Abstract
Nucleus-encoded proteins interact with cis-acting elements in chloroplast transcripts to promote RNA stability and translation. We have analyzed the structure and function of three such elements within the Chlamydomonas petD 5′ untranslated region; petD encodes subunit IV of the cytochrome b6/f complex. These elements were delineated by linker-scanning mutagenesis, and RNA secondary structures were investigated by mapping nuclease-sensitive sites in vitro and by in vivo dimethyl sulfate RNA modification. Element I spans a maximum of 8 nucleotides (nt) at the 5′ end of the mRNA; it is essential for RNA stability and plays a role in translation. This element appears to form a small stem-loop that may interact with a previously described nucleus-encoded factor to block 5′→3′ exoribonucleolytic degradation. Elements II and III, located in the center and near the 3′ end of the 5′ untranslated region, respectively, are essential for translation, but mutations in these elements do not affect mRNA stability. Element II is a maximum of 16 nt in length, does not form an obvious secondary structure, and appears to bind proteins that protect it from dimethyl sulfate modification. Element III spans a maximum of 14 nt and appears to form a stem-loop in vivo, based on dimethyl sulfate modification and the sequences of intragenic suppressors of element III mutations. Furthermore, mutations in element II result in changes in the RNA structure near element III, consistent with a long-range interaction that may promote translation.
Chloroplasts are photosynthetic organelles that contain their own DNA and gene expression machinery. Chloroplast genes are dependent on nucleus-encoded proteins that are imported into chloroplasts and interact with mRNAs to control RNA maturation (5, 6, 11, 34, 46), stability (19, 28, 42, 48, 52, 54), and translation (9, 56, 64, 71). In several cases, these factors have been shown to be required for the expression of a single chloroplast gene, acting through the 5′ untranslated region (UTR) (19, 54, 71). Biochemical studies have identified RNA-binding proteins that associate with 5′ UTRs in vitro, and these proteins have been proposed to have roles in gene expression (17, 30, 39, 72). In general, these RNA-binding proteins do not appear to be sequence specific, and their roles in chloroplast gene expression remain to be confirmed.
Many aspects of chloroplast gene expression reflect their bacterial ancestry (for a review, see reference 65). A significant difference, however, is that nucleus-encoded proteins activate chloroplast translation, whereas Escherichia coli regulatory factors generally repress translation. Furthermore, while in E. coli the Shine-Dalgarno (SD) sequence base pairs with 16S rRNA, many chloroplast transcripts do not contain SD-like sequences. In most (but see references 32 and 47) cases where SD-like sequences have been mutated, translation is impaired mildly or not at all (24, 47, 60). One possibility is that instead of SD sequences, chloroplast 5′ UTRs contain sequence elements that function as binding sites for gene-specific translational activators, which interact with ribosomes to initiate translation, akin to the mechanisms described for yeast mitochondrial translation (for a review, see reference 13).
To study the mechanisms that govern chloroplast RNA stability and translation, we have focused on the petD gene, which encodes subunit IV (SUIV) of the cytochrome b6/f complex. This thylakoid membrane complex functions in photosynthetic electron transport, and nuclear and chloroplast mutants that fail to accumulate SUIV are nonphotosynthetic. petD mRNA is 900 nucleotides (nt) in length, including a 362-nt-long 5′ UTR, and can be transcribed from either the petD promoter or the upstream petA promoter (see Fig. 1) (68). The mature 5′ end of the transcript is generated by RNA processing, an event that may be required for the accumulation of functional mRNA. RNA processing is mediated by sequences located between nt 1 and 25 with respect to the mature RNA 5′ end (60, 62). This part of the 5′ UTR, which we have termed element I, is necessary for the accumulation of the mature mRNA (61, 68) and is sufficient to stabilize chimeric mRNAs (60, 61). Molecular genetic data suggest that it interacts with the product of the nuclear gene MCD1, which stabilizes the transcript by blocking 5′→3′ exoribonuclease activity (19).
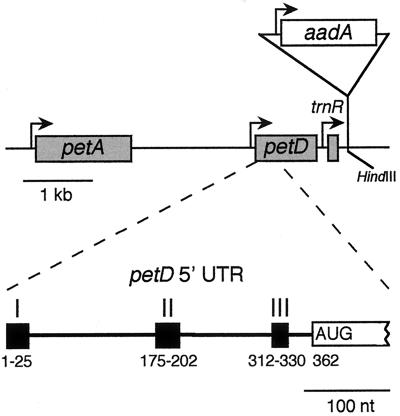
FIG. 1.
The Chlamydomonas chloroplast petD region. The upper diagram shows that petD is located between petA and trnR, with known promoters shown as bent arrows. Shaded boxes indicate transcribed regions. The aadA selectable marker cassette was inserted into a downstream HindIII site to create some transformants (see Materials and Methods). The lower diagram shows the three petD 5′ UTR elements studied in this paper. Nucleotide 1 represents the 5′ end of the mature mRNA, whereas the AUG initiation codon starts at position 362.
Deletion analysis of the petD 5′ UTR identified two additional elements required for petD expression (61). Elements II and III were localized to nt 172 through 202 and 312 through 330, respectively. Deletion of either element resulted in chloroplasts that accumulated petD mRNA but not SUIV, suggesting that both elements are essential for translation. Previously, nucleus-encoded suppressors were identified that overcame an element III mutation, indicating that nucleus-encoded factors are involved in SUIV translation initiation (60).
The mechanisms by which nucleus-encoded proteins activate translation or stabilize mRNAs in chloroplasts are incompletely understood. Relevant cis elements have only been roughly mapped in chloroplast transcripts, and the questions of how they provide specificity and interact with nucleus-encoded proteins have not been addressed. To define the essential nucleotides in the three petD elements more precisely, we introduced mutations into these regions and tested their effects in vivo. To investigate the RNA secondary structures, we used dimethyl sulfate (DMS) to modify petD RNA in vivo and compared these results to RNA modified by DMS or cleaved by specific ribonucleases in vitro, in the absence of proteins. Finally, strains with mutations in element III were used to isolate intragenic suppressors that carry point mutations and support a role for the proposed element III structure.
MATERIALS AND METHODS
Plasmids and chloroplast transformation.
Linker-scanning (LS) mutations were introduced into the wild-type (WT) Chlamydomonas petD pD501 plasmid (60) by site-directed mutagenesis by a traditional method (43) or by PCR. pD501 also contained an engineered BglII restriction endonuclease site at the +25 position of petD, plus flanking chloroplast DNA upstream (600 bp) and downstream (2 kb) of petD. Mutations in elements I and II were made by traditional site-directed mutagenesis with antisense primers with the changes indicated (see Fig. 2 and 3). The tightly linked BglII site at position +25 was used to track the LS mutation in transformants. The 8-base mutations in element II introduced NotI sites, while mutations in element III were made by PCR amplification and introduced either 4-base HaeIII or 6-base NcoI sites.
FIG. 2.
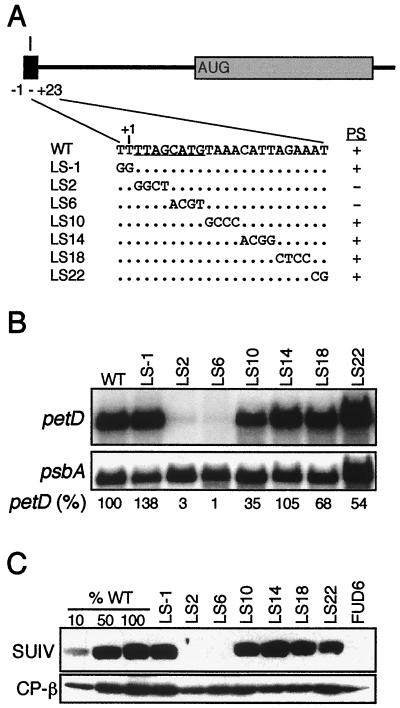
Functional analysis of element I. (A) WT sequences of element I and the seven linker-scanning (LS) mutations, named for the 5′-most changed nucleotide. Unchanged nucleotides are shown as dots. The photosynthetic (PS) phenotypes (+/−) of the transformants are indicated at the right, and the deduced critical nucleotides of element I are underlined in the WT sequence. (B) Filter blots of total RNA (10 μg/lane) from the indicated strains were hybridized with petD or psbA probes. The petD mRNA signal was normalized to psbA; the WT level was set to 100%. The values shown are averages of three independent RNA samples. (C) Immunoblots of total proteins from the indicated strains were challenged with antibodies raised against SUIV or the chloroplast ATP synthase β subunit (CP-β). To estimate quantity, 10, 50, and 100% of WT protein samples were included, along with the negative control FUD6, which fails to accumulate SUIV due to a deletion upstream of petD (68).
FIG. 3.
