Abstract
A highly sensitive enzyme immunoassay (EIA) for the hepatitis C virus (HCV) core antigen (HCVcAg) was developed, and its performance was compared with that of the AMPLICOR HCV test (Roche Molecular Systems). The developed one-step pretreatment method, 30-min incubation of the specimen with a solution containing three different types of detergents (Triton X-100, 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], and sodium dodecyl sulfate), does not require any special device. Because the interfering anti-core antibody in the sample was sufficiently inactivated by the pretreatment, HCVcAg in the sample could be detected. The immunoreactivity on gel filtration was shifted from void fractions to those corresponding to the molecular mass range from 20 to 25 kDa, which is equal to the estimated molecular mass of HCVcAg, after the pretreatment. By the recovery test with HCVcAg-positive serum, the recovery rate was 93.5 to 106.5%. There was no interference with the EIA by anticoagulants or blood components in the serum. When the cutoff value was tentatively set at 0.5 mU/ml based on the distribution of healthy subjects’ sera, the sera of all healthy subjects (n = 125) and patients with hepatitis B (n = 50) were negative. HCVcAg was detected in sera from 57 of 73 individuals (78.1%) with anti-HCV antibody. Similarly, HCV RNA was detected in sera from 59 individuals (80.8%) with the AMPLICOR HCV as the qualitative test (AMPLICOR HCV test) and in sera from 54 individuals (74.0%) by the AMPLICOR HCV Monitor as the quantitative test (AMPLICOR Monitor test). Concentrations of HCVcAg and HCV RNA (measured by the AMPLICOR Monitor test) correlated significantly (r = 0.8, P < 0.001). On seroconversion panels, HCVcAg was detected during the early stage of infection, when anti-HCV antibodies had not been produced. This assay for HCVcAg is simpler than assays for HCV RNA based on gene technology and shows specificity and sensitivity equivalent to those of the AMPLICOR HCV test.
Hepatitis C virus (HCV) is a single-stranded, positive-sense RNA virus with a genome of approximately 9,500 nucleotides coding for a polypeptide with a length of about 3,000 amino acids (aa) (4, 10). HCV is the causative agent of hepatitis C, and it has been clearly shown that the primary routes of infection are through blood and blood products infected with HCV. After the development of an anti-HCV antibody detection test using recombinant HCV antigen (14, 19), it has become possible to identify nearly all persons infected with HCV.
However, the test is unable to confirm viral infections during periods in the early phase of the infection before anti-HCV antibody has been produced (16, 29). Cases of posttransfusion hepatitis C caused by the transfusion of blood that tested negative for anti-HCV antibody, donated by individuals in this early period, have been reported (3, 21, 38). Therefore, the risk of secondary infection caused by blood components still needs to be eliminated. In addition, antibody tests cannot distinguish between persons with anti-HCV antibodies who have recovered and patients exhibiting an active infection, and they are not sufficient for the monitoring of therapy (15, 32). Therefore, a method that is able to detect HCV in samples is required.
The AMPLICOR HCV test and branched-chain DNA signal amplification assay (b-DNA: Quantiplex HCV-RNA assay) to detect HCV genome RNA have been used to detect HCV and to monitor the efficacy of treatment (15, 37, 40), and recently both assay systems have been applied to partially automated systems (22, 26, 41). Even so, there are several problems with the application of these methods to the mass screening of blood donors: the b-DNA assay requires a long incubation period and has a low sensitivity (15); PCR requires considerable skill and has been reported to give a high false-positive rate.
Recently, methods for detecting viral antigen were developed by applying a monoclonal antibody to the HCV core antigen (HCVcAg) (11, 34). The assay, as reported by Takahashi et al. (34), had a low sensitivity for detecting HCVcAg present at a few nanograms per milliliter and required the concentration and fractionation of HCV to detect the antigen. Thus, the performance of this assay was clearly insufficient for clinical application (20). Other methods that detect the presence of HCVcAg in serum were reported to be useful for monitoring interferon therapy (35, 36). However, their low sensitivity (the detection limit is between 104 and 105 equivalent copies of HCV RNA/ml) (11, 25, 36) and the complicated specimen pretreatment process make it difficult to apply them to the mass screening of blood donors.
The present study was aimed at overcoming the problems described above by introducing an efficient specimen pretreatment method into enzyme immunoassay (EIA) for HCVcAg.
MATERIALS AND METHODS
Samples and reagents.
Sera testing positive for anti-HCV antibody were collected from blood donors screened with the Ortho HCV Ab ELISA test III kit (Ortho Clinical Diagnostics Systems, Raritan, N.J.). The presence of anti-HCV antibody was confirmed by the RIBA 3 test kit (Chiron Corporation and Ortho Clinical Diagnostics Systems). Control samples that tested negative for anti-HCV antibody and hepatitis B virus surface antigen were also obtained from blood donors. Serum samples were collected consecutively from patients with chronic hepatitis B at the Shinshu University Hospital in October 1997. The seroconversion panels were purchased from Boston Biomedica Inc. (West Bridgewater, Mass.). All sera were stored at −80°C until tested.
Hemoglobin, bilirubin, lipemic interferents, and rheumatoid factor were purchased from International Reagents Corp. (Kobe, Japan); peroxidase (POD) was obtained from Toyobo (Osaka, Japan); the mouse monoAb-ID kit came from Zymed (San Francisco, Calif.); pepsin and alkaline phosphatase (ALP) were from Boehringer (Mannheim, Germany); and the bicinchoninic acid protein assay kit was from Pierce Chemical Co. (Rockford, Ill.).
Recombinant core antigen.
The fusion protein of the recombinant core antigen (aa 1 through 160 of the HCV genotype 2a isolate [GenBank accession no. I49748]) was expressed in Escherichia coli and was solubilized and purified from inclusion bodies by ion exchange chromatography and hydrophobic chromatography as described previously (28). The amount of recombinant core antigen was determined by using the bicinchoninic acid assay kit according to the manufacturer’s instructions.
