A., Zhang F., Zheng S. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14(12):2083–2103. doi: 10.1080/15548627.2018.1503146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z., Guo M., Shen M., Kong D., Zhang F., Shao J., Tan S., Wang S., Chen A., Cao P., Zheng S. The BRD7-P53-SLC25A28 axis regulates ferroptosis in hepatic stellate cells. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon S.J., Stockwell B.R. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 2014;10(1):9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 16.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamj A.F., Clish C.B., Brown L.M., Girotti A.W., Cornish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang R., Zhu S., Zeh H.J., Klionsky D.J., Tang D. BECN1 is a new driver of ferroptosis. Autophagy. 2018;4(12):2173–2175. doi: 10.1080/15548627.2018.1513758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou W., Xie Y., Song X., Sun X., Lotze M.T., Zeh H.J., 3rd, Kang R., Tang D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12(8):1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng X., Su R., Weng H., Huang H., Li Z., Chen J. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muratori L., Muratori P., Lanzoni G., Ferri S., Lenzi M. Application of the 2010 American Association for the study of liver diseases criteria of remission to a cohort of Italian patients with autoimmune hepatitis. Hepatology. 2010;52(5):1857–1858. doi: 10.1002/hep.23924. [DOI] [PubMed] [Google Scholar]
- 21.Nagai H., Kanekawa T., Kobayashi K., Mukozu T., Matsui D., Matsui T., Kanayama M., Wakui N., Momiyama K., Shinohara M., Ishii K., Igarashi Y., Sumino Y. Changes of cytokines in patients with liver cirrhosis and advanced hepatocellular carcinoma treated by sorafenib. Canc. Chemother. Pharmacol. 2014;73(2):223–229. doi: 10.1007/s00280-013-2344-1. [DOI] [PubMed] [Google Scholar]
- 22.McDaniel K., Meng F., Wu N., Sato K., Venter J., Bernuzzi F., Invernizzi P., Zhou T., Kyritsi K., Wan Y., Huang Q., Onori P., Francis H., Gaudio E., Glaser S., Alpini G. Forkhead box A2 regulates biliary heterogeneity and senescence during cholestatic liver injury in mice. Hepatology. 2017;65(2):544–559. doi: 10.1002/hep.28831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xing P., Niu Y., Mu R., Wang Z., Xie D., Li H., Dong L., Wang C. A pocket-escaping design to prevent the common interference with near-infrared fluorescent probes in vivo. Nat. Commun. 2020;11(1):1573. doi: 10.1038/s41467-020-15323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Liu J., Xiao W., Zeng Q., Bo H., Zhu Y., Gong L., He D., Xing X., Li R., Zhou M., Xiong W., Zhou Y., Zhou J., Li X., Guo F., Xu C., Chen X., Wang X., Wang F., Wang Q., Cao K. SIRT1 regulates N6-methyladenosine RNA modification in hepatocarcinogenesis by inducing RANBP2-dependent FTO SUMOylation. Hepatology. 2020;72(6):2029–2050. doi: 10.1002/hep.31222. [DOI] [PubMed] [Google Scholar]
- 25.Opefi C.A., Tranter D., Smith S.O., Reeves P.J. Construction of stable mammalian cell lines for inducible expression of G protein-coupled receptors. Methods Enzymol. 2015;556:283–305. doi: 10.1016/bs.mie.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y., Luo M., Zhang K., Zhang J., Gao T., Connell D.O., Yao F., Mu C., Cai B., Shang Y., Chen W. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020;11(1):433. doi: 10.1038/s41467-020-14324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song T., Yang Y., Wei H., Xie X., Lu J., Zeng Q., Peng J., Zhou Y., Jiang S., Peng J. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Res. 2019;47(12):6130–6144. doi: 10.1093/nar/gkz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X., Chai G., Wu Y., Li J., Chen F., Liu J., Luo G., Tauler J., Du J., Lin S., He C., Wang H. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019;10(1):2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Sun X., Ou Z., Chen R., Niu X., Chen D., Kang R., Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63(1):173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., Ren B., Pan T., He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy. 2021;17(1):1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., Jin C., Shen M., Wang Z., Tan S., Chen A., Wang S., Shao J., Zhang F., Zhang Z., Zheng S. Iron regulatory protein 2 is required for artemether-mediated anti-hepatic fibrosis through ferroptosis pathway. Free Radic. Biol. Med. 2020;160:845–859. doi: 10.1016/j.freeradbiomed.