In order to identify new players modulating the circadian clock in the model Neurospora crassa, we adopted a reverse genetics strategy. Thus, we focused on transcription factors knockouts and crossed them to strains containing circadian luciferase reporters. Our screen covered close to 60% de the 302 genes encoding for such proteins in Neurospora, identifying that 23 of them appear to modulate period, while none of the tested ones (besides the classic core-clock components) are essential for daily rhythms.
Keywords: circadian clock, Neurospora crassa, transcription factors, luciferase, reverse genetic screen
Abstract
Eukaryotic circadian oscillators share a common circuit architecture, a negative feedback loop in which a positive element activates the transcription of a negative one that then represses the action of the former, inhibiting its own expression. While studies in mammals and insects have revealed additional transcriptional inputs modulating the expression of core clock components, this has been less characterized in the model Neurospora crassa, where the participation of other transcriptional components impacting circadian clock dynamics remains rather unexplored. Thus, we sought to identify additional transcriptional regulators modulating the N. crassa clock, following a reverse genetic screen based on luminescent circadian reporters and a collection of transcription factors (TFs) knockouts, successfully covering close to 60% of them. Besides the canonical core clock components WC-1 and -2, none of the tested transcriptional regulators proved to be essential for rhythmicity. Nevertheless, we identified a set of 23 TFs that when absent lead to discrete, but significant, changes in circadian period. While the current level of analysis does not provide mechanistic information about how these new players modulate circadian parameters, the results of this screen reveal that an important number of light and clock-regulated TFs, involved in a plethora of processes, are capable of modulating the clockworks. This partial reverse genetic clock screen also exemplifies how the N. crassa knockout collection continues to serve as an expedite platform to address broad biological questions.
Introduction
Circadian rhythms are a widespread phenomenon across the tree of life, conferring individuals the capacity to coordinate cellular and organismal metabolism, physiology, and behavior to day/night cycles (Dunlap 1999; Loudon 2012; Montenegro-Montero and Larrondo 2016; Dunlap and Loros 2017). These rhythms exhibit periodic oscillations circa 24 hours, which can be sustained even in the absence of environmental signals, and can be synchronized to external cues such as light and temperature (Lakin-Thomas and Brody 2004; Narasimamurthy and Virshup 2017; Kuhlman et al. 2018). The circadian core oscillator is composed of a topologically conserved transcriptional-translational negative feedback loop (TTFL), although the molecular composition of the core components differs across phyla (Merrow et al. 2010; Zhang and Kay 2010; Zheng and Sehgal 2012). In the model fungus Neurospora crassa, the positive element is the white collar complex (WCC), a heterodimer formed by the TFs white collar 1 (WC-1) and white collar 2 (WC-2). The WCC is responsible for activating transcription of the frequency gene, which encodes for the negative element (FRQ) that feeds back to inhibit the activity of WCC, therefore closing the loop. As FRQ is progressively phosphorylated by several kinases, including CK1, its affinity for the latter as well as for WCC diminishes, therefore no longer inactivating its own expression. As this occurs, hyperphosphorylated FRQ is subjected to proteasomal degradation, all of which can be visualized as daily oscillations in frq mRNA and protein levels (Dunlap 1999; Montenegro-Montero et al. 2015; Diernfellner and Brunner 2020). The changing levels and activities of these two core components allow the entire system to oscillate (Aronson et al. 1994a,b; Brunner and Kaldi 2008; Neiss et al. 2008). However, different aspects of how eukaryotic clocks keep their period constant, and are robust to external perturbations, remain partially unsolved (Ripperger and Brown 2010; Brown et al. 2012; Partch et al. 2014; Kramer 2015; Hurley et al. 2016).
Mechanisms involved in eukaryotic clock regulation comprise several layers of modulation, such as chromatin remodeling, transcriptional control, alternative splicing, antisense transcripts, post-transcriptional regulation and, a high degree of post-translational modifications (phosphorylation) of clock proteins (Brunner and Schafmeier 2006; Ripperger and Brown 2010; Kojima et al. 2011; Hurley et al. 2014, 2016; Montenegro-Montero et al. 2015; Mendoza-Viveros et al. 2017). In plant and animal models, transcriptional regulation of clock components had been profusely addressed, leading to the identification of several transcription factors (TFs) which participate in accessory transcriptional loops interlocked with the central core oscillator (Honma et al. 2002; Kadener et al. 2007; Rossner et al. 2008; Ripperger and Brown 2010; Zhang and Kay 2010; Hardin 2011; Lowrey and Takahashi 2011; Nagel and Kay 2012; Zheng and Sehgal 2012; Ronald and Davis 2017). In N. crassa, the WCC plays a pivotal role in mastering frq expression, although other transcriptional regulators have been found to fine-tune it such as IEC-1 (Gai et al. 2017) and CBF-1 (Cao et al. 2018), or indirectly do so by modulating WCC abundance upon changes in sugar levels, impacting metabolic compensation, as observed for CSP-1 and RCO-1 (Sancar et al. 2012; Olivares-Yanez et al. 2016). Thus, these regulators are capable of affecting the expression of core clock components, having an effect on key circadian parameters (Sancar et al. 2011, 2012; Olivares-Yanez et al. 2016; Gai et al. 2017). Nevertheless, besides this limited list of transcriptional modulators, there is scarce information regarding additional TFs impacting clock function in N. crassa, in contrast to what has been elucidated in other models. (Zhang and Kay 2010; Zheng and Sehgal 2012). Indeed, while there is evidence that in N. crassa the core TTFL is interlocked with a positive feedback loop in which FRQ posttranslationally upregulates WC-1 levels by stabilizing it, there is no information on which TFs may be also contributing to ancillary positive feedback loops that modulate WCC components. Thus, although there is a slight positive direct effect of WCC over wc-1 transcription (Kaldi et al. 2006; Sancar et al. 2012), FRQ favors wc-2 expression through an indirect and still unknown mechanism, involving unidentified TFs (Cheng et al. 2001; Neiss et al. 2008).
The ease in conducting genetic analyses (Beadle and Tatum 1941) and the robust circadian phenotype of daily spore production (banding) (Sargent et al. 1966; Montenegro-Montero et al. 2015), have made N. crassa a key model for unveiling the molecular details of clocks (Dunlap 1993, 2008; Brody 2011). This has been aided by forward genetics analyses of naturally occurring or induced mutations (Feldman and Hoyle 1973; Feldman and Atkinson 1978). However, these banding-based screens can yield confounding results by identifying mutations that affect conidiation per se and not necessarily the core clock (He et al. 2003; Zhou et al. 2018). Nevertheless, in recent years it has been possible to conduct bioluminescence-based studies, utilizing luciferase as a proxy to follow circadian gene expression reporting, unambiguously, the status of the core clock (Gooch et al. 2008; Larrondo et al. 2012; Montenegro-Montero et al. 2015). Importantly, luciferase clock reporters can reveal normal clock function in strains that otherwise may appear arrhythmic, or devoid of circadian banding in race tube assays (He et al. 2003; Shi et al. 2007; Cha et al. 2015; Larrondo et al. 2015; Montenegro-Montero et al. 2015; Olivares-Yanez et al. 2016). Taking all these into account, we adopted a reverse genetic approach, utilizing the N. crassa Knockout (KO) collection (Colot et al. 2006) as well as luminescent reporters to analyze circadian phenotypes in different genetic backgrounds. Herein, we describe such efforts focused on TFs, aiming at identifying the ones that when absent modulate clock period. Future work will provide further understanding on how these TFs regulate circadian properties either by modulating core clock components expression, or by other mechanisms.
Materials and methods
Strains and crosses
For this reverse genetic screen, we analyzed the sexual progeny obtained from crosses between circadian luciferase reporter strains and TF KOs available from the Fungal Genetic Stock Center (FGSC, Kansas City, MO, USA). The KOs were obtained as part of the N. crassa Functional Genomics Project, where individual loci were replaced with the hygromycin B phosphotransferase (hph) gene drug-resistance cassette (Colot et al. 2006; Collopy et al. 2010). We generated a list of putative TF-encoding genes based on the presence of the DNA binding domain sequence described for the N. crassa genome available in the web platform CISBP (cisbp.ccbr.utoronto.ca) (Weirauch et al. 2014). Such list included other genes previously related to transcriptional function (Borkovich et al. 2004; Tian et al. 2011), and which has been recently compiled (Carrillo et al. 2017), yielding a final list of 302 putative TFs loci listed in the Supplementary Table S1.
The reporter utilized in the primary screen was a firefly luciferase gene under the control of a minimal frq clock promoter (frqc-box-luc) integrated into the his-3 locus in LG I: his-3::frqc-box-luc (Gooch et al. 2008; Larrondo et al. 2015). Alternatively, another reporter consisting on a destabilized firefly luciferase (containing a PEST domain), under the control of the same minimal frqc-box promoter was utilized, this time targeted to the csr-1 locus: csr-1::frqc-box-lucPEST, as previously described (Cha et al. 2015; Olivares-Yanez et al. 2016). Both loci are ∼3 million bp apart, improving the number of successful crosses to that linkage group, by diminishing cosegregation of the reporter and hygromycin cassettes (Bardiya and Shiu 2007; Gooch et al. 2008; Honda and Selker 2009; Gooch et al. 2014). To confirm that the observed effects were not due to some spurious factor arising during the cross, and to also test the extent of circadian alterations caused by the missing gene, we also conducted a secondary screen utilizing a circadian output reporter. For this, we used con-10luc (con-10luc-bar) (Lauter and Yanofsky 1993; Olivares-Yanez et al. 2016), which corresponds to a fusion of luciferase to the con-10 ORF at its endogenous locus. And while technically con-10luc is a translational reporter, it faithfully recapitulates core circadian alterations as frqc-box-luc does (Olivares-Yanez 2015) and, therefore, it helps further characterizing the mutants of interest. The reporters were introduced in lab strains derived from crosses between progenies of 87-74 and FGSC #9568 (Larrondo et al. 2015; Olivares-Yanez 2015) which were used as the parental strains for the crosses. They were also utilized as WT controls, along with selected hygromycin sensitive siblings from the progenies derived from the screen crosses.
Out of a list of 302 putative TFs encoding genes in the N. crassa genome (Montenegro-Montero 2014), 45 KO strains were not available in the N. crassa FGSC KO collection (http://www.fgsc.net/) at the time we obtained the arrayed strains from the FGSC.
