Abstract
Proteins in the E2A family of basic helix-loop-helix transcription factors are important in a wide spectrum of physiologic processes as diverse as neurogenesis, myogenesis, lymphopoeisis, and sex determination. In the pancreatic β cell, E2A proteins, in combination with tissue-specific transcription factors, regulate expression of the insulin gene and other genes critical for β-cell function. By yeast two-hybrid screening of a cDNA library prepared from rat insulinoma (INS-1) cells, we identified a novel protein, Bridge-1, that interacts with E2A proteins and functions as a coactivator of gene transcription mediated by E12 and E47. Bridge-1 contains a PDZ-like domain, a domain known to be involved in protein-protein interactions. Bridge-1 is highly expressed in pancreatic islets and islet cell lines and the expression pattern is primarily nuclear. The interaction of Bridge-1 with E2A proteins is further demonstrated by coimmunoprecipitation of in vitro-translated Bridge-1 with E12 or E47 and by mammalian two-hybrid studies. The PDZ-like domain of Bridge-1 is required for interaction with the carboxy terminus of E12. In both yeast and mammalian two-hybrid interaction studies, Bridge-1 mutants lacking an intact PDZ-like domain interact poorly with E12. An E12 mutant (E12ΔC) lacking the carboxy-terminal nine amino acids shows impaired interaction with Bridge-1. Bridge-1 has direct transactivational activity, since a Gal4 DNA-binding domain–Bridge-1 fusion protein transactivates a Gal4CAT reporter. Bridge-1 also functions as a coactivator by enhancing E12- or E47-mediated activation of a rat insulin I gene minienhancer promoter-reporter construct in transient-transfection experiments. Substitution of the mutant E12ΔC for E12 reduces the coactivation of the rat insulin I minienhancer by Bridge-1. Inactivation of endogenous Bridge-1 in insulinoma (INS-1) cells by expression of a Bridge-1 antisense RNA diminishes rat insulin I promoter activity. Bridge-1, by utilizing its PDZ-like domain to interact with E12, may provide a new mechanism for the coactivation and regulation of transcription of the insulin gene.
Basic helix-loop-helix (bHLH) transcription factors regulate a diverse array of physiologic processes in developing and adult organisms. Myogenesis, lymphocyte differentiation, neurogenesis, sex determination, and the development and functions of pancreatic β cells are dependent on the regulated action of both ubiquitous and tissue-specific bHLH proteins (26, 28). The ubiquitous E2A family of proteins, including E12 and E47, function either as homodimers or as heterodimers with tissue-specific class B bHLH proteins to bind and transactivate promoters via conserved sequence elements known as E boxes. Although E2A proteins are best characterized in their interaction with other bHLH proteins, other types of protein-protein interactions have been described (12, 20, 21, 24, 35). The activation of gene transcription by E2A may occur directly by the binding of bHLH dimers to DNA, via synergistic interactions with proteins bound to adjacent DNA, or by interaction with coactivating proteins.
One experimental model system for the transactivation of transcription by E2A is the pancreatic β cell, in which the rat insulin I gene is regulated by glucose-responsive minienhancers consisting of E box binding sites for E2A (30) and A boxes that bind homeoproteins (14). At the E boxes, E2A forms heterodimers with tissue-specific bHLH transcription factors such as Beta-2/NeuroD (29). Homeoproteins, such as PDX-1, Lmx1, and Isl-1, bind to the A boxes and act in synergy with E2A heterodimers on adjacent E boxes to activate transcription of the insulin gene (14, 18, 32).
The importance of these regulatory factors for both the development and function of β cells is illustrated by several lines of investigation. Targeted disruption of Beta-2/NeuroD in mice results in abnormalities of islet development and diabetes mellitus (28). Inactivation of E2a does not impair pancreatic development, but compensation by other members of the E2A family may be responsible for this observation (16, 39). In both mice and humans, homozygous disruption of the pdx-1 gene arrests pancreatic development, whereas heterozygous disruption results in a diabetic phenotype (11, 19, 31, 41, 43).
Additional factors also regulate the transactivational activities of E2A in the pancreatic β cell. Both CBP and p300 may act as E2A coactivators (12, 35). p300 serves as a coactivator for both E2A and Beta-2/NeuroD in insulin gene transcription (35).
PDZ domains, named for three proteins in which the motif was initially noted (postsynaptic density protein PSD-95 [5], Drosophila disc-large tumor suppressor protein DlgA [47], and the mammalian tight junction protein ZO-1 [17]), are conserved domains that mediate protein-protein interactions in a variety of intracellular signaling processes (38). For example, PDZ domains have been implicated in protein-protein interactions required for postsynaptic density ion channel and receptor clustering, signal transduction pathways regulating cell growth, visual signal transduction cascade regulation, and Fas-mediated regulation of apoptosis (38). PDZ domains appear in proteins with a diverse range of functions, including protein tyrosine phosphatases, proteases, ion channels, and signal transduction scaffolding molecules (38).
In studies designed to identify new factors that might regulate insulin gene transcription, we discovered Bridge-1, a novel coactivator for E2A isolated from pancreatic insulinoma cells. We suggest that Bridge-1 represents a novel PDZ-domain coactivator for E2A and further that it participates in the regulation of insulin gene transcription in pancreatic β cells.
MATERIALS AND METHODS
Cloning of rat Bridge-1 by a yeast two-hybrid system.
Standard molecular biology techniques were used (37). A directional INS-1 cDNA library was constructed in the plasmid vector pJG4-5 by using a Stratagene cDNA synthesis kit. The E12 bait was constructed by reverse transcriptase PCR (RT-PCR) amplification of DNA encoding amino acids 521 to 649 of E12 from total RNA isolated from rat 18-day-postcoitus (dpc) pancreas, followed by cloning into the plasmid pEG202 in frame with an upstream LexA DNA-binding domain. Yeast two-hybrid screening was conducted according to standard methods (15). The complete 1.4-kb Bridge-1 cDNA was isolated by the screening of a rat 14-dpc pancreatic library by using the 30-mer oligonucleotide 5′-TCACTCGACATCGCGGACCTAGCCTAAAA-3′. Sequencing was performed by the Sanger dideoxy chain termination method. Protein similarity indices were determined by using the Lipman-Pearson Protein Alignment function of the software package Lasergene (DNASTAR, Inc., Madison, Wis.) and the alignment program BLAST (GenBank). Rat Bridge-1 cDNA sequence was submitted to GenBank under accession number AF067728.
Northern blot analysis.
Total RNAs were extracted by the guanidinium isothiocyanate method (7), and poly(A)+ RNA was prepared by using the PolyATract mRNA isolation system (Promega, Madison, Wis.). RNAs were electrophoresed on 1% agarose-formaldehyde gels and blotted onto nylon membranes (GeneScreen; NEN Life Science Products, Boston, Mass.) prior to a probing with 32P-labeled Bridge-1 cDNA. Membranes were stripped and reprobed with rat γ-actin according to the manufacturer’s instructions. Mouse and human endocrine system multiple tissue Northern blots (Clontech Laboratories, Inc., Palo Alto, Calif.) were probed with 32P-labeled Bridge-1 cDNA by using high-stringency washing conditions as described by the manufacturer.
Plasmid construction.
pBSII–Bridge-1 and pcDNA3–Bridge-1 were constructed by inserting the 1.4-kb Bridge-1 cDNA into an EcoRI site within the multiple cloning region of pBSII or pcDNA3, respectively. AS–Bridge-1–pcDNA3 was generated by inserting the 1.4-kb Bridge-1 cDNA into the EcoRI site of pcDNA3 in the antisense orientation. For the mammalian two-hybrid studies, the Mammalian Matchmaker Two-Hybrid Assay Kit (Clontech Laboratories) vectors pM and PVP16 were used to construct plasmids expressing Gal4 DNA-binding domain–Bridge-1, VP16–Bridge-1, Gal4 DNA-binding domain–E12, VP16-E12, and Gal4 DNA-binding domain–Beta-2 fusion proteins. pM–Bridge-1 and pVP16–Bridge-1 were constructed by inserting a blunt-ended 900-bp BstUI/EcoRI fragment of pBSII–Bridge-1 in frame into a blunt-ended MluI site of the multiple cloning site of the plasmids pM and pVP16, respectively. pM-E12 and pVP16-E12 were constructed by inserting a 2.7-kb blunt-ended NdeI/EcoRI fragment of Pan 2 excised from the vector PARP5 (gift of C. Nelson) in frame into a SmaI site of the pM and pVP16 vectors, respectively. pM-Beta-2 was constructed by inserting a 2.4-kb blunt-ended BamHI/cohesive-ended XbaI fragment from pcDNA1-Beta-2 (gift of J. Seufert) into the vector pM that had been prepared by digesting with BamHI, followed by blunting with Klenow polymerase and digestion with XbaI. pcDNAI-Beta-2 had been previously constructed by inserting a 2.4-kb BamHI/XhoI fragment from pCMV-Beta-2 (gift of F. Naya and M. J. Tsai) into pcDNAI that had been digested with BamHI and XhoI. After the cloning was completed, the matchmaker vector constructs were verified by automated DNA sequencing. pcDNA3-E12 and pcDNA3-E47 were constructed by inserting 2.7-kb BglII/EcoRI fragments of Pan 2 from the vector PARP5 or Pan 1 from the vector PARP5P2, respectively (vectors were gifts of C. Nelson), into pcDNA3 prepared by digestion with BamHI and EcoRI. 5FF1CAT was a gift from J. L. Moss. pcDNA3-E12ΔC and pVP16-E12ΔC were constructed by point mutagenesis with insertion of a premature stop codon in pcDNA3-E12 and pVP16-E12, respectively, by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, Calif.) according to the manufacturer’s instructions with the oligonucleotides 5′-ACCCGGGCCTGGGTTAGGCCCACAAT-3′ and 5′-ATTGTGGGCCTAACCCAGGCCCGGGT-3′. pVP16-E12ΔbHLH was constructed by point mutagenesis with insertion of a premature stop codon in pVP16-E12 by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, Calif.) according to the manufacturer’s instructions with the oligonucleotides 5′-TACCAGCCCAGACTAGGACGAGGACGA-3′ and 5′-TCGTCCTCGTCCTAGTCTGGGCTGGTA-3′. pM–Bridge-1(1-72) and pM–Bridge-1(1-184) were constructed by point mutagenesis with the insertion of premature stop codons in pM–Bridge-1 by utilizing the QuikChange Site-Directed Mutagenesis Kit. The oligonucleotides used for construction of pM–Bridge-1(1-72) were 5′-GGATTTGTATCAGGTCTGAACAGCAAGGCAC-3′ and 5′-GTGCCTTGCTGTTCAGACCTGATACAAATCC-3′ and of pM–Bridge-1(1-184) were 5′-CAGCACAGCGAGGGGTAGCCCCTGAATGTC-3′ and 5′-GACATTCAGGGGCTACCCCTCGCTGTGCTG-3′. pM–Bridge-1(1-133) was constructed by digestion of pM–Bridge-1 with StuI and HindIII, blunting with Klenow polymerase, and religation. Mutants were verified by sequencing, and expression was assessed by Western blotting of transfected cell extracts.
