Abstract
All what we see, touch, hear, taste, or smell must first be detected by the sensory elements of our nervous system. Sensory neurons, therefore, represent a critical component in all neural circuits and their correct function is essential for the generation of behavior and adaptation to the environment. Here, we report that the evolutionarily-conserved microRNA (miRNA) miR-263b plays a key behavioral role in Drosophila melanogaster through effects on the function of larval sensory neurons. Several independent experiments (in 50:50 male:female populations) support this finding: first, miRNA expression analysis, via reporter expression and fluorescent-activated cell sorting (FACS)-quantitative PCR (qPCR) analysis, demonstrate miR-263b expression in larval sensory neurons. Second, behavioral tests in miR-263b null mutants show defects in self-righting, an innate and evolutionarily conserved posture-control behavior that allows larvae to rectify their position if turned upside-down. Third, competitive inhibition of miR-263b in sensory neurons using a miR-263b “sponge” leads to self-righting defects. Fourth, systematic analysis of sensory neurons in miR-263b mutants shows no detectable morphologic defects in their stereotypic pattern, while genetically-encoded calcium sensors expressed in the sensory domain reveal a reduction in neural activity in miR-263b mutants. Fifth, miR-263b null mutants show reduced “touch-response” behavior and a compromised response to sound, both characteristic of larval sensory deficits. Furthermore, bioinformatic miRNA target analysis, gene expression assays, and behavioral phenocopy experiments suggest that miR-263b might exert its effects, at least in part, through repression of the basic helix-loop-helix (bHLH) transcription factor Atonal. Altogether, our study suggests a model in which miRNA-dependent control of transcription factor expression affects sensory function and behavior.
SIGNIFICANCE STATEMENT Sensory neurons are key to neural circuit function, but how these neurons acquire their specific properties is not well understood. Here, we examine this problem, focusing on the roles played by microRNAs (miRNAs). Using Drosophila, we demonstrate that the evolutionarily-conserved miRNA miR-263b controls sensory neuron function allowing the animal to perform an adaptive, elaborate three-dimensional movement. Our work thus shows that microRNAs can control complex motor behaviors by modulating sensory neuron physiology, and suggests that similar miRNA-dependent mechanisms may operate in other species. The work contributes to advance the understanding of the molecular basis of behavior and the biological roles of microRNAs within the nervous system.
Introduction
Although mechanisms of biological communication are fundamental to biological processes across all scales and species (Lerner et al., 2016), the nervous system is probably one of the best examples of high-speed and complex information transmitted within a cellular network.
In neural systems, input information is represented by sensory signals which provide the brain with essential information about the external environment, so that adequate actions can be selected and implemented. Sensory information is encoded in the form of firing patterns of populations of peripheral neurons, collectively known as sensory neurons. To recognise minor, yet potentially crucial, changes in the external world, sensory neurons must be able to detect diverse stimuli, and transform and transmit this information to the rest of the system. This requires that neural network components are “tuned” or aligned by biochemical machineries operating through a common language, so that neurons can talk to one another in an efficient manner, at rates compatible with the speed of the behaviors they control. Several genetic systems modulate neuronal biochemistries, including dynamic quantitative feed-back control devices able to monitor concentrations of gene products and maintain them within a suitable range. These molecular control devices rely on both, transcriptional as well as posttranscriptional processes.
In this article, we focus on the posttranscriptional component, studying the roles played by small regulatory non-coding RNAs termed microRNAs (miRNAs) on the specification of sensory neurons in Drosophila larvae. miRNAs regulate the expression of suites of protein encoding mRNAs by inducing their degradation and/or blocking their translation into protein (Bartel, 2018). Removal of miRNA genes can lead to target de-repression (upregulation), and this may, in certain circumstances, disrupt neural functions critical to physiology and behavioral control.
Our focus on miRNA roles is based on our recent discovery that mutation of a single miRNA gene in Drosophila disrupts a particular larval locomotor behavior termed self-righting: a movement that restores normal position after the animal is placed upside-down (Picao-Osorio et al., 2015). Mapping the “focus” of action (Benzer, 1967; Hotta and Benzer, 1972) of this miRNA (miR-iab4) led to the finding that this gene did not affect neural development, and instead, controls the physiology of a specific set of motor neurons in the larva (Picao-Osorio et al., 2015). Extensions of this work demonstrated that miR-iab4 also controls self-righting in the Drosophila adult (Issa et al., 2019) through actions on a different set of motor neurons, indicating that individual miRNAs may affect equivalent behaviors on systems bearing profoundly different neuroanatomy and biomechanics. Furthermore, a genetic screen (Picao-Osorio et al., 2017) aimed at identifying all miRNAs with impact on larval self-righting, revealed a pervasive influence of miRNA control on this postural behavior.