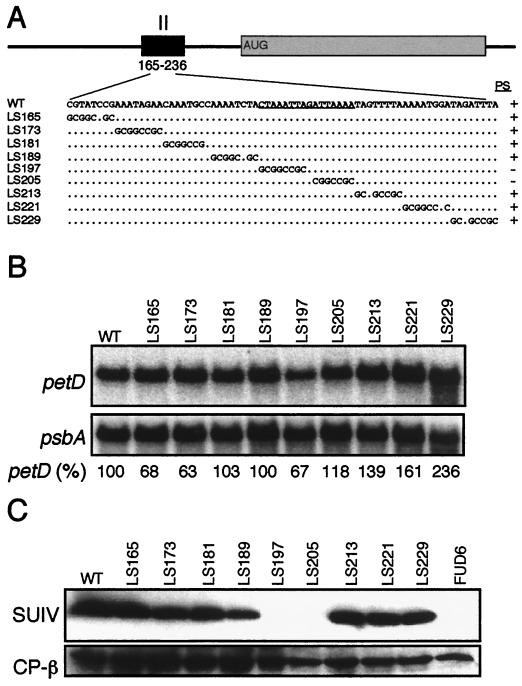
Functional analysis of element II. (A) WT and LS mutant sequences, as described in the legend to Fig. 2A. Total RNA (B) and total protein (C) samples were analyzed as described in the legend to Fig. 2.
The LS309, LS313, and LS317 element III mutants were amplified from pD501 with the sense primer WS13, which anneals at the 5′ end between positions −2 and +24 just upstream of the BglII site, and a downstream antisense primer that contained both the LS mutation (HaeIII site) and the endogenous HindIII site at nt 321 to 326 (see Fig. 4A). The BglII-HindIII fragments, which contained most of the 5′ UTR and the LS mutations, were substituted for the corresponding WT fragments in pD501. LS327 and LS331 of element III were PCR amplified from pD501 with a sense primer that contained both the endogenous HindIII site and LS mutations and the WS4 antisense primer (61), which anneals downstream of the petD gene. The HindIII-PstI fragments, which contained the LS mutations and the 5′ half of the coding region, were substituted for the corresponding fragment in pD501. LS321 was made by a PCR strategy in which the endogenous HindIII site was replaced with an NcoI site. First, the upstream PCR fragment was amplified from pD501 with the sense primer WS13, which anneals at the petD 5′ end, and an antisense primer with the mutations at nt 321 to 326 (NcoI site), replacing the HindIII site. Next, the downstream PCR fragment was amplified from pD501 with a sense primer with the same mutations at nt 321 to 326 and the WS4 antisense primer (61). These intermediate PCR fragments were fused at the NcoI site, and from this fused clone, the BglII-PstI fragment, with most of the 5′ UTR and the 5′ half of the coding region, was isolated and inserted into the BglII-PstI sites of pD501. All final LS mutant clones were confirmed by sequencing. To make the petD-aadA transforming plasmid, the aadA gene cassette (26) was isolated as an EcoRV-SmaI fragment and inserted into a Klenow-filled HindIII site downstream of petD and trnR (see Fig. 1). The aadA cassette was oriented in tandem with petD.
FIG. 4.
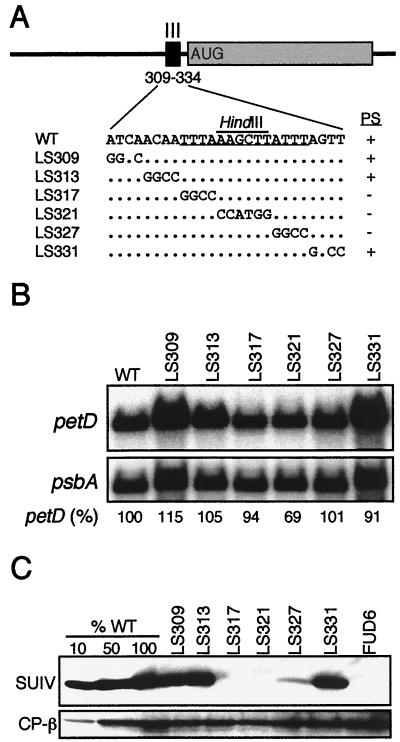
Functional analysis of element III. (A) WT and LS mutant sequences, as described in the legend to Fig. 2A. A HindIII site used for cloning is overlined (see Materials and Methods). Total RNA (B) and total protein (C) samples were analyzed as described in the legend to Fig. 2.
Mutant petD genes were introduced by particle bombardment (38) into the nonphotosynthetic Chlamydomonas strain FUD6, which has a 236-bp deletion at the 5′ end of petD (68). Photoautotrophic transformants were streaked for single colonies on minimal medium (lacking acetate) (29) until homoplasmic strains were recovered. Plasmids in which the LS mutation compromised petD function were unable to rescue the mutation in FUD6. These plasmids, bearing the aadA cassette, were introduced into the WT strain P17. Transformants were initially selected in low light on TAP medium (containing acetate) (29) supplemented with 100 μg of spectinomycin per ml. Between 5 and 25% of the primary transformants contained both the aadA gene and the petD LS mutation. These cotransformants were subsequently streaked for single colonies on medium with 400 μg of spectinomycin per ml until the strains were homoplasmic for the introduced LS mutation, as confirmed by PCR, DNA filter hybridizations, and sequencing. The photosynthetic phenotypes of two independent transformants for each LS mutation were determined by measuring the chlorophyll fluorescence transients of cells grown on TAP medium (74) and by plating strains on minimal medium to determine acetate dependence.
RNA and protein analyses.
Total RNA was isolated from Chlamydomonas liquid cultures as previously described (21). RNA was separated in 1.2% agarose–3% formaldehyde gels, blotted to GeneScreen membranes (Du Pont), and crosslinked to the membrane by exposure to ultraviolet light (UV Stratalinker 1800; Stratagene) (15). RNA filters were hybridized (12) with petD and psbA DNA probes (31). Radioactive bands were visualized and quantified with a PhosphorImager (Molecular Dynamics). Total proteins were isolated and analyzed on immunoblots by enhanced chemiluminescence as previously described (8).
RNA secondary structure analyses.
5′-end-labeled petD 5′ UTR RNA was prepared essentially as previously described (66). For transcription templates, PCR fragments were amplified from WT, LS2, LS197, and LS321 DNAs with a sense primer (5′-CTAATACGACTCACTATAGGGTTTAGCATGTAAACATTAG), which anneals at the petD 5′ end, and an antisense primer (WS11) (61), which anneals at the initiation codon, including 5 bases downstream of AUG. The 5′ half of the sense primer contained the T7 promoter (underlined) (4), which added three Gs to the 5′ end of the RNA, while the 3′ portion annealed to petD beginning at nt +1. Because this primer overlaps the LS2 mutation, a primer with the LS2 sequence and upstream T7 promoter was used for that template. Two femtomoles (6.6 × 103 cpm) of RNA was treated with 5.0 U of RNase T1 (Gibco-BRL) in 10 μl of in vitro nuclease digestion buffer (10 mM HEPES [pH 7.9], 230 mM KCl, 81 mM MgCl2, 0.05 mM EDTA, 41 mM dithiothreitol, 8% glycerol) for 1 min at 25°C. Reactions were stopped by adding 1 μl of yeast tRNA (10 μg/μl; Sigma), 2 μl of 0.1 M aurintricarboxylic acid, and 1.5 μl of 3 M sodium acetate. Digestion products were ethanol precipitated and analyzed in either 15 or 4% sequencing gels; they were compared to a nucleotide ladder generated by boiling 5′-end-labeled petD RNA in 135 mM sodium bicarbonate, 15 mM sodium carbonate, and 3 mM EDTA for 5 min (1).
In vivo DMS treatments were similar to those described for Tetrahymena (73). Ten milliliters of Chlamydomonas cells grown in TAP to a density of 2 × 106 cells/ml was centrifuged for 4 min at 1,500 × g. The cell pellet was resuspended in 1 ml of DMS buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 3 mM CaCl2), and 5 μl of 7.9 M DMS (Aldrich Chemical Co.) was added. The reaction was incubated for 5 min at 25°C, then quenched with 50 μl of β-mercaptoethanol. Cells were collected by centrifugation, and total RNA was extracted as previously described (21). Twenty-five micrograms of this RNA was used in primer extension reactions (62) with gel-purified antisense WS5 (62), D246 (5′-TTAGATCTACAGAATTACATTTTACTTCTG), and WS10 (5′-TTGAGATCTCTGGATCGCTTAAATCAGG) primers. Products were analyzed in 7% sequencing gels and sized by comparison to corresponding petD sequencing ladders.
RNA was treated with DMS in vitro by adding 75 μg of total RNA extracted from untreated cells to 200 μl of in vitro nuclease buffer, adding 1 μl of 7.9 M DMS, incubating for 5 min at 25°C, and then quenching the reaction with 10 μl of β-mercaptoethanol. RNA was precipitated and washed with ethanol to remove the DMS, and 25 μg of RNA was used for the primer extension. The free energies of RNA secondary structures were estimated with the mFold computer software (75, 76) with folding conditions of 25°C and 1 M NaCl.
Isolation and analysis of chloroplast-encoded suppressors.
A total of 109 viable cells of each nonphotosynthetic LS mutant was plated on minimal medium and selected for photosynthetic growth under high light (230 microeinsteins/m2/s). Suppressors (mating type plus [mt+]) were streaked for single colonies on minimal medium four times and then crossed (29) to WT cells (mt−) to identify chloroplast suppressors, for which all tetrad progeny were capable of photoautotrophic growth. The 5′ UTRs from these chloroplast suppressors were PCR amplified with the sense primer WS12 (31) and the antisense primer WS11 (60), and the products were cloned and sequenced. To control for mutations arising during PCR, two independently amplified PCR fragments were sequenced for each suppressor. For reporter gene constructs, the petD 5′ UTRs from LS317 and both suppressors were fused to the DG2 reporter gene which has the bacterial uidA coding region, encoding β-glucuronidase (GUS), and these constructs were introduced into Chlamydomonas chloroplasts as previously described (61). Fluorometric GUS activity assays were conducted on homoplasmic transformants as previously described (61); activities were measured relative to DG2, which contains the WT petD 5′ UTR.
