Abstract
Tandem repeats are inherently unstable and exhibit extensive copy number polymorphisms. Despite mounting evidence for their adaptive potential, the mechanisms associated with regulation of the stability and copy number of tandem repeats remain largely unclear. To study copy number variation at tandem repeats, we used two well-studied repetitive arrays in the budding yeast genome, the ribosomal DNA (rDNA) locus, and the copper-inducible CUP1 gene array. We developed powerful, highly sensitive, and quantitative assays to measure repeat instability and copy number and used them in multiple high-throughput genetic screens to define pathways involved in regulating copy number variation. These screens revealed that rDNA stability and copy number are regulated by DNA replication, transcription, and histone acetylation. Through parallel studies of both arrays, we demonstrate that instability can be induced by DNA replication stress and transcription. Importantly, while changes in stability in response to stress are observed within a few cell divisions, a change in steady state repeat copy number requires selection over time. Further, H3K56 acetylation is required for regulating transcription and transcription-induced instability at the CUP1 array, and restricts transcription-induced amplification. Our work suggests that the modulation of replication and transcription is a direct, reversible strategy to alter stability at tandem repeats in response to environmental stimuli, which provides cells rapid adaptability through copy number variation. Additionally, histone acetylation may function to promote the normal adaptive program in response to transcriptional stress. Given the omnipresence of DNA replication, transcription, and chromatin marks like histone acetylation, the fundamental mechanisms we have uncovered significantly advance our understanding of the plasticity of tandem repeats more generally.
Keywords: qRIN, ddPCR, ribosomal DNA, CUP1, genome instability, copy number variation, replication-transcription conflicts, H3K56 acetylation, Rtt109, adaptation, yeast
INTRODUCTION
Repetitive DNA sequences constitute large fractions of all eukaryotic genomes and copy number polymorphisms at repetitive regions are now recognized as a significant source of genetic diversity. In fact, copy number variations are now recognized as the most significant source of genetic diversity in human populations (Iafrate et al. 2004; Sebat et al. 2004; Redon et al. 2006; Zarrei et al. 2015) and are associated with many human chromosomal syndromes (Wyandt et al. 2017). A recent genome-wide association study of 1011 natural isolates of the budding yeast, Saccharomyces cerevisiae, showed that copy number variations not only constituted the most genetic variation, but also had the most significant effect on phenotype (Peter et al. 2018). Computational studies estimate that the human genome contains at least 25,000 arrays of tandem repeats between 600 bp and 10 kb in length, with 503 arrays larger than 10 kb (Warburton et al. 2008), and these tandem repeats exhibit extreme variability in copy number (Brahmachary et al. 2014). These data suggest that copy number polymorphisms, particularly at tandem repeats, may significantly contribute to genome function. To elucidate the mechanisms underlying the regulation of instability and adaptive copy number changes at tandem repeats, we chose to study the two features of tandem repeats that are key to their ability to undergo extensive copy number variation—instability, and copy number maintenance—using two well-studied repetitive arrays in the yeast genome, the ribosomal DNA (rDNA) locus, and the copper-resistance CUP1 gene array.
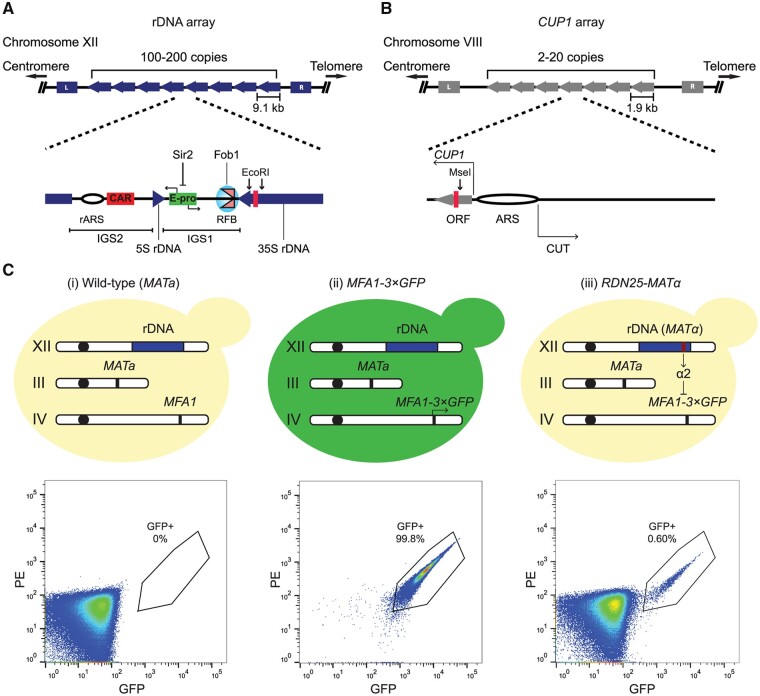
The rDNA genes encode ribosomal RNA (rRNA), and are the most well-characterized tandem repeat family. Given their universally conserved function in ribosome biogenesis, and the conservation of the organization of the genes and key regulatory elements from yeast to humans, the budding yeast rDNA locus has been used as a model to study mechanisms of copy number variation at tandem repeats. The budding yeast rDNA locus is comprised of ∼100–200 copies of a 9.1-kb repeat unit arranged in tandem on the long arm of Chromosome XII. Each 9.1-kb repeat unit contains coding sequences for a pre-35S rRNA, transcribed by RNA polymerase I (RNAPI), that gives rise to the 18S, 25S, and 5.8S rRNA species, and a 5S rRNA, transcribed by RNA polymerase III (RNAPIII). The 35S and 5S rRNA coding sequences are separated by two intergenic spacers (IGS), IGS1 and IGS2, which contain important regulatory elements. IGS2 contains an rDNA origin of replication, rARS, and a cohesin-associating sequence (CAR). IGS1 contains a replication fork barrier (RFB) site bound by the replication fork blocking protein Fob1, and E-pro, a noncoding, bidirectional, RNA polymerase II (RNAPII) promoter whose activity is normally suppressed by Sir2, an NAD+-dependent histone deacetylase (Figure 1A). The rDNA array is the most highly transcribed locus in the yeast genome, and high rates of transcription from multiple copies of rDNA repeats are essential to support rRNA production in actively growing cells. rDNA repeat units are maintained far in excess of the requirement for ribosome biogenesis, with only about half of the 100–200 repeats transcribed in actively growing yeast cells (French et al. 2003). Additionally, while rDNA copy number can be reduced significantly (down to ∼20 copies) with little effect on rRNA output or cell growth (Kobayashi et al. 1998; French et al. 2003), extra, untranscribed rDNA repeats are required to ensure efficient DNA damage repair in the highly transcribed rDNA array (Ide et al. 2010).
Figure 1.
Development of a system to study copy number variation at tandem repeats. (A, B) The rDNA and CUP1 gene arrays share design features. Cartoons showing the structure of the (A) rDNA and (B) CUP1 gene arrays in S. cerevisiae along with key regulatory elements in each array. The rDNA array comprises 100–200 ∼9.1 kb repeat units arranged in tandem at a single locus on Chromosome XII. Direction of blue block arrows indicates direction of rDNA transcription. IGS1 and IGS2, Intergenic spacers 1 and 2; rARS, rDNA origin of replication; CAR, cohesin associated region; E-pro, bidirectional RNAPII promoter whose activity is suppressed by Sir2 binding; RFB, replication fork barrier, bound by Fob1. The CUP1 array comprises 2–20 ∼2kb repeats arranged in tandem at a single locus on Chromosome VIII. Direction of grey block arrows indicates direction of CUP1 ORF transcription. ARS, origin of replication; CUT, cryptic unstable transcript. Red bars within coding sequences of both arrays indicate target regions for ddPCR assays to measure repeat copy number, relevant restriction sites flanking the ddPCR assay targets are also indicated. (C) Basic principle of the quantitative, single cell assay to measure repeat instability (qRIN). MFA1 in a haploid, wild-type MATa strain (i) is tagged with three copies of GFP to generate the MFA1-3×GFP strain (ii). A single copy of MATALPHA is integrated at the locus of interest, e.g., one rDNA repeat (iii). The α2 repressor produced from MATALPHA represses GFP expression. GFP-positive cells produced by loss of the MATALPHA-containing region can be rapidly counted using flow cytometry as shown in representative flow cytometry analyses of the wild-type MATa strain, the MFA1-3×GFP strain, and the RDN25-MATα strains with GFP-positive cells gated by the black polygon.
Normal rDNA copy number is thought to be maintained by a recombination-mediated amplification of rDNA repeats that depends on RNAPI transcription (Kobayashi et al. 1998). Recent work from Mansisidor et al. (2018) and Iida and Kobayashi (2019) suggests that cells depend on multiple RNAPI transcription-dependent mechanisms to (a) monitor rDNA copy number at every cell division, and (b) trigger amplification of the array in cells with critically low rDNA copy number (Mansisidor et al. 2018; Iida and Kobayashi 2019). During S-phase, the binding of Fob1 to the RFB inhibits DNA replication in the direction opposite to 35S rDNA transcription, preventing the head on collision of transcription and replication machinery (Brewer et al. 1992; Kobayashi 2003). DNA replication forks stalled at the RFB are processed into double-stranded breaks (DSBs), which are repaired by homologous recombination-mediated repair pathways (Kobayashi et al. 1998; Kobayashi 2003). The presence of multiple identical tandem repeats that can serve as templates for recombination allows unequal sister chromatid exchange (USCE) events to occur, which frequently result in copy number variations. USCE is suppressed by cohesin binding at the CAR (Kobayashi et al. 2004). Recombination is further suppressed by binding of Sir2 to E-pro (Kobayashi et al. 2004; Kobayashi and Ganley 2005). Despite these known mechanisms to suppress recombination at the rDNA, recombination-mediated repeat loss is relatively high in wild-type cells, even in unperturbed conditions. Additionally, copy number changes at the rDNA have been observed as adaptive responses to mutations and environmental stresses, particularly DNA replication stress (Ide et al. 2007; Kwan et al. 2013; Shyian et al. 2016; Salim et al. 2017) and perturbations in RNAPI transcription (Oakes et al. 1993; Kobayashi et al. 1998; Oakes et al. 1999; Albert et al. 2011). This relatively high instability at the rDNA locus, the paradoxical, stable maintenance of normal repeat copy number, and the apparent lack of correlation between instability and repeat copy number changes have all been areas of active investigation for the last several years. These data suggest that the functions of the rDNA go well beyond ribosome biogenesis. Therefore, characterization of the mechanisms regulating the stability of the rDNA array and its ability to accommodate extensive copy number variation is key to understanding the impact of these variations on genomic adaptation to the environment.
While the yeast rDNA array has served as a major model to study the behavior of tandem repeats, rDNA genes are constitutively transcribed, and their transcription is essential for cell viability. Work from Hull et al. (2017) showed that transcription of the CUP1 gene array induces copy number variation at this locus, suggesting that the inherent instability at the rDNA array is due in part to constitutively high levels of transcription at the array. The requirement of rDNA transcription for cell survival makes it impossible to study transcription-dependent and independent aspects of copy number variation using the rDNA array. The CUP1 array shares design features with the rDNA array. It is comprised of 2–20 copies of a ∼2kb repeat unit, arranged in tandem at a single locus on chromosome VIII. Each repeat unit contains the CUP1 coding sequence and an origin of replication (ARS). The CUP1 promoter is bidirectional, with sense and antisense transcription producing CUP1 mRNA and a cryptic unstable transcript (CUT), respectively (Hull et al. 2017) (Figure 1B). CUP1 encodes a metallothionein that sequesters environmental copper and cadmium. CUP1 is only transcribed in the presence of copper in the medium, and copy number correlates directly with copper resistance, making the CUP1 array a powerful, inducible system to study copy number variation and adaptation. Further, work from Hull et al. (2017) showed that CUP1 copy number variation was regulated by acetylation of the lysine 56 of histone H3 (H3K56), a chromatin mark well known for its role in maintaining rDNA copy number (Ide et al. 2013). These data suggest the conservation of the basic principles of copy number variation at the rDNA and CUP1 arrays.
While the last two decades have witnessed the discovery of many genes involved in the regulation of rDNA copy number variation in budding yeast (Smith et al. 1999; Ide et al. 2013; Saka et al. 2016; Salim et al. 2017), the lack of quantitative, sensitive assays to measure the rapid induction of instability in a high-throughput manner has limited our understanding of how instability aids adaptive copy number variation. The lack of a clear distinction between repeat instability and changes in steady-state copy number has also resulted in several apparently paradoxical findings. Here we report the development and validation of a quantitative, single cell-based assay to measure repeat instability (qRIN), and demonstrate its use in a quantitative and unbiased high-throughput screen to identify genetic factors that regulate rDNA stability. We identified several pathways that impact rDNA instability; notably, in addition to factors that elevate instability, factors that suppress instability were also identified. This suggests that rather than minimizing instability, cells may have evolved to maintain an “optimal rDNA stability” that promotes genome stability while allowing for copy number variations to occur readily in response to genomic stresses. Additionally, to identify factors involved in the maintenance of normal rDNA copy number, we used a droplet digital PCR (ddPCR) based assay to measure rDNA copy number in 279 strains of a yeast conditional temperature-sensitive (ts) mutant collection of essential genes. Our screens, in conjunction with follow-up experiments, revealed that instability and maintenance of copy number of the rDNA and CUP1 arrays are regulated by DNA replication, transcription, and acetylation of the lysine 56 residue of histone H3 (H3K56). Based on these data, we propose that instability at tandem repeats can be rapidly induced by replication and transcription. While changes in instability at both rDNA and CUP1 arrays in response to stress are observed within a few cell divisions, a change in steady-state repeat copy number, or adaptation, requires prolonged propagation under selective conditions. H3K56 acetylation specifically governs transcription-induced array amplification, making it a regulator of the normal process of adaptation. We propose that modulation of replication and transcription is a direct, reversible strategy to alter instability at tandem repeats in response to environmental stimuli, which provides cells rapid adaptability through copy number variation.
Materials and methods
Yeast strains and media
All yeast strains used are listed in Supplementary Table S2. Strains were grown in nonselective synthetic complete (SC) medium [6.7 g/L yeast nitrogen base without amino acids + ammonium sulfate, 20 g/L dextrose, 2 g/L SC supplement], or SD-dropout medium (6.7 g/L yeast nitrogen base without amino acids + ammonium sulfate, 20 g/L dextrose, CSM-dropout supplement) lacking specific amino acids as indicated. Copper and/or hydroxyurea (HU)-treated cells were grown in SC complete medium containing indicated concentrations of CuSO4 and/or HU. Rich medium used was YPD (1% yeast extract, 2% peptone, 2% dextrose). For CUP1-MATα cup2Δ (+PGAL1-CUP1) strains, selective medium used was SRaff-Leu-Ura (6.7 g/L Yeast Nitrogen Base without amino acids + ammonium sulfate, 0.67 g/L CSM-Leu-Ura supplement, 20 g/L raffinose), and nonselective medium used was SRaff+Gal-Ura (6.7 g/L Yeast Nitrogen Base without amino acids + ammonium sulfate, 0.77 g/L CSM-Ura supplement, 20 g/L raffinose, 20 g/L galactose) containing the indicated concentration of CuSO4. All growth was at 30°C. Strain construction was carried out using standard yeast protocols. All strains were verified by replica plating and PCR, followed by ddPCR to measure copy numbers of rDNA, CUP1 as well as the MATα-LEU2 repressor (Supplementary Table S4). Instability reporter strains were also tested for green fluorescent protein (GFP) expression and repression by flow cytometry.
ddPCR
Genomic DNA isolation, quantification, and ddPCR were carried out, as previously described (Salim et al. 2017). Quantification was performed using the Quantasoft software. For copy number measurements, SD for each individual reaction was calculated using the formula:
where (CImax − CImin) is the 95% Confidence Interval for the ratio of absolute copy number of the target of interest and TUB1 in each reaction, with both assays multiplexed in the same well, as generated by Quantasoft. ddPCR primers, probes, and PCR conditions used are listed in Supplementary Table S3.