Cell culture and immunocytochemistry.
HeLa cells (American Type Culture Collection, Manassas, Va., and the gift from R. Stein), BHK-21 (C-13) cells (American Type Culture Collection), and RIN1027-B2 cells (33) were grown in Dulbecco modified Eagle medium (4.5 g of glucose per liter) supplemented with 10% fetal bovine serum, 100 U of penicillin G, and 100 μg of streptomycin sulfate per ml (GIBCO-BRL Life Technologies, Inc., Gaithersburg, Md.). INS-1 cells (gift from C. Wollheim) were grown as previously described (2). For immunocytochemistry, RIN1027-B2 cells were grown on glass slide culture chambers (Nunc, Inc., Naperville, Ill.) prior to staining. Slides were rinsed several times in phosphate-buffered saline (PBS), followed by fixation in 4% paraformaldehyde in PBS for 10 min at room temperature. After several additional rinses in PBS, cells were permeabilized with 100% methanol at −20°C for 5 min, followed by blocking with 1% normal donkey serum for 20 min at room temperature. Slides were then incubated with preimmune or rabbit polyclonal anti-Bridge-1 antisera generated against the peptide immunogen EEALHQLHARDKEKQ at a dilution of 1:500 at 4°C overnight. After several additional rinses in PBS, cells were incubated with donkey anti-rabbit immunoglobulin G (IgG) Cy2 (Jackson ImmunoResearch Laboratories, West Grove, Pa.) at a dilution of 1:500 for 1 h at room temperature in the dark. Slides were then rinsed with PBS and mounted with fluorescence mounting medium (Kirkegaard and Perry Laboratories, Gaithersburg, Md.). Adult murine pancreas was embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Torrance, Calif.) and frozen on dry ice. Tissue was sectioned at 7-μm increments and fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. After four rinses with PBS, permeabilization with methanol, and blocking with 1% donkey serum, sections were incubated overnight at 4°C either with rabbit polyclonal anti-Bridge-1 antiserum (1:500 dilution) and guinea pig anti-insulin antiserum (1:200 dilution) (Linco Research, Inc., St. Charles, Mo.) or with preimmune rabbit serum (1:500 dilution) and guinea pig anti-insulin antiserum (1:200 dilution). Sections were then rinsed in PBS and incubated for 90 min with donkey anti-rabbit IgG Cy3 (1:1,500 dilution) and donkey anti-guinea pig IgG Cy2 (1:500 dilution) (Jackson ImmunoResearch Laboratories). Sections were then rinsed in PBS and mounted in fluorescence mounting medium (Kirkegaard and Perry). Embryonic day 19 mouse pancreas was fixed in 4% paraformaldehyde, followed by incubation in 30% sucrose prior to embedding in paraffin. Sections were cut at 5-μm intervals, and paraffin was extracted with sequential washes in xylene, ethanol solutions, and PBS. Immunostaining was conducted with rabbit polyclonal anti-Bridge-1 antisera (1:500 dilution). Slides were incubated with a biotinylated secondary antibody, followed by an avidin-biotinylated horseradish peroxidase complex (Vectastain ABC System; Vector Laboratories, Burlingame, Calif.). Images were acquired with a Nikon epifluorescence microscope with an Optronics TEC-470 camera (Optronics Engineering, Goleta, Calif.) with an interface to a Power Macintosh 7100 computer. Image analysis was conducted with IP Lab Spectrum software (Signal Analytics Corp., Vienna, Va.) and Adobe Photoshop 4.0 software (Adobe Systems Incorporated, San Jose, Calif.).
Transfections.
In some experiments HeLa cells were transfected with 25 μg of total DNA by the calcium phosphate precipitation method with the CalPhos Maximizer Transfection Kit (Clontech Laboratories) according to the manufacturer’s instructions. In additional studies, INS-1, BHK, or HeLa cells were transfected with 5 μg of total DNA and 5 to 10 μl of Lipofectamine as outlined by the manufacturer (GIBCO-BRL Life Technologies). Cells were harvested 48 h after transfection. Chloramphenicol acetyltransferase (CAT) assays were conducted with the fluorescent substrate assay kit FAST CAT (Molecular Probes, Eugene, Oreg.) and thin-layer chromatography on silica gel plates (Eastman Kodak, Rochester, N.Y.) as previously reported (42). Quantitation was performed with a FluorImager 575 interfaced with ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). Luciferase assays were conducted as previously described (25).
In vitro transcription and translation reactions.
Rat Bridge-1 was synthesized in rabbit reticulocyte extracts by coupled in vitro transcription and translation from the plasmid pcDNA3-Bridge-1 by using the T7 polymerase and the TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer’s instructions. In vitro-translated E12 and E47 were generated by using the same procedure with the plasmids pcDNA3-E12 and pcDNA3-E47, respectively. Reactions were conducted with either cold or 35S-radiolabeled methionine (NEN Life Science Products). To visualize the incorporation of [35S]methionine, reactions were subjected to sodium dodecyl sulfate (SDS)–10% polyacrylamide electrophoresis (PAGE), followed by gel incubation in an autoradiography enhancer (Enlightening; NEN Life Science Products) prior to autoradiography.
Western blot analysis.
In vitro-translated reaction products were fractionated by SDS-PAGE, electroblotted onto Immobilon-P membranes (Millipore, Bedford, Mass.), and incubated with rabbit polyclonal anti-Bridge-1 antisera (1:2,000 dilution). Extracts from transfected cells were fractionated by SDS-PAGE, electroblotted onto Immobilon-P membranes, and incubated with rabbit polyclonal anti-Bridge-1 antisera (1:2,000 dilution), rabbit polyclonal anti-Gal4DBD antiserum (1:1,000 dilution) (sc-577; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), or rabbit polyclonal anti-E12 antisera (1:1,000 dilutions) (sc-349 and sc-762; Santa Cruz Biotechnology). Protein bands were visualized by chemiluminescence with ECL Western blotting detection reagents (Amersham Life Sciences, Arlington Heights, Ill.) with a horseradish peroxidase-conjugated goat anti-rabbit antibody (Bio-Rad Laboratories, Richmond, Calif.).
Immunoprecipitations.
In vitro-translated proteins were preincubated in PBS for 30 min at 4°C prior to preclearing with protein A-Sepharose (Pharmacia Biotech AB, Uppsala, Sweden) for 30 additional minutes. Protein supernatants were incubated for 1 h at 4°C with rabbit polyclonal anti-Bridge-1 antisera or preimmune antisera. Immune complexes were then precipitated with protein A-Sepharose, washed, and separated by SDS-PAGE, followed by autoradiographic enhancement and autoradiography.
RESULTS
Cloning and characterization of Bridge-1.
To identify proteins that might modulate the activity of E2A on target genes in pancreatic β cells, such as the insulin gene, a yeast two-hybrid screening system was developed. A cDNA library derived from the insulinoma cell line, INS-1 (2), was screened by using a bait derived from the carboxy terminus of E12 that included the bHLH DNA-binding and dimerization domains (amino acids 521 to 649). Approximately 0.5 × 106 colonies were screened to identify four clones that interacted strongly with E12 (Table 1). Sequencing and database comparisons identified clones 169 and 6 as rat Twist and Id3, respectively; both proteins are class B bHLH proteins known to dimerize with E12 (23, 40). Their isolation indicated that the E12 bait worked as predicted in the yeast two-hybrid screening system. In addition, two novel sequences were identified. Clone 36 encodes PIN-1, a novel 177-amino-acid open reading frame with homology to PDZ domains (47a). Clone 18 encodes Bridge-1, which is the focus of this report. Interactions of these four clones with unrelated baits, including human interleukin receptor (cytoplasmic domain, amino acids 477 to 527) and Drosophila melanogaster bicoid (pRFHM-1 [15]), were tested as negative controls.
TABLE 1.
Specificity of clone interactions with E12 in yeast two-hybrid screeninga
| Clone no. | Identification | Interaction with:
|
||
|---|---|---|---|---|
| IL-R | Bicoid | E12 | ||
| 169 | Rat Twist | − | − | ++ |
| 6 | Rat Id3 | − | − | ++ |
| 36 | PIN-1 | − | − | ++ |
| 18 | Bridge-1 | − | − | ++ |
Four strong E12 interacting clones identified by yeast two-hybrid screening were tested for their interaction with human interleukin-1 receptor (IL-R), D. melanogaster bicoid, and rat E12 fusion proteins as indicated by semiquantitative yeast two-hybrid interaction assay. The strength of interaction was measured by growth on leucine dropout plates to follow LEU reporter gene activity and by the intensity of blue color on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates to assess lacZ reporter gene activity after a 30°C incubation for 72 h. −, No growth on leucine dropout plates and white colonies on X-Gal plates; ++, growth on leucine dropout plates and intense blue colonies on X-Gal plates.
