Abstract
Mitochondrial dynamics plays an important role in mitochondrial quality control and the adaptation of metabolic activity in response to environmental changes. The disruption of mitochondrial dynamics has detrimental consequences for mitochondrial and cellular homeostasis and leads to the activation of the mitochondrial unfolded protein response (UPRmt), a quality control mechanism that adjusts cellular metabolism and restores homeostasis. To identify genes involved in the induction of UPRmt in response to a block in mitochondrial fusion, we performed a genome-wide RNAi screen in Caenorhabditis elegans mutants lacking the gene fzo-1, which encodes the ortholog of mammalian Mitofusin, and identified 299 suppressors and 86 enhancers. Approximately 90% of these 385 genes are conserved in humans, and one-third of the conserved genes have been implicated in human disease. Furthermore, many have roles in developmental processes, which suggests that mitochondrial function and their response to stress are defined during development and maintained throughout life. Our dataset primarily contains mitochondrial enhancers and non-mitochondrial suppressors of UPRmt, indicating that the maintenance of mitochondrial homeostasis has evolved as a critical cellular function, which, when disrupted, can be compensated for by many different cellular processes. Analysis of the subsets “non-mitochondrial enhancers” and “mitochondrial suppressors” suggests that organellar contact sites, especially between the ER and mitochondria, are of importance for mitochondrial homeostasis. In addition, we identified several genes involved in IP3 signaling that modulate UPRmt in fzo-1 mutants and found a potential link between pre-mRNA splicing and UPRmt activation.
Keywords: mitochondrial unfolded protein response, mitochondrial dynamics, fzo-1, IP3 signaling, Mitoguardin
Introduction
Mitochondria are important for cellular adenosine triphosphate (ATP) production, iron–sulfur-cluster biogenesis, lipid metabolism and apoptosis, and therefore, mitochondrial homeostasis is tightly regulated by several quality control mechanisms (Tatsuta and Langer 2008; Kornmann 2014). Moreover, mitochondria are required to respond to environmental challenges, which are often accompanied by alterations in energy demand (Youle and van der Bliek 2012). Mitochondrial dynamics controls mitochondrial shape and distribution, thus playing a central role in both mitochondrial homeostasis and the adjustment to changing energy demands (Yaffe 1999; van der Bliek et al. 2013). Dynamics of mitochondrial membranes is controlled by large guanosine triphosphate-binding proteins (GTPases) of the dynamin-like family, which are conserved from yeast to humans (Hales and Fuller 1997; Otsuga et al. 1998; Smirnova et al. 1998; Bleazard et al. 1999; Labrousse et al. 1999; Shepard and Yaffe, 1999; Chen et al. 2003; Santel et al. 2003; Ichishita et al. 2008; Kanazawa et al. 2008). In the nematode Caenorhabditis elegans, fusion of the outer and inner mitochondrial membrane (OMM and IMM) is facilitated by FZO-1MFN1,2 (Ichishita et al. 2008) and EAT-3OPA1 (Kanazawa et al. 2008), respectively. Conversely, fission of the OMM and IMM is carried out by DRP-1DRP1 (Labrousse et al. 1999), whose ortholog in Saccharomyces cerevisiae (Dnm1p) has been shown to form constricting spirals around mitochondria (Ingerman et al. 2005). The disruption of mitochondrial dynamics has detrimental consequences for mitochondrial and ultimately cellular homeostasis and is associated with several human diseases. Thus, mitochondrial homeostasis is controlled by several additional protective quality control mechanisms, including the UPRmt and mitophagy (Chen and Chan 2004; Youle and van der Bliek 2012; van der Bliek et al. 2013; Kornmann 2014). How these quality control mechanisms are coordinated with mitochondrial dynamics is not fully understood. Recently, disruption of mitochondrial dynamics has been shown to induce UPRmt (Kim and Sieburth 2018; Zhang et al. 2018; Rolland et al. 2019; Haeussler et al. 2020). UPRmt has been studied extensively in the past decade using genome-wide RNAi screens in C. elegans (Haynes et al. 2007; Runkel et al. 2013; Bennett et al. 2014; Liu et al. 2014; Rolland et al. 2019). Upon mitochondrial stress and the concomitant decrease in mitochondrial membrane potential, the master regulator of UPRmt, “activating transcription factor associated with stress 1” (ATFS-1ATF4,5), instead of being imported into mitochondria, translocates from the cytosol to the nucleus, where it activates a broad transcriptional program (Haynes et al. 2010; Nargund et al. 2012; Rolland et al. 2019). UPRmt activation leads to the expression of a large set of cytoprotective genes including genes encoding chaperones [e.g., hsp-6mtHSP70 and hsp-60HSDP1, whose transcription is commonly used to monitor UPRmt activation (Yoneda et al. 2004)] or proteases, and has been shown to promote mitochondrial biogenesis and coordinate cellular metabolism (Nargund et al. 2012; Rauthan et al. 2013; Liu et al. 2014; Ranji et al. 2014; Nargund et al. 2015; Oks et al. 2018; Haeussler et al. 2020) (All genes that are specifically up- or downregulated upon induction of UPRmt are referred to as UPRmt effectors). Moreover, UPRmt has been shown to act in a cell non-autonomous way, and once activated in a certain tissue can result in a systemic response (Durieux et al. 2011; Shao et al. 2016; Kim and Sieburth 2018; Zhang et al. 2018; Kim and Sieburth 2020).
In this study, we performed a genome-wide RNAi screen to identify regulators of UPRmt in fzo-1(tm1133) mutants and identified 299 suppressors and 86 enhancers. We analyzed this dataset using bioinformatic tools, such as GO enrichment analysis, gene network analysis and analysis of transcription factor (TF) binding sites in promotors of candidate genes. Furthermore, we determined the specificities of the candidates identified with respect to their ability to modulate UPRmt using secondary screens. Finally, we identified the C. elegans ortholog of the mammalian genes Miga1 and Miga2, which have been implicated in mitochondrial fusion, and demonstrate that the loss of the C. elegans ortholog leads to mitochondrial fragmentation and the induction of UPRmt.
Methods
General C. elegans methods and strains
C. elegans strains were cultured as previously described (Brenner 1974). Bristol N2 was used as the wild-type strain. All experiments were carried out at 20°C and all strains were maintained at 20°C. The following alleles and transgenes were used: LGI: spg-7(ad2249) (Zubovych et al. 2010); LGII: fzo-1(tm1133) (National BioResource Project); eat-3(ad426) (Kanazawa et al. 2008); LGIV: drp-1(tm1108) (National BioResource Project); bcSi9 (Phsp-6::gfp::unc-54 3’UTR) (Haeussler et al. 2020); LGV: miga-1(tm3621) (National BioResource Project). In addition, the following multi-copy integrated transgenes were used: zcIs9 (Phsp-60::gfp::unc-54 3’UTR), zcIs13 (Phsp-6::gfp::unc-54 3’UTR) (Yoneda et al. 2004); bcIs78 (Pmyo-3::gfpmt) (Rolland et al. 2013).
RNA-mediated interference
RNAi by feeding was performed using the updated “Ahringer” RNAi library (Kamath and Ahringer 2003), which covers around ∼87% of the currently annotated C. elegans protein-coding genes. For the primary and secondary screens with the multi-copy zcIs13 transgene in the fzo-1(tm1133), drp-1(tm1108), eat-3(ad426), or spg-7(ad2249) background, RNAi clones were cultured overnight in 100 µl of LB containing carbenicillin (100 μg/mL) in a 96 well plate format at 37°C and 200 rpm. 10 µl of each RNAi culture was used to seed one well of a 24 well RNAi plate containing 0.25% Lactose (w/v) as described previously (Rolland et al. 2019). The plates were incubated at 20°C in the dark. 24 hours later, 3 L4 larvae of all strains carrying the fzo-1(tm1133) and spg-7(ad2249) allele, and 2 L4 larvae of drp-1(tm1108) were transferred to each well of the RNAi plates. The F1 generation was scored by eye for fluorescence intensity of the Phsp-6mtHSP70gfp reporter after 4–12 days and compared to worms of the respective genotype on the negative control sorb-1(RNAi).
For double-RNAi experiments (Supplementary Figure S1), RNAi clones were cultured as described above but experiments were conducted in three independent experiments using RNAi plates containing 6 mM IPTG. rps-1(RNAi) was diluted 1:1 with either empty vector RNAi (L4440) or kgb-1(RNAi).
Screening procedure and sequencing of RNAi-clones
For the primary screen, all RNAi clones of the library were tested once. Bacterial RNAi clones that enhanced or suppressed the Phsp-6mtHSP70gfp reporter were picked from the wells and inoculated in 100 µl of LB containing carbenicillin (100 μg/mL) in a 96 well plate format and cultured overnight at 37°C and 200 rpm. Glycerol stocks of these overnight cultures were prepared the following day by adding 100 µl of LB containing 30% glycerol and frozen at −80°C. After all RNAi clones of the library were tested, the 657 identified candidates were retested at least three times in duplicates for verification of the observed phenotype. The RNAi clones that reproduced the suppression or enhancement phenotype at least three out of six times were considered as verified candidates.
The 385 verified RNAi clones were sequenced. For this, colony PCRs were performed directly from the glycerol stocks using the primers L4440F and L4440R. To remove excessive primers and nucleotides, PCR products were treated with ExoSAP-IT™ (Applied Biosystems, Cat.no. 78200.200.UL) according to manufacturer’s protocol. After PCR clean-up, samples were sent for sequencing using L4440F primer.
L4440 F 5′-TGGATAACCGTATTACCGCC-3′
L4440 R 5′-GTTTTCCCAGTCACGACGTT-3′
According to our sequencing results, seven of the RNAi clones covered two genes. These are indicated in column B (“Sequence”) in Supplementary Table S1. These RNAi clones were assigned to the GO group of the gene, which was predominantly covered by our sequencing result and all subsequent analysis were carried out using this gene.
Subsequently, the verified and sequenced clones were rescreened in technical duplicates in three independent experiments in the secondary screens in drp-1(tm1108), eat-3(ad426), and spg-7(ad2249) mutant backgrounds.
Identification of human orthologs
Human orthologs and OMIM data (Amberger et al. 2019) were extracted from wormbase.org using https://intermine.wormbase.org (Harris et al. 2020). Human orthologs were then manually verified using “alliancegenome.org” (The Alliance of Genome Resources, 2019), “orthodb.org” (Kriventseva et al. 2019), “ensembl.org” (Hunt et al. 2018), and “uniprot.org” (UniProt Consortium 2018).
Prediction of mitochondrial localization and mitochondrial targeting sequences
First, https://intermine.wormbase.org (Harris et al. 2020) was used to identify all candidate genes, which are related to any mitochondrial processes/pathways. To that end, we extracted all 698 genes currently associated with at least one of the 404 GO-terms containing “mitochond” and checked how many of our 385 candidate genes are among them. In addition, we used the online platform “MitoProt” (https://ihg.gsf.de/ihg/mitoprot.html) (Claros and Vincens 1996) for computational prediction of mitochondrial targeting sequences. Proteins for which the value of a mitochondrial targeting sequence was ≥0.5 in this analysis were predicted to be mitochondrial.
Gene ontology enrichment analysis using DAVID
In search of enriched gene ontology (GO) terms, we used the DAVID tool (version 6.8, Huang et al. 2009a, 2009b) and ran the list of candidates against all genes of the C. elegans genome as a background list. Using an EASE score from the modified fisher-exact test, the clustering algorithm groups genes based on their association in GO categories and assigns a significance value to the group (Huang et al. 2007). The clustered groups were then plotted using modified functions from the GO plot package (R version 1.0.2, Walter et al. 2015).
TF enrichment analysis
We searched for enriched TFs using the tool g:Profiler [a tool for functional enrichment analysis using over-representation (Raudvere et al. 2019)]. The two input lists [suppressors and enhancers of fzo-1(tm1133)-induced UPRmt) with WBGene-IDs of the identified candidate genes were used to search in the Transfac database [annotations: TRANSFAC Release 2019.1 classes: v2 (Knüppel et al. 1994; Matys et al. 2006)].
