Abstract
The application of recombinant antibodies for the analysis of foods and food contaminants is now a major focus, given their capacity to be engineered to tailor their specificity, enhance their stability, and modify their structural formats to fit the desired analytical platform. In this study, human scFv antibody fragments generated against aflatoxin B1 (AFB1) were selected as the model antibody to explore the effect of antibody formats on their binding activity and to evaluate their potential use as immunoreagents for food contaminant analysis. Four human scFv antibody fragments against aflatoxin B1 (AFB1), previously isolated and engineered by chain shuffling, were converted into various formats, that is, scFv-AP fusions, scFv-Fc, and whole IgG molecules. The result indicated that the effects of the antibody format on the binding property varied, depending on the sequence of scFv. For all of the scFv clones, the scFv-AP fusion format showed the highest sensitivity by competitive ELISA, while the effects on the binding activity after conversion to scFv-Fc or IgG format varied, depending on the amino acid sequence of the antibodies. The sAFH-3e3 antibodies that showed the best performance by competitive ELISA were selected for further investigation. The sAFH-3e3 was converted to the scFv-GFP format and tested by fluorescence-linked immunosorbent assay (FLISA), which showed that its binding property was equivalent to those of scFv-Fc and IgG formats. The potential applications of the sAFH-3e3 in a rapid test kit format based on ELISA (scFv-AP) and in a lateral flow immunochromatography assay (LFIA) (IgG) were demonstrated. A comparison of methods for the extraction of AFB1 from matrices for use with these assay formats indicated that PBS and TBST are better than 70% methanol.
Introduction
Polyclonal and monoclonal antibodies are the most widely used reagents for food immunoanalysis. However, their production is laborious, time-consuming, and costly and requires animal immunization and sacrifice.1 Recombinant antibody has been proven to be highly advantageous in overcoming these limitations. Moreover, recombinant antibodies can be genetically engineered into different formats, thus enhancing their utility for different purposes, including effective clinical applications.2 Recently, there is much more focus on generating recombinant antibodies, given their flexibility and the need to be able to engineer antibodies to perform much more effectively in newly developing assay platforms such as sensors.
Recent trends in antibody engineering are focusing on small antibody fragments, which can be easily expressed in Escherichia coli or other expression systems. These formats include the single-chain variable fragment (scFv), antigen-binding fragment (Fab), and the single-domain antibody (sdAb). However, such antibody fragments may need to be further engineered to suit different diagnostic purposes and, consequently, may or may not lose binding affinity when some of the nonspecificity-associated portions of the antibody are modified.3,4 These issues are explored in this study, where a recombinant antibody against aflatoxin was used as a model to investigate the effect of antibody format on binding activity and to evaluate its use as an immunoreagent for food contaminant analysis.
Results and Discussion
Aflatoxin B1 (AFB1), the model mycotoxin used in this study belongs to a group of mycotoxins that are harmful to human and animal health, due to both acute and chronic effects, including allergic reactions, mutagenicity, immunosuppression, and potent carcinogenicity. It can be found in agricultural commodities and animal feed.5 The maximum level for different types of mycotoxins, especially aflatoxin B1, has been set by many national and international governments and organizations, including the international CODEX standard.6
Among a variety of established methodologies for analyzing mycotoxins, namely, thin layer chromatography, ultrapressured layer chromatography, immunoaffinity chromatography, high-performance liquid chromatography, and near infrared spectroscopy,7−9 immunological methods are among the most rapid, most simple, cheapest, sufficiently sensitive, and most suitable for on-site screening. In the developing countries, which are the major exporters of food and agricultural products such as South-East Asia, Africa, and the Middle East, there is an ever increasing demand for simpler and cheaper methods for the easy detection of mycotoxin contamination.10 The two main immunoassay methods commonly utilized for detecting mycotoxins are ELISA and lateral flow immunoassay (LFIA). These methods require antibody as the vital reagent for detection. Determining the most appropriate recombinant antibody formats for use in these assays is a key step for their optimal exploitation to generate more effective aflatoxin tests.
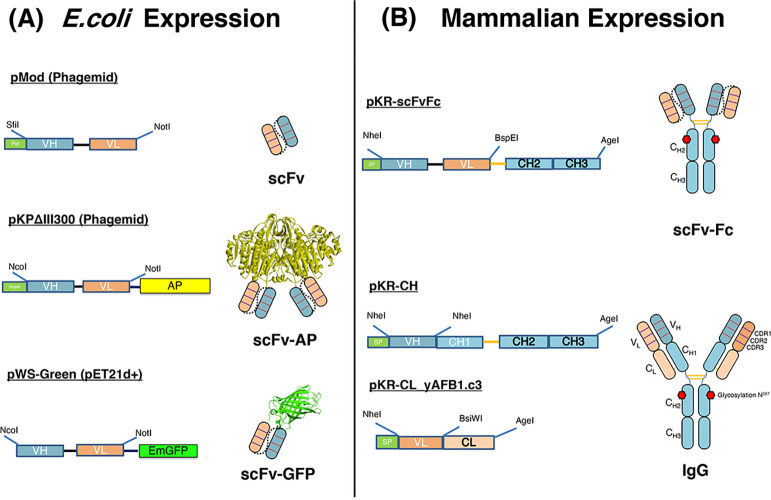
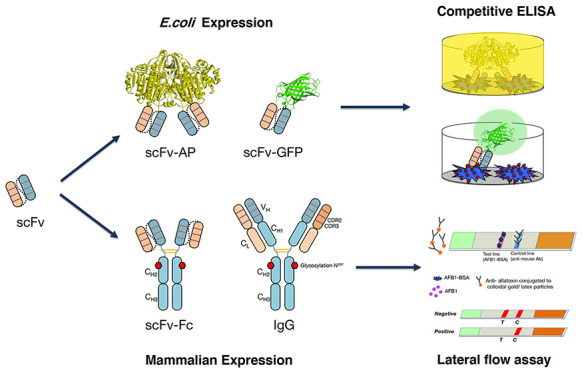
In this study, various formats of anti-aflatoxin B1 antibody were investigated. The expression cassettes and their resulting antibodies are summarized in Figure 1.
Figure 1.
Schematic illustration of phage display and mammalian cassette vectors for the expression scFv, scFv-AP, scFv-GFP, IgG, and scFv-Fc in this study. pMOD, pKPΔIII300 phagemids, and pWs-Green were used to generate scFv, scFv-AP (scFv fused with alkaline phosphatase, AP), and scFv-GFP (scFv fused with green fluorescence protein, GFP), respectively (A). The restriction sites for cloning scFv fragments are shown. Co-transfection of pKR-CH and pKR-CL_yAFB1.c3 in HEK293-6E cells can produce human IgG1 (B). The pKR-scFv-Fc was used to express scFv-Fc. The restriction sites for cloning the V regions and scFv fragment are indicated. The 3D modeling structure of GFP (PDB code: 2gx2) and AP (PDB code: 1urb) was taken from pdb website11 and modified with the program PyMol.12
Expression and Purification of Various Antibody Formats
The four anti-aflatoxin scFvs were converted into IgG and scFv-Fc formats by subcloning into mammalian expression vectors, as depicted in Figure S1. Anti-aflatoxin IgG were produced from 2 monocistronic vectors, that is, pKR-CH (Figure S1A) and pKR-CL_yAFB1.c3 (Figure S1B) by co-transfection of both plasmids into HEK293-6E cells. The mammalian expression vector pKR-scFv-Fc (Figure S1C) was used for the expression of scFv-Fc. Both IgG and scFv-Fc were produced in HEK293-6E cells on a 25 mL scale. These antibody formats were purified by protein A affinity chromatography. Purity and apparent molecular weight of purified antibodies were assessed by SDS–PAGE analysis (Figure S2). Under nonreducing conditions, apparent molecular sizes were approximately 120 and 150 kDa for scFv-Fc and IgG, respectively. Under reducing conditions, both heavy and light chains could be observed, of which doublet bands of the light chain were detected (Figure S2A). This could be because of the heterogeneity in glycosylation that did not interfere with its binding activity as was previously observed.13 Yields of the four different scFv-Fc and IgG antibodies were between 270 and 389 mg L–1 and 277 and 480 mg L–1, respectively. The scFv-Fc formats of the three mutant clones could be purified at a higher yields than those of the parental clones and in IgG formats. These results corresponded with the previous observation that the scFv-Fc generated from scFv expressed in higher yields than those of IgG.14 These yields were much higher than that of scFv-AP format that was expressed in E. coli, under the control of the PhoA promoter and produced in low phosphate media to preserve the alkaline phosphatase activity.1 The scFv-AP bands appeared at the expected size of approximately 80 kDa (Figure S2B). The yields of the four different scFv-AP ranged between 0.7 and 2.7 mg L–1.