Growth conditions
Culture conditions for vegetative growth and asexual reproduction utilized Vogel minimal medium (VM) (Vogel 1956), whereas conditions for sexual development used synthetic crossing medium (SCM) (Westergaard and Mitchell 1947). Sorbose-containing medium (FIGS) was used for colony isolation on plates and ascospore germination and isolation (Davis and de Serres 1970). Picked ascospores were then grown on slants containing VM media supplemented with hygromycin (200 μg/ml; Calbiochem, San Diego, CA, USA) and luciferin (GoldBio) (10 μM), in order to select for progenies carrying knockout cassettes and reporter activity, respectively.
To conduct the circadian analyses (see below), spores from the selected progenies were inoculated in a 96 well plate containing LNN-CCD media (Larrondo et al. 2015; Olivares-Yanez 2015), with 25 μM of Luciferin (GoldBio). Cultures were grown for 24 hours in constant light (LL) at 25°C and then were analyzed under free-running conditions; consisting of constant darkness (DD) and 25°C, for 4–5 days in Percival incubators equipped with CCD PIXIS 1024B cameras (Princeton Instruments). As part of the high-throughput design, several 96 well-plates were run together in a single CCD camera run.
Luciferase data analysis and statistical tests
The resulting images series obtained from CCD camera runs were analyzed with a customized script for ImageJ (Larrondo et al. 2012). The acquired data sets varied in some cases containing 2 or 3 pictures per hour, with exposition times of 10 or 5 minutes respectively, a difference that does not affect the analyses as information can be compared throughout the data sets. Importantly, control wild-type (WT) strains were included in each 96-well plate and each experimental run. The obtained luciferase traces were analyzed as raw as well as normalized data sets (see below).
For the circadian analysis of the recorded time series, the data were uploaded as individual CCD camera runs (each run containing the corresponding WT controls) in BioDare2 (Biological Data Repository 2), a free-available online platform (https://biodare2.ed.ac.uk/) (Moore et al. 2014; Zielinski et al. 2014) which provides a comprehensive analysis of circadian parameters using different algorithms (Zielinski et al. 2014). The data were processed in the following manner for the primary screen: first, data were detrended to remove stationary effects over the time series, as such trends can cause distortions in the data masking circadian information. In addition, we discarded the first and last 12 hours of the data to minimize noise effects associated with the transition from light to darkness and improve subsequent detrending. Periods were calculated using a fast Fourier transform-nonlinear least-squares analysis (FFT-NLLS). Finally, we normalized and aligned every data point respect to the average of all points in their time series to facilitate their visualization (Plautz et al. 1997; Zielinski et al. 2014). The entire data sets produced in this genetic screen are stored in the BioDare2 platform, as an open access repository, with the spirit of propelling further analysis of these and other data by the circadian community. Importantly, the data sets from each CCD camera run are enlisted in the platform as “Neurospora TF Circadian Clock” plus the date of the CCD camera run; these entries also describe the experimental conditions, reporters and the TF KOs analyzed in each run. Period was re-calculated (see below) to compare this circadian parameter for each strain with an internal control (WT) in each 96-experimental run, to minimize the potential noise when comparing different plates in different experiments. The WT strains used in each plate and camera run were the parental reporters used for the crosses, their siblings, along WT siblings (hygromycin sensitive) of the KO crosses. The averaged period calculated for these WT is 21.76 hours, similar to the previous reported value of ∼21.5 utilizing similar WT controls (Larrondo et al. 2015). For the analyses, we calculated period change (Δτ; as PeriodKO—Periodwt.), measured in hours.
We defined the tolerance interval for the WT population using their Δτ in each of the experiments, taking three standard deviation from the mean population (Zhang et al. 2009), after confirming their normal distribution (Shapiro–Wilk test, P < 0.05). With this interval, we covered approximately 99.7% of the WT population and we were able to define TF KOs of interest as the ones that fell outside this range. To reduce the outlier effects, we compared the median of the different selected progenies from each cross.
To analyze the circadian defects between the obtained results using the core and the circadian output reporters we applied a t-test comparing them, discarding samples that showed different results (P < 0.05).
N. crassa knockout complementation
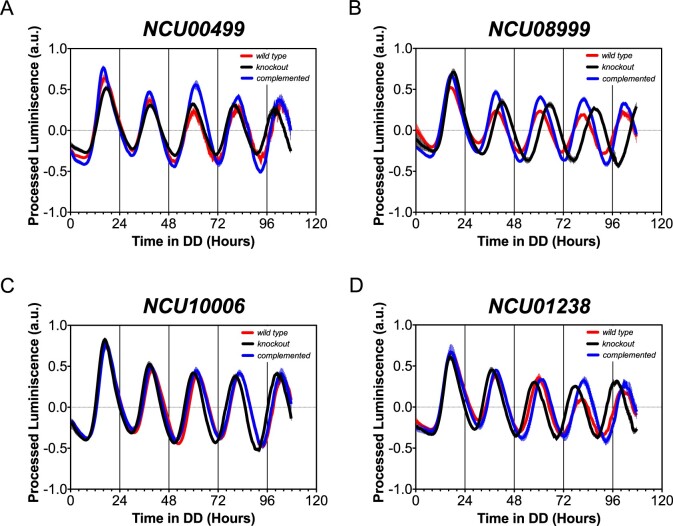
Selected KO strains from the primary genetic screen were complemented by electroporating their conidia with an amplicon containing the corresponding gene, aiming at replacing the hph cassette located in the respective knockout loci (Colot et al. 2006; Collopy et al. 2010). The complementation cassettes were constructed by yeast recombination cloning (Oldenburg et al. 1997; Raymond et al. 1999) containing 5’- and 3’-Flanking regions with the ORF plus a V5-tag and the bar cassette for resistance selection (Collopy et al. 2010). Subsequently, the selection was made through microconidiation to obtain homokayotic strains containing the complemented genes at their endogenous loci (Ebbole and Sachs 1990). Thus, complementation was conducted on a subset of particular KOs derived from the screen, to confirm that the absence of a specific TF encoding gene is the cause of the observed period defect. As the absence of some TFs leads to conidial and growth problems (Carrillo et al. 2017), we focused instead on strains of interest that could be easily subjected to a transformation protocol. Thus, we complemented KOs for NCU01238, NCU00499, NCU10006, and NCU08999, which were transformed with a cassette reconstituting the missing ORF, plus a V5 tag: complementation of the four above-mentioned mutant loci recovered a WT period phenotype.
Data availability
Strains and plasmids are available upon request. Circadian data sets associated with the genetic screen are available at https://biodare2.ed.ac.uk/ (Zielinski et al. 2014) as “Neurospora TF Circadian Clock” and have been also uploaded as supplementary tables to figshare: https://gsajournals.figshare.com/articles/figure/Supplemental_Material_for_Mu_oz-Guzm_n_Caballero_and_Larrondo_2021/14036507?file=26476886.
Results
Primary circadian screen
To identify novel regulators impacting the N. crassa circadian clock we adopted a reverse genetic strategy, focusing on putative TFs encoded in its genome (Borkovich et al. 2004; Tian et al. 2011; Weirauch et al. 2014). The screen consisted of analyzing the behavior of circadian luciferase reporters in the absence of individual TFs. As indicated in the methods section, this was achieved by crossing strains missing a particular TF, available from the N. crassa KO collection, with strains containing a circadian luciferase reporter. Diverse biological and technical issues limited the current extensiveness of this screen: we started our study with a list of 289 genes encoding for putative TFs in the N. crassa genome, defined from previous work from our lab (Weirauch et al. 2014), adding later on other putative TF encoding genes (Borkovich et al. 2004; Tian et al. 2011; Carrillo et al. 2017), yielding a final list of 302 TF possible candidates summarized in Supplementary Table S1. Nevertheless, KOs for 45 of these genes were unavailable in our version of the N. crassa KO collection (Colot et al. 2006) when we started the screen (Supplementary Table S1; KOs not available), while some of the genes were essential and therefore obtaining such mutants as homokaryons were not possible (Giaever and Nislow 2014; Carrillo et al. 2017). Thus, for the available 257 KOs, sexual crosses were conducted with a strain containing a clock-luciferase reporter, in order to analyze the effect of deleting specific TFs. From this long list, 91 crosses failed to yield enough hygR, luc+ offspring for the circadian luciferase analyses.
Genetic linkage issues can explain some of these 91 nonproductive crosses; 41 KO loci are in the same chromosome (LGI) as his-3, implying a genetic linkage between the hph cassette and the luciferase reporter. Therefore, for these unsuccessful crosses (based on a reporter strain where luciferase was inserted at his-3) (Honda and Selker 2009), we conducted new crosses for 18 knockouts, but this time utilizing a frq-c-box reporter located at a different region of LGI; the csr-1 locus (Bardiya and Shiu 2007), which allowed obtaining progeny for 11 additional TF Knockouts, leaving only 80 unsuccessful crosses (Supplementary Table S1; failed offspring) of which 30 can be clearly attributed to genetic linkage. Importantly, among these 80, many of the KOs appeared to be associated with developmental and growth problems (Carrillo et al. 2017), partially explaining the failure to obtain successful progenies. Thus, in toto, this screen circadianly examined a total of 177 TFs knockouts, corresponding to ∼60% of the cohort of putative N. crassa transcriptional regulators (Supplementary Table S1; analyzed strains). The resulting progenies from each cross, and the corresponding WT controls (parental strains and hygromycin sensitive siblings) were monitored for luciferase expression, and analyzed through the Biodare2 platform, focusing on period change (Δτ) (Moore et al. 2014; Zielinski et al. 2014) and plotted accordingly as shown in Supplementary Figure S1. Progeny of two of the 177 crosses were not included in the plots as they were cataloged as arrhythmic: Δwc-1 and Δwc-2, which is expected as they correspond to core clock components (Supplementary Figure S1). Interestingly, the analysis of ΔNCU02666 first suggested that its absence also abrogated rhythms, as it exhibited extremely low and apparently arrhythmic LUC signals. Nevertheless, normalization of the data revealed weak, but rhythmic oscillations for that reporter (c-box-luc), whilst analysis with a different one (con-10luc, see next section) confirmed that in ΔNCU02666 the clock still runs with a WT period (Supplementary Figure S2). The reason why in this mutant the behavior of the c-box reporter is extremely weak, although the core clock still runs, remains to be determined.