Construction of gene networks of FZO-1 and MFN1/2, and the UPRmt
The C. elegans interactomes were compiled for FZO-1 or all 16 genes that are currently associated with the GO-term “mitochondrial unfolded protein response” (GO: 0034514) from scientific literature (Durinck et al. 2009; Simonis et al. 2009) and databases such as mentha (Calderone et al. 2013), BioGRID3.5 (Oughtred et al. 2019), IntAct (Orchard et al. 2014), and STRING (Szklarczyk et al. 2019) (STRING was only used to build the FZOome). The human orthologs of those genes were identified and were searched as well. Whenever possible, the interaction partners were converted back to C. elegans genes using biomaRt (Durinck et al. 2009) and available scientific literature (Shaye and Greenwald 2011; Kim et al. 2018). The complete list of interactions was uploaded to cytoscape (v.3.7.2, Shannon et al. 2003) and a network was calculated, highlighting both enhancers and suppressors from the screening results.
Image acquisition, processing, and analysis
For double-RNAi experiments (Supplementary Figure S1), 10–20 fzo-1(tm1133) mutants carrying bcSi9 (Phsp-6mtHSP70gfp) were immobilized with M9 buffer containing 10 mM levamisole on 2% agarose pads and imaged using a Nikon SMZ18 dissecting microscope and Nikon-Elements software.
For each mutant in Supplementary Figure S2, 10–20 animals were immobilized with M9 buffer containing 150 mM sodium azide on 2% agarose pads and imaged using a Leica GFP dissecting microscope (M205 FA) and Leica Application Suite software (3.2.0.9652).
For the analysis of mitochondrial morphology a strain carrying bcIs78 (Pmyo-3::gfpmt) was imaged using a Zeiss Axioskop 2 with a 63x objective and MetaMorph software (Molecular Devices). Subsequently, mitochondrial morphology was assessed using the deep learning algorithm MitoSegNet (Fischer et al. 2020).
Data availability
Strains are available upon request. Supplementary Figure S1 contains data about involvement of the cSADDs response in suppression of UPRmt upon attenuation of cytosolic translation. Supplementary Figure S2 shows different mutants inducing the UPRmt reporter. Supplementary Figure S3 shows the FZOome. Supplementary Figure S4 contains a subset of the UPRmtome coming from direct interactions in C. elegans. Supplementary Figure S5 depicts a subset of the UPRmtome coming from interactions of human orthologs. Supplementary Figure S6 shows the complete UPRmtome. Supplementary Table S1 contains all suppressors and enhancers of fzo-1(tm1133)-induced UPRmt identified in a genome-wide RNAi screen in C. elegans. Supplementary Table S2 contains the GO enrichment analysis of suppressors and enhancers of fzo-1(tm1133)-induced UPRmt. Supplementary Table S3 contains TF enrichment analysis of suppressors and enhancers of fzo-1(tm1133)-induced UPRmt. Supplementary Table S4 contains the results of the interactome analysis (UPRmtome). The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. The supplemental material is available at figshare: https://doi.org/10.25387/g3.14262425.
Results and Discussion
Genome-wide RNAi screen for suppressors and enhancers of fzo-1(tm1133)-induced UPRmt identifies highly conserved set of genes with relevance to human health
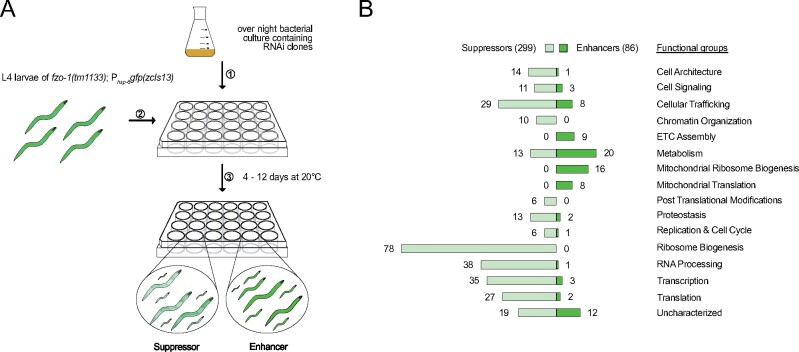
The disruption of mitochondrial dynamics in C. elegans induces the UPRmt (Kim and Sieburth 2018; Zhang et al. 2018; Rolland et al. 2019; Haeussler et al. 2020). To identify genes affecting mitochondrial homeostasis in animals with defects in mitochondrial dynamics, we used a loss-of-function mutation of fzo-1MFN1,2, tm1133, (National BioResource Project) to induce the UPRmt reporter Phsp-6mtHSP70gfp (zcIs13) and screened the C. elegans genome for modifiers. To that end, we used RNA-mediated interference (RNAi) and targeted ∼87% of the currently annotated protein-coding genes (Kamath and Ahringer 2003) (Figure 1A). The moderate induction of the Phsp-6mtHSP70gfp reporter in the fzo-1(tm1133) background allowed the identification of both suppressors and enhancers of the response. Using a protocol in which the F1 generation is scored for Phsp-6mtHSP70gfp expression levels in the fourth larval stage of development (L4), we initially identified 657 candidate genes of which 385 reproduced. Of the 385 candidates identified, 299 act as suppressors upon knock-down and 86 as enhancers (Figure 1B and Supplementary Table S1). In addition, upon knock-down, many candidates result in synthetic slow growth and/or reduced fertility (indicated in the “Overview” sheet in Supplementary Table S1). In order to assess whether the 86 identified enhancers are specific to the fzo-1(tm1133) background or if their depletion induces UPRmt also in the absence of mitochondrial stress, we knocked them down in a wild-type background and tested for induction of the Phsp-6mtHSP70gfp reporter. All except three genes (copd-1ARCN1, F25H9.6PPCDC, and metl-17METTL17) induce Phsp-6mtHSP70gfp expression when knocked-down in wild-type animals, suggesting that the induction of UPRmt by depletion of these candidates is independent of the loss of fzo-1 [Candidates that encode mitochondrial proteins and that induce UPRmt in a wild-type background upon knock-down were included in a recent publication, which reported the systematic identification of mitochondrial inducers of UPRmt (Rolland et al. 2019)].
Figure 1.
Overview of genome-wide RNAi screen for suppressors and enhancers of fzo-1(tm1133)-induced UPRmt. (A) Schematic overview of the RNAi screening procedure using the RNAi feeding library (Kamath and Ahringer 2003) in fzo-1(tm1133) mutants that express the UPRmt reporter Phsp-6mtHSP70gfp (zcIs13). The moderate induction of the reporter in the fzo-1(tm1133) background allowed screening for both suppressors and enhancers of the response. (B) The screen resulted in identification of 299 suppressors and 86 enhancers of fzo-1(tm1133)-induced UPRmt, which were sorted into categories that we defined according to their function. ETC, electron transport chain.
Among the 299 suppressors, only 25 (8%) have previously been found to suppress UPRmt induced by other means upon knock-down (Haynes et al. 2007; Runkel et al. 2013; Liu et al. 2014). Similarly, among the 86 enhancers, only 15 (17%) have previously been shown to induce UPRmt upon knock-down (indicated “Previously identified” in the “Overview” sheet of Supplementary Table S1). This may be due to different genetic backgrounds and to differences in RNAi-protocols. Moreover, false negatives in RNAi screens have been estimated to vary between 10% and 30%, even if the same protocol is used by the same laboratory (Simmer et al. 2003).
Using “alliancegenome.org” (The Alliance of Genome Resources, 2019), “orthodb.org” (Kriventseva et al. 2019), “ensembl.org” (Hunt et al. 2018), “uniprot.org” (UniProt Consortium, 2018), and “wormbase.org” (Harris et al. 2020) databases, we found that approximately 90% of the suppressors and enhancers (348) have at least one ortholog in humans (indicated “Human ortholog” in the “Overview” sheet of Supplementary Table S1). For comparison, the overall conservation of genes from C. elegans to humans is only about 41% (Shaye and Greenwald 2011; Kim et al. 2018). Moreover, we found that the orthologs of 36% (126) of the conserved candidates have previously been associated with human disease and are listed in the “Online Mendelian Inheritance in Man” database (Amberger et al. 2019) (indicated “OMIM” in the “Overview” and “OMIM” sheet of Supplementary Table S1). In summary, we identified a set of predominantly conserved genes, many of them relevant to human health, which when knocked-down affect mitochondrial homeostasis in mutants with defects in mitochondrial fusion.
Genes with functions in development, receptor-mediated endocytosis, and metabolism modulate UPRmt signaling
In order to obtain an overview of the type of processes that affect fzo-1(tm1133)-induced UPRmt, we analyzed the GO terms of all 385 candidates, sorted them into “functional groups” (Figure 1B), and performed a clustered gene enrichment analysis using DAVID (Huang et al. 2009a, 2009b) (Figure 2 and Supplementary Table S2) [Thirty-one suppressors and enhancers could not be assigned to functional groups since these genes are uncharacterized in C. elegans and/or lack orthologs in humans. For this reason, they were assigned to the functional group “uncharacterized” (Figure 1B)].
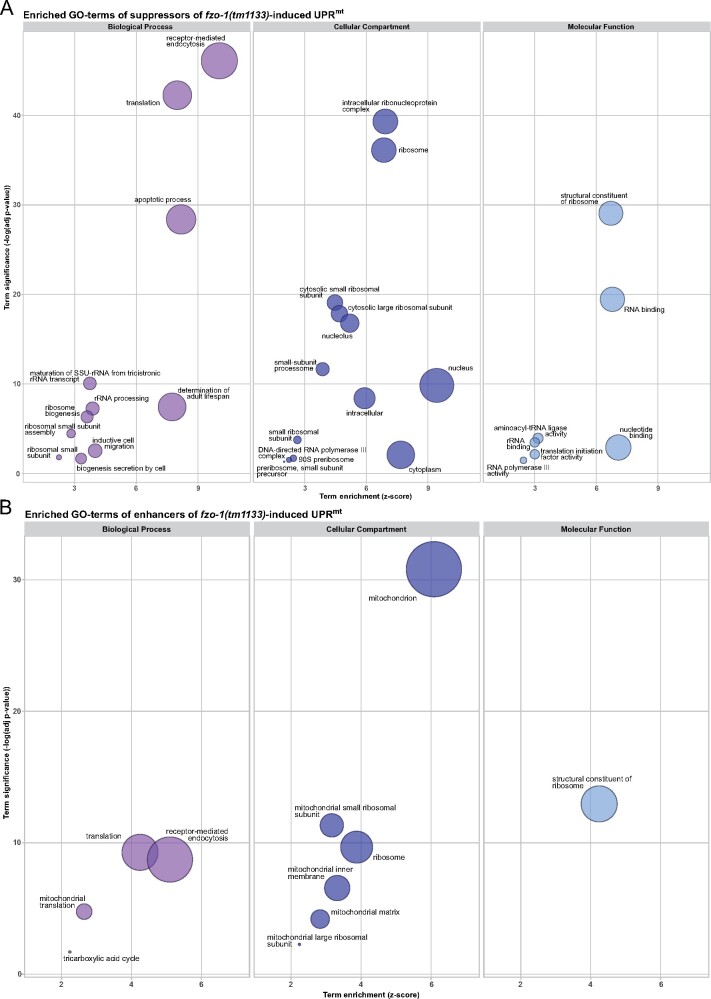
Figure 2.
GO enrichment analysis of suppressors and enhancers of fzo-1(tm1133)-induced UPRmt using DAVID. (A) Results of the clustered GO enrichment analysis of suppressors of fzo-1(tm1133)-induced UPRmt using DAVID (Huang et al. 2009a, 2009b). (B) Results of the clustered GO enrichment analysis of enhancers of fzo-1(tm1133)-induced UPRmt using DAVID. (A and B) Statistically significant (P > 0.05) enriched GO-terms, except the nematode specific GO-terms, of fzo-1(tm1133)-induced UPRmt are depicted. Circle size correlates with the number of genes associated with a specific GO-term.
In the clustered gene enrichment analysis, we found that the majority of both suppressors and enhancers are associated with at least one of the following GO-terms: “nematode larval development,” “embryo development ending in birth or egg hatching” or “reproduction” (Supplementary Table S2). It has been shown that reducing the functions of some genes encoding components of the ETC [e.g., cox-5B(RNAi)] in specific tissues and at specific times during development can lead to both systemic activation of UPRmt and longevity (Dillin et al. 2002; Rea et al. 2007; Durieux et al. 2011). This indicates that the activity levels of mitochondria in an individual animal are “set” at a specific developmental stage and, once set, are maintained throughout development and adult life. Our results demonstrate that disrupting development compromises this process, thereby affecting an animal’s ability to cope with mitochondrial stress and to respond to UPRmt activation, which is expected to indirectly affect processes such as its lifespan. In support of this notion, we found that approximately 20% of the suppressors carry the GO-term “determination of adult lifespan” (Supplementary Table S2).