RESULTS
LS mutations reveal short sequences essential for petD RNA stability and translation.
Figure 1 shows a map of the petD gene with the adjacent petA and trnR genes and a diagram of the petD 5′ UTR, highlighting the three elements roughly defined by our previous studies. To map these elements precisely, a series of LS mutations spanning elements I, II, and III were made by site-directed mutagenesis. Mutated petD genes and a linked selectable marker conferring antibiotic resistance were introduced into Chlamydomonas chloroplasts by particle bombardment (see Materials and Methods). In cases where LS mutations did not significantly alter petD expression, transformants were photosynthetic and could be selected on minimal medium. In cases where petD expression was severely compromised, transformants were selected for antibiotic resistance. These transformants failed to grow on minimal medium and displayed high chlorophyll fluorescence. Homoplasmy of the transformants was confirmed by PCR and DNA filter hybridizations.
For element I, seven LS mutations were made, spanning 24 nt; Fig. 2A shows their DNA sequences and photosynthetic phenotypes. In addition to the indicated sequence changes, a BglII site was added at position +25; this change affects neither petD mRNA abundance nor SUIV accumulation (60). LS mutation 1 (LS1) has a 2-nt mutation beginning at position −1 with respect to the 5′ end of petD mRNA and thus straddles the 5′ processing site. Only two mutants (LS2 and LS6) were nonphotosynthetic, defining the maximum size of element I as 8 nt.
To determine the size and abundance of petD mRNA in these strains, total RNA was blotted to filters and hybridized with a petD probe. Compared to WT cells, a dramatic decrease in the amount of petD mRNA was observed in LS2 and LS6, relative to the abundance of the chloroplast psbA transcript, which encodes the D1 protein of photosystem II (Fig. 2B). LS2 cells accumulated 3% of the WT amount of petD mRNA, while LS6 cells accumulated consistently less, approximately 1% of the WT amount. petD mRNA abundance was reduced to 35% of the WT amount in the adjacent LS10 mutant, but there was no effect on SUIV accumulation or photosynthetic growth (see below). Previously, we showed that deleting nt 1 to 270 did not affect the petD transcription rate but instead caused RNA instability (61). In addition, the redundant upstream petA promoter was shown to be sufficient to synthesize WT amounts of petD mRNA in absence of the petD promoter (68). Taken together, we conclude that petD mRNA decreased in LS2, LS6, and LS10 due to RNA instability.
To measure SUIV accumulation, total protein was used for immunoblot analysis with the chloroplast ATP synthase β subunit as a loading control. Figure 2C shows that LS2 and LS6 accumulated no detectable SUIV. When compared to a dilution series in which 1% of WT SUIV could be detected, no SUIV was seen in these strains (data not shown). Because LS2 and LS6 accumulate trace amounts of petD mRNA, we expected that SUIV would be detectable if RNA stability was the only function of element I. Since SUIV was not detected, our data suggest that element I also functions in translation initiation.
To analyze element II, nine LS mutations were made in which a NotI site replaced 8 bp within a previously identified 72-bp region. Figure 3A shows the DNA sequences and photosynthetic phenotypes of these strains. Two mutants (LS197 and LS205) were nonphotosynthetic, defining the maximum size of element II as 16 nt. RNA filter hybridizations (Fig. 3B) showed that all of the LS mutants in this region accumulated near-WT amounts of petD mRNA. Immunoblot analysis, however, showed that LS197 and LS205 accumulated no detectable SUIV (Fig. 3C), even in gels where 1% of the WT level of SUIV could be detected (data not shown). We conclude that element II is essential for SUIV translation but plays no obvious role in mRNA stability.
To analyze element III, six LS mutations were made that spanned 26 nt. Figure 4A shows the DNA sequences and photosynthetic phenotypes of these mutants, which carry either 4- or 6-bp changes. Three mutants (LS317, LS321, and LS327) were nonphotosynthetic, defining the maximum size of element III as 14 nt. RNA filter hybridizations (Fig. 4B) showed that all of these LS mutants accumulated near-WT amounts of petD mRNA. Immunoblot analyses, however, showed that LS317 and LS321 accumulated no detectable SUIV (Figure 4C), even in gels where 1% of the WT level of SUIV could be detected (data not shown). LS327 accumulated approximately 5% of the WT level of SUIV, a level insufficient to support photosynthesis (8). We conclude that element III is essential for SUIV translation and spans up to 14 nt, although the results with LS327 suggest that changes at the 3′ end of element III have a smaller impact on SUIV translation than changes elsewhere in element III.
RNA secondary structure analysis predicts a small stem-loop for element I.
It has previously been proposed that RNA secondary structures play important roles in chloroplast translation initiation (47, 58), but experimental tests of these hypotheses have been limited. To examine petD mRNA secondary structures and RNA-protein interactions, we used two experimental approaches. First, in vitro-synthesized, 5′-end-labeled petD transcripts were treated with ribonuclease (RNase) T1, which cleaves single-stranded G residues. In vitro RNase V1 treatment, which cleaves double-stranded RNA, was also performed, but the results were inconclusive due to the permissiveness of V1 (55). Second, petD RNA was modified in vivo by treating cells with DMS, which readily crosses cell membranes and methylates the N-1 position of cytosine and the N-3 position of adenine residues, if they are not blocked by base pairing or bound to proteins (51, 73). Residues methylated by DMS stop primer extension 1 nt downstream of the methylated site, yielding bands whose intensities correspond to the amount of methylation. Phenol-extracted total RNA was in vitro-modified with DMS, thus uncovering the influence of chloroplast proteins on RNA secondary structure. This approach has been used to determine the in vivo and in vitro secondary structures of ribosomal and messenger RNAs from both prokaryotes (3, 7, 51) and eukaryotes (18, 63, 69, 73).
To study the petD RNA structure around element I, extension was carried out with a primer which anneals 67 nt downstream of the 5′ end. As expected, the extension of untreated RNA yielded a single major product corresponding to the mature 5′ end (Fig. 5A, lane 2). When RNA from DMS-treated cells was analyzed, the major product also corresponded to the mature 5′ end, but numerous bands corresponding to methylation at A and C residues were also detected (Fig. 5A, lane 3). A4, A7, A11, and A12, which include element I, exhibited very little or no methylation, suggesting they are base paired or protected by tightly bound protein(s). C14 and A18-A22 were intermediately methylated, whereas A13, A15, and A24-A32 were heavily methylated, indicating that they are single stranded. No difference in the methylation pattern was observed whether cells were treated with 10, 40, or 160 mM DMS for 5 min (data not shown). To determine whether bound proteins affected the methylation pattern, phenol-extracted RNA was treated with DMS in vitro (Fig. 5A, lane 4). Overall, methylation was similar within element I, except that A7 and A11 showed small but reproducible increases in methylation, whereas methylation decreased at A24-C26. These data would be consistent with protein binding at A7 and A11 and/or secondary effects of proteins bound elsewhere.
FIG. 5.
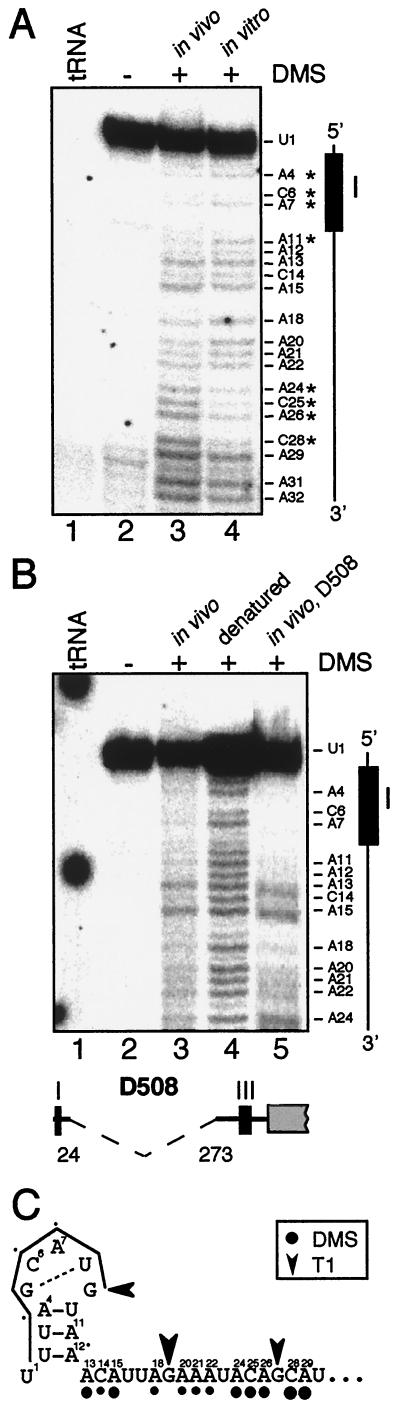
RNA secondary structure analysis of element I. (A) In vivo (lane 3) versus in vitro (lane 4) DMS-treated RNA (25 μg) was analyzed by primer extension with primer WS5 and compared to tRNA (lane 1) and untreated RNA (lane 2). A diagram orienting element I with the corresponding bands in the gel is shown at the right, along with the indicated bases, and bands with different relative levels of in vitro versus in vivo methylation are indicated by asterisks. (B) In vivo DMS-treated RNA (lane 3) was compared to untreated RNA (lane 2), in vitro-treated, heat-denatured RNA (lane 4), RNA from the in vivo-treated D508 deletion mutant (lane 5), and tRNA (lane 1). The 249-bp deletion in D508 is shown at the bottom. (C) Proposed secondary structure for element I, in which nt +1 to +30 are shown and positions are indicated by superscripts. Element I is overlined. Residues methylated by DMS in vivo are identified by filled circles (●), and those cleaved by RNase T1 by arrowheads. The sizes of the symbols represent the amount of modification or cleavage.