Monoclonal antibodies.
BALB/c mice were immunized five times by subcutaneous injection of the recombinant core antigen mixed with TiterMax (CytRx Corporation, Norcross, Ga.). Splenocytes from the immunized mice were fused with myeloma cells (P3X63Ag8) (13). The cell mixture was cultured in RPMI 1640 supplemented with 10% fetal calf serum, and the hybridoma cells were selected by using a hypoxanthine-aminopterin-thymidine medium. Thirty-two hybridoma cell lines were established, and all monoclonal antibodies were prepared from ascite fluids of the mice and were purified by chromatography on a protein-A column (Pharmacia, Uppsala, Sweden). The subclasses of the purified monoclonal antibodies were determined with the mouse monoAb-ID kit. The epitopes of the monoclonal antibodies and polyclonal antibodies were analyzed by an EIA system by using 15 synthesized peptides covering aa 1 to 160 of the recombinant core antigen, as described previously (11).
Preparation of POD-conjugated polyclonal antibody.
New Zealand White rabbits were immunized subcutaneously six times with recombinant core antigen containing Freund’s complete adjuvant (Wako Pure Chemical Industries, Osaka, Japan). The immunoglobulin G (IgG) fraction was separated by protein-A column chromatography. The IgG was digested by pepsin in 0.1 M acetate buffer (pH 4.5), and the F(ab′)2 fragment was isolated by gel filtration on a Superdex 200 pg column (1.6 by 60 cm). The Fab′ fragment was prepared by reducing the F(ab′)2 fragment and was conjugated to POD by the maleimide hinge method (39). The conjugated material was purified by gel filtration on a Superdex 200 pg column (1.6 by 60 cm).
Preparation of ALP-conjugated monoclonal antibody.
Monoclonal antibodies (c11-10 and c11-14) were digested with pepsin, and the F(ab′)2 fragment was isolated by gel filtration on a Superdex 200 pg column (1.6 by 60 cm). The Fab′ fragment was prepared and conjugated to ALP as described above. The conjugated material was fractionated by chromatography on a Superdex 200 pg column (1.6 by 60 cm).
Specimen pretreatment and EIA for HCVcAg.
A well of a microtiter plate (Microlite 2; Dynatech Laboratories, Chantilly, Va.) was coated with 100 μl of an equimolar mixture of the anti-HCVcAg monoclonal antibodies c11-3 and c11-7 at a total concentration of 8 μg/ml and allowed to sit overnight at 4°C. Thereafter, the well was washed twice with phosphate-buffered saline (PBS) (0.15 M NaCl, 10 mM sodium phosphate buffer [pH 7.3]), followed by incubation with a blocking reagent (1% bovine serum albumin [BSA] in PBS [pH 7.3]) at room temperature for 2 h. After removal of the blocking solution, 100 μl of a reaction buffer (1% BSA, 5 mM EDTA, 0.1 M NaCl, 3% mouse serum, 0.3% Triton X-100, 0.1 M phosphate buffer [pH 7.2]) was added to each well. For the absorption test, to confirm the specificity of the immunoreaction, c11-3 and c11-7 monoclonal antibodies at a total concentration of 20 μg/ml were added to this reaction buffer. One hundred microliters of specimen was mixed with 50 μl of pretreatment solution (a mixture of 0.3% Triton X-100, 1.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], and 15% sodium dodecyl sulfate [SDS]) and incubated at 56°C for 30 min. One hundred microliters of pretreated specimen was added to each well prefilled with an equal volume of reaction buffer. The plates were incubated for 90 min at room temperature and then washed five times with washing buffer (0.05% Tween 20 in PBS [pH 7.3]). ALP-conjugated monoclonal antibodies (equimolar mixture of c11-10 and c11-14) were added to each well, and the plates were incubated for 30 min at room temperature. After the plates were washed six times with washing buffer, CDP star (Emerald II, used as an enhancer; Tropix, Bedford, Mass.) was added and incubated for 15 min at room temperature. Then the relative chemiluminescence units (RLU) were measured by using a microplate reader (LUMINOUS CT-9000D; DIA-IATRON, Tokyo, Japan).
Panel serum 50, anti-HCV antibody-positive serum, was used as the standard serum, and its immunoreactive titer was defined as 1 U/ml. This standard serum was serially diluted with normal horse serum and assayed in the same manner as described above. A calibration curve was generated by plotting the obtained RLU against the log of the concentration of the standard serum. Concentrations of HCVcAg in pretreated specimens were obtained by interpolation of their RLU from the calibration curve.
EIA for anti-HCV core antibodies.
Five micrograms of recombinant core antigen/ml was applied to the microtiter plate wells. After overnight incubation at 4°C, the plate was washed twice with PBS, followed by 2 h of incubation with a blocking reagent (0.5% casein in PBS [pH 7.3]). After the blocking solution was removed, 200 μl of reaction buffer (0.5% casein, 0.1 M NaCl, 0.05% Tween 20, 0.1 M phosphate buffer [pH 7.3]) was added to each well. Then, 20 μl of specimen diluted with PBS (1.5-fold concentration) or pretreated specimen was added to each well. The plate was incubated for 60 min at room temperature and then washed five times with washing buffer. POD-conjugated anti-human IgG (Boehringer) was added to each well, and the plate was incubated 30 min at room temperature. After the plate was washed five times with washing buffer, the enzymatic color reaction was effected by the addition of substrate solution (2 mg of o-phenylenediamine/ml, 0.4 μl of 30% hydrogen peroxide/ml, 50 mM citric acid [pH 5.0]). This enzymatic reaction was allowed to proceed for 30 min at room temperature and was stopped by addition of 100 μl of 1 M sulfuric acid. The absorbance of each well was measured at 492 nm, with 630 nm as the reference wavelength, by using a plate reader. The cutoff value was determined based on the distribution of absorbance values of specimens from healthy subjects. Anti-HCV core antibody EIA results are expressed as the ratio of specimen absorbance to the cutoff value (s/co ratio). Ratios over 1.0 are considered to indicate a positive result.