Among the suppressors, the GO-term “receptor-mediated endocytosis” is enriched (Figure 2A and Supplementary Table S2). It contains many genes with roles in vesicular trafficking and vesicle budding. Genes required for vesicular trafficking have been shown to affect mitochondrial morphology and homeostasis when inactivated, and it has been proposed that this is the result of altered contact sites between organelles and altered lipid transfer into mitochondria (Altmann and Westermann 2005). Furthermore, we recently demonstrated that approximately half of the candidates in this GO-category are negative regulators of autophagy. Upon knock-down, these genes suppress fzo-1(tm1133)-induced UPRmt most probably by inducing autophagy thereby causing changes in lipid metabolism (Haeussler et al. 2020). Moreover, many cellular signaling pathways originate at the plasma membrane and, thus, are dependent on endocytosis (Sorkin and von Zastrow 2009; Di Fiore and von Zastrow 2014). Therefore, we speculate that depletion of the genes associated with the GO-term “receptor mediated endocytosis” may either cause changes in lipid metabolism thereby suppressing UPRmt or disrupt cell non-autonomous UPRmt signaling.
The functional group “ribosome biogenesis” contains 78 (26%) of the suppressors (Figure 1B) and includes both small- and large-ribosomal subunits, as well as proteins with roles in the maturation or transport of ribosomal subunits and rRNAs. Accordingly, in all three GO-domains (Biological Process, Cellular Compartment, and Molecular Function), we found that several GO-terms related to the ribosome were significantly enriched (Figure 2A and Supplementary Table S2) (The GO-term “apoptotic process” also contains many ribosomal subunits leading to its enrichment in our analysis).
Moreover, we assigned a substantial part of the suppressors to the groups “RNA processing” (38), “transcription” (35), and “translation” (27) (Figure 1B). Hence, we found five GO-terms related to translation-, two to transcription- and one to RNA-related processes to be enriched in a statistically significant manner in the GO enrichment analysis (Figure 2A and Supplementary Table S2). These results raise the question whether knock-down of the candidates involved in cytosolic translation specifically suppresses UPRmt or simply reduces the expression of the Phsp-6mtHSP70gfp reporter. A previous study also identified many genes related to ribosome biogenesis and cytosolic translation in a screen for suppressors of paraquat-induced UPRmt (Runkel et al. 2013). Runkel and colleagues reported reduced levels of two other reporters (Phsp-16.2 CRYABgfp, Phsp-4 HSPA5gfp) upon attenuation of cytosolic translation by rpl-36(RNAi). In contrast, they showed that Pgst-4 HPGDSgfp was slightly hyperactivated (Runkel et al. 2013), as previously shown for this reporter upon knock-down of several other genes related to cytosolic translation (Melo and Ruvkun 2012). We recently showed that knock-down of the cytosolic tRNA synthetase hars-1HARS1, which we found to suppress Phsp-6mtHSP70gfp expression in fzo-1(tm1133) and which presumably also compromises cytosolic translation, results in reduced expression of a control reporter that is unrelated to other stress responses, Pges-1GES2gfp (Haeussler et al. 2020). Taken together, we cannot exclude the possibility that the knock-down of candidates related to the functional groups of transcription, RNA processing, ribosome biogenesis, and translation may, at least to some extent, interfere with reporter expression per se. In addition, Runkel and colleagues showed that depletion of KGB-1MAPK10, a JNK-like MAP-kinase mediating cellular surveillance-activated detoxification and defenses (cSADDs) in C. elegans (Melo and Ruvkun 2012), derepresses UPRmt induced by paraquat upon attenuation of cytosolic translation (Runkel et al. 2013). Therefore, we tested whether knock-down of kgb-1 also relieves the induction of UPRmt upon knock-down of rps-1 and found that Phsp-6mtHSP70gfp expression was partially restored under these conditions (Supplementary Figure S1). Thus, attenuation of cytosolic translation may activate cSADDs through KGB-1MAPK10, thereby preventing UPRmt induction in fzo-1(tm1133) mutants.
Among the enhancers, we assigned most candidates to the functional groups “metabolism” and “mitochondrial ribosome biogenesis” as well as “cellular trafficking,” “mitochondrial translation,” and “ETC assembly” (Figure 1B). Accordingly, GO analysis of the enhancers shows that the cellular compartments “mitochondrion,” “mitochondrial small ribosomal subunit,” “mitochondrial large ribosomal subunit,” “mitochondrial inner membrane,” “mitochondrial matrix,” and “ribosome” are enriched (Figure 2B and Supplementary Table S2). In addition, the biological processes “translation” (which also includes “mitochondrial translation”), “tricarboxylic acid cycle” and “receptor-mediated endocytosis” are enriched as is the molecular function “structural constituent of ribosome” (Figure 2B and Supplementary Table S2). Among the enhancers carrying the GO-term “receptor-mediated endocytosis,” we identified many subunits of the mitochondrial ribosome and genes required for mitochondrial translation, which are most likely misannotated and therefore led to enrichment of this GO-term. In summary, we showed that disrupting mitochondrial translation and metabolism induces UPRmt in fzo-1(tm1133). Disruption of these processes has also previously been shown to induce UPRmt in wild type (Durieux et al. 2011; Houtkooper et al. 2013). Therefore, we conclude that reducing mitochondrial function induces UPRmt independently of the genetic background.
In summary, the GO enrichment analysis revealed that depletion of the majority of candidates in our dataset may modulate UPRmt due to their role in development. Furthermore, we propose that the suppressors with roles in endocytosis modulate UPRmt signaling indirectly and speculate that cellular signaling and/or alterations in organellar contact sites may influence mitochondrial metabolism and hence, UPRmt signaling. Finally, we find disruption of mitochondrial metabolism and translation to robustly enhance UPRmt signaling in fzo-1(tm1133).
Mitochondrial fitness balances cellular homeostasis
Next, we determined which fraction of the identified enhancers and suppressors encode proteins that have a mitochondrial function or localize to mitochondria. We extracted all 698 genes that are associated with at least one of the 404 GO-terms containing “mitochond” using the “WormMine” database (https://intermine.wormbase.org) (Harris et al. 2020), and then determined how many of our candidate genes are associated with any of these GO-terms. Using this approach, we identified 11 suppressors and 59 enhancers that encode proteins that localize to mitochondria or play a role in mitochondrial metabolism and dynamics, respectively (indicated “GO mitochond” in “Overview” and “Mitochondrial” sheet of Supplementary Table S1). Next, we used the online platform “MitoProt” (https://ihg.gsf.de/ihg/mitoprot.html) (Claros and Vincens 1996) for computational prediction of mitochondrial targeting sequences and identified an additional 5 suppressors and 14 enhancers that are predicted to localize to mitochondria (cut-off value ≥ 0.5) (indicated “MitoProt prediction” in “Mitochondrial” sheet of Supplementary Table S1). Third, by literature searches, we found that the orthologs of 3 enhancers localize to mitochondria (Shafqat et al. 2003; Spaan et al. 2005; Cambier et al. 2012). In summary, 76 out of 86 (88%) enhancers and 16 out of 299 (5%) suppressors encode proteins that have a mitochondrial function. This suggests that only a few processes exist outside of mitochondria that can perturb mitochondrial homeostasis when compromised. Conversely, many processes and mechanisms exist outside of mitochondria that can compensate for mitochondrial dysfunction, thereby ensuring mitochondrial and consequently cellular homeostasis.
Among the 10 “non-mitochondrial” enhancers of UPRmt are three genes (F29B9.8, Y61A9LA.11, C25H3.10) with yet unknown functions, which lack orthologs in other systems. ORC-1ORC1 is a component of the origin recognition complex and plays a role in DNA replication (Gavin et al. 1995; Ohta et al. 2003; Tatsumi et al. 2003). The disruption of DNA replication or cell cycle progression has previously not been reported to lead to UPRmt induction. We speculate that disruption of DNA replication leads to developmental defects and therefore induces UPRmt. F25H9.6PPCDC is the C. elegans ortholog of phosphopantothenoylcysteine decarboxylase, an enzyme required for biosynthesis of coenzyme A (CoA) (Daugherty et al. 2002). Thus, knock-down of F25H9.6PPCDC may interfere with critical biosynthetic and metabolic pathways (including the TCA cycle) and therefore enhance UPRmt. NHR-209HNF4A,G is orthologous to Hepatocyte Nuclear Factor 4α (HNF4A) and belongs to the family of nuclear hormone receptors, a class of cofactor and ligand-inducible TFs that regulate various cellular processes, including metabolism, development, and homeostasis (Aranda and Pascual 2001; Bolotin et al. 2010). Interestingly, long-chain fatty acids are ligands of HNF4A and, depending on their chain length and degree of saturation, activate or repress the transcriptional activity of HNF4A (Hertz et al. 1998; Dhe-Paganon et al. 2002; Wisely et al. 2002; Duda et al. 2004). Furthermore, HNF4A activity has been shown to be required for ß-oxidation of fatty acids both in mice and Drosophila melanogaster (Palanker et al. 2009; Chen et al. 2020). Thus, NHR-209HNF4A,G may have a similar role in C. elegans and act as a metabolic sensor, which when deactivated, enhances UPRmt in fzo-1(tm1133). Moreover, we identified cpna-3CPNE5,8,9, an ortholog of mammalian copine family members, a class of calcium dependent phospholipid binding proteins with roles in intracellular signaling and membrane trafficking (Creutz et al. 1998; Tomsig et al. 2003; 2004; Ramsey et al. 2008). Previously, another gene of the copine family, gem-4CPNE8, has been shown to be upregulated upon UPRmt activation (Nargund et al. 2012). Therefore, we speculate that signaling via copine family members may be important for UPRmt regulation. Another non-mitochondrial enhancer, copd-1ARCN1, encodes a protein orthologous to the delta subunit of coatomer in S. cerevisiae and humans (RET2 and ARCN1, respectively), which is involved in the formation of coat protein complex I (COPI) vesicles. COPI vesicles play a central role in the secretory pathway and are required for the retrieval of lipids and proteins from the Golgi apparatus and the subsequent retrograde transport of these lipids and proteins to the ER (Lee et al. 2004; Beck et al. 2009). Furthermore, the trafficking to their final destination of most non-mitochondrial and non-peroxisomal transmembrane proteins, as well as proteins required for the release of neurotransmitters, such as SNARE proteins, is dependent on COPI-mediated transport (Beck et al. 2009). Thus, disruption of the secretory pathway affects many intra- and intercellular signaling pathways, including the Ras and TOR signaling pathways, as well as signaling via G-protein-coupled receptors (GPCRs) and receptor tyrosine kinases (Farhan and Rabouille 2011). Moreover, disruption of the retrograde transport system has been shown to lead to erroneous secretion of ER resident proteins (e.g., ER chaperones) and, consequently, to the activation of UPR in the ER (UPRER) (Aguilera-Romero et al. 2008; Izumi et al. 2016). Therefore, we speculate that the enhancement of UPRmt induction in fzo-1(tm1133) animals upon copd-1(RNAi) may be due to alterations in one of the above-mentioned signaling pathways. This notion is supported by the finding that phospholipase C (PLC-1PLCE1), a GPCR associated enzyme, is among the non-mitochondrial enhancers, as well as srh-40 (serpentine receptor class H), which is predicted to encode a GPCR. Taken together, we identified many genes among the “non-mitochondrial” enhancers, which regulate intra- and intercellular signaling cascades, and we speculate that these may play a role in signaling of UPRmt, both in a cell autonomous and cell non-autonomous fashion. In addition, we identified “non-mitochondrial” enhancers that directly regulate metabolic homeostasis and, thus, enhance UPRmt in fzo-1(tm1133) mutants.
