Abstract
The European hazelnut (Corylus avellana L.; 2n = 2x = 22) is a worldwide economically important tree nut that is cross-pollinated due to sporophytic incompatibility. Therefore, any individual plant is highly heterozygous. Cultivars are clonally propagated using mound layering, rooted suckers, and micropropagation. In recent years, the interest in this crop has increased, due to a growing demand related to the recognized health benefits of nut consumption. C. avellana cv “Tonda Gentile delle Langhe” (“TGdL”) is well-known for its high kernel quality, and the premium price paid for this cultivar is an economic benefit for producers in northern Italy. Assembly of a high-quality genome is a difficult task in many plant species because of the high level of heterozygosity. We assembled a chromosome-level genome sequence of “TGdL” with a two-step approach. First, 10X Genomics Chromium Technology was used to create a high-quality sequence, which was then assembled into scaffolds with cv “Tombul” genome as the reference. Eleven pseudomolecules were obtained, corresponding to 11 chromosomes. A total of 11,046 scaffolds remained unplaced, representing 11% of the genome (46,504,161 bp). Gene prediction, performed with Maker-P software, identified 27,791 genes (AED ≤0.4 and 92% of BUSCO completeness), whose function was analyzed with BlastP and InterProScan software. To characterize “TGdL” specific genetic mechanisms, Orthofinder was used to detect orthologs between hazelnut and closely related species. The “TGdL” genome sequence is expected to be a powerful tool to understand hazelnut genetics and allow detection of markers/genes for important traits to be used in targeted breeding programs.
Keywords: genomics, NGS, 10X genomics, hazelnut
Introduction
The European hazelnut (Corylus avellana L.) is a woody species belonging to the Betulaceae family. It is an economically important tree nut whose production is mostly destined to the confectionery industry with a demand that has rapidly increased (Molnar 2011). As a consequence, hazelnut harvested areas showed a 58% increase in 2019 (1,000,231 ha) compared to 2014 (FAOSTAT 2019). Hazelnut is cultivated in many countries, including Turkey (65% World production), Italy (12.5%), Azerbaijan (4.6%), USA (3.9%), Chile, China, and Georgia (Botta et al. 2019). In the Piedmont Region of Italy, hazelnut production is mainly based on the cultivar “Tonda Gentile delle Langhe” (syn. “Tonda Gentile,” “Tonda Gentile Trilobata,” hereafter “TGdL”), a small-sized and trilobate shaped kernels command a premium price due to their high quality, especially after roasting (Valentini et al. 2014). In December 1993, the European Union recognized the Protected Geographical Indication (PGI) “Nocciola Piemonte” to “TGdL” produced in the piedmont areas of northern Italy (https://eur-lex.europa.eu). “TGdL” is considered to have a monoclonal origin and it is clonally propagated by mound layering, rooted suckers, and micropropagation (Valentini et al. 2014).
High-quality genome assembly in many fruit tree species is a difficult task due to their high heterozygosity and thus haploid or doubled haploid plants have been often used to accomplish this goal (Jaillon and Aury 2007) or genomes have been assembled with specialized algorithms (e.g., Platanus, Kajitani et al. 2014). Recently, long-read sequencing technologies, such as single-molecule real-time sequencing (SMRT, Pacific Biosciences) or nanopore sequencing (Oxford Nanopore Technologies) have been adopted to face this task. Moreover, scaffolding-like technologies such as optical mapping (Bionano Genomics, Barchi et al. 2019), proximity ligation methods (Hi-C, Dovetail Genomics, Acquadro et al. 2020b), and linked-reads (10X Genomics, Hulse-Kemp 2018) are generally used as companion strategies. The latter, a low-cost approach, can also be used to improve assembly metrics and to reconstruct long-range haplotypes. The 10X Linked-Reads technique amplifies the potential of short-read sequencing to achieve a much more complete genomic analysis. Using this technology, it is possible to discriminate the two haplotypes and also to analyze regions with high repetitiveness.
European hazelnut is a diploid species with 11 chromosomes (2n = 2x = 22), with an estimated genome content (1 C) of 0.43 pg (Pustahija et al. 2013). Being an outbred species (Beltramo et al. 2016) hazelnut shows a high level of heterozygosity. In 2009, a draft genome of the European hazelnut cultivar “Jefferson” was released (https://www.cavellanagenomeportal.com), while a chromosome-scale assembly of the Turkish cv. “Tombul” was recently reported (Lucas et al. 2021). Rowley et al. (2012, 2018) studied the cv “Jefferson” transcriptome (4 tissues), and re-sequenced seven European cultivars (∼20x coverage). More recently, transcriptome sequences were obtained for C. heterophylla Fisch. (Chen et al. 2014) and C. mandshurica Maxim. (Ma et al. 2013). Mehlenbacher et al. (2006) constructed a genetic linkage map of C. avellana based on RAPD and SSR markers, while Torello Marinoni et al. (2018) developed SNP-based genetic maps for “TGdL” × “Merveille de Bollwiller,” and detected QTL regions associated with time of leaf budburst. Many studies in addition to Öztürk et al. (2018) have used SSR markers to study diversity sequencing/assembly.
