Acquired resistance to the antibody–drug conjugate sacituzumab govitecan can arise through genomic alterations affecting either the antigen (TROP2) or the payload target (TOP1).
Abstract
Sacituzumab govitecan (SG), the first antibody–drug conjugate (ADC) approved for triple-negative breast cancer, incorporates the anti-TROP2 antibody hRS7 conjugated to a topoisomerase-1 (TOP1) inhibitor payload. We sought to identify mechanisms of SG resistance through RNA and whole-exome sequencing of pretreatment and postprogression specimens. One patient exhibiting de novo progression lacked TROP2 expression, in contrast to robust TROP2 expression and focal genomic amplification of TACSTD2/TROP2 observed in a patient with a deep, prolonged response to SG. Analysis of acquired genomic resistance in this case revealed one phylogenetic branch harboring a canonical TOP1E418K resistance mutation and subsequent frameshift TOP1 mutation, whereas a distinct branch exhibited a novel TACSTD2/TROP2T256R missense mutation. Reconstitution experiments demonstrated that TROP2T256R confers SG resistance via defective plasma membrane localization and reduced cell-surface binding by hRS7. These findings highlight parallel genomic alterations in both antibody and payload targets associated with resistance to SG.
Significance:
These findings underscore TROP2 as a response determinant and reveal acquired SG resistance mechanisms involving the direct antibody and drug payload targets in distinct metastatic subclones of an individual patient. This study highlights the specificity of SG and illustrates how such mechanisms will inform therapeutic strategies to overcome ADC resistance.
This article is highlighted in the In This Issue feature, p. 2355
Introduction
Metastatic triple-negative breast cancer (mTNBC), defined by absence of estrogen and progesterone receptors (ER and PR) and human epidermal growth factor receptor 2 (HER2) gene amplification, is associated with very poor prognosis (1, 2). Standard chemotherapy, the mainstay of management, is associated with low response rates (5%–10%) and poor median progression-free survival (2–3 months) in the relapsed/refractory setting (3, 4). Similarly, for patients with endocrine-resistant hormone receptor–positive (HR+) metastatic breast cancer (MBC), chemotherapy is the mainstay of management but has limited efficacy and has not been demonstrated to improve overall survival (OS). Antibody–drug conjugates (ADC), generated by linking cancer-directed antibodies to potent payloads, can selectively deliver toxic payloads to cancer cells, thereby maximizing the therapeutic window (5).
Although TNBC lacks the traditional actionable receptors (ER, PR, and HER2), the majority (85%–90%) express TROP2, making it an attractive target for ADCs (6). Sacituzumab govitecan (SG) is a first-in-class TROP2 ADC in which SN-38 (the active metabolite of irinotecan), a topoisomerase I (TOP1) inhibitor, is coupled to the humanized anti-TROP2 monoclonal antibody hRS7 IgG1κ via the hydrolysable CL2A linker. In a phase I/II clinical trial, patients with metastatic TNBC (median of 3 prior regimens) treated with SG had remarkable therapeutic responses (33%), including complete responses (7). The pivotal randomized phase III ASCENT trial demonstrated comparable clinical activity and a near-doubling of OS versus physicians' choice chemotherapy (8). Based on its clinical activity in advanced refractory TNBC, SG received accelerated FDA approval for mTNBC in April 2020 and full approval in April 2021. Substantial clinical efficacy has also been observed in HR+ MBC and other solid tumors that express TROP2 (9).
Despite multiple clinical trials of SG, little is known about predictors of therapeutic response and mechanisms of de novo and acquired resistance. Thus, we carried out genomic and transcriptomic analysis of tumor tissue from patients with mTNBC treated with SG for whom we had access to pretreatment and multisite postprogression (rapid autopsy) specimens.
Results
Association of TROP2 Levels and Gene Copy Number with SG Response/Resistance
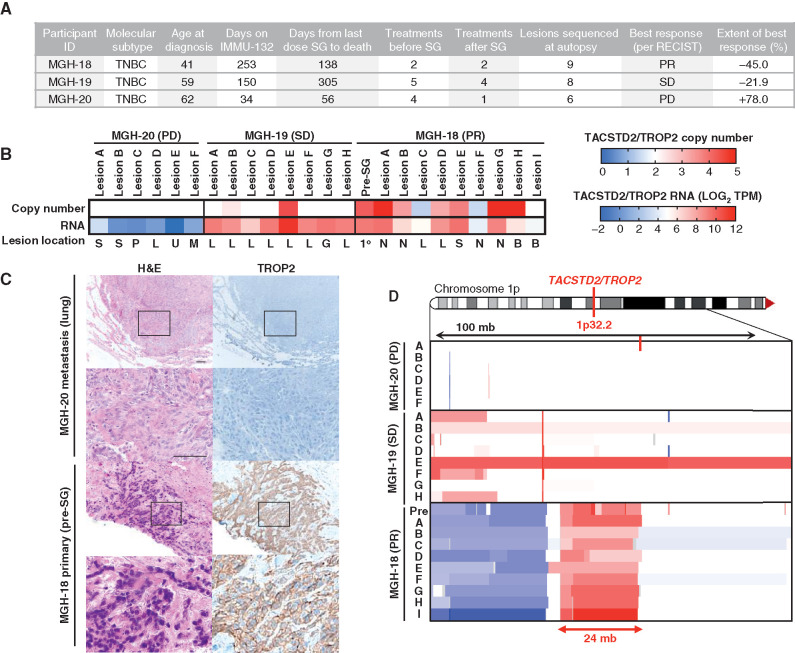
As TROP2 is the protein bound by the antibody moiety of SG, we first analyzed TROP2 gene expression and genomic copy number in pre- and posttreatment specimens from the three-case autopsy series (Fig. 1A; Supplementary Table S1). Each of these patients received SG following progression on at least two prior therapies for mTNBC (Fig. 1A; Supplementary Table S2). Case MGH-20 showed evidence of clinical de novo progression by the time of first interval radiologic assessment, resulting in drug discontinuation. MGH-19 experienced stable disease (SD) by Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 criteria for a period of 5 months, whereas MGH-18 had a substantial and prolonged partial response according to RECIST 1.1 (45% tumor regression, >8 months), prior to multisite progressive disease (PD; Fig. 1A; Supplementary Fig. S1). RNA-sequencing (RNA-seq) analysis of all analyzed tumor specimens from MGH-20 demonstrated essentially undetectable TROP2 RNA expression, and this was corroborated by the absence of TROP2 protein expression on IHC (Fig. 1B and C). MGH-18 and MGH-19 both exhibited detectable TROP2 RNA expression in all specimens analyzed, including in metastatic foci that had progressed on SG treatment (Fig. 1B). MGH-18 was distinguished by the presence of genomic amplification of the TACSTD2 locus (encoding TROP2) in multiple specimens tested, and as anticipated the primary tumor showed robust levels of membrane-localized TROP2 protein (Fig. 1C and D). These findings are in keeping with recent data demonstrating that absence of TROP2 expression is commonly associated with de novo clinical resistance to SG (8).