Two control experiments were carried out to confirm the significance of the results shown in Fig. 5A; the results of these experiments are shown in Fig. 5B. First, to show that the patterns of methylation around element I were due to local rather than long-range interactions, we took advantage of the deletion mutant D508, which carries a deletion between positions +24 and +273, yielding an accumulating but nontranslatable mRNA (60) due to the absence of element II. Around element I, the in vivo DMS methylation of D508 (Fig. 5B, lane 5) appeared identical to that of WT RNA (Fig. 5B, lane 3), suggesting that in vivo DMS protection within the first 12 nt is due either to base pairing or protein binding localized to the first 24 residues. Second, to show that the lack of methylation near the RNA 5′ end (e.g., A4, C6, and A7) was not due to their proximity to the transcript terminus, we treated WT RNA in vitro under denaturing conditions. As shown in Fig. 5B, lane 4, all A and C residues were extensively methylated by DMS, demonstrating that the lack of methylation under other conditions was not an experimental artifact.
In general, the in vivo DMS data showed potential base pairing at least among Cs and As in the first 12 nt, whereas the RNA downstream, at least to position A51, appears to be largely single stranded (Fig. 5 and data not shown). We used the computer modeling program mFold (76), which is based on the algorithm developed by Zuker (75) to predict possible RNA secondary structures within the first 24 nt. We used 24 nt because the data with mutant D508 RNA (Fig. 5B) as well as data from element I reporter gene fusions (61) suggested that this region is autonomous with respect to RNA structure. Of the alternative structures that could be predicted, the one best fitting the data is shown in Fig. 5C and predicts that A4, A11, and A12 would not be methylated, whereas positions downstream of A13 would be methylated. In vitro RNase T1 cleavage sites were determined (data not shown) and are represented by arrowheads in Fig. 5C. With the exception of G5, all Gs were cleaved and therefore predicted to lie in single-stranded regions. This is consistent with the proposed structure, if we assume a G-U base pair within the proposed loop; such intraloop base pairing is well known (2, 36, 45, 69). An inconsistency between the DMS data and the proposed structure is that C6 and A7 should be methylated; however, they are not. One possible explanation, at least for the in vivo data, is protein binding.
If the structure shown in Fig. 5C is biologically significant, it would be expected that LS mutations which affect its function should alter it. Indeed, LS2 and LS6, which destabilize petD mRNA (Fig. 2), would not allow the structure to form. In contrast, LS1, LS14, LS18, and LS22, which do not affect petD expression, would not alter the structure. LS10 however has an intermediate effect on petD mRNA accumulation, with 35% of the WT mRNA amount, yet the structure shown in Fig. 5C could not form. One possible scenario is that while RNA structure is compromised in LS10, protein binding, presumably of the mRNA-stabilizing factor MCD1, is affected only partially. Alternatively, a different and partially functional structure might form.
RNA secondary structures and RNA-binding proteins of translation elements II and III.
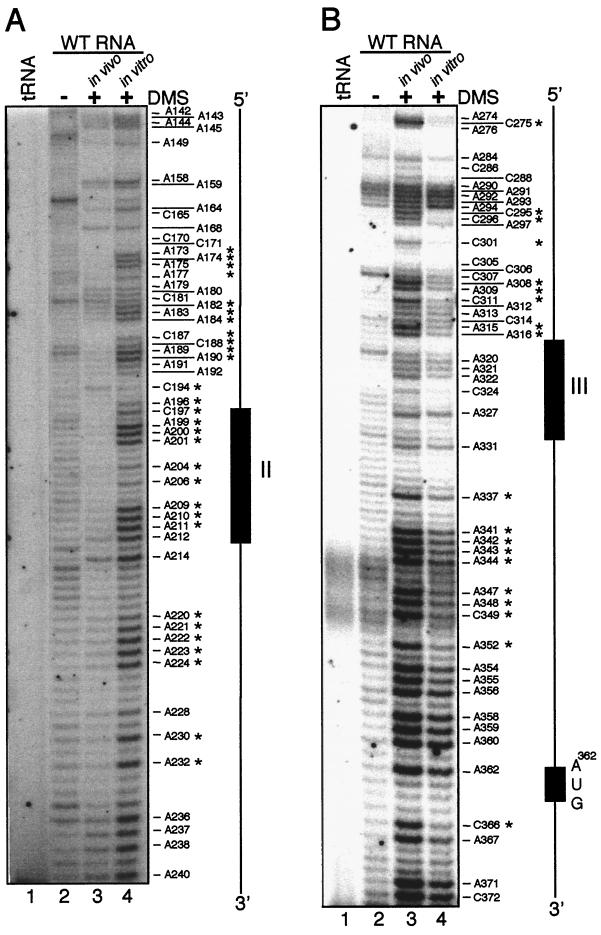
To assess the potential secondary structures of elements II and III, an approach similar to that for element I was used, with in vitro RNase T1 mapping and DMS modification. For element II, primer extension of total RNA from untreated WT cells resulted in a variety of bands due to premature termination (Fig. 6A, lane 2), which is not uncommon in this type of assay. In contrast, extension of in vivo-treated RNA (lane 3) resulted in relatively few bands (e.g., A158-A168 and A214) that were of higher intensity than the background level seen in lane 2. A priori, this would suggest that element II and its flanking regions were in strong secondary structures. However, primer extension of in vitro-treated RNA (lane 4) yielded numerous bands corresponding to heavily methylated A and C residues, suggesting that much of this region was single stranded. In addition, G205 within element II was readily cleaved by RNase T1. Finally, the in vitro pattern was consistent with a computer-predicted structure (data not shown). Of particular interest is that one of the least methylated regions contains element II, namely C197-A212. In summary, the dramatic differences between the in vivo (lane 3) and in vitro (lane 4) patterns implicate RNA-binding proteins as either directly impeding DMS modification or inducing alternate RNA structures that force double-stranded character.
FIG. 6.
In vivo RNA secondary structure analysis of elements II and III. (A) For element II, RNA isolated from DMS-treated cells was analyzed by primer extension with primer D246 (lane 3) and compared to tRNA (lane 1), untreated RNA (lane 2), or phenol-extracted RNA treated with DMS (lane 4). A diagram orienting element II is shown at the right. Bands with different relative levels of methylation in vitro and in vivo are indicated by asterisks. (B) A similar experiment for element III was performed with primer WS10. In addition, the AUG translation initiation codon is shown.
The analysis of element III is shown in Fig. 6B. Primer extension of total RNA from untreated WT cells resulted in faint bands and a few relatively strong bands, due to premature termination (Fig. 6B, lane 2). Using in vivo DMS-treated RNA (lane 3), methylation was considerable between A341 and C372, including the initiation codon at A362, indicating that this region is largely single stranded. Residues A290-A296, A308-C311, and A315-A316 were also heavily methylated, whereas most residues between A320 and C331, which includes element III, were consistently less methylated (Fig. 6B, lane 3, and 7B, lanes 3 and 9). The pattern of in vivo methylation around element III was similar to that of in vitro-treated RNA, while outside of element III several differences were apparent (Fig. 6B, lanes 3 and 4). First, A274-A276 and C295-A316 were much less methylated in vitro, particularly C295-C296. In addition, A308-C311, A315-A316, A337-A352, and C366 were less methylated in vitro. The reduced methylation around element III is consistent with secondary structure and/or protein binding, whereas the change between the in vivo- and in vitro-methylated samples, with the in vitro samples being hypomethylated, is most consistent with altered secondary structures.