Self-righting is a complex evolutionarily conserved, three-dimensional, adaptive, and innate locomotor sequence (Ashe, 1970; Faisal and Matheson, 2001; Jusufi et al., 2011). To trigger this movement, the fruit fly larva must, first, determine that its position is abnormal. This suggests that sensory processes may play a key role in this behavior. Drosophila larvae possess a wide range of sensory organs including multidendritic (md) sensory neurons (SNs) (Bodmer and Jan, 1987; Grueber et al., 2002) and chordotonal organs (Field and Matheson, 1998) capable of detecting chemical and mechanical inputs. These sensory systems are present on the body wall, arranged in highly stereotypical patterns within each segment of the larva, and show complex and largely invariant morphologies (Zawarzin, 1912; Hartenstein, 1988).
Here, we show that the evolutionarily conserved miRNA miR-263b is essential for larval self-righting, through modulatory effects on the sensory system. Cell ablation experiments show that sensory neurons are essential for self-righting, and gene expression analyses demonstrate that miR-263b is expressed across different populations of larval sensory neurons. In addition, genetic manipulations show that normal miR-263b expression in these sensory elements is essential for normal self-righting. Morphologic analyses demonstrate that lack of miR-263b does not disrupt the intricate morphology or array of larval sensory neurons, suggesting miRNA impact on neural function, rather than on structure. Behavioral and optical imaging experiments confirm this, indicating that the functionality and physiology of sensory neurons is abnormal in the absence of miR-263b. Lastly, based on bioinformatic, gene expression and behavioral analyses, we propose a model in which miR-263b exerts its actions on sensory neurons via repression of the basic helix-loop-helix (bHLH) transcription factor encoded by the gene atonal. Our work thus provides evidence that miRNAs can control complex motor behaviors by modulating the physiology of sensory neurons.
Materials and Methods
Drosophila melanogaster strains
Cultures of D. melanogaster stocks were kept under standard conditions with 50–60% relative humidity and a 12/12 h light/dark cycle at 18°C, while working copies were held at 25°C. The following stocks were used in this study: 109(2)80-Gal4 (BDSC #8769), iav-Gal4 (BDSC #52273), ppk-1.9-Gal4 (stock described in Ainsley et al., 2003; a gift from Matthias Landgraf), ato-Gal4 (BDSC # 6480), 109(2)80-Gal4,UAS-mCD8::GFP (BDSC #8768) ΔmiR-263b-Gal4 (stock described in Hilgers et al., 2010; a gift from Sherry Aw), UAS-TeTN Lc (BDSC #28838), UAS-nls-GFP (BDSC #4775), UAS-miR-263b sponge (BDSC #61403), UAS-scramble sponge (BDSC #61501), UAS-GCaMP6m (BDSC #42748), ΔmiR-263b (BDSC #58903), UAS-ato (BDSC #39679); w1118 flies (BDSC #5905) were used as controls.
Immunohistochemistry
Embryo collection, dechorionation, devitellinization, and fixation were conducted as previously described (Picao-Osorio et al., 2015) and samples were stored in 100% methanol at −20°C. To obtain various embryonic stages an overnight collection was used. The samples were gradually rehydrated, rinsed in 1× PBS and 0.3% Triton X-100 (PBTx) and washed with PBTx 4 × 30 min. Subsequently, the samples were incubated with primary antibody (in PBTx) over night at 4°C. Primary antibodies and their concentration used are: 1:10 mouse anti-22C10 (Developmental Studies Hybridoma Bank), 1:2000 chicken anti-GFP (Abacam Probes), 1:100 guinea pig anti-atonal (a gift from Daniel Marenda) and goat anti-HRP-A556. Primary antibody was removed with three rinses and 4 × 30 min PBTx washes. The secondary antibody was added and incubated for 2 h at room temperature. Secondary antibodies and their concentrations used are: 1:500 anti-mouse Alexa Fluor 488 (Invitrogen), 1:500 anti-mouse Alexa Fluor A555 (Invitrogen), 1:500 anti-chicken Alexa Fluor 488 (Invitrogen), 1:500 anti-guinea pig Alexa Fluor A555 or A488. During the incubation with the secondary antibody 1:500–1:1000 Hoechst 33 342 (Life Technologies) was added. Samples were rinsed three times, washed 4 × 30 min with PBTx and transferred to 75% glycerol for mounting. Samples were imaged on a Leica SP8 confocal laser scanning microscope and further processed using ImageJ/FIJI and Adobe Photoshop CS6. Schematic representations and figure arrangement were made with Adobe Illustrator CS6.