Figure 1.
TROP2 expression and gene copy number and response to SG. A, Clinical characteristics, treatment history, and SG response data for the three autopsy series patients. PR, partial response; SD, stable disease; PD, progressive disease. B, TACSTD2/TROP2 copy number (top) and RNA expression (bottom) for all analyzed tumor specimens from each case. Note, white indicates diploid copy number. Key for lesion location: S, subcutaneous; P, pericardium; L, liver; U, lung; M, mediastinum; G, gallbladder; N, lymph node; B, brain; 1o, primary breast tumor. C, Hematoxylin and eosin (H&E) and TROP2 IHC show tumor TROP2 protein expression is absent in MGH-20 but shows intense membrane staining in MGH-18. Scale bars, 50 μm. D, Focal amplification of TACSTD2/TROP2 in pretreatment primary tumor and multiple metastatic lesions from MGH-18. Copy-number scale per B is shown for the indicated region of chromosome 1p of each lesion, indicating up to N = 5 TACSTD2/TROP2 copies. See also Supplementary Fig. S1 and Supplementary Table S1.
Parallel Genomic Alterations of TACSTD1/TROP2 and TOP1 Denote Acquired Resistance to SG
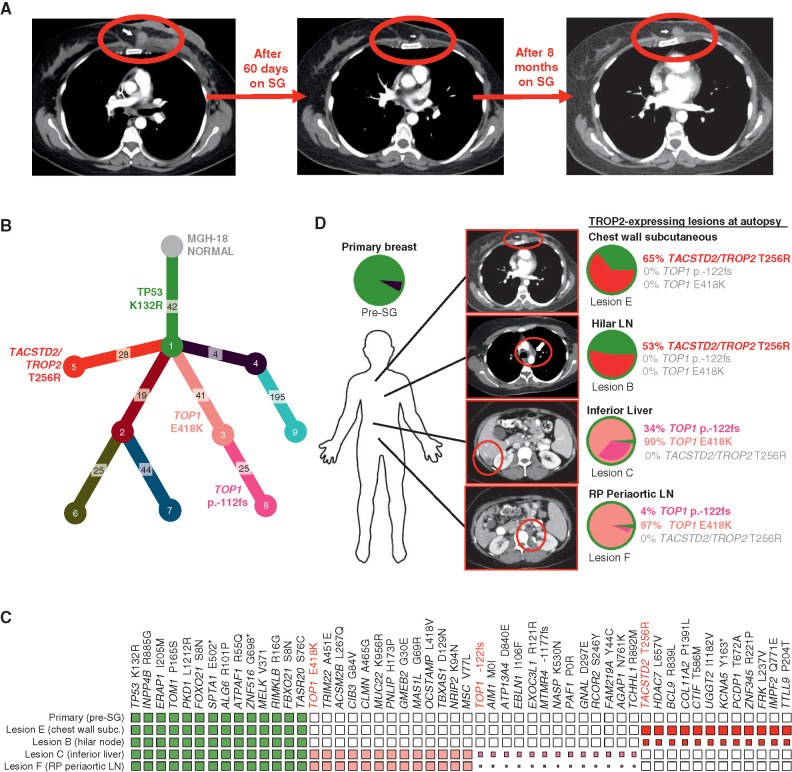
We focused our analysis of acquired resistance on MGH-18 because this case was associated with the most profound and prolonged response, and a relatively short interval (<4 months) between progression on SG and tissue harvest (autopsy). This case involved a 42-year-old female who underwent standard preoperative chemotherapy for primary ER/PR/HER2-negative invasive ductal carcinoma (i.e., TNBC), but within weeks of completion of neoadjuvant therapy was found to have metastatic disease to liver and other organs. The patient underwent palliative mastectomy and then received two sequential investigational therapeutic combinations that were short-lived, and there was no objective response (Supplementary Table S2). The patient then initiated treatment with SG, and restaging scans after two months demonstrated a radiologic partial response with 45% reduction, as per RECIST (Fig. 2A). However, after eight months, restaging scans revealed disease progression at multiple sites, and following brief treatment with standard chemotherapy, the patient expired (Fig. 2A; Supplementary Table S2).
Figure 2.
Parallel and mutually exclusive mutations in TOP1 and TACSTD2/TROP2 in an individual patient with acquired resistance to SG. A, CT radiographs showing deep response and subsequent progression of chest wall lesion (red circle) in MGH-18 under treatment with SG. B, Phylogenetic tree representing the clonal architecture present in primary tumor and metastatic (autopsy) lesions of MGH-18 shown in Fig. 1, using PhylogicNDT (10). Circles indicate numbered clones, and numbers in squares indicate their associated somatic alterations. The primary tumor (green clone) harbors a truncal TP53 mutation, and two major branches harbor TACSTD2/TROP2 and TOP1 mutations. C, Representative clonal and subclonal somatic alterations detected in the indicated tissue specimens. The size of each square represents the estimated tumor proportion of each alteration, with an empty box indicating no detection. D, Clonal composition of primary and metastatic lesions of MGH-18. Layered pie charts represent the likely clonal composition of the indicated specimens, with the color of each subclone matching the color of the respective clone/branch in the phylogenetic tree. The percent of the TACSTD2/TROP2 and TOP1-mutant clones are indicated for each lesion. Note, the TOP1 p.-122fs (frameshift mutation) denotes a subbranch that also harbors the TOP1E418K mutation. CT images show the respective lesions (circled). Note, pie charts and clonality charts for lesions lacking TOP1 and TACSTD2/TROP2-mutant branches are shown in Supplementary Fig. S2. LN, lymph node; RP, retroperitoneal.
