Abstract
Vibrio parahaemolyticus is one of the most important food-borne pathogens in Taiwan, Japan, and other countries with long coastlines. This paper reports on the development of a new random amplified polymorphic DNA (RAPD) method for the molecular typing of this pathogen. The 10-mer primer 284 (5′-CAG GCG CAC A-3′) was selected to generate polymorphic amplification profiles of the genomic DNA at an annealing temperature of 38°C. A total of 308 clinical isolates of V. parahaemolyticus collected during food poisoning outbreaks in Taiwan, mostly occurring between 1993 and 1995, plus 11 environmental and clinical reference strains were analyzed by this RAPD method. A total of 41 polymorphic RAPD patterns were recognized, and these patterns were arbitrarily grouped into 16 types (A to P). Types A, B, C, D, and E were the major types, and subtypes C3, C5, E1, B1, D2, and A2 were the major patterns. The major types were phylogenetically more closely related to each other than to any of the minor types.
Vibrio parahaemolyticus is a halophilic gram-negative bacterium that causes acute gastroenteritis in humans. It is one of the most important food-borne pathogens in Taiwan, Japan, and other countries with long coastlines (1). Isolates of V. parahaemolyticus can be differentiated by serotyping. Commercial serotyping antisera are available in Japan and other countries (e.g., from Denka Seiken, Tokyo, Japan). There are 13 O groups and 71 K types identified by these commercial antisera. Usually the serotyping method cannot differentiate all isolates which originate from different regions or sources. Dependable molecular methods for the typing of strains would greatly aid epidemiological investigations. However, molecular typing methods for the subspecies differentiation of V. parahaemolyticus have not been well developed. Recently, we described the pulsed-field gel electrophoresis (PFGE) method for the subspecies typing of this pathogen (10). This paper reports on the development of another molecular method, random amplified polymorphic DNA (RAPD), for the typing of V. parahaemolyticus. A total of 308 clinical isolates obtained during food poisoning outbreaks, mostly occurring from 1993 to 1995 in Taiwan, and several environmental and clinical reference strains were characterized by this procedure.
MATERIALS AND METHODS
Bacterial strains.
A total of 308 clinical isolates selected from the stool samples of patients involved in food poisoning outbreaks which occurred mostly from 1993 to 1995 in Taiwan were examined in this study. These isolates were identified by API (Montalieu-Vercieu, France) 20E identification strips and also by conventional methods, which included swarming, luminescence, and 29 other biochemical assays (7). Four environmental and seven clinical reference strains from Japan, the United States, and local sources were also examined (Table 1). These cultures were stored at −85°C in tryptic soy broth (Difco, Detroit, Mich.)–3% NaCl containing 20% glycerol. Serotyping of these clinical isolates was performed with commercial K-type antisera (Denka Seiken) following the procedure provided by the supplier.
TABLE 1.
V. parahaemolyticus cultures examined by RAPD in this study
| Category | No. | Designationa | Origin | Remarks |
|---|---|---|---|---|
| Clinical isolate | 308 | Taiwan | 295 collected 1993–1995 | |
| Reference environmental strain | 4 | Laboratory stock 109 | Taiwan | |
| Laboratory stock 226 | ||||
| CCRC12963 | ||||
| CCRC12958 | ||||
| Reference clinical strain | 7 | CCRC10806 | Japan | Type strain ATCC 17802 |
| CCRC12864 | United States | |||
| CCRC12865 | United States | |||
| CCRC13025 | Taiwan | |||
| CCRC12863 | Japan | |||
| CCRC13027 | Taiwan | |||
| ST550 | Japan | Reference 9 |
CCRC, Culture Collection and Research Center, Hsin-chu, Taiwan, Republic of China; ST550 was furnished by T. Arai, Showa College of Pharmaceutical Sciences, Japan.
Preparation of genomic DNA.
