Abstract
The majority of emerging infectious diseases originate in animals. Current routine surveillance is focused on known diseases and clinical syndromes, but the increasing likelihood of emerging disease outbreaks shows the critical importance of early detection of unusual illness or circulation of pathogens - prior to human disease manifestation. In this Viewpoint, we focus on one key pillar of preparedness—the need for early warning surveillance at the human, animal, environmental interface. The COVID-19 pandemic has revolutionized the scale of sequencing of pathogen genomes, and the current investments in global genomic surveillance offer great potential for a novel, truly integrated Disease X (with epidemic or pandemic potential) surveillance arm provided we do not make the mistake of developing them solely for the case at hand. Generic tools include metagenomic sequencing as a catch-all technique, rather than detection and sequencing protocols focusing on what we know. Developing agnostic or more targeted metagenomic sequencing to assess unusual disease in humans and animals, combined with random sampling of environmental samples capturing pathogen circulation is technically challenging, but could provide a true early warning system. Rather than rebuilding and reinforcing the pre-existing silo's, a real step forward would be to take the lessons learned and bring in novel essential partnerships in a One Health approach to preparedness.
1. Introduction
The ongoing COVID-19 pandemic has highlighted how extremely vulnerable we are as a society when unforeseen infectious disease events happen. The WHO R&D blueprint program, launched in 2016, called for action for a list of potential pandemic threats, which subsequently triggered the launch of ambitious vaccine development programs funded through the Coalition for Emerging disease Preparedness Innovations (https://www.who.int/research-observatory/analyses/rd_blueprint/en/ and https://cepi.net), specifically dedicated to emerging infectious diseases (EID).
The rationale was that the majority of emerging diseases come from animals and are first detected by the time human disease outbreaks occur in regions with sufficient health system infrastructure to allow diagnosis of the etiology [1]. With the changing risks of global spread, the timeliness of early detection is becoming a serious factor in our ability to detect outbreaks at the stage when they still can be contained. The use of vaccines and/or non-medical interventions is a second critical pillar in preparedness. This was highlighted from a vaccine trial embedded in outbreak response towards the end of the West African outbreak of Ebola Zaire, in 2015-2016 [2], that was successful, despite the huge challenges of establishing a trial in the midst of an outbreak [3]. Based on the trial outcomes, vaccination of at risk populations was done as part of the response to subsequent outbreaks in Democratic Republic of Congo with some success, although community acceptance is not a given (https://www.msf.org/drc-tenth-ebola-outbreak).
The Ebola example of preparedness is based on combining early diagnosis of known risks, followed by public health measures and risk-group targeted priority vaccination programs. It is a model for prevention of Backspace high human health impact for known EIDs that may cause outbreaks but do not (yet) spread very efficiently among humans. For highly transmissible EIDs, however, the bar is even higher and early detection may not be sufficient. Here, early warning surveillance programs are needed that allow early recognition of unusual disease syndromes, as well as the ability to rapidly identify reservoirs and pre-diagnostic transmission following the identification of the causative agent(s). Such surveillance is a challenging yet crucial element of future preparedness. For diseases with prolonged incubation periods, or complications months after initial infection, this is even more critical. Examples here are the impact on fetal development from Zika virus or Schmallenberg virus infection during pregnancy in humans and animals, respectively, both detected long after the initial introduction and spread of the disease in the Americas and Europe, respectively [4,5].
It is now during the current COVID-19 pandemic that we should decide how we can use the current experiences for future outbreaks and pandemics in the years to come. As the majority of EID come from animal reservoirs, risk-targeted surveillance should include knowledge on the ecology of reservoir hosts and drivers for emergence of outbreaks [6].