In vivo calcium imaging
The calcium sensor GCaMP6m was used to measure the Ca2+ signal in SNs. Calcium imaging was conducted on stage 17 embryos to reduce movement during the recording. Parental lines were raised at 25°C in collection cages bearing apple juice-based medium agar plates, supplemented with yeast paste. Before recording, stage 16 embryos were collected, dechorionated and transferred into a drop (1 µl) of PBS (to prevent dehydration) previously left on a poly-L-lysine coated glass slides. The embryo was placed with its lateral side up, to allow visibility of the majority of SNs. Next, the Ca2+ signal within the SNs was captured during 3 min using Leica DM6000 microscope (Leica Microsystems) and processed with the software Fiji imageJ. GCaMP signals from the soma were analysed. The average signal from the first 10-s was taken as fluorescence baseline F0 to calculate the ΔF/F0 for each recording.
Cell sorting experiments
The fluorescent-activated cell sorting (FACS) dissociation protocol followed was the one described by Harzer and colleagues (Harzer et al., 2013) with some adjustments. Control (w1118) and experimental larvae [109(2)80-Gal4,UAS-mCD8::GFP; 50 h old] were opened anterodorsally and the gut was partly removed. For each genotype larvae were dissected for 30 min, usually obtaining 50–60 larvae, which were pooled in an Eppendorf tube filled with Rinaldini's solution (800 mg NaCl, 20 mg KCl, 5 mg NaH2PO4, 100 mg NaHCO3, and 100 mg glucose in 100 ml H2O). The larvae were washed once with 500-µl Rinaldini's solution. Rinaldini's solution was replaced with dissociation buffer, the mixture was incubated for 1 h at 30°C and gently mixed twice during incubation. Dissociation buffer needs to be prepared fresh, by adding 25-µl collagenase Type I (20 mg/ml, Sigma-Aldrich) and 25-µl papain (20 mg/ml, Sigma-Aldrich) to 200-µl complete Schneider's culture medium (5 ml heat-inactivated fetal bovine serum, 0.1 ml insulin, 1 ml PenStrep, 5 ml L-glutamine, 0.4 ml L-glutathione, and 37.85 ml Schneider's medium). All subsequent washing steps need to be conducted very slowly and carefully to avoid premature dissociation of the larval bodies. After the removal of the dissociation solution, the samples were washed twice with 500-µl Rinaldini's solution first, and then twice with 500-µl complete Schneider's culture medium. All medium was removed and the larval bodies were dissociated using 200-µl complete Schneider's medium, which was pipetted up and down (medium and larvae) with as little foaming as possible. The solution started to appear homogenous after pipetting was repeated ∼50 times. The cell suspension was filtered through a 30-µm mesh into a 5-ml FACS tube, placed on ice, and immediately taken to a FACS system (BD-FACSMelody). One biological replicate comprised around 3000 sorted cells (one sorting event), cells were sorted directly into TRIzol (Invitrogen) and stored at −80°C until further processing.
Quantitative PCR (qPCR) assays
A standard phenol/chloroform protocol was employed to extract RNA from FACS samples using Phase Lock Gel 2 ml Heavy tubes (Fisher Scientific). RNA was treated for 30 min with DNase (TURBO DNA-free, Invitrogen) subsequently. SuperScript III First Strand (Invitrogen) was used for cDNA synthesis. A single qPCR is made of 5 µl 2xSYBR green mix (LightCycler 480 SYBR Green I master, Roche), 2 µl H2O, 1 µl 5 μm forward primer, 1 µl 5 μm reverse primer, 1 µl cDNA (diluted 1:2 with H2O). Every sample was run in technical triplicates. The cycle conditions used were 40× (10 s 95°C, 20 s 60°C, 20 s 72°C) with fluorescent readings during annealing and elongation on a QuantStudio 3 machine (Thermo Fisher Scientific). Three reference genes were tested, Cdc5, RpS9, and eEF1α, and Cdc5 was chosen as appropriate reference gene. Primer sequences used are listed below:miR-263b (forward) 5′-ACTTTGAGTCTTGGCACTGG-3′miR-263b (reverse) 5′-GAAATCGTTGTACAAAGCCGG-3′miR-10 (forward) 5′-GCTTGCCATCAGCAACACTTT-3′miR-10 (reverse) 5′-CGGACTTCATTTCGCCCCAG-3′
GFP (forward) 5′-CATTCATCAGCCGTCTTCCG-3′
GFP (reverse) 5′-GAGTGCCCAAGAAAGCTACC-3′futsch (forward) 5′-TATTAGGGAAGACGCCGACC-3′futsch (reverse) 5′-AGGACTGGAGGCCTTAATGC-3′
Cdc5 (forward) 5′-CGGCAAGATCGAGAAGAAGC-3′
Cdc5 (reverse) 5′-GTTCTGCTCAATCTGGCCG-3′