Competitive ELISA
To determine the sensitivity of various formats of the four antibodies clones (yAFB1-c3, sAFH-3e11, sAFH-3f11, and sAFH-3e3), indirect competitive ELISA was performed. The yAFB1-c3 was the parental antibody clone; while the sAFH-3e11, sAFH-3f11, and sAFH-3e3 are the affinity-matured clones that were obtained using the chain shuffling technique from our previous study.15 For our antibody nomenclature system beginning with sAFH, s stands for shuffled, AF for aflatoxin, and H indicated that the heavy chain was being shuffled. This is then followed by clone number. The graphs were plotted from the absorbance values (expressed as A/A0) and the concentrations of AFB1. The IC50 values for the scFv,15 scFv-AP, scFv-Fc, and IgG are shown in Table 1. The results indicated that the sensitivity of antigen detection by different antibodies was influenced by the conversion into different formats. Overall, the scFv-AP format showed the highest sensitivity to aflatoxin with the IC50 ranging from 0.007 to 0.06 μg mL–1. For clones sAFH-3e11 and sAFH-3f11, the scFv-Fc format had IC50 values close to those of scFv formats, whereas the IgG format showed lower sensitivity. Loss of affinity after conversion of the scFv fragment to IgG was previously reported for human anti-CD30.16 However, the yAFB1-c3 IgG format showed higher sensitivity than the scFv and scFv-Fc formats. For sAFH-3e3 clone, which has the highest sensitivity, the IC50 values of scFv-Fc and IgG formats were similar to that of the scFv format. These results are correlated with several previous studies, which demonstrated that conversion of antibody fragments back to full length IgG or scFv-Fc resulted in similar or improved antigen binding.17−19 Conversion of scFv into IgG can result in similar,18 improved,19 or loss of activity.16 This seems to depend on amino acid sequence, 3D-structure, and the mode of binding of heavy and light variable domains of different scFv molecules.21 Our results confirmed these previous observations. The main reason for the change in their binding property is because the arrangement of VH and VL fragments, which is artificially linked with the linker peptide, in the scFv format, is quite different from the structure of VH and VL in the whole IgG molecule. It has been proposed that affinity maturation in the scFv format may not lead to improved antigen binding properties when converted into IgG because new mutations into scFv antibodies may require the presence of the peptide linker.14 This suggestion has been observed in our study. The affinity-matured-shuffled clones (sAFH-3e11, sAFH-3f11, and sAFH-3e3) showed similar or improved binding sensitivity after conversion into scFv-AP and scFv-Fc formats, where the peptide linker was retained. However, when these shuffled clones were converted into IgG format, the binding sensitivity was lost, probably because the peptide linker was absent in the IgG form. On the contrary, for the parental clone (yAFB1-c3), the binding sensitivity was improved after conversion into IgG.
Table 1. IC50 of Different Formats of Antibody.
| IC50 (μg mL–1)#b |
||||
|---|---|---|---|---|
| scFv (*)a | scFv-AP | scFv-Fc | IgG | |
| yAFB1-c3 | 0.120 ± 0.028 | 0.060 ± 0.006 | 0.230 ± 0.021 | 0.060 ± 0.000 |
| sAFH-3e11 | 0.042 ± 0.005 | 0.009 ± 0.000 | 0.038 ± 0.000 | 0.090 ± 0.000 |
| sAFH-3f11 | 0.055 ± 0.001 | 0.009 ± 0.001 | 0.040 ± 0.000 | 0.070 ± 0.001 |
| sAFH-3e3 | 0.018 ± 0.002 | 0.008 ± 0.002 | 0.022 ± 0.002 | 0.022 ± 0.000 |
*Results from a previous study.15
#Data were obtained from a plot of the mean ± standard deviation (SD) of duplicate wells.
In summary, the results from competitive ELISA showed that the conversion of scFv to IgG and scFv-Fc formats did not help to improve the sensitivity of the antibody. However, the yields of IgG and scFv-Fc formats were much higher when produced in a mammalian system, when compared to those in the E. coli system. The antibody in the scFv-AP format showed the highest sensitivity by ELISA, which could be because AP is a very efficient enzyme,22 and also less washing steps, which could reduce the signal, were required for one-step detection using the scFv-AP format.23 Moreover, because AP is a dimeric enzyme; therefore, scFv-AP is bivalent. An increase in avidity by constructing bivalent scFv-Fc has been reported to increase binding affinity to C-reactive protein (CRP) 50-fold, compared to the monovalent scFv fragment.24 Because in this study, the scFv-AP formats showed the highest sensitivity, it was further used to investigate the biding specificity of each antibody in the next step.
Specificity of scFv-AP Format Against Aflatoxin
Cross-reactivities of four scFv-AP clones were determined against structurally related aflatoxins; namely, aflatoxin B2, G1, G2, and M1 (a metabolite of B1). Different scFv-AP clones showed various degrees of cross-reactivity against different related aflatoxin except for AFM1, as shown in Table 2. For sAFH-3e11, the degree of cross reactivity was in the decreasing order from AFG1 > AFB2 > AFG2, whereas the other two mutant clones showed a decreasing degree of cross-reactivity from AFG1 > AFG2 > AFB2. Interestingly, clone sAFH-3e3, which has the highest sensitivity against AFB1 (sAFH-3e3), showed cross reactivity to AFM1 and a high percentage of cross reactivity against AFG1 in a manner similar to another previously published clone, indicating that the antibody is binding near the C9 position of the toxin molecule (Figure S3).25 None of these clones showed cross reactivity against nonrelated mycotoxins, namely, ochratoxin and zearalenone (see Supporting Information Figure S4). Therefore, these antibodies could be applicable for the detection of total aflatoxins.
Table 2. Results of Cross-Reactivity of Four scFv-AP Against Aflatoxin.
| cross-reactivity (%)*a |
|||||
|---|---|---|---|---|---|
| AFB1 | AFB2 | AFG1 | AFG2 | AFM1 | |
| yAFB1-c3 | 100 | 30.0 | 70.6 | 34.3 | 0.0 |
| sAFH-3e11 | 100 | 33.3 | 83.33 | 28.6 | 0.5 |
| sAFH-3f11 | 100 | 22.5 | 75.0 | 52.9 | 0.3 |
| sAFH-3e3 | 100 | 30.8 | 125.0 | 111.0 | 18.2 |
*data were obtained from a plot of triplicate experiments.
Binding Activity of scFv-GFP to Free Mycotoxin by Competitive FLISA
GFP has been successfully used in biosensors;26 hence, the binding of recombinant anti-aflatoxin antibody in the scFv-GFP format was investigated to explore the possibility for use in mycotoxin biosensor detection. The sAFH-3e3 clone, which showed the highest binding activity, was fused with EmGFP, and the binding sensitivities were determined by competitive fluorescence-linked immunosorbent assay (FLISA).27,28 The yield of the scFv-GFP was 0.44 mg L−1. The optimal concentrations of conjugated mycotoxins and recombinant scFv-GFP proteins were first determined by checkerboard titration (data not shown). Competitive FLISA indicated that the IC50 values of sAFH-3e3-GFP were 26 and 30 ng mL–1 when measured with the excitation/emission wavelengths of 484/509 and 478/506 nm, respectively (Figure 2). The IC50 values of sAFH-3e3-GFP were close to those of scFv-Fc and IgG but not as low as that of scFv-AP format. The limit of detection (LOD) for sAFH-3e3-GFP was approximately 13 ng mL–1.
Figure 2.
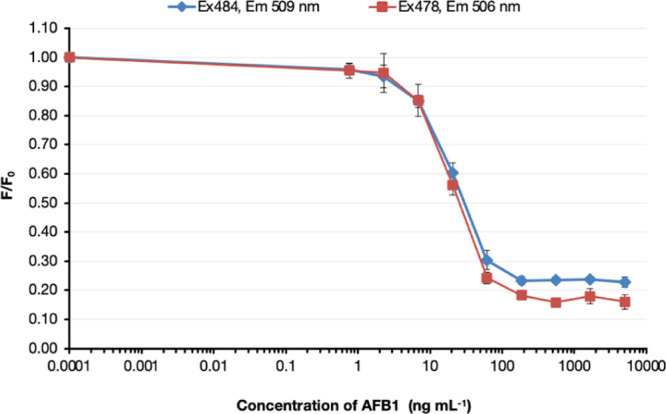
Competitive FLISA of sAFH-3e3 scFv-GFP against AB1-BSA.
