Abstract
The opportunistic fungal pathogen Candida albicans undergoes an unusual parasexual cycle wherein diploid cells mate to form tetraploid cells that can generate genetically diverse progeny via a nonmeiotic program of chromosome loss. The genetic diversity afforded by parasex impacts clinically relevant features including drug resistance and virulence, and yet the factors influencing genome instability in C. albicans are not well defined. To understand how environmental cues impact genome instability, we monitored ploidy change following tetraploid cell growth in a panel of different carbon sources. We found that growth in one carbon source, D-tagatose, led to high levels of genomic instability and chromosome loss in tetraploid cells. This sugar is a stereoisomer of L-sorbose which was previously shown to promote karyotypic changes in C. albicans. However, while expression of the SOU1 gene enabled utilization of L-sorbose, overexpression of this gene did not promote growth in D-tagatose, indicating differences in assimilation of the two sugars. In addition, genome sequencing of multiple progenies recovered from D-tagatose cultures revealed increased relative copy numbers of chromosome 4, suggestive of chromosome-level regulation of D-tagatose metabolism. Together, these studies identify a novel environmental cue that induces genome instability in C. albicans, and further implicate chromosomal changes in supporting metabolic adaptation in this species.
Keywords: parasexual cycle, ploidy change, tetraploid, aneuploidy, stress adaptation
Introduction
Candida albicans is a human commensal fungus that colonizes approximately 70% of healthy adults (Noble et al. 2017). In immunocompromised hosts, the commensal form can seed debilitating mucosal or life-threatening systemic infections, and treatment of these is complicated by an increasing prevalence of drug resistance in clinical isolates (Ksiezopolska and Gabaldon 2018). While C. albicans primarily exists as a diploid species, it has also been observed in haploid, tetraploid, and numerous aneuploid forms that can show altered clinical features including differences in virulence and drug resistance (Olaiya and Sogin 1979; Ibrahim et al. 2005; Selmecki et al.2006, 2008; Hickman et al. 2013; Ford et al. 2015; Hirakawa et al. 2017). Although C. albicans exists in diverse ploidy states, the cues that drive ploidy changes in fungal species are not fully understood.
The most studied mechanism for eliciting ploidy change in C. albicans involves its unusual program of parasexual reproduction. Entry into the parasexual cycle is governed by a phenotypic switch wherein diploid cells undergo a transition from the sterile “white” state to the mating-competent “opaque” state (Slutsky et al. 1987; Miller and Johnson 2002). Diploid opaque cells mate to form tetraploid cells that, rather than a conventional meiosis, can return to a diploid or near-diploid state via reductional divisions in a process known as concerted chromosome loss (CCL) (Bennett and Johnson 2003; Forche et al. 2008; Hickman et al. 2015). CCL promotes genetic diversity as parasexual progeny display shuffled combinations of chromosome homologs, high levels of recombination, and frequent aneuploidy (Forche et al. 2008; Berman and Hadany 2012; Hirakawa et al. 2017; Anderson et al. 2019). The genotypic diversity ultimately afforded by parasex potentiates phenotypic diversity and can alter clinically relevant traits (Hirakawa et al. 2017). Importantly, tetraploid C. albicans cells can also arise independently of the parasexual program, as diploid cells can form a transient tetraploid state due to mitotic collapse in response to treatment with the antifungal drug fluconazole (Harrison et al. 2014).
The mechanisms impacting ploidy change in C. albicans continue to be investigated. Extended passaging in replete or nutrient-depleted media (e.g., low nitrogen or phosphorus) can enable tetraploid cells to return to a diploid state over time (Hickman et al. 2015; Zhang et al. 2015; Gerstein et al. 2017). Moreover, ploidy loss in tetraploid C. albicans cells is facilitated by growth on Saccharomyces cerevisiae pre-sporulation (PRE-SPO) medium, as well as by growth on medium containing the sugar L-sorbose (Bennett and Johnson 2003; Thomson et al. 2019). Genome instability on PRE-SPO medium was recently linked to the high levels of oxidative stress and DNA damage that occur during the growth of tetraploid cells on this medium (Thomson et al. 2019). Growth on L-sorbose is generally toxic to both diploid and tetraploid forms of C. albicans, yet cells can adapt to utilize this sugar by undergoing large-scale genomic changes (Janbon et al. 1998, 1999; Bennett and Johnson 2003; Yang et al. 2013; Hickman et al. 2015). Diploid cells typically lose a copy of chromosome (Chr) 5 during adaptation to L-sorbose, as multiple regions on this chromosome negatively regulate the SOrbose Utilization gene, SOU1, on Chr 4 (Janbon et al. 1998, 1999; Kabir et al. 2005). Increased expression of SOU1 promotes the conversion of L-sorbose to D-sorbitol, which can then be converted into fructose-6-phosphate for entry into glycolysis (Greenberg et al. 2005). It has not been determined how tetraploid cells adapt to assimilate L-sorbose, although progeny recovered from this medium are often diploid with extra copies of Chr 4 (Forche et al. 2008), suggesting that increases in the relative copy number of SOU1 may also enable adaptation of tetraploid cells to this sugar.
In this study, we further examined the environmental conditions that promote genomic instability in C. albicans tetraploid cells. We tested tetraploid cells when grown on a panel of 190 different carbon sources and identified one carbon source, D-tagatose, which frequently led to a reduction in ploidy. D-tagatose is a stereoisomer of L-sorbose, and yet the mechanism for D-tagatose utilization appears distinct from that of L-sorbose as overexpression of SOU1 promoted L-sorbose growth but did not enhance growth on D-tagatose. Interestingly, genomic analysis of cells recovered from D-tagatose cultures (and selected for loss of a genetic marker) revealed that progeny had a reduced ploidy and contained increased relative copy numbers of Chr 4. This suggests that elevated expression of genes on Chr 4 may promote D-tagatose utilization. Overall, our findings identify a novel nutritional cue that leads to high levels of genome instability in tetraploid C. albicans cells and implicate karyotypic changes in adaptation to this nutrient.
Materials and methods
Strains and media
Yeast extract peptone dextrose (YPD) and synthetic complete dextrose (SCD) media were prepared as described (Guthrie and Fink 1991). Liquid minimal media + D-tagatose, L-sorbose, or D-glucose contained 0.7% yeast nitrogen base (without any carbon source or amino acids) supplemented with 2% D-tagatose (NuNaturals), 2% L-sorbose (ACROS Organics), or 2% D-glucose (Sigma-Aldrich), respectively. 2-deoxygalactose (2-DOG) plates contained SC medium with 2% glycerol, 2 mg/ml 2-DOG, and 2% agar.