For each primer set a standard curve was generated using serial dilution of the cDNA (undiluted, 1:2, 1:4, 1:8, 1:16, and 1:32), which was employed to calculate primer efficiency E (E = 10(−1/slope)). The primer efficiency was incorporated into the formula to calculate fold change values (R) as described below:
Sequences, statistics, and miRNA target prediction
Sequences for miR-263b/183/228 were recovered from miRBase.org (release 22) for schematic representation of the phylogenetic tree. Statistical analysis was done using Microsoft Excel. Statistical significance was calculated using Student's t tests with a p value threshold of 0.05 for significance. For atonal expression, fluorescent signal comparisons from wild-type and ΔmiR-263b mutant embryos were processed the same/at the same time and images were collected using the same settings. ImageJ was employed to analyse fluorescent intensity of individual atonal-positive clusters, with the help of the region of interest (ROI) manager. A ROI was manually drawn around an atonal-positive cell cluster as well as a region adjacent to the atonal-positive clusters where atonal is not expressed (to normalize background expression). This was repeated for a minimum of seven clusters (14 cluster-pairs), all located in the thoracic or abdominal region of the embryo. After all ROIs were selected, they were measured using “Measure” button within the ROI manager tool. Per ROI four values related to fluorescence were provided by the program: area, mean, minimum and maximum. First, mean intensity was calculated per area (intensity/area). To calculate the fluorescent intensity of an atonal-positive cluster, the intensity/area of the background expression (atonal-negative area) was subtracted from the intensity/area of the atonal-positive cluster. Per embryo between 7 and 10 atonal-positive clusters were measured and the average was taken. At least 10 embryos were analysed per genotype. For D. melanogaster miR-263b target prediction, we used PITA (Kertesz et al., 2007) and TargetScanFly 7.2 (Agarwal et al., 2018).
Behavioral tests
All embryos/larvae were kept on apple juice agar plates containing a small amount of yeast paste at 25°C. Self-righting and anterior touch response assays were conducted on freshly hatched L1 larvae (<30 min old). For those, late-stage embryos were transferred to a fresh apple juice agar plate (without yeast paste) and monitored for emerging larvae. Newly hatched L1 larvae were transferred to another fresh apple juice agar plate, which was used for testing throughout one session (1 biological repeat). Early L3 larvae (72 h) were used for the sound response assay (startle assay). At least three biological replicates with a minimum of 20 larvae per replicate were analysed for each behavioral test, including self-righting, touch-response, and sound response/startle. A paint brush was used to roll larvae for self-righting tests, which were otherwise performed as described in (Picao-Osorio et al., 2015). An eyelash was employed to deliver a soft stroke to the anterior region (head and thorax only) for anterior touch response assays. The protocol followed those described by Kernan and colleagues (Kernan et al., 1994). Overall response score and hesitation time were analysed. For sound response/startle assays the protocol developed by Zhang and colleagues (Zhang et al., 2013) was followed. In brief, 72 h larvae were washed with PBS and transferred to a testing plate. The plate was put on top of a speaker and videotaped from above. Larval response to sound was assayed by stimulation with a 1-s sound pulse (pure tone, 400 Hz), which was repeated 10 times.
Experimental design and statistical analysis
Time to self-righting, sound response, hesitation time, and atonal fluorescence intensity graphs were generated and compared using GraphPad Prism 6 software. Sample size of n ≥ 50 and n = 6–10 animals were used in total per genotype for behavioral and functional/imaging assays, respectively; each experiment was performed at least three times. Mean and SEM values were calculated for each trial and analysed by Mann–Whitney test, or one-way ANOVA and Kruskal–Wallis tests, when more groups were present (as indicated in each figure). GraphPad Prism 6 was used for all statistical analyses. Significant values in all figures: *p < 0.05, **p < 0.01, ***p < 0.001.