In order to understand the evolution of acquired resistance to SG in this case, we carried out whole-exome sequencing (WES) of the pre-SG treatment and nine postprogression rapid autopsy tumor lesions. From these data, we then reconstructed the phylogenetic relationships and clonal architecture of the resistant lesions using PhylogicNDT, an integrated suite of tools designed for jointly modeling evolution of many samples from the same patient (Fig. 2B; ref. 10). This analysis revealed shared clonal (truncal) mutations common to all samples and private mutations within a given branch (Supplementary Table S3). A truncal TP53 mutation (K132R) was present in the pre-SG primary tumor, as is commonly observed in TNBC, and was present in all postprogression lesions (Fig. 2B and C). In most cases, multiple branches were detected in each metastatic lesion (Fig. 2D; Supplementary Fig. S2A and S2B). Most notable in the phylogenetic tree of resistance were two major branches that demonstrated distinct, branch-restricted mutations of TOP1 (encoding the SN-38 drug target topoisomerase 1) and TACSTD2 (encoding TROP2; Fig. 2B; Supplementary Fig. S2C). These two phylogenetic branches were present in multiple, mutually exclusive metastatic lesions at high frequency. One branch, which was present at high frequency within abdominal lesions in the liver and periaortic lymph nodes, harbored the TOP1 missense mutation E418K (Fig. 2C and D; Supplementary Fig. S2C). This specific TOP1 mutation has been previously described and is established to confer resistance to clinical TOP1 inhibitors, most likely by altering sequence specificity for DNA cleavage by TOP1 (11, 12). Further evolution of this TOP1E418K-containing branch involves an additional subclonal 8-bp frameshift insertion in TOP1 (TOP1p.-122fs), which is present in these same metastases (Fig. 2C and D; Supplementary Fig. S2C). Of note, because the TOP1 frameshift mutation appears to arise from the clone harboring the TOP1E418K mutation, this suggests that the frameshift could represent a “second hit” promoting enhanced resistance to SG. Furthermore, analysis of RNA-seq data demonstrated that both TOP1 mutations were expressed in the metastatic lesions (Supplementary Fig. S2D).
A second major phylogenetic branch of resistance was hallmarked by a missense mutation of TACSTD2/TROP2, T256R. As noted, this mutation occurred in a distinct phylogenetic branch from the TOP1 mutations, and this TACSTD2/TROP2-mutant branch was present at high frequency in multiple thoracic metastatic lesions, including hilar lymph node and chest wall (Fig. 2C and D; Supplementary Fig. S2C). RNA bearing this mutation was also expressed in the respective lesions (Supplementary Fig. S2D). To our knowledge, this mutation of the gene encoding TROP2 has not been previously described in the literature in any context. However, its acquisition in the setting of tumor progression on SG therapy suggested the possibility of parallel mechanisms of resistance to SG involving both the antibody target, TROP2, and its SN-38 payload target, TOP1.
The Membrane-Proximal TACSTD2/TROP2 Mutation T256R Alters Plasma Membrane Localization and Cell-Surface Binding of hRS7
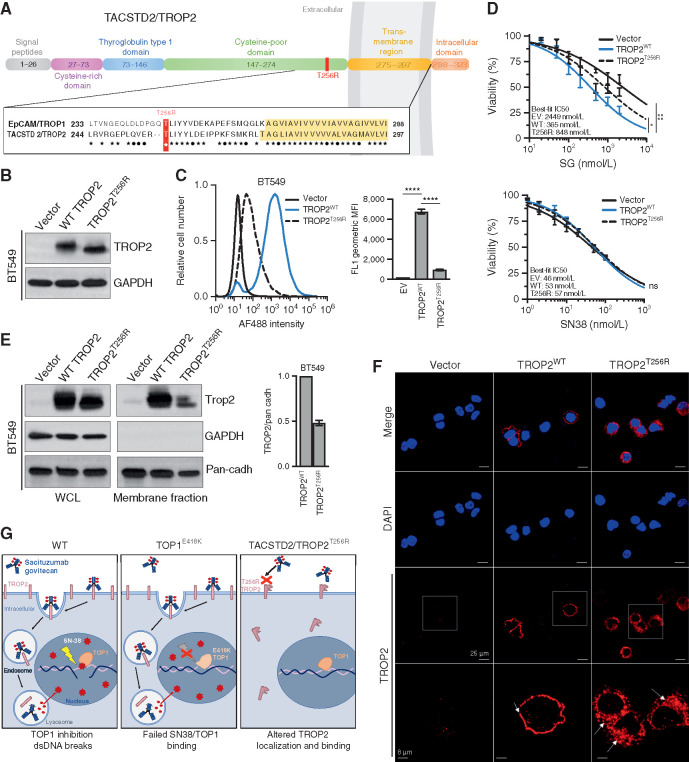
To determine the potential functional significance of the TACSTD2/TROP2T256R mutation for SG treatment, we synthesized cDNAs encoding either the wild-type or mutant TROP2 and reconstituted them into multiple TROP2-negative cell-based models. TROP2 is a cell-surface glycoprotein that exhibits high homology to epithelial cell adhesion molecule (EPCAM), also known as TROP1/TACSTD1 (Fig. 3A). Amino acid T256 of TROP2 resides in a conserved, membrane-proximal region of the extracellular domain, less than 20 amino acids from the transmembrane domain that begins at residue 275. We found that TACSTD2/TROP2T256R encodes a stable protein that could be readily expressed in both TNBC (BT549) cells and nontransformed (NIH 3T3) cells, which both lack endogenous TROP2 expression (Fig. 3B; Supplementary Fig. S3A). We verified equivalent expression levels of wild-type and mutant TROP2 protein and then assayed cell-surface binding to TROP2 in these cells by hRS7, the (humanized) antibody incorporated into SG. Although control transfected cells showed no binding as anticipated, reconstitution of wild-type TROP2 conferred robust cell-surface binding by hRS7. In contrast, in cells reconstituted with the TROP2 mutant, the binding was reduced by >80% compared with wild-type TROP2 in TNBC cells, and similar results were observed in 3T3 cells (Fig. 3C; Supplementary Fig. S3B). Binding of an unrelated anti-TROP2 antibody was also highly significantly reduced in these same cells (Supplementary Fig. S3C and S3D). Thus, TACSTD2/TROP2T256R encodes a stable protein associated with diminished cell-surface binding by multiple antibodies including the SG backbone hRS7.
Figure 3.