To generate strains overexpressing SOU1, we used plasmid pLC605 which contains a Tet-OFF promoter sequence for gene overexpression (Veri et al. 2018). A Tet-OFF overexpression (O/E) cassette for SOU1 was created by PCR amplification of pLC605 using oligonucleotides 4304 and 4305 (see Supplementary Table S2), which was then transformed into SC5314 and RBY18 to generate strains CAY8734 (diploid SOU1 O/E) and CAY8740 (tetraploid SOU1 O/E).
Phenotype microarray plate assays
Tetraploid strain RBY18 was grown in YPD medium and then resuspended in water to an OD600 of 0.2. The cell suspension was diluted 1:48 into inoculating fluid (IFY-0) and 100 µl of the cell suspension was aliquoted into each well of Biolog PM1 and PM2 plates (Biolog Inc., Hayward, CA, USA). The plates were grown at 37°C for 72 h on a shaking platform at 200 RPM. Following incubation, 10 µl of the cultures from each well were spotted undiluted onto SCD and 2-DOG media and incubated at 22°C for 7 days, at which point plates were imaged and analyzed for growth. Experiments were performed with biological duplicates.
Quantification of chromosome loss
Tetraploid strain RBY18 was inoculated into liquid minimal media + 2% D-glucose or 2% D-tagatose and incubated for 7 days at 30 and 37°C. Cells were then collected from each culture and successive 1:100 dilutions were made in PBS. 100 μl of the undiluted and 1:102 dilution cell suspensions were plated onto 2‐DOG medium, and 100 μl of the 1:102 dilution and 1:104 dilution cell suspensions were plated onto YPD medium. These plates were incubated at 30°C for 48 h after which the total number of colonies were counted on each plate. The percent of 2-DOGR progeny was calculated as the (number of colonies on 2-DOG plate × the dilution factor) × 100/(the number of colonies on the YPD plate × the dilution factor). Experiments were performed with biological triplicates.
DNA staining for ploidy analysis
Ploidy analysis was performed on random colonies taken from three independent experiments following growth in either glucose- or tagatose-containing media (at 30 or 37°C) and subsequent selection on YPD or 2-DOG plates. Six-seven independent colonies as well pooled samples (cells from 10 different colonies) were used for each test condition. Tag1-12 isolates represent single colonies originating from three independent experiments (4 colonies selected from each experimental replicate) and were also analyzed for ploidy. Diploid and tetraploid controls were included in each run for comparison of ploidy states.
Colonies were inoculated into liquid YPD and grown overnight at 30°C, at which point cells were collected, fixed with 70% ethanol, and stored overnight at 4°C. Cells were washed twice with 50 mM Tris–Cl, pH 8.0, 5 mM EDTA, resuspended in 2 mg/ml RNase A (Sigma‐Aldrich) in 50 mM Tris–Cl, pH 8.0, 5 mM EDTA, and incubated for 2 h at 37°C. RNase‐treated cells were pelleted and resuspended in 5 mg/ml pepsin (Sigma‐Aldrich) in 55 mM HCl and incubated for 1 h at 37°C. Cells were washed twice with 50 mM Tris–Cl, pH 7.5, 5 mM EDTA, and resuspended in 1 μM SYTOX Green Nucleic Acid Stain (Thermo Fisher Scientific) in 50 mM Tris–Cl, pH 7.5, 5 mM EDTA and stored overnight at 4°C. For ploidy analysis, 104 cells were run on a FACS Celesta (BD Biosciences) and data analyzed using FlowJo 10.7.1 (Ashland, OR, USA). Histograms of SYTOX fluorescence levels were evaluated in FlowJo and the subpopulation of cells in G1 of the cell cycle was gated for analysis of each sample. Mean SYTOX fluorescence intensity levels were calculated for cells under the G1 peak and used to assess ploidy levels. To control for variations between experiments, SYTOX values were normalized to the diploid control.
Growth rate assays
SC5314 [wild type (WT) diploid], RBY18 (WT tetraploid), CAY8734 (diploid SOU1 O/E), CAY8740 (tetraploid SOU1 O/E) and Tag1-12 isolates were grown in liquid YPD medium overnight at 30°C. The next day, 1 μl of each culture was inoculated into 250 μl minimal media + 2% L-sorbose or 2% D-tagatose in a 96-well plate. The plate was incubated at 37°C with continuous shaking using a Biotek Epoch 2 plate reader. OD600 readings were recorded every 24 h for 6–8 days and growth curves were plotted using GraphPad Prism 8. Experiments were performed with 2–4 biological replicates.
Cell wall stress assays
For cell wall sensitivity assays, Congo red (Fisher Scientific) was added to SCD plates at 25, 50, or 100 µg/ml. Strains were grown overnight in liquid YPD at 30°C and further diluted in YPD and grown for an additional 4 h at 30°C to reach exponential phase. Exponentially growing cultures were diluted to 103 cells/ml. Diluted cultures were spotted on SCD or SCD with Congo red and were incubated at 30°C. After 48 h of growth, plates were imaged using a digital camera. Assays were performed with biological duplicates.
Whole-genome sequencing, heterozygosity levels, and ploidy assays
To extract genomic DNA C. albicans isolates were grown overnight in YPD at 30°C and DNA isolated from ∼109 cells using a Qiagen Genomic Buffer Set and a Qiagen Genomic-tip 100/G according to manufacturer’s instructions. Libraries were prepared using the Nextera XT DNA Library preparation kit protocol (Illumina) with an input of 2 ng/μl in 10 μl. Each isolate was sequenced using Illumina HiSeq 2000 generating 101 bp paired reads. The nuclear genome sequences and General Feature Files (GFF) for C. albicans SC5314 reference genome (version A22) were downloaded from the Candida Genome Database (http://www.candidagenome.org/). Reads were aligned to the SC5314 reference genome assembly 22 (haplotype A chromosomes) using Burrows-Wheeler Aligner (BWA) v0.7.4-r385 mem (Li and Durbin 2009), and converted to sorted BAM format using Samtools v0.1.9 (r783) (Li et al. 2009). Reads were trimmed using trimmomatic 0.36 (with default parameters except for SLIDINGWINDOW : 10:25 and MINLEN : 75) (Bolger et al. 2014). Briefly, the Picard tools (http://picard.sourceforge.net/) AddOrReplaceReadGroups, MarkDuplicates, CreateSequenceDictionary, and ReorderSam were used to preprocess the alignments. This resulted in average coverage levels of 138X per sample. To examine ploidy variation across the genome, the Illumina read alignment depth was calculated for 1 kbp windows across the genome using BEDTools 2.28 (Quinlan and Hall 2010) and SAMtools 1.3.1 (Li et al. 2009). The read depth was calculated as the number of bases aligned per window divided by the length of the window (1 kbp) and normalized to the average depth per strain and to GC content using pybedtools 0.8.1 (Quinlan and Hall 2010; Dale et al. 2011). The normalized alignment depth for each 1 kbp was averaged across 10 kbp windows and plotted using GraphPad Prism 8. Relative changes in ploidy were identified, including whole chromosome and segmental aneuploidies larger than 0.1 Mbp.