Results
Sensory and genetic requirements for self-righting behavior
The self-righting response is an innate and evolutionarily conserved movement that involves body rotation when the organism is placed upside-down (Ashe, 1970; Faisal and Matheson, 2001; Jusufi et al., 2011; Picao-Osorio et al., 2015). In Drosophila larvae, self-righting concerns the coordinated three-dimensional motion of multiple larval segments (Fig. 1A,B) and is triggered by the inversion of the position of the larval body in respect to the substrate. A possibility to explain the activation of self-righting is that the response might be triggered by a change in the orientation of the gravitational field; but this is not the case: inversion of the polarity of the gravitational field does not affect larval movement or trigger the self-righting response (Movies 1, 2). An alternative model is that the animal detects an anomaly in the normal pattern of sensory stimuli that informs the status of its contact with substrate and, based on this change, triggers the self-righting sequence. Peripheral sensory inputs are detected by genetically-defined subsets of larval sensory neurons arranged dorsoventrally along the larval body wall (Zawarzin, 1912; Hartenstein, 1988), including md sensory neurons (Bodmer and Jan, 1987; Grueber et al., 2002) and chordotonal organs (Field and Matheson, 1998; Fig. 1C–E). We hypothesised that these peripheral sensors may play a role in conveying the necessary information that triggers self-righting behavior, and, to test this model, we disabled subsets of peripheral SNs using the tetanus toxin (Sweeney et al., 1995) and examined the effects of these perturbations on self-righting. The results of these experiments demonstrate that SNs demarked by expression of the drivers 109(2)80 (multiple dendritic neurons, oenocytes, and chordotonal organs; Gao et al., 1999), ppk (Class IV dendritic arborization neurons and, less strongly, Class III neurons; Grueber et al., 2002, 2007), and iav (chordotonal neurons; Kwon et al., 2010) are essential for normal self-righting (Fig. 1F).
Figure 1.
Self-righting behavior and its sensory requirements. A, The self-righting sequence shown as a series of still images acquired from a video captured during larval self-righting (arrow, self-righting time). B, Self-righting sequence. Schematic representation indicating the individual steps observed during the self-righting behavioral sequence. C, The sensory system of stage 16 Drosophila embryo, in ventro-lateral view. Sensory neurons were labeled with α-22C10 [green; α-HRP (red) provides a general neuronal marker]. D, Sensory organ arrangement of embryonic abdominal segments, ventro-lateral view. Note the complexity and regularity of the arrangement of elements. Schematic representation of the distribution and composition of embryonic/larval sensory organs in the abdomen (E), morphology of chordotonal organs (F), and md neurons (G) is depicted by means of cartoons. H, Self-righting performance is decreased when all components of the sensory system (109(2)80) or some of them (ppk, md SNs; iav, chordotonal organs) are disabled by the tetanus toxin, indicating that information detected by the sensory system is essential for normal larval self-righting (n > 60, ANOVA and Kruskal–Wallis test).
Substrate exploration of a Drosophila first instar larva under normal gravitational conditions. The movie shows substrate exploratory behavior of a wild-type Drosophila first instar larva (∼30 min old) when the gravitational field is in a normal orientation (right-side up). Note that during the sequence, the specimen displays normal forward locomotion and turning behavior, but does not engage in self-righting behavior (Fig. 1).
Substrate exploration of a Drosophila first instar larva under inverted gravitational conditions. The movie shows substrate exploratory behavior of a wild-type Drosophila first instar larva (∼30 min old) when the gravitational field is in an inverted orientation (wrong-side up). Note that, just as in when the experiment is conducted under a normal orientation of the gravitational field, the specimen displays normal forward locomotion and turning behavior but does not engage in self-righting behavior (Fig. 1). These observations demonstrate that an inversion of the polarity of the gravitational field is insufficient to trigger self-righting behavior, indicating that Drosophila larvae must utilize other sensory information than gravity, to trigger the self-righting sequence.
Genetic elements affecting self-righting sensory function: a role for small non-coding RNAs
The physiology of SNs (and that of all neurons) largely relies on biochemistry; and the latter, is defined by the gene expression programs active in the cell. In this context, genetic elements with regulatory roles, i.e., affecting the expression of cohorts of target genes, might play an important function in setting the global biochemical and physiological properties of neurons, allowing for specific contributions to behavior. We have recently identified several genes encoding small non-coding RNAs, i.e., miRNAs, that are essential for a normal self-righting response in Drosophila (Picao-Osorio et al., 2015, 2017; Issa et al., 2019). Several of these miRNAs, collectively termed SR-miRNAs (self-righting miRNAs; Picao-Osorio et al., 2017), are expressed in the central nervous system, but others have been previously reported with expression in the peripheral nervous system (PNS) of larvae and adult (Pierce et al., 2008; Clark et al., 2010; Hilgers et al., 2010; Sun et al., 2012). Among these, we became particularly interested in miR-263b (Pierce et al., 2008; Hilgers et al., 2010) because of its pervasive evolutionary conservation (Pierce et al., 2008; Fig. 2A,B) and roles in multiple sensory and neural processes in the adult (Hilgers et al., 2010; Nian et al., 2019) and set to determine whether miR-263b might play a direct role in the SNs underlying self-righting behavior. For this we first aimed at establishing the expression pattern of miR-263b in larval SNs. FACS experiments coupled to qPCR expression assays (Fig. 2D,E), demonstrate expression of miR-263b in Drosophila larval SNs. Furthermore, spatial expression analysis of a Gal4 insertion located within the miR-263b locus (Hilgers et al., 2010) confirms expression of this genetic element within SNs in the embryo (Fig. 2F) providing independent experimental evidence that supports expression of miR-263b in SNs involved in self-righting control.