Among the 16 identified “mitochondrial suppressors” of UPRmt are candidates, such as TFG-1TFG and GBF-1GBF1, that encode proteins that have been shown to associate with mitochondria but also other organelles. GBF-1GBF1 is a guanine nucleotide exchange factor (GEF) for the small GTPase ARF-1.2ARF1, which in yeast recruits ARF-1.2ARF1,3 to ER-mitochondria contact sites (Ackema et al. 2014). Depletion of GBF-1GBF1 leads to altered ARF-1.2ARF1,3 localization and changes in mitochondrial morphology both in yeast and C. elegans and this appears to be independent of their roles in endosomal transport (Ackema et al. 2014). Ackema and colleagues observed an increase in mitochondrial connectivity upon GBF-1GBF1 depletion, similar to that observed upon knock-down of miro-1MIRO1 and vdac-1VDAC, both of which encode proteins that also localize to ER-mitochondria contact sites. However, the alterations in mitochondrial morphology of FZO-1MFN1,2 depleted animals were shown to be epistatic to the changes in mitochondrial morphology observed upon gbf-1(RNAi) and arf-1.2(RNAi). Therefore, the suppression of UPRmt observed in fzo-1(tm1133) animals upon gbf-1(RNAi) may not be due to a rescue of the mitochondrial morphology defect but rather be the consequence of changes in ER-mitochondria contact sites. This highlights the importance of organellar contact sites for the maintenance of mitochondrial and consequently cellular homeostasis. Furthermore, we identified TFG-1TFG, a component of the secretory pathway via COPII vesicles (Witte et al. 2011), as a suppressor of fzo-1(tm1133)-induced UPRmt. COPII vesicles transport newly synthesized proteins and lipids from specialized ER zones, so-called ER exit sites (ERES), to the Golgi apparatus (Budnik and Stephens 2009; Kurokawa and Nakano 2019). Similar to what we propose for copd-1(RNAi) (see above), we speculate that disruption of the secretory pathway may lead to alterations in cellular signaling, ER-mitochondria contact sites and, depending on the context, either to suppression or enhancement of UPRmt. Taken together, we demonstrate that the perturbation of primarily mitochondrial processes leads to the enhancement of UPRmt. However, the identification of non-mitochondrial enhancers demonstrates that disruption of processes taking place outside of mitochondria can also compromise mitochondrial function and activate or enhance UPRmt. Alterations in cellular signaling pathways and/or organellar contact sites may play a role in this respect. Moreover, we find that the majority of suppressors of fzo-1(tm1133)-induced UPRmt are non-mitochondrial, suggesting that many cellular pathways outside of mitochondria exist that can compensate for mitochondrial stress and, hence, ensure mitochondrial homeostasis. In line with this notion, we identified a few “mitochondrial suppressors,” most of which are involved in the maintenance of contacts to other organelles, especially the ER.
Defects in mitochondrial fusion and fission are suppressed and enhanced by the same pathways
In order to define the specificity of the 299 suppressors and 86 enhancers, we carried out secondary screens. To identify general modifiers of UPRmt, we rescreened the candidates in the background of spg-7(ad2249), which induces UPRmt (Supplementary Figure S2). spg-7AFG3L2 encodes a mitochondrial matrix AAA-protease, which induces UPRmt when depleted and which is commonly used as a positive control for UPRmt activation (Yoneda et al. 2004; Haynes et al. 2007; 2010). To identify genes in our dataset that specifically modify UPRmt induced by defects in mitochondrial membrane fusion, we rescreened all candidates in the eat-3(ad426) background, in which IMM fusion is blocked. Finally, to identify genes that may modulate UPRmt induced by defects in mitochondrial dynamics, we rescreened all candidates in the drp-1(tm1108) background, in which mitochondrial fission is blocked. In the drp-1(tm1108) background, of the 385 candidates, 291 suppress and 59 enhance. In the eat-3(ad426) background, 242 suppress and 49 enhance. Finally, in the spg-7(ad2249) background, 181 suppress and 54 enhance (Supplementary Table S1). (Of note, there is an inverse correlation between the level of Phsp-6mtHSP70gfp expression in the above-mentioned mutant background and the number of candidates that reproduce. Hence, the level of reporter expression may correlate with the number of false negatives in a given dataset of the secondary screens, for both suppressors and enhancers.) Since more suppressors reproduced in drp-1(tm1108) and eat-3(ad426) compared to spg-7(ad2249), we conclude that defects in mitochondrial dynamics, to some extent, are suppressed or enhanced by the same pathways. Moreover, the suppressors of fzo-1(tm1133)-induced UPRmt that were sorted into the functional groups “ribosome biogenesis,” “RNA processing” and “translation,” reproduced comparably well in all secondary screens. Thus, attenuation of cytosolic translation may either be a general mechanism to suppress UPRmt or, as discussed above, interfere with reporter expression. Among the enhancers, genes that sorted into the functional groups “ETC assembly factors,” “mitochondrial ribosome biogenesis” and “mitochondrial translation” showed the highest overlap among the secondary screens (Supplementary Table S1), which demonstrates that disruption of mitochondrial translation robustly enhances UPRmt, independent of genetic background.
While we did not identify any suppressors that act exclusively in the fzo-1(tm1133) background, we found six enhancers (slc-25A26SLC25A26, frh-1FXN, sdha-1SDHA, sucg-1SUCLG2, metl-17METTL17, and K03B4.1) that did not reproduce in any of the secondary screens. Among these, metl-17METTL17, a methyltransferase required for mitochondrial ribosome assembly and mitochondrial translation in mice (Shi et al. 2019), also did not induce UPRmt expression in wild type and, thus, specifically enhances fzo-1(tm1133)-induced UPRmt.
Twelve candidates that suppressed UPRmt in the primary screen using fzo-1(tm1133), enhanced UPRmt in one or more of the secondary screens. Conversely, 10 enhancers of fzo-1(tm1133)-induced UPRmt suppress UPRmt in at least one of the mutants in the secondary screens (listed in the “Opposing UPRmt phenotypes” sheet in Supplementary Table S1). For example, knock-down of icd-1βNAC suppresses Phsp-6mtHSP70gfp in all mitochondrial dynamics-related backgrounds, but enhances spg-7(ad22449)-induced UPRmt. Knock-down of icd-1βNAC in C. elegans has been reported to induce UPRER in wild-type embryos (Arsenovic et al. 2012). Furthermore, icd-1βNAC has been described as a cytosolic stress sensor, which in the absence of stress associates with ribosomes to promote cytosolic translation, and acts as a chaperone in the cytosol upon heat stress (Kirstein-Miles et al. 2013). We recently showed that icd-1βNAC is a negative regulator of autophagy and that increased autophagic flux fuels mitochondria with certain triacylglycerols, thereby suppressing UPRmt in fzo-1(tm1133) and drp-1(tm1108) mutants (Haeussler et al. 2020). Thus, blocking mitochondrial dynamics may reduce the flux of lipids into mitochondria, which can be compensated for by the induction of autophagy and we speculate that this mechanism may also apply to eat-3(ad426) mutants. Conversely, we speculate that defects in mitochondrial homeostasis induced by a point mutation in spg-7, may exert stress to the cytosol and that this is normally compensated for by factors, such as icd-1βNAC. Knocking-down icd-1βNAC may therefore increase cytosolic stress, which in turn enhances UPRmt in spg-7(ad2249) mutants. Taking the candidates into account that have opposing UPRmt phenotypes in the secondary screens, 95% of the suppressors and 66% of the enhancers reproduce in drp-1(tm1108), while 79% of the suppressors and 57% of the enhancers reproduce in eat-3(ad426). We found the lowest overlap of candidate genes in spg-7(ad2249) mutants, with 59% of the suppressors and 60% of the enhancers reproducing in this background. Taken together, the results of the secondary screens show that there are candidates that, when depleted, act to influence UPRmt signaling in general whereas others are specific to a certain type of UPRmt induction, such as the disruption of mitochondrial dynamics.
TF enrichment analysis identifies factors with roles in development, metabolism, and oxidative stress response
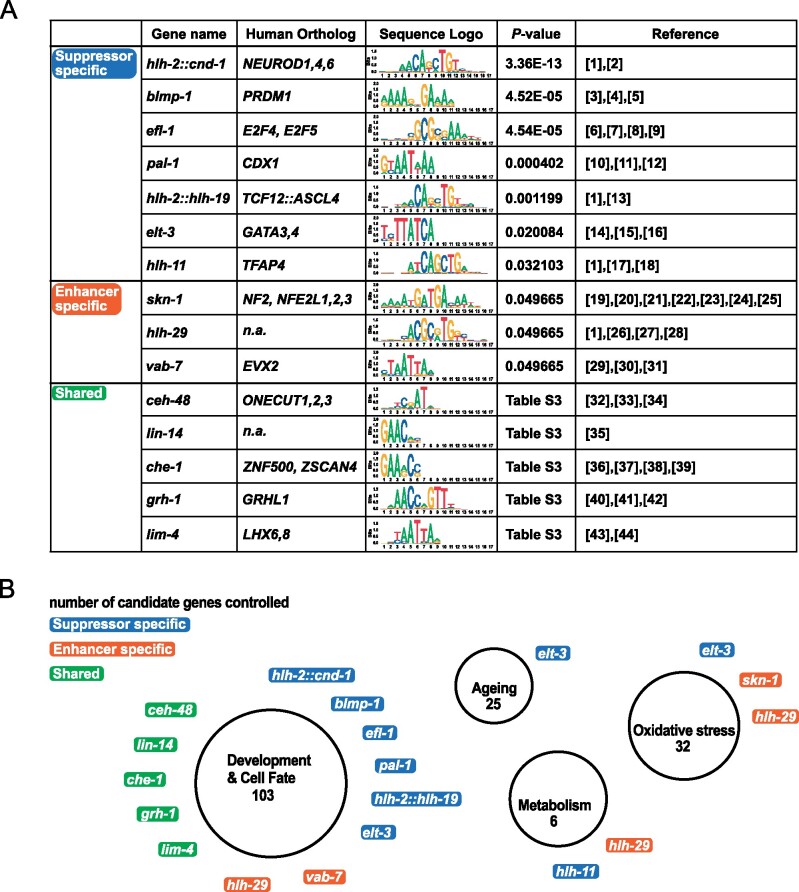
Next, we identified TF binding sites in the promoters of our candidates using ChIP-seq datasets from the modENCODE project (Celniker et al. 2009) in order to test for enrichment of TFs that bind to these sites. To that end, we used g:Profiler, a tool for functional enrichment analysis using over-representation (Raudvere et al. 2019), which utilizes TRANSFAC resources (Knüppel et al. 1994; Matys et al. 2006). Using this approach, we found 15 TFs to be enriched in a statistically significant manner (Figure 3 and Supplementary Table S3). Ten of these TFs only bind promotor regions of suppressors (7) or enhancers (3) (“suppressor- or enhancer-specific”). The remaining five TFs bind to promotor regions of both suppressors and enhancers (“shared”). The “shared” TFs have previously been implicated in cell fate determination or developmental timing. Five out of seven “suppressor specific” TFs have been shown to exclusively control developmental processes. The remaining two “suppressor-specific” TFs are ELT-3GATA3,4 and HLH-11TFAP4, which have been shown to play a role in development, aging, and the response to oxidative stress (Gilleard et al. 1999; Budovskaya et al. 2008; Hu et al. 2017) and to act as a dietary sensor that regulates metabolic gene expression, respectively (Soo-Ung et al. 2009; Watson et al. 2013).
Figure 3.
Enrichment analysis of transcription factors binding to promotors of candidate genes that suppress or enhance fzo-1(tm1133)-induced UPRmt. (A) Transcription factor (TF) binding sites were identified using the modENCODE database (Celniker et al. 2009) and enrichment analysis was performed separately for suppressors and enhancers of fzo-1(tm1133)-induced UPRmt using g:profiler (Knüppel et al. 1994; Raudvere et al. 2019). TFs that are statistically enriched among the candidate genes are shown. References: [1] (Grove et al. 2009), [2] (Hallam et al. 2000), [3] (Horn et al. 2014), [4] (Huang et al. 2014), [5] (Armakola and Ruvkun 2019), [6] (Ceol and Horvitz 2001), [7] (Garbe et al. 2004), [8] (Chi and Reinke 2006), [9] (Miller et al. 2016), [10] (Baugh et al. 2005), [11] (Maduro et al. 2005), [12] (Lei et al. 2009), [13] (Schwarz et al. 2012), [14] (Gilleard et al. 1999), [15] (Budovskaya et al. 2008), [16] (Hu et al. 2017), [17] (Soo-Ung et al. 2009), [18] (Watson et al. 2013), [19] (An and Blackwell, 2003), [20] (An et al. 2005), [21] (Inoue et al. 2005), [22] (Nargund et al. 2012), [23] (Nargund et al. 2015), [24] (Kim and Sieburth 2018), [25] (Wu et al. 2018) [26] (Neves and Priess 2005), [27] (McMiller et al. 2007), [28] (Quach et al. 2013), [29] (Ahringer, 1996), [30] (Esmaeili et al. 2002), [31] (Pocock et al. 2004), [32] (Jacquemin et al. 2003), [33] (Furuno et al. 2008), [34] (Klimova et al. 2015), [35] (Ambros and Horvitz 1984), [36] (Chang et al. 2003), [37] (Uchida et al. 2003), [38] (Etchberger et al. 2007), [39] (Rahe and Hobert, 2019), [40] (Huang et al. 1995), [41] (Wilanowski et al. 2002), [42] (Venkatesan et al. 2003), [43] (Pradel et al. 2007), [44] (Kim et al. 2015). (B) Graphical representation of enriched TFs and the cellular processes they control. “Suppressor specific” TFs are indicated in blue, “enhancer specific” TFs in orange and “shared” TFs in green. The number of candidate genes controlled by a certain group of TFs is indicated in each circle below the functional group name.