To identify changes in heterozygosity, the number of heterozygous positions was calculated per 1 kbp window using vcftools (Danecek et al. 2011) and the genotypes assigned by GATK4 (Brouard et al. 2019). Heterozygosity levels were then averaged across 10 kbp windows and plotted using Prism 8. Large loss of heterozygosity (LOH) tracts were identified and confirmed by visual inspection in IGV (Thorvaldsdottir et al. 2013).
GATK4 outputs were used to calculate allele frequencies for each heterozygous position across all chromosomes. Thus, positions that were heterozygous in either reference strain (SC5314 version A22) or strains of interest were examined and the percent of reads mapping to each chromosome homolog at these positions was determined. Allele frequencies for each chromosome homolog were plotted using GraphPad Prism 8 and heterozygous tracks were manually inspected using IGV.
Statistical analysis
Comparisons between different experimental conditions were performed in GraphPad Prism 8 using two-way ANOVAs and Student’s T-tests.
Data availability
Strains and plasmids are available upon request. Whole-genome sequencing data for D-tagatose-cultured isolates (Tag1–12 and RBY18) is available at NCBI SRA as BioProject PRJNA686496. The whole-genome assembly for SC5314 has been previously published under NCBI BioProject PRJNA193498 (Hirakawa et al. 2015). Supplementary material includes Supplementary Figures S1–S4 and Supplementary Tables S1 and S2, and have been uploaded to figshare: https://doi.org/10.25387/g3.13505709.
Results
Analysis of C. albicans tetraploid cells grown in a diverse set of carbon sources
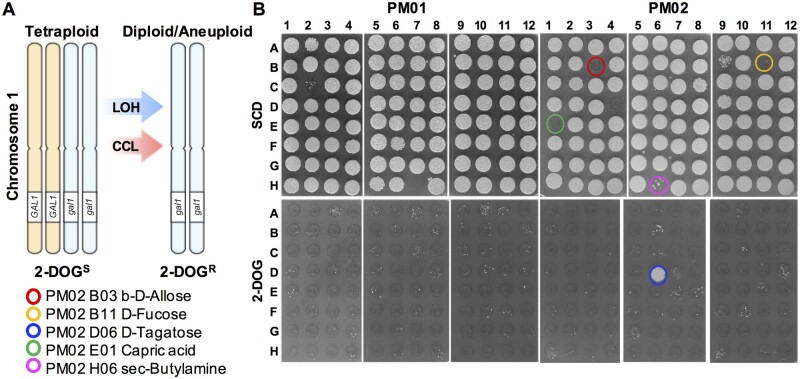
To identify environmental cues that promote genome instability in C. albicans, we examined tetraploid cell behavior when grown in different carbon sources using Biolog Phenotype Microarray (PM) plates. We used PM 96-well plates that contained 190 alternative carbon sources including a variety of carbohydrates, carboxylic acids, and amino acids (Supplementary Table S1). To monitor for ploidy change, we utilized the tetraploid strain RBY18 which is derived from the standard laboratory isolate SC5314 and is heterozygous for GAL1 on Chr 1 (GAL1/GAL1/gal1/gal1). Cells lacking GAL1 function are capable of growing on a medium containing 2-deoxygalactose (2-DOG), and this can be used to select for RBY18-derived progeny that have lost both functional copies of GAL1. 2-DOG-resistant colonies can arise in RBY18 due to LOH events at GAL1 or by loss of both of the GAL1-containing Chr 1 homologs (Figure 1A) (Gorman et al. 1992; Bennett and Johnson 2003).
Figure 1.
C. albicans tetraploid cell response to growth in a diverse set of carbon sources. (A) Assay to monitor for GAL1 loss in tetraploid strain RBY18. Cells containing a wild-type copy of GAL1 do not grow on 2-DOG medium (2-DOGS). Loss of the two functional copies of GAL1, e.g., by loss of the appropriate chromosome homologs, causes cells to become 2-DOG resistant (2-DOGR). (B) Tetraploid cells from strain RBY18 were grown in PM carbon source plates for 3 days at 37°C, at which point cells were transferred onto SCD medium to monitor for viability and onto 2-DOG medium to select for cells that have lost GAL1. Highlighted wells indicate carbon sources that produced few or none CFUs on SCD, as well as D-tagatose, the only condition to result in high numbers of 2-DOG-resistant colonies (PM02 D6; see also Supplementary Figure S1).
Strain RBY18 was grown on PM carbon source plates (37°C for 72 h) and cells replica plated onto SCD and 2-DOG media to monitor for cell viability and loss of GAL1 function, respectively. Several carbon sources did not produce any colony forming units (CFUs), with no viable cells recovered from wells containing β-D-allose (PM2 B3), D-fucose (PM2 B11), capric acid (PM2 E1), or sec-butylamine (PM2 H6; Figure 1B and Supplementary Figure S1). These compounds were not further investigated in this work but may warrant examination for potential roles in inhibiting C. albicans growth. In addition, only rare colonies were recovered from wells containing the sugar L-sorbose, consistent with observations that isolates often require a relatively long incubation period for adapted clones to arise in this nutrient (Janbon et al. 1999).
Notably, only growth in D-tagatose-containing medium (PM2 D6) resulted in a high number of 2-DOGR colonies in the PM screen (Figure 1B). This is indicative of tetraploid RBY18 cells having lost GAL1 function (via recombination or chromosome loss, Figure 1A). To further examine the capacity for D-tagatose to promote GAL1 loss in tetraploid cells, strain RBY18 was grown in minimal media containing 2% D-tagatose or 2% D-glucose at both 30 and 37°C, at which point cells were analyzed by plating on YPD (nonselective) and 2-DOG (selective) media. Irrespective of temperature, growth in D-tagatose consistently resulted in frequencies of 2-DOGR colonies that were more than two orders of magnitude higher than those in D-glucose (Figure 2A).