Concentrations of soluble AFB1 from 0.762 to 5000 ng mL–1 were incubated with purified sAFH-3e3-GFP antibodies at 37 °C for 30 min before addition to wells of Immuno 96 MicroWell plates and coated with 0.25 μg of AFB1-BSA. Bound antibodies were detected by fluorescence intensity. F indicates the fluorescence signal of scFv-GFP that bound to AFB1-BSA in the presence of various concentrations of soluble AFB1. F0 is the fluorescence signal of scFv-GFP at 0 ng mL–1 soluble AFB1. Fluorescence values (expressed as F/F0) were plotted against the logarithm of AFB1 concentration. The IC50 values of sAFH-3e3-GFP were 26 and 30 ng mL–1 when measured with the excitation and emission wavelengths at 484–509 and 478–506 nm, respectively.
Detection of AFB1 from Grain Samples Using scFv-AP
To demonstrate the feasibility of employing recombinant scFv-AP, which showed the best binding performance, for the detection of mycotoxin contamination in agricultural products, competitive ELISA was performed using spiked samples. The IC50 and limit of detection was determined from a graph plotted with A/A0 versus log of concentration of spiked AFB1 (Figure 3). The result indicated that a suitable buffer for extraction of the reference sample was TBST, showing the IC50 value of 9 ng mL–1. Interestingly, the IC50 value was 90 ng mL–1, which was 10 times lower in sensitivity when 70% methanol was used as an extraction buffer. An increase in IC50 is likely due to matrix interferences, which could be decreased by diluting the sample extract with phosphate buffered-tween (PBST) solution.10 The detection limits of scFv-AP when extracted with TBST and 70% methanol were 1.5 and 35 ng mL–1, respectively.
Figure 3.
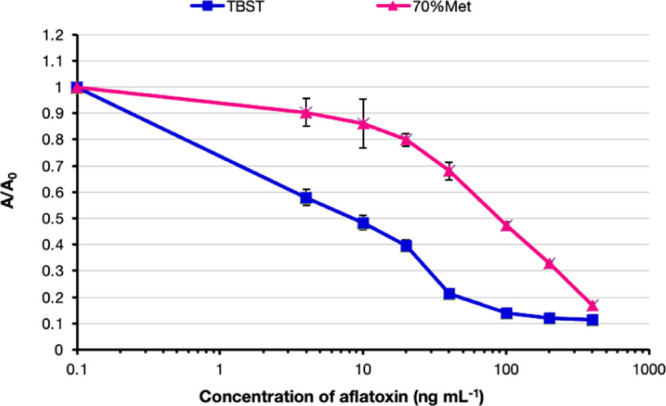
Detection of aflatoxin contamination in the agricultural sample using sAFH-3e3-AP. The extraction buffers [TBST or 70% (v/v) methanol; Met] were compared. The spiked AFB1 at various concentrations (4, 10, 20, 40, 100, 200, and 400 ng mL–1) was incubated with sAFH-3e3-AP. Absorbance values (expressed as A/A0) were plotted against the logarithm of AFB1 concentration. The IC50 values of sAFH-3e3-AP when using TBST or 70% (v/v) methanol as extraction buffer were 9 and 90 ng mL–1, respectively.
While the specificity of our recombinant antibodies is comparable or better than commercial monoclonal or polyclonal antibodies, respectively; the limit of detection (LOD) is still too high for certain regulatory limits and not as good as commercially available polyclonal and monoclonal antibodies. Further improvement of binding affinity could be performed to increase the binding affinity using antibody engineering.29,30 Nevertheless, these results confirmed the previous observation that recombinant antibody in the format of scFv-AP is suitable and can be used to detect mycotoxin contamination in agricultural samples.1 Various methods of affinity maturation can be used to improve antibody limit of detection. For example, the chain shuffling technique has been successfully used to improve the binding sensitivity of human scFv antibody against aflatoxin 7.5-fold over the parental clone.15 A combination of random mutation and yeast display method has been successfully used to improve the binding affinity of mouse scFv antibody against AFB1 9-fold.31 Site-direct mutagenesis in combination with multiple sequence alignment and molecular modeling could lead to 4–20 fold improvement in sensitivity of mouse monoclonal anti-AFB1.32 Recently, computational design of binding antibodies has gained growing interest.33 An automated web server to optimize both antibody affinity and stability by the design of the variable light–heavy chain interface within the antibody core has been created.21 These novel approaches to improve antibody performance are applicable for anti-AFB1 antibody as well.
Recently, several recombinant antibodies have been produced and successfully used for aflatoxin detection in ELISA-based assays on the microtiter plates.6 These included the use of various formats of antibody such as scFv fragment,31,32,35−37 Fab,38 and VHH.39 So far, there have been only a few example of applying recombinant antibody for lateral flow immuno assay (LFIA) or strip test, for example, the detection of morphine by using scFv conjugated with colloidal gold.40 Because there has been no report on using recombinant antibody as an immunoreagent for the detection of aflatoxin by LFIA, in the next step, we further investigated the potential use of recombinant antibody for the detection of AFB1 by LFIA.
LFIA for the Detection of Aflatoxin Using Colloidal Gold-IgG
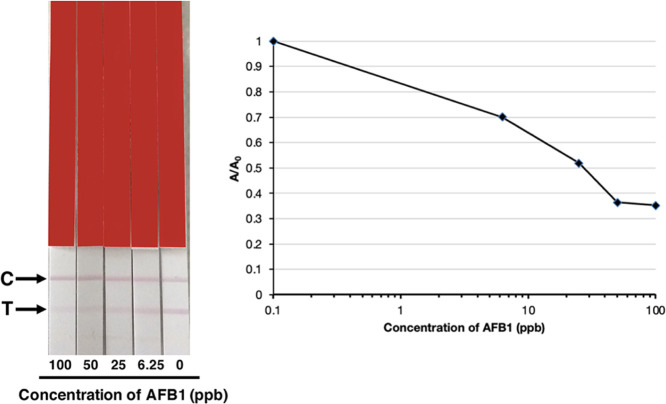
Colloidal gold is commonly used to conjugate with traditional polyclonal and monoclonal antibodies for incorporation into LFIA or test strips. In this study, quantitative detection of AFB1 was investigated using an AgraStrip (Romer Labs). Colloidal gold conjugated to sAFH-3e3 IgG acted as the probe. The anti-mouse mAb and AFB1-BSA were applied to a strip as a control line (C) and test line (T), respectively.
Corn samples were extracted with PBS. Then, different concentrations of AFB1 were spiked into the extracted solution. These spiked solutions were mixed with IgG labeled with colloidal gold and added to microwells. Then, a test strip was dipped into the well. The IgG-colloidal gold complex flowed along the membrane, encountered the coated AFB1-BSA, and was captured. Thus, a red color appeared at the test line (T). When AFB1 was present in the sample, the binding of the IgG-colloidal gold to the AFB1-BSA was inhibited and the color on the test line was reduced. The control line is visible because of the reaction between colloidal gold-sAFH-3e3 IgG and anti-mouse IgG, indicating good functionality of the test. The red color intensity of the test lines decreased with increasing AFB1 concentrations. The intensity of the test line on a strip was evaluated using an AgraStrip reader (Romer Labs, Austria). Graphs were plotted from the peak values. There was an inverse relationship between the signal intensity and aflatoxin concentration. The result showed that the IC50 value was 28 ppb. The limit of detection was estimated from the graph at 0.7 A/A0 as 6 ppb (Figure 4).
Figure 4.
LFIA analysis of the extracted corn sample spiked with AFB1 standard using colloidal gold-sAFH-3e3 IgG antibody. Various concentrations of soluble AFB1 mixed with colloidal gold-antibody conjugated were tested. The red color at the test line (T) decreased when the concentration of AFB1 increased. The graph was plotted from A/A0 against log of the concentration of AFB1; A = peak area value of each concentration of spiked AFB1 and A0 = peak area value at 0 ppb.