Three TFs (SKN-1NFE2, NFE2L1,2,3, HLH-29, and VAB-7EVX2) were identified to be “enhancer-specific” (Figure 3 and Supplementary Table S3). VAB-7EVX2 and HLH-29 are both required for certain aspects of development (Ahringer 1996; Esmaeili et al. 2002; Pocock et al. 2004; Neves and Priess 2005; McMiller et al. 2007; Grove et al. 2009) and HLH-29 has additional roles in fatty acid metabolism and energy homeostasis (McMiller et al. 2007; Quach et al. 2013). Furthermore, HLH-29 and SKN-1NFE2, NFE2L1,2,3 are regulators of the oxidative stress response (An and Blackwell 2003; An et al. 2005; Inoue et al. 2005; Quach et al. 2013), and SKN-1NFE2, NFE2L1,2,3 has previously been implicated in the UPRmt pathway in C. elegans (Nargund et al. 2012; 2015; Wu et al. 2018). In summary, we identified several TFs that bind to promotors of our candidate genes, which have previously been implicated in oxidative stress response, cellular metabolism and development in C. elegans. Interestingly, fzo-1(tm1133) mutants have previously been shown to be slightly sensitive to oxidative stress and have increased levels of carbonylated proteins, a measure for oxidative damage (Yasuda et al. 2011). Moreover, in isp-1(qm150) and clk-1(qm30) mutants, both of which have increased levels of reactive oxygen species (ROS) (Van Raamsdonk et al. 2010; Yang and Hekimi 2010; Dues et al. 2017), UPRmt activation has been shown to lead to ATFS-1ATF4,5-dependent expression of genes required for detoxification of ROS (Wu et al. 2018). This induction is orchestrated by ATFS-1ATF4,5 but may, to some extent, additionally be facilitated through activation of ELT-3GATA3,4 and HLH-29, as it has previously been shown for SKN-1NFE2, NFE2L1,2,3 (Nargund et al. 2012, 2015; Wu et al. 2018). The identification of many TFs controlling developmental processes is in agreement with our finding that GO-terms related to developmental processes are enriched among our dataset. This again highlights that the activity levels of critical cellular processes and responses in somatic tissues appear to be set during development. Finally, we previously found that the induction of autophagy suppresses UPRmt in fzo-1(tm1133) mutants most likely through increased metabolic activity (Haeussler et al. 2020). In our analysis, we identified two TFs, which regulate energy homeostasis and metabolic gene expression. This supports the notion that UPRmt in fzo-1(tm1133) mutants acts to compensate for metabolic defects. In summary, we identified several TFs with roles in development, oxidative stress response and metabolism that previously have not been connected to UPRmt signaling. These TFs may be specific to UPRmt in fzo-1(tm1133) but some may generally be involved in UPRmt signaling.
Interactome of UPRmt reveals potential new regulators
In order to determine whether any of the suppressors or enhancers that we identified have previously been shown to interact with fzo-1MFN1,2 or its mammalian orthologs MFN1 or MFN2, we built a gene network containing all known interactions of fzo-1MFN1,2 and its mammalian orthologs MFN1 and MFN2. Using the interaction databases “string-db.org,” “IntAct,” “BioGRID3.5,” “Genemania,” “CCSB” and “mentha” (Warde-Farley et al. 2010; Calderone et al. 2013; Orchard et al. 2014; Rolland et al. 2014; Oughtred et al. 2019; Szklarczyk et al. 2019), we included genetic and physical interactions (but not predicted interactions or co-expression data) and uploaded them to the cytoscape software (Shannon et al. 2003) to calculate a complete interaction network. The resulting network contains 38 genes and 67 interactions (Supplementary Figure S3). None of the 10 interactors of fzo-1MFN1,2 in C. elegans was identified in our screen (turquois dots in Supplementary Figure S3). Next, we manually annotated the C. elegans orthologs of 24 interactors of Mfn1 or Mfn2 in mammals (except FAF2, MAVS, TCHP, SLC25A38 for which we did not find any orthologs in C. elegans, indicated in dark blue in Supplementary Figure S3) but again did not find any overlap between the gene network and our screen dataset (orange dots in Supplementary Figure S3). In summary, in our screen for modifiers of fzo-1(tm1133)-induced UPRmt, we did not find any previously known interactors of fzo-1MFN1,2. These could either have been missed in the RNAi screen, be essential in the fzo-1(tm1133) background or not have a function in mitochondrial homeostasis and, hence, UPRmt signaling.
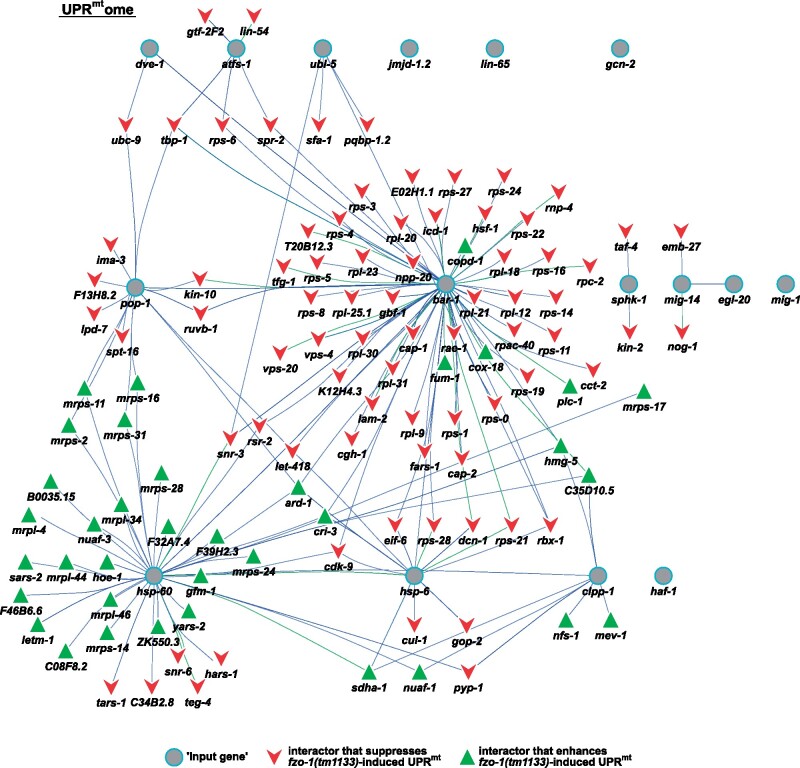
Similar to the approach described above, we used the 16 C. elegans genes currently associated with the GO-term “mitochondrial unfolded protein response” (GO: 0034514) (referred to as “input genes”), identified their human orthologs and included known physical and genetic interactors from the interaction databases “BioGRID3.5,” “IntAct,” and “mentha” (Calderone et al. 2013; Orchard et al. 2014; Oughtred et al. 2019) to calculate an interaction network containing 2603 genes and 4655 interactions (Supplementary Figures S4–S6). In this “UPRmtome,” we identified 129 genes (including the 16 “input genes”), 36 of which are enhancers and 77 of which are suppressors of fzo-1(tm1133)-induced UPRmt, with a total of 213 interactions (Figure 4 and Supplementary Table S4). For the “input gene” atfs-1ATF4,5, we found five interactors (gtf-2F2GTF2F2, lin-54LIN54, rps-6RPS6, spr-2SET, and tbp-1TBP) that suppress fzo-1(tm1133)-induced UPRmt and the gene products of four of these localize to the nucleus (Sopta et al. 1989; Lichtsteiner and Tjian 1993; Wen et al. 2000; Thomas et al. 2003; Harrison et al. 2006; Tabuchi et al. 2011). These could potentially facilitate or directly be involved in the transcription of UPRmt effectors upon activation of the UPRmt response. Moreover, for the “input gene” ubl-5UBL5, we found four interactors that overlap with our dataset of suppressors, three of which are splicing factors (pqbp-1.2PQBP1, sfa-1SF1, snr-3SNRPD1) (Thomas et al. 1988; Krämer, 1992; Arning et al. 1996; Imafuku et al. 1998; Kambach et al. 1999; Mazroui et al. 1999; Waragai et al. 1999). Of note, HUB1, the ortholog of UBL-5UBL5 in Saccharomyces pombe, has been shown to interact with components of the spliceosome. Furthermore, the loss of HUB1 results in reduced splicing efficiency of a variety of mRNAs (Wilkinson et al. 2004). However, in C. elegans, ubl-5(RNAi) has previously been reported to not cause splicing defects (Haynes et al. 2007). Thus, the identification of the splicing factor genes pqbp-1.2PQBP1, sfa-1SF1, snr-3SNRPD1 in our dataset presents an interesting potential link between UPRmt activation and pre-mRNA splicing via UBL-5UBL5. In addition, we identified taf-4TAF4, which encodes an associated factor of transcription factor TFIID, to interact with the “input gene” sphk-1SPHK1,2 and to suppress fzo-1(tm1133)-induced UPRmt upon knock-down. taf-4TAF4 has previously been shown to be required for life span extension in isp-1(qm150), clk-1(qm30) and tpk-1(qm162) mutants, (Walker et al. 2001, 2004; Khan et al. 2013). Finally, we identified many genes interacting with the “input gene” bar-1JUP, CTNNB1, which has previously been shown to be involved in cell non-autonomous propagation of UPRmt signaling (Zhang et al. 2018). Among these interactors is phospholipase C (plc-1PLCE), which enhances fzo-1(tm1133)-induced UPRmt and plays a central role in the inositol triphosphate (IP3) signaling pathway (Clandinin et al. 1998; Kariya et al. 2004). In summary, we identified several genes in our dataset using gene network analysis that have previously not been identified to play a role in UPRmt signaling in C. elegans. The genes with roles in pre-mRNA splicing and IP3 signaling may be particularly interesting in this respect. Furthermore, we propose that these genes may directly influence UPRmt signaling through interactions with known players of the UPRmt pathway.
Figure 4.
Analysis of a gene network—the UPRmtome. Interactors of all genes that are currently associated with the GO-term “mitochondrial unfolded protein response” and of their human orthologs were identified to build the complete UPRmtome using “IntAct,” “BioGRID3.5” and “mentha” databases (Calderone et al. 2013; Orchard et al. 2014; Oughtred et al. 2019). One hundred and twenty-nine genes are depicted, which overlapped between the complete UPRmtome and the candidate list of our screen in fzo-1(tm1133) mutants. Turquois circles: “input genes” currently associated with GO-term “mitochondrial unfolded protein response,” red arrowheads: suppressors of fzo-1(tm1133)-induced UPRmt that overlap with the complete UPRmtome, green triangles: enhancers of fzo-1(tm1133)-induced UPRmt that overlap with the complete UPRmtome. Interactions of two genes that were identified for C. elegans genes are indicated with green lines, interactions that were identified in human orthologs are indicated with blue lines.