1.1. One Health definition
The need for a holistic perspective on infectious diseases has long been recognized and there is no shortage of definitions of the OneHealth concept. Following several decades of focus mainly on human medicine this was revitalized in the early years of the 21st century, most prominently at a meeting by the three global institutes with a focus on human health, food- and environment, and animal health, respectively (WHO, FAO and OIE) in 2004. The parties convened a global think tank that concluded that the threat of EIDs was increasing, that there were shortfalls in the organisation of surveillance, and that new mechanisms of surveillance and response were required, "using new approaches (e.g. syndromic surveillance), using new tools (e.g. geographic information systems, remote sensing data and molecular epidemiology) and bringing together different sectors and disciplines (e.g. medical, veterinary, population biology, information technology, economics, social science and diagnostics)". https://apps.who.int/iris/bitstream/handle/10665/68899/WHO_CDS_CPE_ZFK_2004.9.pdf. This definition clearly was focussed on early detection of zoonotic diseases. A report by the World Bank also made the case for One Health surveillance based on economic assessment of the net benefits of prevention [7].
Since then the concept has been widely used and many people and groups have claimed to be doing OneHealth research. The World Health Organization today uses the following definition: 'One Health' is an approach to designing and implementing programmes, policies, legislation and research in which multiple sectors communicate and work together to achieve better public health outcomes (https://www.who.int/news-room/q-a-detail/one-health). However, despite the meetings, policy statements, and research projects, some of the key recommendations from the 2004 report were not implemented. Also, one can argue whether this anthropocentric definition of One Health is sufficiently balanced with threats to animal health and ecosystem health from the same global processes driving disease emergence [8]. Recently, a formal expert panel was established to advise the tripartite (WHO, FAO, OIE), and the UN environmental program on the future needs of One Health surveillance. This group assumes a wider definition of One Health, and is currently reviewing regional examples of integrated One Health surveillance to assess scalability of these systems to other regions starting from a systematic review listing examples for West Nile, avian influenza, rabies, Rift valley fever and others [9].
In May 2021, in preparation of the World Health Assembly, leaders of the EU, the G7, and the G20 emphasized the need for future preparedness. The European commission has launched an initiative to develop infrastructure for preparedness for SARS COV 2 variants and future pandemic threats. This involves significant investments in development of infrastructure for detection, and for development and production of vaccines (https://ec.europa.eu/commission/presscorner/detail/en/fs_21_650). This also offers a huge opportunity to establish the integrated infrastructure recommended 17 years ago.
1.2. The breakthrough of genomic surveillance
Prior to the pandemic, genomic pathogen surveillance was slowly but gradually making its way into public health and clinical diagnostics. This was arguably most advanced in bacterial food safety where next generation sequencing and near-real time sharing of data between human clinical diagnostic, food and veterinary microbiology has been implemented in several countries; most noteworthy in the USA with the GenomeTrakr programme that was initiated already in 2011 [10].
The pandemic has clearly catapulted genomic surveillance to center stage. The scale of sequencing of SARS-CoV-2 genomes has outpaced all other surveillance systems, and the number of available genomes after 18 months of SARS-CoV-2 is higher than those for influenza, HIV and all foodborne bacterial species. Genomic sequencing has been embraced as part of the essential public health toolbox, driving massive investments in further development of sequencing capacity. Given the roll-out of vaccines, it is likely that this capacity will no longer be needed to the same scale in the near future. Therefore, it is pertinent to consider what it would take to make the developing infrastructure and tools applicable for preparedness for other future outbreaks and pandemic threats. This includes a critical appraisal of the data sharing infrastructures. Currently, pathogen specific options are developed. For instance, for SARS-CoV-2, the database model for influenza virus surveillance was adopted [11]. This was developed in response to criticism from low and middle income countries (LMICs) that wanted some governance over who could do what with data on pathogens from specific countries. Sharing of pathogen genomic data across domains is further complicated, and solutions chosen may differ regionally [12].