Measurement of repeat loss rates
Growth and sample collection:
Single colonies (four each) of freshly revived reporter strains were inoculated into 5 mL SD-Leu medium and grown overnight (up to 24 h) at 30°C. The next day, each overnight culture was used to inoculate cells into 5 mL SC complete such that the starting cell density of this culture was OD600≥0.05. Actual OD600 following inoculation into SC complete medium (t = 0), OD600-0, was also measured and recorded, following which cells were allowed to grow in SC complete medium for ∼24 h (∼10–12 doublings, t = 24h) at 30°C. Additionally, 100–200µL of each overnight culture (SD-Leu) was also harvested and fixed for cytometric analysis of the fraction of GFP-positive cells at t = 0. The next day, the cell density of each SC Complete culture (t = 24h), OD600-24, was measured and recorded. Additionally, 100–200µL of each culture was harvested and fixed for cytometric analysis of the fraction of GFP-positive cells at t = 24h.
Preparing samples for cytometry:
To prepare cells for cytometry, cells were harvested from an appropriate volume of the culture by centrifuging at <3000 rpm for 5 min. The medium was aspirated, and cells were washed once in 1× phosphate-buffered saline (PBS) (<3000 rpm, 5 min). Following removal of 1×PBS, cells were resuspended in 100–200 µL 4% paraformaldehyde solution [Per 40 mL: 10 mL 16% paraformaldehyde (Ted Pella), 1.36 g sucrose] and incubated at room temperature, in the dark, for 15 min. Fixed cells were centrifuged at 3000 rpm for 5 min to remove the paraformaldehyde, washed once with 1×PBS (3000 rpm, 5 min), and resuspended in 1 mL 1×PBS. Fixed samples were then stored at 4°C in the dark for up to a week until needed for cytometry.
Flow cytometry:
Fixed samples were analyzed on a MACSQuant Analyzer (Miltenyi Biotec) or a ZE5 Cell Analyzer (Bio-Rad). At least 200,000 single cells were counted for each sample. Data analysis was performed using FlowJo v10 to obtain the fraction of GFP-positive cells. The FlowJo analysis template used can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1576.
Calculation of repeat loss rates:
The fraction of GFP-positive cells and OD600 measurements for each sample at t = 0 and t = 24 h were recorded in Microsoft Excel. Equations (5) and (6) in Supplementary File S1 were applied to these data to calculate repeat loss rates using Mathematica 10 (Wolfram Research 2014).
Testing for loss of the MATα-LEU2 repressor
Single colonies of freshly revived MFA1-3×GFP, RDN25-MATα, and CUP1-MATα strains were inoculated into 5 mL SC Complete and SD-Leu, respectively, and allowed to grow to overnight at 30°C. The next day, a small aliquot of each overnight culture (t = 0) was used for fluorescence-activated cell sorting (FACS) sorting GFP-positive and GFP-negative cells from each sample. The overnight culture was also diluted into 5 mL SC Complete medium, and allowed to grow overnight at 30°C. The following day, an aliquot from this culture (t = 24h) was also used for FACS.
Samples from t = 0 and t = 24h were sorted using an S3 Cell Sorter (Bio-Rad), MoFlo Legacy cell sorter (Beckman Coulter), or an Influx cell sorter (BD Biosciences) depending on instrument availability. 100,000 GFP-negative cells and 1000–100,000 GFP-positive cells (depending on the frequency of GFP-positive cells) were sorted from each sample into ∼500µL YPD. The sorted cells were diluted and plated at single cell density on to YPD plates. The plates were incubated at 30°C for ∼2 days until colonies appeared. The plates were photographed, following which each plate was replica plated on to both, YPD and SD-Leu plates. The plates were incubated at 30°C for 1 day, until colonies appeared and photographed.
Twelve single colonies were then picked at random from the YPD replica plates, resuspended in 200 µL ddH2O, and stored at −80°C prior to genomic DNA isolation for ddPCR. Four additional single colonies were also picked from the replica plates to generate the growth curves in Figure 2B. Single colonies were used to inoculate 175 µL SC Complete at a starting OD600 of 0.05 in a 96-well plate. Each colony was inoculated in triplicate, and growth curves were generated from average (of 3) measurements of OD600 taken every 15 min using a TECAN Infinite M200 plate reader for growth at 30°C for 24 h.
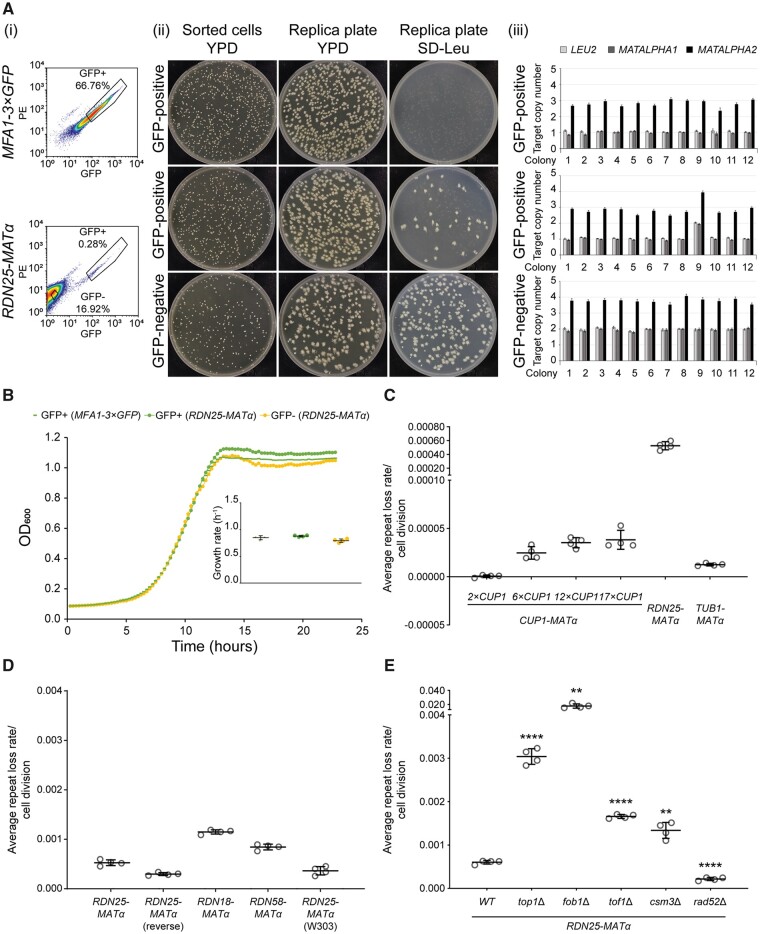
Figure 2.
Validation of the qRIN assay. (A) GFP-positive cells in the RDN25-MATα strain at t = 24h are generated predominantly by a complete loss of MATALPHA-LEU2. (i) Flow cytometry data showing GFP-positive and GFP-negative cells sorted from the MFA1-3×GFP and RDN25-MATα strains grown in nonselective medium (t = 24h). (ii) Sorted GFP-positive and GFP-negative cells plated on YPD plates, and replica plated on to YPD and SD-Leu plates. 91% (364/400 colonies) GFP-positive cells from RDN25-MATα strain are Leu−. (iii) Copy number of MATALPHA-LEU2 in 12 colonies picked from the YPD replica plate for each sorted population measured by ddPCR. Error bars represent standard deviation for each individual reaction. 91.67% (11/12 colonies) GFP-positive cells from the RDN25-MATα strain have lost the MATALPHA-LEU2 construct. (B) Loss of MATALPHA-LEU2 does not confer a growth advantage. Representative growth curves of single colonies (1 each) derived from sorted GFP-positive cells from RDN25-MATα and MFA1-3×GFP strains and sorted GFP-negative cells from RDN25-MATα. Growth curves were plotted based on average of triplicate measurements of OD600 taken at 15-min intervals for 24 h. Inset: growth rates for four independent colonies derived from each population. Error bars represent standard deviation. (C) Average repeat loss rates from three different genomic regions—CUP1 gene arrays containing 2, 6, 12, or 17 copies of CUP1, the rDNA array, and a “stable” intergenic region downstream of TUB1. (D) Similar rDNA repeat loss rates obtained from a variety of reporter strains. (E) Altered rDNA repeat loss rates in mutants known to affect rDNA stability. For (C–E), error bars represent standard deviation based on four biological replicates. Statistical significance was calculated using a standard two-tailed t-test. **P < 0.01, ****P < 0.0001.
High-throughput screens
Plasmid preparation and transformation into WT reporter strain:
The freshly revived RDN25-MATα strain was inoculated into 200 mL SD-Leu and incubated overnight, with shaking, at 30°C. In the following morning, the culture was spun down and the medium decanted. The cell pellet was then washed twice with ddH2O and once with 0.1 M lithium acetate. The washed pellet was resuspended in 1.5 mL 1 M lithium acetate, 0.5 mL ddH2O, and 2 mL 2 mg/mL sheared salmon sperm DNA. 50 µL of this mix was aliquoted into all wells of a 96-well PCR plate. 200 ng of each MoBY plasmid was then added to each well of the PCR plate and vortexed prior to the addition of 100 µL 50% PEG3350. The plate was sealed, vortexed, and briefly spun down before heat shocking at 42°C for 1 h. The plate was then spun down, and the supernatant was aspirated off. 200 µL SD-Leu-Ura was then added to each well, and the cultures were transferred to a flat-bottom 96-well plate and incubated overnight, with shaking, at 30°C. Cultures were then spotted onto SD-Leu-Ura PlusPlates and grown for two nights at 30°C. Cells from each spot were then inoculated into 150 µL SD-Leu-Ura broth and grown overnight at 30°C. 65 µL 50% glycerol was added to each well of the plate, mixed, and frozen at −80°C.
Growth and preparation for cytometry:
30 µL of the glycerol stock of each strain from RDN25-MATα + MoBY library was inoculated into 1.5 mL SD-Leu-Ura in a 96-deepwell plate. These plates were incubated overnight, with shaking, at 30°C. The next morning, 100 µL of the overnight cultures were fixed for cytometric analysis. In parallel, the overnight cultures were also diluted 1:50 into 1.5 mL SD-Ura in 96-deepwell plates. OD600 following inoculation into nonselective medium (t = 0), OD600-0, was measured immediately after inoculation using a TECAN Infinite M200 plate reader and recorded. This set of plates was incubated overnight, with shaking, at 30°C. The following morning, 100 µL of the overnight cultures in nonselective media were fixed for cytometric analysis. Additionally, the cultures were diluted 1:10 and used to obtain OD600 following growth in nonselective medium (t = 24h), OD600-24, using a TECAN Infinite M200 plate reader. Cells were fixed for cytometry as described above, with minor modifications. Briefly, cells were pelleted, resuspended in 100 µL 4% paraformaldehyde, and incubated at room temperature for 15 min. Fixed cells were then washed twice with 200 µL 1×PBS and resuspended in 250 µL 1×PBS. Flow cytometry to estimate the fraction of GFP-positive cells, and calculation of rDNA repeat loss rates was carried out as described above.
Hit validation:
The top ∼200 hits with high rDNA instability and the top ∼100 hits with low rDNA instability were cherry-picked and re-arrayed into 96-well plates, as previously described (Salim et al. 2017). rDNA repeat loss rates were measured in these strains two additional times (three independent measurements including initial screen) as described above. Genomic DNA was isolated from these hits for ddPCR, as previously described (Salim et al. 2017).
Screen to identify genes involved in rDNA copy number maintenance:
Genomic DNA isolation from the yeast ts mutant collection of essential genes (Ben-Aroya et al. 2008) and ddPCR was performed, as previously described (Salim et al. 2017). rDNA copy number measurements for eight colonies of wild-type BY4741 (Supplementary Table S6) were used to set thresholds as follows: mean rDNA copy number ± 2SD – no change. rDNA copy number > mean rDNA copy number + 2SD OR rDNA copy number < mean rDNA copy number – 2SD – significant change.
GO enrichment analysis:
GO enrichment analyses were performed using GOrilla (Eden et al. 2007, 2009).
Subculturing experiments
All subculturing experiments were performed, as described previously (Salim et al. 2017).
H3 point mutants and plasmid shuffle
Plasmids containing hht2(K56A)-HHF2, hht2(K9A)-HHF2, hht2(K14A)-HHF2, and HHT2-HHF2 (wild-type) were obtained from the SHIMA library (Nakanishi et al. 2008). Plasmids containing hht2(K56R)-HHF2 and hht2(K56Q)-HHF2 were generated by site-directed mutagenesis as described in Nakanishi et al. (2008). Each of these plasmids also contain the TRP1 selectable marker. Each plasmid was transformed into the RDN25-MATα (YBL574) reporter strain, and transformants selected by plating on SD-Leu-Trp medium. Cells were subjected to two rounds of selection on SD-Leu-Trp + 5-FOA to eliminate the plasmid containing wild-type HHT2-HHF2 (URA3) to obtain the RDN25-MATα (YBL574) + HHT2-HHF2 or H3K56A or H3K56R or H3K56Q or H3K9A or H3K14A strains. rDNA repeat loss rates in these strains were measured as described above. Selective medium used was SD-Leu-Trp, and nonselective medium used was SD-Trp.
CUP1 mRNA measurements
Freshly revived CUP1-MATα reporter strains were grown overnight at 30°C in 5 mL selective medium (SD-Leu). In the morning, 100 µL of this overnight culture was diluted into 10 mL of SC complete or SC complete containing 1 mM (for 6×CUP1 and 17×CUP1 strains) CuSO4. These cultures were incubated at 30°C and allowed to grow to an OD600 of ∼0.6–1.0. Cells were harvested, and pellets were flash frozen in liquid nitrogen and stored at −80°C until further use. RNA was isolated from frozen cell pellets using the hot acid phenol method. Briefly, the frozen cell pellet was resuspended in 800 µL of cold AE Buffer (50 mM NaOAc (pH 5.2), 10 mM EDTA in RNase free water), 80 μL of 10% SDS and 800 µL acid phenol (pH 4.3) (Ambion #AM9720) and incubated at 65°C for 10 min. The cell suspension was incubated on ice for 5 min and centrifuged for 20 min at 14,000 rpm, 4°C. The supernatant was transferred to a fresh tube, mixed with 800 µL of chloroform, and centrifuged for 15 min at 14,000 rpm, 4°C. The chloroform extraction was repeated one more time. The supernatant was transferred to a fresh tube, and 10 μg of linear acrylamide, 3 M NaOAc, pH 5.2–5.6 (1/10th of total volume), and isopropanol (volume equal to total volume) were added and centrifuged for 20 min at 14,000 rpm, 4°C. The pellet was washed with 1 mL 70% ethanol and resuspended in 100 μL TE buffer (10 mM Tris (pH 8.0), 0.1 mM EDTA in RNase-free water). The RNA was chilled on ice for 5 min, then incubated for ∼30 s at 65°C. It was mixed by gentle vortexing and chilled on ice for 5 min before storing at −80°C until further use.
