Abstract
We present a genome assembly from an individual male Rattus norvegicus (the Norway rat; Chordata; Mammalia; Rodentia; Muridae). The genome sequence is 2.44 gigabases in span. The majority of the assembly is scaffolded into 20 chromosomal pseudomolecules, with both X and Y sex chromosomes assembled. This genome assembly, mRatBN7.2, represents the new reference genome for R. norvegicus and has been adopted by the Genome Reference Consortium.
Keywords: Rattus norvegicus, Norway rat, genome sequence, chromosomal, reference genome
Species taxonomy
Eukaryota; Metazoa; Chordata; Mammalia; Rodentia; Muridae; Rattus; Rattus norvegicus Berkenhout 1769 (NCBI:txid10116).
Introduction
Rattus norvegicus is one of the most well-established experimental model organisms, with use of the species dating back to the mid-19th century ( Modlinska & Pisula, 2020). The longstanding use of R. norvegicus in the laboratory as a model organism has led to a multitude of discoveries, providing insight into human physiology, behaviour and disease. The complexity of R. norvegicus relative to many other model organisms, in addition to its well-characterised physiology, means that it is frequently used in cancer research, behavioral neuroscience, and the pharmaceutical industry.
We present the reference genome mRatBN7.2 for the Norway rat, Rattus norvegicus. This genome assembly represents a substantial improvement on the previous assemblies, correcting areas of potential mis-assembly in the 2014 reference assembly, Rnor_6.0 ( Ramdas et al., 2019). The new reference has a mean genome coverage of ~92x for a single male individual of the BN/NHsdMcwi strain, which was obtained from the same colony as the original “Eve” rat that was sampled 18 years ago for use in previous rat reference genome assemblies (Eve was a female rat of generation F14, the index male described here is generation F61). The new assembly contains no gaps between scaffolds and has a scaffold N50 an order of magnitude higher than the previous reference assembly; with just 756 contigs (N50 >29 Mb), its contiguity is comparable to that of reference assemblies for humans and mice.