There is a large gap between the tools available for other fruit species and the existing knowledge on hazelnut. This study aims to fill this gap, considering that hazelnut is a strategic crop for Italy. The European hazelnut genome sequencing will allow the study of the Corylus pan-genome, the identification of variants for traceability, or the implementation of genome-wide association studies.
For this reason, here we report the chromosome-scale assembly of the European hazelnut cultivar “TGdL” established through a two-tiered approach: (i) 10X Genomics sequencing/assembling and (ii) scaffolding using the RaGOO pipeline (Alonge et al. 2019) and the “Tombul” genome as a guide.
Materials and methods
Plant materials and DNA sequencing
Young fresh leaves were collected from the “TGdL” UNITO-AD17 clone. DNA extraction was performed by Novogene (Genome Sequencing Company, Hong Kong) and used to construct 10X Genomics Chromium technology (Weisenfeld et al. 2017) libraries. Sequencing was then performed on an Illumina NovaSeq 6000 System.
De novo genome assembly and reference-guided scaffolding
The “TGdL” genome was de novo assembled using Supernova Assembler v 2.1.1 (Weisenfeld et al. 2017) software (10X Genomics) using 10X linked-reads as input. The Supernova Assembler was run directly on raw data derived from the sequencing process without any read cleaning process. The output format chosen for the subsequent analyses was “pseudohap.” The gap-closing process was performed using GapCloser script from SOAPdenovo2 pipeline (Luo et al. 2012).
The reference-guided scaffolding was performed using the RaGOO v1.1 (Alonge et al. 2019) scaffolder with the “Tombul” genome as reference (PRJEB31933, https://www.ebi.ac.uk/ena), with default parameters. The gap-closing process was repeated to further decrease the rate of indeterminate bases (N). Quality assessment of the genome assemblies was obtained using the QUAST tool (http://quast.sourceforge.net/). SNP/Indels were counted and analyzed using custom bash scripts. The estimation of the genome heterozygosity level was calculated by considering the ratio between the number of SNP/Indels (called in heterozygous state) and the size of the assembled genome after removal of Ns (404.097.498 bp) as previously reported (Acquadro et al. 2020a).
Genome annotation, integrity, and completeness
The de novo assembly was masked using RepeatMasker (Smit et al. 2013–2015) and the gene prediction used Maker-P (Campbell et al. 2014). The prediction process was made using Augustus (Stanke et al. 2006) Hidden Markov Models and SNAP (Bromberg and Rost 2007) algorithms aided by a set of NCBI available hazelnut proteins and transcripts. All the genes detected were evaluated considering AED values and only genes with AED ≤0.4 were maintained. The AED values measure the concordance between the predicted gene and a transcript, mRNA-seq, and protein homology library data. In a case of perfect concordance, the score is 0, in the opposite case 1. To measure the quality and completeness of the predicted proteomes, a quantitative assessment was carried out based on evolutionary informed expectations of gene content known as Benchmarking Universal Single-Copy Orthologs (BUSCO Simão et al. 2015; v3.0.2., Embryophyta odb 10).
Gene function was attributed using BLASTP (Altschul et al. 1990) comparing data with the Uniprot/Swissprot Viridiplantae database. Default parameters, except for the e-value (<1e-5) were applied. InterProScan (v. 5.33-72.0; Jones et al. 2014) was also introduced for domain inspection using all the available databases (ProSitePro les-20.119, PANTHER-10.0, Coils-2.2.1, PIRSF-3.01, Hamap-201511.02, Pfam29.0, ProSitePatterns-20.119, SUPERFAMILY-1.75, ProDom-2006.1, SMART-7.1, Gene3D-3.5.0, and TIGRFAM-15.0).
OrthoFinder
OrthoFinder software was used for the detection of putative orthologs and orthology groups. The comparisons were made among three C. avellana cultivars (“TGdL,” “Jefferson,” and “Tombul”), Quercus suber, Betula pendula, and Carpinus fangiana. Gene ontology (GO) term enrichment was carried out with AGRIGOv2 (http://systemsbiology.cau.edu.cn/agriGOv2) to find a representative subset of the GO terms previously identified with the Interproscan pipeline.
Resistance genes analogs
Candidate resistance genes were identified using RGAugury (Li et al. 2016). Resistance genes analogs (RGA) candidates were classified into four major families based on the presence of combinations of these RGA domains and motifs: NBS-encoding [subsequently divided into subgroups according to their domain architecture, namely NBS (NBS domain), CNL (CC-NBS-LRR domains), TNL (TIR-NBS-LRR), TN (TIR-NBS), CN (CC-NBS), NL (NBS-LRR), and TX (TIR-unknown domain and other)], TM-CC, and membrane-associated RLP and RLK. MAFFT v7.450 (https://mafft.cbrc.jp) was used for protein alignment with the following parameters: –ep 0 –reorder –maxiterate 1000 –genafpair. Genetic relationships were described by constructing a phylogenetic tree by maximum likelihood by using the IQ-TREE software (v.1.6.12, http://www.iqtree.org); branch supports were obtained with the ultrafast bootstrap with 1000 replicates. Trees were visualized using interactive Tree of Life (iTOL v3, https://itol.embl.de).