Gel filtration analysis of HCVcAg in serum.
Panel serum 13 (1.5 ml), anti-HCV antibody-positive serum, was subjected to gel filtration on a Sepharose CL-4B column (1.6 by 70 cm; Pharmacia), and then the total amount of HCVcAg in each fraction was measured by EIA as described above. To analyze the immunoreactive form of HCVcAg in a pretreated specimen, 1 ml of panel serum 13, which was pretreated as described above, was applied to a Superdex 200 pg column (1.6 by 60 cm) and the amount of HCVcAg in each fraction was measured by EIA. The molecular mass of each fraction was calibrated with ribonuclease A (13.7 kDa), recombinant core antigen (21 kDa), chymotrypsinogen (25 kDa), BSA (68 kDa), aldolase (158 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa).
AMPLICOR HCV test.
The AMPLICOR HCV test (Roche Molecular Systems, Branchburg, N.J.) (40) was performed according to the manufacturer’s instructions. The cutoff value of the AMPLICOR HCV for the qualitative test (AMPLICOR HCV test) was set at 200 copies/ml, and that of the AMPLICOR HCV Monitor for the quantitative test (AMPLICOR Monitor test) was set at 1,000 copies/ml.
In-house RT-PCR.
The nested cDNA reverse transcription (RT)-PCR assay was used for detecting HCV RNA in 100-μl samples of serum, as described previously (28). The primer set used for the in-house PCR assay was selected from the 5′ untranslated region of the HCV genome.
Determination of HCV genotype.
The HCV genotype was determined by the method reported by Okamoto et al. (23, 24). A line probe assay (INNO-LiPA HCV II test; INNOGENETICS, Zwijndrecht, Belgium) was performed according to the manufacturer’s instructions (31).
Liver function test.
Serum alanine aminotransferase (ALT) levels were measured with a Hitachi 7450 analyzer at 37°C, according to the manufacturer’s protocol (the reagent used was Clinimate GPT; Daiichi Pure Chemical, Tokyo, Japan).
RESULTS
To evaluate the specimen pretreatment method, a sandwich EIA for the detection of HCVcAg was established by using anti-recombinant core polypeptide rabbit polyclonal antibody, which was blended so as to recognize almost all peptide fragments covering the HCV core region. A prototype of the specimen pretreatment method using a solution containing Triton X-100, CHAPS, and SDS was selected. Then, the combinations of monoclonal antibodies were investigated among 32 clones for ability to detect HCVcAg in serum pretreated by the method. Consequently, we selected c11-3 and c11-7 as immobilized monoclonal antibodies to capture HCVcAg and c11-10 and c11-14 as secondary monoclonal antibodies. The combination of these antibodies exhibited the lowest detection limit for a certain number of the pretreated specimens (detailed data not shown). The epitopes of each monoclonal antibody (c11-3, c11-7, c11-10, and c11-14), which were determined with 20-residue-long peptides with 10-aa overlap covering the core region, were located at aa residues 100 to 120, 120 to 130, 21 to 40, and 41 to 50, respectively. Various conditions of the specimen pretreatment and the sandwich EIA for HCVcAg were optimized for this combination of monoclonal antibodies; for example, the concentrations of ingredients in the various solutions, the incubation temperature, and the length of incubation time were studied.
Effect of specimen pretreatment.
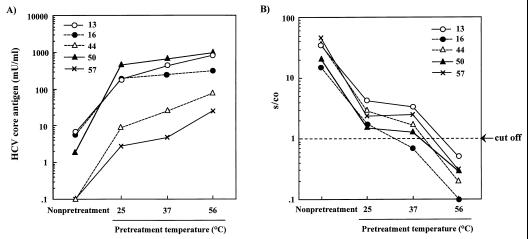
Fig. 1 shows the effect of pretreatment incubation temperature on the immunoreactivity of HCVcAg and the titer of anti-HCV core antibodies in anti-HCV antibody-positive sera. It was clear that the activity of the anti-HCV core antibodies in all the sera was reduced to an undetectable level by incubation at 56°C (Fig. 1B). For panel sera 44 and 57, in which it was impossible to detect HCVcAg without this pretreatment, HCVcAg became detectable after pretreatment (Fig. 1A). The gradual increase in sensitivity of the HCVcAg assay and decrease in titer of the anti-HCV core antibodies in these samples with high titers of anti-HCV antibodies (s/co ratio, over 35) indicated that the anti-HCV core antibodies in serum would interfere with the binding of monoclonal antibodies to HCVcAg in this EIA.
FIG. 1.
The effect of pretreatment on the immunoreactivity of HCV core antigen (A) and the titer of anti-HCV core antibodies in anti-HCV antibody-positive sera (B) at various incubation temperatures. EIA results for anti-HCV core antibodies are means of duplicate assays, expressed as ratios of specimen absorbance to cutoff value (s/co ratios). Ratios over 1.0 are considered to show reactivity.
EIA for HCVcAg.
Serum 50, anti-HCV antibody-positive serum with HCV genotype 1a, was used as the standard. The HCVcAg concentration of serum 50 was defined as 1.0 U/ml. The HCV RNA titer of this serum, measured by the AMPLICOR HCV Monitor test, was 1.0 million copies/ml.