The production of a high-quality reference genome assembly for R. norvegicus allows researchers using rats for research, as a model organism for human diseases, and for determining drug interactions to have as complete and reliable a genome as possible. The result is a greater depth and certainty in data interpretation and species comparison, which will have numerous benefits for biological understanding and health.
Genome sequence report
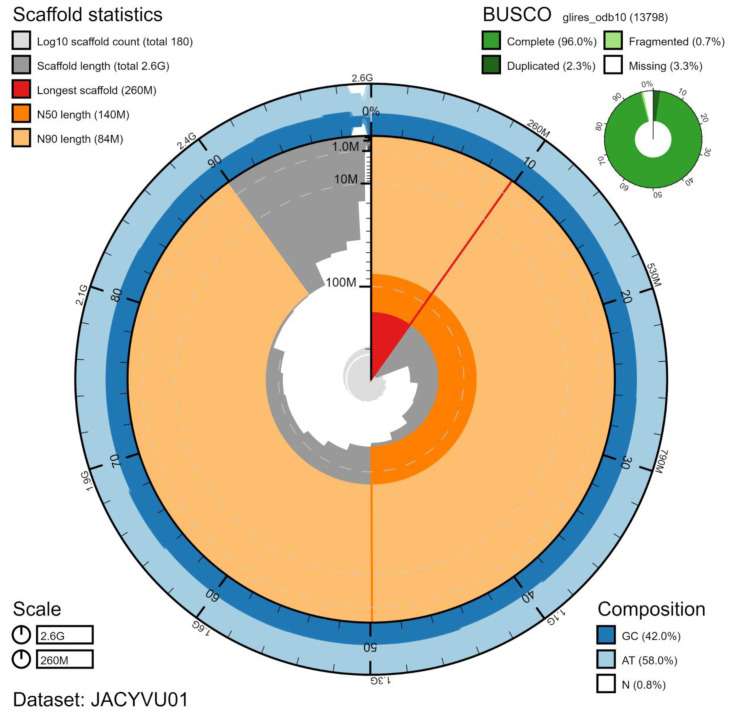
The genome was sequenced from the kidney tissue of a single male R. norvegicus (strain BN/NHsdMcwi, generation F61) housed at the Medical College of Wisconsin, Milwaukee, Wisconsin, USA. A total of 80-fold coverage in Pacific Biosciences single-molecule long reads (N50, 37 kb) and 31-fold coverage in 10X Genomics read clouds (from molecules with an estimated N50 of 26 kb) were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data (29-fold coverage). Manual assembly curation corrected 234 missing/misjoins and removed 34 haplotypic duplications, reducing the scaffold number by 4.8%, increasing the scaffold N50 by 0.04% and decreasing the assembly length by 0.9%. The final assembly has a total length of 2.65 Gb in 219 sequence scaffolds with a scaffold N50 of 135.0 Mb ( Table 1). The majority, 99.7%, of the assembly sequence was assigned to 20 chromosomal-level scaffolds representing 20 autosomes and the X and Y sex chromosomes ( Figure 1– Figure 4; Table 2). The assembly has a BUSCO ( Simão et al., 2015) completeness of 96.2% using the mammalia_odb10 reference set. The primary assembly is a large-scale mosaic of both haplotypes (i.e. is not fully phased) and we have therefore also deposited the contigs corresponding to the alternate haplotype.
Figure 1. Genome assembly of Rattus norvegicus, mRatBN7.2: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/Rattus%20norvegicus/dataset/JACYVU01/snail.
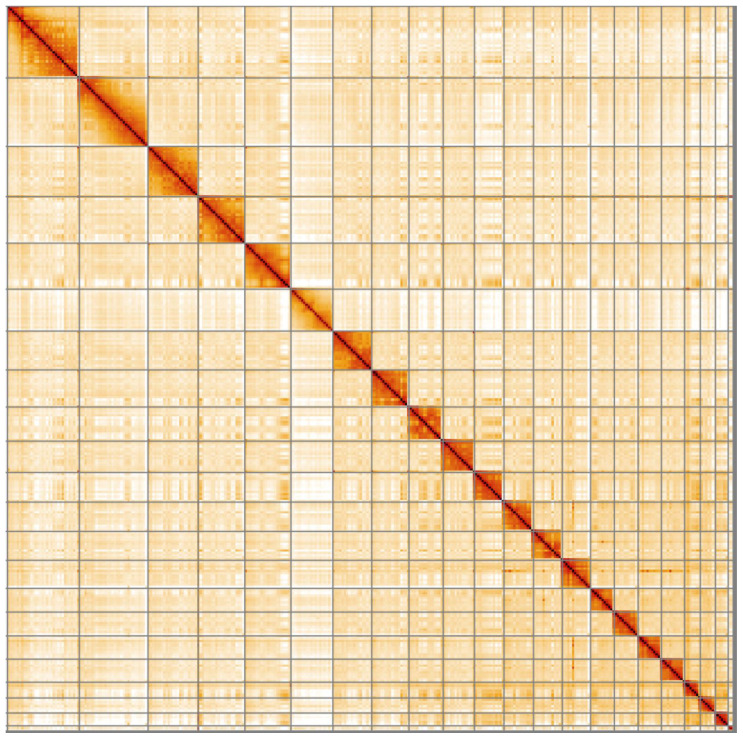
Figure 4. Genome assembly of Rattus noevegicus, mRatBN7.2: Hi-C contact map.
Hi-C contact map of the mRatBN7.2 assembly, visualised in HiGlass.