To select for the strongest circadian phenotypes in our screen, we defined upper and lower thresholds based on a tolerance interval of three standard deviations of the WT mean values for Δτ (see Materials and Methods; Supplementary Figure S3); this tolerance interval corresponds to ±0.49 hours. With this approach, we covered ∼99.7% of the WT group (assuming a normal distribution, Shapiro–Wilk test P < 0.05). The selection of TF KOs of interest comprise strains with calculated median outside this tolerance interval (Zhang et al. 2009). For a better visualization of the scattered Δτ data emerging from the plotted 175 crosses, it was separated in two groups: strains exhibiting shorter (Figure 1) and longer (Figure 2) periods. The results of our primary screen analyses are summarized in Supplementary Table S2, indicating the experimental identifiers, calculated period change (Δτ), number of biological replicates, and the descriptive statistics for each of the studied TF KOs; also, we listed in a secondary supplementary table (Supplementary Table S3), the raw results from each individual strain retrieved from the BioDare2 platform for simpler and faster access.
Figure 1.
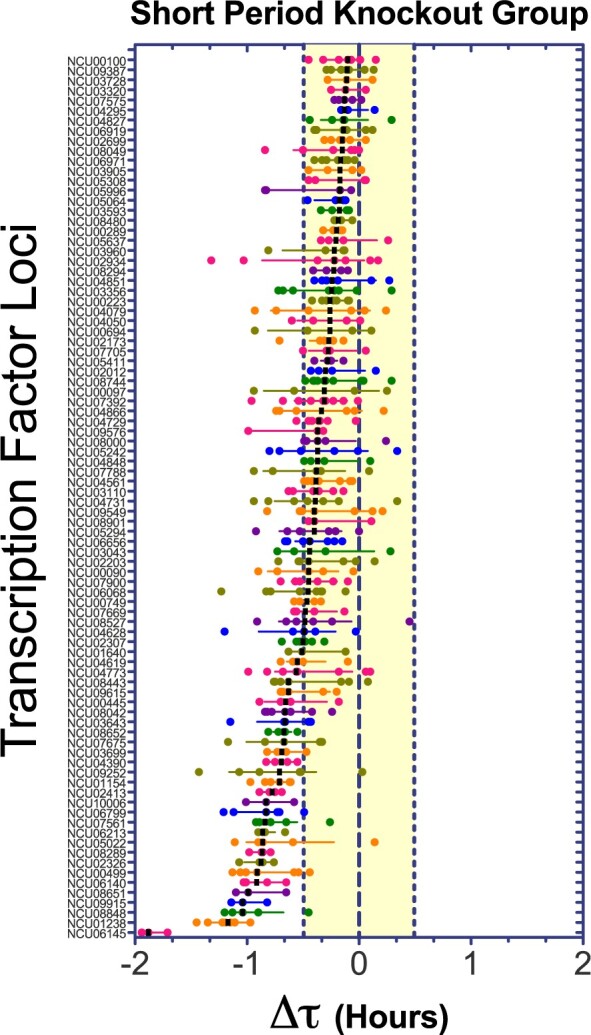
Primary circadian screen of TFs KO strains (short period group). The plot of the short period KO group depicts the Δτ values for 88 KOs crosses. Each dot reflects the value for a replicate, the black bar is the median for each KO population and the error bars are the interquartile range. Transcription factor loci are ordered by median values and the yellow filled range reflect the behavior of the 99.7% of the WT population. Twenty-nine strains are outside the range and are selected as TF KO candidates with shorter periods.
Figure 2.
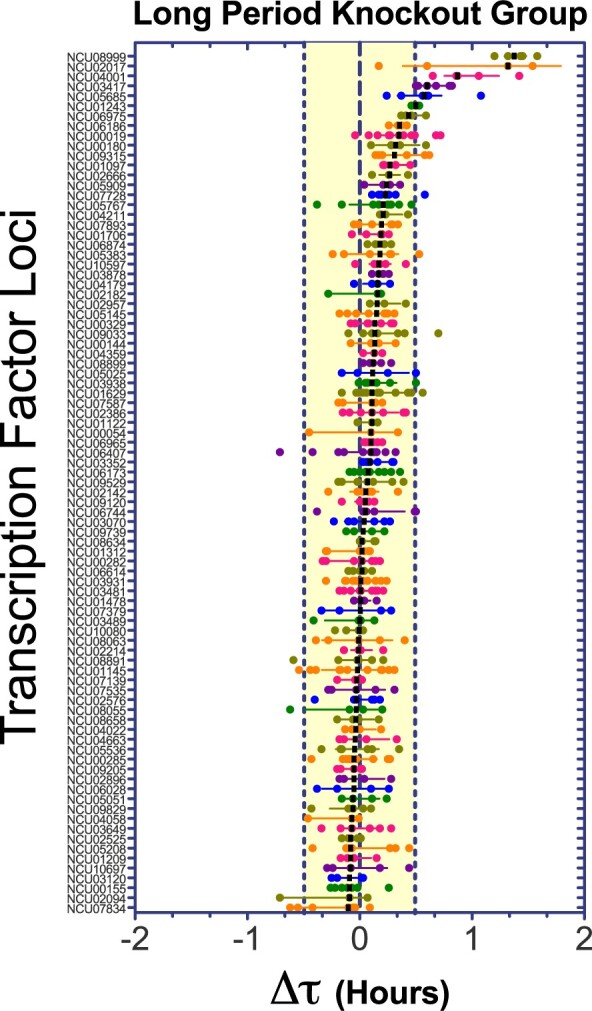
Primary circadian screen of TF KO strains (long period group). The plot of the long period KO group depicts the Δτ values for 87 KOs crosses. Each dot reflects the value for a replicate, the black bar is the median for each KO population and the error bars are the interquartile range. The transcription factor loci are ordered by median values, where the yellow filled range reflect the behavior of the 99.7% of the WT population. Based on this, seven KO strains were selected as candidates with a longer period.
Out of this analysis, only 36 KOs showed statistically significant differences in the primary screen, based on the above-mentioned criteria of three standard deviation from the wild-type population mean (Zhang et al. 2009). Notably, 30 out of 36 TF KOs displayed shorter period, results that contrast with what has been observed in similar screens conducted in other circadian models, where most of the identified genes affecting the clock yield longer periods (Matsumoto et al. 2007; Hirota et al. 2008; Zhang et al. 2009; Agrawal and Hardin 2016). To ensure that the circadian change in these 36 candidates was caused by the removal of the specific locus of interest, we also analyzed WT siblings for each selected KO. This additional evaluation helped reducing the deviation associated with technical noise derived from the analysis of different plates and/or camera runs, or for potential unlinked spontaneous mutations present in the KO or emerging during the sexual crosses (Watters and Stadler 1995).
Confirmation of circadian phenotype by output reporters
To confirm that the clock defects observed in our screen with a minimal core clock reporter (c-box-luc) were actually due to circadian alterations, and not to unidentified technical issues, such as low reporter expression or other reasons (see Supplementary Figure S2), we evaluated the behavior of a different reporter in the selected TF KOs. For this, we utilized the output gene con-10 (NCU07325), which exhibits robust circadian expression (Lauter and Yanofsky 1993; Hurley et al. 2014). con-10 is a vastly studied gene, expressed in late stages of conidial differentiation (Roberts et al. 1988; Olmedo et al. 2010a), responds to light (Olmedo et al. 2010b; Wu et al. 2014), and is highly expressed during carbon starvation, similar to our experimental conditions, (Xiong et al. 2014, 2017) among others regulations (Kays et al. 2000; Thompson et al. 2008; Wang et al. 2012; Pengkit et al. 2016; Dekhang et al. 2017). We created a con-10luc reporter by integrating luc at the corresponding locus obtaining a fusion between the latter and the con-10 ORF (Olivares-Yanez et al. 2016).
This secondary analysis (Figure 3) reduced the number of TFs of interest derived from the primary screen. Four of the 36 candidate TFs were disregarded in the following analysis for experimental issues; three of them (NCU04390, NCU08289, and NCU08651) failed to produce successful offspring with the con-10luc reporter, whereas the KO for NCU02017 severely affected the expression of the circadian output reporter; leaving further confirmation of these KOs pending. Nine strains were also left out of the refined list of TFs of interest as in the con-10luc analyses since period differences did not statistically recapitulate what had been observed with the core clock reporter (Figure 3).
Figure 3.
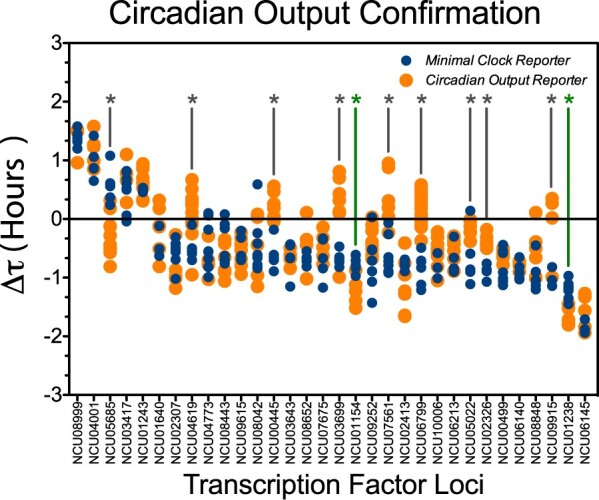
Confirmation of TF candidates using a Circadian Output Reporter. Thirty-two TF candidates were successfully analyzed utilizing the circadian output reporter con-10luc (orange dots) and frqc-box-luc (blue dots), where each dot is a biological replicate. Eight candidates were statistically different between both reporters (gray asterisk), showing a Δτ value for the circadian output reporters close to zero. Two strains (Δsub-1 and ada-9) also showed significant differences between both reporters (green asterisk), but with a shorter period for con-10luc compared to frqc-box-luc. All things considered, the intersection of both screens allowed identifying twenty-three TF as significantly impacting circadian period in Neurospora.