An important question to consider is—what a future infrastructure could look like. The next pandemic could be caused by a completely different pathogen and it would be logical to utilize a platform with a broad focus, targeting all potential infectious agents. The world has an established, global, neutral data-sharing platform in place, namely http://www.insdc.org/. This resource is currently mostly focused on research data [13], and separate databases have been developed to share time-sensitive data for surveillance [14], [15], [16], [17]. A disadvantage of this approach is that there is no public repository of this data, and that publicly released data are incomplete. There is an increasing call for open sharing of data for the research community, and the European Commission has invested in the European Open Science cloud, building on the long-established infrastructures for biodata [18]. While custom-built fit for purpose databases may still be the norm, in years to come, a longer term strategy for data-sharing that compensates for the fragmentation and that focuses on improving the already existing global data infrastructures is necessary [19]. The willingness to move towards full open sharing of data also has not been fully endorsed. In particular, scientists from LMICs have expressed that this was a "first world" way of thinking, and that there is a need for controlled access solutions to avoid a further deepening of the gap in research contributions from LMIC [20]. In addition, while FAIR data is advocated widely, the reality is much more challenging. The interpretation of what is allowed under the General Data Protection Regulation differs between institutes, countries, and continents, making sharing of patient related information a formidable challenge. Similarly, the Nagoya protocol, developed to protect national resources, has turned out to be a considerable barrier to international sharing of samples and even sequence data. As the Nagoya protocol also specifies that decisions on data exchange can no longer be made by researchers or research institutes, a new burocracy is developing with the risk of increasing barriers to data sharing. Thirdly, sharing of data in a structured manner requires the agreement on standards, another issue that is far from solved. These issues have been recognized at the level of international organizations and the European commission, and will need to be resolved to be able to fully capitalize on the potential of the rapidly increasing data for health [21].
1.3. Early detection
Whether it is risk-targeted monitoring, syndromic surveillance, or clinical diagnostic evaluation, engagement of citizens, care providers, veterinarians, farmers, and others involved in collection of essential samples is critical to the success of such programs. Studies have clearly shown that capacity building only happens when the individuals that need to build a novel capacity also feel they are contributing and have something to offer [22]. A potential way of promoting this is to provide frontline settings with a capacity to analyse their own data, and contribute to conclusions and/or publications prior to sharing them publicly. Thus, we can for example focus resources on open science, non-commercial web-based solutions [23], [24], [25] that will allow relatively un-experienced scientists in the clinical and veterinary frontline to complete simple analyses of their data and thereby take part in the research on global issues and not simply see their data being exploited by others. Furthermore, collaboration can also be improved through international clinical trial networks set up to rapidly respond to new infections. For instance, REMAP-CAP is an adaptive platform trial evaluating potential treatments for COVID-19 in 321 clinical sites in five continents. VEO is a project that aims to develop smart strategies for surveillance and is sampling the virome in large cities to assess how this methodology could be employed cease, cease to exist, as in stop existing.
However, improving capacity in the frontline is not sufficient. Easier access to reference testing and verification is needed, particularly when symptoms are seen and a known causative agent can not immediately be identified. Establishing international networks between clinical laboratories, public health institutions and academia, and the ability to provide reference testing without repercussions for the requesting individual would be key to make this happen. And while the metagenomic sequencing of samples does hold promise, it is unlikely to provide early warning surveillance unless it is embedded in well thought through sampling strategies, guided by ecological and epidemiological knowledge to avoid generation of massive amounts of data that do not provide meaningful surveillance information [26]. Finally, cost of using genomic technologies may be a barrier. Genome-based surveillance and metagenomic sequencing are more expensive than traditional pathogen targeted detection, although there is economy of scale when combining sequencing capacity for multiple purposes [27]. A recent study found that the costs per sample varied greatly, depending on how the sequencing effort and data analytics were organized, ranging from 15 to > 700 euro per sample. There was an inverse relationship between per sample costs, sample volume, and batch sizes, which in turn were determined by the breadth of applications for which the sequencing capacity had been established (single pathogen to multipurpose). Therefore, with proper preparation, genome-based surveillance can be made cost effective.