Clone 18 contained a cDNA insert of 934 bp. Because Northern blot analysis of INS-1 RNA revealed two larger Bridge-1 transcripts of 1.3 and 1.0 kb (see Fig. 3A), a 14 dpc rat pancreatic cDNA library was screened by using a 30-mer oligonucleotide from the clone 18 sequence. A single clone (Bridge-1) with an insert of 1.4 kb was isolated (Fig. 1). DNA sequence analysis revealed an open reading frame of 222 amino acids. The start codon for this open reading frame lies within the yeast two-hybrid clone 18.
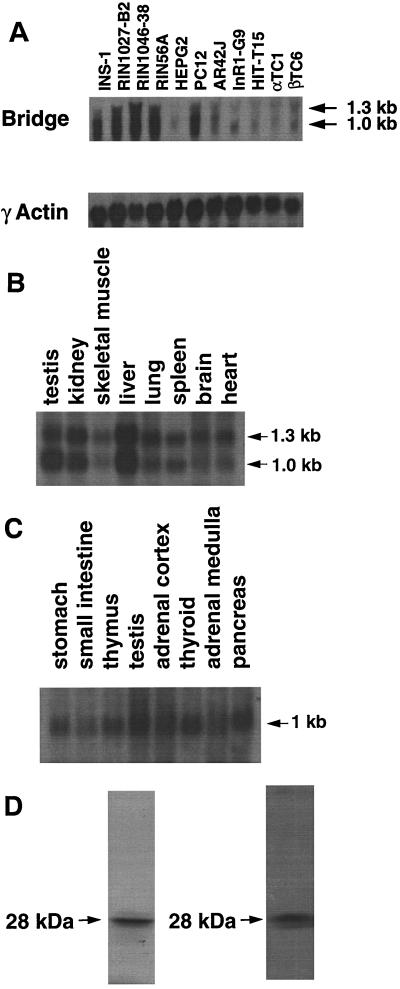
FIG. 3.
Characterization of Bridge-1 expression. (A) Northern blot of Bridge-1 transcript in total RNA from rodent cell lines (upper panel). For each lane, 20 μg of total RNA derived from the following cell lines was hybridized with 32P-labeled Bridge-1 cDNA: INS-1, rat insulinoma cells; RIN1027-B2, rat islet tumor somatostatin-secreting cells; RIN1046-38, rat insulinoma cells; RIN56A, rat insulinoma cells; HepG2, human hepatoblastoma cells; PC12, rat pheochromocytoma cells; AR42J, rat exocrine pancreatic tumor cells; InR1-G9, hamster islet tumor glucagon-secreting cells; HIT-T15, hamster insulinoma cells; αTC1, mouse islet tumor glucagon-secreting cells; βTC6, mouse islet tumor insulin-secreting cells. The blot was stripped and reprobed with a gamma-actin cDNA as a loading control (lower panel). (B) Northern blot of Bridge-1 transcript in mouse tissues. A commercially generated Northern blot (Clontech) containing 2 μg of poly(A)+ RNA per lane from the murine tissues indicated was hybridized with 32P-labeled Bridge-1 cDNA. (C) Northern blot of Bridge-1 transcript in human endocrine tissues. A commercially generated Northern blot (Clontech) containing 2 μg of poly(A)+ RNA per lane from the human tissues indicated was hybridized with 32P-labeled Bridge-1 cDNA. (D) Autoradiogram after SDS-PAGE fractionation of 35S-labeled in vitro-translated Bridge-1 protein (left panel). Western blot analysis of in vitro-translated Bridge-1 after SDS-PAGE fractionation (right panel).
FIG. 1.
Sequence of rat Bridge-1 cDNA and encoded protein. Positions of in-frame stop codons are designated by asterisks under the corresponding nucleotide sequences. The arrowhead is placed below the corresponding nucleotide sequence to indicate the starting position of the two-hybrid clone 18. The PDZ-like domain is underlined.
The Bridge-1 sequence of 222 amino acids predicts a protein with a molecular mass of 24.8 kDa and a pI of 6.70. When Bridge-1 cDNA was introduced into a coupled in vitro transcription-translation system under the control of a T7 RNA polymerase promoter, a single radiolabeled protein was produced that migrated at approximately 28 kDa (Fig. 3D, left panel).
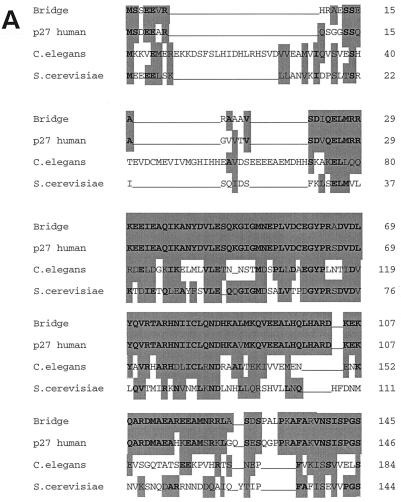
Comparison of the rat Bridge-1 protein sequence with the GenBank database via BLAST analysis revealed that Bridge-1 is highly conserved across species, including Clostridium elegans, and Saccharomyces cerevisiae (Fig. 2a). Bridge-1 cDNA is homologous to murine and human expressed sequence tag sequences from a variety of embryonic and adult tissues as well as to a human sequence with the designation proteasomal modulator subunit p27 (GenBank accession number AB003177). Rat Bridge-1 and human p27 are highly homologous, with 70% identity (156 of 222 amino acids) and 82% similarity at the protein level. The two sequences diverge at the carboxy termini of the proteins. Comparison of the first 184 amino acids of rat Bridge-1 and p27 proteins yields 84% identity and 98% similarity. Of note, the predicted translations of several human expressed sequence tagged sequences in the GenBank database diverge from the p27 sequence at the carboxy terminus and more closely resemble the carboxy terminus of rat Bridge-1 (data not shown) (44). Homologies between Bridge-1 and proteins from S. cerevisiae and C. elegans are weaker, with 37 and 35% identities, respectively.
FIG. 2.
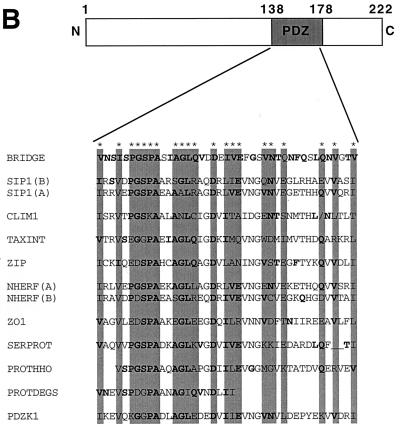
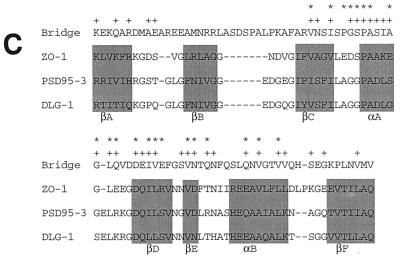
(A) Homologies between Bridge-1 protein sequence and sequences from other species. Sequences used for alignment are as follows: Bridge, rat Bridge-1 sequence; p27 human, human proteasomal modulator subunit p27 (GenBank number AB003177); C. elegans, sequence from chromosome III of C. elegans (46) (GenBank number U23453); S. cerevisiae, hypothetical 24.8-kDa protein in FAA3-BET1 intergenic region from S. cerevisiae (GenBank number P40555). Amino acid similarities as determined by BLAST analysis are shown as gray boxes, and identities are in boldface. (B) PDZ-like domain homologies between Bridge-1 and other PDZ domain-containing proteins. The Bridge-1 protein is schematically depicted to illustrate the PDZ-like domain (PDZ) identified by homology with other proteins. Alignment positions with conserved similar or identical residues by BLAST analysis in at least 50% of the aligned sequences are indicated with asterisks and gray boxes. Identical amino acids are in boldface. Protein sequences represented are as follows: BRIDGE, rat Bridge-1, amino acids 138 to 178; SIP1 A and B, interacting protein with human SRY (34) (GenBank number U82108), PDZ-domain A, amino acids 36 to 76, and PDZ-domain B, amino acids 176 to 216; CLIM1, human carboxyl-terminal LIM domain protein (GenBank number U90878), amino acids 30 to 65 and 76 to 80; TAXINT, human Tax interaction protein 1 (36) (GenBank number AF028823), amino acids 49 to 89; ZIP, human zipper containing protein (9) (GenBank number 631508), amino acids 76 to 116; NHERF, rabbit protein cofactor that mediates protein kinase A regulation of the renal brush border membrane Na+-H+ exchanger (45, 48) (GenBank number U19815) PDZ-domain A, amino acids 39 to 79, and PDZ-domain B, amino acids 179 to 219; ZO-1, mouse tight junction protein ZO-1 (17) (GenBank number P39447), amino acids 447 to 487; SERPROT, Aquifex aeolicus periplasmic serine protease (8) (GenBank number AE000741), amino acids 280 to 318; PROTHHO, Synechocystis sp. protease HhoB (GenBank number D90911), amino acids 346 to 384; PROTDEGS, E. coli protease DEGS precursor (GenBank number P31137), amino acids 284 to 305; and PDZK1, human PDZ domain containing-protein (22) (GenBank number AF012281), amino acids 403 to 443. (C) Comparison of the Bridge-1 PDZ-like domain with typical PDZ domain sequences. Alignment of typical PDZ domains and designation of regions of secondary structure (boxed) are depicted as described by Doyle et al. (10). A segment of Bridge-1 sequence is aligned for comparison. Amino acids within the Bridge-1 sequence identified as conserved among sequences aligned in Fig. 2B are designated with an asterisk for reference. For each aligned position, similarities or identities determined by BLAST analysis between the Bridge-1 sequence and any of the three ZO-1, PSD95-3, or DLG-1 sequences are indicated (+). Amino acid sequences illustrated are as follows: Bridge-1, amino acids 105 to 190; ZO-1, mouse tight junction protein ZO-1 (17) (GenBank number P39447), amino acids 423 to 501; PSD95-3, rat presynaptic density protein 95, PDZ domain 3 (5) (GenBank number P31016), amino acids 312 to 391; and DLG-1, D. melanogaster lethal (1) discs-Large-1 tumor suppressor protein (47) (GenBank number P31007), amino acids 485 to 564.