TACSTD2/TROP2T256R confers resistance to SG via altered localization and decreased binding to hRS7. A, Domain structure and homology between TROP1/EPCAM and TROP2/TACSTD2. EPCAM and TROP2 share 49% sequence identity and 67% sequence similarity. Homology is highest in the membrane-proximal region and transmembrane region (yellow highlight). ★, identical amino acids; •, conservative differences. T256R mutation site indicated in red. B, Western blot of lentivirally expressed control vector or TROP2 cDNAs in TROP2-negative BT549 cells, showing equivalent expression levels of wild-type (WT) and mutant proteins. C, Live-cell flow cytometry of BT549 cells shown in B stained with hRS7, the anti-TROP2 antibody backbone of SG, reveals a marked decrease in signal intensity for cells harboring TROP2T256R compared with WT. Bar graphs to the right represent geometric mean fluorescent intensity (MFI). Data are representative of three independent experiments, and error bars show SD. ****, P < 0.0001. D, TROP2T256R-mutant confers resistance to SG but not SN38. BT549 cells expressing empty vector (EV), WT TROP2, or T256R-mutant TROP2 were treated with SG or SN-38 at the indicated doses for 4 hours and then cultured in fresh medium for 96 hours before assessing cell viability by using CellTiter-Glo. Values plotted represent the mean of quadruplicate wells and an average of three independent experiments. Error bars indicate SD. P value calculated by two-way ANOVA (cell line, concentration). *, P < 0.05; ns, nonsignificant. E, TROP2T256R has significantly decreased plasma membrane association, shown by comparing whole-cell lysate (WCL) and plasma membrane fraction of BT549 cells expressing the vector, WT TROP2, or T256R-mutant TROP2. GAPDH and pan-cadherin serve as controls for cytosol and membrane fractions, respectively. Bar graphs at right represent the mean of at least two independent experiments. Error bars indicate range. F, Confocal immunofluorescence using rabbit monoclonal clone EPR20043 reveals plasma membrane staining of WT TROP2 and intracellular staining of TROP2T256R. Vector, WT, or T256R TROP2-expressing BT549 cells were stained for TROP2 and then examined at 40× using a Nikon A1R confocal microscope. Bottom, magnified images corresponding to white boxes. Scale bars, 25 μm and 8 μm (magnified). G, Model for parallel polyclonal acquired resistance to SG. Left, SG induces tumor cell killing through TROP2-dependent delivery of SN-38, which binds at the TOP1/DNA interface and induces double-strand DNA (dsDNA) breaks. Middle, TOP1E418K has altered DNA binding and sequence specificity, resulting in failure of SN-38 binding and SN-38–induced DNA damage. Right, the novel TROP2T256R mutant shows attenuated cell membrane localization and decreased binding to hRS7, impeding SG binding and SN-38 delivery. See also Supplementary Figs. S3 and S4.
We next tested directly whether attenuated binding of the mutant TROP2 protein by hRS7 was associated with decreased sensitivity to SG. As expected, reconstitution of wild-type TROP2 into TROP2-negative cells conferred highly significant increases in sensitivity to SG, in both TNBC and 3T3 cells (Fig. 3D; Supplementary Fig. S3E). In contrast, reconstitution with mutant TROP2 conferred only minimal sensitivity that was only marginally distinguishable from that of nonreconstituted cells. As a control, we tested sensitivity to the SG payload SN-38 itself in these cells. As anticipated, reconstitution of either wild-type or mutant TROP2 had no impact on SN-38 sensitivity in either TNBC or 3T3 cells (Fig. 3D; Supplementary Fig. S3E). Thus, expression of the mutant TROP2 compared with the wild-type protein is associated with decreased cellular sensitivity to SG.
Given attenuated cell-surface binding of TROP2T256R to hRS7 despite expression levels comparable to wild-type protein, we hypothesized that the mutant protein might exhibit altered subcellular localization. We therefore isolated plasma membrane fractions from TROP2-expressing TNBC and 3T3 cells and compared localization of mutant and wild-type TROP2 proteins. In both cell types, TROP2T256R consistently displayed decreased plasma membrane localization compared with the wild-type protein (Fig. 3E; Supplementary Fig. S3F). We then used confocal immunofluorescence (IF) to corroborate these findings. Staining TNBC cells with a rabbit monoclonal antibody, we observed nearly exclusive plasma membrane staining of wild-type TROP2. In contrast, TROP2T256R showed substantial localization to the cytosol (Fig. 3F). Furthermore, confocal IF in these same cells using the hRS7 antibody revealed both mislocalized and decreased staining, possibly suggesting a selective decrease in binding of the mutant protein to hRS7 (Supplementary Fig. S3G). Finally, we carried out IHC for TROP2 in parallel on the patient MGH-18 hilar lymph node metastasis containing the mutant TROP2T256R and a brain metastasis harboring only wild-type TROP2. Consistent with IF analysis, the hilar lesion showed a largely diffuse cytoplasmic staining pattern for TROP2, whereas the brain metastasis showed a membrane pattern similar to the primary tumor (Supplementary Fig. S4A and S4B). Altogether, this study suggests the emergence of parallel genetic mechanisms of acquired resistance under selective pressure from SG treatment in this patient. A subset of metastatic lesions harbor an established resistance mutation to the SG TOP1 inhibitor (SN-38) payload, TOP1E418K, whereas a nonoverlapping set of lesions harbor the TACSTD2/TROP2T256R mutation, which encodes a protein with altered subcellular localization, attenuated cell-surface binding to the therapeutic antibody backbone, and ability to confer SG resistance (Fig. 3G).
Discussion
To our knowledge, this is the first report to describe the emergence of acquired resistance to SG hallmarked by mutually exclusive somatic mutations in genes encoding both TROP2 (target of antibody) and TOP1 (target of payload) in distinct metastatic lesions of the same patient. We use the term “parallel” to describe this pattern of polyclonal acquired genetic resistance, in contrast to multiple recently described “convergent” mechanisms that mediate resistance to a single pathway inhibitor (13, 14). Parallel resistance seems naturally applicable to ADCs, given they have dual molecular targets for the antibody and drug payload. The parallel resistance mechanisms we describe highlight the specificity of this ADC, and they may have direct implications for the choice of subsequent therapies, particularly as additional ADCs with potentially overlapping antibody and payload targets become available for this and other patient populations (5, 6).
Comparing three autopsy cases with multiple tumor lesions available for analysis, we find an association between absence of TROP2 expression and a lack of clinical response to SG. In general, reduced expression of an ADC target antigen could affect both binding of the antibody as well as intracellular uptake and payload release (15, 16). In the pivotal EMILIA trial, which led to approval of the HER2-directed ADC trastuzumab emtansine (T-DM1) for metastatic HER2+ breast cancer, the survival benefit with T-DM1 compared with control arm was greater in patients whose tumors had high HER2 mRNA levels (hazard ratio = 0.53; median OS, 34.1 vs. 26.5 months), compared with lower HER2 mRNA levels (hazard ratio, 0.80; median OS, 24.8 vs. 23.7 months; ref. 17). Similarly, in the confirmatory phase III clinical trial with SG (ASCENT), patients with mTNBC who had high or medium TROP2 expression had higher objective response rates (44% and 39%, respectively), as compared with low TROP2 expression (22%; refs. 8, 18). Furthermore, out of seven patients who had tumors with complete absence of TROP2 expression in ASCENT, only one had a response (8). These observations are consistent with our findings and highlight how lower expression of TROP2 could modulate therapeutic efficacy and ultimately might influence patient selection for SG.