The isolate was cultured in 5 ml of tryptic soy broth–3% NaCl medium and incubated in a rotary shaking incubator at 37°C and 160 rpm for 16 h. Bacterial cells were collected by centrifugation. Genomic DNA from these isolates was prepared by following the small-scale preparation method of Sambrook et al. (5), suspended in TE buffer (10 mM Tris hydrochloride buffer, 1 mM EDTA, pH 7.5), and stored at −20°C until required.
DNA amplification.
For the development of the RAPD method for V. parahaemolyticus, the 100 different 10-mer primers (UBC RAPD primer set 100/3) (Finnzymes Oy, Espoo, Finland) were screened for the amplification of template DNA from reference strain ST550 and domestic clinical isolate DOH548. Since PCR in the RAPD method is affected by the experimental parameters (6), all of the screening steps in this study were performed under the conditions described below, except that the annealing temperature was 36°C.
DNA amplifications for the screening of primers or final typing were carried out in buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris HCl, pH 8.8) containing 200 μM (each) dATP, dCTP, dGTP, and dTTP; 0.25 μM primer; 2.0 U of DyNAZyme II thermostable DNA polymerase (Finnzymes); and 100 ng of template DNA in a final volume of 100 μl. Primer 284 (5′-CAG GCG CAC A-3′) was selected for final RAPD typing. Amplification was performed in a thermal cycler (Personal Cycler 20; Biometra biomedizinische analytik Gmbh, Gottingen, Germany). All manipulations were carried out with dedicated DNA-free pipettes in a sterile field to minimize the risk of contamination. The reaction mixture was overlaid with 50 μl of sterile mineral oil and incubated in the thermal cycler at 95°C for 5 min. Then, thermostable DNA polymerase was added and amplification was carried out for 45 cycles, each of which went as follows: 94°C for 1 min, 38°C for 1 min, 72°C for 2 min, and finally, an additional 72°C for 10 min.
Gel electrophoresis.
The amplified products were electrophoresed at 75 V in a horizontal 10- by 15-cm 1.5% agarose gel in Tris-borate buffer for about 4.5 h. The amplified DNA bands were visualized after ethidium bromide staining and photographed under UV light. A 100-bp ladder (Pharmacia Biotech, Uppsala, Sweden) was used as a marker in determining the sizes of the amplification products.
Similarities among patterns.
The size of each band was determined by Stratascan 7000 densitometry with one-dimensional analysis software (Stratagene, La Jolla, Calif.). Data were coded as 0 (negative) or 1 (positive). Following the method described by Martin-Kearley et al. (3), hierarchical cluster analysis was done by the average linkage method with the squared Euclidean distance measure. The dendrogram was produced with the SPSS for Windows release 6.0 program (SPSS Inc., Chicago, Ill.) (3).
RESULTS AND DISCUSSION
Recently, a PFGE method was developed for epidemiological examination of V. parahaemolyticus by molecular means. Genomic DNA was digested with SfiI and resolved on 1% agarose by a contour-clamped homogeneous electric field apparatus. A total of 130 selected isolates were grouped into 14 PFGE types, which consisted of 1 to 6 patterns, and a total of 39 patterns were identified. Most of these domestic clinical isolates could be clustered into several major groups (A, B, C, and G) (10). PFGE is a reliable method with high discrimination efficiency. However, the whole process takes several days to complete. In comparison, RAPD has the advantages of being less labor-intensive and easier to standardize between laboratories. It has been proven that short primers of arbitrary nucleotide sequences can be used to amplify segments of genomic DNA reproducibly from a wide variety of species. Polymorphisms among the amplification products are detected frequently and are useful as genetic markers (8).