Thus, interestingly, the use of two different circadian reporters allowed us to observe differences in the degree of circadian alteration of several TF KOs strains (Supplementary Figure S2). These differences could relate to a direct effect of the candidate TF in both pathways: alterations on the N. crassa clock and in output pathways (con-10), versus a major effect only in the former and compensatory mechanisms in the latter (albeit this would be less likely). A clear example of a KO impacting both core clock function and the output pathways while severely compromising the quality of con-10 rhythms is Δada-2 (NCU02017), where the expression of con-10luc appears severely affected. This TF KO shows defects in both sexual and asexual development (Sun et al. 2019) and differential gene expression upon carbon source differences (Reilly et al. 2015), conditions where con-10 regulation is affected (Wang et al. 2012; Xiong et al. 2014, 2017).
In terms of period, Δsub-1 (NCU01154) and Δada-9 (NCU01238), where significantly shorter when examining output compared to the core clock reporter (Figure 3). Both of these TFs had been previously associated with con-10 regulation: in Δsub-1, light induction of con-10 is reduced (Sancar et al. 2015a), and in the case of ADA-9, this TF is capable of interacting with RCO-1 (Olivares-Yanez et al. 2016), a transcriptional co-repressor that drastically affects con-10 expression (Yamashiro et al. 1996). In addition, both KOs show alterations on sexual and asexual development (Carrillo et al. 2017), conditions where con-10 expression is differentially regulated, as commented above. Yet, it is not obvious to explain that period for a particular KO would yield so different results with the core-clock and output reporters.
Thus, applying a conservative criterion restricting the list to those which absence showed significant period changes in both screens, we identified 23 TF encoding genes, which corresponds to ∼7.6% of the total number of putative TFs in N. crassa (and a ∼13% of the successfully analyzed for luc expression). Importantly, other clock screens based on similar reverse genetics approaches have shown variable results. For example, while in a broad screen using human cells the rate of genes of interest was near 1% for a total of 22,468 genes, or a smaller subgroup (Maier et al. 2009; Zhang et al. 2009), depending on the size of the screen or the category of the analyzed genes such rates can go up. Thus, genes flagged as of interest were ∼3.8% of 133 circadianly expressed genes screened in Drosophila (Matsumoto et al. 2007), compared to ∼22% of 86 phosphatase encoding genes in the same organism (Agrawal and Hardin 2016). To our knowledge, there are no published studies exclusively focusing on the circadian impact of TFs, although TFs with a clock-related function have been already identified in unbiased screens (Matsumoto et al. 2007).
The TFs of interest are associated with multiple processes
Thus, based on the strict criteria of showing period alterations when assessing with both reporters, we have identified 23 TFs with strong and reproducible circadian defects, which appear to be involved in a broad range of cellular processes, as it is indicated in the next paragraphs for each one. The absence of the corresponding ORFs in the progeny of these 23 KOs was confirmed by PCR, and for four particular KOs (that will be further pursued for mechanistic studies) we conducted complementation assays (see below and Supplementary Table S4).
NCU08999: (Δτ = +1.38 h) this locus encodes for a bHLH TF, which is an ortholog of the yeast centromere binding factor 1 (cbf-1) (Stoyan et al. 2001). This gene is known to be expressed late after phytosphingosine treatment (Videira et al. 2009). Recently this N. crassa TF was described to have a similar circadian phenotype (∼2 h lengthened period), as determined by race tube assays (Cao et al. 2018). Interestingly, the authors failed to observe circadian expression of luciferase in their characterization of this mutant, which could be partially explained by their experimental settings (see Discussion), a discrepancy we have seen with other mutants like Δrco-1 (Zhou et al. 2013; Olivares-Yanez et al. 2016). The period phenotype associated with ΔNCU08999 was restored to WT upon complementation (Figure 4B).
Figure 4.
Validation of TFs of interest by complementation. Four KO strains from the selected candidates were individually transformed, as explained in methods, with a construct that allowed reinserting the missing TF locus. KOs for the TF encoding genes: A, ada-1 (NCU00499); B, cbf-1 (NCU08999); C, sgr-30 (NCU10006), and D, ada-9 (NCU01238) were complemented. The red lines, depict the oscillations of the wild types; black, the traces of the KO strains, whereas the blue lines, correspond to the complemented KO strains. For each strain, we used at least three biological replicates. All complementations successfully restored the wild-type circadian phenotype.
NCU04001: (Δτ = +0.87 h) identified as female fertility 7 (ff-7), this zinc-finger TF was found to interact with SUB-1 (NCU01154) in regulating the expression of several genes in LL and DD conditions. This transient complex is able to interact with WCC upon light exposure, modulating several genes (Sancar et al. 2015a). ADV-1 (NCU07392) regulates NCU04001/ff-7 expression in a light dependent manner (Dekhang et al. 2017). ff-7 is subjected to metabolic regulation and its expression is decreased in a Δcol-26 (NCU07788) strain in amylose (Xiong et al. 2017), and it is also reduced in a strain overexpressing CSP-1 (NCU02713) in light conditions (Sancar et al. 2011).
NCU03417: (Δτ = +0.52 h) encodes for a hypothetical C6 zinc-finger containing protein. Its expression in DD is altered in the absence of MAK-1 (Mitogen-Activated Protein Kinase) (Bennett et al. 2013).
NCU01243: (Δτ = +0.50 h) corresponds to zinc-finger 41 (znf-41), a TF highly conserved in many pathogenic fungi. In N. crassa, it is induced by menadione, and its absence leads to enhanced ROS sensitivity (Zhu et al. 2013). It is a light-responsive gene, directly regulated by WCC (Smith et al. 2010), while also displaying rhythmic expression (Hurley et al. 2014).
NCU01640: (Δτ = −0.51 h) corresponds to a C2H2 TF named regulatory particle non-ATPase-like-4 (rpn-4). rpn-4 is a light-responsive gene, whose expression is up-regulated by ADV-1 (Dekhang et al. 2017) and down-regulated by CSP-1 (Sancar et al. 2011). Both of these TFs are rhythmically expressed (Hurley et al. 2014; Dekhang et al. 2017), and are related to responses to particular sugar availability, implying a possible metabolic control over rpn-4. Thus, expression of the latter and of adv-1, are decreased in the presence of amylose in a Δcol-26 strain (Xiong et al. 2017). Also, the expression of this gene is strongly activated under conditions which challenge cell integrity; such as phytosphingosine treatment, an inducer of programed cell death in N. crassa, (Videira et al. 2009), and conditions that favor cell fusion and cell-to-cell communication, regulated by ADV-1 and PP-1 (NCU00340) (Fischer et al. 2018). This gene exhibits rhythmic expression (Hurley et al. 2014).
NCU02307: (Δτ = −0.51 h) is a light-responsive gene encoding for a zinc-finger TFs, up-regulated indirectly by ADV-1 (Dekhang et al. 2017), with reduced expression in a csp-1 overexpressing strain (Sancar et al. 2011). It varies differentially between new and old vegetative tissue (Tian et al. 2011), and it is mostly expressed in aerial hyphae compared to mycelia (Greenwald et al. 2010). Albeit this TF has not been well characterized yet, its expression is strongly tied to plant cell-wall degradation, being highly expressed under such conditions (Tian et al. 2009), depending on XLR-1, a well-known TF involved in hemicellulose degradation (Sun et al. 2012a). Its expression is also differentially regulated in starch, where it is repressed by the TF COL-26 (Xiong et al. 2017). NCU02307 has been reported as exhibiting rhythmic expression, with a peak in the evening (Hurley et al. 2014).
NCU04773: (Δτ = −0.56 h) encodes for a conserved fungal hypothetical protein containing a copper ion-binding domain. A related ortholog is GRISEA, a copper-dependent TF in Podospora anserina or yeast MAC1 (Borghouts and Osiewacz 1998). Podospora lacking GRISEA shows an increased lifespan by reduction of ROS and ATP, this through switching from a copper-dependent to an iron-dependent respiration system (Borghouts et al. 1997; 2001; Gredilla et al. 2006).
NCU08443: (Δτ = −0.63 h) This gene encodes for a zinc ion-binding TF. It has been shown to display rhythmic expression by RNA-Seq, with a morning peak of expression (Hurley et al. 2014).
NCU09615: (Δτ = −0.63 h) encodes for a zinc TF named vegetative asexual development 14 (vad-14) (Carrillo et al. 2017) and is a light-responsive gene, with a late-light expression pattern—between 60 and 120 min—(Wu et al. 2014), directly regulated by WCC (Smith et al. 2010) and indirectly by ADV-1 (Dekhang et al. 2017). It has been described to have rhythmic expression, with a morning peak (Hurley et al. 2014). Also, it is induced by starch-related carbon sources and not by simple sugars such as maltose (Xiong et al. 2017). In cell viability assays its levels are increased (Hutchison et al. 2009).
NCU08042: (Δτ = −0.64 h) this locus encodes for the C6 zinc-finger TF cellulose degradation regulator 2 (CLR-2). Identified by its severe growth defect on crystalized cellulose (Avicel) (Coradetti et al. 2012). CLR-2 regulates the expression of several cellulolytic genes (Coradetti et al. 2013), and itself is induced by cellulose in a CLR-1 (NCU07705) dependent manner (Coradetti et al. 2012; Znameroski et al. 2012; Xiong et al. 2017).
NCU03643: (Δτ = −0.66 h) Encodes for a zinc-finger TF ortholog of cutinase transcription factor 1 beta (ctf1β) (Li et al. 2002; Tang et al. 2011). It is a light-responsive gene (Dong et al. 2008; Chen et al. 2009) whose expression appears to be regulated by FF-7 (Sancar et al. 2015a), and it is also affected by menadione (Zhu et al. 2013). It is downregulated in the absence of col-26, under amylose conditions (Xiong et al. 2017), and it exhibits a delayed up-regulation to quinic acid (QA), likely mediated by QA-1F (NCU06028), the main controller of the QA cluster in N. crassa (Patel and Giles 1985; Tang et al. 2011). It is positively regulated by direct binding of ADV-1, providing its basal expression (Dekhang et al. 2017). CTF1β has been predicted as an “activator” of the delayed group of metabolic genes up-regulated after QA addition (Tang et al. 2011), and it has also been reported as exhibiting rhythmic expression (Hurley et al. 2014).
NCU08652: (Δτ = −0.67 h) this locus encodes for hypothetical C6 zinc-finger TF, described based on its knockout phenotype as slower growth rate 31 (sgr-31) (Carrillo et al. 2017). It has a differential expression when grown under maltose compared with other carbon source or sucrose (Xiong et al. 2017).