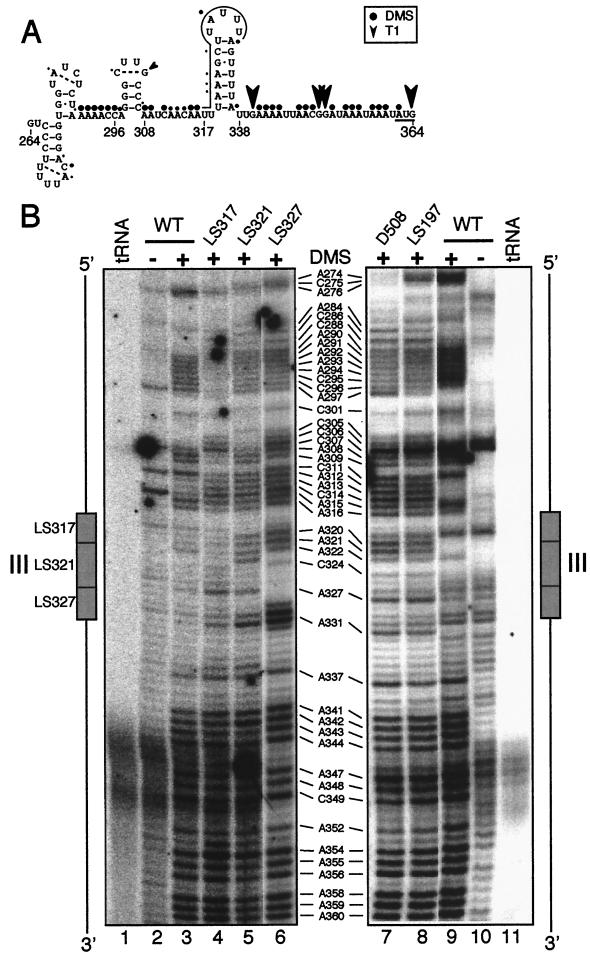
For the region around element III, a model containing four stem-loops could be derived by using mFold with constraints based upon the RNase T1 (not shown) and in vivo DMS (Fig. 6B) data; this is shown in Fig. 7A. The model proposes that element III (overlined) (Fig. 7A) exists within a stem-loop, and that the initiation codon (underlined) is within a single-stranded region. Although U319-A337 is shown as the first pair in the stem, the partial methylation of A337 suggests that it has some single-stranded character.
FIG. 7.
Element III in vivo RNA secondary structures in LS mutants and in D508 (Fig. 5A). (A) Predicted RNA secondary structures between nt 264 and 364. Element III is overlined, nucleotide modifications are described in the legend to Fig. 5C, and the initiation codon is underlined. (B) Primer extension of in vivo DMS-modified RNA from the WT (lanes 3 and 9) and the indicated mutants (lanes 4 to 8) with primer WS10. The left panel shows the WT and three LS element III mutants (lanes 4 to 6) and a diagram to orient the fragments. The right panel shows the primer extension of element III with RNA from in vivo DMS-treated D508 (Fig. 5B, lane 5) and LS197 mutants.
As one approach to test whether this potential secondary structure is biologically significant, nonfunctional element III LS mutants were analyzed in vivo with DMS. Figure 7B, lanes 3 to 6, shows that these mutations resulted in altered methylation patterns, both within and upstream of element III. Some of the alterations within element III can be ascribed to sequence changes; for example, position 323 is A in LS321 but a nonmethylatable G in the other strains. However, most of the increase in methylation within element III cannot be explained in this way, leading to the hypothesis that the LS mutations destabilize the proposed secondary structure and/or compromise protein binding.
Two types of changes occurred upstream of element III; in LS317, hypomethylation relative to the WT was seen at A292-C296 and A308-A309, whereas hypermethylation occurred at C305-C307 in both LS317 and LS327. Taken together, these results imply structural interdependence over a nearly 50-nt region roughly defined by A290-A337. In stark contrast to the changes between A290-A337 was the downstream region, including the initiation codon, which remained single stranded in all the LS mutants tested. This suggests that steric blockage of the initiation codon was not causing the loss of translation.
Because element II and III mutants had similar phenotypes (Fig. 3 and 4), we considered the possibility that they interact with each other directly or via possible trans factors. Because primer extension analysis of element II is technically difficult, we chose to examine element III structure in two element II mutants: LS197, in which its sequence is changed, and D508 (Fig. 5B), in which it is deleted. The methylation patterns of the mutants were compared to those of the WT, as shown in Fig. 7, lanes 7 to 9. Remarkably, the methylation patterns in D508 and LS197 around element III did not resemble that of the WT, but instead resembled that of LS327 (Fig. 7, lane 6, positions 329 and 330 are methylatable Cs in this strain) although the element III sequence is WT in both D508 and LS197. These data thus suggest an interaction between elements II and III. Because long-range interactions are difficult to predict with chemical probing (23), and possible structures formed by base pairing between element II and element III are neither consistent with the chemical probing data nor very strong, precisely defining this interaction will require further experimentation.
Intragenic mutations can suppress element III mutations.
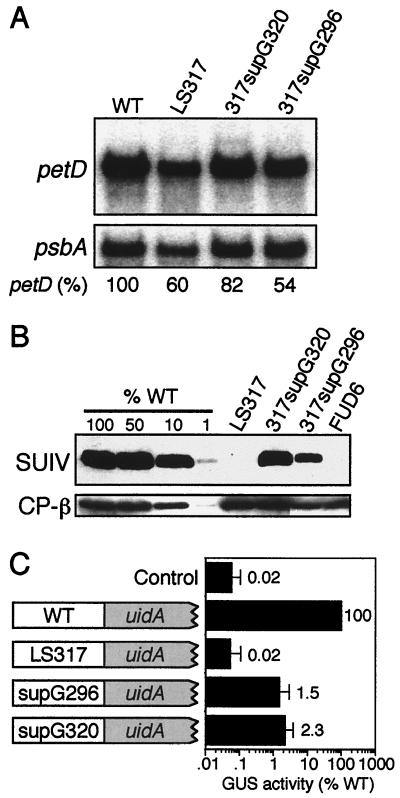
To gain additional information regarding sequence requirements around element III, we took a genetic approach. To do this, rare photosynthetic revertants were selected from large numbers of LS317 cells plated on minimal medium. Four spontaneous photoautotrophic colonies were obtained, and all were shown genetically to be due to chloroplast DNA mutations or reversions (see Materials and Methods). The petD 5′ UTR was amplified and sequenced, and it was found that in one case (317supG296) the original LS mutation was still present but an upstream base change had occurred, whereas the other strains had identical mutations within element III (see below). A basic molecular analysis of the LS317 suppressors is shown in Fig. 8. Panel A shows that both LS317 and the suppressors accumulate near-WT amounts of petD mRNA, whereas panel B shows that there was an increase from no detectable SUIV in LS317 to approximately 10 to 25% of the WT amount in the suppressors. We tentatively concluded that the new petD 5′ UTR mutations overcame the original LS317 mutation by increasing SUIV translation.
FIG. 8.
Intragenic suppressors of LS317. Total RNA (A) and total protein (B) were analyzed as described in the legend to Fig. 2. To help estimate SUIV accumulation, a dilution series of WT protein was included, along with a negative control (FUD6). (C) Fluorometric analysis of GUS enzyme activity from petD-uidA reporter genes. The petD WT, LS317, 317supG296 (supG296), and 317supG320 (supG320) 5′ UTRs were translationally fused to the uidA coding region and introduced into chloroplasts. GUS assays were conducted on these and a nontransformed control strain (control). GUS activity (in picomoles of methyumbelliferone/minute/milligram of protein) is presented on a log scale as the percentage of the WT positive control that has the WT petD 5′ UTR, and values were standardized to total protein. Error bars indicate standard errors of the mean.
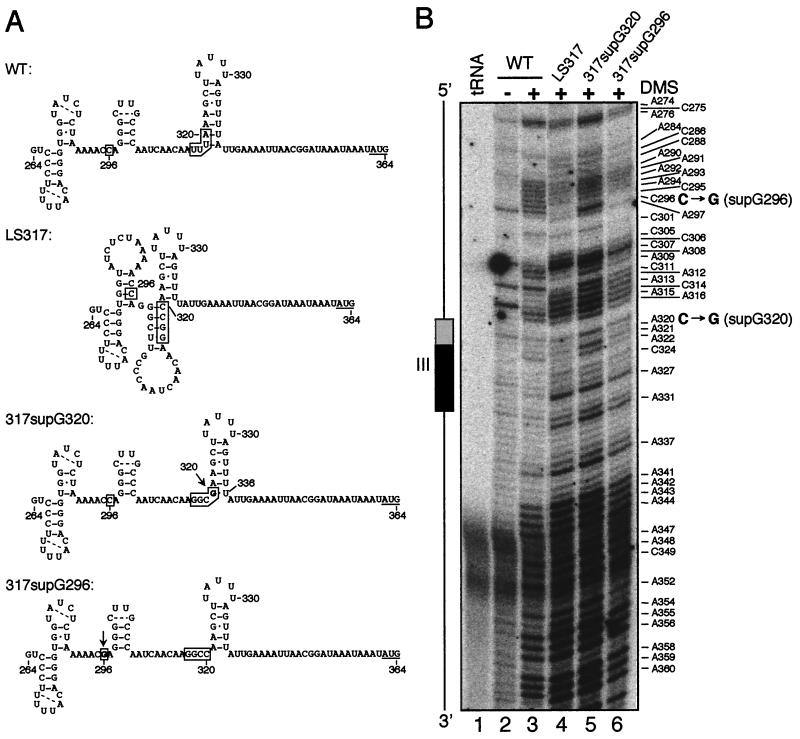
As noted above, two sequence classes of LS317 suppressors were isolated. 317supG296 has a C-to-G transversion at nt 296, 21 nt upstream of the original LS mutation, whereas 317supG320 has a C-to-G transversion at nt 320. To confirm that these intragenic point mutations, rather than undetected mutations elsewhere in the chloroplast genome, restored SUIV accumulation, WT and mutant petD 5′ UTRs were fused to the bacterial uidA coding region and the Chlamydomonas chloroplast rbcL 3′ UTR. These constructs, which also contained a functional atpB gene, were introduced into Chlamydomonas chloroplasts of an atpB deletion mutant, and expression of GUS, the product of uidA, was measured. The results of fluorometric GUS assays are shown in Fig. 8C and reveal that GUS activity in both suppressor-uidA fusions was more than 25 times that of the LS317-uidA fusion and the nontransformed control (note the log scale), yet all transformants accumulated similar amounts of uidA mRNA (data not shown). These data confirm that the identified point mutations are responsible for the suppressor phenotype. Interestingly, the ratio of GUS activity in WT-uidA versus suppressor-uidA fusions (∼50:1) was much greater than the corresponding SUIV ratio (∼4:1) (Fig. 4B). Our prior experience with uidA fusions leads us to suspect that this is a result of poor translation of the mutant 5′ UTR-uidA fusions. For example, while an AUG-to-AUU initiation codon mutation in petD reduced SUIV translation to 10% of the WT amount (8), the same mutant petD 5′ UTR fused to uidA produced less than 1% of the WT petD 5′ UTR-uidA GUS activity (10).