The concentration of MgCl2 used in the reaction buffer was 1.5 mM, and this concentration enhanced the specificity of the PCR and produced informative arrays in this study (2). In the first round of screening, amplification by 19 of the primers resulted in several clear DNA bands in both strains (data not shown). Eighteen of these primers, all of them from 50 to 90% G+C in composition and containing no palindromic sequences (8), were chosen for further verification and selection. Amplification of 20 other domestic clinical isolates by using these 18 selected primers individually was examined. Four primers, namely, 241, 243, 284, and 290, with 90, 70, 70, and 90% G+C composition, respectively, were selected for final-round examination. Furthermore, the effect of different annealing temperatures (36 or 38°C) on the amplification results of these primers was also tested for four domestic clinical isolates (DOH155, DOH355, DOH548, and DOH584) and then for another eight strains (ST550, CCRC12958, DOH304, DOH616, DOH646, DOH650, DOH665, and DOH687). The amplifications of these strains with these four primers were repeated three times. Judging from the number of clearly discernible amplified DNA bands and the reproducibility, primer 284 (5′-CAG GCG CAC A-3′) was selected for the typing of the rest of the isolates by the RAPD method at 38°C annealing temperature (data not shown).
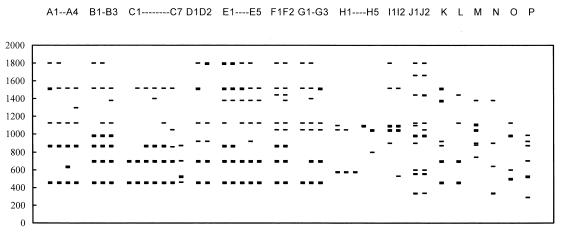
A total of 308 domestic isolates of V. parahaemolyticus and 11 reference strains were analyzed by this RAPD method. One to 10 amplified DNA bands were resolved by agarose gel electrophoresis in these isolates. Among the amplified bands, 460-, 700-, 870-, 1,130-, 1,560-, and 1,800-bp bands were relatively conserved in most of the isolates (Fig. 1 and 2). A total of 41 different RAPD patterns were discerned. After hierarchical cluster analysis, those polymorphic patterns with dissimilarity values of less than 7 were arbitrarily grouped into 16 types (A to P), while type J consisted of patterns with a dissimilarity of about 11 (Fig. 2 and 3). Types A, B, C, D, and E are the major groups, comprising 7.79, 9.74, 45.81, 6.17, and 21.75% of the isolates, respectively. The rest of the RAPD types (F to P) together made up only 8.75% of the isolates. A2, B1, C3, C5, D2, and E1 are the major patterns, containing 4.22, 7.79, 20.45, 21.75, 5.84, and 14.93% of the isolates, respectively (Table 2).
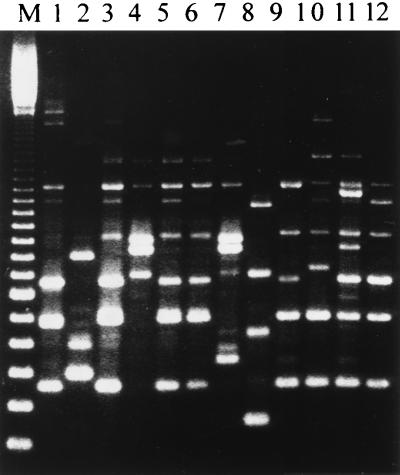
FIG. 1.
Amplified DNA polymorphisms of V. parahaemolyticus isolates with primer 284. Lane M, 100-bp ladder marker; each band represents a 100-bp increment, with 300 bp at the low end. Lanes: 1, isolate DOH702 (pattern C4); 2, DOH714 (0); 3, DOH718 (E1); 4, DOH719 (I1); 5, DOH720 (E1); 6, DOH730 (C5); 7, DOH733 (I2); 8, DOH738 (N); 9, DOH740 (C5); 10, DOH741 (E4); 11, DOH747 (F1); 12, DOH755 (E1).
FIG. 2.
Diagram of RAPD types and patterns of V. parahaemolyticus. Numbers on the left are molecular size markers in base pairs.
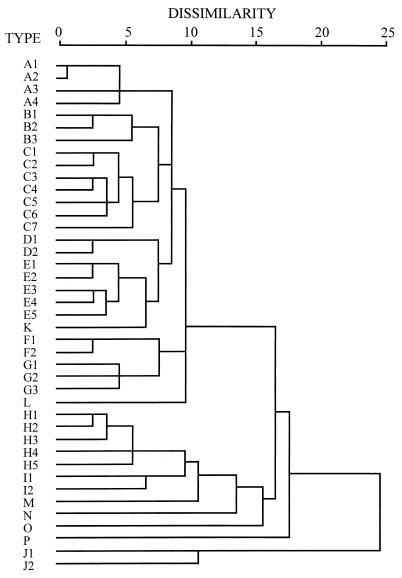
FIG. 3.