Figure 2.
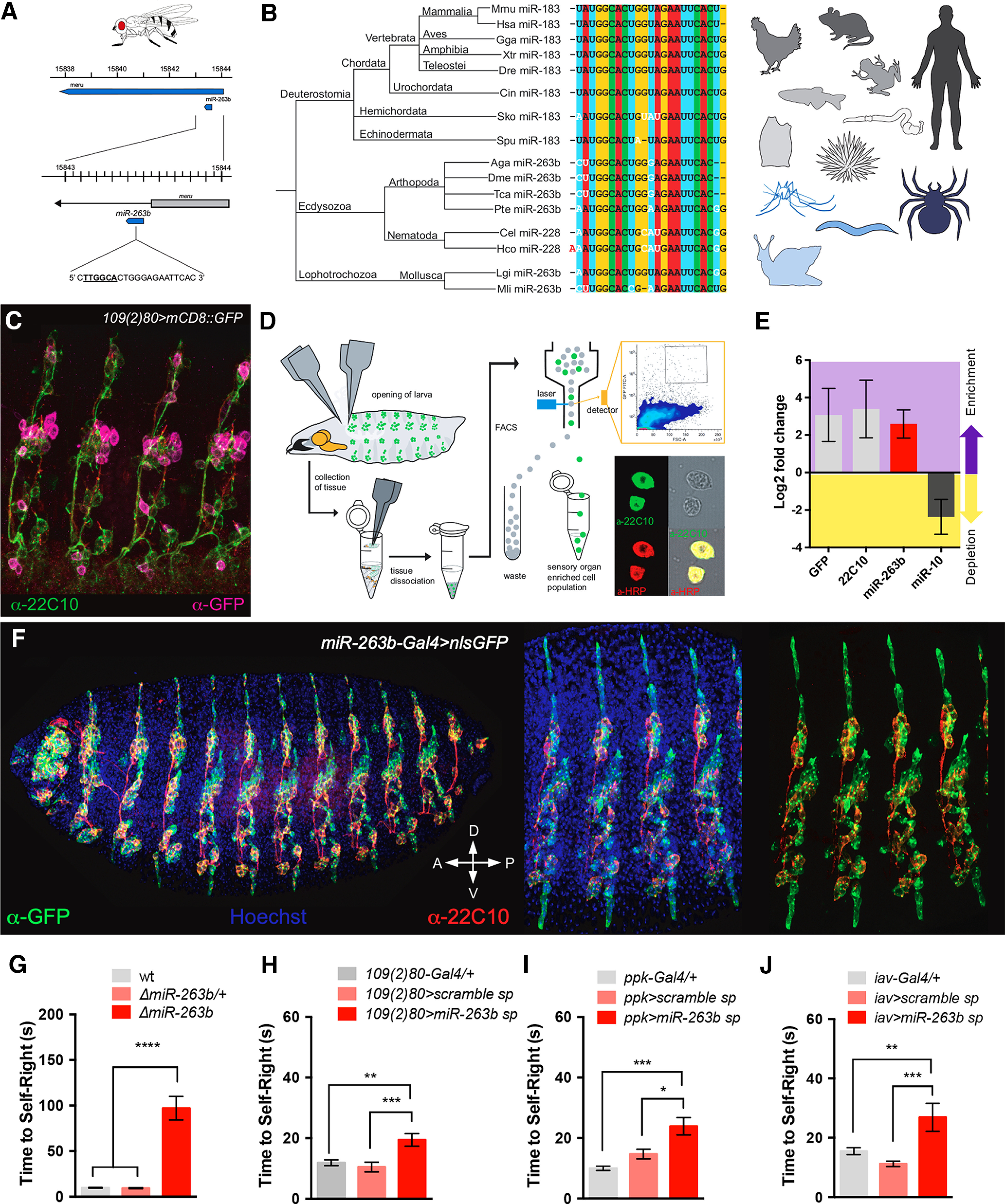
Expression and behavioral roles of miR-263b. A, Genomic location of miR-263b in Drosophila. B, Evolutionary conservation of miR-263b throughout the animal kingdom. The sequence encoding miR-263b is highly conserved across large phylogenetic distances; there are several names for this miRNA: miR-263b in arthropods and molluscs; miR-183 in deuterostomes; miR-228 in nematodes. C, The driver line 109(2)80-Gal4 was used in cell sorting (FACS) experiments; its coupling to the UAS-mCD8::GFP construct labels larval sensory neurons (magenta). D, Schematic representation of the FACS protocol used to study miRNA expression in the sensory system. The process involved the dissection of larval tissue (top left), and cell type validation via antibody staining (bottom right). E, Relative gene expression of GFP and futsch (labeled as “22C10”) in sensory organ enriched cell populations. F, miR-263b expression using a the miR-263b driver confirms miRNA expression in sensory organ-associated cells in the Drosophila embryo. Sensory neurons are visualized in red (α-22C10) and expression of the miR-263b driver is depicted via GFP signal, in green (α-GFP). G, miR-263b homozygote mutants (red) exhibit a pronounced delay in self-righting. H, Reduction of miR-263b function via competitive inhibition (miRNA sponges) in md neurons and chordotonal neurons results in a very significant increase of self-righting time. I, Functional inhibition of miR-263b in md (Class IV) neurons, or in (J) chordotonal organs, leads to significant delays in self-righting time (for panels G–J, we used sample size n > 50, ANOVA and Kruskal–Wallis tests). Altogether these results indicate that normal expression of miR-263b in the sensory system is necessary for normal larval behavior.