NCU07675: (Δτ = −0.67 h) Encodes for a C6 zinc-finger TF defined as tall aerial hyphae 4 (tah-10). Its expression is affected by amylose, showing decreased levels in the absence of col-26 (Xiong et al. 2017), with high expression in asexual reproduction conditions (Wang et al. 2019). Its expression is reduced in a CSP-1 overexpression strain, albeit it is not a direct target of this TF (Sancar et al. 2011).
NCU01154: (Δτ = −0.71 h) submerged protoperithecia 1 (sub-1), a GATA TF, is an early light-responsive gene, directly regulated by WCC (Chen et al. 2010; Smith et al. 2010; Wu et al. 2014), exhibiting a rhythmic expression with a morning pattern (Hurley et al. 2014). Functionally, it is able to dynamically interact with FF-7, and regulate the expression of several genes in light and darkness, also acting synergistically with WCC in light-responsive genes (Sancar et al. 2015a), connecting light responses and fungal development (Kasuga et al. 2005; Wang et al. 2019). It is differentially expressed in young versus old tissue (Tian et al. 2011), favored in sexual reproduction and down-regulated in asexual development (Wang et al. 2019). It is also subjected to metabolic regulation under different carbon sources, such as maltose and amylose in a col-26 dependent manner (Xiong et al. 2017).
NCU09252: (Δτ = −0.71 h) Encodes for a hypothetical C2H2 TF whose expression in darkness depends on SUB-1 and FF-7 at basal levels (Sancar et al. 2015a), as well as on the TF vad-5 (NCU06799) (Sun et al. 2012b). It has been described as clock-controlled and temperature-regulated (Nowrousian et al. 2003), and is member of the over-expressed genes of the starch-regulon in N. crassa, dependent on COL-26 (Xiong et al. 2017), and preferentially expressed in young versus old tissue (Tian et al. 2011).
NCU02413: (Δτ = −0.78 h) defined as response regulator 2 (rrg-2), is part of a two-component regulatory system for stress response in N. crassa (Catlett et al. 2003; Froehlich et al. 2005; Jones et al. 2007); it has been described as containing a truncated HSF DNA-binding domain and it is involved in ROS responses (Banno et al. 2007; Thompson et al. 2008). In addition, it exhibits a repressive effect on the secretion of lignocellulases, based on its requirement in the ER stress response (Fan et al. 2015). It is also downregulated during cell viability assays (Hutchison et al. 2009), being more expressed in mycelia than in aerial hyphae (Greenwald et al. 2010).
NCU10006: (Δτ = −0.83 h) Named as slow growth rate 30 (sgr-30) (Carrillo et al. 2017), is a gene coding for a C2H2 TF with an increased expression under high level of phosphate in the media (Gras et al. 2013), with no additional function or process associated with this gene. Complementation of the mutant back with the NCU10006 gene recovered WT period (Figure 4C).
NCU06213: (Δτ = −0.85 h) zinc-finger transcription factor 9 (znf-9). This gene has shown to display rhythmic expression, with an evening peak (Hurley et al. 2014).
NCU00499: (Δτ = −0.91 h) corresponds to all development altered 1 (ada-1), a bZIP, of which KO yields a strong growth phenotype (Carrillo et al. 2017). This TF has an increased expression in young versus old tissue, and mostly in aerial hyphae (Greenwald et al. 2010; Tian et al. 2011), and displays rhythmic expression (Hurley et al. 2014). The period phenotype observed in Δada-1 was reverted by complementation (Figure 4A).
NCU06140: (Δτ = −0.92 h) vegetative and sexual development (vsd-8) (Carrillo et al. 2017) encodes for a MYB TF. Under light exposure, it is up-regulated by SUB-1 and modulated by ADV-1 (Sancar et al. 2015a; Dekhang et al. 2017); in darkness, its basal levels are diminished in the absence of ff-7 and sub-1, being also a direct target of FF-7 (Sancar et al. 2015a). It exhibits metabolic regulation, having an increased expression under amylose in a Δcol-26 strain (Xiong et al. 2017), and is a downstream target of the MAP kinase signaling through the regulation by the TF PP-1 (Gras et al. 2013; Leeder et al. 2013).
NCU08848: (Δτ = −1.04 h) is a hypothetical protein with a zinc-ion binding domain. Its expression varies during conidial germination (Kasuga et al. 2005).
NCU01238: (Δτ = −1.17 h) is a PHD TF named as all developmental alteration 9 (ada-9). It is capable of interacting with the co-repressor RCO-1, a transcriptional regulator, devoid of a DNA binding domain, known to impact clock regulation and which absence leads to lengthened period (Olivares-Yanez et al. 2016). Complementation of ΔNCU01238 recovered WT period (Figure 4D).
NCU06145: (Δτ = −1.88 h) Encodes for a C2H2 TF named as really interesting gene 6 (ring-6). It is also a light down-regulated gene, bound by ADV-1 (Dekhang et al. 2017), with an expression that is affected by the presence of maltose (Xiong et al. 2017). This KO strain does not show the characteristic displays of apical branching when is exposed to cold shock (Watters et al. 2018). In our hands, ΔNCU06145 yielded the shortest period among the mutants identified in this screen.
Discussion
Our study constitutes, so far, one of the most extensive reverse genetic analyses concentrating on the N. crassa clock utilizing luciferase as a proxy for circadian molecular phenotypes. Such an approach is a major advance compared to previous N. crassa circadian forward genetic screens based on race tubes (Feldman and Hoyle 1973; Feldman and Atkinson 1978), due its fine spatiotemporal resolution, ideal for measuring key clock parameters as period, and facilitating high-throughput analyses by monitoring multiple strains and replicas simultaneously (Gooch et al. 2008; Larrondo et al. 2012; Cha et al. 2015). Undeniably, the race tube assay is a robust method for circadian screenings, which led to the identification of frequency (Feldman and Hoyle 1973; Loros and Feldman 1986; Aronson et al. 1994a) and several other clock affecting loci (Lakin-Thomas et al. 1990; Loros and Dunlap 2001; Morgan et al. 2001; Lakin-Thomas et al. 2011), some of which were subsequently functionally characterized such as prd-1, prd-4, and prd-6 (Pregueiro et al. 2006; Emerson et al. 2015; Adhvaryu et al. 2016). However, race tubes may overlook mutations of interest, as any alterations which impact conidiation per se would show overt arrhythmicity, although core circadian function may remain intact, or even with distorted parameters. Indeed, KOs of TFs associated with asexual growth (Colot et al. 2006) could yield confusing or inconclusive results on race tube assays, as conidiation banding would be obscured (Baker et al. 2012).
Out of all the screened TFs (∼60% of 302 in N. crassa), WC-1 and WC-2 continue to be the only ones essential for the clockworks. Nevertheless, extensive analysis of the remaining TFs is needed in order to identify whether another regulator plays a critical role, most likely commanding the expression of clock components other than FRQ, such as the WCC, CK1, or FRH encoding genes. It is noteworthy, though, that none of the screened TF causes major clock alterations and that, although a limited number of TFs affect period, they do so within a rather narrow range of hours. The low number of TFs strongly impacting the clockworks could be explained by two possibilities: a “TF” or a “circadian system” interpretation. The former is the most parsimonious interpretation and implies that the circadian core oscillator is not critically regulated by a significant number of TFs, as opposed to what occurs in other regulatory systems. On the other hand, we have the “circadian system explanation,” which argues of a pervasive robustness of circadian systems, taking the clock phenotype as a robust structure resilient to individual genetic perturbations (Kitano 2002; Félix and Barkoulas 2015). A corollary of the latter premise is that some genetic perturbations may require a particular environmental stimulus, or combination of stimuli, to reveal an important role in the clockworks (Kitano 2002; Félix and Barkoulas 2015), this due the dynamic reprogramming of regulatory networks, which enables cells to modify network topology to adapt to complex environmental perturbations (Hu et al. 2016; Swift and Coruzzi 2017). Thus, a given regulator may not be identified as important in metabolic or temperature compensation unless defined sugar levels or temperatures are tested (Sancar et al. 2012; Olivares-Yanez et al. 2016). Indeed, supporting this idea are additional pieces of evidence to discuss. For example, CSP-1 (NCU02713), a TF implicated in metabolic compensation of the clock (Sancar et al. 2011, 2012), has been shown to modulate circadian period only under high glucose conditions, since under low glucose levels, no circadian alterations are appreciated (as we saw herein for Δcsp-1 in our screen conditions). Likewise, we have also reported that Δrco-1 is a mutant which exhibits a longer period, but is shortened to almost WT levels as glucose concentration is increased (Olivares-Yanez et al. 2016). Herein, we were able to observe clear luciferase rhythms in Δcbf-1, contrary to what was reported for this mutant (Cao et al. 2018); such difference could be due to the culture conditions used by the authors to monitor luciferase (GAI et al. 2017), which are similar to ours, except that theirs (FGS, consisting of 0.05% fructose, 0.05% glucose and 2% sorbose), contains other sugars and promotes colonial growth (ours is only 0.03% glucose). Importantly, the molecular rhythms we detected by bioluminescence confirmed the increase in period, which was described for this mutant based on race tube assays. Indeed, by parsimony it is expected that a robust overt conidiation rhythm, as seen for Δcbf-1 (Cao et al. 2018), should be accompanied by molecular rhythms in luc expression, as we have reported herein. Importantly, these case studies exemplify the advantage of having a minimal clock promoter reporter (frqc-box-luc) that can be easily crossed to mutants of interest, with no concerns for ripping as it would occur for full frq promoter-reporters (Gooch et al. 2008). Likewise, the con-10luc reporter, which we also utilized in this screen, can be easily crossed as it corresponds to a luc knock-in at the con-10 locus, creating a translational fusion reporter (Olivares-Yanez et al. 2016). Such resources are extremely useful considering the existing N. crassa KO collection (Colot et al. 2006), as well as the great assortment of historical mutants at the FGSC (McCluskey et al. 2011), which may be hard to screen by the conventional race tube assay, since many exhibit growth problems.