Data availability
Raw reads are publicly available in the NCBI sequence read archive under the bioproject: PRJNA694440. The reference assembly and annotation data are also available for downloading from https://zenodo.org/deposit/4454484. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14502048.
Results and discussion
Genome sequencing and assembly
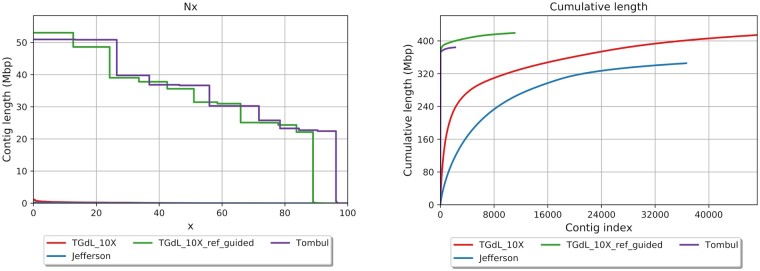
The chromosome-scale “TGdL” genome was developed using two-tiered approach. The 10X Genomics Chromium Technology was firstly used to obtain a high-quality preliminary assembly. Reference-guided scaffolding was then implemented using the “Tombul” genome as the reference. The 10X genomic library was sequenced with Illumina technology and 138.56 million raw pair-end reads were generated (52X coverage). The average read length was 138.50 bp, with 86.06% of them having Q > 30. These data are comparable to the optimal standard values suggested by Supernova Assembler software manufacturer (Table 1). In details, Supernova assembled 47,216 scaffolds having a total length of 414.38 Mb and an N50 of 51,567 bp. The results were similar (Table 2) to the other genome assemblies at a contig level (“Jefferson,” Rowley et al. 2018; “Tombul,” Lucas et al. 2021; Table 2). A single library (10X Genomics Chromium Technology) produces a more optimized “TGdL” genome assembly compared to the “Jefferson” assembly (Table 2), the latter being obtained using three different Illumina libraries, 250-bp and 350-bp Illumina paired-end (PE) libraries and a 4.5-Kb mate-pair (MP) library. Moreover, the 10X strategy proved to be a cost-effective route being the “TGdL” assembly highly comparable to the “Tombul” (contigs) draft made with a higher coverage (108X, short reads) and 9.3X of long Nanopore reads (Figure 1) prior to scaffolding.
Table 1.
Summary of the metrics of the “TGdL” 10X Genomics Chromium Technology
| “TGdL” Results | Optimal standard values | |
|---|---|---|
| Reads number | 138.56 M | — |
| Reads average length (bp) | 138.50 | 140 |
| Coverage | 52X | 56X |
| % reads with Q30 quality | 86.06 | 75–85 |
Table 2.
Metrics of the genome assemblies of the “TGdL,” “Tombul,” and “Jefferson” cultivar
| “TGdL” (contigs) | “Tombul” (contigs) | “Jefferson” | “TGdL” | “Tombul” | |
|---|---|---|---|---|---|
| Scaffold number (#) | 47,216 | 12,557 | 36,641 | 11,059 | 2,207 |
| Total length (Mb) | 414.38 | 383.1 | 345.54 | 419.46 | 384.20 |
| N50 | 51,567 | 1,299 | 21,508 | 35,598,485 | 36,653,616 |
| L50 | 1,457 | 78,800 | 4,253 | 5 | 5 |
| Largest contig (bp) | 1,152,936 | — | 274,525 | 53,036,447 | 50,950,907 |
| # N’s/100 Kb | 3,811.59 | 180 | 12,054.75 | 3,514.85 | 468.09 |
| GC (%) | 36.87 | — | 36.30 | 36.84 | 35.91 |
Figure 1.
Contiguity statistics performed on “TGdL” (contigs), “TGdL” (pseudomolecules plus unplaced scaffolds), “Tombul” (pseudomolecules), and “Jefferson” genomes. Left picture: Nx statistics (Nx is the largest contig length, L, such that using contigs of length ≥ L accounts for at least x% of the bases of the assembly) with x varying between 1 and 100. Right picture represents the cumulative length increment of the genome through the scaffold/contig addition.
The reference-guided scaffolding was made using the RaGOO pipeline, which was able to optimize the “TGdL” assembly (contigs) adopting the “Tombul” genome (PRJEB31933), previously obtained with proximity ligation technology (Dovetail Genomics using Chicago & HiC protocols), as reference. It produced 11 pseudomolecules (11 chromosomes) and 11,046 scaffolds belonging to chromosome 0, which represent 11% of the genome (46,504,161 bp). The resulting assembly was a complete “TGdL” chromosome-scale genome (Table 2), whose total length (without chr0) resembled that on the “Tombul” assembly. Following the scaffolding process, we renamed the super scaffolds based on Torello Marinoni et al. (2018) linkage groups (Table 3).
Table 3.