When the suppressor mutations were viewed in the context of the predicted element III secondary structure, it can be seen that the LS317 mutation disrupted the two basal pairs in the element III stem-loop, resulting in a less stable stem while favoring an alternative structure predicted by mFold (Fig. 9A). The C-to-G mutation in 317supG320 could result in a G-U wobble pair with U336, thus favoring the original structure while destabilizing the alternative one. Support for this hypothesis comes from the in vivo DMS methylation data shown in Fig. 9B. For example, C296 is single stranded in the WT (Fig. 9B, lane 3) but base paired in LS317 (lane 4) (see also C295), consistent with the alternative structure for LS317. C296 reverted to single-stranded character in 317supG320, in keeping with the proposed structures. Potential methylation at some putatively double-stranded positions (e.g., A295 and A298) suggests that these structures may not be completely stable, consistent with the partial translatability of the suppressor 5′ UTRs.
FIG. 9.
Intragenic suppressors cause changes in the in vivo RNA secondary structure upstream of element III. (A) Possible secondary structures in the petD 5′ UTR nt 264 to 364 regions of the indicated strains. Positions 296 and 317 to 320 (the location of LS317) are boxed, and base changes in suppressors are shown in boldface type and marked by arrows. Putative intraloop base pairs are marked by dashed lines. (B) Primer extension reactions of unmodified (lane 2) and in vivo DMS-modified WT RNA (lane 3) were compared to those of RNA from in vivo DMS-treated LS317 (lane 4) and suppressors (lanes 5 and 6). The WT lanes are the same as those shown in Fig. 7B (lanes 2 and 3) for reference. A diagram orienting element III is shown, with the location of the LS317 mutation indicated by a shaded box and that of other essential nucleotides by a filled box. Base changes in the suppressors are marked in large boldface letters.
317supG296 appeared to be more enigmatic due to its location 21 nt upstream of the LS mutation. However, it can be hypothesized that the C-to-G change at nt 296 would destabilize the alternative structure shown for LS317, and thus predicts that G296 in 317supG296 would be single stranded, as compared to the double-stranded C296 in its progenitor. This could not be directly tested, however, since DMS does not methylate G residues. We conclude that secondary structures around element III are likely to be important for translation, including the amount of single-stranded character at C295-C296.
DISCUSSION
Here we have defined three petD 5′ UTR cis elements in detail by using LS mutagenesis and genetic suppressors. Element I is critical for petD mRNA stability and SUIV translation, whereas elements II and III function only in translation. In addition, we made extensive use of chemical (DMS) probing as a first step in describing their in vivo higher-order structures. While previous studies of organellar transcripts have largely relied upon computer folding or in vitro analysis to interpret mutagenic data and predict essential secondary structures (17, 40, 58, 64, 66, 67), the approaches used in this study have allowed us to experimentally arrive at structure-function models that will direct future investigations.
Element I includes a maximum of 8 nt and is located at the immediate 5′ end of mature petD mRNA (Fig. 2A). Element I mutations reduced petD mRNA accumulation to less than 5% of the WT amount; based on previous work, we conclude that this results from RNA destabilization and not a decrease in transcription (61, 62, 68). Because all the element I LS mutants accumulated only mature-sized petD mRNA, element I does not appear to have a direct role in petD 5′ end formation. However, changes of a few nucleotides cannot be ruled out for LS2 and LS6, since the low accumulation of petD mRNA precluded primer extension analysis (data not shown). This is in contrast to the ΔP deletion, which has an 81-bp deletion upstream of the mature 5′ end of petD, causes inefficient processing, and results in the accumulation of a dicistronic petA-petD transcript (62, 68).
The 5′ terminal location and RNA stability function of element I appears to be typical of organellar mRNAs. For example, a deletion of 81 nt near the 5′ end of the yeast mitochondrial COX3 mRNA caused an 85% decrease in its steady-state abundance, although the residual message was translatable (70). Deleting the first 10 nt of the yeast mitochondrial COB transcript also destabilized it; again, the residual mRNA was translated (49). Deletions near the 5′ ends of Chlamydomonas chloroplast psbA (47) and psbD (53) mRNAs caused 95 and 100% reductions in RNA accumulation, respectively, and the reduced psbA mRNA did not appear to be translated (47). We proposed earlier that petD element I protects the RNA from 5′→3′ exoribonuclease in concert with the product of the MCD1 gene (19); it is conceivable and likely, in the case of psbD (53), that the other 5′ terminal stability elements have similar functions.
Like Chlamydomonas psbA, petD mRNA carrying element I mutations did not appear to be translatable. In repeated experiments, we found that LS2 and LS6 accumulated 1 to 3% of the WT mRNA level. Since 1 to 3% of WT SUIV was easily detectable on immunoblots (Fig. 8B), translation appears to be impaired by these mutations. However, if the RNAs were inefficiently translated, SUIV might have been below the level of detection. Another way of interpreting the data for element I is to argue that compromised translation leads to RNA instability. The relationship between the two processes varies in Chlamydomonas chloroplasts, and mutations that eliminate translation can result in increased, decreased, or unaltered accumulation of mRNA (for a review, see reference 57). The petD element II and element III mutations described here, for example, completely abolish translation but do not markedly affect RNA abundance. These observations, however, do not rule out a causal relationship between impaired translation and RNA instability in element I mutants.
The in vivo DMS methylation data suggest that element I exists in a higher-order structure which is confined to the first 24 nt. Differences in the pattern of in vitro versus in vivo DMS methylation suggest that the structure forms but is less stable in the absence of proteins. The simplest interpretation of these results is that element I exists in vivo within the small stem-loop structure shown in Fig. 5C. The LS10 mutation is predicted to destabilize this structure and indeed, LS10 accumulated only 35% of the WT amount of petD mRNA. This observation suggests that the stem-loop structure promotes RNA stability but may not be essential.
As determined by nuclear magnetic resonance spectroscopy and chemical probing, stem-loop structures commonly contain stabilizing tetra- and pentaloops, similar to that proposed for element I, in which there can be pairing across the loop (2, 7, 35, 36); also included in these examples is the spinach chloroplast 5S rRNA (69). Loops commonly contain extruded nucleotides that can enable ribonucleoprotein formation for small RNA elements (44). Similar to petD, E. coli ompA mRNA harbors a short stem-loop structure at its 5′ end. In this case a stem as short as 4 bp is sufficient to confer RNA stability in vivo, although its primary sequence appears to be unimportant (3). In the case of petD, the primary sequence of element I likely is important, since previous work has shown that the product of the nuclear MCD1 gene interacts specifically with it (19). MCD1 itself appears to be a translation factor in addition to promoting RNA stability (20).
Translation elements II and III are located approximately 150 and 40 nt upstream of the initiation codon, respectively, and have no obvious complementarity to the 3′ end of the chloroplast 16S rRNA nor homology to other known chloroplast translation-activating sequences, such as those in psbC (71) and psaB (64). Element II was defined by two LS mutations spanning 16 nt, and element III was defined by three LS mutations spanning 14 nt. Deletions and/or insertions in these regions were previously shown to block SUIV translation (60). By analogy to well-characterized translation activation mechanisms for yeast mitochondrial mRNAs such as COX2 (27), COX3 (70), and COB (50, 59), we expect that elements II and III interact either together or separately with gene-specific translation activators, which in turn facilitate recognition of the start codon by ribosomes. In Chlamydomonas as in yeast, numerous nuclear mutants defining gene-specific translation activators have been isolated (for a review, see reference 65). Although SUIV translation-specific mutants have not yet been isolated, it is unlikely that genetic screens have already identified all such loci.