Dendrogram showing the clustering of RAPD patterns for V. parahaemolyticus. The dendrogram was made based on the squared Euclidean distance measure and average linkage clustering method by the SPSS for Windows release 6.0 program. The dissimilarity units are arbitrary, based on the squared Euclidean distance measurement. The strains were arbitrarily grouped into different types. The letters in the column on the left represent the RAPD patterns.
TABLE 2.
RAPD analysis of the 308 clinical isolates of V. parahaemolyticus isolated in Taiwan from food poisoning outbreaks
| Type | No. of strains (% of total) | Pattern | No. of strains (% of total) | Reference straina |
|---|---|---|---|---|
| A | 24 (7.79) | A1 | 9 (2.92) | |
| A2 | 13 (4.22) | |||
| A3 | 1 (0.32) | |||
| A4 | 1 (0.32) | |||
| B | 30 (9.74) | B1 | 24 (7.79) | |
| B2 | 5 (1.62) | ST550, laboratory no. 226 | ||
| B3 | 1 (0.32) | |||
| C | 133 (45.81) | C1 | 1 (0.32) | |
| C2 | 6 (1.95) | CCRC12864, CCRC13027 | ||
| C3 | 63 (20.45) | CCRC12958 | ||
| C4 | 2 (0.65) | CCRC10806, | ||
| C5 | 67 (21.75) | CCRC12863 | ||
| C6 | 1 (0.32) | |||
| C7 | 1 (0.32) | Laboratory no. 109 | ||
| D | 19 (6.17) | D1 | 1 (0.32) | |
| D2 | 18 (5.84) | |||
| E | 67 (21.75) | E1 | 46 (14.93) | |
| E2 | 0 | CCRC12963 | ||
| E3 | 7 (2.27) | |||
| E4 | 10 (3.24) | |||
| E5 | 4 (1.29) | |||
| F | 3 (0.97) | F1 | 2 (0.65) | |
| F2 | 1 (0.32) | |||
| G | 3 (0.97) | G1 | 1 (0.32) | |
| G2 | 1 (0.32) | |||
| G3 | 1 (0.32) | |||
| H | 10 (3.25) | H1 | 3 (0.97) | |
| H2 | 4 (1.30) | |||
| H3 | 1 (0.32) | |||
| H4 | 1 (0.32) | |||
| H5 | 1 (0.32) | |||
| I | 4 (1.30) | I1 | 1 (0.32) | |
| I2 | 3 (0.97) | |||
| J | 2 (0.65) | J1 | 1 (0.32) | |
| J2 | 1 (0.32) | |||
| K | 0 | K | 0 | CCRC13025 |
| L | 0 | L | 0 | CCRC12865 |
| M | 1 (0.32) | M | 1 (0.32) | |
| N | 1 (0.32) | N | 1 (0.32) | |
| O | 1 (0.32) | O | 1 (0.32) | |
| P | 2 (0.65) | P | 2 (0.65) |
The reference strains were typed by the RAPD method but were not counted in the distribution of the 308 local clinical isolates. The four environmental strains were subtyped as B2, C2, C5, and E2.
The major types (A to E), which together accounted for about 91.25% of the domestic clinical isolates examined, were closely related and showed low degrees of similarity to the minor types (F to P) (Fig. 3). Type K consisted of a single domestic clinical reference strain and was also close to the major types (Fig. 3). The foreign and environmental reference strains were subtyped as B, C, E, K, and L by the RAPD method, putting some into the major groups and some into the minor groups (Table 2). Thus, the different geographic or environmental origins of these strains did not have any significant effect on the clustering of the RAPD types.