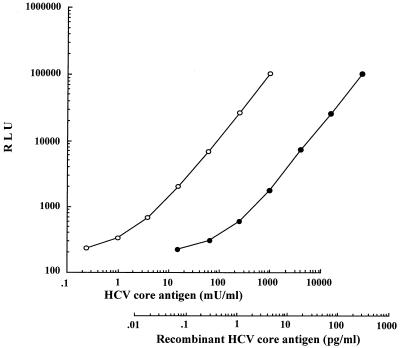
The calibration curve of the serial dilutions of serum 50 with normal horse serum was determined (Fig. 2). The analytical lower detection limit of this assay was 0.2 mU/ml: the point did not overlap with the concentration corresponding to 2 standard deviations above the mean RLU of the zero calibrator (n = 6). The dilution curves of the two sera in our panel (data not shown) and the recombinant core antigen (Fig. 2) were parallel to that of the standard serum. The recovery rates of various concentrations of HCVcAg-positive serum or the recombinant core antigen, which were determined by the addition of these samples to three reference sera, ranged from 93.5 to 106.5%. Intraassay precision was assessed by making eight measurements of four specimens. The mean HCVcAg values for the specimens were 1.3, 16.5, 59.3, and 198.2 mU/ml, and the coefficients of variation (CVs) ranged from 2.7 to 12.7%. The interday reproducibility was assessed for three specimens. The mean HCVcAg values for the specimens were 13.5, 54.5, and 232.6 mU/ml for 3 days of measurements, and the CVs ranged from 4.0 to 7.9%.
FIG. 2.
Standard serum and recombinant HCV core antigen dilution curve. Standard serum (○) and recombinant HCV core antigen (●) were serially diluted four times with normal horse serum.
The influence of anticoagulants and blood elements in serum on EIA results was assessed. The anticoagulants EDTA (<2.0 mg/ml), heparin (<100 U/ml), and sodium citrate (<10 mg/ml) showed no interference with the assay. Assessment of the influence of hemoglobin (<4.9 mg/ml), bilirubin (<0.19 mg/ml), lipemia (turbidity, with lipid concentration as high as 16.5 mg/ml), and rheumatoid factor (<500 U/ml) also revealed that these did not interfere with this assay.
Gel filtration analysis of HCVcAg in pretreated serum.
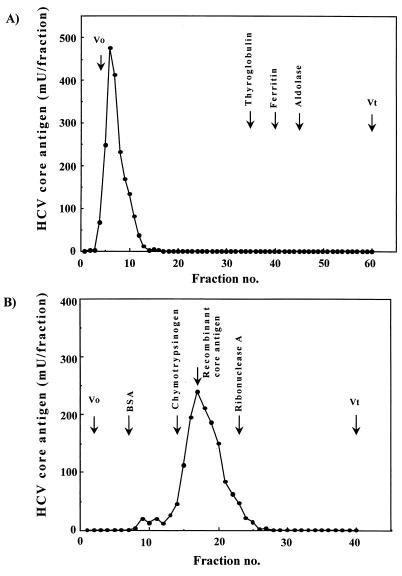
To analyze the immunoreactivity of HCVcAg detected after this pretreatment, a gel filtration study of nonpretreated or pretreated anti-HCV antibody-positive serum (panel serum 13) was performed. The results are shown in Fig. 3. The peak of the immunoreactivity of serum samples without pretreatment was detected in void fractions (Fig. 3A). When the pretreated serum was analyzed, the immunoreactivity was eluted at a position of approximately 20 to 25 kDa, and the molecular mass of this peak corresponded with that of the recombinant core antigen and also with the molecular mass of HCVcAg estimated based on an in vitro study (9). No other immunoreactive peak fractions were detectable (Fig. 3B).
FIG. 3.
(A) Gel filtration patterns of immunoreactive HCV core antigen in nonpretreated anti-HCV antibody-positive serum. Each fraction after fractionation on a Sepharose CL-4B column was pretreated and then measured by EIA. (B) Gel filtration patterns of immunoreactive HCV core antigen in pretreated anti-HCV antibody-positive serum on a Superdex 200 pg column. The total amount of HCV core antigen in each fraction after the chromatography of pretreated serum was measured by the EIA. The elution points for molecular markers are identified at the top. Vo, void volume; Vt, total column volume.
Sera from healthy subjects and hepatitis B patients.
In this assay, 125 serum samples from healthy subjects were measured, and the values were below 0.35 mU/ml. Based on this data, the cutoff value of this assay was tentatively set at 0.5 mU/ml. The immunoreactivities of sera from all healthy subjects and hepatitis B patients were below this cutoff value.
Detection of HCVcAg in anti-HCV antibody-positive sera.
HCVcAg for 73 human sera with anti-HCV antibodies was determined. The concentration of HCV RNA was also measured by using two AMPLICOR HCV tests. The results are summarized in Table 1. The positive rates for the HCVcAg assay, the AMPLICOR HCV test, and AMPLICOR Monitor test were 78.1% (57 of 73 sera), 80.8% (59 of 73 sera), and 74.0% (54 of 73 sera), respectively. All positive sera detected by the AMPLICOR Monitor test were judged to be positive based on both the AMPLICOR HCV test and the HCVcAg assay. The positive rate of the in-house RT-PCR was 79.5% (58 of 73 sera). The 58 positive sera identified by the in-house RT-PCR showed HCV genotypes 1a (35 sera), 1b (7 sera), 2b (6 sera), 3a (3 sera), and 3c (1 serum). The genotypes for three sera could not be determined by either genotyping method. The remaining three sera were not tested, because the volume of each was less than that required for the test.
TABLE 1.