Table 1. Genome data for R. norvegicus.
| Project accession data | |
|---|---|
| Assembly identifier | mRatBN7.2 |
| Species | Rattus norvegicus |
| Specimen | mRatNor1 |
| NCBI taxonomy ID | 10116 |
| BioProject | PRJNA662962 |
| BioSample ID | SAMN16261960,
SAMEA5928170 |
| Isolate information | Laboratory animal, male,
kidney tissue |
| Raw data accessions | |
| PacificBiosciences SEQUEL II | ERR5310326-ERR5310327 |
| 10X Genomics Illumina | ERR5309015-ERR5309022 |
| Hi-C Illumina | ERR5309023, ERR5309024 |
| BioNano | ERZ1741012 |
| Genome assembly | |
| Assembly accession | GCA_015227675.2 |
| Accession of alternate haplotype | GCA_015244455.1 |
| Span (Mb) | 2,648 |
| Number of contigs | 738 |
| Contig N50 length (Mb) | 34 |
| Number of scaffolds | 219 |
| Scaffold N50 length (Mb) | 135 |
| Longest scaffold (Mb) | 260 |
| BUSCO * genome score | C:96.2%[S:94.0,D:2.2%],F:0.
9%,M:2.8%,n:9226 |
*BUSCO scores based on the mammalia_odb10 BUSCO set using v5.0.0. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/Rattus%20norvegicus/dataset/JACYVU01/busco.
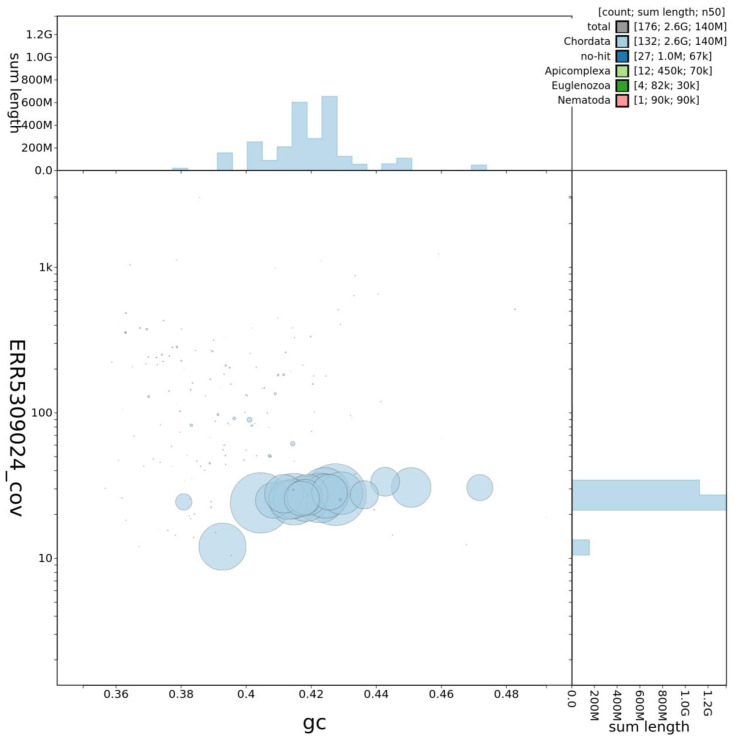
Figure 2. Genome assembly of Rattus norvegicus, mRatBN7.2: GC coverage.
BlobToolKit GC-coverage plot. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/Rattus%20norvegicus/dataset/JACYVU01/blob?plotShape=circle.
Table 2. Chromosomal pseudomolecules in the primary genome assembly of Rattus norvegicus mRatBN7.2.
| Accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| CM026974.1 | 1 | 260.52 | 42.8 |
| CM026975.1 | 2 | 249.05 | 40.5 |
| CM026976.1 | 3 | 169.03 | 42.5 |
| CM026977.1 | 4 | 182.69 | 41.6 |
| CM026978.1 | 5 | 166.88 | 42.3 |
| CM026979.1 | 6 | 140.99 | 41.9 |
| CM026980.1 | 7 | 135.01 | 42.4 |
| CM026981.1 | 8 | 123.90 | 43 |
| CM026982.1 | 9 | 114.18 | 41.9 |
| CM026983.1 | 10 | 107.21 | 45.1 |
| CM026984.1 | 11 | 86.24 | 40.8 |
| CM026985.1 | 12 | 46.67 | 47.2 |
| CM026986.1 | 13 | 106.81 | 41.5 |
| CM026987.1 | 14 | 104.89 | 41.3 |
| CM026988.1 | 15 | 101.77 | 41.2 |
| CM026989.1 | 16 | 84.73 | 41.8 |
| CM026990.1 | 17 | 86.53 | 42.6 |
| CM026997.1 | 18 | 83.83 | 41.7 |
| CM026992.1 | 19 | 57.34 | 44.3 |
| CM026993.1 | 20 | 54.44 | 43.7 |
| CM026994.1 | X | 152.45 | 39.5 |
| CM026995.1 | Y | 18.32 | 42.2 |
Methods
The Norway rat specimen (strain BN/NHsdMcwi, generation F61) was a male individual housed in a standard rodent microisolator cage at the Medical College of Wisconsin, Milwaukee, Wisconsin, USA. The animal was euthanised by CO 2 inhalation. This procedure was approved by the Medical College of Wisconsin Institutional Animal Care and Use Committee.