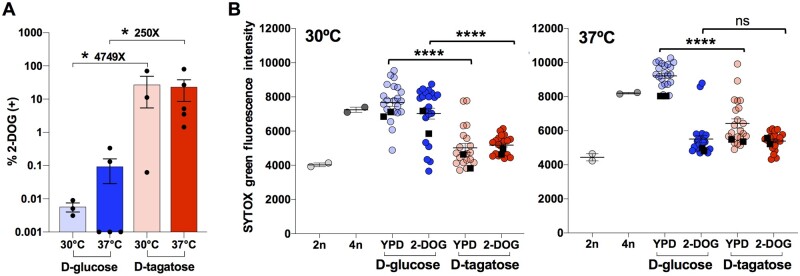
Figure 2.
Growth in D-tagatose promotes GAL1 marker loss and ploidy reduction in tetraploid cells. (A) Analysis of GAL1 loss (2-DOG-resistant colonies) in tetraploid strain RBY18 after growth in minimal media + 2% D-glucose or 2% D-tagatose for 7 days at 30 and 37°C. Asterisks denote a significant difference in the percentage of 2-DOGR colonies between the indicated conditions (P < 0.05, Student’s T-test, n = 3–5, error bars represent ± SEM). (B) RBY18 was grown in minimal media + 2% D-glucose or 2% D-tagatose at 30 or 37°C and colonies recovered on YPD and 2-DOG media. DNA content of progeny was analyzed via flow cytometry in comparison with diploid (2n) and tetraploid (4n) controls. Lines indicate mean values ± SEM. Data points reflect SYTOX green mean fluorescence intensity levels for 19–21 individual colonies and two sets of pooled isolates from two independent experiments for each condition. Pooled isolates were obtained by combining cells from 10 different colonies (black squares). Asterisks denote significant differences between D-glucose and D-tagatose conditions (two-way ANOVAs, ****P < 0.0001, df = 22).
The ploidy of cells following growth in D-glucose or D-tagatose media and recovered on YPD or 2-DOG media was directly evaluated by DNA staining and flow cytometry. In total, ∼20 individual colonies and 2 pooled samples (each pool a combination of 10 individual colonies) were assessed for DNA content for each condition. The most striking observation from this data was that the majority of colonies derived from D-tagatose cultures were closer to diploid than to tetraploid in DNA content, indicative of having undergone a substantial reduction in ploidy, and this occurred at both 30 and 37°C (Figure 2B). Similar results were obtained whether D-tagatose-cultured cells had been recovered on YPD or 2-DOG media, indicating that ploidy reduction occurred independent of selection for gal1- cells. In contrast, tetraploid cells grown in D-glucose and recovered on YPD showed ploidy levels close to that of the tetraploid (4n) control (Figure 2B). Selection of D-glucose-cultured cells on 2-DOG medium gave rise to a range of ploidies; experiments performed at 37°C resulted in ploidies close to diploid, while those performed at 30°C produced ploidies that were closer to tetraploid (Figure 2B).
Taken together, these experiments establish that tetraploid cells cultured in D-tagatose consistently undergo large-scale reductions in ploidy, most frequently to levels between that of diploid and triploid, and that this occurs at both 30 and 37°C. In contrast, culture in D-glucose rarely results in reduced ploidy, although karyotypic changes still occur and can be enriched by selection of cells that have lost a genetic marker.
Examination of the role of SOU1 in adaptation to D-tagatose
D-tagatose is a stereoisomer of L-sorbose (Figure 3A), a sugar previously shown to promote ploidy reduction in tetraploid C. albicans cells, as well as chromosomal-level changes in diploid cells (Bennett and Johnson 2003; Rustchenko 2007). L-sorbose utilization requires expression of the SOU1 gene on Chr 4, which encodes for a sorbose reductase that converts L-sorbose to D-sorbitol (Greenberg et al. 2005). Diploid cells can adapt to L-sorbose by increasing the relative copy number of the SOU1 gene (e.g., by becoming trisomic for Chr 4 which encodes SOU1) and/or by losing a copy of Chr 5, as the latter contains multiple elements that negatively regulate SOU1 expression (Janbon et al. 1998, 1999; Kabir et al. 2005; Rustchenko 2007).
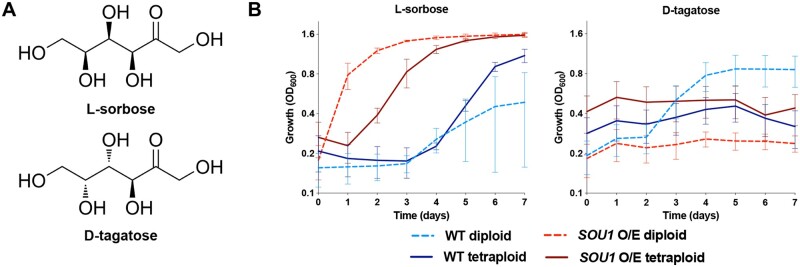
Figure 3.
Utilization of L-sorbose, but not D-tagatose, is promoted by overexpression of the SOU1 gene. (A) Skeletal formulas of L-sorbose and D-tagatose. (B) Growth (measured by OD600) of wild type (WT) and SOU1 overexpression (O/E) in diploid and tetraploid strains in minimal media supplemented with 2% L-sorbose or 2% D-tagatose at 37°C (n = 4).
To examine whether or not D-tagatose utilization involves SOU1, diploid and tetraploid C. albicans strains were constructed that overexpress SOU1 using a regulatable Tet-Off system (Veri et al. 2018). As anticipated, SOU1-overexpressing strains more readily utilized L-sorbose than wild-type diploid and tetraploid control strains (Figure 3B). Thus, diploid and tetraploid cells overexpressing SOU1 showed substantial growth in L-sorbose by 1 and 2 days, respectively, whereas control strains did not show detectable growth until 3–4 days (Figure 3B). In contrast, however, SOU1 overexpression did not provide any growth advantage in D-tagatose cultures (Figure 3B). D-tagatose, therefore, appears to have distinct metabolic requirements from those used to assimilate L-sorbose.