The results obtained with anti-HCV antibody-positive sera by the HCV core antigen assay and the AMPLICOR HCV test
| AMPLICOR HCV test result | No. of serum samples with HCV core antigen assay result
|
Correlation (%) | |
|---|---|---|---|
| Positive | Negative | ||
| Qualitative test | |||
| Positive | 56 | 3 | |
| 94.5 | |||
| Negative | 1 | 13 | |
| Monitor test | |||
| Positive | 54 | 0 | |
| 95.9 | |||
| Negative | 3 | 16 | |
Samples for which there was a discrepancy between the results obtained by the HCVcAg assay and those obtained by the AMPLICOR HCV test were analyzed by using the in-house RT-PCR assay and RIBA 3 test (Table 2). Serum 76 was an indeterminate sample with only an NS5 band identified by the RIBA 3 test. The results of the HCVcAg assay for three samples (sera 4, 76, and 83) coincided with those of the in-house RT-PCR assay. For serum 69 the results of the AMPLICOR HCV test were the same as those of the in-house RT-PCR. We confirmed the immunoreactivity of serum 4 in the HCVcAg assay using the absorption test by the addition of anti-HCV core monoclonal antibodies, c11-3 and c11-7, to the reaction buffer. The positive signal was diminished by addition of monoclonal antibodies.
TABLE 2.
Resolution of specimens with discordant results obtained by HCV core antigen assay and by AMPLICOR HCV test
| Test | Sample no.
|
|||
|---|---|---|---|---|
| 4 | 69 | 76 | 83 | |
| AMPLICOR HCV test | − | + | + | + |
| AMPLICOR Monitor test (k copies/ml) | <1.0 | <1.0 | <1.0 | <1.0 |
| HCV core antigen (mU/ml) | 1.31 | <0.5 | <0.5 | <0.5 |
| ALT (U/liter) | 27 | 32 | 6 | 28 |
| In-house RT-PCR | + | + | − | − |
| HCV genotype | 1a | N.D.a | − | − |
| RIBA 3 | + | + | +/− | + |
| C100p | 4+ | 1+ | − | +/− |
| C33C | 4+ | 4+ | − | 4+ |
| C22P | 4+ | − | − | 1+ |
| NS5 | +/− | 4+ | 4+ | +/− |
N.D., not determined.
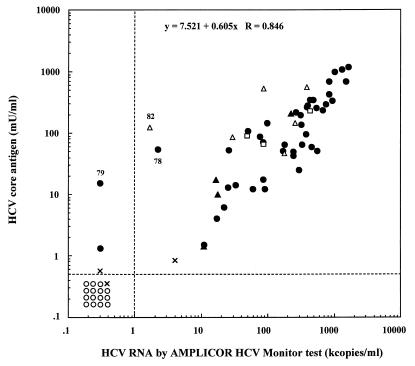
The correlation between the concentrations of HCVcAg and HCV RNA as detected by the AMPLICOR Monitor test is shown in Fig. 4. The coefficient of correlation of the values obtained by the HCVcAg assay with those obtained by the AMPLICOR Monitor test was over 0.8 (P < 0.001), indicating that the results of the two assays were well correlated. There were some samples with a negative or low positive value by the AMPLICOR Monitor test and a highly positive value by the HCVcAg assay (sera 78, 79, and 82). The immunoreaction in these samples was also confirmed to be specific by the absorption test described above. The genotypes of HCV in sera 78, 79, and 82 were 1a, 1b, and 2b, respectively.
FIG. 4.
Correlation between the concentrations of HCV core antigen and HCV RNA as detected by the AMPLICOR Monitor test. ○, negative sample as determined by the in-house RT-PCR; ●, sera with HCV of genotype 1; ▵, sera with HCV of genotype 2; ▴, sera with HCV of genotype 3; ×, sera with HCV of undetermined genotype; and □, sera for which HCV genotype was not tested.
Detection of HCVcAg in the early period of HCV infection.
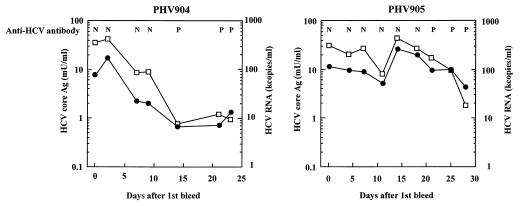
The values of various HCV markers in commercially available seroconversion panel sera, which are a group of serial samples obtained from an individual plasma donor during a period of anti-HCV antibody seroconversion, are shown in Fig. 5. The data for the anti-HCV antibody are provided by the supplier. For the PHV904 panel, anti-HCV antibodies were detected on the 14th day after the first bleed, but HCVcAg and HCV-RNA were detected at the time of the first bleed. The patterns of changes in the concentrations of HCVcAg and HCV-RNA resembled each other. For the PHV905 panel, though anti-HCV antibodies were detected on the 21st day after the first bleed, the HCVcAg assay showed performance equivalent to that of the AMPLICOR Monitor test, as was found with the PHV904 panel. For the PHV907 panel, anti-HCV antibodies, HCVcAg, and HCV RNA were detected on the 21st day, first day, and first day after the first bleed, respectively. For PHV901 and PHV908 panel sera as well, HCVcAg and HCV-RNA could be detected at some point before the anti-HCV antibodies were detected.
FIG. 5.
Patterns of various HCV markers in commercially available seroconversion panels. The data for anti-HCV antibody were obtained by Ortho EIA (3.0) by using the supplier’s data sheet. The concentrations of HCV core antigen (□) and HCV RNA as measured by AMPLICOR Monitor test (●) were measured at the points indicated. N, negative; P, positive.
DISCUSSION
The conventional EIA, which captures the antigen specifically using antibodies, has several merits when compared with nucleic acid detection systems. EIAs generally can detect quantities with lower variance and are low cost. Since additional amplification procedures are not used in this HCVcAg assay system, this system is expected to be resistant to contamination, in contrast to the high prevalence of false-positive results shown by PCR. In addition, we expect that it should not be necessary to take the extra precaution of storing specimens because this assay does detect a stable substance, HCV protein (1a), in contrast to RNA, which was reported to be unstable (6, 7).