The domestic isolates examined in this study were selected from the stool samples of patients involved in food poisoning outbreaks which occurred mostly from 1993 to 1995 in Taiwan. One hundred and twenty-eight isolates examined in this study had also been typed by the PFGE method (10). The epidemiological information about these isolates and outbreaks were provided in our previous publication (10). The corresponding PFGE typing results for the major RAPD patterns are summarized in Table 3. These major RAPD patterns each consisted of 1 to 10 PFGE patterns or one to six PFGE types. However, a major PFGE pattern or type was found in most of the RAPD patterns; for instance, RAPD patterns A1, A2, B1, C3, C5, D2, and E1 contained major PFGE types F, J, A, C, C, B, and G, respectively. The typings of isolates from different outbreaks by these two methods were also examined separately. Similar results were observed for most of the outbreaks. For some outbreaks, the isolates were typed into a single pattern by both methods; for instance, isolates from outbreaks 10, 35, and 37 were typed solely as RAPD patterns A1, A2, and B2 and PFGE patterns F1, J2, and A1, respectively (10). For some outbreaks, the isolates were typed into different patterns by the two methods (for instance, in outbreaks 9, 11, 15, and 17), although most of them could be grouped into closely related types. The discriminatory ability of RAPD was lower than that of the PFGE method. Isolates from outbreaks 39 and 40, which had been typed into several PFGE patterns (10), were all grouped into pattern E1 by RAPD.
TABLE 3.
Comparison of the typing of clinical isolates of V. parahaemolyticus by RAPD and PFGE
| Major RAPD pattern | PFGE (no. of isolates)a
|
|
|---|---|---|
| Pattern | Type | |
| A1 | F1 (7), F2 (1), C2 (1) | F (8), C (1) |
| A2 | J2 (11) | J (11) |
| B1 | A1 (4), A2 (2), A3 (1), E1 (1), M1 (1) | A (7), E (1), M (1) |
| B2 | A1 (1), B6 (1) | A (1), B (1) |
| C2 | K (1) | K (1) |
| C3 | C2 (1), C6 (12), D1 (2), E1 (2), H1 (1), I1 (1), I2 (1) | C (13), D (2), E (2), H (1), I (2) |
| C4 | C6 (1) | C (1) |
| C5 | C2 (4), C3 (1), C4 (1), C5 (7), C6 (7), D1 (3), D2 (2), G2 (2), L1 (1), L2 (1) | C (20), D (5), G (2), L (2) |
| C7 | H1 (1) | H (1) |
| D2 | B1 (2), B2 (3), B3 (3), B5 (2) | B (10) |
| E1 | A1 (1), C2 (1), C4 (1), F2 (1), G2 (1), G3 (1), G4 (6), M1 (2), M2 (1), M3 (2), N (2) | A (1), C (2), F (1), G (8), M (5), N (2) |
| E3 | J1 (1) | J (1) |
One hundred and thirteen of the clinical isolates were typed by both RAPD and PFGE (10). The major RAPD patterns and their corresponding PFGE typing results were compared.
The RAPD method can be used in the molecular subspecies typing of V. parahaemolyticus independently or as a supplement to other typing methods when very fine typing is needed. The most frequent serovars isolated from food poisoning outbreaks between 1993 and 1995 were K15, K8, K29, K56, and K12, with frequencies of 19.23, 13.94, 12.98, 8.65, and 6.25%, respectively. These major serovars could be further subdivided into two to seven patterns by the RAPD method, while three to seven patterns were discernible by the PFGE method (Table 4). In fact, 13 of 18 serovars of this pathogen could be subtyped into at least two RAPD patterns. In 1996, the O3:K6 serovar accounted for 50 to 80% of the V. parahaemolyticus outbreaks in India. This serovar also accounted for 80% of the outbreaks in 1997 in Taiwan. As examined by the PFGE method, these domestic O3:K6 cultures are phylogenetically identical or closely related to the strains isolated in India in 1996 (4) but are not close to those isolated before 1996 (data not shown). The present RAPD method may be also useful in differentiating strains of the same serovars (Table 4).