Optimization of Antibody-Latex Particle Conjugation
About 0.5 mg mL–1 concentrations of IgG and scFv-Fc antibodies were conjugated with 70 nm and 169 nm latex particles using the methods as described in the Materials and Methods section. As illustrated in Figure S5, when 0.05 mg mL–1 IgG was conjugated with 70 nm latex particles, both control and test lines appeared as green lines, indicating optimal conjugation conditions. Other conditions using 169 nm latex particles or scFv-Fc did not show the right appearance of the green lines. Therefore, 0.05 mg mL–1 IgG and 70 nm latex particles were used for LFIA detection in this study.
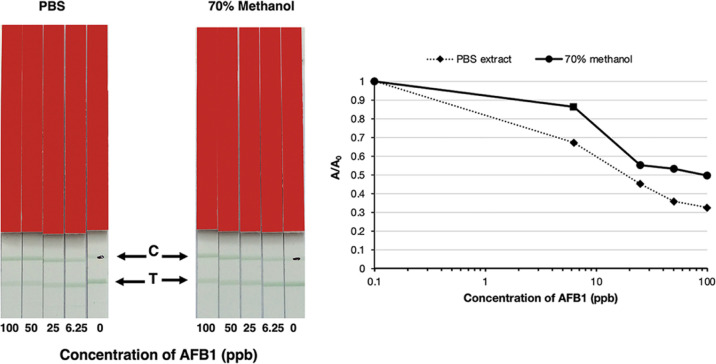
LFIA for the Detection of Aflatoxin Using Latex-IgG
Colloidal gold is widely used as a label for LFIA, but appears mainly as a red color. Latex particles offer a wider range of colors, which will be useful for multiplexing LIFA for the detection of multiple mycotoxins. Therefore, the sAFH-3e3 IgG–latex was generated to investigate its potential use for the detection of mycotoxin contamination in LFIA. The procedure was the same as that used for the colloidal goal-conjugated antibody, except that the IgG-latex conjugation gave a green color on the test line and control line. As illustrated in Figure 5, the result showed that the IC50 value was 18 ppb. When 70% methanol was used as the extraction buffer, the IC50 value was 70 ppb. This result and a previous observation41 indicated that water-based buffer is a suitable extraction buffer for LFIA. The limit of detection when using PBS as an extraction buffer was at 5 ppb. In this study, the extraction solution was diluted 1:3, as a multifold dilution has been shown to reduce matrix interference and improve sensitivity.42
Figure 5.
LFIA analysis of the extracted corn sample spiked with AFB1 standard using latex microsphere-sAFH-3e3 IgG antibody. The corn sample was extracted with PBS buffer or 70% (v/v) methanol and spiked with various concentrations of AFB1. The green color in the test line (T) decreased when the concentration of AFB1 was increased. The graph was plotted from A/A0 against log of concentration of aflatoxin; A = peak area of each concentration of spiked AFB1 and A0 = peak area at 0 ppb.
This is the first preliminary study on using recombinant antibody-based conjugation with latex particles as a detection probe for LFIA. The results showed that both gold and latex can be used to conjugate with recombinant IgG for LFIA. To improve the performance of LFIA, various parameters can be optimized, for example, the conjugation methods, types of strip utilized, and concentrations of reagents on the test and control lines.43
A summary of the LOD of different formats of antibody clone sAFH-3e3 in this study is shown in Table 3. These values were obtained from spike experiments of corn extracts except for the data of scFv and scFv-GFP, which were obtained from a standard competitive ELISA because their performances were inferior to those of scFv-AP and not selected for further analysis.
Table 3. Summary of LOD Values of Antibody Clone sAFH-3e3 in Different Formats.
| antibody format | detection method | extraction buffer | LOD (ng mL–1) |
|---|---|---|---|
| scFv | ELISA | PBST*a | 9*a |
| scFv-GFP | FLISA | PBST*a | 13*a |
| scFv-AP | ELISA | 70% methanol in TBST | 35 |
| TBST | 1.5 | ||
| IgG-Gold | LFIA | PBST | 6 |
| IgG-Latex | LFIA | 70% methanol in PBST | 13 |
| PBST | 5 |
*LOD values of scFv and scFv-GFP were obtained from standard competitive ELISA using indicated buffer. All other LOD values were from spiked experiment of corn standard, extracted with different extraction buffers.
Because aflatoxin contamination is a worldwide problem, different countries, regions, and international agencies have created regulatory limits based on both socioeconomics and scientific factors.44 Currently, aflatoxin regulations have been set in approximately 120 countries around the word either in the form of total aflatoxins or AFB1.45 While European Union (EU) regulation has the strictest regulations, the Codex standard, which has been established by Food and Agricultural Organization (FAO) and the World Health Organization (WHO), is globally acknowledged. The maximum limits of aflatoxin varied from 2 to 35 μg/kg (ppb), depending on the type of aflatoxins (total or AFB1), country, organization, and type of food or agricultural products. Maximum limit of AFB1 has been set for the EU (2 μg/kg for peanut products), China (20 μg/kg for peanut and corn), India (30 μg/kg for all food), Indonesia (15 μg/kg for peanut and corn), and South Korea (10 μg/kg for grains and cereal products).46 Based on this information, the recombinant antibody from this study could be applicable for point-of-demand detection (POD) of contamination in the raw material in several countries. Nevertheless, the real application of recombinant antibody for aflatoxin detection requires several additional critical steps, ranging from sampling and extraction methods to enrichment, detection, and sensor platforms.44 Because different samples may require different methods; therefore, further investigation on the application of recombinant antibody for on-site determination of aflatoxin in various raw materials or food products must be performed.
Conclusions
In this study, the effects of recombinant antibody formats on their performance were investigated by using an antibody against AFB1 as a model. Four recombinant antibodies against AFB1 were engineered into four different formats, that is, scFv-AP, scFv-Fc, IgG, and scFv-EmGFP. The binding properties of these antibodies were compared. The results showed that when scFv was converted to different formats; it can retain, increase, or lose binding activity, depending on the variable sequence of the clones. The scFv-AP format was found to be highly suitable for the detection by ELISA; however, the production yield is relatively low, when compared to scFv-Fc or IgG constructs. The IgG format, conjugated to either gold or latex particles, was highly applicable for the detection of mycotoxin by LFIA.
Materials and Methods
Materials and Chemicals
All reagents were of molecular grade or analytical grade. Anti-aflatoxin scFv antibody yAFB1-c3 (parental clone) and three mutant clones (sAFH-3e11, sAFH-3f11, and sAFH-3e3) were produced in our laboratory.15,36 Standard aflatoxin B1 (AFB1), B2, G1, G2, M1, and AFB1-BSA conjugates for ELISA and lateral flow assays were obtained from Aokin, Germany and Romer Labs, Austria, respectively. The TMB substrate (3, 3′, 5, 5′-tetramethylbenzidine) and PNPP substrate (p-nitrophenyl phosphate) were obtained from Sigma (St. Louis, MO). E. coli TG1 was obtained from the MRC, Cambridge, UK, and used for the production scFv-AP. E. coli SHuffle T7 Express was obtained from New England Biolabs (NEB, Massachusetts, USA). A HiTrap protein A HP and HisTrap HP affinity column were purchased from GE Healthcare (Chicago, USA). The peroxidase AffiniPure F(ab′)2 fragment (goat anti-human IgG (H + L) was purchased from Jackson ImmunoResearch Inc. (PA, USA). The HEK293-6E and pTT28 vectors were originally developed at the NRC-BRI in Montreal, Canada. The 25 kDa PEI was purchased from Polysciences (Polysciences Europe GmbH, Germany). Serum-free Freestyle 17 (F17) expression medium was obtained from Life Technologies Inc. (Burlington, Ontario, CA). l-glutamine and Pluronic F-68 were purchased from Sigma-Aldrich (Oakville, Ontario, CA). Colloidal gold was purchased from BBI solutions (Crumlin, UK). Estapor Microspheres (Green latex microspheres) size 70 nm (K007) and 169 nm (K3-020) were obtained from Merk Millipore (Darmstadt, Germany).