By focusing on a single patient with a deep and prolonged response to SG who subsequently progressed, we were able to identify multiple potentially important mechanisms of acquired genetic resistance to this ADC. The point mutation in TOP1 we describe, E418K, is one of several reported missense mutations that are known to induce resistance to clinical TOP1 inhibitors but do not alter intrinsic enzyme catalytic activity (11, 19). Instead, this mutation is believed to alter the DNA sequence specificity and/or DNA binding affinity of the enzyme and thereby hinder the stereotactic binding of drug to the enzyme/DNA interface (12). Although such mutations are established to induce resistance in the heterozygous state, the potential relevance of the mutation in this case is further underscored by the development of a coexisting, subclonal frameshift TOP1 mutation in the same metastatic lesions. We are not certain that the frameshift mutation exists in trans with TOP1E418K, though the resulting loss of the wild-type protein would seem potentially advantageous under the selective pressure of SG. In theory, these genetic events would be expected to induce cross-resistance to multiple emerging ADCs that use TOP1 inhibitors as payloads. However, future treatment strategies using different payloads such as maytansine (microtubule inhibitor), or possibly a more potent TOP1 inhibitor payload such as deruxtecan (>10 times potent than SN-38), could potentially overcome therapeutic resistance induced by such TOP1 mutations (20, 21).
Although germline loss-of-function mutations of TACSTD2/TROP2 are associated with a hereditary human corneal dystrophy, to our knowledge no prior somatic mutations of this gene have been described in the context of acquired resistance to SG or in any other context (22). We find that TACSTD2/TROP2T256R encodes a protein that can readily be expressed at equivalent total cellular levels to the wild-type protein but is dramatically attenuated in its cell-surface binding to the SG backbone antibody hRS7. Furthermore, TROP2T256R is mislocalized from the plasma membrane and instead appears to be primarily intracellular. Of note, we consistently observe altered migration of the mutant TROP2 protein on Western blot analysis, potentially suggesting a posttranslational modification that may contribute to altered localization and/or attenuated hRS7 binding. Although it remains to be seen how common such mutations will prove to be as a mechanism of acquired resistance to SG, our results provide strong proof-of-principle that genetic alteration of the ADC target TROP2 is a relevant resistance mechanism. In contrast, resistance to T-DM1 has been shown to occur through downregulation of HER2 expression in cell culture models, but the acquisition of missense mutations that confer resistance has not been well documented (23). Whether TROP2 alteration is a preferred mechanism of cellular resistance to SG could depend on its expression level and its functional contribution in tumor cells that may vary, both between TNBCs of different patients and between different tumor types for which SG is in clinical development (6, 24).
This study utilized genomic analyses of pretreatment and postprogression lesions to identify acquired alterations, a strategy that is increasingly being used to identify novel alterations mediating therapeutic resistance. Comparison of molecular alterations from pre- and posttreatment biopsies led to discovery of acquired ESR1 mutations emerging under selective pressure from aromatase inhibitors (25,26,27), acquired PTEN loss in response to PI3K inhibition (13), acquired RB1 mutations in response to CDK4/6 inhibition (28, 29), and acquired T790M EGFR mutations that circumvent inhibition from first- and second-generation EGFR inhibitors (30). Whether the emergence of such alterations represents newly acquired mutations versus expansion of a preexisting, low-frequency subclone is a long-standing question. Future improvement in sequencing strategies, including ultra-deep sequencing, will further illuminate this issue.
Currently, there are nine ADCs approved by the FDA in the field of oncology, and more than 100 being investigated in clinical trials. Our study highlights that molecular alterations in both antibody binding target and payload target may mediate resistance to ADCs, a finding that is likely to have implications for therapeutic sequencing of ADCs. For example, resistance mechanisms to SG due to drug payload target alterations would likely confer cross-resistance to other ADCs with similar (TOP1 inhibitor) payloads, such as trastuzumab deruxtecan and datopotomab deruxtecan (DS1062). In such a scenario, therapeutic sequencing with the same antibody but a different payload could provide therapeutic benefit. On the other hand, resistance mechanisms due to TROP2 alteration would likely confer cross-resistance to other TROP2 ADCs such as DS1062, but not to non–TROP2-directed ADCs regardless of payload. Another important consideration related to antibody target–associated therapeutic resistance is the bystander effect, which describes the ability of the ADC to kill adjacent non–target-expressing cells. Intratumoral heterogeneity in expression of the target would be predicted to affect ADCs with limited bystander effect more than those with bystander effect, as seen with T-DM1 versus trastuzumab deruxtecan (21). Finally, combination therapies that could increase ADC internalization via effects on the antibody target could potentially overcome resistance and provide synergistic effects, such as seen with T-DM1 and neratinib in the setting of HER2-mutant tumors (31).
The study has a few limitations. First, the molecular alterations were seen in a single patient, and the findings and implications for clinical management need to be evaluated in larger studies. Second, patient MGH-18 received two additional therapies after SG (vinorelbine and capecitabine). However, these are not TOP1 inhibitors, and it is therefore unlikely that the TOP1 mutations emerged due to selective pressure from these agents. Third, MGH-18 had a tumor with focal TACSTD2/TROP2 amplification, which may have increased the chance of developing resistance via acquisition of a mutation in TACSTD2/TROP2 itself. Whether such mutations would also be seen in tumors without TACSTD2/TROP2 amplification is unclear and requires further investigation.
In conclusion, our findings highlight the role of parallel genetic alterations in both antibody and payload targets in mediating resistance to ADCs such as SG. Future studies are needed to determine the prevalence of the key resistance mutations we have identified across large numbers of patients and to establish their detailed mechanisms in vivo. Additional work will also be required to identify how these mutations temporally emerge under selective pressure from ADCs, in order to enable development of rational, mechanism-based sequential strategies to improve outcomes of patients treated with these agents.
Methods
Patients and Specimen Collection
All biopsies and tumor specimens were collected in accordance with institutional review board (IRB)–approved protocols. Patients provided written informed consent, and all studies were conducted in accordance with the Declaration of Helsinki. Rapid autopsies were performed within the first 3 hours postmortem with informed consent of patient and/or designee under IRB-approved protocol 13-416. Imaging studies, including CT scans, were obtained as part of routine clinical care and/or clinical trial participation. Imaging response was determined by independent radiology review using RECIST version 1.1. Sex, gender, age, and weight were not used as selection or exclusion factors for this work.