Pseudomolecules reconstructed in “TGdL” and nomenclature according to the genetic map by Torello Marinoni et al. (2018)
| “TGdL” pseudomolecules | “Tombul” nomenclature | Size (bp) | Ns | SNP/indels | SNP/indels frequency (%) |
|---|---|---|---|---|---|
| 1 | 1 | 53,036,447 | 2,194,688 | 430,039 | 0.85 |
| 2 | 2 | 48,611,531 | 1,656,681 | 390,798 | 0.83 |
| 3 | 8 | 25,054,957 | 974,784 | 216,892 | 0.90 |
| 4 | 3 | 39,027,746 | 1,377,931 | 322,996 | 0.86 |
| 5 | 5 | 35,598,485 | 1,323,708 | 299,132 | 0.87 |
| 6 | 10 | 24,339,126 | 955,975 | 211,629 | 0.91 |
| 7 | 6 | 30,979,729 | 1,185,588 | 244,134 | 0.82 |
| 8 | 11 | 22,131,124 | 858,998 | 182,492 | 0.86 |
| 9 | 4 | 37,785,217 | 1,350,719 | 299,648 | 0.82 |
| 10 | 9 | 25,093,934 | 1,084,840 | 201,790 | 0.84 |
| 11 | 7 | 31,441,515 | 1,359,091 | 262,222 | 0.87 |
| Unplaced scaffolds | — | 45,736,493 | 415,803 | 272,272 | 0.60 |
| Whole genome | — | 418,836,304 | 14,738,806 | 3,334,091 | 0.84 |
The nomenclature of “Tombul” pseudomolecules is reported as in Lucas et al. (2021). Observed SNP/indel in heterozygous state and their frequency are calculated using the size of the assembled genome, after removal of Ns (404,097,498 bp).
The rate of heterozygosity of the hazelnut genome was calculated as 0.84% and was similar for all the pseudomolecules, ranging from 0.91 (chromosome 6) to 0.82% (chromosome 7). Unplaced scaffolds showed a lower rate of heterozygosity (0.60%). These data were expected due to the allogamous behavior of hazelnut and are comparable to those of other outbred species (∼1%; Velasco et al. 2007; Acquadro et al. 2017), and higher than the ones obtained in inbred species (∼0.1%; Barchi et al. 2019; Acquadro et al. 2020a).
Genome annotation and Orthofinder analysis
Globally, ∼41.5% of the assembled genome were repeated, 17% of which consisted of LTR elements (Table 4). The assembled genome was then structurally annotated with the Maker-P suite and the total number of genes identified was 27,791 (AED 0.4). The proteome was validated using BUSCO; overall, more than 92% of 1614 expected embryophyta genes were identified in the “TGdL” genome annotations as the complete and partial BUSCO profiles. The number of predicted genes is similar to the one predicted in “Tombul” (27,270) and in “Jefferson” (28,167); a similar number of genes were also identified in the close species C. fangiana (27,384) and B. pendula (28,153), while fewer genes were predicted in Q. suber (25,808).
Table 4.
Masking statistics for the “TGdL” hazelnut genome
| Class | Superfamily | Count | Masked (bp) | Masked (%) |
|---|---|---|---|---|
| DNA | — | — | — | — |
| hAT | 46,506 | 9,536,392 | 2.30 | |
| CACTA | 66,513 | 10,364,758 | 2.50 | |
| PIF/Harbinger | 33,649 | 5,713,915 | 1.38 | |
| Mutator | 208,743 | 34,956,690 | 8.43 | |
| Tcl/Mariner | 9,668 | 1,607,102 | 0.39 | |
| Helitron | 71,851 | 14,190,408 | 3.42 | |
| LTR | — | — | — | — |
| Copia | 46,495 | 19,214,573 | 4.63 | |
| Gypsy | 48,852 | 26,434,445 | 6.37 | |
| Unknown | 95,170 | 26,065,393 | 6.28 | |
| MITE | — | — | — | — |
| hAT | 9,724 | 1,295,113 | 0.31 | |
| CACTA | 1,080 | 128,610 | 0.03 | |
| PIF/Harbinger | 10,503 | 1,717,093 | 0.41 | |
| Mutator | 60,562 | 6,926,393 | 1.67 | |
| Tcl/Mariner | 388 | 33,917 | 0.01 | |
| Unspecified | — | 22,038 | 4,627,197 | 1.12 |
| Total interspersed | 731,742 | 162,811,999 | 39.24 | |
| Low_complexity | — | 30,853 | 1,453,286 | 0.35 |
| Simple_repeat | — | 224,449 | 7,743,022 | 1.87 |
| Total | — | 987,044 | 172,008,307 | 41.46 |
The function attributed to each predicted protein was based on the results of the BLASTP (SwissProt) and the InterProScan domain inspection. InterProScan analyses highlighted about 80% of the predicted proteins with at least one IPR domain. Among the top 20 SUPERFAMILY domains (Table 5), the most abundant in all the genomes was SSF56112 (protein Kinase-like domain), which acts on signaling and regulatory processes in the eukaryotic cell. The other most abundant Superfamilies were: SSF52540 (P-loop containing nucleoside triphosphate hydrolase), which is involved in several UniPathways, including chlorophyll or coenzyme A biosynthesis and SSF52058 (Leucine-rich repeat domain, L domain-like), which is related to resistance to pathogens.