Interestingly, most of the 23 TFs candidates fall in two well-studied biological processes in N. crassa: light responses and carbon availability (Supplementary Table S4). Light triggers well-defined transcriptional responses in this fungus, mediated by the photoreceptor and TF WC-1, being also one of the main inputs to the clock, able to reset and entrain it (Montenegro-Montero et al. 2015). Several of the 23 TFs of interest appear to be subjected to both light- and circadian-regulation. Starting from WCC direct targets, we observed the case of sub-1, znf-41, and vad-14, where the three of them have been both reported to be clock-controlled and light-responsive genes, probably members of the second-tier of TFs able to control downstream circadian effectors (Smith et al. 2010; Hurley et al. 2014). Based on previous works (Smith et al. 2010; Sancar et al. 2011; Dekhang et al. 2017), we know that adv-1 and csp-1 are members of this second-tier, implying a plausible role on the clock-controlled and light-response of rpn-4, ctf1β, and NCU02307 genes, being these last three part of a third-tier of transcriptional regulation which could include a transcriptional feedback to the clock (Sancar et al. 2011; Dekhang et al. 2017). From the 23 TFs of interest, three of them have been described as having circadian expression, but no regulation by light: znf-9, ada-1, and NCU09252 (Hurley et al. 2014), being the latter the only known connected to SUB-1, a second-tier TF (Sancar et al. 2015a). The TFs of interest ring-6, vsd-8, and ff-7, have not been reported to have circadian expression, but are light-responsive genes regulated by ADV-1 (Dekhang et al. 2017). It is outstanding that out of the 23 TFs we identified as modulating period, 9 appear to be under clock-regulation themselves, intertwining aspects of output control back to core clock dynamics. Previous studies (Hurley et al. 2014; Sancar et al. 2015b), have indicated that ∼10% of the N. crassa TF genes are circadianly expressed; our results provide additional hints unveiling potential rhythmic transcriptional networks around the N. crassa circadian clock (Zhang and Kay 2010). Thus, an overrepresentation of the TFs of interest which are not only rhythmic but also, through yet unclear mechanisms, feedback to modulate circadian period, blurring the already fine line and directionality between clock output and core mechanisms. Moreover, we predict that by altering the growth conditions (i.e., varying nitrogen/sugar source or content) several of the clock phenotypes observed for a given set of TFs will change, potentially uncovering new aspects of the intricate mechanisms undelaying metabolic compensation, in the case of CSP-1 (see above) ( Sancar et al. 2011; 2012), or in developmental control, exemplified by SUB-1 and its role in the asexual-sexual switch (Wang et al. 2014, 2018).
N. crassa is a fungus that can be found growing on different type of plant tissues, and therefore, sensing nutrient availability is one a key aspect of its biology, involving a series of fast and accurate responses upon substrate recognition (Horta et al. 2019). Two of the genes of interest, clr-2 and NCU02307 are highly expressed upon exposure to complex substrates, like cellulose and hemicellulose respectively, each of them under the control of the main TFs regulating the expression on these substrates, CLR-1 and XLR-1 (Tian et al. 2009; Sun et al. 2012a). CLR-2 is a TF involved in the decomposition of the plant cell wall, but mainly controlling the expression of cellulases, and having no connection to the circadian clock components in cellulose conditions (Coradetti et al. 2012; Znameroski et al. 2012), whereas recent studies have shown how the clock nicely functions when N. crassa grows on plant material (Diaz and Larrondo 2020). Regarding complex carbon sources, for starch there is a defined starch-regulon commanded by the TF COL-26, with highly expressed genes under amylose conditions, where we can find the candidate gene NCU09252. Not part of the starch-regulon, but regulated by COL-26, are ff-7, rpn-4, sub-1, ctf1β, tah-10, and vsd-8, whereas vad-14 responds to amylose but no depending on COL-26 (Xiong et al. 2017). Simpler carbon source like maltose or glucose show effects in the expression of some of the genes of interest: for maltose there is an increase in the levels of ring-6, srg-31, and sub-1 (Xiong et al. 2017). Related to glucose metabolic compensation of the N. crassa clock, both the co-repressor RCO-1 and the transcriptional repressor CSP-1 have been implicated (Sancar et al. 2012; Olivares-Yanez et al. 2016), appearing to modulate an ancillary feedback loop to the clock under high glucose conditions. While these candidate genes are not glucose regulated, they are regulated by CSP-1 in response to light, like rpn-4 and NCU02307, the latter a plausible connection between light and metabolic regulation (Sancar et al. 2011).
Conclusions
Our screen, focused on TFs, and utilizing two different clock-reporters, has allowed identification of 23 TFs which absence leads to discrete period changes, most of them associated with period shortening. Although the underlying mechanisms of these circadian phenotypes still remain obscure, it is noteworthy that several of the TFs of interest are, themselves, light- or-clock regulated.
On the other hand, this screen has allowed confirmation of period defects for recently characterized mutants such as Δcbf-1, providing clear evidence of molecular rhythms in TF KO strains where they had been hard to observe using luciferase.
Finally, during this study, we have generated a ΔTF frqcbox-luc collection, spanning 177 different TFs, which constitute a valuable resource that will expedite new genetic screens contemplating particular single and combined environmental perturbations such as nutrients, pH, and temperature. This will help further unveiling the existence of a dynamic transcriptional network supporting robust and compensated clock function.
Acknowledgments
The authors thank Dr. Andrew Millar and Dr. Tomasz Zielinski (University of Edinburgh), for their valuable help in the use of the BioDare2 platform. They thank Alejandro Stevens for his technical assistance.
Funding
This research was funded by ANID—Millennium Science Initiative Program—Millennium Institute for Integrative Biology (iBio ICN17_022), ANID/FONDECYT 1171151, and the International Research Scholar program of the Howard Hughes Medical Institute.
Conflicts of interest
None declared.
Contributor Information
Felipe Muñoz-Guzmán, ANID—Millennium Science Initiative Program—Millennium Institute for Integrative Biology (iBio), Santiago 8331150, Chile; Departamento de Genética Molecular y Microbiología, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, Santiago 8331150, Chile.
Valeria Caballero, Departamento de Genética Molecular y Microbiología, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, Santiago 8331150, Chile.
Luis F Larrondo, ANID—Millennium Science Initiative Program—Millennium Institute for Integrative Biology (iBio), Santiago 8331150, Chile; Departamento de Genética Molecular y Microbiología, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, Santiago 8331150, Chile.
Literature cited
- Adhvaryu K, Firoozi G, Motavaze K, Lakin-Thomas P. 2016. PRD-1, a component of the circadian system of Neurospora crassa, is a member of the dead-box RNA helicase family. J Biol Rhythms 31:258–271. [DOI] [PubMed] [Google Scholar]
- Agrawal P, Hardin PE. 2016. An RNAi screen to identify protein phosphatases that function within the Drosophila circadian clock. G3 (Bethesda). 6:4227–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson BD, Johnson KA, Dunlap JC. 1994a. Circadian clock locus frequency: protein encoded by a single open reading frame defines period length and temperature compensation. Proc Natl Acad Sci USA. 91:7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson B Johnson K Loros J Dunlap J. 1994. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 263:1578–1584. 10.1126/science.8128244 [DOI] [PubMed] [Google Scholar]
- Baker CL, Loros JJ, Dunlap JC. 2012. The circadian clock of Neurospora crassa. FEMS Microbiol Rev. 36:95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno S, Noguchi R, Yamashita K, Fukumori F, Kimura M, et al. 2007. Roles of putative his-to-asp signaling modules HPT-1 and rrRRG-2, on viability and sensitivity to osmotic and oxidative stresses in Neurospora crassa. Curr Genet. 51:197–208. [DOI] [PubMed] [Google Scholar]
- Bardiya N, Shiu PK. 2007. Cyclosporin a-resistance based gene placement system for Neurospora crassa. Fungal Genet Biol. 44:307–314. [DOI] [PubMed] [Google Scholar]
- Beadle GW, Tatum EL. 1941. Genetic control of biochemical reactions in Neurospora. Proc Natl Acad Sci USA. 27:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LD, Beremand P, Thomas TL, Bell-Pedersen D. 2013. Circadian activation of the mitogen-activated protein kinase MAK-1 facilitates rhythms in clock-controlled genes in Neurospora crassa. Eukaryot Cell 12:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C, Kimpel E, Osiewacz HD. 1997. Mitochondrial DNA rearrangements of Podospora anserina are under the control of the nuclear gene grisea. Proc Natl Acad Sci USA. 94:10768–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C, Osiewacz HD. 1998. GRISEA, a copper-modulated transcription factor from Podospora anserina involved in senescence and morphogenesis, is an ortholog of MAC1 in Saccharomyces cerevisiae. Mol Gen Genet. 260:492–502. [DOI] [PubMed] [Google Scholar]
- Borghouts C, Werner A, Elthon T, Osiewacz HD. 2001. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol Cell Biol. 21:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, et al. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev. 68:1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody S. 2011. The genetics of circadian rhythms. Introduction. Adv Genet. 74:1–12. [DOI] [PubMed] [Google Scholar]
- Brown SA, Kowalska E, Dallmann R. 2012. (Re)inventing the circadian feedback loop. Dev Cell 22:477–487. [DOI] [PubMed] [Google Scholar]
- Brunner M, Kaldi K. 2008. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol Microbiol. 68:255–262. [DOI] [PubMed] [Google Scholar]
- Brunner M, Schafmeier T. 2006. Transcriptional and post-transcriptional regulation of the circadian clock of cyanobacteria and Neurospora. Genes Dev. 20:1061–1074. [DOI] [PubMed] [Google Scholar]
- Cao X, Liu X, Li H, Fan Y, Duan J, et al. 2018. Transcription factor CBF-1 is critical for circadian gene expression by modulating white collar complex recruitment to the frq locus. PLoS Genet. 14:e1007570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo AJ, Schacht P, Cabrera IE, Blahut J, Prudhomme L, et al. 2017. Functional profiling of transcription factor genes in Neurospora crassa. G3 (Bethesda). 7:2945–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett NL, Yoder OC, Turgeon BG. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell 2:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J, Zhou M, Liu Y. 2015. Methods to study molecular mechanisms of the Neurospora circadian clock. Methods Enzymol. 551:137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, DeMay BS, Gladfelter AS, Dunlap JC, Loros JJ. 2010. Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc Natl Acad Sci USA. 107:16715–16720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. 2009. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 28:1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Liu Y. 2001. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc Natl Acad Sci USA. 98:7408–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collopy PD, Colot HV, Park G, Ringelberg C, Crew CM, et al. 2010. High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods Mol Biol. 638:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, et al. 2006. A high-throughput gene knockout procedure for neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 103:10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, et al. 2012. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci USA. 109:7397–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coradetti ST, Xiong Y, Glass NL. 2013. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen 2:595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH, de Serres FJ. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods in Enzymol. 17:79–143. [Google Scholar]