Purity of each RNA sample was analyzed on a NanoDrop ND-1000 spectrophotometer. RNA integrity was analyzed using a 2100 Bioanalyzer (Agilent Technologies). After ensuring purity and integrity, RNA concentration was measured on a Qubit 2.0 Fluorometer using the Qubit RNA BR assay (Invitrogen). For each sample, 1 μg total RNA was used to set up DNase treatment reactions (to remove genomic DNA contamination) followed by cDNA synthesis reactions using the iScript gDNA Clear cDNA Synthesis Kit (Bio-rad). All reactions were set up in triplicate. Additionally, a “no-RT control” reaction (identical in composition to the three cDNA synthesis reactions, but lacking reverse transcriptase) was also set up for each sample. Following DNase treatment and cDNA synthesis, the cDNA was serially diluted using RNase-free water, and CUP1 mRNA immediately measured using ddPCR.
ddPCR was performed as described previously (Salim et al. 2017), with some modifications. Primers and conditions used are listed in Supplementary Table S3. 5 μL serially diluted cDNA was used per 20 μL reaction. ddPCR was performed according to the manufacturer’s protocol (Bio-Rad). Briefly, master mixes containing the dsDNA binding dye EvaGreen, primers for CUP1 or reference genes, cDNA, and the restriction endonuclease MseI (New England Biolabs, Inc.) were prepared and aliquoted into Eppendorf twin.tec plates. Reaction mixtures were incubated at room temperature for 15 min to allow restriction digestion of cDNA prior to droplet generation. Droplets were cycled to endpoint and subsequently read using the QX200 droplet reader. Quantification was performed using the Quantasoft software to obtain the absolute concentration of the target of interest (copies/μL). Expression of eight reference genes (TUB1, ACT1, CDC28, MUD1, SER2, SPT15, TRP1, and ZWF1) was also measured in each sample, but not used for normalization of CUP1 mRNA levels owing to changes in their expression in the presence of copper, and in mutant backgrounds. The strains tested carry an auxotrophic mutation in the TRP1 gene, and do not express TRP1. Therefore, TRP1 expression was used as a control. Average [CUP1 mRNA] (copies/ng of total RNA) and standard deviation were calculated for each sample based on data from triplicate cDNA samples (at the same dilution) synthesized from the same RNA sample.
Data availability
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1576. Strains and plasmids are available upon request. Supplementary Figure S1 contains additional validation of the qRIN assay. Supplementary Figure S2 contains analysis of rDNA stability in fob1Δ mutants. Supplementary Figure S3 contains analysis of the loss of MATα-LEU2 in fob1Δ mutants. Supplementary Figure S4 contains analysis of rDNA stability in NAM. Supplementary Figure S5 contains validation of the qRIN assay for the CUP1-MATα reporter strains. Supplementary Figure S6 contains validation of hits with altered rDNA stability. Supplementary Figure S7 contains functional validation and additional supporting information with regard to deletion of CUP2 in CUP1-MATα reporter strains. Supplementary Figure S8 contains stress and locus-specific effects of copper treatment. Supplementary Figure S9 contains data regarding how deletion of RTT109, HST3, and HST4 affect stability of TUB1. Supplementary Figure S10 contains data regarding how H3K56 acetylation restricts transcription-induced amplification of the CUP1 array in a 17 copy strain. Supplementary File S1 describes how to calculate repeat loss rates. Supplementary File S2 describes the method for determining and the location of the MATα-LEU2 casette in reporter strains. Supplementary Table S1 summarizes rDNA and CUP1 instability measurements from the literature. Supplementary Table S2 is a list of yeast strains. Supplementary Table S3 is a list of primers. Supplementary Table S4 contains validation copy number measurements for 25S rDNA, CUP1, LEU2, and MATALPHA1 in various yeast strains. Supplementary Table S5 contains the rDNA repeat loss rates and copy number measurements from the screens. Supplementary Table S6 contains rDNA copy number measurements from the temperature-sensitive mutant collection. Supplementary Table S7 contains copy number measurements of subcultured CUP1-MATα strains.
Supplementary material is available at figshare DOI: https://doi.org/10.25387/g3.14195492.
Results
Development and validation of qRIN, a quantitative, single-cell assay to measure rapid induction of repeat instability
The budding yeast rDNA locus has been used to model copy number variation at tandem repeats for several decades. Since recombination-mediated repeat copy number variation is the major source of instability at the rDNA locus, various measures of rDNA copy number variation, and rDNA repeat loss rates have been used as indicators of rDNA stability. In some studies, pulsed-field gel electrophoresis has been used to observe gross changes in rDNA copy number by the size of chromosome XII (Saka et al. 2016; Horigome et al. 2019). However, this method is labor-intensive, and only provides a qualitative estimate of rDNA copy number variation. Studies on recombination at the rDNA in yeast typically involved estimation of rDNA repeat loss rates by measuring the frequency of loss of a selectable marker (such as URA3, ADE2, LEU2) integrated into a single rDNA repeat (Petes 1980; Szostak and Wu 1980; Wagstaff et al. 1985; Gottlieb and Esposito 1989). This method involves plating and counting thousands of colonies, and is therefore not amenable to high-throughput analysis. A more recent study involved the insertion of a small targeted mutation in the IGS of a single rDNA repeat and monitoring the rate of loss of this single unit and its frequency of duplication over several generations (Ganley and Kobayashi 2011). However, this method demands PCR of >200 colonies per time point to estimate the fraction of cells that have lost or gained the tagged repeat, making it unsuitable for high-throughput analysis. Moreover, the use of different parameters by different groups to represent their estimates of rDNA instability makes direct comparison of data from different studies challenging.
To measure rDNA instability in a quantitative, highly sensitive, and simple manner that is amenable to high-throughput studies, we developed a single-cell, fluorescence-based assay that combines the basic principles of traditional marker-loss assays and those of the quantitative, single-cell, cytometry based assay for measurement of chromosome transmission fidelity (qCTF) in yeast developed by Zhu et al. (2015). We call our assay qRIN. To construct the reporter strain to measure rDNA instability, we first tagged the most highly expressed MATa-specific gene, MFA1 (Ghaemmaghami et al. 2003) with three copies of GFP in a haploid S288C yeast strain of MATa mating type. The MATα-LEU2 cassette, containing the MATα locus and the selectable marker, LEU2, was then introduced into a single rDNA repeat unit. The α2 transcriptional repressor produced from the MATα locus strongly represses MATa-specific genes, such as MFA1-3×GFP. Thus, when the MATα-containing rDNA repeat is present, the expression of Mfa1-3×GFP is strongly repressed. However, if this repeat is lost, Mfa1-3×GFP will be expressed and the cell will become highly fluorescent within one cell cycle after the loss event due to rapid proteasome degradation of the α2 repressor (Laney et al. 2006) (Figure 1C).
To measure rDNA repeat loss rates, single colonies of freshly revived reporter strains are inoculated into Leucine-dropout medium to select for the retention of the MATα-LEU2 repressor construct and allowed to grow overnight. The cultures are then diluted into nonselective medium (time t = 0) to allow for the loss of the MATα-containing rDNA repeat. The cell density and fraction of GFP-positive cells are measured at the start of the experiment. Following growth in nonselective medium for ∼24 h (time t = 24h, ∼10–12 doublings), cell density, and fraction of GFP-positive cells in the culture are measured. Optical density at 600 nm (OD600) is used as a measure of cell density, and the fraction of fluorescent cells can be rapidly measured using flow cytometry in low or high-throughput formats, and subsequently used to calculate the rate of loss of rDNA repeats using a simple mathematical formula derived based on the methods in Zhu et al. (2015) and described in Supplementary File S1.
To validate the qRIN assay, we first constructed a reporter strain, hereafter referred to as RDN25-MATα, where the MATα-LEU2 repressor construct was integrated near the 3ʹ-end of the 25S rRNA coding sequence of a single rDNA repeat. We grew this strain overnight in leucine-dropout medium to select for MATα, diluted into complete, nonselective medium (t = 0), and allowed growth for 10–12 generations (t = 24h). We collected and analyzed samples from t = 0 and t = 24 h by flow cytometry. As expected, a vast majority of cells were GFP-negative at both time points (Supplementary Figure S1A). A small fraction of cells (0.436 ± 0.08%, n = 8 experiments) exhibited GFP fluorescence 10- to 100-fold higher than that of the GFP-negative population at t = 24h, and a small, but much lower fraction of cells (0.118 ± 0.02%, n = 8 experiments) also exhibited similar GFP fluorescence at t = 0 (Supplementary Figure S1A). The presence of a small number of GFP-positive cells at t = 0 following growth in selective leucine-dropout medium could be due to silencing of the MATα-LEU2 repressor and the imperfect nature of selection in dropout media. In contrast, this highly fluorescent GFP-positive population was the predominant population (97.06 ± 2.70% at t = 24 h and 94.78 ± 3.79% at t = 0, n = 8 experiments) in samples collected at both time points from the parent strain, hereafter referred to as MFA1-3×GFP, that contains the 3×GFP tagged Mfa1, but lacks the MATα-LEU2 repressor (Supplementary Figure S1A). Additionally, this highly fluorescent GFP-positive population was absent at both time points in a control strain, BY4741, that does not contain the 3×GFP tagged Mfa1 (Supplementary Figure S1A). Altogether these data demonstrate that highly fluorescent cells detected by cytometry derive from the expression of 3×GFP from the MFA1 promoter.
We then used FACS to sort both, the GFP-positive and GFP-negative populations, in the RDN25-MATα reporter strain grown in complete medium to test for the presence of the MATα-LEU2 repressor construct. To do this, we collected samples at t = 0 and t = 24 h from the RDN25-MATα strain grown as described above (Figure 2A(i), Supplementary Figure S1B(i)). As a control, we also sorted GFP-positive cells from the MFA1-3×GFP strain grown in complete medium at both t = 0 and t = 24 h (Figure 2A(i), Supplementary Figure S1B(i)). We then plated the FACS-sorted cells at single-cell density on to rich medium, followed by replica plating on to leucine-dropout medium. We found that all the sorted GFP-negative cells from the RDN25-MATα strain were Leu+ at both time points (Figure 2A(ii), Supplementary Figure S1B(ii)). As expected, 100% of the sorted GFP-positive cells from the MFA1-3×GFP strain at both time points were Leu− (Figure 2A(ii), Supplementary Figure S1B(ii)). Only 78.25% (331/423 colonies) of the GFP-positive cells sorted from RDN25-MATα at t = 0 were Leu- (Supplementary Figure S1B(ii)). Given the (very low) presence of GFP-positive cells at t = 0 following growth in leucine-dropout medium, this is a reasonable fraction of Leu− cells and further highlights the imperfect nature of selection in dropout media. However, 91% (364/400 colonies) of the GFP-positive cells sorted from RDN25-MATα at t = 24h were Leu−, suggesting that most of the GFP-positive cells generated during the course of growth in nonselective medium had lost MATα-LEU2 (Figure 2A(ii)).
To further confirm that most of the GFP-positive cells generated after growth in nonselective medium had in fact lost MATα-LEU2, we used PCR genotyping. We picked 12 colonies at random from the different sorted populations that had been plated on rich medium for both, the MFA1-3×GFP and the RDN25-MATα strains, and measured the copy number of LEU2 and the two genes that constitute the MATα locus, MATα1, and MATα2, using ddPCR assays designed to target these genes. The MFA1-3×GFP strain contains one copy of leu2-3,112, which confers leucine auxotrophy, one copy of MATα1 (at the silenced HML locus), and three copies of MATα2 (1 copy of MATα2 at the silenced HML locus, one copy of MATa2 identical in sequence to MATα2 at the mating type MAT locus, and one copy of MATa2 at the silenced HMR locus). The RDN25-MATα strain derived from this strain contains one additional copy each of LEU2, MATα1, and MATα2. Therefore, the GFP-positive cells sorted from MFA1-3×GFP at t = 0 and t = 24 h should contain one copy each of LEU2 and MATα1, and three copies of MATα2, which is what our ddPCR copy number measurements show (Figure 2A(iii), Supplementary Figure S1B(iii)). Similarly, the GFP-negative cells sorted from RDN25-MATα contain two copies each of LEU2 and MATα1 and four copies of MATα2 as expected (Figure 2A(iii), Supplementary Figure S1B(iii)). Finally, 5/12 (41.67%) of the sorted GFP-positive cells from t = 0 and 11/12 (91.67%) of the sorted GFP-positive cells from t = 24h of the RDN25-MATα strain contain only one copy each of LEU2 and MATα1, and three copies of MATα2, which are the expected copy number measurements of MATα and LEU2 for cells that have lost the MATα-LEU2 repressor (Figure 2A(iii), Supplementary Figure S1B(iii)). Some of these GFP-positive cells could be generated by silencing of the MATα-LEU2 within the rDNA, especially at t = 0, and could account for a small fraction of the GFP-positive cells that still contain the MATα-LEU2 repressor. Silencing of RNAPII transcribed reporters at the rDNA can be altered by several factors including the integration site within a single repeat and relative location within the array, and levels of silencing factors like Sir2 (Smith et al. 1998; Huang and Moazed 2003; Wang et al. 2016); a significant change in silencing of the MATα-LEU2 repressor is expected to result in an underestimation of rDNA repeat loss rates. This is a caveat inherent to all marker loss assays, underscoring the need for validation and follow-up experiments, as for any screening method. Our validation experiments confirm that GFP-positive cells in the wild-type reporter strain grown in nonselective medium are mostly generated by a complete loss of the MATα-LEU2 repressor construct. We also compared growth rates of cultures derived from GFP-positive and GFP-negative cells sorted from samples collected at t = 24h for RDN25-MATα and found no significant differences (Figure 2B), which is critical for downstream rate calculations as described in SupplementaryFile S1.
To test the quantitative performance of our assay, we calculated rDNA repeat loss rates per cell division in the RDN25-MATα reporter strain grown in nonselective medium for 10–12 generations, as described above. While average repeat loss rates ranged from ∼0.0003 to 0.001 per cell division across all our experiments, biological replicates within any single experiment showed low variability, as shown in Figure 2C. The rates of rDNA repeat loss estimated using qRIN are similar to estimates of mitotic intrachromosomal recombination rates at the rDNA obtained using traditional marker loss assays (Supplementary Table S1).