Bridge-1 has a PDZ-like domain.
Further analysis of the Bridge-1 protein sequence reveals a 41-amino-acid segment of Bridge-1, extending from amino acids 138 to 178, that is homologous to protein-protein interaction domains of the PDZ type within several other proteins (Fig. 2B). Homologies with the aligned proteins within this segment range from 27 to 54% identity and 46 to 77% similarity. This segment of homologous sequence in Bridge-1 is shorter than prototypical PDZ domains of approximately 80 to 90 amino acids that form 5 or 6 beta sheets and two alpha helices as determined by crystal structure analysis (4, 10). The high degree of sequence similarity in this region suggests that Bridge-1 contains a PDZ-like domain. A comparison of a longer segment of Bridge-1 sequence with typical PDZ domains from the proteins PSD-95, DLG-1, and ZO-1 demonstrates that the 41-amino-acid segment identified within Bridge-1 corresponds to three beta sheets (βC-βE) and two alpha helices (αA and αB) of PDZ domains (Fig. 2C). The distance between beta sheets B and C varies among PDZ domains within different proteins (4, 10), raising the possibility that additional sequences within Bridge-1 may contribute to forming a complete PDZ domain. Bridge-1 sequence is less similar to the amino-terminal portions of typical PDZ domains. Bridge-1, like ZO-1, lacks the conserved sequence GLGF, between beta sheets A and B, that has been identified as the substrate binding site within the third PDZ domain of PSD-95 (10). Although secondary structure predictions suggest that multiple alpha helices and beta sheets may form within the Bridge-1 protein (data not shown), they do not predict a secondary structure pattern that resembles the crystal structures of PDZ domains within PSD-95 or DLG-1 (4, 10).
Tissue distribution of Bridge-1 expression.
To study the tissue distribution of Bridge-1 at the RNA level, Bridge-1 cDNA was used as a probe for Northern analysis of RNAs from cells and tissues derived from rodents and humans (Fig. 3A to C). Two transcripts of approximately 1.0 kb and 1.3 to 1.4 kb in size are consistently noted in RNA from rodent tissues (Fig. 3A and B); however, only a single transcript of approximately 1.0 kb is observed in human tissues (Fig. 3C). Bridge-1 RNA is highly expressed in a variety of cell lines derived from pancreatic islets, including the insulinoma line INS-1 (2), from which the cDNA was cloned, and the somatostatin-producing cell line RIN1027-B2 (33). Lower levels of expression of Bridge-1 are observed in the glucagon-producing cell lines InR1-G9 and αTC1 and the hepatoma cell line HepG2 (Fig. 3A). Although the expression of Bridge-1 RNA is highest in pancreas, testis, kidney, and liver, expression is detectable in all rodent and human RNA sources tested. Consistent with a widely distributed pattern of Bridge-1 expression are the existence in the GenBank databases of several homologous murine and human expressed sequence-tagged cDNA sequences derived from a wide range of tissues, including whole embryos, placenta, brain, central nervous system, heart, and uterus.
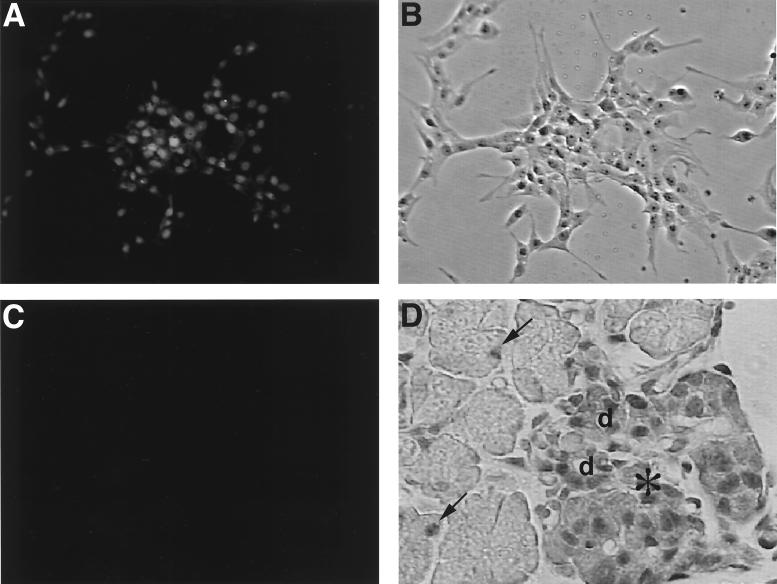
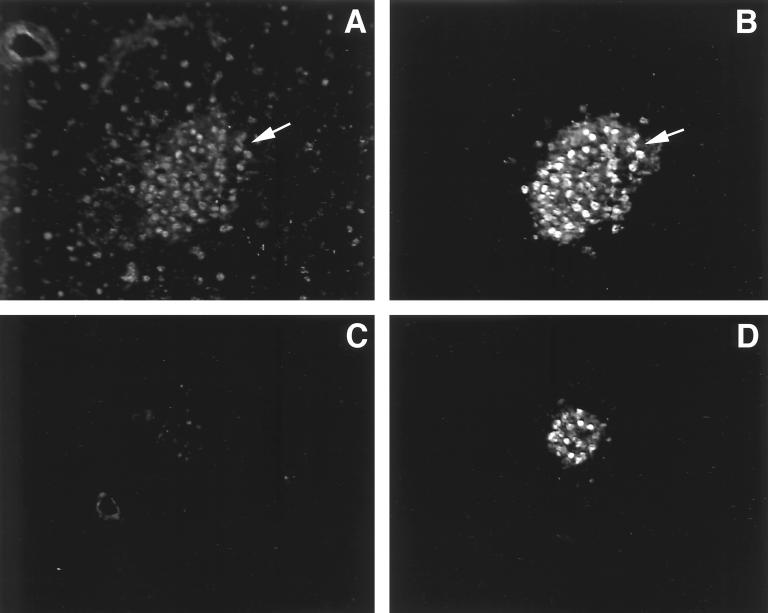
To assess the protein expression pattern of Bridge-1, rabbit polyclonal antisera were generated against an internal peptide sequence within Bridge-1. These antisera distinguish the in vitro-translated Bridge-1 on Western blots from other proteins present in the rabbit reticulocyte lysate (Fig. 3D, right panel). Assessment of Bridge-1 expression in RIN1027-B2 cells by immunocytochemistry reveals that Bridge-1 is predominantly located in the nucleus, although lower levels of cytoplasmic staining are observed in some cells (Fig. 4A). Bridge-1 expression does not appear to localize to the cytoplasmic projections extended by the RIN1027-B2 cells growing in culture. In embryonic day 19 mouse pancreas, nuclear Bridge-1 immunostaining is prominent within the cells of pancreatic islets, in ductal cells, and in a few scattered nuclei of pancreatic exocrine cells (Fig. 4D). A lower level of cytoplasmic Bridge-1 staining is seen in islets but not in the exocrine pancreas. In the adult murine pancreas, Bridge-1 is expressed in pancreatic β cells, as demonstrated by the coexpression with insulin (Fig. 5A and B). The observed pattern of Bridge-1 protein expression by immunostaining, with higher levels of endocrine relative to exocrine pancreatic expression, as well as expression in pancreatic β cells, mimics the RNA expression patterns observed in pancreatic cell lines (Fig. 3A).
FIG. 4.
Immunocytochemical staining of Bridge-1 in RIN1027-B2 cells and mouse pancreas. Fluorescent immunostaining of RIN1027-B2 cells was conducted with rabbit polyclonal anti-Bridge-1 antisera (A) or preimmune antisera (C) and photographed under identical conditions. (B) A phase-contrast view of the field of cells stained with anti-Bridge-1 antisera is shown for comparison. (D) Mouse embryonic pancreas at day 19 was stained with rabbit polyclonal anti-Bridge-1 antisera. An islet (∗), ducts (d), and an adjacent exocrine pancreas are shown. Examples of exocrine cell nuclei with positive Bridge-1 immunostaining are indicated with arrows.
FIG. 5.
Bridge-1 is coexpressed with insulin in murine pancreas. Fluorescent immunostaining of adult murine pancreas was conducted by costaining with rabbit polyclonal anti-Bridge-1 antiserum (A) and guinea pig anti-insulin antiserum (B). Costaining of an adjacent section of murine pancreas with rabbit preimmune antiserum (C) and guinea pig anti-insulin antiserum (D), photographed under identical conditions, is shown for comparison. Arrows point to β cells that coexpress Bridge-1 and insulin within an Islet of Langerhans.
Bridge-1 interaction with E12 and E47.
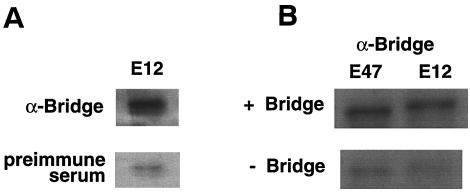
To confirm the observation of the interaction between Bridge-1 and E12 seen by yeast two-hybrid analysis, additional studies were conducted. Antisera directed against Bridge-1 effectively coimmunoprecipitate in vitro-translated 35S-radiolabeled E12 and in vitro-translated Bridge-1, compared with preimmune antisera (Fig. 6A). Both 35S-radiolabeled in vitro-translated E12 and E47 proteins coimmunoprecipitate with in vitro-translated Bridge-1. Immunoprecipitations with anti-Bridge-1 antisera conducted in the presence or absence of in vitro-translated Bridge-1 demonstrate the requirement for Bridge-1 for efficient immunoprecipitation of radiolabeled E12 or E47 (Fig. 6B).