1.4. The need for One Health surveillance
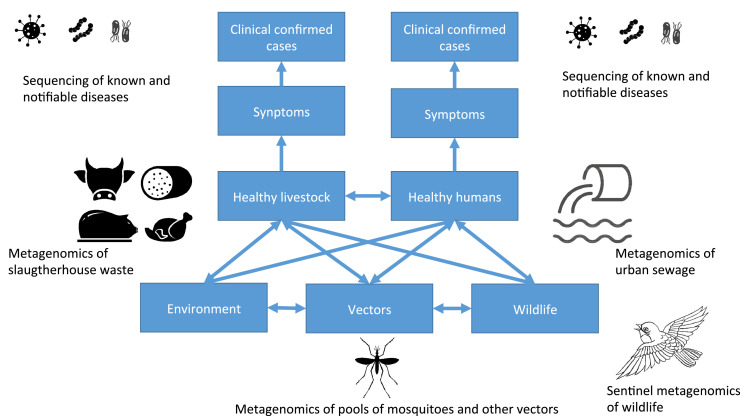
A future catch-all infrastructure would not be limited to surveillance based solely on human clinical cases, but also need to improve the generation and access to data also from other reservoirs (Fig. 1). While the human clinical diagnosis in many countries has been either fully paid or supported by governmental funding and/or heavy investment from large international funding organizations, this has not been the same for livestock clinical diagnostics, which basically has to be paid by the farmers. More control on limiting disease transmission between livestock is in place compared to humans, but host jumps into and between livestock also can occur; emergence of novel diseases in livestock will both have effect on livestock production and be a potential reservoir of diseases that might jump further to humans.
Fig. 1.
Examples for catch-all surveillance, combining disease surveillance in animals and humans targeting known diseases (top row), with catch-all metagenomic surveillance capturing circulation on other pathogens in livestock or humans (in this example depicted as wastewater metagenomics in slaughterhouses and urban sewage, respectively). Additional metagenomic sequencing could include other environmental samples, samples from wild-life and from vectors.
Most novel diseases likely circulate for some time in animals and humans before they eventually are detected in clinical cases. There is thus a need to collect standardised samples that represent the human and animal microbiome (bacteriome and virome) in a comparable way over time and between countries. Samples should be stored in a way that would allow for immediate re-analyses if new diseases emerge. Potential sampling sites could be long distance planes, urban metro-stations or urban sewage followed by metagenomic analyses, as previous piloted [28], [29], [30], [31]. This could potentially be combined with similar surveillance of animal waste (Fig. 1). While having the potential to continuously identify trends of endemic diseases and antimicrobial resistance, such a metagenomics approach has the clear advantage of allowing for rapid re-analyses of the data if a potential novel pathogen is detected and could thereby facilitate a rapid global overview of potential transmission and reservoir. Sampling of sewage also overcomes concerns regarding ethical and GDPR concerns because the samples are already anonymized. For any future design, it is crucial to allow for a mixture of lower cost solutions, in which the collection of samples for metagenomic agnostic testing could be embedded if needed. Also, while promising, the true added value of adding metagenomics to surveillance can only be decided by carefully designed studies. A recent successful example of this model is the discovery of SARS-CoV-2, that was identified through metagenomic sequencing in samples from patients that had been sent to specialised laboratories as part of the Chinese metagenomic disease detection program [32].
We can also improve the environment, wildlife and vector surveillance. Sentinel surveillance of wild birds is already in place for avian flu in some countries and in an informal international collaboration [33]; similarly, there are examples of regional initiatives and projects targeting mosquitos and ticks for surveillance of specific vector borne diseases. Such initiatives could be expanded to not only focus on a single disease, but for detection of a combination of infectious agents of importance and agnostic metagenomics.