Further, we also constructed three additional reporter strains where the MATα-LEU2 repressor was integrated into different parts of a single rDNA repeat. RDN18-MATα and RDN58-MATα contain MATα-LEU2 integrated into the 18S rRNA and the 5.8S rRNA coding sequences of a single rDNA repeat, respectively, and RDN25-MATα (reverse) contains MATα-LEU2 integrated near the 3ʹ-end of the 25S rDNA gene on the noncoding strand. The rDNA repeat loss rates calculated using these different reporter strains were similar to one another in any given experiment, and all within the range of 0.0003–0.001/cell division across all experiments (Figure 2D). We also determined the position of integration of the MATα-LEU2 repressor in the rDNA array in these reporter strains using Southern blotting as described in Supplementary File S2. We found that both, the RDN25-MATα and RDN18-MATα reporter strains each have one MATα-LEU2 unit inserted ∼5 repeats into one end of the rDNA array. RDN58-MATα has one MATα-LEU2 unit inserted ∼15 repeats into one end of the rDNA array, and RDN25-MATα (reverse) has one MATα-LEU2 unit inserted in the middle of the array. The similarity in rDNA repeat loss rates across these reporter strains with different integration sites within a single rDNA repeat and across the rDNA array suggests that the effect of the position of MATα-LEU2 on the estimation of rDNA instability using the qRIN assay is minimal.
We constructed a fourth reporter strain, TUB1-MATα, where the MATα-LEU2 repressor was inserted into the intergenic region downstream of the essential gene TUB1, a unique genomic region that should be “stable” relative to the rDNA. As shown in Figure 2C, the loss of MATα is negligibly low for this region, as expected.
Next, we chose a small set of genes well-known for their effects on rDNA stability—TOP1, which encodes Topoisomerase I, RAD52, which is required for homologous recombination-mediated DSB repair, and FOB1, which encodes the rDNA RFB binding protein. We deleted each of these genes in the RDN25-MATα reporter strain to test the ability of our assay to detect changes in rDNA stability. The top1Δ mutants showed increased rDNA repeat loss rates (Figure 2E), consistent with previous reports of increased marker loss rates and extensive copy number variation at the rDNA in top1 mutants (Houseley et al. 2007; Andersen et al. 2015). The rad52Δ mutants showed decreased rDNA repeat loss rates, as expected, supporting the idea that homologous recombination-mediated repeat loss is a major source of instability at the rDNA.
The RDN25-MATα fob1Δ mutants, on the other hand, showed a significant increase in rDNA repeat loss rates relative to wild-type controls in our assay (Figure 2E), contrary to previous reports of lower marker loss and marker duplication frequencies in these mutants (Kobayashi et al. 1998; Johzuka and Horiuchi 2002; Kobayashi 2003). We observed a consistent increase in rDNA repeat loss rates in fob1Δ mutants in multiple independent isolates of fob1Δ in the RDN25-MATα background across multiple experiments. We also observed elevated repeat loss rates in fob1Δ mutants in the RDN18-MATα and RDN58-MATα backgrounds (Supplementary Figure S2A). To rule out the contribution of the genetic background of our reporter strains to this phenotype, we generated fob1Δ mutants in the RDN25-MATα (W303) strain background and measured rDNA repeat loss rates as before. We found that rDNA repeat loss rates in the fob1Δ mutants were higher than those in the wild-type RDN25-MATα (W303) strain (Supplementary Figure S2A). We confirmed that there were no changes in rDNA copy number, and that there was only one copy of the MATα-LEU2 repressor in the fob1Δ mutants (Supplementary Figure S2B). Given the reported roles of Fob1 in the silencing of RNAPII transcribed genes at the rDNA (Buck et al. 2016; Di Felice et al. 2019) and the lack of clear separation between GFP-positive and GFP-negative cells in this mutant (Supplementary Figure S3), we considered the possibility that the expression of the MATα-LEU2 repressor may be altered in a fob1Δ mutant background. To test for the presence of the MATα-LEU2 repressor construct, we sorted GFP-positive and GFP-negative cells from t = 0 and t = 24h for wild-type and fob1Δ RDN25-MATα strains grown in complete medium. As described above, we plated the sorted cells at single-cell density on to rich medium, followed by replica plating on to leucine-dropout medium. We found that 100% of the GFP-positive cells sorted from the RDN25-MATα fob1Δ strain were Leu+, suggesting retention of the MATα-LEU2 repressor and the generation of GFP-positive cells likely through silencing of the repressor (Supplementary Figure S3).
Tof1 and Csm3, like Fob1, are required for replication fork pausing within the rDNA and RFB activity. However, loss of these proteins does not affect reporter silencing within the rDNA (Bando et al. 2009; Mohanty et al. 2009). To test the effects of reporter silencing-independent loss of RFB activity on rDNA stability, we deleted TOF1 and CSM3 in the RDN25-MATα strain, and measured rDNA stability in the mutant strains. We found that both tof1Δ and csm3Δ mutants had increased rDNA repeat loss rates (Figure 2E), suggesting that the loss of RFB activity decreases rDNA stability, presumably because of increased head-on collisions of the replisome with RNAPI transcription machinery.
The role of Sir2 in suppressing recombination at the rDNA array and the increase in rDNA instability in sir2Δ mutants are well-known (Gottlieb and Esposito 1989). However, Sir2 is important for silencing the HML and HMR loci, which contain full, silenced copies of the MATα and MATa loci (Rine and Herskowitz 1987). Loss of Sir2 is therefore incompatible with our reporter system, which relies on the native HML and HMR loci remaining silenced, so that GFP-repression is dependent solely on the MATα-LEU2 construct inserted in the rDNA. In a sir2Δ mutant strain, the HML and HMR loci may be de-silenced, resulting in constitutive expression of MATα, and consequent repression of Mfa1-3×GFP irrespective of the presence of the MATα-LEU2 repressor construct. To test this, we measured rDNA repeat loss rates in the RDN25-MATα reporter strain grown in nicotinamide (NAM), an inhibitor of sirtuins including Sir2. As predicted, in NAM, the fraction of GFP-positive cells at t = 0 is lower than in untreated controls, and remains low even after growth in nonselective medium, and therefore the calculated repeat loss rates were also low (Supplementary Figure S4). While our qRIN assay depends on normal function and silencing of the HML and HMR loci, the quantitative power and scalability offered by this assay far outweighs this caveat.
Finally, we demonstrate the versatility of the qRIN assay by adapting it to measure repeat loss rates at a second tandem array in the yeast genome, the CUP1 gene array. Estimates of CUP1 instability and copy number variation from previous studies are also summarized in Supplementary Table S1. To construct the CUP1-MATα reporter strains, we integrated the MATα-LEU2 repressor construct at the 3ʹ-end of the CUP1 ORF in a single CUP1 repeat unit in the MFA1-3×GFP strain. This transformation resulted in CUP1-MATα isolates that had CUP1 gene arrays ranging in size from 2 to 17 repeats. As expected, copper resistance in the reporter strains was directly related to CUP1 copy number, and required the CUP1 transcription factor Cup2 (Ace1) (Supplementary Figure S5A). As with the RDN25-MATα strain, we validated that GFP-positive cells in the CUP1-MATα reporter strains were generated mostly by the loss of the MATα-LEU2 repressor (Supplementary Figure S5B). We then measured transcription-independent CUP1 repeat loss rates in each of the reporter strains with 2–17 copies of CUP1. Strains were grown in nonselective medium, without any copper, and repeat loss rates calculated using the formulae described in Supplementary File S1 . We found that transcription-independent repeat loss rates at the CUP1 array correlated positively with CUP1 copy number, and ranged from 1 × 10−6 per cell division in a two-copy array to 5 × 10−5 per cell division in a 17-copy array (Figure 2C). This suggests that transcription-independent repeat instability may be directly related to the size of the tandem array. Further, in the absence of transcription, the instability at the CUP1 array was at least 10-fold lower than that of the constitutively transcribed rDNA (Figure 2C), suggesting that the larger size of the rDNA array and its constitutive transcription are significant sources of instability. This further affirms the power of using an inducible array in the study of mechanisms involved in the regulation of copy number variation at tandem repeats. Notably, repeat loss rates at both tandem arrays are orders of magnitude higher than the rate of single base substitutions in the yeast genome (Supplementary Table S1), or instability at TUB1, consistent with the potential for copy number to profoundly impact genome diversity and evolution [reviewed in (Press et al. 2019)].
High-throughput screens to identify factors that regulate rDNA copy number variation
Saka et al. (2016) reported the first high-throughput screen to identify nonessential genes involved in the maintenance of rDNA stability in yeast. This group used PFGE to screen a yeast deletion mutant collection of nonessential genes for alterations in the extent of rDNA copy number variation, as indicated by the size of chromosome XII (Saka et al. 2016). This approach is, at best, a qualitative estimate of the variability in rDNA copy number. While marker loss assays have been used to carry out screens, these have been focused on the identification of factors involved in silencing of reporters at the rDNA (Smith et al. 1999). Further, several reports suggest that rDNA copy number is maintained by the fundamental processes of DNA replication and RNAPI transcription, but many components of these processes are essential to cell viability and are missing from high-throughput screens using the yeast knockout collection of nonessential genes. In order to identify pathways that regulate rDNA instability in an unbiased manner, while achieving maximum coverage of the genome, we chose to use our qRIN assay to measure rDNA repeat loss rates in a library of strains in which gene dosage was moderately increased. To achieve this, we used the MoBY-ORF library, composed of 4956 uniquely barcoded yeast ORFs, each cloned into a Uracil-selectable plasmid along with their endogenous promoter and 3ʹ-UTR sequences to ensure normal expression patterns (Ho et al. 2009). The plasmids also contain a yeast centromeric sequence, which ensures that the plasmid is maintained at a low copy number (∼1–3 copies/cell), thereby only moderately increasing the gene dosage. This library represents ∼90% of all nondubious ORFs annotated in the Saccharomyces Genome Database (SGD) and contains over 250 essential genes. We isolated the individual MoBY-ORF plasmids and transformed them each into the RDN25-MATα reporter strain to generate the RDN25-MATα (+MoBY-ORF) strain library comprising 4763 strains (representing 96.1% of the 4956 MoBY-ORF plasmids) (Figure 3A). Additionally, the MoBY empty vector, p5472, was also transformed into the RDN25-MATα reporter strain to generate the RDN25-MATα (+p5472) control strain. The RDN25-MATα (+MoBY-ORF) strain library was arrayed into 60 96-well plates, with the following control strains added to random empty wells in each plate: BY4741 (wild-type, MATa strain with no 3×GFP tag), MFA1-3×GFP, RDN25-MATα, and RDN25-MATα (+p5472) (Supplementary Table S5).
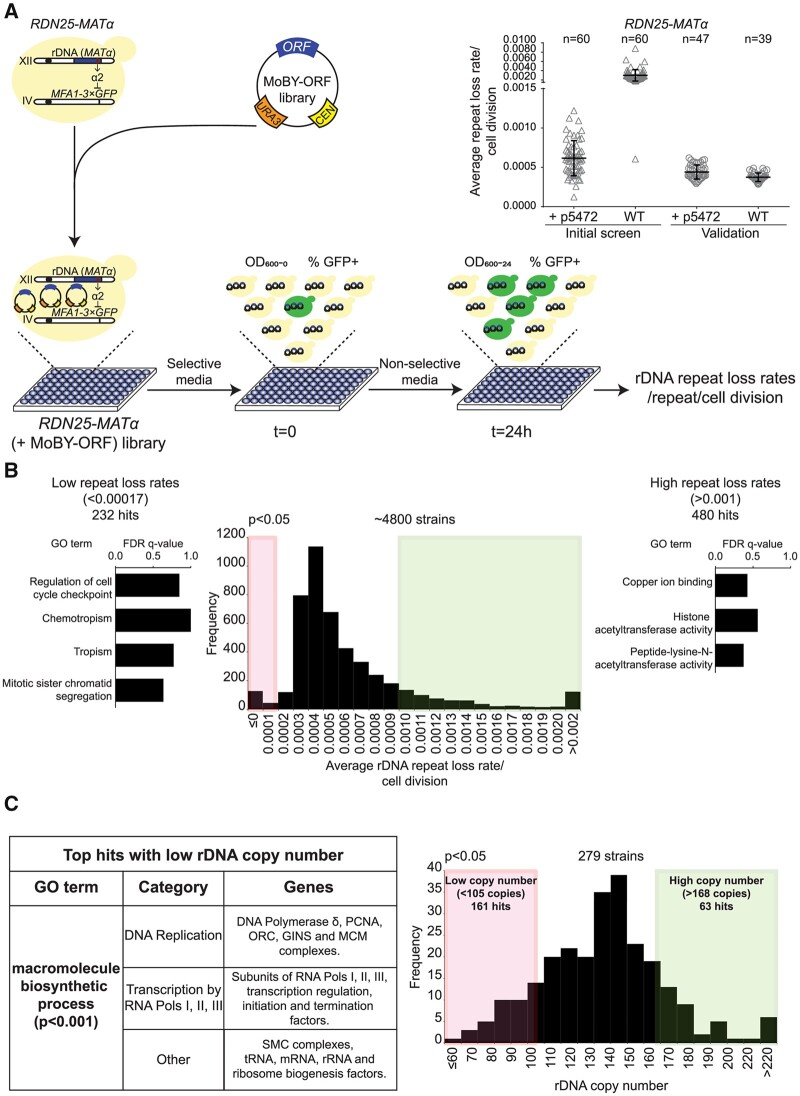
Figure 3.
Screens to identify genes that regulate copy number variation at the rDNA. (A) Cartoon showing the design of the overexpression screen to identify genes that regulate rDNA stability. Inset: rDNA repeat loss rates in wild-type RDN25-MATα and RDN25-MATα (+p5472) strains. Error bars represent standard deviation based on the indicated number of biological replicates. (B) Distribution of rDNA repeat loss rates across the ∼4800 strains of the RDN25-MATα (+MoBY-ORF) library. 232 and 480 strains had significantly lower (<0.00017/cell division) and higher (>0.001/cell division) rDNA repeat loss rates than the empty vector controls, respectively (P < 0.05). FDR q-values for significantly enriched (P < 0.001) GO terms (sorted in order of increasing P-values from top to bottom) for hits with high and low repeat loss rates are also shown. (C) Distribution of rDNA copy number in a second yeast ts mutant collection. 161 and 63 strains had significantly lower (<105 copies) and higher copy number (>168 copies) than wild-type controls, respectively (P < 0.05). The table summarizes the top hits with low rDNA copy number.
We then used our qRIN assay in high-throughput format to measure rDNA repeat loss rates in the RDN25-MATα (+MoBY-ORF) strain library, as illustrated in Figure 3A. As with batch cultures, we first grew the RDN25-MATα (+MoBY-ORF) strain library overnight in leucine and uracil-dropout medium to select for retention of the MATα-LEU2 repressor and the MoBY-ORF plasmid. We diluted the cultures into uracil-dropout medium (t = 0) to select for retention of the MoBY-ORF plasmid while allowing for loss of the MATα-LEU2 repressor, and grew cells for ∼24 h (t = 24h, ∼10–12 doublings). The control strain RDN25-MATα was initially grown in leucine-dropout medium, followed by dilution into nonselective medium at t = 0, whereas the control strains BY4741 and MFA1-3×GFP were always grown in nonselective medium. We collected samples at t = 0 and t = 24h for measuring the fraction of GFP-positive cells by cytometry. We also measured OD600 values at both time-points, and calculated rDNA repeat loss rates/cell division for the RDN25-MATα (+MoBY-ORF) strain library using the formula described in detail in Supplementary File S1.