Genomic analysis of C. albicans progeny from D-tagatose cultures
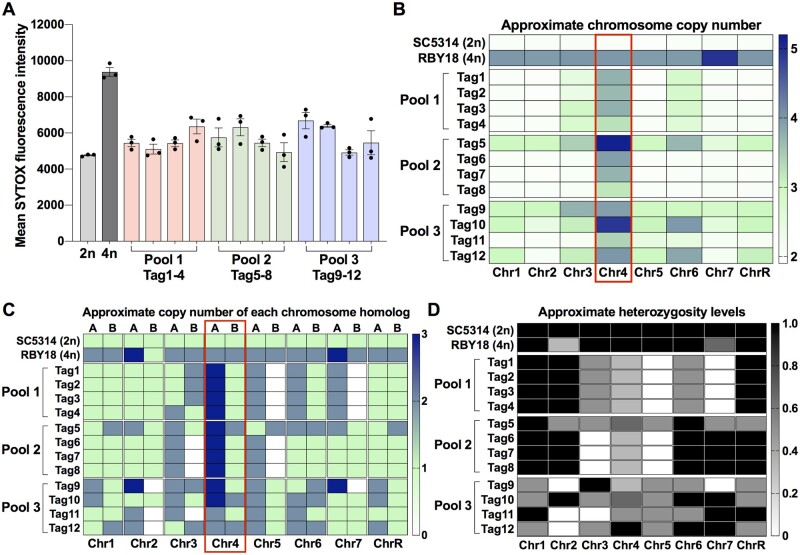
To examine the genotypes formed when tetraploid cells are cultured in D-tagatose, ploidy analysis and genome sequencing were performed on a subset of the recovered progeny. Tetraploid strain RBY18 was inoculated into three separate D-tagatose cultures at 37°C and progeny cells recovered on 2-DOG plates. Twelve isolates (Tag1-12) from the three different cultures (four isolates per culture) were examined for DNA content using flow cytometry. We found that all 12 isolates displayed near diploid ploidy levels or were between diploid and triploid (Figure 4A), indicative of having undergone ploidy reduction.
Figure 4.
Karyotypic analysis of progeny following growth of tetraploid cells in D-tagatose and selection for 2-DOGR colonies. (A) Flow cytometry analysis of DNA content for diploid strain SC5314, tetraploid strain RBY18 and twelve 2-DOGR isolates derived from RBY18 cells that had been grown in D-tagatose-containing medium (Tag1–12). Tag1–12 isolates represent single colonies obtained from three separate pools and are grouped according to the pool of origin. Lines indicate mean SYTOX green fluorescence intensity ± SEM. (B, C) Heatmaps indicate estimated copy numbers for each chromosome (B) and each chromosome homolog (C) across all eight chromosomes. (D) Heatmap shows heterozygosity levels across chromosomes, with 0 indicating whole chromosome LOH and 1 indicating no LOH.
Whole-genome sequencing (∼138X coverage) was performed on Tag1–12 isolates along with the parental tetraploid isolate RBY18. Relative read depth was compared across all eight chromosomes and revealed that most tagatose-derived isolates were aneuploid having a nonequal number of chromosomes (Figure 4B). Eleven of the 12 isolates were confirmed to be lacking an intact GAL1 allele, as expected for cells that had undergone selection on 2-DOG medium. Of these isolates, eight isolates were disomic for Chr 1 (Tag1–4, Tag6–8, and Tag11) and had lost the two chromosome homologs carrying the intact GAL1 allele. Three isolates (Tag9, Tag10, and Tag12) were trisomic for Chr 1 and were also gal1-, which suggests they lost both GAL1-containing homologs as well as underwent LOH at GAL1. Surprisingly, one isolate, Tag5, retained three copies of Chr 1, two of which carried the gal1- allele and one of which carried the GAL1 allele (Figure 4, B and C). It is not clear if expression of GAL1 is defective in this strain and that this allowed growth on 2-DOG medium.
Notably, each of the 12 sequenced isolates showed an increase in Chr 4 copy number relative to the other chromosomes (Figure 4, B and C), suggesting that this may provide an adaptive advantage to growth in D-tagatose. Estimates indicate that Chr 4 was present at trisomic levels in Tag 11, at tetrasomic levels for Tag1–4, Tag6–9, and Tag12, and at pentasomic levels in Tag5 and Tag10 (Figure 4, B and C and Supplementary Figure S2). In addition to increased relative copies of Chr 4, we note that only certain homologs were retained for Chr 2, 3, 5, and 7 in some isolates. This resulted in LOH across the entire chromosome in these examples, and the same patterns of LOH were often present in isolates from the same experimental pool (Figure 4, C and D and Supplementary Figure S2). However, given that different patterns were observed between isolates from independent experiments, we propose that these LOH events are not closely associated with fitness in D-tagatose.
The karyotypes described above were supported by examining the allele frequencies across all heterozygous positions for each chromosome. The parental strain used for these experiments, SC5314, is diploid and heterozygous for the eight disomic chromosomes so median allele frequencies are ∼50%, as chromosome homologs are present in equal proportions (Supplementary Figure S3). Exceptions were noted for the right arms of Chr 3 and Chr R which had undergone LOH and were therefore homozygous even in the parental SC5314 lineage. The 12 isolates recovered from D-tagatose had median allele frequencies of ∼50% for most chromosomes (Supplementary Figure S3), consistent with these chromosomes retaining equal copies of each homolog (Figure 4C). In contrast, strains that had undergone differential retention of the two chromosome homologs showed skewed median allele frequencies. For example, strains carrying two copies of homolog A and one copy of homolog B (AAB configuration) had median allele frequencies close to 66 and 33%, respectively (Supplementary Figure S3A). We note there was a significant bias for retention of the A homolog over the B homolog for both Chr 4 [P < 0.001, two-way ANOVA, F(1,11) = 59.22, df = 11] and Chr 5 [P < 0.001, two-way ANOVA, F(1,11) = 32.27, df = 11] in the 12 progeny (Figure 4C and Supplementary Figure S3B).
Taken together, these results indicate that the most striking commonalities between the 2-DOGR isolates obtained from D-tagatose cultures are that they carry extra copies of Chr 4 relative to the rest of the genome and show a biased retention of homolog A over homolog B for both Chr 4 and 5.
Phenotypic analysis of C. albicans progeny from D-tagatose cultures
To determine whether isolates recovered from D-tagatose cultures show improved growth in this sugar relative to the tetraploid parent, we performed growth curves of Tag1–12 isolates and RBY18 in minimal media supplemented with D-glucose or D-tagatose. Growth in D-glucose did not reveal any differences between the parental RBY18 strain and D-tagatose-derived isolates, with all isolates reaching maximum biomass within the first day of growth (Supplementary Figure S4A). However, Tag1–12 showed improved growth in D-tagatose relative to RBY18 (Supplementary Figure S4A), as these isolates displayed shorter lag times and reached maximum biomass approximately 2 days earlier than the parental strain. These results indicate that the 12 isolates are better adapted for growth in D-tagatose compared to the parental strain.