Interactome analysis reveals involvement of IP3 signaling pathway in UPRmt regulation in fzo-1(tm1133)
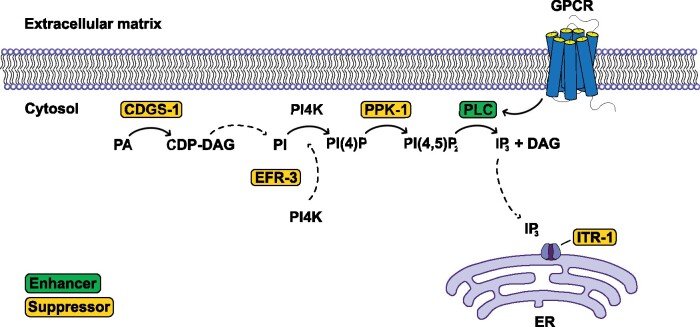
In our gene network analysis, we identified plc-1PLCE, which encodes phospholipase C, as an interactor of bar-1β-catenin (Byrne et al. 2007). Interestingly, we and others found several genes that play a role in inositol triphosphate (IP3) signaling (Figure 5) (Liu et al. 2014). The IP3 pathway is well known for its role in the regulation of intracellular calcium levels and transmits signals from the extracellular space via GPCRs and second messengers to the ER (Berridge 2009). Thus, this signaling pathway may have a role in cell non-autonomous propagation of UPRmt. We identified the enzyme CDGS-1CDS1, which is essential for the production of phosphatidylinositol (PI) (Wu et al. 1995; Vance, 1998), and EFR-3EFR3B, which targets PI-4-kinase (PI4K) to the plasma membrane (Nakatsu et al. 2012). Furthermore, we identified the sole type I PIP kinase in C. elegans, PPK-1PIP5K1A (Weinkove et al. 2008), which phosphorylates PI4P to form PI(4,5)P2 (Ishihara et al. 1996; Loijens and Anderson 1996). PLC-1PLCE is activated via GPCR and hydrolyzes PI(4,5)P2 to generate the second messengers DAG and IP3, known regulators of several signal transduction pathways (Clandinin et al. 1998; Kariya et al. 2004). One mechanism that is dependent on IP3-signaling is the release of calcium from the ER (Clandinin et al. 1998; Kariya et al. 2004; Kovacevic et al. 2013). Interestingly, the IP3 receptor at the ER, ITR-1ITPR1, has previously also been identified as a suppressor of antimycin-induced UPRmt (Liu et al. 2014). Thus, it is tempting to speculate that altering IP3 signaling influences cellular calcium signaling in fzo-1(tm1133), thereby affecting mitochondrial homeostasis and consequently UPRmt signaling. Moreover, we propose that the effect on UPRmt signaling may be indirect since we previously showed that knock-down of mitochondrial genes controlling calcium homeostasis does not induce UPRmt in wild type (Rolland et al. 2019). Furthermore, we propose that fzo-(tm1133) mutants may be more prone to changes in IP3 signaling and, consequently, calcium signaling since these mutants may have altered ER-mitochondria contact sites, as shown in tissue culture cells lacking the mammalian ortholog MFN2 (de Brito and Scorrano 2008; Cosson et al. 2012; Filadi et al. 2015, 2016; Leal et al. 2016; Naon et al. 2016; Basso et al. 2018).
Figure 5.
Candidate genes with roles in IP3 signaling. We identified four genes in our dataset that either play a direct role in the IP3 signaling pathway or are crucial for the synthesis of phosphatidylinositol-4,5-biphosphate [PI(4,5)P2]. The IP3 receptor has previously been identified (Liu et al. 2014). Suppressors are shown in yellow boxes, enhancers in green boxes. PA, phosphatidic acid; CDP-DAG, cytidine biphosphate-diacylglycerol; PI, phosphatidylinositol; PI(4), P phosphatidylinositol-4-phosphate; IP3, inositol triphosphate; ER, endoplasmic reticulum; GPCR, G-protein coupled receptor.
miga-1(tm3621) mutants show mitochondrial fragmentation and induce UPRmt
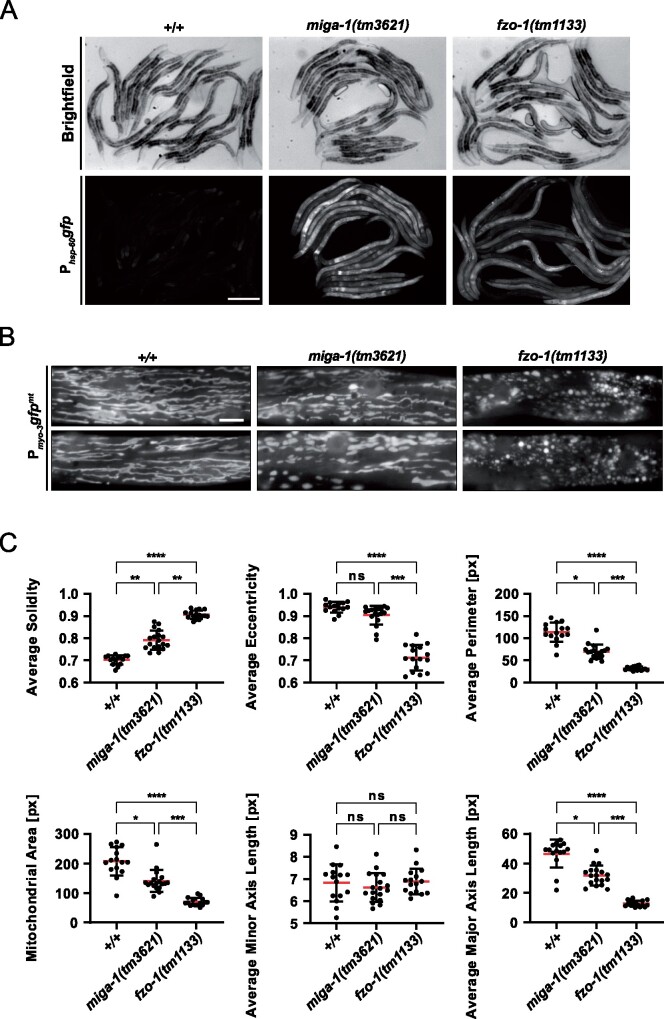
One of the enhancers we identified is K01D12.6, which is conserved from C. elegans to humans. The D. melanogaster ortholog of this gene has previously been identified in a screen for genes, which when knocked-down induce photoreceptor cell neurodegeneration. Furthermore, it was shown to be required for the maintenance of mitochondrial morphology and hence, named “Mitoguardin” (Zhang et al. 2016). Moreover, the two orthologs of this gene in mammals (MIGA1, MIGA2) were found to regulate mitochondrial fusion and to be critical for mitochondrial function in human tissue culture cells and in mice (Liu et al. 2016; Zhang et al. 2016; Liu et al. 2017). Therefore, we named K01D12.6 “mitoguardin homolog-1 (miga-1).” We verified UPRmt induction using the Phsp-60HSPD1gfp (zcIs9) reporter in the miga-1(tm3621) mutant background (Figure 6A). On average, the induction of Phsp-60HSPD1gfp is higher in miga-1(tm3621) animals compared to fzo-1(tm1133) animals. Moreover, we tested the effects of miga-1(tm3621) on steady-state mitochondrial morphology, which, in C. elegans, is carried out using a mitochondrial matrix-targeted GFP under a promoter that expresses the transgene in body wall muscle cells (Pmyo-3MYHgfpmt) (Labrousse et al. 1999; Ichishita et al. 2008; Rolland et al. 2013). While wild-type worms show a tubular network of mitochondria, miga-1(tm3621) mutants have a “fragmented mitochondria” phenotype, which is less severe than that caused by the loss of fzo-1 (Figure 6B). In addition, we analyzed mitochondrial morphology using the MitoSegNet algorithm (Fischer et al. 2020) and confirmed the “fragmented mitochondria” phenotype of miga-1(tm3621) mutants. Specifically, for most of the shape descriptors analyzed, miga-1(tm3621) mutants were statistically different from wild type but distinct from fzo-1(tm1133) mutants (Figure 6C). In summary and in line with previous observations in other organisms, we see drastic changes in mitochondrial morphology in miga-1(tm3621) mutants, which are accompanied by the induction of UPRmt.
Figure 6.
miga-1(tm3621) mutants induce UPRmt and have altered mitochondrial morphology. (A) Fluorescence images of L4 larvae expressing Phsp-60mtHSPD1gfp (zcIs9) in wild type (+/+), miga-1(tm3621) or fzo-1(tm1133) mutants. Scale bar: 200 µm. (B) Fluorescence images of L4 larvae expressing mitochondrial targeted gfp (Pmyo-3gfpmt) in wild type (+/+), miga-1(tm3621) or fzo-1(tm1133) mutants. Representative images are shown. Scale bar: 10 µm. (C) Fluorescence images of L4 larvae expressing mitochondrial targeted gfp (Pmyo-3gfpmt) in wild-type (+/+), miga-1(tm3621), or fzo-1(tm1133) mutants were quantified using the MitoSegNet algorithm (Fischer et al. 2020). ns: not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 using Kruskal–Wallis test with Dunn’s post hoc test for multiple comparison among all three genotypes, n ≥ 15. px: pixel.
Acknowledgments
The authors thank members of the Conradt lab and the “Mito Club” for lively discussions. We thank M. Bauer, L. Jocham, N. Lebedeva, and M. Schwarz for excellent technical support and S. Mitani (National BioResource Project, Tokyo, Japan) for fzo-1(tm1133), drp-1(tm1108), and miga-1(tm3621). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding
This work was supported by funding from the Deutsche Forschungsgemeinschaft (CO204/6-1 and CO204/9-1 to B.C. and RO5352/1-1 to S.G.R.), the Institute for Basic Science (IBS-R022-A2-2020 to S.G.R) and a Royal Society Wolfson Fellowship (RSWF\R1\180008 to B.C.).
Conflicts of interest
The authors declare no conflicts of interest.