Conversion of Anti-Aflatoxin scFv to scFv-Fc and IgG
The pTT28 (NRC-BRI, Canada) was used as a framework for the generation of monocistronic IgG and scFv-Fc expression vectors. The vector consisted of a bacterial origin of replication (ori), ampicillin resistance gene (AmpR) for plasmid amplification and selection in E. coli, and a CMV promoter, which is suitable for expression in HEK 293-6E cells and had the synthetic and codon-optimized human signal peptide for protein secretion.47
A schematic diagram for the construction of expression vectors used in this study is shown in Figure S1. To construct the heavy-chain expression vector, pKR-CH (Figure S1A), the constant heavy-chain regions of human IgG1 from the pRom108-3D6H chain (gift from Kunert’s lab, BOKU, Austria) was amplified with primers containing the NheI site at 5′ end (CH-NheIFw) and AgeI site at 3′ end (CH-AgeIRv) of the constant heavy-chain gene and cloned into pTT28 that was cut with respective restriction enzymes (NEB, USA). Cloning of four anti-aflatoxin VH clones (sAFH-3e11, sAFH-3f11, and sAFH-3e3 and yAFB1-c3)15 was performed in a similar manner via the NheI/NheI site of the pTT28 vector.
For light-chain expression vector, the kappa (κ) light chain was amplified from the MIS104 vector (pTT5-3D6 L chain, a gift from Florain’s lab, BOKU, Austria) with primers introducing the BsiWI site at the 5′ end and AgeI site at the 3′ end of the constant light-chain gene. The anti-aflatoxin VL (yAFB1-c3) was amplified with primers introducing the NheI site at the 5′ end and BsiWI site at the 3′ end of VL gene. The kappa (κ) light chain and VL genes were cloned into pTT28 between NheI/AgeI sites, resulting in the kappa light-chain expression vector pKR-CL_yAFB1.c3 (Figure S1B).
For the conversion of scFv to scFv-Fc, the Fc region was amplified from pRom108 by PCR with a primer introducing the BspEI site at the 5′ end (Fc-BspEIFw) and AgeI site at 3′ end (CH-AgeIRv); while the 4 anti-aflatoxin scFv genes were amplified with a primer introducing the NheI site at the 5′ end and BspEI site at the 3′ end of the scFv gene. Then, the scFv and Fc regions were subcloned into pTT28 via NheI and AgeI sites. This scFv-Fc vector was designated as KR-scFv-Fc- (yAFB1-c3, sAFH-3e11, sAFH-3f11, and sAFH-3e3) vectors (Figure S1C).
All of the PCR reactions were composed of 0.2 μM each of primers, 0.2 mM dNTP, 0.25 μL (1.25 U) of One Taq polymerase (NEB, USA), 0.5 μL of the vector template and made up to 50 μL with distilled water. Primers for vector construction are listed in Table 4. After digestion of the insert and vector with corresponding restriction enzymes, the vector and insert were joined using the T4 DNA ligase (400 U/μL, NEB, USA) at 25 °C for 1 h before transformation into E. coli TOP 10 (Thermo Fisher Scientific, USA). The individual colonies were picked and cultured overnight for plasmid preparation using Nucleospin plasmid (Macherey-Nagel, Germany), according to the manufacturer’s protocol. The integrity of the plasmids was confirmed by DNA sequencing. Large-scale plasmid preparation for antibody expression was performed by using Nucleobond extra Midi (Macherey-Nagel Germany) from 100 mL culture volume of E. coli TOP 10.
Table 4. Primers for the Construction scFv-Fc and IgG Expression Vectors Used in This Studya.
| primer | sequence |
|---|---|
| Fc-BspEIFw | 5′AGT CTC CGG AGA GCC CAA GAG CTG CGA C3′ |
| CH-NheIFw | 5′ATG CGC TAG CAC CAA GGG CCC CAG CGT GTT CC3′ |
| CH-AgeIRv | 5′GCC ACC GGT TCA CTT GCC GGG GGA CAG GCT3′ |
| C3 VLNheIFw | 5′AGT GCC GCT AGC GAC ACC GTG ATG ACC CAG TCT3′ |
| LC conAgeIRv | 5′AGT GCC ACC GGT CTA ACA CTC TCC CCT GTT3′ |
| C3 VHNheIFw | 5′GCC GCT AGC CAG GTG CAG CTG GTG CAG TC3′ |
| 3E3 VHNheIFw | 5′AGT GCC GCT AGC GGG GTG CAG CTG GTG GAG TC3′ |
| VL BspEIRv | 5′GCA CAG TCC GGA ACG TTT GAT CTC CAC CTT GGT3′ |
| C3 VHNheIRv | 5′TTG GTG CTA GCT GAG GAG ACG GTG ACC AGG G3′ |
| C3 VLNheIFw | 5′AGT GCC GCT AGC GAC ACC GTG ATG ACC CAG TCT3′ |
Sequences corresponding to NheI, BspEI, BsiWI, and AgeI restriction sites are underlined.
Construction of the scFv-AP Fusion
The three anti-aflatoxin scFv genes from the phagemid vector15 were sub-cloned into the pKP300 ΔIII vector48 between the NcoI and NotI sites. The integrity of the construct was confirmed by automated DNA sequencing.
Expression and Purification of scFv and scFv-AP
The expression and purification of scFv were done as previously described.15 The scFv-APs were expressed according to a previously published protocol.36 For purification, after 18–20 h, the cultures were centrifuged at 7455g for 10 min at 4 °C. The pellet was resuspended in 8 mL of ice-cold periplasmic buffer (1 × PBS, 1 M NaCl and 1 mM EDTA) and left on ice for 20 min. The resuspended solution was centrifuged at 3000g for 10 min at 4 °C. The supernatants, composed of the periplasmic fractions containing scFv fragments, were collected, and then, MgCl2 was added to a final concentration of 1 mM. The periplasmic fraction was purified with the ÄKTA start protein purification system using immobilized metal affinity chromatography (IMAC) according to the manufacturer’s instructions (GE Healthcare, USA). Before purification, cell debris was removed from cell lysate by filtration (0.45 μm; Corning, New York, USA). A 1 ml His-Trap column (GE Healthcare, USA) was equilibrated with 10 column bed volume (CV) binding buffer (20 mM Tris–HCl, 500 mM NaCl, 20 mM imidazole, pH 7.9) before the sample was loaded. Then, the column was washed with 20 CV of the same buffer. The scFv was eluted with 20 CV of a linear gradient (0–100%) of imidazole by mixing binding buffer and elution buffer (20 mM Tris–HCl, 500 mM NaCl, 500 mM imidazole, pH 7.9). Then, the antibody fraction was dialyzed with 2 L of TBS buffer containing 1 mM MgCl2 using 10 kDa snakeskin dialysis tubing (Thermo Scientific, USA) to remove imidazole.
Expression and Purification of IgG and scFv-Fc
IgG and scFv-Fc expression vectors were expressed in HEK293-6E cells (NRC-BRI, Canada). Before transfection, the cells was passaged and cultured in F17 media (Invitrogen, USA) supplemented with 4 mM l-glutamine, 0.1% (v/v) Pluronic F68, and 25 μg mL–1 G418, at 37 °C, under 5% CO2 in a hydrated atmosphere with shaking at 130 rpm until the cell density reached 1.7–2.0 × 106 cells/mL. Then, the cells were transfected with 1 μg mL–1 of plasmid DNA by slowly adding a 3 mL complex solution of plasmid DNA and PEI (polyethylenimine) (Polysciences, USA) at a ratio of 1:2.
For IgG expression, 25 mL HEK-293-6E cells were transfected with 12.5 μg mL–1 of each heavy-chain and light-chain plasmid DNA. The cells were fed with 0.5% (v/v) TN1 medium (Tekniscience, Canada) 48 h after transfection. The scFv-Fc or IgG was harvested from the culture supernatant after another 72 h of cultivation by centrifugation at 1300g for 5 min. The supernatant was passed through a 0.45 μm filter before purification.
Human IgG and scFv-Fc were purified using a 1 mL HiTrap Protein-A HP column (GE Healthcare, USA). The column was equilibrated and washed with PBS buffer. The scFv-Fc or IgG was eluted in 0.1 M glycine-HCl, pH 3.5. The elution fractions were neutralized with 2 M Tris–HCl, pH 8.0 and dialyzed with PBS buffer. The samples were kept at 4 °C until further analysis.
Competitive ELISA Using Different Antibody Formats
The purified antibodies were used to test binding sensitivity (scFv-AP, scFv-Fc, and IgG) and specificity (scFv-AP) by competitive ELISA, as previously described.36 Briefly, 1 μg of the AFB1-BSA conjugate (Aokin, Germany) was immobilized on the wells of an ELISA plate and blocked with 2% (w/v) skimmed milk. The optimal concentration of scFv-AP that showed a linear relationship by indirect ELISA was preincubated with increasing amounts of soluble AFB1, ranging from 0.028 to 5000 ng mL–1 at 37 °C for 30 min before adding into the previously coated wells of the ELISA plates. To develop color, the PNPP substrate was added and the absorbance measured at 405 nm by a Sunrise Tecan ELISA reader (Männedorf, Switzerland). The data obtained were used to plot the inhibition curve as A/A0 versus logarithm of analyte concentration. A half-maximum inhibition (IC50) was estimated at 50% A/A0.