HCV sequence variability has been reported, and at least six genotypes of HCV have been classified (30). Several studies have shown that differences in sensitivity among HCV genotypes by the AMPLICOR HCV test and b-DNA exist (8, 25). To minimize the effect of sequence variability on the sensitivity of the assay, we used monoclonal antibodies recognizing amino acid sequences rather well conserved among the six genotypes (2). However, amino acid sequence alignment of the epitope peptide with the published sequences of HCV isolates (GenBank) revealed that several isolates had sequences different from our isolate at several residues. Therefore, we should examine the effects of the sequence heterogeneity on the sensitivity of this EIA using a large pool of samples consisting of sera with various HCV genotypes.
The pretreatment method described in a previous report, which consisted of a six-step process, took about 2 h (11). Our pretreatment method is a single-step process and requires as little as 30 min. Furthermore, it requires less time and labor, and procedures, such as centrifugation, used in the previous study have been eliminated. The reduction of the number of steps should contribute to improving the sensitivity and specificity of EIA for HCVcAg.
It is generally accepted that a high concentration of SDS destroys the structure of the polypeptide and thereby weakens the affinity of the antigen-antibody binding. The first reaction in the HCVcAg EIA, the binding of free HCVcAg to the immobilized monoclonal antibody, is carried out in buffer containing a high concentration of SDS. We speculate that there are several reasons that the anti-HCV core monoclonal antibodies efficiently capture HCVcAg under such a stringent condition. First, we selected monoclonal antibodies that were able to capture recombinant HCV core protein in the presence of SDS. Second, the efficient inactivation of anti-HCV core antibodies in specimens required high concentration of SDS and heat treatment (Fig. 1 and Materials and Methods); in addition, the first reaction was carried out at room temperature and SDS was diluted to the concentration of 2.5%. Third, when the standard serum was diluted with PBS containing 1% BSA instead of normal horse serum, the sensitivity was decreased (data not shown), suggesting that the SDS would be absorbed by the materials in serum and that the amount of free SDS would be reduced before application for the first reaction.
The gel filtration study revealed that the HCVcAg in serum was detected as a high-molecular-weight substance, suggesting that HCVcAg should exist in free virion or virus particles associated with immunoglobulins or low-density lipoproteins (5, 27). Therefore, the pretreated HCVcAg needed to be freed from all associated materials by the destruction of the complex containing the virus particles and the inactivation of the anti-HCV core antibody present in the serum. The molecular mass of HCVcAg estimated based on gel filtration analysis and SDS-polyacrylamide gel electrophoresis analysis (33) suggested that the molecular mass of HCVcAg in serum should be between 20 and 25 kDa.
The detection limit (0.2 mU/ml) corresponded to 0.06 pg of recombinant core antigen/ml. The cutoff value of 0.5 mU/ml, based on the values obtained for the sera from the healthy subjects tested, was approximately equivalent to the value of 500 copies of HCV RNA/ml which was determined by the AMPLICOR Monitor test. The sensitivity of this HCVcAg assay was nearly equal to that of the AMPLICOR HCV test and higher than that of the AMPLICOR Monitor test (Table 1). These data indicate that the present assay is highly sensitive to HCVcAg and exhibits better performance than the previous EIAs for HCVcAg (11, 25, 36).
False-negative results of the PCR method were reported to occur when heparin had been added to the specimen. The inhibitory effects of heparin on the detection of HCV RNA were often noted when the HCV copy number was low (1, 17). In contrast, the performance of the EIA in the present study was not hampered by the addition of blood elements found in serum or anticoagulants, including heparin. This is a point that makes our EIA system superior to PCR.
For this HCVcAg assay, the good results of the recovery test performed after the addition of HCVcAg-positive serum or recombinant core antigen to reference sera indicated that the specificity of the EIA for HCVcAg is high. Regarding HCVcAg released from virions or replicating cells in sera that have been stored under various conditions or are present in the circulation of infected hosts, the good results of the recovery test after the addition of recombinant core antigen suggest that the released HCVcAg would be detected as sensitively as the HCVcAg in virions.
The confirmed correlation between the concentration of core antigen detected by the HCVcAg assay and the amount of HCV RNA detected by the AMPLICOR Monitor test supports the previous finding that the HCV core antigenic activity is internally associated with HCV RNA, in the form of a nucleocapsid (34). It was shown that the sensitivity of AMPLICOR Monitor test to the HCV genotypes 2 and 3 was lower than that to genotype 1 (8). In this study, the frequency of the HCV genotypes 2 and 3 combined in test sera was only 17.2% (10 of 58 sera). The good correlation of the results of these two tests reflects bias as a result of the distribution of HCV genotypes in the sera in this study. There were some samples that had negative or low positive values in the AMPLICOR Monitor test and strongly positive values in the HCVcAg assay (sera 78, 79, and 82). These samples with discrepant results were confirmed to have specific immunoreactivity by the absorption test. We speculate that these discrepancies may result from the differences in reactivity among genotypes when tested by the AMPLICOR HCV test (8, 25), the spread of data obtained by the PCR method as a standard (18), the stability of specimens during storage (6, 7), or possible inclusion of HCVcAg without genomic RNA, as noninfectious particles.
In this study, the EIA for HCVcAg could detect HCV viremia at an earlier point than the anti-HCV antibody test when applied to several commercially available seroconversion panels (Fig. 5). The amounts of HCVcAg and HCV RNA in plasma also varied with nearly the same pattern. Recently, Kobayashi et al. reported that the measurement of HCVcAg was useful for the early diagnosis of acute hepatitis C (12). Currently, it is reported that the risk of infectious HCV-positive donor blood entering the blood supply is on the order of 9.70 per million donations, which is approximately five times greater than that for blood containing human immunodeficiency virus or human T-cell lymphotropic virus; if the HCV RNA test is introduced into the screening test, the risk will be reduced to 2.72 per million donations (29). The present EIA system has better performance than the assay used by Kobayashi et al. (12), and the sensitivity is nearly equal to that of the AMPLICOR HCV test; therefore, we think that this HCVcAg assay will contribute to the early diagnosis of hepatitis C and reduction of the risk of contracting HCV infection from donated blood. To confirm these points, we will measure the HCVcAg values in many clinical seroconversion specimens.