Literature cited
- Ackema KB, Hench J, Böckler S, Wang SC, Sauder U, et al. 2014. The small GTPase Arf1 modulates mitochondrial morphology and function. EMBO J. 33:2659–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Romero A, Kaminska J, Spang A, Riezman H, Muñiz M.. 2008. The yeast p24 complex is required for the formation of COPI retrograde transport vesicles from the Golgi apparatus. J Cell Biol. 180:713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahringer J. 1996. Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes Dev. 10:1120–1130. [DOI] [PubMed] [Google Scholar]
- Altmann K, Westermann B.. 2005. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 16:5410–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Bocchini CA, Scott AF, Hamosh A.. 2019. OMIM.org: leveraging knowledge across phenotype–gene relationships. Nucleic Acids Res. 47:D1038–D1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR.. 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science . 226:409–416. [DOI] [PubMed] [Google Scholar]
- An JH, Blackwell TK.. 2003. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17:1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, et al. 2005. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci USA. 102:16275–16280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda A, Pascual A.. 2001. Nuclear hormone receptors and gene expression. Physiol Rev. 81:1269–1304. [DOI] [PubMed] [Google Scholar]
- Armakola M, Ruvkun G.. 2019. Regulation of Caenorhabditis elegans neuronal polarity by heterochronic genes. Proc Natl Acad Sci USA. 116:12327–12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arning S, Grüter P, Bilbe G, Krämer A.. 1996. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 2:794–810. [PMC free article] [PubMed] [Google Scholar]
- Arsenovic PT, Maldonado AT, Colleluori VD, Bloss TA.. 2012. Depletion of the C. elegans NAC engages the unfolded protein response, resulting in increased chaperone expression and apoptosis. PLoS One. 7:e44038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso V, Marchesan E, Peggion C, Chakraborty J, von Stockum S, et al. 2018. Regulation of ER-mitochondria contacts by Parkin via Mfn2. Pharmacol Res. 138:43–56. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, et al. 2005. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development. 132:1843–1854. [DOI] [PubMed] [Google Scholar]
- Beck R, Rawet M, Ravet M, Wieland FT, Cassel D.. 2009. The COPI system: molecular mechanisms and function. FEBS Lett. 583:2701–2709. [DOI] [PubMed] [Google Scholar]
- Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, et al. 2014. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat Commun. 5:3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. 2009. Inositol trisphosphate and calcium signalling mechanisms. Biochim Biophys Acta. 1793:933–940. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, et al. 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1:298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, et al. 2010. Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology. 51:642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The Genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik A, Stephens DJ.. 2009. ER exit sites – Localization and control of COPII vesicle formation. FEBS Lett. 583:3796–3803. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, et al. 2008. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 134:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne AB, Weirauch MT, Wong V, Koeva M, Dixon SJ, et al. 2007. A global analysis of genetic interactions in Caenorhabditis elegans. J Biol. 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone A, Castagnoli L, Cesareni G.. 2013. mentha: a resource for browsing integrated protein-interaction networks. Nat Methods. 10:690–691. [DOI] [PubMed] [Google Scholar]
- Cambier L, Rassam P, Chabi B, Mezghenna K, Gross R, et al. 2012. M19 modulates skeletal muscle differentiation and insulin secretion in pancreatic β-cells through modulation of respiratory chain activity. PLoS One. 7:e31815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, et al. 2009. Unlocking the secrets of the genome. Nature. 459:927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR.. 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize ras signaling in C. elegans vulval development. Mol Cell. 7:461–473. [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Hobert O.. 2003. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17:2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC.. 2004. Mitochondrial dynamics in mammals. Curr Top Dev Biol. 59:119–144. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, et al. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 160:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Vasoya RP, Toke NH, Parthasarathy A, Luo S, et al. (2020). HNF4 regulates fatty acid oxidation and is required for renewal of intestinal stem cells in mice. Gastroenterology. 158, 985–999.e989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Reinke V.. 2006. Promotion of oogenesis and embryogenesis in the “C. elegans” gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB). Development. 133:3147–3157. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW.. 1998. Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell. 92:523–533. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P.. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 241:779–786. [DOI] [PubMed] [Google Scholar]
- Cosson P, Marchetti A, Ravazzola M, Orci L.. 2012. Mitofusin-2 Independent juxtaposition of endoplasmic reticulum and mitochondria: an ultrastructural study. PLoS One. 7:e46293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutz CE, Tomsig JL, Snyder SL, Gautier MC, Skouri F, et al. 1998. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J Biol Chem. 273:1393–1402. [DOI] [PubMed] [Google Scholar]
- Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, et al. 2002. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J Biol Chem. 277:21431–21439. [DOI] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L.. 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 456:605–610. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Duda K, Iwamoto M, Chi Y-I, Shoelson SE.. 2002. Crystal structure of the HNF4α ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 277:37973–37976. [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, von Zastrow M.. 2014. Endocytosis, signaling, and beyond. Cold Spring Harb Perspect Biol. 6:a016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu A-L, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, et al. 2002. Rates of behavior and aging specified by mitochondrial function during development. Science. 298:2398–2401. [DOI] [PubMed] [Google Scholar]
- Duda K, Chi Y-I, Shoelson SE.. 2004. Structural basis for HNF-4α activation by ligand and coactivator binding. J Biol Chem. 279:23311–23316. [DOI] [PubMed] [Google Scholar]
- Dues DJ, Schaar CE, Johnson BK, Bowman MJ, Winn ME, et al. 2017. Uncoupling of oxidative stress resistance and lifespan in long-lived isp-1 mitochondrial mutants in Caenorhabditis elegans. Free Radic Biol Med. 108:362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A.. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 144:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W.. 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili B, Ross JM, Neades C, Miller DM, Ahringer J.. 2002. The C. elegans even-skipped homologue vab-7 specifies DB motoneurone identity and axon trajectory. Development. 129:853. [DOI] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, et al. 2007. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21:1653–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H, Rabouille C.. 2011. Signalling to and from the secretory pathway. J Cell Sci. 124:171–180. [DOI] [PubMed] [Google Scholar]
- Filadi R, Greotti E, Turacchio G, Luini A, Pozzan T, et al. 2015. Mitofusin 2 ablation increases endoplasmic reticulum–mitochondria coupling. Proc Natl Acad Sci USA. 112:E2174–E2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filadi R, Greotti E, Turacchio G, Luini A, Pozzan T, et al. 2016. Presenilin 2 modulates endoplasmic reticulum-mitochondria coupling by tuning the antagonistic effect of mitofusin 2. Cell Rep. 15:2226–2238. [DOI] [PubMed] [Google Scholar]
- Fischer CA, Besora-Casals L, Rolland SG, Haeussler S, Singh K, et al. 2020. MitoSegNet: easy-to-use deep learning segmentation for analyzing mitochondrial morphology. iScience. 23:101601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno K, Ikeda K, Hamano S, Fukuyama K, Sonoda M, et al. 2008. Onecut transcription factor OC2 is a direct target of T-bet in type-1 T-helper cells. Genes Immun. 9:302–308. [DOI] [PubMed] [Google Scholar]
- Garbe D, Doto JB, Sundaram MV.. 2004. Caenorhabditis elegans lin-35/Rb, efl-1/E2F and other synthetic multivulva genes negatively regulate the anaphase-promoting complex gene mat-3/APC8. Genetics. 167:663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin KA, Hidaka M, Stillman B.. 1995. Conserved initiator proteins in eukaryotes. Science. 270:1667–1671. [DOI] [PubMed] [Google Scholar]
- Gilleard JS, Shafi Y, Barry JD, McGhee JD.. 1999. ELT-3: A Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Dev Biol. 208:265–280. [DOI] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, et al. 2009. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 138:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler S, Köhler F, Witting M, Premm MF, Rolland SG, et al. 2020. Autophagy compensates for defects in mitochondrial dynamics. PLoS Genet. 16:e1008638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales KG, Fuller MT.. 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 90:121–129. [DOI] [PubMed] [Google Scholar]
- Hallam S, Singer E, Waring D, Jin Y.. 2000. The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development. 127:4239. [DOI] [PubMed] [Google Scholar]
- Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, et al. 2020. WormBase: a modern Model Organism Information Resource. Nucleic Acids Res. 48:D762–D767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Ceol CJ, Lu X, Horvitz HR.. 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci USA. 103:16782–16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D.. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 13:467–480. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D.. 2010. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 37:529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz R, Magenheim J, Berman I, Bar-Tana J.. 1998. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4α. Nature. 392:512–516. [DOI] [PubMed] [Google Scholar]
- Horn M, Geisen C, Cermak L, Becker B, Nakamura S, et al. 2014. DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Dev Cell. 28:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, et al. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497:451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, D'Amora DR, MacNeil LT, Walhout AJM, Kubiseski TJ.. 2017. The oxidative stress response in Caenorhabditis elegans requires the GATA transcription factor ELT-3 and SKN-1/Nrf2. Genetics. 206:1909–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, et al. 2007. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JD, Dubnicoff T, Liaw GJ, Bai Y, Valentine SA, et al. 1995. Binding sites for transcription factor NTF-1/Elf-1 contribute to the ventral repression of decapentaplegic. Genes Dev. 9:3177–3189. [DOI] [PubMed] [Google Scholar]
- Huang T-F, Cho C-Y, Cheng Y-T, Huang J-W, Wu Y-Z, et al. 2014. BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 10:e1004428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SE, McLaren W, Gil L, Thormann A, Schuilenburg H, et al. 2018. Ensembl Variation Resources. Database 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichishita R, Tanaka K, Sugiura Y, Sayano T, Mihara K, et al. 2008. An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. J Biochem. 143:449–454. [DOI] [PubMed] [Google Scholar]
- Imafuku I, Waragai M, Takeuchi S, Kanazawa I, Kawabata M, et al. 1998. Polar Amino acid-rich sequences bind to polyglutamine tracts. Biochem Biophys Res Commun. 253:16–20. [DOI] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, et al. 2005. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 170:1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, et al. 2005. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 19:2278–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, Shibasaki Y, Kizuki N, Katagiri H, Yazaki Y, et al. 1996. Cloning of cDNAs Encoding Two Isoforms of 68-kDa Type I Phosphatidylinositol4-phosphate 5-Kinase. J Biol Chem. 271:23611–23614. [DOI] [PubMed] [Google Scholar]
- Izumi K, Brett M, Nishi E, Drunat S, Tan E-S, et al. 2016. ARCN1 mutations cause a recognizable craniofacial syndrome due to COPI-mediated transport defects. Am J Hum Genet. 99:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P, Lemaigre FP, Rousseau GG.. 2003. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev Biol. 258:105–116. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods .30:313–321. [DOI] [PubMed] [Google Scholar]
- Kambach C, Walke S, Nagai K.. 1999. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr Opin Struct Biol. 9:222–230. [DOI] [PubMed] [Google Scholar]
- Kanazawa T, Zappaterra MD, Hasegawa A, Wright AP, Newman-Smith ED, et al. 2008. The C. elegans opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 4:e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya K-I, Kim Bui Y, Gao X, Sternberg PW, Kataoka T.. 2004. Phospholipase Cɛ regulates ovulation in Caenorhabditis elegans. Dev Biol. 274:201–210. [DOI] [PubMed] [Google Scholar]
- Khan MH, Ligon M, Hussey LR, Hufnal B, Farber R, et al. 2013. TAF-4 is required for the life extension of isp-1, clk-1 and tpk-1 Mit mutants. Aging (Albany NY). 5:741–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yeon J, Choi S-K, Huh YH, Fang Z, et al. 2015. The evolutionarily conserved LIM homeodomain protein LIM-4/LHX6 specifies the terminal identity of a cholinergic and peptidergic C. elegans Sensory/Inter/Motor Neuron-Type. PLoS Genet. 11:e1005480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sieburth D.. 2018. Sphingosine kinase activates the mitochondrial unfolded protein response and is targeted to mitochondria by stress. Cell Reports. 24, 2932–2945.e2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sieburth D.. 2020. FSHR-1/GPCR regulates the mitochondrial unfolded protein response in Caenorhabditis elegans. Genetics. 214:409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Underwood RS, Greenwald I, Shaye DD.. 2018. OrthoList 2: a new comparative genomic analysis of human and Caenorhabditis elegans genes. Genetics. 210:445–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein-Miles J, Scior A, Deuerling E, Morimoto RI.. 2013. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 32:1451–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimova L, Antosova B, Kuzelova A, Strnad H, Kozmik Z.. 2015. Onecut1 and Onecut2 transcription factors operate downstream of Pax6 to regulate horizontal cell development. Dev Biol. 402:48–60. [DOI] [PubMed] [Google Scholar]
- Knüppel R, Dietze P, Lehnberg W, Frech K, Wingender E.. 1994. TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J Comput Biol. 1:191–198. [DOI] [PubMed] [Google Scholar]
- Kornmann B. 2014. Quality control in mitochondria: use it, break it, fix it, trash it. F1000Prime Rep. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic I, Orozco JM, Cram EJ.. 2013. Filamin and Phospholipase C-ε are required for calcium signaling in the Caenorhabditis elegans Spermatheca. PLoS Genet. 9:e1003510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. 1992. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol Cell Biol. 12:4545–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, et al. 2019. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47:D807–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Nakano A.. 2019. The ER exit sites are specialized ER zones for the transport of cargo proteins from the ER to the Golgi apparatus. J Biochem. 165:109–114. [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM.. 1999. C. elegans Dynamin-Related Protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell 4:815–826. [DOI] [PubMed] [Google Scholar]
- Leal NS, Schreiner B, Pinho CM, Filadi R, Wiehager B, et al. 2016. Mitofusin-2 knockdown increases ER–mitochondria contact and decreases amyloid β-peptide production. J Cell Mol Med. 20:1686–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R.. 2004. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 20:87–123. [DOI] [PubMed] [Google Scholar]