For element II, in vitro DMS analysis was consistent with a computer-predicted stem-loop structure, but this structure was not consistent with the in vivo RNA structural data, nor was its importance supported by the phenotypes of relevant LS mutants. The striking reduction in DMS methylation of element II residues in vivo was consistent with the hypothesis that proteins interact with this element and either bind directly to element II, cause it to form secondary structures, or both. It was not possible to determine whether element II mutations restored methylation by compromising protein binding, since this region is refractory to primer extension (the data shown in Fig. 6A could be reproduced but most experiments failed). Somewhat surprisingly, we found that at least some element II mutations affect in vivo RNA structure near element III (Fig. 7B, lanes 7 and 8). We cannot rule out, therefore, that element II LS mutations act indirectly by altering element III structures. Nonetheless, element II would still be defined as an essential translation element. This type of long-range interaction would be typical of many functional RNA structures including those of introns, rRNAs, and ribozymes.
Element III appears to exist in a stem-loop (Fig. 7A) which forms in vitro and in vivo. Mutations in the 5′ half of this stem block translation and destabilize the stem in vivo, consistent with the notion that this structure is necessary for translation. Although the LS331 mutation alters the sequence of the 3′ half of the stem, it did not affect SUIV accumulation. The sequence of LS331, however, still allows 5 of 7 bases to pair, giving a structure with a thermodynamic stability comparable to that of the WT sequence.
Additional stem-loops upstream of element III were predicted by computer modeling and shown to be mostly consistent with DMS methylation data. These structures contained tetra-, penta-, and hexaloops, which are known to be stabilizing (35, 36). Tetraloops commonly have intraloop pairing of the first and fourth bases, leaving only two unpaired bases. An example is the CUUG tetraloop sequence in which the C-G intraloop base pair forms to stabilize the stem-loop (35). The closing base pairs of stems, just below the loop, are conserved and are frequently G-C pairs. Consistent with these tetraloop features, the predicted small stem-loop immediately upstream of element III has a G300-C305 closing pair, and C301 in the loop was only slightly methylated by DMS, suggesting that it may form an intraloop pair with G304.
Four spontaneous intragenic suppressors of LS317 were isolated that comprised two different single-base changes. Three had C-to-G transversions within the LS mutation, while one had a mutation further upstream. The effects of the LS mutations and suppressors on translation could be rationalized in terms of competing secondary structures, as shown in Fig. 9 and discussed in the text. Some of the predicted changes are subtle but mirror the observation that a 5-bp but not a 4-bp stem is functional in the 5′ UTR of yeast mitochondrial COX2 mRNA (22). In general, the single-strandedness of C295 and C296, which are upstream of element III, correlates with SUIV translation. A similar phenomenon has been reported for yeast mitochondrial COX3, where upstream insertions, deletions, or base changes can suppress a large 5′ UTR deletion (14, 70). We suggest that the LS317 mutation promotes the formation of an alternative, inhibitory structure (Fig. 9A) which affects both element III and nt 295 and 296. The suppressor mutations appear to destabilize the alternative structure, allowing a limited amount of translation. A similar hypothesis was proposed for Chlamydomonas chloroplast psbC, based on computer modeling (58).
Our methylation data suggest that the petD initiation codon resides in a long single-stranded region. Various models have been proposed to explain the relationship between RNA secondary structure at the initiation codon and chloroplast translation (6, 16, 33, 37, 40, 41, 47, 64, 65). Some models propose that stem-loop structures at the initiation codon must be destabilized to allow translation (6, 47, 64), while others propose that the initiation codons reside in relatively unstructured A+U-rich regions. The data presented here are most consistent with the second model, in which the initiation codon remains single stranded. We also have provided limited evidence for protein binding sites, especially at element II, supporting the idea that gene-specific translation activators may facilitate the interaction between the ribosome and mRNA. Genetic approaches that are feasible in Chlamydomonas should allow the nature of these interactions to be elucidated and the nuclear genes encoding these trans-acting factors to be isolated. Of considerable interest is whether there will be any relationship to previously characterized yeast mitochondrial proteins of similar function, as has recently been shown in maize chloroplasts (25).
ACKNOWLEDGMENTS
We thank the members of the Stern and Kindle laboratories, in particular the late Robert G. Drager, for stimulating discussions.
This work was supported by National Science Foundation grant MCB-9723274 to D.B.S. and K.L.K. D.C.H. was supported by Individual National Research Award 5-F32-GM17938-2 from the National Institutes of Health, and R.S.S. was supported by a Howard Hughes Undergraduate Research Scholarship.
REFERENCES
- 1.Adams C C, Stern D B. Control of mRNA stability in chloroplasts by 3′ inverted repeats: effects of stem loop mutations on degradation of psbA mRNA in vitro. Nucleic Acids Res. 1990;18:6003–6010. doi: 10.1093/nar/18.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addess K J, Basilion J P, Klausner R D, Rouault T A, Pardi A. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins. J Mol Biol. 1997;274:72–83. doi: 10.1006/jmbi.1997.1377. [DOI] [PubMed] [Google Scholar]
- 3.Arnold T E, Yu J, Belasco J G. mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA. 1998;4:319–330. [PMC free article] [PubMed] [Google Scholar]
- 4.Baklanov M M, Golikova L N, Malygin E G. Effect on DNA transcription of nucleotide sequences upstream of T7 promoter. Nucleic Acids Res. 1996;24:3659–3660. doi: 10.1093/nar/24.18.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell. 1993;5:389–402. doi: 10.1105/tpc.5.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barkan A, Walker M, Nolasco M, Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunel C, Romby P, Westhof E, Ehresmann C, Ehresmann B. Three-dimensional model of E. coli ribosomal 5S RNA as deduced from structure probing in solution and computer modeling. J Mol Biol. 1991;221:293–308. doi: 10.1016/0022-2836(91)80220-o. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Kindle K, Stern D. Initiation codon mutations in the Chlamydomonas chloroplast petD gene result in temperature-sensitive photosynthetic growth. EMBO J. 1993;12:3627–3635. doi: 10.1002/j.1460-2075.1993.tb06036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Simpson C L, Kindle K L, Stern D B. A dominant mutation in the Chlamydomonas reinhardtii nuclear gene SIM30 suppresses translational defects caused by initiation codon mutations in chloroplast genes. Genetics. 1997;145:935–943. doi: 10.1093/genetics/145.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, X., and D. B. Stern. Unpublished data.
- 11.Choquet Y, Goldschmidt-Clermont M, Girard-Bascou J, Kuck U, Bennoun P, Rochaix J-D. Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C. reinhardtii chloroplast. Cell. 1988;52:903–913. doi: 10.1016/0092-8674(88)90432-1. [DOI] [PubMed] [Google Scholar]
- 12.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costanzo M C, Fox T D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- 14.Costanzo M C, Fox T D. Suppression of a defect in the 5′ untranslated leader of mitochondrial COX3 mRNA by a mutation affecting an mRNA-specific translational activator protein. Mol Cell Biol. 1993;13:4806–4813. doi: 10.1128/mcb.13.8.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotton J L S, Ross C W, Byrne D H, Colbert J T. Down regulation of phytochrome mRNA abundance by red light and benzyladenine in etiolated cucumber cotyledons. Plant Mol Biol. 1990;14:707–714. doi: 10.1007/BF00016503. [DOI] [PubMed] [Google Scholar]
- 16.Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danon A, Mayfield S P Y. Light regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO J. 1991;10:3993–4002. doi: 10.1002/j.1460-2075.1991.tb04974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doktycz M, Larimer F W, Pastrnak M, Stevens A. Comparative analyses of the secondary structures of synthetic and intracellular yeast MFA2 mRNAs. Proc Natl Acad Sci USA. 1998;95:14614–14621. doi: 10.1073/pnas.95.25.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drager R G, Girard-Bascou J, Choquet Y, Kindle K L, Stern D B. In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 1998;13:85–96. doi: 10.1046/j.1365-313x.1998.00016.x. [DOI] [PubMed] [Google Scholar]
- 20.Drager R G, Higgs D C, Kindle K L, Stern D B. 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J. 1999;19:521–532. doi: 10.1046/j.1365-313x.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- 21.Drager R G, Zeidler M, Simpson C L, Stern D B. A chloroplast transcript lacking the 3′ inverted repeat is degraded by 3′→5′ exoribonuclease activity. RNA. 1996;2:652–663. [PMC free article] [PubMed] [Google Scholar]
- 22.Dunstan H M, Green-Willms N S, Fox T D. In vivo analysis of Saccharomyces cerevisiae COX2 mRNA 5′-untranslated leader functions in mitochondrial translation initiation and translational activation. Genetics. 1997;147:87–100. doi: 10.1093/genetics/147.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fargo D C, Zhang M, Gillham N W, Boynton J E. Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet. 1998;257:271–282. doi: 10.1007/s004380050648. [DOI] [PubMed] [Google Scholar]