By the development of a simple procedure for specimen pretreatment and a sensitive EIA for HCVcAg, a detection system for HCVcAg with sensitivity equivalent to that of the AMPLICOR HCV test was established. A study of mass screening using this EIA to reduce the risk of secondary infection from blood components is needed, and the determination of the precise amount of HCV in serum will be very useful for the clinical observation of hepatitis C patients.
ACKNOWLEDGMENTS
We express our gratitude to Kenjiro Yamaguchi and Yuka Matsunaga for their technical assistance with the in-house RT-PCR assay.
REFERENCES
- 1a.Aoyagi, K., C. Ohue, K. Iida, and S. Yagi. Unpublished data.
- 1.Beutler E, Gelbart T, Kuhl W. Interference of heparin with the polymerase chain reaction. BioTechniques. 1990;9:166. [PubMed] [Google Scholar]
- 2.Bukh J, Purcell R H, Miller R H. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc Natl Acad Sci USA. 1994;91:8239–8243. doi: 10.1073/pnas.91.17.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busch M P, Korelitz J J, Kleinman S H, Lee S R, AuBuchon J P, Schreiber G B. Declining value of alanine aminotransferase in screening of blood donors to prevent posttransfusion hepatitis B and C virus infection. The Retrovirus Epidemiology Donor Study. Transfusion. 1995;35:903–910. doi: 10.1046/j.1537-2995.1995.351196110893.x. [DOI] [PubMed] [Google Scholar]
- 4.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 5.Choo S H, So H S, Cho J M, Ryu W S. Association of hepatitis C virus particles with immunoglobulin: a mechanism for persistent infection. J Gen Virol. 1995;76:2337–2341. doi: 10.1099/0022-1317-76-9-2337. [DOI] [PubMed] [Google Scholar]
- 6.Davis G L, Lau J Y, Urdea M S, Neuwald P D, Wilber J C, Lindsay K, Perrillo R P, Albrecht J. Quantitative detection of hepatitis C virus RNA with a solid-phase signal amplification method: definition of optimal conditions for specimen collection and clinical application in interferon-treated patients. Hepatology. 1994;19:1337–1341. [PubMed] [Google Scholar]
- 7.Halfon P, Khiri H, Gerolami V, Bourliere M, Feryn J M, Reynier P, Gauthier A, Cartouzou G. Impact of various handling and storage conditions on quantitative detection of hepatitis C virus RNA. J Hepatol. 1996;25:307–311. doi: 10.1016/s0168-8278(96)80116-4. [DOI] [PubMed] [Google Scholar]
- 8.Hawkins A, Davidson F, Simmonds P. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by Quantiplex HCV RNA assay versions 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J Clin Microbiol. 1997;35:187–192. doi: 10.1128/jcm.35.1.187-192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houghton M, Weiner A, Han J, Kuo G, Choo Q-L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991;14:381–388. [PubMed] [Google Scholar]
- 11.Kashiwakuma T, Hasegawa A, Kajita T, Takata A, Mori H, Ohta Y, Tanaka E, Kiyosawa K, Tanaka T, Tanaka S, Hattori N, Kohara M. Detection of hepatitis C virus specific core protein in serum of patients by a sensitive fluorescence enzyme immunoassay (FEIA) J Immunol Methods. 1996;190:79–89. doi: 10.1016/0022-1759(95)00261-8. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi M, Tanaka E, Matsumoto A, Yoshizawa K, Imai H, Sodeyama T, Kiyosawa K. Clinical application of hepatitis C virus core protein in early diagnosis of acute hepatitis C. J Gastroenterol. 1998;33:508–511. doi: 10.1007/s005350050123. [DOI] [PubMed] [Google Scholar]
- 13.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 14.Kuo G, Choo Q-L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, Tegtmeier G E, Bonino F, Colombo M, Lee W-S, Kuo C, Berger K, Shuster J R, Overby L R, Bradley D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 15.Lau J Y, Davis G L, Kniffen J, Qian K P, Urdea M S, Chan C S, Mizokami M, Neuwald P D, Wilber J C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993;341:1501–1504. doi: 10.1016/0140-6736(93)90635-t. . (Erratum, 342:504, 1993.) [DOI] [PubMed] [Google Scholar]
- 16.Lelie P N, Cuypers H T, Reesink H W, van der Poel C L, Winkel I, Bakker E, van Exel-Oehlers P J, Vallari D, Allain J P, Mimms L. Patterns of serological markers in transfusion-transmitted hepatitis C virus infection using second-generation HCV assays. J Med Virol. 1992;37:203–209. doi: 10.1002/jmv.1890370310. [DOI] [PubMed] [Google Scholar]
- 17.Masukawa A, Miyachi H, Ohshima T, Fusegawa H, Ando Y. Monitoring of inhibitors of the polymerase chain reaction for the detection of hepatitis C virus using the positive internal control. Jpn J Clin Pathol. 1997;45:673–678. [PubMed] [Google Scholar]
- 18.Miskovsky E P, Carrella A V, Gutekunst K, Sun C A, Quinn T C, Thomas D L. Clinical characterization of a competitive PCR assay for quantitative testing of hepatitis C virus. J Clin Microbiol. 1996;34:1975–1979. doi: 10.1128/jcm.34.8.1975-1979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamura T, Saito I, Katayama T, Kikuchi S, Tateda A, Houghton M, Choo Q L, Kuo G. Detection of antibody against antigen expressed by molecularly cloned hepatitis C virus cDNA: application to diagnosis and blood screening for posttransfusion hepatitis. Proc Natl Acad Sci USA. 1990;87:983–987. doi: 10.1073/pnas.87.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriya T, Sasaki F, Tanaka J, Mizui M-A, Nakanishi T, Takahashi K, Yoshizawa H. Comparison of HCV core antigen activity by ELISA and amount of HCV RNA by branched DNA assay. Int Hepatol Commun. 1994;2:175–177. [Google Scholar]