The fact that miR-263b is expressed in larval SNs is consistent with a potential role of this miRNA in SNs, but falls short from demonstrating functional roles. A more direct way to test the involvement of miR-263b in the biology of SNs is genetic removal: if miR-263b is essential for SN function, elimination of this genetic element from larvae is predicted to affect behaviors that rely on normal SNs, including self-righting. Experiments in Figure 2G show that homozygote as miR-263b mutant larvae display severe self-righting phenotypes demonstrating that this miRNA gene is essential for normal self-righting. Furthermore, functional attenuation of miR-263b by means of miRNA-specific sponges (Fulga et al., 2015) applied to the whole sensory domain or within specific sensory subsets (Fig. 2H–J) further confirms that miR-263b-mediated activities are necessary for normal self-righting behavior to take place.
miR-263b affects the physiology of sensory neurons
The regulatory nature of miRNAs makes them suitable for regulation of multiple functions within the organism, including developmental as well as physiological roles. To examine the point of action of miR-263b in the sensory system, we first set to establish whether this miRNA controls the development of SNs. For this we looked at the morphology of the sensory system in miR-263b null mutant larvae and wild-type specimens. The complex and stereotyped morphology of the larval sensory field makes it particularly suitable as a system to examine effects on the developmental process. Figure 3A,B shows that detailed and systematic characterization of the individual sub-components of the sensory system using confocal microscopy, in both, wild-type and miR-263b null mutant larvae, reveals no detectable differences among these two genotypes (Fig. 3B).
Figure 3.
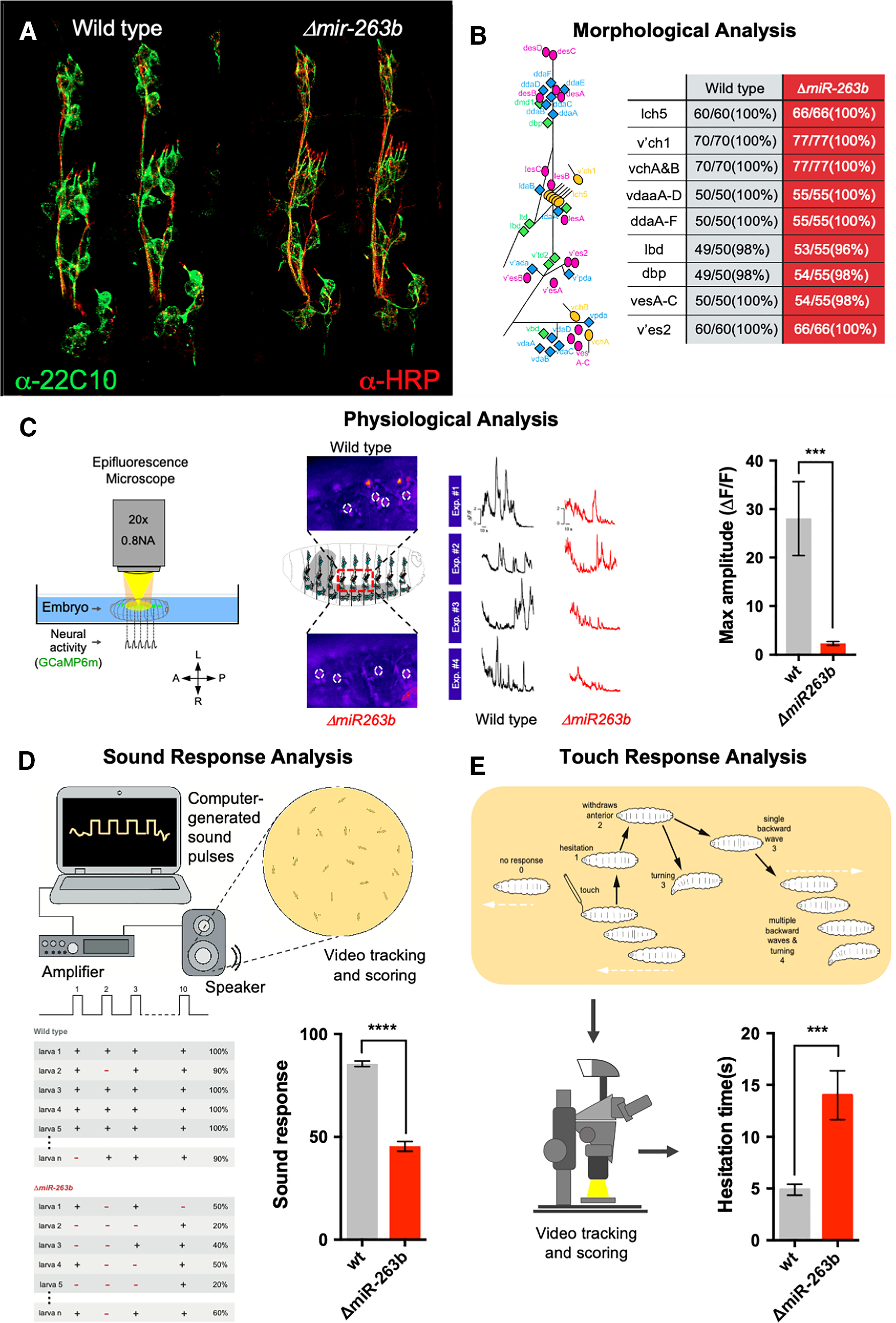
Effects of miR-263b on neural morphology, physiology and behavior. A, Labeling of sensory neurons (green, α-22C10) in stage 16 embryos in wild-type and miR-263b mutant specimens reveals the detailed morphology of the sensory system. B, Comparison of the integrity and arrangements of a comprehensive series of sensory organ subtypes (see diagram on the left) in wild-type and miR-263b mutants; there are no morphologic anomalies detected in miR-263b mutants. C, Calcium sensor analysis (GCaMP6m) shows reduced neural activity in miR-263b mutants when compared with wild type (n = 6, Mann–Whitney test). D, Experimental set up for sound response experiments (top). Diagram describing a representative set of sound response experiments in both genotypes (bottom