FIG. 6.
Coimmunoprecipitation of Bridge-1 and E2A proteins. (A) 35S-labeled in vitro-translated E12 was incubated with cold in vitro-translated Bridge-1 prior to immunoprecipitation with anti-Bridge-1 rabbit polyclonal antisera (upper panel) or preimmune antisera (lower panel). (B) Immunoprecipitation reactions with anti-Bridge-1 antisera were conducted with 35S-labeled in vitro-translated E12 or E47 in the presence (upper panel) or absence (lower panel) of cold in vitro-translated Bridge-1. Autoradiograms of the immunoprecipitation reactions after SDS-PAGE fractionation are shown. In vitro-translated E12 and E47 migrated on SDS-PAGE at approximately 69 and 68 kDa, respectively.
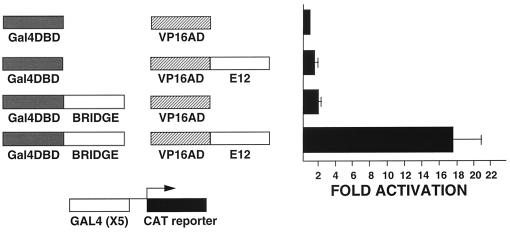
In mammalian two-hybrid studies, fusion protein constructs were transiently transfected into HeLa cells and tested for their ability to activate a Gal4CAT reporter (Fig. 7). Fusion protein constructs were generated with the Gal4DNA-binding domain or the activation domain of the VP16 protein from the herpes simplex virus. Empty vectors or the VP16-E12 fusion protein alone have little activity in this system. The Gal4 DNA-binding domain–Bridge-1 fusion construct has a slight but detectable level of activation of the reporter. However, in the presence of both the VP16-E12 and the Gal4 DNA-binding domain–Bridge-1 fusion constructs, an 18-fold increase in CAT activity is seen. This activity, in excess of the sum of the activation observed for either fusion construct alone, demonstrates that Bridge-1 interacts with E12 in living mammalian cells to bring the Gal4 DNA-binding domain into proximity to the VP16 activation domain to activate the transcriptional reporter.
FIG. 7.
Bridge-1 interacts with E12 in a mammalian two-hybrid system. HeLa cells were transiently transfected with 5 μg of Gal4CAT reporter and 10 μg of pM (Gal4DBD), pM-Bridge (Gal4DBD–Bridge-1), pVP16 (VP16AD), or pVP16-E12 (VP16AD-E12) as indicated. Results shown are the means ± the standard error of the mean (SEM) of five transfections (n = 5), each conducted in duplicate.
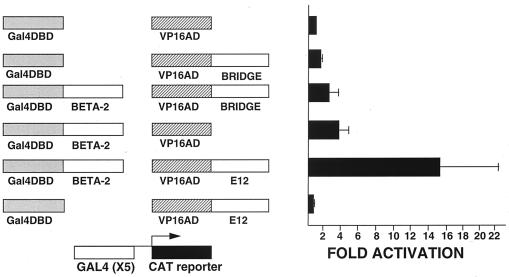
Bridge-1 does not interact with Beta-2/NeuroD.
To determine whether Bridge-1 might be interacting in a nonspecific manner with other bHLH proteins, the pancreas-specific transcription factor Beta-2/NeuroD was analyzed in the mammalian two-hybrid system. Beta-2/NeuroD is known to directly interact with E2A proteins via heterodimerization of bHLH domains (27, 29). As a positive control, Beta-2/NeuroD and E12 interactions were assessed (Fig. 8). The VP16-E12 and Gal4 DNA-binding domain–Beta-2 fusion proteins together activate the Gal4CAT reporter by 15-fold, a substantially higher activation than that seen for either fusion construct alone. These findings demonstrate and confirm Beta-2/NeuroD interaction with E12. In contrast, the combination of VP16–Bridge-1 and Gal4 DNA-binding domain–Beta-2 fusion constructs did not activate the Gal4CAT reporter compared with either of these two fusion constructs tested individually. Neither mammalian two-hybrid studies nor coimmunoprecipitation studies with in vitro-translated proteins (data not shown) demonstrate any interactions between Bridge-1 and Beta-2/NeuroD.
FIG. 8.
Bridge-1 and Beta-2/NeuroD do not interact in a mammalian two-hybrid system. HeLa cells were transiently transfected with 5 μg of Gal4CAT reporter and 10 μg of pM (Gal4DBD), pM–Beta-2 (Gal4DBD BETA-2), pVP16 (VP16AD), pVP16-E12 (VP16AD E12), or pVP16–Bridge-1 (VP16AD BRIDGE) as indicated. Results shown are the means ± the SEM of two transfections conducted in duplicate.
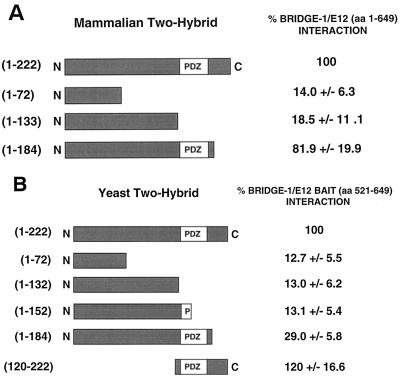
The PDZ-like domain of Bridge-1 is required for interaction with E12.
To identify the domains within Bridge-1 that mediate its interaction with E2A proteins, Bridge-1 deletion mutants were constructed as Gal4 DNA-binding domain fusion proteins for analysis of interaction with the VP16-E12 fusion protein construct in mammalian two-hybrid studies. Bridge-1 fusion constructs containing the PDZ domain interacted efficiently with the full-length E12 fusion protein, whereas Bridge-1 constructs without the PDZ domain were weak interactors relative to full-length Bridge-1 (Fig. 9A). Marked overexpression of a mutant Bridge-1 fusion protein retaining amino acids 1 to 133, but lacking the PDZ domain could produce an interaction with the VP16-E12 fusion construct, raising the possibility that a second weaker cryptic interaction domain may exist within the amino-terminal portion of Bridge-1 (data not shown).
FIG. 9.
The Bridge-1–E12 interaction requires the PDZ-like domain of Bridge-1. (A) In a mammalian two-hybrid system, HeLa cells were transiently transfected with Gal4CAT reporter, pM, pM-Bridge-1, pVP16, or pVP16-E12, as in Figure 7. pM–Bridge-1(1-72), pM–Bridge-1(1-133), and pM–Bridge-1(1-184) were substituted for pM–Bridge-1. The interaction of each mutant with pVP16-E12 was assessed and normalized to the interaction seen for the full-length pM–Bridge-1 and pVP16-E12. Aliquots of the transfected cell extracts were assessed by Western blotting to confirm comparable expression of the pM–Bridge-1 constructs tested. Results shown are the mean ± the SEM of four transfections (n = 4), conducted in duplicate. (B) By yeast two-hybrid screening, Bridge-1 mutant fusion proteins (amino acids 1 to 222, amino acids 1 to 72, amino acids 1 to 132, amino acids 1 to 152, amino acids 1 to 184, and amino acids 120 to 122) were tested for the strength of their interaction with rat E12(521-649) fusion proteins by measurement of β-galactosidase levels extracted from transformants in a yeast two-hybrid interaction assay. Results shown are the mean ± the SEM of three independent experiments (n = 3), conducted with two yeast transformants per construct. Expression levels of each of the Bridge-1 fusion constructs were comparable, as assessed by Western blots.
In an independent series of experiments by yeast two-hybrid analysis, the original E12 carboxy-terminal bait (amino acids 521 to 649) was used to assess the strength of interaction of a series of Bridge-1 deletion mutants (Fig. 9B). In a pattern analogous to that seen in mammalian cells, Bridge-1 mutants lacking a complete PDZ-like domain failed to efficiently interact with the E12 bait. In contrast, a mutant lacking the amino-terminal portion of Bridge-1 but retaining the PDZ-like domain (amino acids 120 to 222) demonstrated a potent interaction with E12. In both mammalian and yeast cells, Bridge-1 requires an intact PDZ-like domain in order to interact with E12.
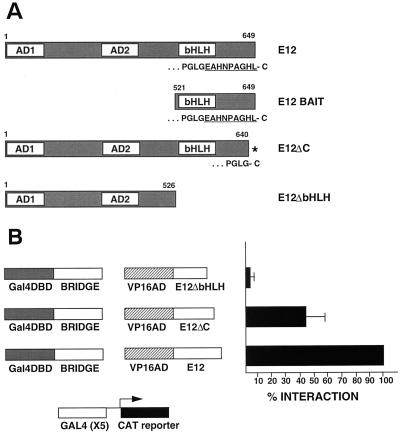
The carboxy terminus of E12 participates in the interaction with Bridge-1.
Although the PDZ domain within Bridge-1 has some atypical structural features, it may function as an acceptor site for carboxy-terminal residues of interacting proteins analogous to more typical PDZ domains (38). To test whether the carboxy-terminal amino acids of E12 contribute to the Bridge-1–E12 interaction, a mutant (E12ΔC) was generated to prematurely truncate E12 by removing the carboxy-terminal nine amino acids but retaining the two activation domains and the bHLH domain (Fig. 10A). With this mutation, E12 no longer terminates in a hydrophobic residue. In mammalian two-hybrid analysis, the introduction of this mutation into the VP16-E12 fusion construct significantly impairs interaction with the Gal4 DNA-binding domain–Bridge-1 fusion construct, decreasing the strength of the E12ΔC interaction with Bridge-1 to 45% of that with full-length E12 (Fig. 10B). These data indicate that the carboxy terminus of E12 participates in the interaction with Bridge-1.
FIG. 10.