Table 5.
Top 20 SUPERFAMILY domains in the “TGdL” hazelnut genome
| Superfamily | Description | Count |
|---|---|---|
| SSF56112 | Protein kinase-like domain | 1577 |
| SSF52540 | P-loop containing nucleoside triphosphate hydrolase | 1565 |
| SSF52058 | L domain-like | 935 |
| SSF51735 | NAD(P)-binding domain | 484 |
| SSF57850 | RING/U-box | 468 |
| SSF48371 | Armadillo-type fold | 428 |
| SSF48452 | Tetratricopeptide-like helical domain | 413 |
| SSF57889 | Cysteine-rich domain | 409 |
| SSF52047 | RNI-like | 380 |
| SSF48264 | Cytochrome P450 | 376 |
| SSF53474 | Alpha/Beta hydrolase fold | 362 |
| SSF53756 | UDP-Glycosyltransferase/glycogen phosphorylase | 362 |
| SSF46689 | Homeobox-like domain | 336 |
| SSF53335 | S-adenosyl-L-methionine-dependent methyltransferase | 335 |
| SSF48403 | Ankyrin repeat-containing domain | 324 |
| SSF81383 | F-box-like domain | 322 |
| SSF54928 | RNA-binding domain | 321 |
| SSF51445 | Glycoside hydrolase | 313 |
| SSF50978 | WD40-repeat-containing domain | 291 |
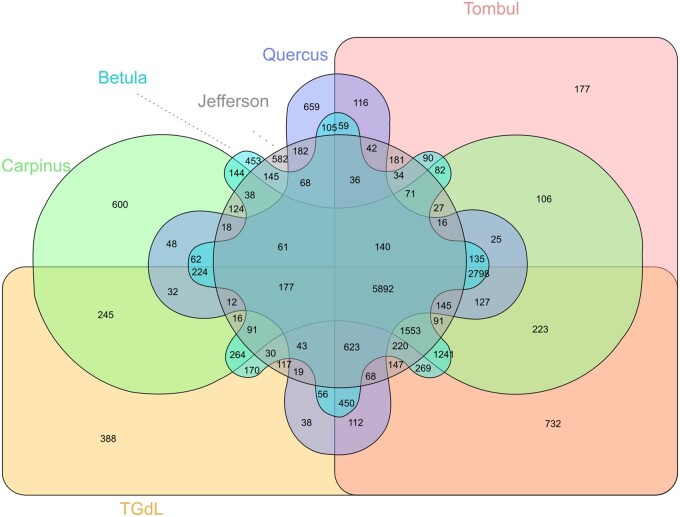
Clustering by Orthofinder the proteomes (164,573 sequences) of the three hazelnut genomes together with the ones from B. pendula, C. fangiana, and Q. suber, produced a set of 21,239 gene families (plus 24,639 unassigned genes), of which 5892 (including 59,597 genes) were shared (Figure 2). Focusing on hazelnut, the “Jefferson” proteome showed the highest percentage of unassigned genes (41.6%), presumably due to the fragmented assembly which limited the annotation procedure. On the other hand, the “TGdL” and “Tombul” assemblies showed a high percentage of assigned genes to orthogroups (93.1 and 96.3%, respectively). The “TGdL” proteome contained 388 genome-specific orthogroups (1279 genes), while 732 (with 2040 genes) were shared between “TGdL” and “Tombul,” but not the other genomes. For the former, the analysis revealed significant gene enrichment for some GO terms (Supplementary Table S1), including GO:0042908 (xenobiotic transport) as well as GO terms related to nuclease activity [GO:0016891 (endoribonuclease activity, producing 5'-phosphomonoesters), GO:0004540 (ribonuclease activity)] and transport [GO:0008559 (xenobiotic-transporting ATPase activity) and GO:0090484 (drug transporter activity)]. For genes shared by “TGdL” and “Tombul,” enriched GO terms included GO:0044092 (negative regulation of molecular function), GO:0043086 (negative regulation of catalytic activity) and GO:0050790 (regulation of catalytic activity), as well as several nuclease related terms [as GO:0004523 (RNA-DNA hybrid ribonuclease activity) GO:0004521 (endoribonuclease activity) and GO:0004540 (ribonuclease activity)] (Supplementary Table S2).
Figure 2.
Orthofinder analysis performed using “TGdL,” “Jefferson,” “Tombul,” C. fangiana, B. pendula, and Q. suber genomes.