- Lei H, Liu J, Fukushige T, Fire A, Krause M.. 2009. Caudal-like PAL-1 directly activates the bodywall muscle module regulator hlh-1 in C. elegans to initiate the embryonic muscle gene regulatory network. Development. 136:1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtsteiner S, Tjian R.. 1993. Cloning and properties of the Caenorhabditis elegans TATA-box-binding protein. Proc Natl Acad Sci USA. 90:9673–9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-M, Zhang Y-P, Ji S-Y, Li B-T, Tian X, et al. 2016. Mitoguardin-1 and -2 promote maturation and the developmental potential of mouse oocytes by maintaining mitochondrial dynamics and functions. Oncotarget. 7:1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-M, Zhang Y-L, Ji S-Y, Zhao L-W, Shang W-N, et al. 2017. Mitochondrial function regulated by mitoguardin-1/2 is crucial for ovarian endocrine functions and ovulation. Endocrinology. 158:3988–3999. [DOI] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, Ruvkun G.. 2014. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 508:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loijens JC, Anderson RA.. 1996. Type I Phosphatidylinositol-4-phosphate 5-Kinases are distinct members of this novel lipid kinase family. J Biol Chem. 271:32937–32943. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Kasmir JJ, Zhu J, Rothman JH.. 2005. The WNT effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev Biol. 285:510–523. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, et al. 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34:D108–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Puoti A, KrÄMer A.. 1999. Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA. 5:1615–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMiller TL, Sims D, Lee T, Williams T, Johnson CM.. 2007. Molecular characterization of the Caenorhabditis elegans REF-1 family member, hlh-29/hlh-28. Biochim Biophys Acta. 1769:5–19. [DOI] [PubMed] [Google Scholar]
- Melo JA, Ruvkun G.. 2012. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 149:452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JG, Liu Y, Williams CW, Smith HE, O’Connell KF.. 2016. The E2F-DP1 transcription factor complex regulates centriole duplication in Caenorhabditis elegans. G3 (Bethesda). 6:709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, et al. 2012. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol. 199:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naon D, Zaninello M, Giacomello M, Varanita T, Grespi F, et al. 2016. Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum–mitochondria tether. Proc Natl Acad Sci USA. 113:11249–11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM.. 2015. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPRmt. Mol Cell 58:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM.. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 337:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves A, Priess JR.. 2005. The REF-1 Family of bHLH transcription factors pattern C. elegans embryos through Notch-Dependent and Notch-Independent Pathways. Dev Cell 8:867–879. [DOI] [PubMed] [Google Scholar]
- Ohta S, Tatsumi Y, Fujita M, Tsurimoto T, Obuse C.. 2003. The ORC1 cycle in human cells: II. Dynamic changes in the human ORC complex during the cell cycle. J Biol Chem. 278:41535–41540. [DOI] [PubMed] [Google Scholar]
- Oks O, Lewin S, Goncalves IL, Sapir A.. 2018. The UPRmt Protects Caenorhabditis elegans from mitochondrial dysfunction by upregulating specific enzymes of the mevalonate pathway. Genetics. 209:457–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, et al. 2014. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42:D358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, et al. 1998. The Dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 143:333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred R, Stark C, Breitkreutz B-J, Rust J, Boucher L, et al. 2019. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47:D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS.. 2009. Drosophila HNF4 regulates lipid mobilization and β-Oxidation. Cell Metab. 9:228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocock R, Ahringer J, Mitsch M, Maxwell S, Woollard A.. 2004. A regulatory network of T-box genes and the even-skipped homologue vab-7 controls patterning and morphogenesis in C. elegans. Development. 131:2373–2385. [DOI] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, et al. 2007. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci USA. 104:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach TK, Chou HT, Wang K, Milledge GZ, Johnson CM.. 2013. Genome-wide microarrray analysis reveals roles for the REF-1 family member HLH-29 in ferritin synthesis and peroxide stress response. PLoS One. 8:e59719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahe DP, Hobert O.. 2019. Restriction of cellular plasticity of differentiated cells mediated by chromatin modifiers, transcription factors and protein kinases. G3 (Bethesda). 9:2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey CS, Yeung F, Stoddard PB, Li D, Creutz CE, et al. 2008. Copine-I represses NF-kappaB transcription by endoproteolysis of p65. Oncogene. 27:3516–3526. [DOI] [PubMed] [Google Scholar]
- Ranji P, Rauthan M, Pitot C, Pilon M.. 2014. Loss of HMG-CoA Reductase in C. elegans causes defects in protein prenylation and muscle mitochondria. PLoS One. 9:e100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, et al. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M, Ranji P, Aguilera Pradenas N, Pitot C, Pilon M.. 2013. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc Natl Acad Sci USA. 110:5981–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE.. 2007. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 5:e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland SG, Motori E, Memar N, Hench J, Frank S, et al. 2013. Impaired complex IV activity in response to loss of LRPPRC function can be compensated by mitochondrial hyperfusion. Proc Natl Acad Sci USA. 110:E2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland SG, Schneid S, Schwarz M, Rackles E, Fischer C, et al. 2019. Compromised mitochondrial protein import acts as a signal for UPRmt. Cell Rep 28, 1659–1669.e1655. [DOI] [PubMed] [Google Scholar]
- Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, et al. 2014. A proteome-scale map of the human interactome network. Cell. 159:1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel ED, Liu S, Baumeister R, Schulze E.. 2013. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 9:e1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Frank S, Gaume B, Herrler M, Youle RJ, et al. 2003. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 116:2763–2774. [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Kato M, Sternberg PW.. 2012. Functional transcriptomics of a migrating cell in Caenorhabditis elegans. Proc Natl Acad Sci USA. 109:16246–16251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafqat N, Marschall H-U, Filling C, Nordling E, Wu X-Q, et al. 2003. Expanded substrate screenings of human and Drosophila type 10 17β-hydroxysteroid dehydrogenases (HSDs) reveal multiple specificities in bile acid and steroid hormone metabolism: characterization of multifunctional 3α/7α/7β/17β/20β/21-HSD. Biochem J. 376:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L-W, Niu R, Liu Y.. 2016. Neuropeptide signals cell non-autonomous mitochondrial unfolded protein response. Cell Res. 26:1182–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I.. 2011. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One. 6:e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Yaffe MP.. 1999. The yeast dynamin-like Protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J Cell Biol. 144:711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Xu S, Xing S, Yao K, Zhang L, et al. 2019. Mettl17, a regulator of mitochondrial ribosomal RNA modifications, is required for the translation of mitochondrial coding genes. FASEB J. 33:13040–13050. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PVE, et al. 2003. Genome-Wide RNAi of C. elegans Using the Hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis N, Rual J-F, Carvunis A-R, Tasan M, Lemmens I, et al. 2009. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 6:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland D-L, Ryazantsev SN, van der Bliek AM.. 1998. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 143:351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo-Ung L, Hyun-Ok S, Wonhae L, Gunasekaran S, Jae-Ran Y, et al. 2009. Identification and Characterization of a Putative Basic Helix-Loop-Helix (bHLH) Transcription Factor Interacting with Calcineurin in C. elegans. Mol Cells 28:455–461. [DOI] [PubMed] [Google Scholar]
- Sopta M, Burton ZF, Greenblatt J.. 1989. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 341:410–414. [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M.. 2009. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 10:609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan AN, Ijlst L, van Roermund CWT, Wijburg FA, Wanders RJA, et al. 2005. Identification of the human mitochondrial FAD transporter and its potential role in multiple acyl-CoA dehydrogenase deficiency. Mol Genet Metab. 86:441–447. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, et al. 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi TM, Deplancke B, Osato N, Zhu LJ, Barrasa MI, et al. 2011. Chromosome-Biased Binding and Gene Regulation by the Caenorhabditis elegans DRM Complex. PLoS Genet. 7:e1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C.. 2003. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J Biol Chem. 278:41528–41534. [DOI] [PubMed] [Google Scholar]
- Tatsuta T, Langer T.. 2008. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 27:306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Alliance of Genome Resources 2019. Alliance of genome resources portal: unified model organism research platform. Nucleic Acids Res. 48:D650–D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Conrad RC, Blumenthal T.. 1988. The C. elegans Trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell. 54:533–539. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Ceol CJ, Schwartz HT, Horvitz HR.. 2003. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics. 164:135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsig JL, Snyder SL, Creutz CE.. 2003. Identification of targets for calcium signaling through the copine family of proteins. Characterization of a coiled-coil copine-binding motif. J Biol Chem. 278:10048–10054. [DOI] [PubMed] [Google Scholar]
- Tomsig JL, Sohma H, Creutz CE.. 2004. Calcium-dependent regulation of tumour necrosis factor-alpha receptor signalling by copine. Biochem J. 378:1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida O, Nakano H, Koga M, Ohshima Y.. 2003. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 130:1215–1224. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium 2018. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM, Shen Q, Kawajiri S.. 2013. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 5:a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Meng Y, Camp D, Yang W, Jia X, et al. 2010. Decreased energy metabolism extends life span in Caenorhabditis elegans without reducing oxidative damage. Genetics. 185:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE. 1998. Eukaryotic lipid-biosynthetic enzymes: the same but not the same. Trends Biochem Sci. 23:423–428. [DOI] [PubMed] [Google Scholar]
- Venkatesan K, McManus HR, Mello CC, Smith TF, Hansen U.. 2003. Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res. 31:4304–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Rothman JH, Shi Y, Blackwell TK.. 2001. Distinct requirements for C. elegans TAFIIs in early embryonic transcription. EMBO J. 20:5269–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Shi Y, Blackwell TK.. 2004. An extensive requirement for transcription factor IID-specific TAF-1 in Caenorhabditis elegans embryonic transcription. J Biol Chem. 279:15339–15347. [DOI] [PubMed] [Google Scholar]
- Walter W, Sánchez-Cabo F, Ricote M.. 2015. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 31:2912–2914. [DOI] [PubMed] [Google Scholar]
- Waragai M, Lammers CH, Takeuchi S, Imafuku I, Udagawa Y, et al. 1999. PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum Mol Genet. 8:977–987. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, et al. 2010. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38:W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Arda HE, Zhu LJ, Walhout AJM.. 2013. Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response. Cell. 153:253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinkove D, Bastiani M, Chessa TAM, Joshi D, Hauth L, et al. 2008. Overexpression of PPK-1, the Caenorhabditis elegans Type I PIP kinase, inhibits growth cone collapse in the developing nervous system and causes axonal degeneration in adults. Dev Biol. 313:384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C, Levitan D, Li X, Greenwald I.. 2000. spr-2, a suppressor of the egg-laying defect caused by loss of sel-12 presenilin in Caenorhabditis elegans, is a member of the SET protein subfamily. Proc Natl Acad Sci USA. 97:14524–14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilanowski T, Tuckfield A, Cerruti L, O'Connell S, Saint R, et al. 2002. A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainy head. Mech Dev. 114:37–50. [DOI] [PubMed] [Google Scholar]
- Wilkinson CRM, Dittmar GAG, Ohi MD, Uetz P, Jones N, et al. 2004. Ubiquitin-like protein Hub1 is required for Pre-mRNA splicing and localization of an essential splicing factor in fission yeast. Curr Biol. 14:2283–2288. [DOI] [PubMed] [Google Scholar]
- Wisely GB, Miller AB, Davis RG, Thornquest AD, Johnson R, et al. 2002. Hepatocyte nuclear factor 4 is a Transcription Factor that constitutively binds fatty acids. Structure. 10:1225–1234. [DOI] [PubMed] [Google Scholar]
- Witte K, Schuh AL, Hegermann J, Sarkeshik A, Mayers JR, et al. 2011. TFG-1 function in protein secretion and oncogenesis. Nat Cell Biol. 13:550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Niemeyer B, Colley N, Socolich M, Zuker CS.. 1995. Regulation of PLC-mediated signalling in vivo by CDP-diacylglycerol synthase. Nature. 373:216–222. [DOI] [PubMed] [Google Scholar]
- Wu Z, Senchuk MM, Dues DJ, Johnson BK, Cooper JF, et al. 2018. Mitochondrial unfolded protein response transcription factor ATFS-1 promotes longevity in a long-lived mitochondrial mutant through activation of stress response pathways. BMC Biol. 16:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP. 1999. The machinery of mitochondrial inheritance and behavior. Science. 283:1493–1497. [DOI] [PubMed] [Google Scholar]
- Yang W, Hekimi S.. 2010. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 8:e1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Hartman PS, Ishii T, Suda H, Akatsuka A, et al. 2011. Interrelationships between mitochondrial fusion, energy metabolism and oxidative stress during development in Caenorhabditis elegans. Biochem Biophys Res Commun. 404:751–755. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, et al. 2004. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 117:4055–4066. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM.. 2012. Mitochondrial fission, fusion, and stress. Science. 337:1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wu X, Chen P, Liu L, Xin N, et al. (2018). The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent WNT Signaling. Cell. 174, 870–883.e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu X, Bai J, Tian X, Zhao X, et al. 2016. Mitoguardin regulates mitochondrial fusion through MitoPLD and is required for neuronal homeostasis. Mol Cell 61:111–124. [DOI] [PubMed] [Google Scholar]
- Zubovych IO, Straud S, Roth MG, Newmeyer DD.. 2010. Mitochondrial dysfunction confers resistance to multiple drugs in Caenorhabditis elegans. Mol Biol Cell 21:956–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. Supplementary Figure S1 contains data about involvement of the cSADDs response in suppression of UPRmt upon attenuation of cytosolic translation. Supplementary Figure S2 shows different mutants inducing the UPRmt reporter. Supplementary Figure S3 shows the FZOome. Supplementary Figure S4 contains a subset of the UPRmtome coming from direct interactions in C. elegans. Supplementary Figure S5 depicts a subset of the UPRmtome coming from interactions of human orthologs. Supplementary Figure S6 shows the complete UPRmtome. Supplementary Table S1 contains all suppressors and enhancers of fzo-1(tm1133)-induced UPRmt identified in a genome-wide RNAi screen in C. elegans. Supplementary Table S2 contains the GO enrichment analysis of suppressors and enhancers of fzo-1(tm1133)-induced UPRmt. Supplementary Table S3 contains TF enrichment analysis of suppressors and enhancers of fzo-1(tm1133)-induced UPRmt. Supplementary Table S4 contains the results of the interactome analysis (UPRmtome). The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. The supplemental material is available at figshare: https://doi.org/10.25387/g3.14262425.