To evaluate binding specificity, the scFv-AP antibody constructs were assayed against a range of soluble aflatoxins, that is, B1, B2, G1, G2, M1, ochratoxin A (OTA), and zearalenone (ZEN). Stock solutions were diluted using TBST. The assays were performed following the competitive ELISA protocol, as described above. The percentage cross-reactivity (% CR50) was determined by the IC50 value of aflatoxin B1 divided by the IC50 value of other aflatoxins, multiplied by 100.
For IgG and scFv-Fc, an HRP-conjugated goat anti-human IgG F(ab′)2 secondary antibody was added and incubated for 1 h. After that, the plate was washed with PBST three times and PBS two times. Then, the TMB substrate was added. The reaction was stopped with 10% (v/v) HCl after incubation for 10–30 min. Triplicate measurements of absorbance values at 450 nm were performed, and the mean values with SD (standard deviations) were reported.
Cloning and Expression of scFv-EmGFP
The gene of the sAFH-3e3 anti-aflatoxin scFv, which showed the best sensitivity,15 was sub-cloned from the phagemid vector into the pWS-Green vector (constructed in our laboratory), between the NcoI and NotI sites. The integrity of the construct was confirmed by automated DNA sequencing.
To express scFv-EmGFP, the sAFH-3e3-EmGFP vector was transformed into E. coli SHuffle T7 Express. A single colony of E. coli harboring recombinant plasmid was inoculated into 5 mL of LB media containing 100 μg mL–1 ampicillin and cultured at 30 °C with shaking at 250 rpm overnight. 4 mL of this overnight culture was used to inoculate 400 mL of LB medium containing 100 μg mL–1 ampicillin. Cells were cultured at 30 °C until an OD600 of 0.9 was reached. Then, the culture was induced with 0.4 mM IPTG and grown with shaking at 25 °C, 250 rpm for 16 h.
The cell pellet was harvested by centrifugation at 7455g for 10 min and resuspended in binding buffer (20 mM Tris–HCl, 500 mM NaCl, and 20 mM imidazole, pH 7.4) containing 1 mg mL–1 lysozyme. After adding 1 mM phenylmethylsulfonyl fluoride (PMSF), cells were disrupted by intermittent sonication at 40% amplitude for 5 min on ice using 30 s pulse and 30 s break for cooling. The cell debris was removed by centrifugation at 10,000g for 30 min at 4 °C, followed by filtering through a 0.45 μm filter. The antibody was purified by the ÄKTA start protein purification system (GE Healthcare, USA), using a 1 mL His-Trap column (GE Healthcare, USA) pre-equilibrated with 10 column bed volume (CV) of binding buffer (20 mM Tris–HCl, 500 mM NaCl, 20 mM imidazole, pH 7.4) before the clear supernatant containing the crude antibody was loaded. Then, the column was washed with 20 CV of the same buffer. The scFv-EmGFP was eluted with 20 CV of a linear gradient (0–100%) of imidazole by mixing binding buffer with elution buffer (20 mM Tris–HCl, 500 mM NaCl, 500 mM imidazole, pH 7.4 buffer). Fractions containing the scFv-EmGFP fusion protein were pooled and exchanged by dialysis into PBS buffer at 4 °C, using 10 kDa MWCO Snakeskin dialysis tubing (ThermoFisher Scientific, USA) at 4 °C.
Fluorescence-Linked Immunosorbent Assay
Binding activity of sAFH-3e3-EmGFP antibodies was determined by FLISA. Black Nunc-Immuno 96 well plates (Nunc, Denmark) were coated with 0.25 μg of AFB1-BSA in 100 μL 1× PBS. After incubation at 4 °C overnight, the plates were blocked with 2% (w/v) MPBS at room temperature for 1 h. The plate was washed three times with PBS. The optimal dilution of scFv-emGFP that showed a linear relationship by indirect ELISA was pre-incubated with increasing amounts of soluble AFB1 ranging from 0.762 to 5,000 ng mL–1 at 37 °C for 30 min before adding into previously coated wells of the FLISA plates. After that, the plate was washed three times with 0.05% (v/v) of Tween-20 in PBS (PBST), followed by twice washing with PBS. Finally, 100 μL of PBS was added into the plate wells. The fluorescence intensity was measured by a fluorescence-based microplate reader (ThermoFisher Scientific, USA). The excitation–emission wavelengths were set at 484–509 and 478–506 nm, respectively. Standard curves were plotted as F/F0 versus logarithm of analyte concentration. A half-maximum inhibition (IC50) was estimated at 50% F/F0. A detection limit was estimated at 70% F/F0.
Passive Adsorption of Antibodies to Colloidal Gold
Colloidal gold at a particle size of 40 nm (BBI solutions, UK) was used in this study. For conjugation, 5 μg mL–1 sAFH-3e3 IgG and 5 μg mL–1 BSA in 100 μL of 0.2 M borate buffer, pH 8.55 were slowly mixed with 10 mL of colloidal gold solution. After 30 min, 100 μL of 1% (w/v) BSA was added to block the surface of colloidal gold particles and incubated for 30 min at room temperature. Then, the reaction was centrifuged at 9000g at 4 °C for 20 min before the supernatant was discarded. The pellet was resuspended in 20 mL of deionized water. The centrifugation step was repeated. Finally, 600 μL of deionized water containing 1.2% (w/v) BSA and 0.08% NaN3 (v/v) was used to resuspend the pellet for stabilizing the conjugate. The antibody–gold complex was diluted with conjugation buffer to 5–10% (v/v) before use.
Passive Adsorption of Antibodies to Latex Microspheres
The adsorption of purified scFv-Fc and IgG to latex microspheres was modified from a previously described method.9 Different sizes of green latex microspheres (70 and 169 nm; Merk Millipore, Germany) were tested for conjugation with scFv-Fc and IgG. Fifty microliters of green latex microspheres were diluted in 450 μL of borate buffer (10 mM borate buffer, pH 8.7) and mixed by vortexing. After centrifugation at 10,000g for 5 min, the supernatant was carefully discarded. The pellet was resuspended in 250 μL of 10 mM borate buffer. Then, an equal volume of 0.05 mg mL–1 of sAFH-3e3 antibody in the same borate buffer was added and incubated for 90 min at room temperature with rocking. After incubation, 10% (w/v) BSA was added to block nonspecific binding and further incubated for 45 min. Then, the particles were washed with 10 mM borate buffer and centrifuged at 10000g, 20 °C for 5 min. After that, the pellet was resuspended in 500 μL of 100 mM borate buffer, pH 8.7. The antibody-latex particle complex was diluted at 1:3 with 10 mM borate buffer before use.
Analysis of Corn Samples Spiked with AFB1 Using scFv-AP by Competitive ELISA
Two grams of the corn sample containing 0 ppb AFB1 (Trilogy, USA) were mixed with 20 mL of TBST or 70% (v/v) methanol in a glass bottle. This was then vortexed for 3 min and allowed to stand to let the matrix settle for 10–15 min. After that the solution was filtered through Whatman filter paper no. 1. Various concentrations of AFB1 standard (4, 10, 20, 40, 100, 200, and 400 ng mL–1) were spiked into different tubes of the filtered solution. These were diluted in TBST or 70% (v/v) methanol at a ratio of 1:3. Analysis was performed by competitive ELISA, using scFv-AP as described above, except that the spiked AFB1 samples were used instead of AFB1 standards. ELISA plate wells were coated with 0.25 μg of AFB1-BSA, and the scFv-AP at a ratio of 1:20 was used. A competitive curve between A/A0 and concentration of spiked AFB1 was plotted. The IC50 and the limit of detection was estimated at 50% A/A0 and a detection limit was estimated at 70% A/A0, respectively.