2. Conclusion
The close interactions between humans, animals and the environment do hold a risk of emergence of infectious diseases, and their likelihood is increasing. Therefore, preparing for future epidemics and pandemics needs special focus on understanding complexity of the interactions between humans, animals and the environment, the different farming systems with their levels of biosecurity, prevention of disease introduction - for instance through vaccines-, and rapid control of outbreaks by containment measures. The pandemic has shown the critical importance of collaboration beyond the "normal" infrastructures, networks, and disciplines. This also applies to the use of genomics and open data infrastructures. The current investments in global genomic surveillance offer great potential, precluded we do not make the same old mistake of developing them solely for the case at hand. Rather than rebuilding and reinforcing the pre-existing silo'’, a real step forward would be to bring these novel essential partnerships together [34].
Contributors
Frank Aarestrup and Marion Koopmans drafted the first version. Marc Bonten reviewed and edited substantially. All authors approved the final version of the manuscript.
Declaration of Interests
Funding to the institution. No involvement in the writing. None other to declare.
Acknowledgements
This work was supported by the European Union's Horizon H2020 grants VEO (874735), RECOVER (101003589), ECRAID (965313) and Novo Nordisk Foundation (NNF16OC0021856).
References
- 1.Machalaba CM, Karesh WB. Emerging infectious disease risk: shared drivers with environmental change. Rev Sci Tech. 2017;36:435–444. doi: 10.20506/rst.36.2.2664. [DOI] [PubMed] [Google Scholar]
- 2.Henao-Restrepo AM, Camacho A, Longini IM. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!) Lancet. 2017;389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walldorf JA, Cloessner EA, Hyde TB, MacNeil A, CDC Emergency Ebola Vaccine Taskforce Considerations for use of Ebola vaccine during an emergency response. Vaccine. 2019;37:7190–7200. doi: 10.1016/j.vaccine.2017.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faria NR, Quick J, Claro IM. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. 2017;546:406–410. doi: 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann B, Scheuch M, Höper D. Novel Orthobunyavirus in Cattle, Europe, 2011. Emerg Infect Dis. 2012;18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sikkema RS, Koopmans MPG. Preparing for Emerging Zoonotic Viruses. Encyclopedia Virol. 2021:256–266. [Google Scholar]
- 7.World Bank . 2012. People, Pathogens and Our Planet: The Economics of One Health.https://openknowledge.worldbank.org/handle/10986/11892 Washington, DC. [Google Scholar]
- 8.Zinsstag J, Schelling E, Waltner-Toews D, Tanner M. From "one medicine" to "one health" and systemic approaches to health and well-being. Prev Vet Med. 2011;101:148–156. doi: 10.1016/j.prevetmed.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordier M, Uea-Anuwong T, Binot A, Hendrikx P, Goutard FL. Characteristics of ssalth surveillance systems: a systematic literature review. Prev Vet Med. 2020;181 doi: 10.1016/j.prevetmed.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Timme RE, Sanchez Leon M, Allard MW. Utilizing the Public GenomeTrakr Database for Foodborne Pathogen Traceback. Methods Mol Biol. 2019;1918:201–212. doi: 10.1007/978-1-4939-9000-9_17. [DOI] [PubMed] [Google Scholar]
- 11.Shu Y, McCauley J.GISAID. Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dos S Ribeiro C, Koopmans MP, Haringhuizen GB. Threats to timely sharing of pathogen sequence data. Science. 2018;362:404–406. doi: 10.1126/science.aau5229. [DOI] [PubMed] [Google Scholar]