The average of rDNA repeat loss rates/cell division obtained across all 60 96-well plates was 0.0029 ± 0.0012 (n = 60) in RDN25-MATα, and 0.0006 ± 0.0002 (n = 60) in RDN25-MATα (+p5472) (Figure 3A). Surprisingly, the average rDNA repeat loss rates/cell division for the RDN25-MATα control strain was approximately fivefold higher than the rate obtained for the RDN25-MATα (+p5472) control strain. However, the rDNA repeat loss rates for the RDN25-MATα (+p5472) strain is within the range of 0.0003–0.001/cell division we observed for the RDN25-MATα strain in batch cultures. A closer examination of the cytometry data for the RDN25-MATα strain revealed that the fraction of GFP-positive cells was higher than usual at both, t = 0 and t = 24 h, suggesting a jackpot event, a loss of the MATα-LEU2 repressor early in the establishment of the culture that was used to inoculate all 60 plates used in the initial screen. We measured rDNA repeat loss rates in 47 colonies each of freshly revived RDN25-MATα and RDN25-MATα (+p5472) strains grown in a 96-well plate as in the initial screen. 8/47 colonies of the RDN25-MATα strain showed higher fractions of GFP-positive cells at t = 0 and t = 24 h, characteristic of jackpot events, and data from these colonies were excluded from the analyses. Based on the analysis of rDNA repeat loss rates for the remaining colonies, we found that the two reporter strains had similar rDNA repeat loss rates that were indistinguishable from rates calculated for the RDN25-MATα (+p5472) strain in the initial screen (Figure 3A).
A distribution of rDNA repeat loss rates/cell division for the entire RDN25-MATα (+MoBY-ORF) strain library is shown in Figure 3B. rDNA repeat loss rates ranged from −0.003 to +0.01/cell division, with the distribution centered around ∼0.0004/cell division. We used the variation in rDNA repeat loss rates for the RDN25-MATα (+p5472) strain across all 60 96-well plates from the initial screen to set thresholds to identify RDN25-MATα (+MoBY-ORF) strains that had significantly altered rDNA repeat loss rates. Of the ∼4800 RDN25-MATα (+MoBY-ORF) strains screened, 712 strains had significantly altered rDNA repeat loss rates relative to the empty vector control strain (P < 0.05). Of these, 480 strains had significantly higher (>0.001/cell division) rDNA repeat loss rates, and 232 strains had significantly lower (<0.0001/cell division) rDNA repeat loss rates than the empty vector control strain. We then performed a gene ontology (GO) enrichment analysis on the initial 712 hits to identify pathways involved in regulating rDNA stability to guide our validation experiments. We found that the hits with lower rDNA repeat loss rates were significantly enriched for genes involved in nucleic-acid metabolism and DNA repair and/or recombination pathways (e.g., SGS1, HCA4, MLH1, ADE6), mitotic cell cycle checkpoint regulation (e.g., MAD1, RTT107, SLX4, CDH1), and sister chromatid segregation (e.g., SMC4, ECO1, CIN8, NPA3) (P < 0.001) (Figure 3B, Supplementary Table S5). In contrast, the hits with elevated rDNA repeat loss rates were significantly enriched for genes involved in acetylation of histones, specifically at lysine residues (e.g., RTT109, GCN5, SPT10, HFI1, SAS3, SAS4, SAS5, NAT4) (P < 0.001) (Figure 3B, Supplementary Table S5). Interestingly, the hits with elevated rDNA repeat loss rates were also significantly enriched for genes involved in copper transport and/or homeostasis (P < 0.001) (Figure 3B, Supplementary Table S5). This suggests that rDNA stability could be modulated by environmental stresses such as high concentrations of copper. However, the copper response genes in yeast (e.g. SOD1) are also known to be activated by DNA damage (Dong et al. 2013), and could regulate rDNA stability through their role in the DNA damage response. Further, the identification of hits with both increased and decreased rDNA repeat loss rates also suggests that cells may have evolved to optimize rDNA instability rather than minimize it so as to allow for rDNA copy number variation in response to genomic stresses.
As a preliminary validation of the results of our screen, we chose to focus on the genes involved in sister chromatid segregation, specifically sister chromatid cohesion. We first performed sequence validation and then manually transformed low copy plasmids containing various genes involved in sister chromatid cohesion into the RDN25-MATα strain and measured rDNA repeat loss rates. In agreement with the results from our initial screen, these experiments showed that a moderate increase in the dosage of SMC1, SMC3, ECO1, SCC2, MCD1, and RAD61 increased rDNA stability (Supplementary Figure S6A). Consistent with these data, overexpression of ESP1, which encodes separase that disrupts cohesion, made the rDNA more unstable (Supplementary Figure S6A).
Next, we selected ∼200 hits with elevated rDNA repeat loss rates, and ∼100 hits with reduced rDNA repeat loss rates and subjected them to additional validation. We re-arrayed these hits into three 96-well plates, along with control strains BY4741, MFA1-3×GFP, RDN25-MATα, RDN25-MATα (+p5472), and RDN25-MATα top1Δ (which has elevated rDNA repeat loss rates, Figure 2E). We measured rDNA repeat loss rates in high-throughput format in these hits two more times to obtain three independent measurements of rDNA repeat loss rates (including the initial screen) for each of these strains. The rDNA repeat loss rates from these three independent measurements are summarized in (Supplementary Table S5). Sixty-five of the ∼200 hits with high instability and 73 of the ∼100 hits with low instability showed similarly high or low rDNA instability rates in at least one additional validation run. We also isolated genomic DNA from these hits and measured rDNA copy number as well as MATα-LEU2 copy number. These copy number measurements are also summarized in (Supplementary Table S5). We wanted to ensure that the reduction in rDNA repeat loss rates were not due to amplifications of the MATα-LEU2 repressor. In fact, almost all the hits had only one copy of MATα-LEU2 repressor, and rDNA copy number similar to that of the RDN25-MATα (+p5472) strain. The only exception was RDN25-MATα (+MoBY-YCR035C), which had reduced rDNA repeat loss rates, and four copies of the MATα-LEU2 repressor, suggesting duplications of the MATα-LEU2 repressor, likely during strain construction. This strain also had a significantly lower rDNA copy number (∼69 copies), and so we excluded this strain from further analyses. These data suggest that the results of our screen were not confounded significantly by copy number changes at the rDNA or amplifications of the MATα-LEU2 repressor.
Finally, to gain a comprehensive understanding of the pathways involved in the regulation of copy number variation at the rDNA, we supplemented our results from the screen for pathways that regulate instability with pathways that are involved in the maintenance of rDNA copy number. We had reported previously, through an unbiased screen of 787 mutants of a yeast conditional temperature-sensitive (ts) mutant collection covering ∼45% of essential yeast genes, that mutations in DNA replication machinery, particularly, subunits of DNA polymerases α, δ, and ε, and various replication initiation complexes, such as the Mini chromosome maintenance 2–7 (Mcm2–7) complex, the Origin Recognition Complex (ORC), and the Cdc7–Dbf4 complex, were associated with a loss of rDNA repeats (Salim et al. 2017). However, this collection was missing mutants representing RNAPI and RNAPIII transcription, and many additional DNA replication processes, all of which have the potential to regulate rDNA copy number maintenance. Therefore, in this study, we extended this screen to nearly 75% of essential yeast genes by screening an additional yeast ts mutant collection of 279 strains (Ben-Aroya et al. 2008). We measured rDNA copy number in this yeast ts mutant collection using established ddPCR based assays (Salim et al. 2017). The mutant strains, along with wild-type controls, were grown at the permissive temperature (room temperature), and then shifted to the restrictive temperature (37˚C) for 3 h. Following this, genomic DNA was isolated, and copy number measured using ddPCR.
A distribution of rDNA copy number across the 279 strains screened is shown in Figure 3C, and also summarized in Supplementary Table S6. The mean rDNA copy number of wild-type strains was 136.58 ± 15.8 (n = 8). Mutants with significantly higher or lower rDNA copy number were identified based on thresholds set by variation in rDNA copy number in wild-type controls. Of the 279 strains screened, 161 strains had a significantly lower copy number (<105 copies), and 63 strains had significantly higher copy number (>168 copies) than wild-type controls (P < 0.05) (Figure 3C). GO enrichment analyses of the 63 hits with high copy number did not yield any significantly enriched GO terms. The hits with low rDNA copy number, however, were significantly enriched for genes involved in macromolecular biosynthesis (P < 0.001) (Figure 3C). A closer look at the genes comprising this significantly enriched GO term revealed that these were genes involved in DNA replication (subunits of DNA polymerase δ, the Origin Recognition Complex, MCM and GINS complexes, Proliferating cell nuclear antigen (PCNA), encoded by POL30), transcription (subunits of RNAPI, RNAPII, and RNAPIII, transcription initiation, elongation, and termination factors), rRNA and tRNA synthesis factors, and SMC complex subunits (cohesin and SMC5/6 complex subunits) (Supplementary Table S6). These data suggest modifications in rDNA copy number are a common strategy to adapt to the loss of essential functions related to DNA replication and transcription. Combining multiple high-throughput genetic screens revealed that rDNA stability and repeat copy number are regulated by the fundamental processes of DNA replication, transcription, and histone acetylation.
Repeat instability at the rDNA and CUP1 arrays is induced by DNA replication stress and transcription
Seminal work in bacteria has shown that replication-transcription conflicts promote instability [reviewed in (Lang and Merrikh 2018)], and elegant work in yeast demonstrated the role of transcription in promoting copy number variation at the CUP1 array (Hull et al. 2017). Taken together with our results, we postulate that copy number variation at tandem repeats like the rDNA is regulated by modulation of replication and/or transcription. Conflicts between the DNA replication machinery and transcription machinery operating on the same DNA template have now been established as a significant source of genomic instability. These replication–transcription conflicts occur frequently genome-wide and result installed or collapsed DNA replication forks which can be processed into DSBs and repaired by one of many recombination-mediated repair pathways. When this occurs at tandem repeats, recombination can result in copy number variation at every cell division. While average repeat copy number of a population remains stable in unperturbed conditions, under stress, selection of advantageous copy number variants can facilitate adaptation [reviewed in (Salim and Gerton 2019)]. Based on this model, we hypothesized that instability at tandem repeats like the rDNA and CUP1 arrays could be regulated by modulation of DNA replication, transcription, or even the downstream processes of recombination-mediated DSB repair.
To test this hypothesis, we measured the effect of DNA replication stress on instability at both, the rDNA and CUP1 arrays. We measured rDNA and CUP1 repeat loss rates in RDN25-MATα or CUP1-MATα (17×CUP1) treated with 0–200 mM HU, a ribonucleotide reductase inhibitor that depletes cellular dNTP pools. We found that HU induced repeat instability at both the rDNA and CUP1 gene arrays in a dose-dependent manner. Importantly, the induction of instability at CUP1 was transcription-independent (Figure 4A). As a control, we also treated the TUB1-MATα strains with 0–200 mM HU and found that instability at this nonrepetitive genomic locus remained low and unchanged irrespective of the dose of HU (Figure 4A). Therefore, DNA replication stress can increase instability specifically at tandem repeats.
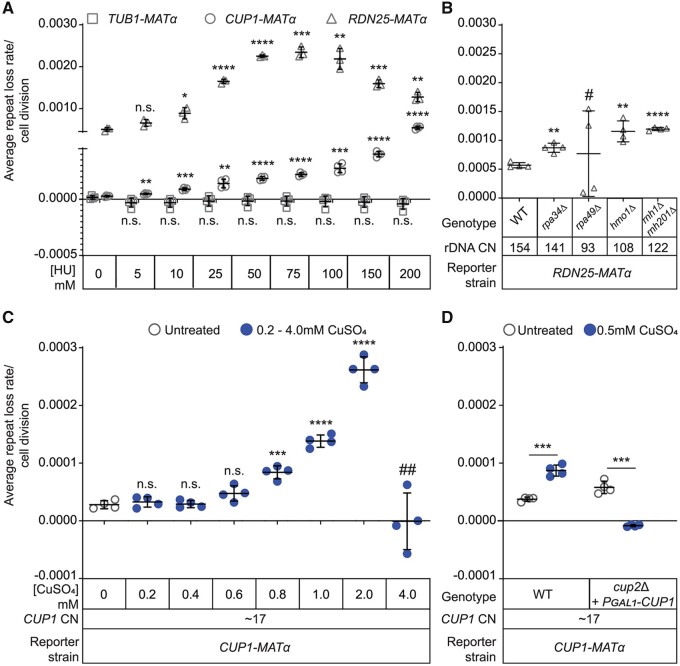
Figure 4.
Transcription and replication stress induce repeat instability. (A) Repeat instability is induced by Hydroxyurea (HU) induced replication stress in a dose-dependent manner at both, the rDNA and CUP1 gene arrays, but not at TUB1. (B) rDNA repeat loss rates in mutants that affect RNAPI transcription. # High variability in repeat loss rates between biological replicates. (C) Dose-dependent induction of instability at the CUP1 gene array by copper. ## No growth/cell divisions in the duration of the experiment. (D) Copper induces CUP1 instability through Cup2-mediated transcription of CUP1. (A–D) Error bars represent standard deviation based on four biological replicates. Statistical significance was calculated using a standard two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n. s., not significant.
Next, we perturbed transcription at the rDNA by deleting several genes known to play critical roles in RNAPI transcription. Loss of Rpa34, a subunit of RNAPI involved in transcription elongation, resulted in increased instability at the rDNA (Figure 4B). Loss of the Rpa49 subunit of RNAPI also altered rDNA stability, however we observed significant variability in the direction and magnitude of change in rDNA stability between biological replicates (Figure 4B). We speculate that this could be due to suppressor mutations that are known to arise in the rpa49Δ background that could result in altered RNAPI transcription (Darriere et al. 2019). Loss of Hmo1, an HMG-box protein involved in regulation of RNAPI transcription, induced instability at the rDNA (Figure 4B), consistent with a previous report of increased marker loss rates at the rDNA in hmo1Δ mutants (Mansisidor et al. 2018). We also generated mutants lacking Rnh1 and Rnh201, subunits of the functionally redundant RNases H1 and H2, respectively, that are required to process R-loops generated at the highly transcribed rDNA genes. Loss of Rnh1 and Rnh201 should result in an accumulation of R-loops at the rDNA and create more replication-transcription conflicts. Our model predicts that this should induce instability at the rDNA, which is exactly what we observed in rnh1Δrnh201Δ mutants (Figure 4B). HMO1 and RNH201 were also validated hits from our screen; overexpression of each of these genes increased rDNA instability (Supplementary Table S5), suggesting both loss and increased dosage compromise stability.
While these results suggest that transcription at the rDNA induces rDNA instability, the requirement of rDNA transcription for cell viability makes it impossible to test this directly. To complement these data, and directly test the effects of transcription on repeat stability, we used the CUP1-MATα reporter strains and measured CUP1 instability in the presence of copper in the medium. We observed that copper-induced CUP1 instability in a dose-dependent manner (Figure 4C). Since copper is also known to induce DNA damage, and copper response genes are induced by and involved in the DNA damage response, we wanted to verify that it is the copper-induced transcription of the array that induces CUP1 instability. To this end, we constructed a strain lacking Cup2, the transcription factor required for copper-induced CUP1 transcription (Welch et al. 1989). Since cup2Δ strains cannot grow in copper, we introduced into this strain a high-copy plasmid containing the CUP1 ORF under the control of a galactose inducible promoter, resulting in CUP1-MATα cup2Δ (+PGAL1-CUP1) strains that do not transcribe the native CUP1 array in the presence of copper in the medium, but are copper resistant when galactose is present in the medium (Supplementary Figure S7A). We found that copper no longer induced CUP1 instability in this strain (Figure 4D, Supplementary Figure S7B), suggesting that it is the copper-induced transcription, and not DNA damage, that induces CUP1 instability. These data suggest that instability at both the rDNA and CUP1 gene arrays can be rapidly induced (∼5–10 cell divisions) by modulation of replication or transcription.