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Kuo C.Y., Chiu V., Hsieh P.C., Huang C.Y., Huang S.J., Tzeng I.S., Tsai F.M., Chen M.L., Liu C.T., Chen Y.R. Chrysophanol attenuates hepatitis B virus X protein-induced hepatic stellate cell fibrosis by regulating endoplasmic reticulum stress and ferroptosis. J. Pharmacol. Sci. 2020;144(3):172–182. doi: 10.1016/j.jphs.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Gao J., Zhang D., Zhang J., Ma J., Jiang H. New insights into the antifibrotic effects of sorafenib on hepatic stellate cells and liver fibrosis. J. Hepatol. 2010;53(1):132–144. doi: 10.1016/j.jhep.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 37.Huang H., Weng H., Zhou K., Wu T., Zhao B.S., Sun M., Chen Z., Deng X., Xiao G., Auer F., Klemm L., Wu H., Zuo Z., Qin X., Dong Y., Zhou Y., Qin H., Tao S., Du J., Liu J., Lu Z., Yin H., Mesquita A., Yuan C.L., Hu Y.C., Sun W., Su R., Dong L., Shen C., Li C., Qing Y., Jiang X., Wu X., Sun M., Guan J.L., Qu L., Wei M., Müschen M., Huang G., He C., Yang J., Chen J. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature. 2019;567(7748):414–419. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Wu R., Liu Y., Zhao Y., Bi Z., Yao Y., Liu Q., Shi H., Wang F., Wang Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16(7):1221–1235. doi: 10.1080/15548627.2019.1659617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu Y., Lin Z., Wan A., Chen H., Liang H., Sun L., Wang Y., Li X., Xiong X.F., Wei B., Wu X., Wan G. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Canc. 2019;18(1):46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Z., Geng Y., Lu X., Shi Y., Wu G., Zhang M., Shan B., Pan H., Yuan J. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc. Natl. Acad. Sci. U. S. A. 2019;116(8):2996–3005. doi: 10.1073/pnas.1819728116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao M., Monian P., Pan Q., Zhang W., Xiang J., Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26(9):1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schroeder B., Schulze R.J., Weller S.G., Sletten A.C., Casey C.A., McNiven M.A. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 2015;61(6):1896–1907. doi: 10.1002/hep.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang M., Chen P., Liu J., Zhu S., Kroemer G., Klionsky D.J., Lotze M.T., Zeh H.J., Kang R., Tang D. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci. Adv. 2019;5(7) doi: 10.1126/sciadv.aaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C., Zhang Y., Cheng X., Yuan H., Zhu S., Liu J., Wen Q., Xie Y., Liu J., Kroemer G., Klionsky D.J., Lotze M.T., Zeh H.J., Kang R., Tang D. PINK1 and PARK2 suppress pancreatic tumorigenesis through control of mitochondrial iron-mediated immunometabolism. Dev. Cell. 2018;46(4):441–455. doi: 10.1016/j.devcel.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang X., Liu B., Nie Z., Duan L., Xiong Q., Jin Z., Yang C., Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021;6(1):74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q., Ni Y., Zhang L., Jiang R., Xu J., Yang H., Hu Y., Qiu J., Pu L., Tang J., Wang X. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target. Ther. 2021;6(1):76. doi: 10.1038/s41392-020-00453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y., Fan S., Wu M., Zuo Z., Li X., Jiang L., Shen Q., Xu P., Zeng L., Zhou Y., Huang Y., Yang Z., Zhou J., Gao J., Zhou H., Xu S., Ji H., Shi P., Wu D.D., Yang C., Chen Y. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019;10(1):4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi H., Zhang X., Weng Y.L., Lu Z., Liu Y., Lu Z., Li J., Hao P., Zhang Y., Zhang F., Wu Y., Delgado J.Y., Su Y., Patel M.J., Cao X., Shen B., Huang X., Ming G.L., Zhuang X., Song H., He C., Zhou T. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563(7730):249–253. doi: 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.