Growth on L-sorbose represses the synthesis of cell wall enzymes in Neurospora crassa (Mishra and Tatum 1972) and L-sorbose-adapted C. albicans cells (that are Chr 5 monosomic) exhibit reduced levels of 1,3-β-glucan in the cell wall (Yang et al. 2013). It therefore seemed possible that D-tagatose-adapted isolates might also show altered cell wall properties, particularly in those with reduced copy numbers of Chr 5. To test this, we assessed the susceptibility of D-tagatose-adapted isolates to Congo red which targets cell wall β-glucan (Kopecka and Gabriel 1992). The Tag1-12 isolates, along with diploid and tetraploid controls, were spotted in 10-fold serial dilutions on SCD plates with or without Congo red. The tetraploid isolate showed a slight increase in Congo red sensitivity compared to the diploid control. Strikingly, all D-tagatose isolates except for Tag5 showed increased sensitivity to this cell wall agent (Supplementary Figure S4B). This analysis reveals that adaptation to growth on D-tagatose is associated with increased sensitivity to β-glucan-targeting agents, potentially indicative of changes to the cell wall.
Discussion
Several studies have examined the environmental conditions and genetic factors that influence ploidy change in fungal species (for recent reviews see Wertheimer et al. 2016; Todd et al. 2017; Gerstein and Sharp 2021). In Cryptococcus neoformans for example, polyploid Titan cells have been identified that show altered interactions with the mammalian host (Okagaki et al. 2010; Zaragoza et al. 2010; Okagaki and Nielsen 2012), and polyploid cells can subsequently give rise to haploid or aneuploid progeny in a process that is not well understood but is dependent on meiotic genes (Gerstein et al. 2015; Zhao et al. 2020). Studies in S. cerevisiae have also revealed that there are mitotic mechanisms by which triploid or tetraploid cells can shift toward the diploid state during passaging (Gerstein et al. 2006, 2008; Selmecki et al. 2015). Overall, it is evident that polyploid states frequently arise in nature, both in fungi and in higher eukaryotes, that these states exhibit elevated genomic instability, and that this instability can be further accentuated by environmental cues (Bennett et al. 2014; Storchova 2014; Todd et al. 2017).
Here, we focus on ploidy change in C. albicans, a diploid yeast that undergoes a parasexual cycle wherein mating generates tetraploid cells that can then reduce their ploidy by a nonmeiotic mechanism back to a diploid or aneuploid state (Bennett and Johnson 2003; Forche et al. 2008; Berman and Hadany 2012; Hickman et al. 2015). This parasexual cycle enables C. albicans to generate genotypic diversity that impacts multiple phenotypes including drug resistance and virulence (Forche et al. 2008; Hickman et al. 2015; Hirakawa et al. 2017). While a limited set of in vitro conditions are known to promote chromosome instability in tetraploid cells, key questions remain as to where and how parasex (mating and ploidy reduction) occur in nature. Here, we examined how C. albicans tetraploid cells respond to growth in a diverse set of carbon sources and identified one sugar, D-tagatose, that promotes genomic instability, and showed that its assimilation has distinct requirements from that of the related sugar, L-sorbose.
Screening of C. albicans tetraploid cells on 190 different carbon sources revealed several that failed to support growth, including known fungistatic or fungicidal agents such as sec-butylamine and capric acid (Yoshikawa and Oda 1976; Tzatzarakis et al. 2000; Bergsson et al. 2001). We also note that few colonies were recovered following growth in L-sorbose which could indicate that the adaptive changes necessary for L-sorbose utilization (Janbon et al. 1999) did not arise during the 3-day growth period used in this study. We are not aware of any documented antifungal activity of the other compounds that failed to support growth, and these may warrant further investigation for potential inhibitory effects on C. albicans.
Our screen identified only one carbon source, D-tagatose, that led to high frequencies of 2-DOGR progeny from tetraploid strain RBY18, an indication that cells had lost GAL1 function and had potentially undergone chromosome loss or recombination. Growth in D-tagatose resulted in a 2–3 orders of magnitude increase in the frequency of 2-DOGR colonies compared to growth in D-glucose, and assays established that the majority of recovered progeny existed in a state between that of diploid and triploid. Interestingly, D-tagatose is a stereoisomer of L-sorbose which was previously shown to promote ploidy reduction in tetraploid cells (Bennett and Johnson 2003; Hickman et al. 2015). Sorbose utilization in C. albicans is dependent on SOU1 expression which resides on Chr 4 and is repressed by multiple factors encoded on Chr 5, and thus a monosomic Chr 5, either with or without a Chr 4 trisomy, enables sorbose assimilation by diploid cells (Rustchenko et al. 1994; Janbon et al. 1998, 1999; Kabir et al. 2005; Kravets et al. 2014).
Given that SOU1 expression enables utilization of L-sorbose, we were surprised that SOU1 overexpression did not promote growth in D-tagatose, indicating differences in how C. albicans utilizes these sugars. Despite this difference, an examination of 12 isolates recovered from D-tagatose cultures revealed the consistent presence of supernumerary copies of Chr 4. This suggests that increased expression of genes on Chr 4 may promote tagatose utilization, potentially due to the genes on a supernumerary chromosome being expressed in line with their copy number (Tucker et al. 2018). We also observed a bias in the homologs retained for Chr 4 and 5 following tagatose culture, with the A homolog retained more frequently than the B homolog for both chromosomes. Thus, changes in both chromosome copy number and allelic balance may promote adaptation to tagatose, although analysis of a larger set of independently cultured samples will be necessary to establish whether these differences are enabling adaptation. In addition, we note that most D-tagatose-adapted isolates exhibited increased sensitivity to Congo red which targets β-glucan in the cell wall. This is in line with cell wall remodeling changes that occur following adaption to L-sorbose and that aneuploid cells can exhibit altered stress profiles (Yang et al. 2013; Tsai et al. 2019).