- 13.Harrison PW, Ahamed A, Aslam R. The European Nucleotide Archive in 2020. Nucleic Acids Res. 2021;49:D82–D85. doi: 10.1093/nar/gkaa1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulder AC, Kroneman A, Franz E. HEVnet: a One Health, collaborative, interdisciplinary network and sequence data repository for enhanced hepatitis E virus molecular typing, characterisation and epidemiological investigations. Euro Surveill. 2019;24 doi: 10.2807/1560-7917.ES.2019.24.10.1800407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Félix B, Danan C, Van Walle I. Building a molecular Listeria monocytogenes database to centralize and share PFGE typing data from food, environmental and animal strains throughout Europe. J Microbiol Methods. 2014;104:1–8. doi: 10.1016/j.mimet.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Harvala H, Jasir A, Penttinen P, Pastore Celentano L, Greco D, Broberg E. Surveillance and laboratory detection for non-polio enteroviruses in the European Union/European Economic Area, 2016. Euro Surveill. 2017;22:16–00807. doi: 10.2807/1560-7917.ES.2017.22.45.16-00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Agama Study Group. Achtman M. The EnteroBase user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anon European Open Science Cloud. Nat Genet. 2016;48:821. doi: 10.1038/ng.3642. [DOI] [PubMed] [Google Scholar]
- 19.van Noorden R. Scientists call for fully open sharing of coronavirus genome data. Nature. 2021;590:195–196. doi: 10.1038/d41586-021-00305-7. [DOI] [PubMed] [Google Scholar]
- 20.Maxmen A. Why some researchers oppose unrestricted sharing of coronavirus genome data. Nature. 2021;593:176–177. doi: 10.1038/d41586-021-01194-6. [DOI] [PubMed] [Google Scholar]
- 21.Dos S Ribeiro C, Koopmans MP, Haringhuizen GB. Threats to timely sharing of pathogen sequence data. Science. 2018;362:404–406. doi: 10.1126/science.aau5229. [DOI] [PubMed] [Google Scholar]
- 22.Faure MC, Munung NS, Ntusi NAB, Pratt B, de Vries J. Mapping experiences and perspectives of equity in international health collaborations: a scoping review. Int J Equity Health. 2021;20:28. doi: 10.1186/s12939-020-01350-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen MV, Cosentino S, Rasmussen S. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Beek J, de Graaf M, Al-Hello H. NoroNet. Molecular surveillance of norovirus, 2005-16: an epidemiological analysis of data collected from the NoroNet network. Lancet Infect Dis. 2018;18:545–553. doi: 10.1016/S1473-3099(18)30059-8. [DOI] [PubMed] [Google Scholar]
- 25.Bortolaia V, Kaas RS, Ruppe E. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray GC, Robie ER, Studstill CJ, Nunn CL. Mitigating Future Respiratory Virus Pandemics: New Threats and Approaches to Consider. Viruses. 2021;13:637. doi: 10.3390/v13040637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alleweldt F, Kara Ş, Best K. Economic evaluation of whole genome sequencing for pathogen identification and surveillance - results of case studies in Europe and the Americas 2016 to 2019. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.9.1900606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordahl Petersen T, Rasmussen S, Hasman H. Meta-genomic analysis of toilet waste from long distance flights; a step towards global surveillance of infectious diseases and antimicrobial resistance. Sci Rep. 2015;5:11444. doi: 10.1038/srep11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendriksen RS, Munk P, Njage P. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat Commun. 2019;10:1124. doi: 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieuwenhuijse DF, Oude Munnink BB, Phan MVT. Setting a baseline for global urban virome surveillance in sewage. Sci Rep. 2020;10:13748. doi: 10.1038/s41598-020-69869-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danko D, Bezdan D, Afshin EE. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell. 2021;184:3376–3393. doi: 10.1016/j.cell.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu F, Zhao S, Yu B. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lycett SJ, Pohlmann A, Staubach C. Genesis and spread of multiple reassortants during the 2016/2017 H5 avian influenza epidemic in Eurasia. Proc Natl Acad Sci USA. 2020;117:20814–20825. doi: 10.1073/pnas.2001813117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bedford J, Farrar J, Ihekweazu C, Kang G, Koopmans M, Nkengasong J. A new twenty-first century science for effective epidemic response. Nature. 2019;575:130–136. doi: 10.1038/s41586-019-1717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]