- Dekhang R, Wu C, Smith KM, Lamb TM, Peterson M, et al. 2017. The Neurospora transcription factor ADV-1 transduces light signals and temporal information to control rhythmic expression of genes involved in cell fusion. G3 (Bethesda). 7:129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz RD, Larrondo LF. 2020. A circadian clock in Neurospora crassa functions during plant cell wall deconstruction. Fungal Biol. 124:501–508. [DOI] [PubMed] [Google Scholar]
- Diernfellner ACR, Brunner M. 2020. Phosphorylation timers in the Neurospora crassa circadian clock. J Mol Biol. 432:3449–3465. [10.1016/j.jmb.2020.04.004] [DOI] [PubMed] [Google Scholar]
- Dong W, Tang X, Yu Y, Nilsen R, Kim R, et al. 2008. Systems biology of the clock in Neurospora . PLoS ONE 3:e3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. 1993. Genetic analysis of circadian clocks. Annu Rev Physiol. 55:683–728. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. 1999. Molecular bases for circadian clocks. Cell 96:271–290. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. 2008. Salad days in the rhythms trade. Genetics 178:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ. 2017. Making time: conservation of biological clocks from fungi to animals. Microbiol Spectr. 5. [10.1128/microbiolspec.FUNK-0039-2016] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole DJ, Sachs MS. 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genetics Reports 37: [Google Scholar]
- Emerson JM, Bartholomai BM, Ringelberg CS, Baker SE, Loros JJ, et al. 2015. period-1 encodes an atp-dependent RNA helicase that influences nutritional compensation of the Neurospora circadian clock. Proc Natl Acad Sci USA. 112:15707–15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Ma G, Li J, Liu Q, Benz JP, et al. 2015. Genome-wide analysis of the endoplasmic reticulum stress response during lignocellulase production in Neurospora crassa. Biotechnol Biofuels 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JF, Atkinson CA. 1978. Genetic and physiological characteristics of a slow-growing circadian clock mutant of Neurospora crassa. Genetics 88:255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JF, Hoyle MN. 1973. Isolation of circadian clock mutants of Neurospora crassa. Genetics 75:605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M-A, Barkoulas M. 2015. Pervasive robustness in biological systems. Nat Rev Genet. 16:483–496. [DOI] [PubMed] [Google Scholar]
- Fischer MS, Wu VW, Lee JE, O'Malley RC, Glass NL. 2018. Regulation of cell-to-cell communication and cell wall integrity by a network of map kinase pathways and transcription factors in Neurospora crassa. Genetics 209:489–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich AC, Noh B, Vierstra RD, Loros J, Dunlap JC. 2005. Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot Cell 4:2140–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai K, Cao X, Dong Q, Ding Z, Wei Y, et al. 2017. Transcriptional repression of frequency by the IEC-1-INO80 complex is required for normal Neurospora circadian clock function. PLoS Genet. 13:e1006732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Nislow C. 2014. The yeast deletion collection: a decade of functional genomics. Genetics 197:451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooch VD, Johnson AE, Bourne BJ, Nix BT, Maas JA, et al. 2014. A kinetic study of the effects of light on circadian rhythmicity of the frq promoter of Neurospora crassa. J Biol Rhythms 29:38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, et al. 2008. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell 7:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gras DE, Persinoti GF, Peres NT, Martinez-Rossi NM, Tahira AC, et al. 2013. Transcriptional profiling of Neurospora crassa deltamak-2 reveals that mitogen-activated protein kinase MAK-2 participates in the phosphate signaling pathway. Fungal Genet Biol. 60:140–149. [DOI] [PubMed] [Google Scholar]
- Gredilla R, Grief J, Osiewacz HD. 2006. Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina. Exp Gerontol. 41:439–447. [DOI] [PubMed] [Google Scholar]
- Greenwald CJ, Kasuga T, Glass NL, Shaw BD, Ebbole DJ, et al. 2010. Temporal and spatial regulation of gene expression during asexual development of Neurospora crassa. Genetics 186:1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. 2011. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 74:141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, He Q, Yu H, et al. 2003. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J. 22:4421–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, et al. 2008. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci USA. 105:20746–20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Selker EU. 2009. Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, et al. 2002. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419:841–844. [DOI] [PubMed] [Google Scholar]
- Horta MAC, Thieme N, Gao Y, Burnum-Johnson KE, Nicora CD, et al. 2019. Broad substrate-specific phosphorylation events are associated with the initial stage of plant cell wall recognition in Neurospora crassa. Front Microbiol. 10:2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JX, Thomas CE, Brunak S. 2016. Network biology concepts in complex disease comorbidities. Nat Rev Genet. 17:615–629. [DOI] [PubMed] [Google Scholar]
- Hurley JM, Dasgupta A, Emerson JM, Zhou X, Ringelberg CS, et al. 2014. Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proc Natl Acad Sci USA. 111:16995–17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Loros JJ, Dunlap JC. 2016. Circadian oscillators: around the transcription-translation feedback loop and on to output. Trends Biochem Sci. 41:834–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison E, Brown S, Tian C, Glass NL. 2009. Transcriptional profiling and functional analysis of heterokaryon incompatibility in Neurospora crassa reveals that reactive oxygen species, but not metacaspases, are associated with programmed cell death. Microbiology 155:3957–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CA, Greer-Phillips SE, Borkovich KA. 2007. The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female fertility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol Biol Cell 18:2123–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. 2007. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 21:1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldi K, Gonzalez BH, Brunner M. 2006. Transcriptional regulation of the Neurospora circadian clock gene wc-1 affects the phase of circadian output. EMBO Rep. 7:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T, Townsend JP, Tian C, Gilbert LB, Mannhaupt G, et al. 2005. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 33:6469–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays AM, Rowley PS, Baasiri RA, Borkovich KA. 2000. Regulation of conidiation and adenylyl cyclase levels by the galpha protein GNA-3 in Neurospora crassa. Mol Cell Biol. 20:7693–7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. 2002. Systems biology: a brief overview. Science 295:1662–1664. [DOI] [PubMed] [Google Scholar]
- Kojima S, Shingle DL, Green CB. 2011. Post-transcriptional control of circadian rhythms. J Cell Sci. 124:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A. 2015. Circadian rhythms. When the circadian clock becomes blind. Science. 347:476–477. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Craig LM, Duffy JF. 2018. Introduction to chronobiology. Cold Spring Harb Perspect Biol. 10:a033613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin-Thomas PL, Bell-Pedersen D, Brody S. 2011. The genetics of circadian rhythms in Neurospora. Adv Genet. 74:55–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin-Thomas PL, Brody S. 2004. Circadian rhythms in microorganisms: new complexities. Annu Rev Microbiol. 58:489–519. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas PL, Coté GG, Brody S. 1990. Circadian rhythms in Neurospora crassa: biochemistry and genetics. Crit Rev Microbiol. 17:365–416. [DOI] [PubMed] [Google Scholar]
- Larrondo LF, Loros JJ, Dunlap JC. 2012. High-resolution spatiotemporal analysis of gene expression in real time: in vivo analysis of circadian rhythms in Neurospora crassa using a FREQUENCY-luciferase translational reporter. Fungal Genet Biol. 49:681–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrondo LF, Olivares-Yanez C, Baker CL, Loros JJ, Dunlap JC. 2015. Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination. Science 347:1257277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter FR, Yanofsky C. 1993. Day/night and circadian rhythm control of con gene expression in Neurospora. Proc Natl Acad Sci USA. 90:8249–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeder AC, Jonkers W, Li J, Glass NL. 2013. Early colony establishment in Neurospora crassa requires a map kinase regulatory network. Genetics 195:883–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Sirakova T, Rogers L, Ettinger WF, Kolattukudy PE. 2002. Regulation of constitutively expressed and induced cutinase genes by different zinc finger transcription factors in Fusarium solani f. Sp. Pisi (Nectria haematococca). J Biol Chem. 277:7905–7912. [DOI] [PubMed] [Google Scholar]
- Loros JJ, Dunlap JC. 2001. Genetic and molecular analysis of circadian rhythms in Neurospora. Annu Rev Physiol. 63:757–794. [DOI] [PubMed] [Google Scholar]
- Loros JJ, Feldman JF. 1986. Loss of temperature compensation of circadian period length in the frq-9 mutant of Neurospora crassa. J Biol Rhythms 1:187–198. [DOI] [PubMed] [Google Scholar]
- Loudon AS. 2012. Circadian biology: a 2.5 billion year old clock. Curr Biol. 22:R570–571. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. 2011. Genetics of circadian rhythms in mammalian model organisms. Adv Genet. 74:175–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, et al. 2009. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, et al. 2007. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 21:1687–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey K, Wiest AE, Grigoriev IV, Lipzen A, Martin J, et al. 2011. Rediscovery by whole genome sequencing: classical mutations and genome polymorphisms in Neurospora crassa. G3. G3 (Bethesda). 1:303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Viveros L, Bouchard-Cannon P, Hegazi S, Cheng AH, Pastore S, et al. 2017. Molecular modulators of the circadian clock: lessons from flies and mice. Cell Mol Life Sci. 74:1035–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M, Lenssen D, Roenneberg T. 2010. Comparative clocks. Protein Rev. 12:157–177. [Google Scholar]
- Montenegro-Montero A. 2014. Ph.Thesis. Bzip transcription factors and transcriptional regulatory networks in the Neurospora circadian system. Pontificia Universidad Católica de Chile, Santiago, Chile. [Google Scholar]
- Montenegro-Montero A, Canessa P, Larrondo LF. 2015. Around the fungal clock: recent advances in the molecular study of circadian clocks in Neurospora and other fungi. Adv Genet. 92:107–184. [DOI] [PubMed] [Google Scholar]
- Montenegro-Montero A, Larrondo LF. 2016. In the driver's seat: the case for transcriptional regulation and coupling as relevant determinants of the circadian transcriptome and proteome in eukaryotes. J Biol Rhythms 31:37–47. [DOI] [PubMed] [Google Scholar]