RNA-seq and WES Analysis
For WES, the AllPrep DNA/RNA Mini Kit (Qiagen) was used for dual extraction of both genomic DNA and RNA. DNA was quantified in triplicate using a standardized PicoGreen dsDNA Quantitation Reagent (Invitrogen) assay. The quality control identification check was performed using fingerprint genotyping of 95 common single-nucleotide polymorphisms by Fluidigm Genotyping (Fluidigm). Library construction from double-stranded DNA was performed using the KAPA Library Prep Kit with palindromic forked adapters from Integrated DNA Technologies. Libraries were pooled before hybridization. Hybridization and capture were performed using the relevant components of Illumina's Rapid Capture Enrichment Kit, with a 37-Mb target. All library construction, hybridization, and capture steps were automated on the Agilent Bravo liquid handling system. After postcapture enrichment, library pools were denatured using 0.1 N NaOH on the Hamilton STARlet. Cluster amplification of DNA libraries was performed according to the manufacturer's protocol (Illumina) using HiSeq 4000 exclusion amplification chemistry and HiSeq 4000 flow cells. Flow cells were sequenced utilizing sequencing-by-synthesis chemistry for HiSeq 4000 flow cells. The flow cells were then analyzed using RTA version 2.7.3 or later. Each pool of whole-exome libraries was sequenced on paired 76-cycle runs with two eight-cycle index reads across the number of lanes needed to meet coverage for all libraries in the pool.
Phylogenetic Analysis of Multiple Samples from the Same Patient
WES data were analyzed on the FireCloud cloud-based analysis platform (https://portal.firecloud.org/). Somatic mutations for tumor/normal pairs were detected using the Cancer Genome Analysis WES Characterization Pipeline available on FireCloud. The WES Characterization Pipeline includes multiple steps, including MuTect (for detection of somatic single-nucleotide variants; ref. 32), Strelka (for detecting small insertions and deletions; ref. 33), deTiN (estimates potential tumor-in-normal contamination; ref. 34), ContEst (for detecting cross-patient contamination; ref. 35), AllelicCapSeg (for assessing allele specific copy-number alterations; ref. 36), and ABSOLUTE (for estimating tumor purity, ploidy, absolute allelic copy number, and cancer cell fractions; ref. 36). The data for this study consisted of multiple samples collected from each patient (pre- and posttreatment), as well as autopsy samples. Phylogenetic analysis, subclonal reconstruction, and tree building were done with the PhylogicNDT package (10). Purity of samples was inferred using the established ABSOLUTE method (36).
Cell Lines, Cell Culture, and Drug Sensitivity Studies
All cell lines were obtained from the MGH Center for Molecular Therapeutics cell bank and underwent high-density SNP typing to confirm their identity. All experiments shown were performed within less than 6 months' passage of all lines since acquisition. Cells were maintained at 37°C in 5% CO2. BT549 (ATCC) cells were grown in RPMI (Lonza) supplemented with 10% FBS (SAFC), 1% penicillin (Gibco), and streptomycin (Gibco). NIH3T3 (ATCC) cells were grown in DMEM (Lonza) supplemented with 10% FBS (SAFC), 1% penicillin (Gibco), and streptomycin (Gibco).
pReceiver-based wild-type TROP2 lentiviral vector and empty control vector were purchased from GeneCopoeia. T256R-mutant TROP2 was constructed by introducing the mutation using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent). The primers for site-directed mutagenesis is: 5′-CTGCAGGTGGAGCGCAGGCTCATCTATTACCTG-3′.
The lentiviral constructs were transfected into HEK293T cells with LV-MAX Lentiviral Packaging Mix (Gibco) by using the CalPhos Mammalian Transfection Kit (Clontech Laboratories) according to the manufacturer's instructions. The conditioned media containing lentiviral particles were collected 36 hours after transfection and filtered using 0.45-μm pore filter (Millipore). The filtered media were then used to infect target cells. Polybrene (Sigma) was added into filtered media at a final concentration of 10 μg/mL to increase the infection efficiency. The infected cells were selected with 1 μg/mL puromycin (Sigma) 72 hours after infection. Expression of wild-type and T256R-mutant TROP2 was confirmed via Western blotting.
For SG and SN-38 sensitivity studies, cells were seeded in 96-well plates (5,000 cells per well) and exposed to varying doses of SG or SN-38 for 4 hours before changing to fresh medium. Cell viability was assayed after 96 hours. To determine cell viability upon drug treatment, CellTiter-Glo Luminescent Cell Viability Assay (Promega) was used according to the manufacturer's recommendations.
Protein Extraction, Membrane Fractionation, and Western Blot Analysis
For total protein extraction, cells were lysed in RIPA buffer [10 mmol/L Tris–HCl pH 7.5, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 1% (v/v) NP40, proteinase inhibitor cocktail, phosphatase inhibitor cocktail] for 30 minutes at 4°C.
Cells cultured in 10-cm dish were collected with 500 μL fractionation buffer [250 mmol/L sucrose, 20 mmol/L HEPES (pH 7.4), 10 mmol/L KCl, 2 mmol/L MgCl2, 1 mmol/L EDTA, 1 mA EGTA, 1 mmol/L DTT, proteinase inhibitor cocktail] and passed through a 25-gauge needle 10 times. Cell suspension was centrifuged at 720 × g for 5 minutes, and the supernatant was then centrifuged at 10,000 × g for 5 minutes. The supernatant was then centrifuged at 100,000 × g for 1 hour. Pellets were washed by adding 500 μL of fractionation buffer and resuspended by pipetting and passing through a 25-G needle. The sample was centrifuged for 1 hour. The membrane pellets were resuspended with SDS sample buffer.
Protein samples were mixed with SDS sample buffer and boiled for 10 minutes before being subjected to SDS-PAGE. The protein samples on the SDS-PAGE gel were then transferred onto PVDF membrane (Millipore), which was blocked by 5% nonfat milk in PBST (PBS plus 0.02% Tween 20) at room temperature for 1 hour. Then, the PVDF membrane was incubated with primary antibodies diluted in 3% BSA in PBST at 4°C overnight and horseradish peroxidase–conjugated secondary antibodies (Sigma) diluted in 3% BSA in PBST at room temperature for 2 hours. The signal was detected by enhanced chemiluminescence solution (PerkinElmer).
Live-Cell Flow Cytometry
BT549 or NIH3T3 cells were plated, allowed to attach overnight, and then harvested by trypsinization the following day. Cell pellets were washed with 1 mL of ice-cold 1× PBS and maintained at 4°C for further processing. Cells were blocked for 30 minutes with 1% BSA + 5% normal goat serum (Sigma) in 1× PBS, incubated with 5 μg/mL hRs7 (Immunomedics/Gilead, Immunomedics, Inc.), and then incubated with a goat anti-human F(ab)2 immunoglobulin secondary (Jackson ImmunoResearch). Samples were examined using a FACSAria flow cytometer. Analysis was conducted with FlowJo using Cell Quest software (Becton Dickinson). Data were representative of three biological repeats with at least triple technical replicates.