While this study identifies D-tagatose as a carbon source that promotes tetraploid cell instability, it is currently unclear how C. albicans assimilates this sugar. Chr 4 could harbor a tagatose utilization gene that modifies D-tagatose into an intermediate that can be utilized for glycolysis, such as a “tagatose reductase” analogous to the sorbose reductase encoded by SOU1 (Greenberg et al. 2005). It is also possible that D-tagatose becomes phosphorylated and is then utilized by catabolic pathways similar to those in bacteria (Brinkkotter et al. 2002; Shakeri-Garakani et al. 2004; Van der Heiden et al. 2013). Additional studies are therefore required to identify C. albicans pathways that support the metabolism of this sugar.
Finally, it is worth noting that D-tagatose is a naturally occurring sugar that has been isolated from milk and mycobacteria (Adachi 1958; Szumilo and Russa 1982; Kim 2004). D-tagatose has gained popularity as a sugar substitute due to advancements in its production (Bober and Nair 2019) and because it has 38% of the calories of sucrose, as well as other benefits including prebiotic and digestive properties (Bertelsen et al. 1999; Levin 2002; Liang et al. 2019; Torrico et al. 2019). Dietary consumption of D-tagatose is also capable of increasing the diversity of the microbiome in mice (Liang et al. 2019), although effects on the mycobiome or C. albicans colonization of the gastrointestinal tract have yet to be examined. This therefore represents an interesting avenue for future research.
In summary, this study establishes that tetraploid C. albicans cells undergo ploidy reduction when grown on D-tagatose. Utilization of this sugar is not facilitated by SOU1 overexpression, unlike L-sorbose assimilation, although growth in both sugars generates progeny with increased relative copy numbers of Chr 4. Future studies will characterize how C. albicans assimilates D-tagatose and whether or not it involves processes similar to those characterized in bacteria. As D-tagatose is increasingly popular as a dietary sugar, studies examining the effects of D-tagatose consumption on the gut mycobiome and on C. albicans colonization are also warranted.
Acknowledgments
The authors thank Dr. Leah Cowen (U. Toronto) for plasmids and members of the Bennett lab for feedback. They also thank the Computational Biology Core at Brown University for bioinformatic support.
Funding
This work was supported by NIH grants AI081704 and AI141893 to R.J.B., a Burroughs Welcome Fund PATH award to R.J.B., NIH grant AI139592 to I.V.E., and an NIH IDeA award (P20GM109035) to I.V.E.
Conflicts of interest: The authors do not have any conflicts of interest.
Literature cited
- Adachi S. 1958. Formation of lactulose and tagatose from lactose in strongly heated milk. Nature. 181:840–841. [DOI] [PubMed] [Google Scholar]
- Anderson MZ, Thomson GJ, Hirakawa MP, Bennett RJ.. 2019. A ‘parameiosis’ drives depolyploidization and homologous recombination in Candida albicans. Nat Commun. 10:4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Forche A, Berman J.. 2014. Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb Perspect Med. 4:a019604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD.. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22:2505–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsson G, Arnfinnsson J, Steingrimsson O, Thormar H.. 2001. In vitro killing of Candida albicans by fatty acids and monoglycerides. Antimicrob Agents Chemother. 45:3209–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J, Hadany L.. 2012. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends Genet. 28:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen H, Jensen BB, Buemann B.. 1999. D-tagatose–a novel low-calorie bulk sweetener with prebiotic properties. World Rev Nut Diet. 85:98–109. [DOI] [PubMed] [Google Scholar]
- Bober JR, Nair NU.. 2019. Galactose to tagatose isomerization at moderate temperatures with high conversion and productivity. Nat Commun. 10:4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkkotter A, Shakeri-Garakani A, Lengeler JW.. 2002. Two class II D-tagatose-bisphosphate aldolases from enteric bacteria. Arch Microbiol. 177:410–419. [DOI] [PubMed] [Google Scholar]
- Brouard JS, Schenkel F, Marete A, Bissonnette N.. 2019. The GATK joint genotyping workflow is appropriate for calling variants in RNA-seq experiments. J Anim Sci Biotechnol. 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale RK, Pedersen BS, Quinlan AR.. 2011. Pybedtools: a flexible python library for manipulating genomic datasets and annotations. Bioinformatics. 27:3423–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, et al. 2011. The variant call format and vcftools. Bioinformatics. 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Alby K, Schaefer D, Johnson AD, Berman J, et al. 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CB, Funt JM, Abbey D, Issi L, Guiducci C, et al. 2015. The evolution of drug resistance in clinical isolates of Candida albicans. Elife. 4:e00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Chun HJ, Grant A, Otto SP.. 2006. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2:e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Fu MS, Mukaremera L, Li Z, Ormerod KL, et al. 2015. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio. 6:e01340–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Lim H, Berman J, Hickman MA.. 2017. Ploidy tug-of-war: evolutionary and genetic environments influence the rate of ploidy drive in a human fungal pathogen. Evolution. 71:1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, McBride RM, Otto SP.. 2008. Ploidy reduction in Saccharomyces cerevisiae. Biol Lett. 4:91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein AC, Sharp NP.. 2021. The population genetics of ploidy change in unicellular fungi. FEMS Microbiol Rev. 10.1093/femsre/fuab006. [DOI] [PubMed] [Google Scholar]
- Gorman JA, Gorman JW, Koltin Y.. 1992. Direct selection of galactokinase-negative mutants of Candida albicans using 2-deoxy-galactose. Curr Genet. 21:203–206. [DOI] [PubMed] [Google Scholar]
- Greenberg JR, Price NP, Oliver RP, Sherman F, Rustchenko E.. 2005. Candida albicans SOU1 encodes a sorbose reductase required for L-sorbose utilization. Yeast. 22:957–969. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR.. 1991. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press. [Google Scholar]
- Harrison BD, Hashemi J, Bibi M, Pulver R, Bavli D, et al. 2014. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 12:e1001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MA, Paulson C, Dudley AM, Berman J.. 2015. Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans. Genetics. 200:781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, et al. 2013. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature. 494:55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa MP, Chyou DE, Huang D, Slan AR, Bennett RJ.. 2017. Parasex generates phenotypic diversity de novo and impacts drug resistance and virulence in Candida albicans. Genetics. 207:1195–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa MP, Martinez DA, Sakthikumar S, Anderson MZ, Berlin A, et al. 2015. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 25:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim AS, Magee BB, Sheppard DC, Yang M, Kauffman S, et al. 2005. Effects of ploidy and mating type on virulence of Candida albicans. Infect Immun. 73:7366–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Sherman F, Rustchenko E.. 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc Natl Acad Sci USA. 95:5150–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Sherman F, Rustchenko E.. 1999. Appearance and properties of L-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics. 153:653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir MA, Ahmad A, Greenberg JR, Wang YK, Rustchenko E.. 2005. Loss and gain of chromosome 5 controls growth of Candida albicans on sorbose due to dispersed redundant negative regulators. Proc Natl Acad Sci USA. 102:12147–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P. 2004. Current studies on biological tagatose production using L-arabinose isomerase: A review and future perspective. Appl Microbiol Biotechnol. 65:243–249. [DOI] [PubMed] [Google Scholar]