DNA was extracted using an agarose plug extraction from kidney tissue following the Bionano Prep Animal Tissue DNA Isolation Soft Tissue Protocol. Pacific Biosciences CLR long read and 10X Genomics read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Hi-C data were generated using the Arima v2 Hi-C kit. Sequencing was performed by the Scientific Operations DNA Pipelines at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL II and Illumina HiSeq X instruments. DNA was labeled for Bionano Genomics optical mapping following the Bionano Prep Direct Label and Stain (DLS) Protocol and run on one Saphyr instrument chip flowcell.
Assembly was carried out following the Vertebrate Genome Project pipeline v1.6 ( Rhie et al., 2020) with Falcon-unzip ( Chin et al., 2016), haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020) and a first round of scaffolding carried out with 10X Genomics read clouds using scaff10x (see Table 3 for software versions and sources). Hybrid scaffolding was performed using the BioNano DLE-1 data and BioNano Solve. Scaffolding with Hi-C data ( Rao et al., 2014) was carried out with SALSA2 ( Ghurye et al., 2019). The Hi-C scaffolded assembly was polished with arrow using the PacBio data, then polished with the 10X Genomics Illumina data by aligning to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012) and applying homozygous non-reference edits using bcftools consensus. Two rounds of the Illumina polishing were applied. The assembly was checked for contamination and analysed using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation was performed using gEVAL, Bionano Access, HiGlass and Pretext. In addition, we used 10X longranger and genetic mapping data provided by LS, RWW, HC and AK to identify and resolve regions of concern. Figure 1– Figure 3 were generated using BlobToolKit ( Challis et al., 2020).
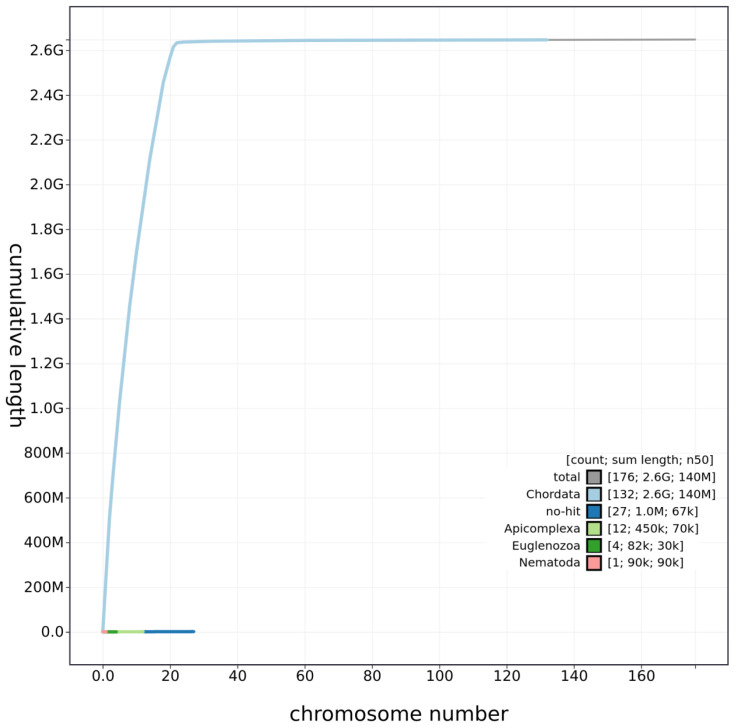
Figure 3. Genome assembly of Rattus norvegicus, mRatBN7.2: cumulative sequence.