Immunofluorescence and Confocal Microscopy
BT549 or NIH3T3 cells were lifted and plated at subconfluent densities into 96-well plates having #1.5 glass coverslips to facilitate confocal microscopy (Cellvis, P96-1.5H-N). Cells were washed with 1× PBS and then fixed at room temperature using 4% PFA (ChemCruz) for 15 minutes at a gentle shake. Thereafter, cells were washed thrice with 1× PBS and then incubated with 0.5% Triton X-100 (Sigma) for 5 minutes to permeabilize. Blocking then used 1% BSA with 5% normal goat serum and 0.1% Triton X-100, followed by one of two anti-TROP2 antibodies (as indicated) for 1 hour at room temperature. Cells were then washed with 1% BSA in 1× PBS thrice for 20 minutes and incubated with a fluorescent FITC secondary antibody (Jackson ImmunoResearch, 1:500) for 1 hour. After incubation with secondary, cells were again washed thrice for 20 minutes with 1% BSA, 5% normal goat serum, and 0.1% Triton X-100 in 1× PBS. Nuclei were counterstained with 0.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; Sigma) and maintained in 200 μL PBS for imaging. Images were acquired using a Nikon A1R confocal microscope with a 60× oil immersion objective, a physical magnification of 1.5×, and a lens magnification of 1.3×, the latter of which was to eliminate distal warping. Images were converted and compiled using ImageJ software.
Antibodies used are listed in Supplementary Table S4.
Statistical Analysis
Tests to determine significance are detailed in the relevant figure legends. Briefly, t tests or one-way ANOVAs were used to determine statistical significance of treatment conditions and two-way ANOVA to discriminate between the contributions of both dose and transformation.
Upload of data from WES and RNA-seq is available in dbGaP, accession number phs2555.v1.
Supplementary Material
Acknowledgments
We gratefully acknowledge Immunomedics/Gilead for the gift of sacituzumab govitecan and hRS7. This work was supported by DOD/CDMRP/BCRP grant BC200927 (to A. Bardia and L.W. Ellisen), by NIH/NCI K99CA259447-01, the Terri Brodeur Breast Cancer Foundation Fellowship, and the MGH ECOR Fund for Medical Discovery Fellowship (to S. Sun), by the Susan Eid Tumor Heterogeneity Initiative (to D. Juric), by the Tracey Davis Breast Cancer Research Fund (to L.W. Ellisen), and by the Broad/IBM Cancer Resistance Research Project (G. Getz and L. Parida). Confocal microscopy core facility is supported by NIH grant 1S10 RR031563-01.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
#J.T. Coates and S. Sun contributed equally to this article.
Cancer Discov 2021;11:2436–45
Authors' Disclosures
S. Sun reports grants from NCI, Terri Brodeur Breast Cancer Foundation, and Massachusetts General Hospital during the conduct of the study. I. Leshchiner reports personal fees from PACT Pharma, Inc and Enov1, LLC outside the submitted work. A. Kambadakone reports grants from Philips Healthcare, GE Healthcare, PanCAN, personal fees from Bayer, and other support from Siemens Healthcare outside the submitted work. S.J. Isakoff reports personal fees from Immunomedics, Mylan, Myriad, Puma, Novartis, Seattle Genetics, grants and personal fees from OncoPep and AbbVie, grants from Merck, AstraZeneca, and Genentech outside the submitted work. D. Juric reports grants and personal fees from Novartis, Genentech, Syros, grants from Pfizer, Takeda, personal fees from Relay, PIC Therapeutics, Mapkure, Vibliome, grants and personal fees from Eisai, grants from InventisBio, Arvinas, Ribon Therapeutics, and Infinity Pharmaceuticals outside the submitted work. G. Getz reports grants from IBM during the conduct of the study; and receives research funds from Pharmacyclics. G. Getz is a founder, consultant, and holds privately held equity in Scorpion Therapeutics. A. Bardia reports grants from Genentech, Novartis, Pfizer, Merck, Sanofi, Radius Health, Immunomedics, and Diiachi Pharma/AstraZeneca, and personal fees from Pfizer, Novartis, Genentech, Merck, Radius Health, Immunomedics, Taiho, Sanofi, Diiachi Pharma/AstraZeneca, Puma; Biothernostics Inc., Phillips, Eli Lilly, Foundation Medicine, during the conduct of the study; personal fees from Pfizer, Novartis, Genentech, Merck, Radius Health, Immunomedics, Taiho, Sanofi, Diiachi Pharma/AstraZeneca, Puma; Biothernostics Inc., Phillips, Eli Lilly, Foundation Medicine, and grants from Genentech, Novartis, Pfizer, Merck, Sanofi, Radius Health, Immunomedics, and Diiachi Pharma/AstraZeneca outside the submitted work. L.W. Ellisen reports grants from DOD/CDMRP and other support from Tracey Davis Memorial Fund during the conduct of the study; and sacituzumab (drug only) provided by the sponsor for in vitro experiments as acknowledged in the manuscript. No disclosures were reported by the other authors.