Stress-induced instability facilitates adaptation through environment and locus-specific copy number changes
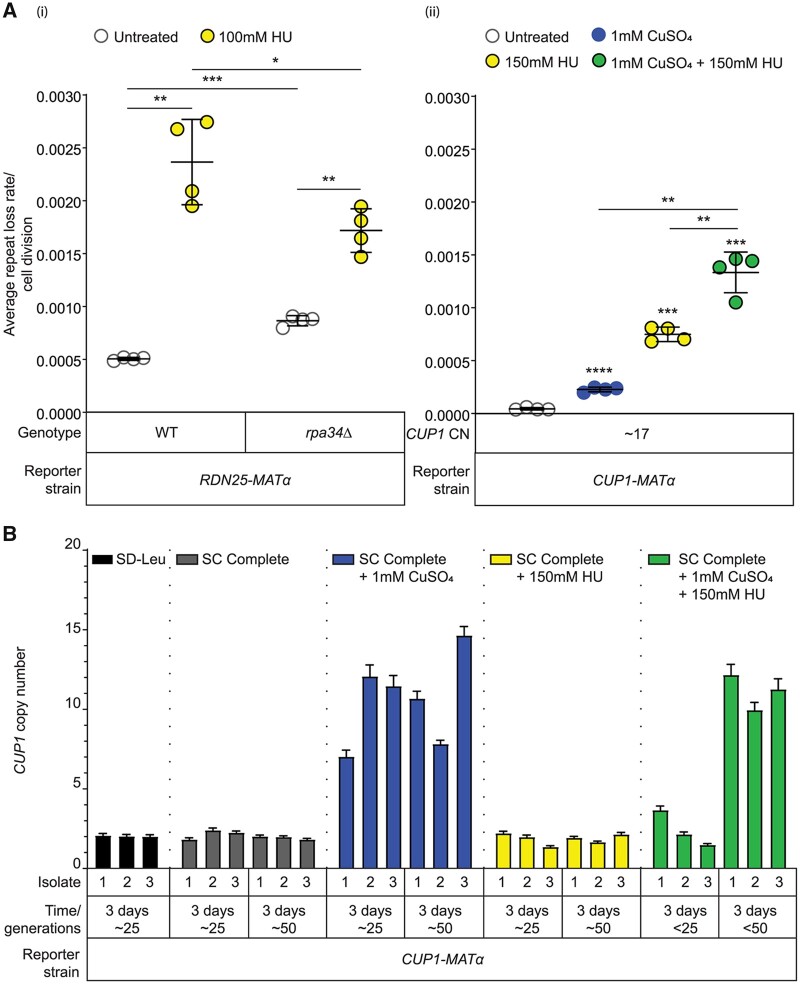
Since replication stress and transcription induce repeat instability, we wondered if there would be synergy between the effects of DNA replication and transcription on repeat instability. To test this, we grew the RDN25-MATα rpa34Δ strain in the presence of HU. Interestingly, HU-induced rDNA instability was lower in the rpa34Δ strain relative to the wild-type RDN25-MATα strain (Figure 5A(i)), suggesting that slowing DNA replication and transcription may promote stability at the rDNA. At the CUP1 array, transcription and replication stress had an additive effect on instability (Figure 5A(ii)). These data suggest that repeat instability is regulated by the relative balance between DNA replication and transcription at the array. Perturbation of one or both can rapidly induce instability within a few generations, presumably by increasing the frequency of replication–transcription conflicts.
Figure 5.
Stress-induced instability facilitates adaptation through stress and locus-specific copy number changes. (A) Synergistic effect of transcription and Hydroxyurea (HU) induced replication stress on instability at (i) rDNA and (ii) CUP1 arrays. Error bars represent standard deviation based on four biological replicates. Statistical significance was calculated using a standard two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (B) The wild-type CUP1-MATα strain (2×CUP1) was subcultured in the indicated medium for ∼6 days (approximately 50 generations). After every 3 days, three independent isolates were used to measure CUP1 copy number by ddPCR. Error bars represent standard deviation for each individual reaction.
We had previously reported that replication stress induced by high levels of HU selects for a loss of rDNA repeats; cells that had lost rDNA repeats survived better under conditions of replication stress, suggesting that the loss of rDNA repeats facilitated adaptation to DNA replication stress (Salim et al. 2017). However, the loss of repeats required propagation under conditions of DNA replication stress (≥150 mM HU) for at least 50 generations (Salim et al. 2017). Repeat instability, as measured by our assay, on the other hand, is rapidly induced within a few generations in a variety of conditions including low doses of HU. Therefore, we wanted to examine changes in steady-state copy number in response to prolonged propagation under these stresses.
To study transcription-dependent adaptation via copy number variation, we chose to monitor CUP1 copy number in a CUP1-MATα reporter strain subcultured for ∼50 generations in complete medium, or complete medium containing copper, or HU. We found that copper-resistant cells emerged after ∼25–50 generations in high concentrations of copper, and these cells had amplified CUP1 arrays (Supplementary Figure 5B). High concentrations of copper that select for amplified CUP1 arrays also induce instability at the rDNA array (Supplementary Figure S8A). However, rDNA copy number remains unaltered in these cells (Supplementary Figure S8B). Importantly, while instability at the CUP1 array is also induced by low concentrations of copper that cells are resistant to, CUP1 copy number remains unchanged in these conditions, even after 50 generations of growth (Supplementary Figure S8C). Further, no changes in CUP1 array size were observed in HU (Figure 5B). These experiments highlight the distinction between instability and copy number changes.
Finally, given the synergistic effect of HU and copper on CUP1 instability, we also subcultured the CUP1-MATα reporter strain used in Figure 5B in medium containing both copper and HU to test whether the additive effect on instability might accelerate adaptation to high concentrations of copper. We found that copper resistant cells with amplified CUP1 arrays emerged, however our ability to qualitatively estimate adaptation rates was confounded by the slow growth of cells in medium containing both copper and HU. Altogether, these data strongly support our hypothesis that repeat instability can be rapidly induced by DNA replication stress and transcription in a dose-dependent manner, but a change in steady-state repeat copy number requires selection for locus-specific advantageous copy number variants that facilitate adaptation.
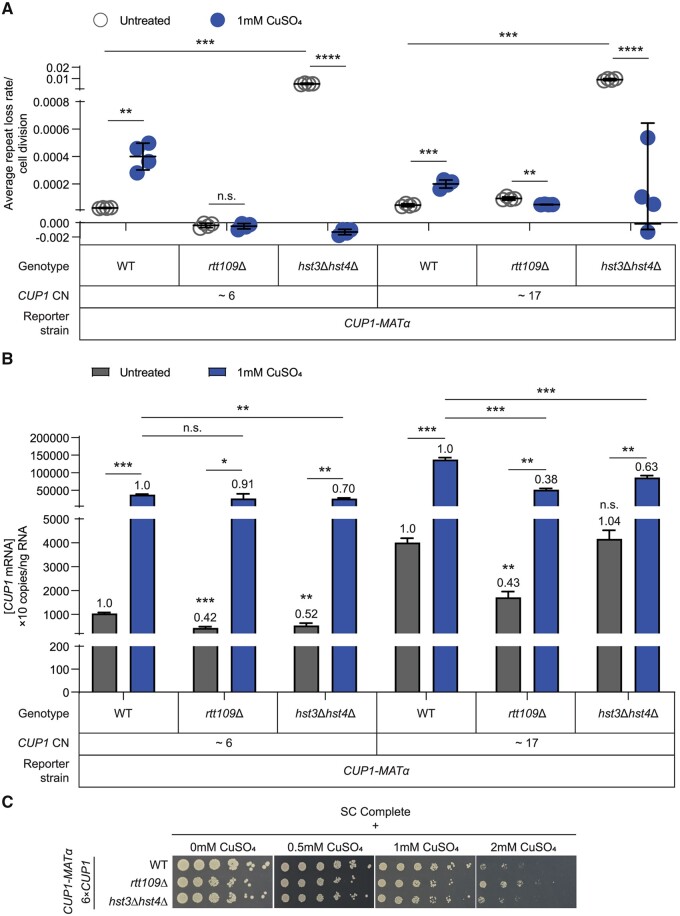
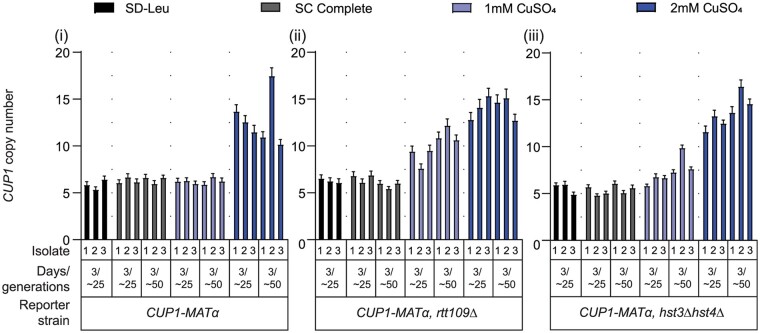
H3K56 acetylation regulates rDNA stability and copy number
Histone acetylation is a chromatin mark well known for its roles in promoting transcription (Yang et al. 2008; Varv et al. 2010), maintaining DNA replication timing and origin firing (Vogelauer et al. 2002; Aparicio et al. 2004; Unnikrishnan et al. 2010; Casas-Delucchi et al. 2012), and even influencing the choice of break repair pathway (Munoz-Galvan et al. 2013; Che et al. 2015), and therefore has the potential to have profound impacts on rDNA stability and copy number variation. Hst3 and Hst4 are key global H3K56 deacetylases, (Celic et al. 2006; Maas et al. 2006), and Rtt109 is the global H3K56 acetyltransferase (Han et al. 2007). Loss of Rtt109, or Hst3 and Hst4 were both shown to result in amplification of rDNA repeats (Ide et al. 2013), suggesting a key role for H3K56 acetylation in steady-state copy number. To validate the results from our screens and directly demonstrate the role of H3K56 acetylation in the regulation of rDNA stability, we generated rtt109Δ mutants and hst3Δhst4Δ double mutants in the RDN25-MATα background. We first measured rDNA repeat loss rates in these mutants. Interestingly, rtt109Δ mutants had significantly lower rDNA repeat loss rates than wild-type RDN25-MATα (Supplementary Figure S6B). On the other hand, the hst3Δhst4Δ double mutants had higher rDNA repeat loss rates than wild-type RDN25-MATα (Supplementary Figure S6B). We also measured the loss of MATα from a unique intergenic region downstream of TUB1 and found that while loss of Rtt109 did not affect the stability of this region, loss of Hst3 and Hst4 increased instability at this region (Supplementary Figure S9). This suggests that perturbing H3K56 acetylation affects global genome stability, although the effects vary. While Hst3 and Hst4 mainly target acetylated H3K56, Rtt109 acetylates both H3K56 and H3K9 residues. Additionally, given the role of Sir2 in regulating rDNA stability, and the role of its multiple targets, particularly H3K14, in regulating silencing at the rDNA, we sought to identify the key lysine residue of histone H3 involved in regulation of rDNA stability and copy number variation.
To characterize the role of the H3K56 residue in regulating rDNA stability, we generated three H3K56 point mutants, an alanine substitution mutant, H3K56A (Nakanishi et al. 2008), and H3K56R (which mimics the unacetylated form of H3K56) and H3K56Q (which mimics the acetylated form of H3K56). We also generated alanine substitution mutants in H3K9 and H3K14, H3K9A and H3K14A (Nakanishi et al. 2008). We transformed each of these plasmids into the RDN25-MATα (YBL574) reporter strain, which was constructed exactly as described above in the yeast histone shuffle strain, YBL574 (Nakanishi et al. 2008). YBL574 has the native histone H3/H4 loci (HHT1-HHF1 and HHT2-HHF2) deleted and carries wild-type HHT2-HHF2 on a Uracil-selectable plasmid. The H3K56A, H3K56R, H3K56Q, H3K9A, and H3K14A mutations were each carried on a tryptophan-selectable plasmid. We transformed each of the mutant plasmids, as well as a tryptophan-selectable plasmid carrying wild-type HHT2-HHF2 into RDN25-MATα (YBL574). We grew transformants on Tryptophan-dropout medium containing 5-fluorooroticacid (5-FOA) to select for the Tryptophan-selectable mutant HHT2-HHF2 plasmids and simultaneously remove the Uracil-selectable wild-type HHT2-HHF2 plasmids. We then measured rDNA repeat loss rates in the resulting strains.
The “wild-type” RDN25-MATα and RDN25-MATα (YBL574) reporter strains as well as the RDN25-MATα (YBL574) strain containing the wild-type HHT2-HHF2 plasmid had comparable rDNA repeat loss rates (Supplementary Figure S6C). We found that the H3K9A and H3K14A mutations did not alter rDNA repeat loss rates. All three H3K56 mutations altered rDNA repeat loss rates, however, unexpectedly, H3K56A, H3K56R, and H3K56Q, all increased rDNA repeat loss rates (Supplementary Figure S6C). Next, we measured rDNA copy number in multiple isolates of each of these strains. Despite isolate to isolate variability in rDNA copy number, the rtt109Δ and hst3Δhst4Δ double mutants, and all three H3K56 mutant strains had amplified rDNA arrays in comparison to the corresponding parent strains, as previously reported (Ide et al. 2013) (Supplementary Figure S6D). These data suggest that the H3K56 acetylation pathway plays a key role in the regulation of rDNA stability and maintenance of normal rDNA copy number. Furthermore, these data suggest the H3K56 acetylation/deacetylation cycle regulates stability more than any particular state of acetylation.
H3K56 acetylation regulates CUP1 transcription and stability
H3K56 acetylation has been demonstrated to directly affect rDNA transcription; while total 18S and 25S rRNA levels in H3K56A mutants are comparable to wild-type strains, these mutants have decreased RNA polymerase I (RNAPI) binding at the rDNA and accumulate unprocessed rRNA precursors, both indicative of reduced rDNA transcription efficiency (Chen et al. 2012). Further, the normal H3K56 acetylation/deacetylation cycle has been shown to promote DNA damage repair via homologous recombination with the sister chromatid (Munoz-Galvan et al. 2013; Che et al. 2015). Consistent with this, control of rDNA amplification by the TOR signaling pathway has been shown to be mediated by H3K56 acetylation through break-induced replication (BIR) mediated repair pathways (Chen et al. 2012; Jack et al. 2015). These data suggest that rDNA amplification in mutants with perturbed H3K56 acetylation is due at least in part to constitutive and perturbed transcription at the locus, which may in turn influence the choice of the repair pathways in a manner that results in a net gain of repeats.