BlobToolKit cumulative sequence plot. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/Rattus%20norvegicus/dataset/JACYVU01/cumulative.
Table 3. Software tools used.
| Software tool | Version | Source |
|---|---|---|
| Falcon-unzip | falcon-kit 1.8.0 | ( Chin et al., 2016) |
| purge_dups | 1.0.0 | ( Guan et al., 2020) |
| Bionano Solve | Solve3.4.1_09262019 | https://bionanogenomics.com/downloads/bionano-solve/ |
| SALSA2 | 2.1 | ( Ghurye et al., 2019) |
| scaff10x | 4.2 | https://github.com/wtsi-hpag/Scaff10X |
| arrow | GCpp-1.9.0 | https://github.com/PacificBiosciences/GenomicConsensus |
| longranger align | longranger align (2.2.2) |
https://support.10xgenomics.com/genome-exome/software/
pipelines/latest/advanced/other-pipelines |
| freebayes | v1.3.1-17-gaa2ace8 | ( Garrison & Marth, 2012) |
| bcftools consensus | 1.11-88-g71d744f8 | http://samtools.github.io/bcftools/bcftools.html |
| HiGlass | 1.11.6 | ( Kerpedjiev et al., 2018) |
| PretextView | 0.0.4 | https://github.com/wtsi-hpag/PretextView |
| gEVAL | N/A | ( Chow et al., 2016) |
| BlobToolKit | 1.2 | ( Challis et al., 2020) |
The mitochondrial genome was assembled as part of assembly mRatBN7.1, but was replaced with the pre-existing mitochondrial assembly MT AY172581.1, which is identical. This replacement occurred as annotation already existed for the pre-existing assembly. As such, the primary assembly is now mRatBN7.2.
Data availability
Underlying data
NCBI BioProject: Rattus norvegicus (Norway rat) genome assembly, mRatBN7, Accession number PRJNA662962: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA662962/
NCBI Assembly: mRatBN7.2 primary assembly, Accession number GCA_015227675.2: https://www.ncbi.nlm.nih.gov/assembly/GCF_015227675.2
NCBI Assembly: mRatBN7.1 alternate haplotype, Accession number GCA_015244455.1: https://www.ncbi.nlm.nih.gov/assembly/GCA_015244455.1
The genome sequence is released openly for reuse. The R. norvegicus genome sequencing initiative is part of the Darwin Tree of Life (DToL) project and the Vertebrate Genome Project (VGP) ordinal references programme. All raw data and the assemblies have been deposited in INSDC databases under BioProject PRJNA662962. Raw data and assembly accession identifiers are reported in Table 1.
Funding Statement
This work was supported by the Wellcome through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328). Maintenance of the BN/NHsdMcwi colony is supported by funding from the National Institutes of Health (NIH grants R24OD024617, DA044223) and the UTHSC Center for Integrative and Translational Genomics. SAM is supported by Wellcome (207492). Genetic marker data are available from the Rat Genome Database (NIH grant R01HL064541).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
References
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, et al. : Phased Diploid Genome Assembly with Single-Molecule Real-Time Sequencing. Nat Methods. 2016;13(12):1050–54. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL - a Web-Based Browser for Evaluating Genome Assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing.arXiv:1207.3907.2012. Reference Source [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. Gigascience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlinska K, Pisula W: The Norway Rat, from an Obnoxious Pest to a Laboratory Pet. eLife. 2020;9:e50651. 10.7554/eLife.50651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas S, Ozel AB, Treutelaar MK, et al. : Extended Regions of Suspected Mis-Assembly in the Rat Reference Genome. Sci Data. 2019;6(1):39. 10.1038/s41597-019-0041-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, et al. : Towards Complete and Error-Free Genome Assemblies of All Vertebrate Species. bioRxiv. 2020; 2020.05.22.110833. 10.1101/2020.05.22.110833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics. 2015;31(19):3210–12. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]