Detection of AFB1 Contaminated in Corn Samples Using Lateral Flow Immunoassay
AgraStrip Aflatoxin WATEX Quantitative Tests were provided by Romer Labs Division Holding GmbH (Tulln, Austria). The antibodies conjugated with colloidal gold or latex particles were used instead of the conjugated antibody from the company. The detection of aflatoxin was done according to the instructions from the test kit. Here, the LFIA is based on a competitive ELISA. The test strip contained mouse monoclonal antibody and aflatoxin-conjugated BSA on the control line and test line, respectively (Figure S6). Gold and latex particle complexes were diluted with their conjugation buffer before use as described above. For LFIA, 10 g of the corn sample (Romer labs, Austria) was weighed into a bag (provided by Romer labs) and 20 mL of PBS buffer or 70% (v/v) methanol was added. The bags were shaken for 2 min in PBS extraction buffer or 70% (v/v) methanol. After that, the corn samples were settled by standing the bag for 2 min. Before testing with the lateral flow assay, the extraction sample solution from the bag was diluted in 1:10 with PBS buffer. Then, 50 μL of different concentrations of AFB1 (100, 50, 25, and 6.25 ppb; Romer Labs) was mixed with 50 μL of antibody-latex complex or colloidal gold conjugated-antibody in the microwell by pipetting up and down. One test strip was put into one microwell for 3 min. The signal was measured using an AgraVision Reader (Romer labs, Tulln, Austria). A competitive curve between A/A0 and logarithm of concentration of spiked AFB1 was plotted. A half-maximum inhibition (IC50) was estimated at 50% A/A0, and a detection limit was estimated at 70% A/A0.
Acknowledgments
This work was supported by Suranaree University of Technology (SUT), grant number SUT3-304-62-12-17; Thailand Science Research and Innovation (TSRI), grant number RTA6180012; Ministry of Higher Education, Science, Research and Innovation (MHESI), Thailand, grant no. CRP550701085; and Ernst Mach Grant, Nachbetreuungsstipendium/EZA from Federal Ministry of Education, Science and Research (BMBWF), grant no. ICM-2019-14954. K.R. were supported by SUT Full-time Doctoral Research [grants no.61/13/2561]. We would like to thank Dr. Milica Sevo for her help with the design and construction of the IgG vector. We are grateful to Romer Laboratories Co., especially Julia Mayer, in Tulln, Austria, for the facility and expertise that involve aflatoxin analysis and lateral flow assay. We also greatly appreciate Dr. Jenny Fitzgerald, Dr. Paul Leonard, and Dr Hui Ma from Dublin City University for their excellent technical support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c03044.
Diagram of the construction of IgG and scFv expression vectors, SDS–PAGE analysis of purified antibody, cross-reactivity of scFv-AP to aflatoxins and nonrelated mycotoxin, the optimization of antibody-latex conjugation, and a cartoon of lateral flow assay for aflatoxins detection (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Sompunga P.; Pruksametanan N.; Rangnoi K.; Choowongkomon K.; Yamabhai M. Generation of human and rabbit recombinant antibodies for the detection of Zearalenone by phage display antibody technology. Talanta 2019, 201, 397–405. 10.1016/j.talanta.2019.04.034. [DOI] [PubMed] [Google Scholar]
- Wan L.; Zhu S.; Zhu J.; Yang H.; Li S.; Li Y.; Cheng J.; Lu X. Production and characterization of a CD25-specific scFv-Fc antibody secreted from Pichia pastoris. Appl. Microbiol. Biotechnol. 2013, 97, 3855–3863. 10.1007/s00253-012-4632-9. [DOI] [PubMed] [Google Scholar]
- Wang S.-H.; Du X.-Y.; Huang Y.-M.; Lin D.-S.; Hart P. L.; Wang Z.-H. Detection of deoxynivalenol based on a single-chain fragment variable of the antideoxynivalenol antibody. FEMS Microbiol. Lett. 2007, 272, 214–219. 10.1111/j.1574-6968.2007.00765.x. [DOI] [PubMed] [Google Scholar]
- Yuan Q.; Clarke J. R.; Zhou H. R.; Linz J. E.; Pestka J. J.; Hart L. P. Molecular cloning, expression, and characterization of a functional single-chain Fv antibody to the mycotoxin zearalenone. Appl. Environ. Microbiol. 1997, 63, 263–269. 10.1128/aem.63.1.263-269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shephard G. S. Impact of mycotoxins on human health in developing countries. Food Addit. Contam. 2008, 25, 146–151. 10.1080/02652030701567442. [DOI] [PubMed] [Google Scholar]
- Yamabhai M.; Rangnoi K.; Sompunga P.; O’Kennedy R.. Novel Recombinant Antibody and Protein-based Approaches for Analysis of Food and Food Contaminants with Particular Relevance to Asia. Rapid Antibody-based Technologies in Food Analysis; The Royal Society of Chemistry, 2019; Chapter 10, pp 195–222. [Google Scholar]
- Goryacheva I. Y.; Saeger S. d.; Eremin S. A.; Peteghem C. V. Immunochemical methods for rapid mycotoxin detection: Evolution from single to multiple analyte screening: A review. Food Addit. Contam. 2007, 24, 1169–1183. 10.1080/02652030701557179. [DOI] [PubMed] [Google Scholar]
- Li P.; Zhang Q.; Zhang D.; Guan D.; Liu D. X.; Fang S.; Wang X.; Zhang W.. Aflatoxin measurement and analysis. Aflatoxins-Detection, Measurement and Control; IntechOpen, 2011. [Google Scholar]
- Zheng M. Z.; Richard J. L.; Binder J. A review of rapid methods for the analysis of mycotoxins. Mycopathologia 2006, 161, 261–273. 10.1007/s11046-006-0215-6. [DOI] [PubMed] [Google Scholar]
- Masinde L. A.; Sheng W.; Xu X.; Zhang Y.; Yuan M.; Kennedy I. R.; Wang S. Colloidal gold based immunochromatographic strip for the simple and sensitive determination of aflatoxin B1 and B2 in corn and rice. Microchim. Acta 2013, 180, 921–928. 10.1007/s00604-013-1008-5. [DOI] [Google Scholar]
- Berman H.; Henrick K.; Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003, 10, 980. 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Schrödinger L.The PyMOL molecular graphics system, version 1.8, November: 2015.
- Chan C. E.; Chan A. H.; Lim A. P.; Hanson B. J. Comparison of the efficiency of antibody selection from semi-synthetic scFv and non-immune Fab phage display libraries against protein targets for rapid development of diagnostic immunoassays. J. Immunol. Methods 2011, 373, 79–88. 10.1016/j.jim.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinwand M.; Droste P.; Frenzel A.; Hust M.; Dübel S.; Schirrmann T. The influence of antibody fragment format on phage display based affinity maturation of IgG. MAbs 2014, 6, 204–218. Taylor & Francis. 10.4161/mabs.27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangnoi K.; Choowongkomon K.; O’Kennedy R.; Rüker F.; Yamabhai M. Enhancement and analysis of human antiaflatoxin b1 (AFB1) scFv antibody–ligand interaction using chain shuffling. J. Agric. Food Chem. 2018, 66, 5713–5722. 10.1021/acs.jafc.8b01141. [DOI] [PubMed] [Google Scholar]
- Menzel C.; Schirrmann T.; Konthur Z.; Jostock T.; Dübel S. Human antibody RNase fusion protein targeting CD30+ lymphomas. Blood 2008, 111, 3830–3837. 10.1182/blood-2007-04-082768. [DOI] [PubMed] [Google Scholar]
- Ames R. S.; Tornetta M. A.; Deen K.; Jones C. S.; Swift A. M.; Ganguly S. Conversion of murine Fabs isolated from a combinatorial phage display library to full length immunoglobulins. J. Immunol. Methods 1995, 184, 177–186. 10.1016/0022-1759(95)00086-p. [DOI] [PubMed] [Google Scholar]
- Huls G. A.; Heijnen I. A. F. M.; Cuomo M. E.; Koningsberger J. C.; Wiegman L.; Boel E.; van der Vuurst de Vries A.-R.; Loyson S. A. J.; Helfrich W.; van Berge Henegouwen G. P.; van Meijer M.; de Kruif J.; Logtenberg T. A recombinant, fully human monoclonal antibody with antitumor activity constructed from phage-displayed antibody fragments. Nat. Biotechnol. 1999, 17, 276–281. 10.1038/7023. [DOI] [PubMed] [Google Scholar]