The carboxy terminus of E12 mediates the Bridge-1–E12 interaction. (A) Schematic diagrams of E12 (amino acids 1 to 649), the E12 fragment utilized as bait in the yeast two-hybrid analysis that identified Bridge-1 (amino acids 521 to 649), the E12 mutant E12ΔC (amino acids 1 to 640), and the E12 mutant E12ΔbHLH (amino acids 1 to 526). The two activation domains are designated as AD1 and AD2 and the bHLH domain as indicated (modeled after a published schematic diagram [39]). The carboxy-terminal sequences of E12, E12 bait, and E12ΔC are shown below the respective schematic diagrams. The asterisk designates the carboxy terminus of E12ΔC, truncated by nine amino acids (EAHNPAGHL), due to the introduction of a TAG stop codon corresponding to amino acid position 641 in E12. (B) HeLa cells were transiently transfected with Gal4CAT reporter, pM, pM–Bridge-1, pVP16 (VP16AD), or pVP16-E12 (VP16AD E12), as in Fig. 7. pVP16-E12ΔC or pVP16-E12ΔbHLH were substituted for pVP16-E12 and assessed for interaction with pM–Bridge-1. The observed interaction was normalized to that seen for pVP16-E12. Aliquots of transfected cell extracts were assessed by Western blotting to confirm comparable expression of E12 and E12 mutant fusion proteins. Results shown are the means ± the SEM of three to five transfections (n = 3 to 5) conducted in duplicate.
In contrast, the E12ΔC mutant retains the ability to interact with Beta-2/NeuroD. In mammalian two-hybrid studies, the VP16-E12ΔC fusion construct interaction with the Gal4 DNA-binding domain–Beta-2 fusion construct is not impaired relative to the VP16/E12 construct interaction with the Gal4 DNA-binding domain–Beta-2 construct (data not shown). These results are consistent with Beta-2/NeuroD heterodimerization with E12 via its bHLH domain, a domain left intact in the E12ΔC mutant.
Deletion of the bHLH domain and carboxy terminus of E12, by introduction of a stop codon at amino acid position 527, results in an E12 mutant (E12ΔbHLH) that is no longer able to interact with Bridge-1 (Fig. 10B). This mutant retains both of the activation domains within E12 (Fig. 10A). These data suggest that the E12 bait used for yeast two-hybrid screening (amino acids 521 to 649) encompasses all of the E12 domains that participate in the interaction of E12 and Bridge-1.
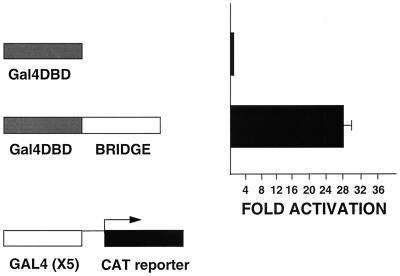
Bridge-1 has intrinsic transactivation potential.
A small but detectable level of activity was observed for the Gal4 DNA-binding domain–Bridge-1 fusion construct alone in transient transfections in HeLa cells (Fig. 7), supporting the idea that Bridge-1 might have intrinsic transactivation activity independent of interactions with E2A. The activity of this construct is considerably higher in transient transfections of BHK cells (Fig. 11), suggesting that Bridge-1 activation may be a regulated function that varies with differences in intracellular signaling. In BHK cells, the Gal4 DNA-binding domain–Bridge-1 fusion construct activates the Gal4CAT reporter 28-fold compared to activation of the reporter by the empty vector containing the Gal4 DNA-binding domain alone.
FIG. 11.
Bridge-1 has intrinsic transactivation potential. BHK cells were transiently transfected with 1 μg of Gal4CAT reporter and 4 μg of pM (Gal4DBD) or 4 μg of pM–Bridge-1 (Gal4DBD-Bridge), as indicated. Results shown are the means ± the SEM of three transfections (n = 3) conducted in triplicate.
Bridge-1 coactivates insulin promoter elements with E12 and E47.
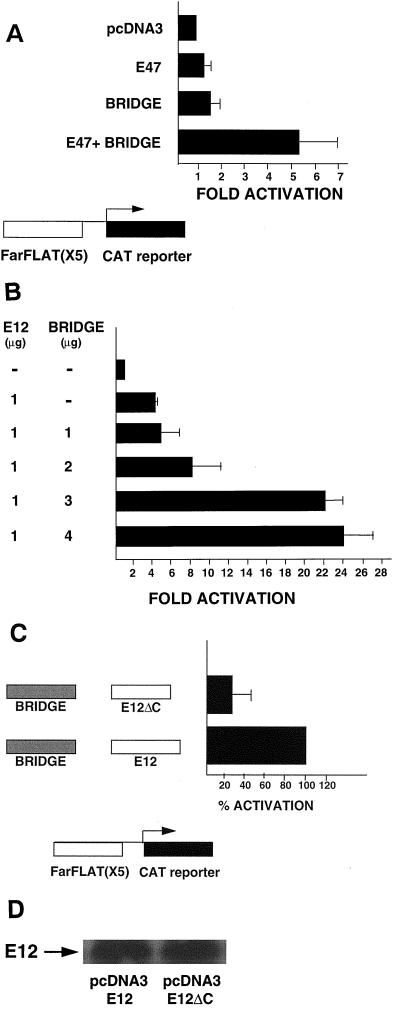
To test the activity of Bridge-1 on mammalian promoter regulatory elements, we utilized a CAT reporter regulated by upstream multimerized E and A box (FarFlat) minienhancers of the rat insulin I gene (5FF1CAT). In transient transfections of HeLa cells with equal amounts of expression vectors for Bridge-1 and E47, activation of the 5FF1CAT reporter was assessed (Fig. 12A). Transfection of E47 alone minimally activates the reporter 1.4-fold, and transfection of Bridge-1 alone results in a 1.5-fold activation. However, transfection of the two constructs in combination results in a 5.2-fold activation of the reporter, suggesting that Bridge-1 enhances E47 transactivation. Transfection of increasing amounts of Bridge-1 expression vector also increases E12-mediated activation of the FarFlat reporter in a dose-dependent manner, up to 24-fold (Fig. 12B). The E12ΔC mutation that impairs Bridge-1–E12 interaction decreases, by 65%, the combined activity of Bridge-1 and E12 on the FarFlat reporter (Fig. 12C). The strength of Bridge-1 interaction with E12 appears to modulate the level of the observed coactivation. Western blotting of extracts from these transfected cells demonstrates that the differences in coactivation are not a result of significant differences in expression of E12 and E12ΔC (Fig. 12D). No significant difference was observed in the activation of 5FF1CAT by E12ΔC versus E12 in transfections conducted without the addition of Bridge-1 (data not shown).
FIG. 12.
Bridge-1 coactivation of rat insulin I minienhancer FarFlat reporter with E47 or E12. (A) Bridge-1 coactivation of FarFlat with E47. HeLa cells were transiently transfected with 2 μg of 5FF1CAT reporter and 2 μg of pcDNA3-E47, and/or 2 μg of pcDNA3-Bridge-1 as indicated. pcDNA3 was added to normalize the amount of pcDNA3 vectors across all transfections. pBluescript was included to provide a total of 25 μg of DNA per transfection. Results shown are the means ± the SEM of six transfections conducted in triplicate (n = 5) or duplicate (n = 1). (B) Dose-dependent Bridge-1 coactivation of FarFlat with E12. HeLa cells were transiently transfected with 2 μg of 5FF1CAT reporter in the presence or absence of 1 μg of pcDNA3-E12 and/or 1 to 4 μg of pcDNA3-Bridge-1 as indicated. pcDNA3 was added to normalize the amount of pcDNA3 vectors across all transfections. pBluescript was included to provide a total of 25 μg of DNA per transfection. Results shown are the means ± the SEM of triplicate samples. (C) The E12ΔC mutant diminishes Bridge-1 coactivation of FarFlat. HeLa cells were transiently transfected with 2 μg of 5FF1CAT reporter in the presence of 4 μg of pcDNA3–Bridge-1 and 1 μg of pcDNA3-E12 or 1 μg of pcDNA3-E12ΔC. pBluescript was included to provide a total of 25 μg of DNA per transfection. Results shown are the means ± the SEM of four transfections (n = 4) conducted in duplicate and normalized to the activity of pcDNA3–Bridge-1 with pcDNA3-E12. (D) Representative Western blot demonstrating comparable expression of pcDNA3-E12 and pcDNA3-E12ΔC. Aliquots of extracts from cells transfected in a representative experiment as described in panel C were subjected to SDS-PAGE, followed by Western blotting with rabbit polyclonal antisera directed against E12.
In selected DNA-binding assays (not shown) with a FarFlat oligonucleotide probe, no direct binding of either in vitro-translated or recombinant Bridge-1 was observed. In similar studies, anti-Bridge-1 antisera did not attenuate DNA-binding of protein complexes from insulinoma cell nuclear extracts, although attenuation of protein-binding was observed with anti-E2A antiserum (Santa Cruz Biotechnology).
Endogenous Bridge-1 inactivation impairs insulin promoter activity in INS-1 cells.
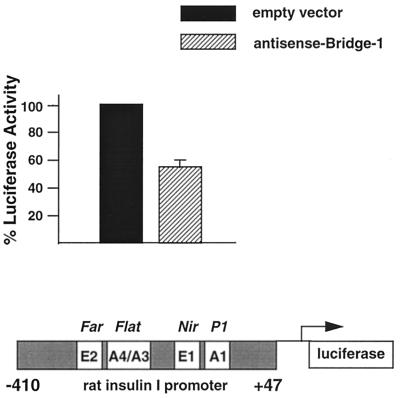
To determine whether endogenous Bridge-1 levels are important in the regulation of insulin promoter activity, an antisense Bridge-1 cDNA construct, AS–Bridge-1–pcDNA3, was employed in transient transfections of the INS-1 rat insulinoma cell line. A promoter-reporter construct consisting of −410 to +47 rat insulin I promoter sequences, −410INS-LUC (25), was utilized to assess the effect of expression of the antisense Bridge-1 construct. Expression of the AS–Bridge-1–pcDNA3 construct decreased insulin promoter activity by 45% (Fig. 13). These results indicate that endogenous Bridge-1 contributes to insulin promoter activation in insulin-producing cells.