- 25.Fisk D G, Walker M B, Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4090. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green-Willms N S, Fox T D, Costanzo M C. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol Cell Biol. 1998;18:1826–1834. doi: 10.1128/mcb.18.4.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gumpel N J, Ralley L, Girard-Bascou J, Wollman F-A, Nugent J H A, Purton S. Nuclear mutants of Chlamydomonas reinhardtii defective in the biogenesis of the cytochrome b6/f complex. Plant Mol Biol. 1995;29:921–932. doi: 10.1007/BF00014966. [DOI] [PubMed] [Google Scholar]
- 29.Harris E H. The Chlamydomonas sourcebook. San Diego, Calif: Academic Press, Inc.; 1989. [Google Scholar]
- 30.Hauser C R, Gillham N W, Boynton J E. Translational regulation of chloroplast genes. J Biol Chem. 1996;271:1486–1497. doi: 10.1074/jbc.271.3.1486. [DOI] [PubMed] [Google Scholar]
- 31.Higgs D C, Kuras R, Kindle K L, Wollman F-A, Stern D B. Inversions in the Chlamydomonas chloroplast genome suppress a petD 5′ untranslated region deletion by creating functional chimeric mRNAs. Plant J. 1998;14:663–671. doi: 10.1046/j.1365-313x.1998.00165.x. [DOI] [PubMed] [Google Scholar]
- 32.Hirose T, Kusumegi T, Sugiura M. Translation of tobacco chloroplast rps14 mRNA depends on a Shine-Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett. 1998;430:257–260. doi: 10.1016/s0014-5793(98)00673-5. [DOI] [PubMed] [Google Scholar]
- 33.Hirose T, Sugiura M. Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J. 1996;15:1687–1695. [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkins B D, Kulhanek D J, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jucker F M, Pardi A. Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry. 1995;34:14416–14427. doi: 10.1021/bi00044a019. [DOI] [PubMed] [Google Scholar]
- 36.Kajava A, Rüterjans H. Molecular modelling of the 3-D structure of RNA tetraloops with different nucleotide sequences. Nucleic Acids Res. 1993;21:4556–4562. doi: 10.1093/nar/21.19.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J, Mullet J E. Ribosome-binding sites on chloroplast rbcL and psbA mRNAs and light-induced initiation of D1 translation. Plant Mol Biol. 1994;25:437–448. doi: 10.1007/BF00043872. [DOI] [PubMed] [Google Scholar]
- 38.Kindle K L, Richards K L, Stern D B. Engineering the chloroplast genome: techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1991;88:1721–1725. doi: 10.1073/pnas.88.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klaff P, Gruissem W. A 43 kD light-regulated chloroplast RNA-binding protein interacts with the psbA 5′ non-translated leader RNA. Photosynth Res. 1995;46:235–248. doi: 10.1007/BF00020436. [DOI] [PubMed] [Google Scholar]
- 40.Klaff P, Mundt S M, Steger G. Complex formation of the spinach chloroplast psbA mRNA 5′ untranslated region with proteins is dependent on the RNA structure. RNA. 1997;3:1468–1479. [PMC free article] [PubMed] [Google Scholar]
- 41.Koo J S, Spremulli L L. Effect of the secondary structure in the Euglena gracilis chloroplast ribulose-bisphosphate carboxylase/oxygenase messenger RNA on translational initiation. J Biol Chem. 1994;269:7501–7508. [PubMed] [Google Scholar]
- 42.Kuchka M R, Goldschmidt-Clermont M, van Dillewijn J, Rochaix J-D. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PSII. Cell. 1989;58:869–876. doi: 10.1016/0092-8674(89)90939-2. [DOI] [PubMed] [Google Scholar]
- 43.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Legault P, Li J, Mogridge J, Kay L E, Greenblatt J. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 45.Leontis N B, Westhof E. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA. 1998;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy H, Kindle K L, Stern D B. A nuclear mutation that affects the 3′ processing of several mRNAs in Chlamydomonas chloroplasts. Plant Cell. 1997;9:825–836. doi: 10.1105/tpc.9.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayfield S P, Cohen A, Danon A, Yohn C B. Translation of the psbA mRNA from Chlamydomonas reinhardtii requires a structured RNA element contained within the 5′ untranslated region. J Cell Biol. 1994;127:1537–1545. doi: 10.1083/jcb.127.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meurer J, Berger A, Westhoff P. A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell. 1996;8:1193–1207. doi: 10.1105/tpc.8.7.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mittelmeier T M, Dieckmann C L. In vivo analysis of sequences necessary for CBP1-dependent accumulation of cytochrome b transcripts in yeast mitochondria. Mol Cell Biol. 1993;13:4203–4213. doi: 10.1128/mcb.13.7.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mittelmeier T M, Dieckmann C L. In vivo analysis of sequences required for translation of cytochrome b transcripts in yeast mitochondria. Mol Cell Biol. 1995;15:780–789. doi: 10.1128/mcb.15.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moazed D, Stern S, Noller H F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 52.Monod C, Goldschmidt-Clermont M, Rochaix J-D. Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol Gen Genet. 1992;231:449–459. doi: 10.1007/BF00292715. [DOI] [PubMed] [Google Scholar]
- 53.Nickelsen J, Fleischmann M, Boudreau E, Rahire M, Rochaix J-D. Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell. 1999;11:957–970. doi: 10.1105/tpc.11.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nickelsen J, van Dillewijn J, Rahire M, Rochaix J-D. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 1994;13:3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puglisi J D, Wyatt J R. Biochemical and NMR studies of RNA conformation with an emphasis on RNA pseudoknots. Methods Enzymol. 1995;261:323–350. doi: 10.1016/s0076-6879(95)61016-2. [DOI] [PubMed] [Google Scholar]
- 56.Rattanachaikunsopon P, Rosch C, Kuchka M R. Cloning and characterization of the nuclear AC115 gene of Chlamydomonas reinhardtii. Plant Mol Biol. 1999;39:1–10. doi: 10.1023/a:1006108203580. [DOI] [PubMed] [Google Scholar]
- 57.Rochaix J-D. Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- 58.Rochaix J-D, Kuchka M, Mayfield S, Schirmer-Rahire M, Girard-Bascou J, Bennoun P. Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 1989;8:1013–1021. doi: 10.1002/j.1460-2075.1989.tb03468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rödel G, Fox T D. The yeast nuclear gene CBS1 is required for translation of mitochondrial mRNAs bearing the cob 5′ untranslated leader. Mol Gen Genet. 1987;206:45–50. doi: 10.1007/BF00326534. [DOI] [PubMed] [Google Scholar]
- 60.Sakamoto W, Chen X, Kindle K L, Stern D B. Function of the Chlamydomonas reinhardtii petD 5′ untranslated region in regulating the accumulation of subunit IV of the cytochrome b6/f complex. Plant J. 1994;6:503–512. doi: 10.1046/j.1365-313x.1994.6040503.x. [DOI] [PubMed] [Google Scholar]
- 61.Sakamoto W, Kindle K L, Stern D B. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of β-glucuronidase translational fusions. Proc Natl Acad Sci USA. 1993;90:497–501. doi: 10.1073/pnas.90.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakamoto W, Sturm N R, Kindle K L, Stern D B. petD mRNA maturation in Chlamydomonas reinhardtii chloroplasts: role of 5′ endonucleolytic processing. Mol Cell Biol. 1994;14:6180–6186. doi: 10.1128/mcb.14.9.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Senecoff J F, Meagher R B. In vivo analysis of plant RNA structure: soybean 18S ribosomal and ribulose-1,5-bisphosphate carboxylase small subunit RNA. Plant Mol Biol. 1992;18:219–234. doi: 10.1007/BF00034951. [DOI] [PubMed] [Google Scholar]
- 64.Stampacchia O, Girard-Bascou J, Zanasco J-L, Zerges W, Bennoun P, Rochaix J-D. A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell. 1997;9:773–782. doi: 10.1105/tpc.9.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stern D B, Higgs D C, Yang J. Transcriptional and translational activities of chloroplasts. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- 66.Stern D B, Jones H, Gruissem W. Function of plastid mRNA 3′ inverted repeats: RNA stabilization and gene-specific protein binding. J Biol Chem. 1989;264:18742–18750. [PubMed] [Google Scholar]
- 67.Stern D B, Radwanski E R, Kindle K L. A 3′ stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991;3:285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sturm N R, Kuras R, Büschlen S, Sakamoto W, Kindle K L, Stern D B, Wollman F-A. The petD gene is transcribed by functionally redundant promoters in Chlamydomonas reinhardtii chloroplasts. Mol Cell Biol. 1994;14:6171–6179. doi: 10.1128/mcb.14.9.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westhof E, Romby P, Romaniuk P J, Ebel J-P, Ehresmann C, Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5S rRNAs. J Mol Biol. 1989;207:417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- 70.Wiesenberger G, Costanzo M C, Fox T D. Analysis of the Saccharomyces cerevisiae mitochondrial COX3 mRNA 5′ untranslated leader: translational activation and mRNA processing. Mol Cell Biol. 1995;15:3291–3300. doi: 10.1128/mcb.15.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zerges W, Girard-Bascou J, Rochaix J-D. Translation of the chloroplast psbC mRNA is controlled by interactions between its 5′ leader and the nuclear loci TBC1 and TBC3 in Chlamydomonas reinhardtii. Mol Cell Biol. 1997;17:3440–3448. doi: 10.1128/mcb.17.6.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zerges W, Rochaix J-D. Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol. 1998;140:101–110. doi: 10.1083/jcb.140.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhaug A J, Cech T R. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]
- 74.Zito F, Kuras R, Choquet Y, Kossel H, Wollman F-A. Mutations in cytochrome b6 in Chlamydomonas disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol Biol. 1997;33:79–86. doi: 10.1023/a:1005734809834. [DOI] [PubMed] [Google Scholar]
- 75.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 76.Zuker, M. 1995–1999, copyright date. [Online.] http://mfold2.wustl.edu/⨍mfold/rna/form1. [13 October 1999, last date accessed.]