Resistance genes
Many plant-pathogen interactions are determined by the presence of resistance (R) genes/alleles, which enable plants to recognize pathogen effectors and subsequently activate effector-triggered immunity (ETI) (Sekhwal et al. 2015), followed by a defense response often leading to cell death or a hypersensitive response (HR) (Zaidi et al. 2018). Most intracellular immune receptors in plants belong to the nucleotide-binding site and leucine-rich repeat (NLR, also known as NB-LRR) superfamilies (Eitas and Dangl 2010; Lee and Yeom 2015). The NLR superfamily proteins include two classes on the basis of the presence of a toll and interleukin-1 receptor domain in the N-terminus (TIR-NLR or TNL) or its absence (non-TIR-NLR or non-TNL). Some non-TNL proteins have a coiled-coil motif (CC-NLR or CNL).
The RGAugury pipeline detected between 86 and 2017 resistance gene analogs (RGAs) among the species/genotypes analyzed (Table 6). The highest percentage of RGAs compared to the total number of genes was found in Q. suber (7.82%), while the lowest was detected in “Jefferson” (0.31%, presumably as a consequence of the low-quality genome annotation) and Carpinus (1.91). In the “TGdL” assembly we identified a total of 810 RGAs. Furthermore, clustering of RLKs, RLPs, NBS-encoding, and TM-CC genes in some chromosomes were detected (Table 7), in agreement with classical genetics and analysis from large-scale sequencing data in plant genomes (Rody et al. 2019). The chromosome 2 was the richest in RGAs followed by 5, 3, and 4, while chromosome 10 was the poorest. The majority of RLK genes was found on chromosomes 2, 5, and 6, while the majority of RLP on chromosomes 2, 9, and 7.
Table 6.
Resistance (R) genes in the “TGdL” hazelnut genome compared with the genomes of “Jefferson,” and “Tombul” and those of B. pendula, C. fangiana, and Q. suber
| Species/Genotype | NBS | CNL | TNL | CN | TN | NL | TX | Others | RLP | RLK | TM-CC | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| “TGdL” | 18 (0.06%) | 32 (0.12%) | 1 (0%) | 23 (0.08%) | 1 (0%) | 33 (0.12%) | 6 (0.02%) | 1 (0%) | 93 (0.33%) | 547 (1.97%) | 55 (0.2%) | 810 (2.91%) |
| “Jefferson” | 2 (0.01%) | 0 (0%) | 0 (0%) | 1 (0%) | 0 (0%) | 2 (0.01%) | 2 (0.01%) | 0 (0%) | 11 (0.04%) | 67 (0.24%) | 1 (0%) | 86 (0.31%) |
| “Tombul” | 14 (0.05%) | 43 (0.15%) | 14 (0.05%) | 12 (0.04%) | 5 (0.02%) | 28 (0.1%) | 23 (0.08%) | 6 (0.02%) | 123 (0.44%) | 673 (2.42%) | 190 (0.68%) | 1131 (4.07%) |
| B. pendula | 20 (0.07%) | 10 (0.04%) | 48 (0.17%) | 0 (0%) | 16 (0.06%) | 31 (0.11%) | 70 (0.25%) | 5 (0.02%) | 30 (0.11%) | 662 (2.38%) | 50 (0.18%) | 942 (3.39%) |
| C. fangiana | 3 (0.01%) | 23 (0.08%) | 0 (0%) | 4 (0.01%) | 1 (0%) | 30 (0.11%) | 4 (0.01%) | 0 (0%) | 55 (0.2%) | 291 (1.05%) | 51 (0.18%) | 462 (1.66%) |
| Q. suber | 47 (0.17%) | 240 (0.86%) | 174 (0.63%) | 30 (0.11%) | 16 (0.06%) | 309 (1.11%) | 118 (0.42%) | 26 (0.09%) | 286 (1.03%) | 736 (2.65%) | 35 (0.13%) | 2017 (7.26%) |
For each resistance gene class, the number as well as the percentage over the total number of genes is reported. Resistance genes abbreviations (from Li et al. 2016): NBS, nucleotide-binding site; CNL, CC (coiled-coil)-NBS-LRR; TNL, TIR (Toll/Interleukin-1 receptor)-NBS-LRR; CN, CC-NBS; TN, TIR-NBS; NL, NBS-LRR; TX, TIR-unknown domain; RLK, receptor-like kinase; RLP, receptor-like protein; TM (transmembrane)-CC.
Table 7.