- Kopecka M, Gabriel M.. 1992. The influence of congo red on the cell wall and (1-3)-beta-D-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch Microbiol. 158:115–126. [DOI] [PubMed] [Google Scholar]
- Kravets A, Yang F, Bethlendy G, Cao Y, Sherman F, et al. 2014. Adaptation of Candida albicans to growth on sorbose via monosomy of chromosome 5 accompanied by duplication of another chromosome carrying a gene responsible for sorbose utilization. FEMS Yeast Res. 14:708–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiezopolska E, Gabaldon T.. 2018. Evolutionary emergence of drug resistance in Candida opportunistic pathogens. Genes (Basel). 9:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin GV. 2002. Tagatose, the new GRAS sweetener and health product. J Med Food. 5:23–36. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. 2009. The sequence alignment/map format and samtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YX, Wen P, Wang Y, OuYang DM, Wang D, et al. 2019. The constipation-relieving property of D-tagatose by modulating the composition of gut microbiota. Int J Mol Sci. 20:5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD.. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 110:293–302. [DOI] [PubMed] [Google Scholar]
- Mishra NC, Tatum EL.. 1972. Effect of L-sorbose on polysaccharide synthetases of Neurospora crassa. Proc Natl Acad Sci USA. 69:313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Gianetti BA, Witchley JN.. 2017. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol. 15:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Nielsen K.. 2012. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell. 11:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, et al. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6:e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaiya AF, Sogin SJ.. 1979. Ploidy determination of Candida albicans. J Bacteriol. 140:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. Bedtools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko E. 2007. Chromosome instability in Candida albicans. FEMS Yeast Res. 7:2–11. [DOI] [PubMed] [Google Scholar]
- Rustchenko EP, Howard DH, Sherman F.. 1994. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J Bacteriol. 176:3231–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J.. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 313:367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J.. 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 68:624–641. [DOI] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, et al. 2015. Polyploidy can drive rapid adaptation in yeast. Nature. 519:349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakeri-Garakani A, Brinkkotter A, Schmid K, Turgut S, Lengeler JW.. 2004. The genes and enzymes for the catabolism of galactitol, D-tagatose, and related carbohydrates in Klebsiella oxytoca M5a1 and other enteric bacteria display convergent evolution. Mol Genet Genomics. 271:717–728. [DOI] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, et al. 1987. "White-opaque transition": a second high-frequency switching system in Candida albicans. J Bacteriol. 169:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z. 2014. Ploidy changes and genome stability in yeast. Yeast. 31:421–430. [DOI] [PubMed] [Google Scholar]
- Szumilo T, Russa R.. 1982. Accumulation of D-tagatose by D-galactose-grown mycobacteria. Ann Univ Mariae Curie Sklodowska Med. 37:11–18. [PubMed] [Google Scholar]
- Thomson GJ, Hernon C, Austriaco N, Shapiro RS, Belenky P, et al. 2019. Metabolism-induced oxidative stress and DNA damage selectively trigger genome instability in polyploid fungal cells. EMBO J. 38:e101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP.. 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RT, Forche A, Selmecki A.. 2017. Ploidy variation in fungi: polyploidy, aneuploidy, and genome evolution. Microbiol Spectr. 5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrico DD, Tam J, Fuentes S, Gonzalez VC, Dunshea FR.. 2019. D-tagatose as a sucrose substitute and its effect on the physico-chemical properties and acceptability of strawberry-flavored yogurt. Foods. 8:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HJ, Nelliat AR, Choudhury MI, Kucharavy A, Bradford WD, et al. 2019. Hypo-osmotic-like stress underlies general cellular defects of aneuploidy. Nature. 570:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker C, Bhattacharya S, Wakabayashi H, Bellaousov S, Kravets A, et al. 2018. Transcriptional regulation on aneuploid chromosomes in diverse Candida albicans mutants. Sci Rep. 8:1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatzarakis M, Tsatsakis AM, Liakou A, Vakalounakis DJ.. 2000. Effect of common food preservatives on mycelial growth and spore germination of Fusarium oxysporum. J Environ Sci Health B. 35:527–537. [DOI] [PubMed] [Google Scholar]
- Van der Heiden E, Delmarcelle M, Lebrun S, Freichels R, Brans A, et al. 2013. A pathway closely related to the D-tagatose pathway of gram-negative enterobacteria identified in the gram-positive bacterium Bacillus licheniformis. Appl Environ Microbiol. 79:3511–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veri AO, Miao Z, Shapiro RS, Tebbji F, O'Meara TR, et al. 2018. Tuning HSF1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS Genet. 14:e1007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer NB, Stone N, Berman J.. 2016. Ploidy dynamics and evolvability in fungi. Philos Trans R Soc Lond B Biol Sci. 371:20150461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Kravets A, Bethlendy G, Welle S, Rustchenko E.. 2013. Chromosome 5 monosomy of Candida albicans controls susceptibility to various toxic agents, including major antifungals. Antimicrob Agents Chemother. 57:5026–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Oda T.. 1976. Metabolism of maltose during surgery in patients with diabetes mellitus under general anesthesia. Res Exp Med (Berl). 167:127–138. [DOI] [PubMed] [Google Scholar]
- Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, et al. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog. 6:e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Magee BB, Magee PT, Holland BR, Rodrigues E, et al. 2015. Selective advantages of a parasexual cycle for the yeast Candida albicans. Genetics. 200:1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang Y, Upadhyay S, Xue C, Lin X.. 2020. Activation of meiotic genes mediates ploidy reduction during cryptococcal infection. Curr Biol. 30:1387–1396. e1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Whole-genome sequencing data for D-tagatose-cultured isolates (Tag1–12 and RBY18) is available at NCBI SRA as BioProject PRJNA686496. The whole-genome assembly for SC5314 has been previously published under NCBI BioProject PRJNA193498 (Hirakawa et al. 2015). Supplementary material includes Supplementary Figures S1–S4 and Supplementary Tables S1 and S2, and have been uploaded to figshare: https://doi.org/10.25387/g3.13505709.