- Liu B.; Conrad F.; Roth A.; Drummond D. C.; Simko J. P.; Marks J. D. Recombinant full-length human IgG1s targeting hormone-refractory prostate cancer. J. Mol. Med. (Berl.) 2007, 85, 1113–1123. 10.1007/s00109-007-0208-z. [DOI] [PubMed] [Google Scholar]
- Warszawski S.; Borenstein Katz A.; Lipsh R.; Khmelnitsky L.; Ben Nissan G.; Javitt G.; Dym O.; Unger T.; Knop O.; Albeck S.; Diskin R.; Fass D.; Sharon M.; Fleishman S. J. Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces. PLoS Comput. Biol. 2019, 15, e1007207 10.1371/journal.pcbi.1007207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabhai M.; Kay B. K. Mapping protein-protein interactions with alkaline phosphatase fusion proteins. Methods Enzymol. 2001, 332, 88–102. 10.1016/s0076-6879(01)32194-8. [DOI] [PubMed] [Google Scholar]
- Yamabhai M.BAP-fusion: A versatile molecular probe for biotechnology research. Biotechnology: Research, technology and applications; Nova Science Publishers, 2008, pp 327–345. [Google Scholar]
- Pohl S. C.; Schwarz S.; Frenzel A.; Schirrmann T. A cassette vector system for the rapid cloning and production of bispecific tetravalent antibodies. Antibodies 2012, 1, 19–38. 10.3390/antib1010019. [DOI] [Google Scholar]
- Moghaddam A.; Løbersli I.; Gebhardt K.; Braunagel M.; Marvik O. J. Selection and characterisation of recombinant single-chain antibodies to the hapten Aflatoxin-B1 from naive recombinant antibody libraries. J. Immunol. Methods 2001, 254, 169–181. 10.1016/s0022-1759(01)00413-6. [DOI] [PubMed] [Google Scholar]
- Crone D. E.; Huang Y.-M.; Pitman D. J.; Schenkelberg C.; Fraser K.; Macari S.; Bystroff C.. GFP-based biosensors. State of the Art in Biosensors-General Aspects; IntechOpen, 2013. [Google Scholar]
- Magnusson K.-E.; Bartonek E.; Nordkvist E.; Sundqvist T.; Asbrink E. Fluorescence-linked immunosorbent assay (FLISA) for quantification of antibodies to food antigens. Immunol. Invest. 1987, 16, 227–240. 10.3109/08820138709030578. [DOI] [PubMed] [Google Scholar]
- Chen M.; Ding S.; Wen K.; Xie S.; Wang Q.; Pei X.; Xie J.; Wang Z.; Jiang H. Development of a fluorescence-linked immunosorbent assay for detection of avermectins using a fluorescent single-domain antibody. Anal. Methods 2015, 7, 3728–3734. 10.1039/c5ay00305a. [DOI] [Google Scholar]
- Ducancel F.; Muller B. H. Molecular engineering of antibodies for therapeutic and diagnostic purposes. MAbs 2012, 4, 445–457. Taylor & Francis, 10.4161/mabs.20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller K. E.; Tessier P. M. Advances in antibody design. Annu. Rev. Biomed. Eng. 2015, 17, 191–216. 10.1146/annurev-bioeng-071114-040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W.-K.; Kim S.-G.; Seo J.-H. Affinity maturation of single-chain variable fragment specific for aflatoxin B(1) using yeast surface display. Food Chem. 2015, 188, 604–611. 10.1016/j.foodchem.2015.04.117. [DOI] [PubMed] [Google Scholar]
- Li X.; Li P.; Zhang Q.; Li Y.; Zhang W.; Ding X. Molecular characterization of monoclonal antibodies against aflatoxins: a possible explanation for the highest sensitivity. Anal. Chem. 2012, 84, 5229–5235. 10.1021/ac202747u. [DOI] [PubMed] [Google Scholar]
- Baran D.; Pszolla M. G.; Lapidoth G. D.; Norn C.; Dym O.; Unger T.; Albeck S.; Tyka M. D.; Fleishman S. J. Principles for computational design of binding antibodies. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 10900–10905. 10.1073/pnas.1707171114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Ding H.; Gu Z.; Zhao J.; Chen H.; Tian F.; Chen Y. Q.; Zhang H.; Chen W. Selection of single chain fragment variables with direct coating of aflatoxin B1 to enzyme-linked immunosorbent assay plates. J. Agric. Food Chem. 2009, 57, 8927–8932. 10.1021/jf9019536. [DOI] [PubMed] [Google Scholar]
- Rangnoi K.; Jaruseranee N.; O’Kennedy R.; Pansri P.; Yamabhai M. One-step detection of Aflatoxin-B 1 using scFv-alkaline phosphatase-fusion selected from human phage display antibody library. Mol. Biotechnol. 2011, 49, 240–249. 10.1007/s12033-011-9398-2. [DOI] [PubMed] [Google Scholar]
- Li X.; Li P.; Lei J.; Zhang Q.; Zhang W.; Li C. A simple strategy to obtain ultra-sensitive single-chain fragment variable antibodies for aflatoxin detection. RSC Adv. 2013, 3, 22367–22372. 10.1039/c3ra42706d. [DOI] [Google Scholar]
- Edupuganti S. R.; Edupuganti O. P.; Hearty S.; O’Kennedy R. A highly stable, sensitive, regenerable and rapid immunoassay for detecting aflatoxin B1 in corn incorporating covalent AFB1 immobilization and a recombinant Fab antibody. Talanta 2013, 115, 329–335. 10.1016/j.talanta.2013.05.012. [DOI] [PubMed] [Google Scholar]
- He T.; Wang Y.; Li P.; Zhang Q.; Lei J.; Zhang Z.; Ding X.; Zhou H.; Zhang W. Nanobody-based enzyme immunoassay for aflatoxin in agro-products with high tolerance to cosolvent methanol. Anal. Chem. 2014, 86, 8873–8880. 10.1021/ac502390c. [DOI] [PubMed] [Google Scholar]
- Gandhi S.; Banga I.; Maurya P. K.; Eremin S. A. A gold nanoparticle-single-chain fragment variable antibody as an immunoprobe for rapid detection of morphine by dipstick. RSC Adv. 2018, 8, 1511–1518. 10.1039/c7ra12810j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfossi L.; D’Arco G.; Calderara M.; Baggiani C.; Giovannoli C.; Giraudi G. Development of a quantitative lateral flow immunoassay for the detection of aflatoxins in maize. Food Addit. Contam. 2011, 28, 226–234. 10.1080/19440049.2010.540763. [DOI] [PubMed] [Google Scholar]
- Xie Y.-J.; Yang Y.; Kong W.-J.; Yang S.-H.; Yang M.-H. Application of nanoparticle probe-based lateral flow immunochromatographic assay in mycotoxins detection. Chin. J. Anal. Chem. 2015, 43, 618–628. 10.1016/S1872-2040(15)60821-0. [DOI] [Google Scholar]
- Garcia V. S.; Guerrero S. A.; Gugliotta L. M.; Gonzalez V. D. G. A lateral flow immunoassay based on colored latex particles for detection of canine visceral leishmaniasis. Acta Trop. 2020, 212, 105643. 10.1016/j.actatropica.2020.105643. [DOI] [PubMed] [Google Scholar]
- Jallow A.; Xie H.; Tang X.; Qi Z.; Li P. Worldwide aflatoxin contamination of agricultural products and foods: From occurrence to control. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2332–2381. 10.1111/1541-4337.12734. [DOI] [PubMed] [Google Scholar]
- Bui-Klimke T. R.; Guclu H.; Kensler T. W.; Yuan J.-M.; Wu F. Aflatoxin regulations and global pistachio trade: insights from social network analysis. PloS One 2014, 9, e92149 10.1371/journal.pone.0092149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norlia M.; Jinap S.; Nor-Khaizura M. A. R.; Radu S.; Samsudin N. I. P.; Azri F. A. Aspergillus section Flavi and Aflatoxins: Occurrence, Detection, and Identification in Raw Peanuts and Peanut-Based Products Along the Supply Chain. Front. Microbiol. 2019, 10, 2602. 10.3389/fmicb.2019.02602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tole S.; Mukovozov I. M.; Huang Y.-W.; Magalhaes M. A. O.; Yan M.; Crow M. R.; Liu G. Y.; Sun C. X.; Durocher Y.; Glogauer M.; Robinson L. A. The axonal repellent, Slit2, inhibits directional migration of circulating neutrophils. J. Leukoc. Biol. 2009, 86, 1403–1415. 10.1189/jlb.0609391. [DOI] [PubMed] [Google Scholar]
- Pershad K.; Sullivan M. A.; Kay B. K. Drop-out phagemid vector for switching from phage displayed affinity reagents to expression formats. Anal. Biochem. 2011, 412, 210–216. 10.1016/j.ab.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.