Distribution of the resistance R genes among the 11 pseudomolecules of the “TGdL” hazelnut genome
| “TGdL” pseudomolecules | CN | CNL | NBS | NL | Other | RLK | RLP | TM-CC | TN | TNL | TX | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | — | 1 | — | — | — | 45 | 10 | 7 | — | — | — | 63 |
| 2 | 6 | 9 | 9 | 18 | — | 96 | 18 | 2 | — | — | 3 | 161 |
| 3 | 2 | 4 | 2 | 3 | — | 51 | 9 | 12 | 1 | — | 1 | 85 |
| 4 | 2 | 3 | — | 2 | 1 | 53 | 8 | 6 | — | 1 | — | 76 |
| 5 | 1 | 5 | — | — | — | 67 | 4 | 7 | — | — | 2 | 86 |
| 6 | 1 | — | — | 4 | — | 58 | 6 | — | — | — | — | 69 |
| 7 | – | 2 | 1 | 1 | — | 44 | 9 | 8 | — | — | — | 65 |
| 8 | 9 | 4 | 3 | 1 | — | 33 | 7 | 3 | — | — | — | 60 |
| 9 | — | 1 | — | 2 | — | 31 | 11 | 2 | — | — | — | 47 |
| 10 | 1 | 3 | 2 | 2 | — | 20 | 5 | 1 | — | — | — | 34 |
| 11 | — | — | — | — | — | 43 | 5 | 4 | — | — | — | 52 |
| Unplaced scaffolds | 1 | — | 1 | — | — | 6 | 1 | 3 | — | — | — | 12 |
| Total | 23 | 32 | 18 | 33 | 1 | 547 | 93 | 55 | 1 | 1 | 6 | 810 |
The majority of RGAs were receptor-like kinases (RLKs), followed by receptor-like proteins (RLPs), while only few RGAs contain at least one NB-ARC domain. Similarly, in other members of the order Fagales, i.e., Juglans microcarpa and J. regia, the most represented RGAs belong to RLK while few TNLs were identified (Zhu et al. 2019). Comparable results have been obtained in other non-woody species, like Solanaceae species such as Capsicum annuum, Solanum melongena, Solanum lycopersicum, and Solanum tuberosum (Barchi et al. 2019; Acquadro et al. 2020a). Furthermore, Kim et al. (2012) highlighted that some Asterids contain functional TNLs, whereas others do not. This resulted in the identification of only 19 and 13 full length CNLs in sunflower and lettuce respectively, but no full length TNLs. Recently, Acquadro et al. (2017) reported that in the Cynara cardunculus genome, the RGAs belong almost exclusively to the RLK/RLP families, while no TNLs and few CNLs were identified. This species-specific RGAs distribution was also observed in Brassica oleracea, B. rapa, Arabidopsis thaliana, and Theobroma cacao, where the number of TNL was higher than CNL, while an opposite situation was found for Populus trichocarpa, Vitis vinifera, and Medicago truncatula (Yu et al. 2014).
The alignments of the amino acid sequences and subsequent IQ-TREE analyses generated phylogenetic trees for CNL-TNL, RLP, and RLK RGA classes (Supplementary Figure S1, A–C).
It has been reported that several R-genes (Triticum aestivum Pm3, Arabidopsis thaliana RPP13, Linum usitatissimum, and Capsicum annuum eIF4E) seem to have evolved following a co-evolutionary relationship with pathogens and thus environment. The difference in terms of number and phylogenetic relationship represent a valuable information for conducting future in-depth studies on particular genes that are associated with their local environment (Rose et al. 2004; Charron et al. 2008).
Conclusions
We performed a whole-genome assembly, using a combination of 10X Chromium linked-read technology and accurate 150 bp paired-end short-read Illumina sequencing, to generate the genome of the European hazelnut cv. “TGdL,” one of the best cultivars for processing due to its high kernel quality. A chromosome-scale assembly of “TGdL” was built and will facilitate the detection of genomic variants, including copy number variations and large insertions/deletions. About 28.000 genes were identified and annotated with known homology. Since the European hazelnut “TGdL” has excellent kernel quality and its genome sequences will be useful for studying important traits, predicting genes, and developing markers for use in breeding programs.
Funding
The research was carried out without financial support.
Conflicts of interest
The authors declare no conflict of interest.
Literature cited
- Acquadro A, Barchi L, Portis E, Nourdine M, Carli C, et al. 2020a. Whole genome resequencing of four Italian sweet pepper landraces provides insights on sequence variation in genes of agronomic value. Sci Rep. 10:9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquadro A, Portis E, Valentino D, Barchi L, Lanteri S.. 2020b. “Mind the Gap”: Hi-C technology boosts contiguity of the globe artichoke genome in low-recombination regions. G3 (Bethesda). 10:3557–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquadro A, Barchi L, Portis E, Mangino G, Valentino D, et al. 2017. Genome reconstruction in Cynara cardunculus taxa gains access to chromosome-scale DNA variation. Sci Rep. 7:5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, et al. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Barchi L, Pietrella M, Venturini L, Minio A, Toppino L, et al. 2019. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci Rep. 9:11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramo C, Valentini N, Portis E, Torello Marinoni D, Boccacci P, et al. 2016. Genetic mapping and QTL analysis in European hazelnut (Corylus avellana L.). Mol Breed. 36:27. [Google Scholar]
- Botta R, Molnar TJ, Erdogan V, Valentini N, Marinoni DT, et al. 2019. Hazelnut (Corylus spp.) Breeding: 157-219. Chapter 6. In: Al-Khayri JM, Jain SM, Johnson DV, editors. Advances in Plant Breeding Strategies, Vol 4: Nut and Beverage Crops. Springer Nature. p. 607. [Google Scholar]
- Bromberg Y, Rost B.. 2007. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 35:3823–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Law M, Holt C, Stein JC, Moghe GD, et al. 2014. MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164:513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron C, Nicolaï M, Gallois JL, Robaglia C, Moury B, et al. 2008. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 54:56–68. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang J, Liu Q, Guo W, Zhao T, et al. 2014. Transcriptome sequencing and identification of cold tolerance genes in hardy Corylus species (C. heterophylla Fisch) floral buds. PLoS One. 9:e108604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL.. 2010. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 13:472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT. 2019. Food and Agriculture Organization of the United Nations (2019). (Accessed: 2020 April 20). http://www.fao.org/faostat/.