FIG. 13.
Expression of antisense Bridge-1 in transfected INS-1 cells decreases rat insulin I promoter activity. INS-1 cells were transiently transfected with 1 μg of AS–Bridge-1–pcDNA3 (antisense Bridge-1), a construct encoding the 1.4-kb Bridge-1 cDNA in the antisense orientation, or 1 μg of pcDNA3 (empty vector) and 1 μg of −410INS-LUC, a rat insulin I promoter-reporter construct spanning residues −410 to +47 of the rat insulin I promoter sequence (25). This portion of the rat insulin I promoter contains two sets of tandem E and A boxes as designated in the schematic diagram, including the minienhancer FarFlat. pBluescript was included to provide a total of 5 μg of DNA per transfection. Results shown are the means ± the SEM of two transfections (n = 2) conducted in duplicate and normalized for cellular extract protein concentrations. Data are expressed relative to the activity of pcDNA3 and −410INS-LUC.
DISCUSSION
We have identified a novel protein that interacts with E12 by yeast two-hybrid analysis. The protein is designated Bridge-1 to reflect its probable role as a coactivator that functions via protein-protein interactions. Consistent with this nomenclature is the localization within Bridge-1 of a truncated PDZ-like domain that is required for its interaction with E12. Whereas most characterized PDZ domain-containing proteins have been localized to membrane or cytoplasmic compartments, a small number of nuclear proteins with PDZ domains have been identified. The PDZ-like domain within Bridge-1 may be a subtype that functions within the nucleus, since it is similar to PDZ domains within the nuclear protein SIP-1. SIP-1 interacts with the testis determining factor SRY (34) and has a sequence identical to that of proteins designated TKA-1 (tyrosine kinase activator-1, GenBank accession number Z50150) and E3KARP (NH3 kinase A regulatory protein [48]). Other proteins with PDZ domains homologous to Bridge-1 include a Tax-binding protein (36), and human proteins of unknown function with leucine zipper or LIM domain motifs usually found in transcription factors (Fig. 2B). The PDZ-like domain within Bridge-1 also shares homology with cytoplasmic proteins, including a regulator of renal Na+-H+ exchange, proteases of the DEGS type, and the tight junction protein ZO-1.
The PDZ-like domain of Bridge-1 has some atypical structural features. The stretch of PDZ homology within Bridge-1 is 41 amino acids, approximately one-half the size of typical PDZ domains (4, 10). It is possible that additional regions within Bridge-1 may contribute secondary structure to complete the PDZ domain. The absence of conservation within Bridge-1 of the peptide binding site described in the third PDZ domain of PSD-95 (10) is of interest. However, alterations in substrate binding site configurations within PDZ domains are likely needed to provide specificity for protein-protein interactions. Bridge-1 may contain a distinct type of PDZ domain with different structural determinants.
The identification, by yeast two-hybrid screening with the same E12 carboxy-terminal bait, of a second novel PDZ domain-containing protein PIN-1 (clone 36) (47a), raises the possibility that Bridge-1 is a part of a larger signaling network involving E2A-PDZ domain communication. Future studies of Bridge-1 and PIN-1 function should provide opportunities to define signaling pathways that may regulate E2A function.
E12 and E47 interact with other bHLH proteins through heterodimerization via HLH domains (26). The yeast two-hybrid bait used to clone Bridge-1 was derived from carboxy-terminal E12 sequences, including the bHLH domain and the carboxy terminus (Fig. 10A). Although the Bridge-1 protein sequence includes regions of hydrophobicity, homologies with bHLH proteins were not observed. In addition, the failure of Bridge-1 to interact with Beta-2/NeuroD, under experimental conditions in which Bridge-1–E12 and Beta-2–E12 interaction occur, indicates the specificity of Bridge-1 and E2A protein interactions. In general, PDZ domains interact with carboxy-terminal sequences within target proteins (38). Truncation of the E12 carboxy terminus markedly reduced its interaction with Bridge-1, suggesting that the model of PDZ domain interaction with carboxy-terminal sequences also applies to the Bridge-1–E12 interaction. In contrast, deletion of nine carboxy-terminal amino acids from E12 did not interfere with the interaction of Beta-2/NeuroD with E12. It is probable that other regions within E12, such as the bHLH domain, stabilize the interaction with Bridge-1, since deletion of both the bHLH domain and the carboxy terminus of E12 abolishes interaction with Bridge-1.
The E12 carboxy terminus is important both for interaction with Bridge-1 and for Bridge-1 coactivation of E12. Bridge-1 coactivation of E12-mediated transactivation of the insulin promoter enhancer sequence FarFlat was impaired by approximately 65% in the absence of the nine carboxy-terminal amino acids of E12. These data support a model in which interaction of the carboxy-terminal domain of E12 with the PDZ-like domain of Bridge-1 results in increased transcriptional activation of E12 targets. This Bridge-1–E12 interaction model is distinct from typical heterodimerization models thought to be employed by most E12 interacting proteins.
Other models of protein-protein interaction for E2A proteins that do not involve heterodimerization via HLH domains are supported by reports of E2A interactions with the polymyositis-scleroderma autoantigen and the ubiquitin-conjugating enzyme, UbcE2A/mUBC9 (20, 21, 24). These proteins interact with internal E12 sequences distinct from the carboxy terminus. The interaction of E2A proteins with UBcE2A-mUBC9 is intriguing in light of the designation, human proteasomal modulator subunit p27, attributed to the human homologue of Bridge-1 (44). Additional evidence for the potential involvement of components of a regulated protein degradation pathway in E2A signaling was provided by a report that the proteasomal subunit S5a regulates the binding of E12 and MyoD to DNA by direct interactions with Id1 (1). Further studies are required to determine whether Bridge-1 may function both as an E2A coactivator and in some additional capacity as a regulator of E2A degradation.
Our original intent in screening an insulinoma cell cDNA library was to identify factors within the endocrine pancreas that may modulate E2A function on β-cell genes. However, the wide distribution of Bridge-1 expression resembles that of the E2A proteins and suggests that it may play a broader role in the modulation of E2A activity. Furthermore, the level of protein conservation of Bridge-1 across species implies a fundamental function of biological importance. It is tempting to speculate that Bridge-1 functions in developing tissues, in light of the embryonic expression of cDNAs homologous to Bridge-1 reported in databases of expressed sequence tags derived from embryonic tissues and of Bridge-1 expression in the 14 dpc rat pancreatic cDNA library. Because E2A null mice have abnormalities of lymphocyte development (3, 49), and mice nullizygous for Beta-2/NeuroD, the pancreas-specific dimerization partner of E2A, demonstrate abnormalities in pancreas development (28), proteins that modulate the transactivation functions of E2A are candidate developmental regulators.
Bridge-1 functions as a strong activator in the context of its fusion to the Gal4 DNA-binding domain. The difference in the activity of this construct in the two cell types tested suggests that this transactivational activity may be a regulated function. Possible explanations for these differences in activity include different posttranslational modifications of Bridge-1 protein that alter its conformation and function or differences in the expression patterns of protein-binding partners for Bridge-1. The transactivation data are consistent with either an intrinsic transactivation domain within Bridge-1 or a recruitment function that attracts additional transactivating proteins.
Bridge-1 may be important in the pancreas in regulating the insulin and other islet genes. The expression of Bridge-1 in a nuclear pattern within the insulin-producing β cells of mouse islets is consistent with its proposed function as a coactivator. Inactivation of endogenous Bridge-1 by expression of Bridge-1 antisense RNA diminishes insulin promoter activity in transient-transfection studies of insulinoma cells, suggesting that Bridge-1 functions to enhance transactivation of the insulin promoter.
The action of Bridge-1 on the insulin promoter appears to be mediated, at least in part, through interaction with E2A proteins. E2A transcription factors regulate the promoter activities of islet-specific genes, including the insulin and glucagon genes, via binding to E boxes within the promoters of these genes (6). In the insulin gene, E boxes in the promoter are partially responsible for glucose-responsive transcription (13). Previous investigators have demonstrated that E2A proteins heterodimerize with the tissue-specific bHLH transcription factor Beta-2/NeuroD in binding to the E boxes of the insulin promoter (29). These heterodimers work in synergy with homeodomain proteins, including PDX-1, to activate the insulin promoter via minienhancers such as FarFlat (14, 32). Bridge-1 enhances the activation of this minienhancer by either E12 or E47. The lack of DNA-binding activity of Bridge-1 on FarFlat oligonucleotides is consistent with a role for Bridge-1 in this context as a coactivator rather than as a direct transactivator. The absence of any obvious DNA-binding domain within the Bridge-1 sequence supports this model, although it is possible that, with other target DNA sequences or under other conditions, Bridge-1 might have a cryptic DNA-binding function. Impairment of this coactivation by deletion of nine carboxy-terminal amino acids of E12, residues implicated in mediating the Bridge-1–E12 interaction, suggests that this transcriptional coactivation results from the interaction of Bridge-1 and E12. The coactivator p300 is proposed to stimulate insulin gene transcription via direct interactions with E47 and Beta-2/NeuroD (35). Our data suggest that Bridge-1 coactivation of E12 is likely to occur through a mechanism independent of Beta-2/NeuroD.
The importance of the regulation of the glucose-responsive regions of the insulin promoter is underscored by the recent reports that inactivating mutations in proteins that regulate this response, such as PDX-1, are linked to diabetes mellitus in humans (41). The observation that Bridge-1 acts with E12 and E47 to increase the activation of a critical enhancer in the insulin promoter provides an alternative model for E2A activation and may permit the elucidation of novel mechanisms of insulin gene regulation.