Authors' Contributions
J.T. Coates: Data curation, supervision, validation, investigation, visualization, methodology, writing–review and editing. S. Sun: Data curation, supervision, validation, investigation, methodology, writing–review and editing. I. Leshchiner: Formal analysis, supervision, investigation, writing–review and editing. N. Thimmiah: Investigation, writing–review and editing. E.E. Martin: Formal analysis, writing–review and editing. D. McLoughlin: Supervision, project administration, writing–review and editing. B.P. Danysh: Software, project administration, writing–review and editing. K. Slowik: Software and project administration. R.A. Jacobs: Formal analysis, project administration, writing–review and editing. K. Rhrissorrakrai: Funding acquisition. F. Utro: Funding acquisition and writing–review and editing. C. Levovitz: Funding acquisition. E. Denault: Project administration. C.S. Walmsley: Conceptualization, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing, and specimen acquisition. A. Kambadakone: Investigation. J.R. Stone: Conceptualization, software, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing, and contributed patients. S.J. Isakoff: Conceptualization, software, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing, contributed patients. L. Parida: Conceptualization, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. D. Juric: Conceptualization, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. G. Getz: Conceptualization, software, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. A. Bardia: Conceptualization, software, funding acquisition, writing–original draft, project administration, writing–review and editing. L.W. Ellisen: Conceptualization, software, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;13:674–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 2010;7:683–92. [DOI] [PubMed] [Google Scholar]
- 3. Anders CK, Zagar TM, Carey LA. The management of early-stage and metastatic triple-negative breast cancer: a review. Hematol Oncol Clin North Am 2013;27:737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gradishar WJ, Anderson BO, Abraham J, Aft R, Agnese D, Allison KH, et al. Breast cancer, version 3.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2020;18:452–78. [DOI] [PubMed] [Google Scholar]
- 5. Nicolo E, Zagami P, Curigliano G. Antibody-drug conjugates in breast cancer: the chemotherapy of the future? Curr Opin Oncol 2020;32:494–502. [DOI] [PubMed] [Google Scholar]
- 6. Nagayama A, Vidula N, Ellisen L, Bardia A. Novel antibody-drug conjugates for triple negative breast cancer. Ther Adv Med Oncol 2020;12:1758835920915980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bardia A, Mayer IA, Vahdat LT, Tolaney SM, Isakoff SJ, Diamond JR, et al. Sacituzumab govitecan-hziy in refractory metastatic triple-negative breast cancer. N Engl J Med 2019;380:741–51. [DOI] [PubMed] [Google Scholar]
- 8. Bardia A, Hurvitz SA, Tolaney SM, Loirat D, Punie K, Oliveira M, et al. Sacituzumab govitecan in metastatic triple-negative breast cancer. N Engl J Med 2021;384:1529–41. [DOI] [PubMed] [Google Scholar]
- 9. Kalinsky K, Diamond JR, Vahdat LT, Tolaney SM, Juric D, O'Shaughnessy J, et al. Sacituzumab govitecan in previously treated hormone receptor-positive/HER2-negative metastatic breast cancer: final results from a phase I/II, single-arm, basket trial. Ann Oncol 2020;31:1709–18. [DOI] [PubMed] [Google Scholar]
- 10. Leshchiner I, Livitz D, Gainor JF, Rosebrock D, Spiro O, Martinez A, et al. Comprehensive analysis of tumour initiation, spatial and temporal progression under multiple lines of treatment. bioRxiv 2018. [Google Scholar]
- 11. Chang JY, Liu JF, Juang SH, Liu TW, Chen LT. Novel mutation of topoisomerase I in rendering cells resistant to camptothecin. Cancer Res 2002;62:3716–21. [PubMed] [Google Scholar]
- 12. Fiorani P, Chillemi G, Losasso C, Castelli S, Desideri A. The different cleavage DNA sequence specificity explains the camptothecin resistance of the human topoisomerase I Glu418Lys mutant. Nucleic Acids Res 2006;34:5093–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature 2015;518:240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanaka N, Lin JJ, Li C, Ryan MB, Zhang J, Kiedrowski LA, et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov 2021;11:1913–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bon G, Pizzuti L, Laquintana V, Loria R, Porru M, Marchio C, et al. Loss of HER2 and decreased T-DM1 efficacy in HER2 positive advanced breast cancer treated with dual HER2 blockade: the SePHER study. J Exp Clin Cancer Res 2020;39:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia-Alonso S, Ocana A, Pandiella A. Resistance to antibody-drug conjugates. Cancer Res 2018;78:2159–65. [DOI] [PubMed] [Google Scholar]
- 17. Baselga J, Lewis Phillips GD, Verma S, Ro J, Huober J, Guardino AE, et al. Relationship between tumor biomarkers and efficacy in EMILIA, a phase III study of trastuzumab emtansine in HER2-positive metastatic breast cancer. Clin Cancer Res 2016;22:3755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bardia A, Tolaney SM, Punie K, Loirat D, Oliveira M, Kalinsky K, et al. Biomarker analyses in the phase III ASCENT study of sacituzumab govitecan versus chemotherapy in patients with metastatic triple-negative breast cancer. Ann Oncol 2021;32:1148–56. [DOI] [PubMed] [Google Scholar]
- 19. Urasaki Y, Laco GS, Pourquier P, Takebayashi Y, Kohlhagen G, Gioffre C, et al. Characterization of a novel topoisomerase I mutation from a camptothecin-resistant human prostate cancer cell line. Cancer Res 2001;61:1964–9. [PubMed] [Google Scholar]
- 20. Ogitani Y, Aida T, Hagihara K, Yamaguchi J, Ishii C, Harada N, et al. DS-8201a, a novel HER2-targeting ADC with a novel DNA topoisomerase I inhibitor, demonstrates a promising antitumor efficacy with differentiation from T-DM1. Clin Cancer Res 2016;22:5097–108. [DOI] [PubMed] [Google Scholar]
- 21. Yver A, Agatsuma T, Soria JC. The art of innovation: clinical development of trastuzumab deruxtecan and redefining how antibody-drug conjugates target HER2-positive cancers. Ann Oncol 2020;31:430–4. [DOI] [PubMed] [Google Scholar]
- 22. McDougall AR, Tolcos M, Hooper SB, Cole TJ, Wallace MJ. Trop2: from development to disease. Dev Dyn 2015;244:99–109. [DOI] [PubMed] [Google Scholar]
- 23. Hunter FW, Barker HR, Lipert B, Rothe F, Gebhart G, Piccart-Gebhart MJ, et al. Mechanisms of resistance to trastuzumab emtansine (T-DM1) in HER2-positive breast cancer. Br J Cancer 2020;122:603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weiss J, Glode A, Messersmith WA, Diamond J. Sacituzumab govitecan: breakthrough targeted therapy for triple-negative breast cancer. Expert Rev Anticancer Ther 2019;19:673–9. [DOI] [PubMed] [Google Scholar]
- 25. Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 2013;45:1446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 2013;45:1439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Cancer therapy: ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014;345:216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Condorelli R, Spring L, O'Shaughnessy J, Lacroix L, Bailleux C, Scott V, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol 2018;29:640–5. [DOI] [PubMed] [Google Scholar]
- 29. O'Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov 2018;8:1390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786–92. [DOI] [PubMed] [Google Scholar]
- 31. Li BT, Michelini F, Misale S, Cocco E, Baldino L, Cai Y, et al. HER2-mediated internalization of cytotoxic agents in ERBB2 amplified or mutant lung cancers. Cancer Discov 2020;10:674–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Källberg M, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591–4. [DOI] [PubMed] [Google Scholar]
- 34. Taylor-Weiner A, Stewart C, Giordano T, Miller M, Rosenberg M, Macbeth A, et al. DeTiN: overcoming tumor-in-normal contamination. Nat Methods 2018;15:531–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 2011;27:2601–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.