- Hulse-Kemp AM, Maheshwari S, Stoffel K, Hill TA, Jaffe D, et al. 2018. Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic Res. 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM.. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 449:463–467. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics. 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lim CJ, Lee BW, Choi JP, Oh SK, et al. 2012. A genome-wide comparison of NB-LRR type of resistance gene analogs (RGA) in the plant kingdom. Mol Cells. 33:385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HA, Yeom SI.. 2015. Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief Funct Genom. 14:233–242. [DOI] [PubMed] [Google Scholar]
- Li P, Quan X, Jia G, Xiao J, Cloutier S, et al. 2016. RGAugury: a pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genomics. 17:852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas SJ, Kahraman K, Avşar B, Buggs RJA, Bilge I.. 2021. A chromosome-scale genome assembly of European Hazel (Corylus avellana L.) reveals targets for crop improvement. Plant J. 105:1413–1430. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Lu Z, Liu B, Qiu Q, Liu J.. 2013. Transcriptome analyses of a Chinese hazelnut species Corylus mandshurica. BMC Plant Biol. 13:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlenbacher SA, Brown RN, Nouhra ER, Gökirmak T, Bassil NV, et al. 2006. A genetic linkage map for hazelnut (Corylus avellana L.) based on RAPD and SSR markers. Genome. 49:122–133. [DOI] [PubMed] [Google Scholar]
- Molnar TJ. 2011. Corylus. In: Kole C, editor. Wild Crop Relatives: Genomic and Breeding Resources. Berlin, Heidelberg: Springer. 2: 15–48. [Google Scholar]
- Öztürk SC, Göktay M, Allmer J, Doğanlar S, Frary A.. 2018. Development of simple sequence repeat markers in hazelnut (Corylus avellana L.) by next-generation sequencing and discrimination of Turkish hazelnut cultivars. Plant Mol Biol Rep. 36:800–811. [Google Scholar]
- Pustahija F, Brown SC, Bogunic F, Basic N, Muratovic E, et al. 2013. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant Soil. 373:427–453. [Google Scholar]
- Rody HVS, Bombardelli RGH, Creste S, Camargo LEA, Van Sluys MA, et al. 2019. Genome survey of resistance gene analogs in sugarcane: genomic features and differential expression of the innate immune system from a smut-resistant genotype. BMC Genomics. 20:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose LE, Bittner-Eddy PD, Langley CH, Holub EB, Michelmore RW, et al. 2004. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics. 166:1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley ER, Fox SE, Bryant DW, Sullivan CM, Givan SA, et al. 2012. Assembly and characterization of the European hazelnut (Corylus avellana L.) ‘Jefferson’ transcriptome. Crop Sci. 52:2679–2686. [Google Scholar]
- Rowley ER, Vanburen R, Bryant DW, Priest HD, Mehlenbacher SA, et al. 2018. A draft genome and high-density genetic map of European hazelnut (Corylus avellana L.). bioArxiv. 1–25. doi:10.1101/469015 [Google Scholar]
- Sekhwal MK, Li P, Lam I, Wang X, Cloutier S, et al. 2015. Disease resistance gene analogs (RGAs) in plants. Int J Mol Sci. 16:19248–19290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, , Waterhouse RM, , Ioannidis P, , Kriventseva EV, , Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31:3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P.. 2013. –2015. RepeatMasker Open-4.0. http://www.repeatmasker.org/faq.html (Accessed: 2020 December 16).
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torello Marinoni D, Valentini N, Portis E, Acquadro A, Beltramo C, et al. 2018. High density SNP mapping and QTL analysis for time of leaf budburst in Corylus avellana L. PLoS One. 13:e0195408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini N, Calizzano F, Boccacci P, Botta R.. 2014. Investigation on clonal variants within the hazelnut (Corylus avellana L.) cultivar ‘Tonda Gentile delle Langhe’. Scientia Hortic. 165:303–310. [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, et al. 2007. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One. 2:e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar P, Shah P, Church DM, Jaffe DB.. 2017. Direct determination of diploid genome sequences. Genome Res. 27:757–767. doi:10.1101/gr.214874.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Tehrim S, Zhang F, Tong C, Huang J, et al. 2014. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics. 15:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SS, Mukhtar MS, Mansoor S.. 2018. Genome Editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36:898–906. [DOI] [PubMed] [Google Scholar]
- Zhu T, Wang L, You FM, Rodriguez JC, Deal KR, et al. 2019. Sequencing a Juglans regia × J. microcarpa hybrid yields high-quality genome assemblies of parental species. Hortic Res. 6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw reads are publicly available in the NCBI sequence read archive under the bioproject: PRJNA694440. The reference assembly and annotation data are also available for downloading from https://zenodo.org/deposit/4454484. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14502048.