TABLE 4.
Distribution of RAPD patterns among different serotypes of the clinical isolates of V. parahaemolyticus
| Sero-typea | Pattern (no. of isolates)
|
|
|---|---|---|
| RAPD | PFGEb | |
| K3 | C3 (8), C5 (6) | D1 (4), D2 (2), E1 (2) |
| K4 | E1 (3), E3 (1), E4 (1), E5 (1) | N (2) |
| K5 | A1 (8) | C2 (1), F1 (7) |
| K6 | C5 (1), E1 (1) | ND |
| K7 | E5 (2) | ND |
| K8 | C5 (15), I2 (1) | A1 (1), C4 (1), C5 (4), G2 (1), L1 (1), L2 (1) |
| K10 | C3 (3), E3 (3), E5 (1), M (1) | C6 (1), J1 (1) |
| K11 | C3 (1) | ND |
| K12 | B1 (11), C5 (1) | A2 (1), C6 (1), M1 (1) |
| K15 | C3 (16), C5 (20), C7 (1), E1 (1) | C2 (5), C3 (1), C6 (8), D1 (1), M1 (1) |
| K29 | B3 (1), C2 (1), C3 (3), C5 (1), D1 (1), D2 (16), E1 (3) | B1 (2), B2 (2), B3 (3), B4 (1), B5 (2), G2 (1), M2 (1) |
| K38 | F1 (1), F2 (1) | ND |
| K41 | C5 (1), E1 (8), I (1), I2 (2) | M3 (4) |
| K56 | A2 (9), C3 (1), E1 (8) | C2 (1), C4 (1), G4 (4), J2 (9), M1 (2) |
| K57 | C2 (1) | K (1) |
| K60 | E1 (7), E3 (1) | F2 (1), G2 (1), G3 (1), G4 (2) |
| K63 | C3 (3), C5 (1), E1 (1) | H1 (1) |
| K68 | E1 (8) | ND |
Only those isolates that were serotyped are included.
Only some of the isolates were examined by PFGE (10); ND, not determined.
ACKNOWLEDGMENTS
This research was supported by the Department of Health, Republic of China (DOH85-TD-090 and DOH86-TD-106).
We also thank Carlos Javier for editing the manuscript.
REFERENCES
- 1.Chiou A, Chen L-H, Chen S-K. Foodborne illness in Taiwan, 1981–1989. Food Aust. 1991;43:70–71. [Google Scholar]
- 2.Makino S, Okada Y, Maruyama T, Kaneko S, Sasakawa C. PCR-based random amplified polymorphic DNA fingerprinting of Yersinia pseudotuberculosis and its practical applications. J Clin Microbiol. 1994;32:65–69. doi: 10.1128/jcm.32.1.65-69.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin-Kearley J, Gow J A, Peloquin M, Greer C W. Numerical analysis and the application of random amplified polymorphic DNA polymerase chain reaction to the differentiation of Vibrio strains from a seasonally cold ocean. Can J Microbiol. 1994;40:446–455. doi: 10.1139/m94-073. [DOI] [PubMed] [Google Scholar]
- 4.Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay A K, Garg S, Bhattacharya S K, Nair G B, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 6.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West P A, Colwell R R. Identification and classification of vibrionaceae—an overview. In: Colwell R R, editor. Vibrios in the environment. New York, N.Y: John Wiley & Sons; 1984. pp. 285–363. [Google Scholar]
- 8.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong H C, Liu C C, Yu C M, Lee Y S. Utilization of iron sources and its possible roles in the pathogenesis of Vibrio parahaemolyticus. Microbiol Immunol. 1996;40:791–798. doi: 10.1111/j.1348-0421.1996.tb01144.x. [DOI] [PubMed] [Google Scholar]
- 10.Wong H C, Lu K-T, Pan T-M, Lee C-L, Shih D Y-C. Subspecies typing of Vibrio parahaemolyticus by pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:1535–1539. doi: 10.1128/jcm.34.6.1535-1539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]