High levels of constitutive rDNA transcription are essential for cell viability. This makes it impossible to directly test transcription-dependent and independent functions of H3K56 acetylation in the maintenance of rDNA stability and copy number. We chose to study this in the copper-inducible CUP1 gene array because Hull et al. (2017) showed that transcription induces copy number variation at the CUP1 array in an Rtt109-dependent manner (Hull et al. 2017). Based on their observation of low CUP1 copy number variation in rtt109Δ mutants, this group had predicted that adaptation and CUP1 amplification in response to copper would require Rtt109. This prediction is confounding given the rDNA hyper-amplification observed in rtt109Δ mutants. However, their results are consistent with our observations of decreased rDNA repeat loss rates in rtt109Δ mutants, and high rDNA repeat loss rates in hst3Δhst4Δ mutants (Supplementary Figure S6B). We also previously showed that instability at both, the rDNA and CUP1 arrays is regulated by the fundamental processes of DNA replication and transcription (Figure 4). Taken together, these data suggest that the use of the inducible CUP1 array can help identify conserved general principles that govern the regulation of these two tandem repeats by H3K56 acetylation.
We found that disrupting the H3K56 acetylation cycle prevented transcription-induced instability at CUP1 in CUP1-MATα reporter strains with 6 or 17 copies of CUP1. We showed previously that copper-induced transcription increases CUP1 instability in a dose-dependent manner in CUP1-MATα reporter strains (Figure 4, C and D). In contrast, we find that transcription does not increase instability in the rtt109Δ or the hst3Δhst4Δ mutants (Figure 6A). Further, while transcription-independent CUP1 instability in the rtt109Δ mutants is comparable to that of corresponding wild-type strains, transcription-independent CUP1 instability in hst3Δhst4Δ mutants is significantly elevated (Figure 6A). These data suggest that both transcription-dependent and independent stability at the CUP1 array are affected by the H3K56 acetylation/deacetylation pathway.
Figure 6.
H3K56 acetylation regulates CUP1 instability and transcription. (A) Transcription dependent and independent CUP1 repeat loss rates in rtt109Δ and hst3Δhst4Δ mutants. Error bars represent standard deviation based on 4 biological replicates. (B) Absolute CUP1 mRNA concentration (×10 copies/ng of total RNA) in rtt109Δ and hst3Δhst4Δ mutants measured by RT-ddPCR. Error bars represent standard deviation based on three technical replicates for cDNA synthesis from the same RNA sample. Fold-change relative to the corresponding wild-type strain is indicated. For (A) and (B), the 6×CUP1 strains and the 17×CUP1 strains were grown in 1 mM CuSO4. Statistical significance was calculated using a standard two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n. s., not significant. (C) Growth assays showing copper resistance of rtt109Δ and hst3Δhst4Δ mutants.
Given the role of H3K56 acetylation in promoting transcription at the rDNA, and the induction of instability at the CUP1 array by transcription in a dose-dependent manner, we wondered whether the changes in CUP1 instability in the rtt109Δ and hst3Δhst4Δ mutants were due in part to altered CUP1 transcription. To test this, we measured basal CUP1 mRNA levels in wild-type, rtt109Δ and hst3Δhst4Δ mutant strains in CUP1-MATα reporter strains with 6 or 17 copies of CUP1. We also measured CUP1 mRNA levels in each of these strains following induction of the CUP1 array with the same concentrations of copper used for the instability measurements in Figure 6A. The decrease in CUP1 mRNA levels is modest in the copper-treated 6×CUP1 strains (91% and 70% of wild-type levels in rtt109Δ and hst3Δhst4Δ, respectively), and significant in the 17×CUP1 strains (38 and 63% of wild-type levels in rtt109Δ and hst3Δhst4Δ, respectively) (Figure 6B). Nevertheless, the rtt109Δ and hst3Δhst4Δ strains do not exhibit altered copper sensitivity compared to corresponding wild-type strains (Figure 6C). These data suggest that the H3K56 acetylation/deacetylation cycle is important for copper-induced CUP1 transcription at ≥6×CUP1 arrays. Given the higher transcriptional output and the more pronounced decrease in CUP1 transcript levels in the mutant 17×CUP1 strains, we speculate that disruption of H3K56 acetylation/deacetylation becomes increasingly detrimental to transcription as array size and transcriptional load on the array increases.
Basal CUP1 mRNA levels in untreated cells were also significantly affected in the rtt109Δ and hst3Δhst4Δ mutants in all three CUP1-MATα reporter strains tested. We found that the basal transcript abundance was significantly reduced in both rtt109Δ and hst3Δhst4Δ mutants in strains with ≥6×CUP1. The 6×CUP1 rtt109Δ and hst3Δhst4Δ strains exhibited a ∼2.4-fold and ∼1.9-fold decrease in basal CUP1 mRNA levels, respectively (Figure 6B). While the 17×CUP1 rtt109Δ strain exhibited a ∼2.3-fold decrease in CUP1 mRNA levels, transcript levels were unchanged in the 17×CUP1 hst3Δhst4Δ strains (Figure 6B). These data show that the H3K56 acetylation/deacetylation pathway is critical to maintain normal basal CUP1 transcription irrespective of array size.
The decrease in both, transcriptional output as well as transcription-dependent instability, suggests that the H3K56 acetylation/deacetylation pathway regulates instability at the CUP1 array partly through its effects on transcription. The differences in the changes in CUP1 transcript levels between the mutant 6×CUP1 and 17×CUP1 strains also suggest that the size of the tandem array may be critical in determining its response to transcription and perturbed H3K56 acetylation. Since transcription-induced CUP1 instability facilitates adaptation to high concentrations of copper (Figure 5), the dependence of transcription and stability of the CUP1 array on the H3K56 acetylation pathway suggests that the typical response of this array to transcription is critically dependent on its H3K56 acetylation status.
H3K56 acetylation restricts transcription-induced CUP1 amplification
Our study showed that in wild-type CUP1-MATα cells, CUP1 amplification facilitates adaptation to high concentrations of copper after prolonged propagation (25–50 generations) in selective conditions (Figure 5). While all mutations that affect H3K56 acetylation are known to be associated with amplifications of the rDNA array, we noticed that in uninduced conditions, copy number of the CUP1 array was stably maintained in both, rtt109Δ and hst3Δhst4Δ mutants irrespective of starting array size (Figure 7, Supplementary Figure S10). Because both rtt109Δ and hst3Δhst4Δ mutants with ≥6×CUP1 had low copper-induced instability and CUP1 mRNA levels, like Hull et al. (2017), we also predicted that these mutants may be defective in adapting to high concentrations of copper. However, when we subcultured various rtt109Δ and hst3Δhst4Δ reporter strains with ≥6×CUP1 in high concentrations of copper, we were surprised to find that copper resistant cells that had amplified CUP1 arrays readily emerged after 25–50 generations as in the corresponding wild-type strains (Figure 7, Supplementary Figure S10). Since this was reminiscent of the amplification of the constitutively transcribed rDNA array observed in rtt109Δ and hst3Δhst4Δ mutants, we hypothesized that in rtt109Δ and hst3Δhst4Δ mutants, transcription of the locus may induce amplification of the CUP1 array independent of growth defects. If this were the case, we would expect CUP1 amplifications to occur even in low concentrations of copper that do not affect cell growth.
Figure 7.
H3K56 acetylation restricts transcription-induced amplification of the CUP1 array. (i) Wild-type, (ii) rtt109Δ or (iii) hst3Δhst4Δ CUP1-MATα strains (6×CUP1) were subcultured in the indicated medium for ∼6 days (approximately 50 generations). After every 3 days, three independent isolates were used to measure CUP1 copy number by ddPCR. Error bars represent standard deviation for each individual reaction.
To test this hypothesis, we subcultured ≥6×CUP1 rtt109Δ and hst3Δhst4Δ strains in low levels of copper that do not impair growth. Importantly, these concentrations of copper induce instability at the CUP1 array in wild-type cells, but not in rtt109Δ or hst3Δhst4Δ mutants (Figure 6A). Interestingly, despite low rates of repeat loss, we found that after 25-50 generations, CUP1 amplification still occurred in all the ≥6×CUP1 rtt109Δ strains, but not in the corresponding wild-type strains (Figure 7, Supplementary Figure S10). This result demonstrates that stability and copy number maintenance need not be directly correlated, and highlights the importance of distinguishing between changes in instability and steady-state copy number.
While the hst3Δhst4Δ mutants did not exhibit net CUP1 amplification after ∼50 generations in these nonselective conditions, we found that there was more isolate to isolate variability in CUP1 copy number in these strains (Figure 7(iii)). Given the high transcription-independent repeat loss rates in this strain, it is possible that CUP1 amplification is countered by high rates of repeat loss, and requires propagation longer than 50 generations in these conditions. These data suggest that H3K56 acetylation is required for regulating transcription and transcription-induced instability at the CUP1 array, and importantly, it also functions to restrict transcription-induced amplification. Therefore, the H3K56 acetylation/deacetylation cycle is required to maintain the characteristic adaptive response to transcriptional stress.
Discussion
Eukaryotic genomes contain large stretches of tandem repeats. These repeats are repositories for copy number variation and can facilitate rapid adaptation to stress. Stress-induced adaptation through copy number variation could facilitate genome evolution in a variety of conditions like cancers, and treatment with anticancer drugs or antibiotics. This underscores the need to characterize the program of adaptation at tandem repeats. Here, we carried out a genetic screen with a new quantitative method to measure instability at two tandem arrays in budding yeast, the rDNA array, and the CUP1 gene array, to define the program of repeat stability and adaptation in response to transcriptional stress. We also used previously established ddPCR-based assays to measure rDNA copy number in a yeast ts mutant collection of 279 essential genes and identify additional essential factors involved in rDNA copy number maintenance. Altogether, our screens revealed that rDNA copy number and stability are regulated by the ubiquitous processes of DNA replication, transcription, and histone acetylation. Our assay was based on repeat loss; additional regulatory factors may be discovered if repeat gain was also monitored.
The traditional view of evolution states that adaptive mutations occur at random under stress and are selected for during growth under that stress. However, several studies in bacteria have shown that the genome may be designed to direct mutations to loci that require rapid evolution and tune the rates of mutation at relevant loci in the face of stress [reviewed in (Salim and Gerton 2019)]. Directing mutations to specific genomic regions is not restricted to bacteria; elegant work by Hull et al. (2017) demonstrated that in budding yeast, mutagenesis could be directed to the CUP1 array in a transcription-dependent manner (Hull et al. 2017). We extend our understanding of repeat stability by demonstrating its dependence not only on transcription itself, but also on transcription-associated consequences, such as R-loops. Furthermore, we demonstrate synergy for instability between DNA replication and transcription. Selection can act on the variation resulting from induced instability, which can produce a new “adapted” steady-state copy number over time. Therefore, stress-induced instability at tandem repeats can accelerate adaptation through copy number variation. These data support the model that the sensitivity of tandem repeats to transcription and DNA replication stress makes them unstable by design, enabling them to accommodate copy number variation and facilitate adaptation to genomic stresses.
Finally, we also define the contributions of histone acetylation to the process of adaptation at tandem repeats. Through studies using the CUP1 array, we show that H3K56 acetylation (a) regulates transcription, (b) supports transcription-dependent repeat instability, and (c) restricts transcription-induced repeat amplification. Importantly, we demonstrate that the responses to perturbation of H3K56 acetylation are dependent on the size of the array and its transcriptional status. While Hull et al. (2017) had proposed the importance of H3K56 acetylation in transcription-induced copy number variation, our studies provide experimental evidence for the role of this chromatin mark in governing transcription-induced instability at the CUP1 array and subsequent adaptation to copper. Our findings also highlight the importance of the characterization of instability and distinguishing it from changes in steady-state copy number upon prolonged propagation under transcriptional stress. Our studies suggest that loss of H3K56 acetylation may bias recombination events at the array toward amplification under transcriptional stress, which manifests as low repeat loss rates, but this requires further investigation. Parallel measurements of repeat stability and copy number changes at the rDNA suggest that the rDNA hyperamplification phenotype in these mutants may also be due to constitutive, perturbed transcription, and homologous recombination at the locus. Altogether, we have demonstrated that under conditions of transcriptional stress, H3K56 acetylation normally restricts amplification.
The fundamental principles underlying copy number variation at tandem repeats in budding yeast may be instructive to understand the behavior of other tandem repeats. One interesting application of our findings is in understanding the molecular evolution of cancer genomes because cancer cells are frequently characterized by transcriptional stress and copy number amplifications. Altered transcription may induce copy number variation at tandem repeats, facilitating adaptive copy number changes to occur, both, during the course of disease establishment and in response to therapy. There are several tandem repeat arrays on the X-chromosome (e.g., CT45, CT47) that are only expressed in the testis and in cancer (Ross et al. 2005); our results suggest transcription would trigger their instability in cancer. The observations that (a) the rDNA array is highly susceptible to recombination-mediated rearrangements in solid tumors (Stults et al. 2009), (b) rRNA transcription is frequently dysregulated in cancers (Lu et al. 2009; Xu et al. 2017; Udugama et al. 2018, (c) RNAPI transcription has emerged as an effective therapeutic target in a variety of cancers (Hannan et al. 2013), and (d) 45S rDNA repeats are lost in many cancer genomes (Wang and Lemos 2017; Xu et al. 2017; Udugama et al. 2018) all support the idea that 45S rDNA arrays are unstable in cancer and their instability is governed by the same foundational principles described in yeast.
Our findings have profound impacts for understanding the general principles underlying the evolution of genomes under stress. The TOR pathway regulates rDNA transcription by targeting the H3K56 acetylation pathway. Interestingly, in cells with low rDNA copy number, TOR-induced rDNA transcription as well as canonical HR-independent rDNA amplification by inhibiting key H3K56 deacetylases (Chen et al. 2012; Jack et al. 2015). Our data show that the regulation of repeat stability and copy number by transcription and histone acetylation is not limited to recovery or maintenance of normal rDNA copy number. Our studies of the CUP1 array suggest that transcription and histone acetylation may govern the behavior of tandem repeats in general and dictate adaptive outcomes. These data also have implications for understanding the molecular evolution of diseases like cancer, which are characterized by global dysregulation of transcription and histone acetylation, and copy number amplifications (Santarius et al. 2010; Audia and Campbell 2016). We speculate that altered transcription in the context of perturbed histone acetylation may underlie at least some of these amplifications. Further, because histone acetylation is often a target of anticancer drugs, understanding how acetylation impacts the stability and copy number of tandem arrays could be relevant to understanding how cancer cells adapt in response to therapy. Taken together, our studies of the yeast rDNA and CUP1 arrays reveal the unifying principles that govern tandem repeat plasticity and adaptation.
Acknowledgments
We thank Jin Zhu, Dominic Heinecke, and Rong Li for generously sharing qCTF strains, plasmids, reagents, and their expertise during the development of the qRIN assay. We also thank Andrew Box, Jungeun Park, and Laura Holmes (Stowers Institute) for assistance with the development of methods for cytometric analysis of samples and analysis of cytometry data, and the Cytometry core facility (Stowers Institute), particularly Dustin DeGraffenreid for assistance with FACS sorting cells and analyzing sorting data. We are also grateful to Sue-Jinks Robertson (Duke University) for strains, and Jerry Workman (Stowers Institute) for the YBL574 strain. We would like to thank the Molecular Biology core facility (Stowers Institute) for construction of H3K56 point mutants and for access to the TECAN plate reader. This work was done to fulfill, in part, requirements for Devika Salim’s PhD thesis research as a student registered with the Open University.