ACKNOWLEDGMENTS
We thank the following individuals for their contributions: Christopher Miller, Mario Vallejo, and the members of the Laboratory of Molecular Endocrinology for valuable discussions and reagents; Heather Hermann, Jee Lee, Wai-Ying Leung, Xiaolin Li, and Josephine Ngai for expert technical assistance; Ming Jer-Tsai, Frank Naya, Roland Stein, Claus Wollheim, J. Larry Moss, and Christian Nelson for valuable reagents; and Townley Budde for assistance in the preparation of the manuscript.
This work was supported by grants from the National Institute of Diabetes, Digestive, and Kidney Diseases (J.F.H. and M.K.T.) and Hong Kong University CRCG (K-M.Y.). M.K.T. is a recipient of a Howard Hughes Medical Institute Postdoctoral Research Fellowship for Physicians. J.F.H. is an Investigator with the Howard Hughes Medical Institute.
M.K.T. and K.-M.Y. contributed equally to this work.
REFERENCES
- 1.Anand G, Yin X, Shahidi A K, Grove L, Prochownik E V. Novel regulation of the helix-loop-helix protein Id1 by S5a, a subunit of the 26S proteasome. J Biol Chem. 1997;272:19140–19151. doi: 10.1074/jbc.272.31.19140. [DOI] [PubMed] [Google Scholar]
- 2.Asfari M, Janjic D, Meda P, Li G, Halban P A, Wollheim C B. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 3.Bain G, Maandag E C, Izon D J, Amsen D, Kruisbeek A M, Weintraub B C, Krop I, Schlisssel M S, Feeney A J, van Room M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 4.Cabral J H M, Petosa C, Sutcliffe M J, Raza S, Byron O, Poy F, Marfatia S M, Chishti A H, Liddington R C. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- 5.Cho K-O, Hunt C A, Kennedy M B. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 6.Cordier-Bussat M, Morel C, Philippe J. Homologous DNA sequences and cellular factors are implicated in the control of glucagon and insulin gene expression. Mol Cell Biol. 1995;15:3904–3916. doi: 10.1128/mcb.15.7.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis L G, Dibner M D, Battey J F. Basic methods in molecular biology. New York, N.Y: Elsevier; 1986. [Google Scholar]
- 8.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 9.Dixon B, Sahely B, Liu L, Pohajdak B. Cloning a cDNA from human NK/T cells which codes for an unusual leucine zipper containing protein. Biochim Biophys Acta. 1993;1216:321–324. doi: 10.1016/0167-4781(93)90165-a. [DOI] [PubMed] [Google Scholar]
- 10.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 11.Dutta S, Bonner-Weir S, Montminy M, Wright C. Regulatory factor linked to late-onset diabetes? Nature. 1998;392:560. doi: 10.1038/33311. [DOI] [PubMed] [Google Scholar]
- 12.Eckner R, Yao T-P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 13.German M S, Wang J. The insulin gene contains multiple transcriptional elements that respond to glucose. Mol Cell Biol. 1994;14:4067–4075. doi: 10.1128/mcb.14.6.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.German M S, Wang J, Chadwick R B, Rutter W J. Synergistic activation of the insulin gene by a LIM-homeodomain protein and a basic helix-loop-helix protein: building a functional insulin minienhancer complex. Genes Dev. 1992;6:2165–2176. doi: 10.1101/gad.6.11.2165. [DOI] [PubMed] [Google Scholar]
- 15.Golemis E A, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins, unit 13.14. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley and Sons; 1994. pp. 13.14.1–13.14.17. [Google Scholar]
- 16.Itkin-Ansari P, Bain G, Beattie G M, Murre C, Hayek A, Levine F. E2A gene products are not required for insulin gene expression. Endocrinology. 1996;137:3540–3543. doi: 10.1210/endo.137.8.8754784. [DOI] [PubMed] [Google Scholar]
- 17.Itoh M, Nagafuchi A, Yonemura S, Kitani-Yasuda T, Tsukita S, Tsukita S. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson J D, Zhang W, Rudnick A, Rutter W J, German M S. Transcriptional synergy between LIM-homeodomain proteins and basic helix-loop-helix proteins: the LIM2 domain determines specificity. Mol Cell Biol. 1997;17:3488–3496. doi: 10.1128/mcb.17.7.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 20.Kho C-J, Huggins G S, Endege W O, Hsieh C-M, Lee M-E, Haber E. Degradation of E2A proteins through a ubiquitin-conjugating enzyme, UbcE2A. J Biol Chem. 1997;272:3845–3851. doi: 10.1074/jbc.272.6.3845. [DOI] [PubMed] [Google Scholar]
- 21.Kho C-J, Huggins G S, Endege W O, Patterson C, Jain M K, Lee M-E, Haber E. The polymyositis-scleroderma autoantigen interacts with the helix-loop-helix proteins E12 and E47. J Biol Chem. 1997;272:13426–13431. doi: 10.1074/jbc.272.20.13426. [DOI] [PubMed] [Google Scholar]
- 22.Kocher O, Comella N, Tognazzi K, Brown L F. Identification and partial characterization of PDZK1: a novel protein containing PDZ interaction domains. Lab Investig. 1998;78:117–125. [PubMed] [Google Scholar]
- 23.Loveys D A, Streiff M B, Kato G J. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucleic Acids Res. 1996;24:2813–2820. doi: 10.1093/nar/24.14.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loveys D A, Streiff M B, Schaefer T S, Kato G J. The mUBC9 murine ubiquitin conjugating enzyme interacts with the E2A transcription factors. Gene. 1997;201:169–177. doi: 10.1016/s0378-1119(97)00444-7. [DOI] [PubMed] [Google Scholar]
- 25.Lu M, Seufert J, Habener J F. Pancreatic beta-cell-specific repression of insulin gene transcription by CCAAT/enhancer-binding protein beta. J Biol Chem. 1997;272:28349–28359. doi: 10.1074/jbc.272.45.28349. [DOI] [PubMed] [Google Scholar]
- 26.Murre C, Bain G, van Dijk M A, Engel I, Furnari B A, Massari M E, Matthews J R, Quong M W, Rivera R R, Stuiver M H. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 27.Mutoh H, Fung B P, Naya F J, Tsai M-J, Nishitani J, Leiter A B. The basic helix-loop-helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc Natl Acad Sci USA. 1997;94:3560–3564. doi: 10.1073/pnas.94.8.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naya F J, Huang H-P, Qiu Y, Mutoh H, DeMayo F J, Leiter A B, Tsai M-J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naya F J, Stellrecht C M, Tsai M-J. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 30.Nelson C, Shen L-P, Meister A, Fodor E, Rutter W J. Pan: a transcriptional regulator that binds chymotrypsin, insulin, and AP-4 enhancer motifs. Genes Dev. 1990;4:1035–1043. doi: 10.1101/gad.4.6.1035. [DOI] [PubMed] [Google Scholar]
- 31.Offield M F, Jetton T L, Labosky P A, Ray M, Stein R W, Magnuson M A, Hogan B L M, Wright C V E. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 32.Peers B, Leonard J, Sharma S, Teitelman G, Montminy M R. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol Endocrinol. 1994;8:1798–1806. doi: 10.1210/mend.8.12.7708065. [DOI] [PubMed] [Google Scholar]
- 33.Philippe J, Chick W L, Habener J F. Multipotential phenotypic expression of genes encoding peptide hormones in rat insulinoma cell lines. J Clin Investig. 1987;79:351–358. doi: 10.1172/JCI112819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poulat F, de Santa Barbara P, Desclozeaux M, Soullier S, Moniot B, Bonneaud N, Boizet B, Berta P. The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J Biol Chem. 1997;272:7167–7172. doi: 10.1074/jbc.272.11.7167. [DOI] [PubMed] [Google Scholar]
- 35.Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rousset R, Fabre S, Desbois C, Bantignies F, Jalinot P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene. 1998;16:643–654. doi: 10.1038/sj.onc.1201567. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Saras J, Heldin C-H. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem Sci. 1996;21:455–458. doi: 10.1016/s0968-0004(96)30044-3. [DOI] [PubMed] [Google Scholar]
- 39.Sharma A, Henderson E, Gamer L, Zhuang Y, Stein R. Analysis of the role of E2A-encoded proteins in insulin gene transcription. Mol Endocrinol. 1997;11:1608–1617. doi: 10.1210/mend.11.11.0004. [DOI] [PubMed] [Google Scholar]
- 40.Spicer D B, Rhee J, Cheung W L, Lassar A B. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 41.Stoffers D A, Ferrer J, Clarke W L, Habener J F. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 42.Stoffers D A, Stanojevic V, Habener J F. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant negative isoprotein. J Clin Investig. 1998;102:232–241. doi: 10.1172/JCI2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoffers D A, Zinkin N T, Stanojevic V, Clarke W L, Habener J F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe T K, Saito A, Suzuki M, Fujiwara T, Takahashi E, Slaughter C A, DeMartino G N, Hendil K B, Chung C H, Tanahashi N, Tanaka K. cDNA cloning and characterization of a human proteasomal modulator subunit, p27. Genomics. 1998;50:241–250. doi: 10.1006/geno.1998.5301. [DOI] [PubMed] [Google Scholar]
- 45.Weinman E J, Steplock D, Wang Y, Shenolikar S. Characterization of a protein cofactor that mediates protein kinase A regulation of the renal brush border membrane Na+-H+ exchanger. J Clin Investig. 1995;95:2143–2149. doi: 10.1172/JCI117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 47.Woods D F, Bryant P J. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 47a.Yao, K.-M., and G. C. Wong. Unpublished data.
- 48.Yun C H C, Oh S, Zizak M, Steplock D, Tsao S, Tse C-M, Weinman E J, Donowitz M. cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein. Proc Natl Acad Sci USA. 1997;94:3010–3015. doi: 10.1073/pnas.94.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]