- Moore A, Zielinski T, Millar AJ. 2014. Online period estimation and determination of rhythmicity in circadian data, using the biodare data infrastructure. Methods Mol Biol. 1158:13–44. [DOI] [PubMed] [Google Scholar]
- Morgan LW, Feldman JF, Bell-Pedersen D. 2001. Genetic interactions between clock mutations in Neurospora crassa: can they help us to understand complexity? Philos Trans R Soc Lond B Biol Sci. 356:1717–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Kay SA. 2012. Complexity in the wiring and regulation of plant circadian networks. Curr Biol. 22:R648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimamurthy R, Virshup DM. 2017. Molecular mechanisms regulating temperature compensation of the circadian clock. Front Neurol. 8:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiss A, Schafmeier T, Brunner M. 2008. Transcriptional regulation and function of the Neurospora clock gene white collar 2 and its isoforms. EMBO Rep. 9:788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M, Duffield GE, Loros JJ, Dunlap JC. 2003. The frequency gene is required for temperature-dependent regulation of many clock-controlled genes in Neurospora crassa. Genetics 164:923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Yanez C. 2015. Ph.D. Thesis. Role of RCO-1 in the control of circadian gene expression and metabolic compensation of the Neurospora crassa circadian clock. Pontificia Universidad Católica de Chile, Santiago, Chile. [Google Scholar]
- Olivares-Yanez C, Emerson J, Kettenbach A, Loros JJ, Dunlap JC, et al. 2016. Modulation of circadian gene expression and metabolic compensation by the RCO-1 corepressor of Neurospora crassa. Genetics 204:163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo M, Ruger-Herreros C, Corrochano LM. 2010a. Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa. Genetics 184:651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo M, Ruger-Herreros C, Luque EM, Corrochano LM. 2010b. A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet Biol. 47:352–363. [DOI] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VB, Giles NH. 1985. Autogenous regulation of the positive regulatory qa-1F gene in Neurospora crassa. Mol Cell Biol. 5:3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengkit A, Jeon SS, Son SJ, Shin JH, Baik KY, Choi EH, et al. 2016. Identification and functional analysis of endogenous nitric oxide in a filamentous fungus. Sci Rep. 6:30037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, et al. 1997. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12:204–217. [DOI] [PubMed] [Google Scholar]
- Pregueiro AM, Liu Q, Baker CL, Dunlap JC, Loros JJ. 2006. The Neurospora Checkpoint Kinase 2: a regulatory link between the circadian and cell cycles. Science. 313:644–649. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Pownder TA, Sexson SL. 1999. General method for plasmid construction using homologous recombination. Biotechniques 26:134–138, 140–1. [DOI] [PubMed] [Google Scholar]
- Reilly MC, Qin L, Craig JP, Starr TL, Glass NL. 2015. Deletion of homologs of the SREBP pathway results in hyper-production of cellulases in Neurospora crassa and Trichoderma reesei. Biotechnol Biofuels 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Brown SA. 2010. Transcriptional regulation of circadian clocks. Protein Rev. 12:37–78. [Google Scholar]
- Roberts AN, Berlin V, Hager KM, Yanofsky C. 1988. Molecular analysis of a Neurospora crassa gene expressed during conidiation. Mol Cell Biol. 8:2411–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J, Davis SJ. 2017. Making the clock tick: the transcriptional landscape of the plant circadian clock. F1000Res. 6:951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner MJ, Oster H, Wichert SP, Reinecke L, Wehr MC, et al. 2008. Disturbed clockwork resetting in Sharp-1 and Sharp-2 single and double mutant mice. PLoS ONE 3:e2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar C, Ha N, Yilmaz R, Tesorero R, Fisher T, et al. 2015a. Combinatorial control of light induced chromatin remodeling and gene activation in Neurospora. PLoS Genet. 11:e1005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar C, Sancar G, Ha N, Cesbron F, Brunner M. 2015b. Dawn- and dusk-phased circadian transcription rhythms coordinate anabolic and catabolic functions in Neurospora. BMC Biol. 13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G, Sancar C, Brugger B, Ha N, Sachsenheimer T, et al. 2011. A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol Cell 44:687–697. [DOI] [PubMed] [Google Scholar]
- Sancar G, Sancar C, Brunner M. 2012. Metabolic compensation of the Neurospora clock by a glucose-dependent feedback of the circadian repressor CSP1 on the core oscillator. Genes Dev. 26:2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent ML, Briggs WR, Woodward DO. 1966. Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiol. 41:1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Larrondo LF, Loros JJ, Dunlap JC. 2007. A developmental cycle masks output from the circadian oscillator under conditions of choline deficiency in Neurospora. Proc Natl Acad Sci USA. 104:20102–20107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Sancar G, Dekhang R, Sullivan CM, Li S, et al. 2010. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for Neurospora White Collar Complex. Eukaryot Cell 9:1549–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyan T, Gloeckner G, Diekmann S, Carbon J. 2001. Multifunctional centromere binding factor 1 is essential for chromosome segregation in the human pathogenic yeast Candida glabrata. Mol Cell Biol. 21:4875–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Tian C, Diamond S, Glass NL. 2012a. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot Cell 11:482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang F, Lan N, Liu B, Hu C, et al. 2019. The Zn(II)2Cys6-type transcription factor ADA-6 regulates conidiation, sexual development, and oxidative stress response in Neurospora crassa. Front Microbiol. 10:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Yu L, Lan N, Wei S, Yu Y, et al. 2012b. Analysis of the role of transcription factor VAD-5 in conidiation of Neurospora crassa. Fungal Genet Biol. 49:379–387. [DOI] [PubMed] [Google Scholar]
- Swift J, Coruzzi GM. 2017. A matter of time - how transient transcription factor interactions create dynamic gene regulatory networks. Biochim Biophys Acta Gene Regul Mech. 1860:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Dong W, Griffith J, Nilsen R, Matthes A, et al. 2011. Systems biology of the qa gene cluster in Neurospora crassa. PLoS One 6:e20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S, Croft NJ, Sotiriou A, Piggins HD, Crosthwaite SK. 2008. Neurospora crassa heat shock factor 1 is an essential gene; a second heat shock factor-like gene, hsf2, is required for asexual spore formation. Eukaryot Cell 7:1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, et al. 2009. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA. 106:22157–22162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Li J, Glass NL. 2011. Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology (Reading) 157:747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira A, Kasuga T, Tian C, Lemos C, Castro A, et al. 2009. Transcriptional analysis of the response of Neurospora crassa to phytosphingosine reveals links to mitochondrial function. Microbiology (Reading) 155:3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ. 1956. A convenient growth medium for Neurospora crassa. Microb Genet Bull. 13:42–47. [Google Scholar]
- Wang Z, Kin K, López-Giráldez F, Johannesson H, Townsend JP. 2012. Sex-specific gene expression during asexual development of Neurospora crassa. Fungal Genet Biol. 49:533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lopez-Giraldez F, Lehr N, Farré M, Common R, et al. 2014. Global gene expression and focused knockout analysis reveals genes associated with fungal fruiting body development in Neurospora crassa. Eukaryot Cell 13:154–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Miguel-Rojas C, Lopez-Giraldez F, Yarden O, Trail F, et al. 2019. Metabolism and development during conidial germination in response to a carbon-nitrogen-rich synthetic or a natural source of nutrition in Neurospora crassa. mBio. 10: e00192-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang J, Li N, Li J, Trail F, et al. 2018. Light sensing by opsins and fungal ecology: NOP-1 modulates entry into sexual reproduction in response to environmental cues. Mol Ecol. 27:216–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters MK, Manzanilla V, Howell H, Mehreteab A, Rose E, et al. 2018. Cold shock as a screen for genes involved in cold acclimatization in Neurospora crassa. G3 (Bethesda). 8:1439–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters MK, Stadler DR. 1995. Spontaneous mutation during the sexual cycle of Neurospora crassa. Genetics 139:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M, Mitchell HK. 1947. A synthetic medium favoring sexual reproduction. Am J Bot. 34:573–577. [Google Scholar]
- Wu C, Yang F, Smith KM, Peterson M, Dekhang R, et al. 2014. Genome-wide characterization of light-regulated genes in Neurospora crassa. G3 (Bethesda). 4:1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Sun J, Glass NL. 2014. VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa. PLoS Genet. 10:e1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Wu VW, Lubbe A, Qin L, Deng S, et al. 2017. A fungal transcription factor essential for starch degradation affects integration of carbon and nitrogen metabolism. PLoS Genet. 13:e1006737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro CT, Ebbole DJ, Lee BU, Brown RE, Bourland C, et al. 1996. Characterization of rco-1 of Neurospora crassa, a pleiotropic gene affecting growth and development that encodes a homolog of Tup1 of Saccharomyces cerevisiae. Mol Cell Biol. 16:6218–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. 2010. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 11:764–776. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, et al. 2009. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. 2012. Speed control: cogs and gears that drive the circadian clock. Trends Neurosci. 35:574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang B, Emerson JM, Ringelberg CS, Gerber SA, et al. 2018. A had family phosphatase CSP-6 regulates the circadian output pathway in Neurospora crassa. PLoS Genet. 14:e1007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Liu X, Hu Q, Zhang N, Sun G, et al. 2013. Suppression of WC-independent frequency transcription by RCO-1 is essential for Neurospora circadian clock. Proc Natl Acad Sci USA. 110:E4867–E4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yu X, Xie B, Gu X, Zhang Z, et al. 2013. Transcriptomic profiling-based mutant screen reveals three new transcription factors mediating menadione resistance in Neurospora crassa. Fungal Biol. 117:422–430. [DOI] [PubMed] [Google Scholar]
- Zielinski T, Moore AM, Troup E, Halliday KJ, Millar AJ. 2014. Strengths and limitations of period estimation methods for circadian data. PLoS ONE 9:e96462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znameroski EA, Coradetti ST, Roche CM, Tsai JC, Iavarone AT, et al. 2012. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc Natl Acad Sci USA. 109:6012–6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Circadian data sets associated with the genetic screen are available at https://biodare2.ed.ac.uk/ (Zielinski et al. 2014) as “Neurospora TF Circadian Clock” and have been also uploaded as supplementary tables to figshare: https://gsajournals.figshare.com/articles/figure/Supplemental_Material_for_Mu_oz-Guzm_n_Caballero_and_Larrondo_2021/14036507?file=26476886.