Funding
This work was funded by the Stowers Institute for Medical Research.
Conflicts of interest: None declared.
Literature cited
- Albert B, Leger-Silvestre I, Normand C, Ostermaier MK, Perez-Fernandez J, et al. 2011. RNA polymerase i-specific subunits promote polymerase clustering to enhance the rrna gene transcription cycle. J Cell Biol. 192:277–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Sloan RS, Petes TD, Jinks-Robertson S.. 2015. Genome-destabilizing effects associated with top1 loss or accumulation of top1 cleavage complexes in yeast. PLoS Genet. 11:e1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM.. 2004. The rpd3-sin3 histone deacetylase regulates replication timing and enables intra-s origin control in Saccharomyces cerevisiae. Mol Cell Biol. 24:4769–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audia JE, Campbell RM.. 2016. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 8:a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M, Katou Y, Komata M, Tanaka H, Itoh T, et al. 2009. Csm3, tof1, and mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem. 284:34355–34365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Coombes C, Kwok T, O'Donnell KA, Boeke JD, et al. 2008. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell. 30:248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachary M, Guilmatre A, Quilez J, Hasson D, Borel C, et al. 2014. Digital genotyping of macrosatellites and multicopy genes reveals novel biological functions associated with copy number variation of large tandem repeats. PLoS Genet. 10:e1004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Lockshon D, Fangman WL.. 1992. The arrest of replication forks in the rDna of yeast occurs independently of transcription. Cell. 71:267–276. [DOI] [PubMed] [Google Scholar]
- Buck SW, Maqani N, Matecic M, Hontz RD, Fine RD, et al. 2016. Rna polymerase i and fob1 contributions to transcriptional silencing at the yeast rdna locus. Nucleic Acids Res. 44:6173–6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Delucchi CS, van Bemmel JG, Haase S, Herce HD, Nowak D, et al. 2012. Histone hypoacetylation is required to maintain late replication timing of constitutive heterochromatin. Nucleic Acids Res. 40:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, et al. 2006. The sirtuins hst3 and hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 16:1280–1289. [DOI] [PubMed] [Google Scholar]
- Che J, Smith S, Kim YJ, Shim EY, Myung K, et al. 2015. Hyper-acetylation of histone h3k56 limits break-induced replication by inhibiting extensive repair synthesis. PLoS Genet. 11:e1004990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fan M, Pfeffer LM, Laribee RN.. 2012. The histone h3 lysine 56 acetylation pathway is regulated by target of rapamycin (tor) signaling and functions directly in ribosomal rna biogenesis. Nucleic Acids Res. 40:6534–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriere T, Pilsl M, Sarthou MK, Chauvier A, Genty T, et al. 2019. Genetic analyses led to the discovery of a super-active mutant of the RNA polymerase i. PLoS Genet. 15:e1008157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Felice F, Egidi A, D'Alfonso A, Camilloni G.. 2019. Fob1p recruits DNA topoisomerase i to ribosomal genes locus and contributes to its transcriptional silencing maintenance. Int J Biochem Cell Biol. 110:143–148. [DOI] [PubMed] [Google Scholar]
- Dong K, Addinall SG, Lydall D, Rutherford JC.. 2013. The yeast copper response is regulated by DNA damage. Mol Cell Biol. 33:4041–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z.. 2007. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 3:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z.. 2009. Gorilla: a tool for discovery and visualization of enriched go terms in ranked gene lists. BMC Bioinformatics. 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL.. 2003. In exponentially growing Saccharomyces cerevisiae cells, rrna synthesis is determined by the summed rna polymerase i loading rate rather than by the number of active genes. Mol Cell Biol. 23:1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley AR, Kobayashi T.. 2011. Monitoring the rate and dynamics of concerted evolution in the ribosomal DNA repeats of Saccharomyces cerevisiae using experimental evolution. Mol Biol Evol. 28:2883–2891. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, et al. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE.. 1989. A new role for a yeast transcriptional silencer gene, sir2, in regulation of recombination in ribosomal DNA. Cell. 56:771–776. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, et al. 2007. Rtt109 acetylates histone h3 lysine 56 and functions in DNA replication. Science. 315:653–655. [DOI] [PubMed] [Google Scholar]
- Hannan RD, Drygin D, Pearson RB.. 2013. Targeting rna polymerase i transcription and the nucleolus for cancer therapy. Expert Opin Ther Targets. 17:873–878. [DOI] [PubMed] [Google Scholar]
- Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, et al. 2009. A molecular barcoded yeast orf library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol. 27:369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigome C, Unozawa E, Ooki T, Kobayashi T.. 2019. Ribosomal rna gene repeats associate with the nuclear pore complex for maintenance after DNA damage. PLoS Genet. 15:e1008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D.. 2007. Trf4 targets ncrnas from telomeric and rdna spacer regions and functions in rdna copy number control. Embo J. 26:4996–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Moazed D.. 2003. Association of the rent complex with nontranscribed and coding regions of rdna and a regional requirement for the replication fork block protein fob1 in rdna silencing. Genes Dev. 17:2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull RM, Cruz C, Jack CV, Houseley J.. 2017. Environmental change drives accelerated adaptation through stimulated copy number variation. PLoS Biol. 15:e2001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. 2004. Detection of large-scale variation in the human genome. Nat Genet. 36:949–951. [DOI] [PubMed] [Google Scholar]
- Ide S, Miyazaki T, Maki H, Kobayashi T.. 2010. Abundance of ribosomal rna gene copies maintains genome integrity. Science. 327:693–696. [DOI] [PubMed] [Google Scholar]
- Ide S, Saka K, Kobayashi T.. 2013. Rtt109 prevents hyper-amplification of ribosomal rna genes through histone modification in budding yeast. PLoS Genet. e1003410.9: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Watanabe K, Watanabe H, Shirahige K, Kobayashi T, et al. 2007. Abnormality in initiation program of DNA replication is monitored by the highly repetitive rrna gene array on chromosome xii in budding yeast. Mol Cell Biol. 27:568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Kobayashi T.. 2019. Rna polymerase i activators count and adjust ribosomal rna gene copy number. Mol Cell. 73:645–654 e613. [DOI] [PubMed] [Google Scholar]
- Jack CV, Cruz C, Hull RM, Keller MA, Ralser M, et al. 2015. Regulation of ribosomal DNA amplification by the tor pathway. Proc Natl Acad Sci USA. 112:9674–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T.. 2002. Replication fork block protein, fob1, acts as an rdna region specific recombinator in S. cerevisiae. Genes Cells. 7:99–113. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ganley AR.. 2005. Recombination regulation by transcription-induced cohesin dissociation in rdna repeats. Science. 309:1581–1584. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T.. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (fob1) protein and the role of rna polymerase i. Genes Dev. 12:3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M.. 2004. Sir2 regulates recombination between different rdna repeats, but not recombination within individual rrna genes in yeast. Cell. 117:441–453. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. 2003. The replication fork barrier site forms a unique structure with fob1p and inhibits the replication fork. Mol Cell Biol. 23:9178–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan EX, Foss EJ, Tsuchiyama S, Alvino GM, Kruglyak L, Kaeberlein M, et al. 2013. A natural polymorphism in rdna replication origins links origin activation with calorie restriction and lifespan. PLoS Genet. 9:e1003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney JD, Mobley EF, Hochstrasser M.. 2006. The short-lived matalpha2 transcriptional repressor is protected from degradation in vivo by interactions with its corepressors tup1 and ssn6. Mol Cell Biol. 26:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KS, Merrikh H.. 2018. The clash of macromolecular titans: replication-transcription conflicts in bacteria. Annu Rev Microbiol. 72:71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Chang Q, Zhang Y, Beezhold K, Rojanasakul Y, et al. 2009. Lung cancer-associated jmjc domain protein mdig suppresses formation of tri-methyl lysine 9 of histone h3. Cell Cycle. 8:2101–2109. [DOI] [PubMed] [Google Scholar]
- Maas NL, Miller KM, DeFazio LG, Toczyski DP.. 2006. Cell cycle and checkpoint regulation of histone h3 k56 acetylation by hst3 and hst4. Mol Cell. 23:109–119. [DOI] [PubMed] [Google Scholar]
- Mansisidor A, Molinar T Jr., Srivastava P, Dartis DD, Pino Delgado A, et al. 2018. Genomic copy-number loss is rescued by self-limiting production of DNA circles. Mol Cell. 72:583–593 e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D.. 2009. Contrasting roles of checkpoint proteins as recombination modulators at fob1-ter complexes with or without fork arrest. Eukaryot Cell. 8:487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Galvan S, Jimeno S, Rothstein R, Aguilera A.. 2013. Histone h3k56 acetylation, rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet. 9:e1003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A.. 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for h3k4 methylation. Nat Struct Mol Biol. 15:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M, Nogi Y, Clark MW, Nomura M.. 1993. Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in rna polymerase i. Mol Cell Biol. 13:2441–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M, Siddiqi I, Vu L, Aris J, Nomura M.. 1999. Transcription factor uaf, expansion and contraction of ribosomal DNA (rdna) repeats, and rna polymerase switch in transcription of yeast rdna. Mol Cell Biol. 19:8559–8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, De Chiara M, Friedrich A, Yue JX, Pflieger D, et al. 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 556:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. 1980. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 19:765–774. [DOI] [PubMed] [Google Scholar]
- Press MO, Hall AN, Morton EA, Queitsch C.. 2019. Substitutions are boring: some arguments about parallel mutations and high mutation rates. Trends Genet. 35:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. 2006. Global variation in copy number in the human genome. Nature. 444:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Herskowitz I.. 1987. Four genes responsible for a position effect on expression from hml and hmr in Saccharomyces cerevisiae. Genetics. 116:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, et al. 2005. The DNA sequence of the human x chromosome. Nature. 434:325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka K, Takahashi A, Sasaki M, Kobayashi T.. 2016. More than 10% of yeast genes are related to genome stability and influence cellular senescence via rdna maintenance. Nucleic Acids Res. 44:4211–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim D, Bradford WD, Freeland A, Cady G, Wang J, et al. 2017. DNA replication stress restricts ribosomal DNA copy number. PLoS Genet. 13:e1007006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim D, Gerton JL.. 2019. Ribosomal DNA instability and genome adaptability. Chromosome Res. 27:73–87. [DOI] [PubMed] [Google Scholar]
- Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS.. 2010. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 10:59–64. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. 2004. Large-scale copy number polymorphism in the human genome. Science. 305:525–528. [DOI] [PubMed] [Google Scholar]
- Shyian M, Mattarocci S, Albert B, Hafner L, Lezaja A, et al. 2016. Budding yeast rif1 controls genome integrity by inhibiting rdna replication. PLoS Genet. 12:e1006414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Pillus L, Boeke JD.. 1998. Distribution of a limited sir2 protein pool regulates the strength of yeast rdna silencing and is modulated by sir4p. Genetics. 149:1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Caputo E, Boeke JD.. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 19:3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, et al. 2009. Human rrna gene clusters are recombinational hotspots in cancer. Cancer Res. 69:9096–9104. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Wu R.. 1980. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 284:426–430. [DOI] [PubMed] [Google Scholar]
- Udugama M, Sanij E, Voon HPJ, Son J, Hii L, et al. 2018. Ribosomal DNA copy loss and repeat instability in atrx-mutated cancers. Proc Natl Acad Sci USA. 115:4737–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T.. 2010. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 17:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varv S, Kristjuhan K, Peil K, Looke M, Mahlakoiv T, et al. 2010. Acetylation of h3 k56 is required for rna polymerase ii transcript elongation through heterochromatin in yeast. Mol Cell Biol. 30:1467–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M.. 2002. Histone acetylation regulates the time of replication origin firing. Mol Cell. 10:1223–1233. [DOI] [PubMed] [Google Scholar]
- Wagstaff JE, Klapholz S, Waddell CS, Jensen L, Esposito RE.. 1985. Meiotic exchange within and between chromosomes requires a common rec function in Saccharomyces cerevisiae. Mol Cell Biol. 5:3532–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Mansisidor A, Prabhakar G, Hochwagen A.. 2016. Condensin and hmo1 mediate a starvation-induced transcriptional position effect within the ribosomal DNA array. Cell Rep. 14:1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Lemos B.. 2017. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 13:e1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Hasson D, Guillem F, Lescale C, Jin X, et al. 2008. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics. 9:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J, Fogel S, Buchman C, Karin M.. 1989. The cup2 gene product regulates the expression of the cup1 gene, coding for yeast metallothionein. Embo J. 8:255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram Research. 2014. Mathematica. Champaign, IL: Wolfram Research, Inc. [Google Scholar]
- Wyandt HE, Wilson GN, Tonk VS.. 2017. Human chromosome variation: heteromorphism, polymorphism and pathogenesis.
- Xu B, Li H, Perry JM, Singh VP, Unruh J, et al. 2017. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 13:e1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Miller A, Kirchmaier AL.. 2008. Hst3/hst4-dependent deacetylation of lysine 56 of histone h3 in silent chromatin. Mol Biol Cell. 19:4993–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, MacDonald JR, Merico D, Scherer SW.. 2015. A copy number variation map of the human genome. Nat Rev Genet. 16:172–183. [DOI] [PubMed] [Google Scholar]
- Zhu J, Heinecke D, Mulla WA, Bradford WD, Rubinstein B, et al. 2015. Single-cell based quantitative assay of chromosome transmission fidelity. G3 (Bethesda). 5:1043–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1576. Strains and plasmids are available upon request. Supplementary Figure S1 contains additional validation of the qRIN assay. Supplementary Figure S2 contains analysis of rDNA stability in fob1Δ mutants. Supplementary Figure S3 contains analysis of the loss of MATα-LEU2 in fob1Δ mutants. Supplementary Figure S4 contains analysis of rDNA stability in NAM. Supplementary Figure S5 contains validation of the qRIN assay for the CUP1-MATα reporter strains. Supplementary Figure S6 contains validation of hits with altered rDNA stability. Supplementary Figure S7 contains functional validation and additional supporting information with regard to deletion of CUP2 in CUP1-MATα reporter strains. Supplementary Figure S8 contains stress and locus-specific effects of copper treatment. Supplementary Figure S9 contains data regarding how deletion of RTT109, HST3, and HST4 affect stability of TUB1. Supplementary Figure S10 contains data regarding how H3K56 acetylation restricts transcription-induced amplification of the CUP1 array in a 17 copy strain. Supplementary File S1 describes how to calculate repeat loss rates. Supplementary File S2 describes the method for determining and the location of the MATα-LEU2 casette in reporter strains. Supplementary Table S1 summarizes rDNA and CUP1 instability measurements from the literature. Supplementary Table S2 is a list of yeast strains. Supplementary Table S3 is a list of primers. Supplementary Table S4 contains validation copy number measurements for 25S rDNA, CUP1, LEU2, and MATALPHA1 in various yeast strains. Supplementary Table S5 contains the rDNA repeat loss rates and copy number measurements from the screens. Supplementary Table S6 contains rDNA copy number measurements from the temperature-sensitive mutant collection. Supplementary Table S7 contains copy number measurements of subcultured CUP1-MATα strains.
Supplementary material is available at figshare DOI: https://doi.org/10.25387/g3.14195492.