- 21.Nagayama R, Miyake K, Takikawa H, Yamanaka M, Tadokoro K, Takahashi Y. A case of posttransfusion hepatitis C caused by a blood donated before an appearance of HCV antibody in early period of hepatitis C virus infection. Acta Hepatol Jpn. 1998;17:73–76. [Google Scholar]
- 22.Nolte F S, Thurmond C, Fried M W. Preclinical evaluation of AMPLICOR hepatitis C virus test for detection of hepatitis C virus RNA. J Clin Microbiol. 1995;33:1775–1778. doi: 10.1128/jcm.33.7.1775-1778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y, Mayumi M. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto H, Tokita H, Sakamoto M, Horikita M, Kojima M, Iizuka H, Mishiro S. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J Gen Virol. 1993;74:2385–2390. doi: 10.1099/0022-1317-74-11-2385. [DOI] [PubMed] [Google Scholar]
- 25.Orito E, Mizokami M, Tanaka T, Lau J Y, Suzuki K, Yamauchi M, Ohta Y, Hasegawa A, Tanaka S, Kohara M. Quantification of serum hepatitis C virus core protein level in patients chronically infected with different hepatitis C virus genotypes. Gut. 1996;39:876–880. doi: 10.1136/gut.39.6.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poljak M, Seme K, Koren S. Evaluation of the automated COBAS AMPLICOR hepatitis C virus PCR system. J Clin Microbiol. 1997;35:2983–2984. doi: 10.1128/jcm.35.11.2983-2984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince A M, Huima-Byron T, Parker T S, Levine D M. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J Viral Hepat. 1996;3:11–17. doi: 10.1111/j.1365-2893.1996.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 28.Saito M, Hasegawa A, Kashiwakuma T, Kohara M, Sugi M, Miki K, Yamamoto T, Mori H, Ohta Y, Tanaka E, Kiyosawa K, Furuta S, Wakashima M, Tanaka S, Hattori N. Performance of an enzyme-linked immunosorbent assay system for antibodies to hepatitis C virus with two new antigens (c11/c7) Clin Chem. 1992;38:2434–2439. [PubMed] [Google Scholar]
- 29.Schreiber G B, Busch M P, Kleinman S H, Korelitz J J. The risk of transfusion-transmitted viral infections. The Retrovirus Epidemiology Donor Study. N Engl J Med. 1996;334:1685–1690. doi: 10.1056/NEJM199606273342601. [DOI] [PubMed] [Google Scholar]
- 30.Simmonds P, Holmes E C, Cha T A, Chan S-W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 31.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Heuverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Tanaka E, Matsumoto A, Urushihara A, Sodeyama T. Usefulness of simple assays for serum concentration of hepatitis C virus RNA and HCV genotype in predicting the response of patients with chronic hepatitis C to interferon alpha 2a therapy. J Med Virol. 1995;46:162–168. doi: 10.1002/jmv.1890460215. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi K, Kishimoto S, Yoshizawa H, Okamoto H, Yoshikawa A, Mishiro S. p26 protein and 33-nm particle associated with nucleocapsid of hepatitis C virus recovered from the circulation of infected hosts. Virology. 1992;191:431–434. doi: 10.1016/0042-6822(92)90204-3. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi K, Okamoto H, Kishimoto S, Munekata E, Tachibana K, Akahane Y, Yoshizawa H, Mishiro S. Demonstration of a hepatitis C virus-specific antigen predicted from the putative core gene in the circulation of infected hosts. J Gen Virol. 1992;73:667–672. doi: 10.1099/0022-1317-73-3-667. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka E, Kiyosawa K, Matsumoto A, Kashiwakuma T, Hasegawa A, Mori H, Yanagihara O, Ohta Y. Serum levels of hepatitis C virus core protein in patients with chronic hepatitis C treated with interferon alfa. Hepatology. 1996;23:1330–1333. doi: 10.1053/jhep.1996.v23.pm0008675147. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka T, Lau J Y, Mizokami M, Orito E, Tanaka E, Kiyosawa K, Yasui K, Ohta Y, Hasegawa A, Tanaka S, Kohara M. Simple fluorescent enzyme immunoassay for detection and quantification of hepatitis C viremia. J Hepatol. 1995;23:742–745. doi: 10.1016/0168-8278(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 37.Urdea M S. Synthesis and characterization of branched DNA (bDNA) for the direct and quantitative detection of CMV, HBV, HCV, and HIV. Clin Chem. 1993;39:725–726. [Google Scholar]
- 38.Vrielink H, van der Poel C L, Reesink H W, Zaaijer H L, Lelie P N. Transmission of hepatitis C virus by anti-HCV-negative blood transfusion. Vox Sang. 1995;68:55–56. doi: 10.1111/j.1423-0410.1995.tb02546.x. [DOI] [PubMed] [Google Scholar]
- 39.Yoshitake S, Imagawa M, Ishikawa E, Niitsu Y, Urushizaki I, Nishiura M, Kanazawa R, Kurosaki H, Tachibana S, Nakazawa N, Ogawa H. Mild and efficient conjugation of rabbit Fab′ and horseradish peroxidase using a maleimide compound and its use for enzyme immunoassay. J Biochem. 1982;92:1413–1424. doi: 10.1093/oxfordjournals.jbchem.a134065. [DOI] [PubMed] [Google Scholar]
- 40.Young K K, Resnick R M, Myers T W. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeuzem S, Ruster B, Roth W K. Clinical evaluation of a new polymerase chain reaction assay (Amplicor HCV) for detection of hepatitis C virus. Z Gastroenterol. 1994;32:342–347. [PubMed] [Google Scholar]