left); quantification of responsiveness to sound (bottom right) is reduced in miR-236b mutants (n > 50, Mann–Whitney test). E, Experimental setup and scoring system for anterior touch-response experiments; video tracking and quantification of individual responses in normal and miRNA mutant larvae shows that miR-236b mutants exhibit an increased hesitation time (bottom right; n > 60, Mann–Whitney test). These experiments suggest that miR-263b plays a role in the physiological control of sensory function with impact on behavior.
The absence of morphological defects in the sensory system of miR-263b null mutants might indicate that this miRNA may impinge on the function (physiology) rather than the development of SNs. To test this possibility directly, we decided to quantify the spontaneous patterns of neural activity produced by SNs in miRNA mutants and compare these results to those obtained in normal specimens. For this we expressed a genetically-encoded calcium sensor (GCaMP6m) within the sensory system and visualized the resulting patterns of activity in live recordings. Figure 3C shows the results of these experiments where a significant decrease in neural activity patterns can be observed in miR-263b samples (Fig. 3C).
We reasoned that if miR-263b affects the general physiology of SNs, then other larval behaviors that rely on sensory input could also be affected in the absence of this miRNA. Two independent series of experiments confirm this. First, quantitative assessment of larval response to sound, previously shown to depend on sensory neurons, particularly, on chordotonal organs (Zhang et al., 2013), reveal that miR-263b mutants display a marked decrease in sound responsiveness when compared with their wild-type counterparts (Fig. 3D). Second, evaluation of the ability of larva to respond to touch (“anterior touch-response”; Kernan et al., 1994) shows that miR-263b mutants display a reduced response with markedly pronounced “hesitation time” (Fig. 3G,H). Altogether, the combination of morphologic, physiological and behavioral analyses of miR-263b mutants strongly indicates that this miRNA is required for normal sensory function in Drosophila larvae.
A molecular model for miR-263b action in sensory neurons and self-righting behavior
To advance the mechanistic understanding of the effects of miR-263b in the sensory system we decided to explore the potential points of action of this miRNA within SNs. Given that miRNAs are regulatory molecules (Bartel, 2018) their biological roles in the cell are likely to emerge indirectly, via effects on so-called “miRNA target” genes. Bioinformatic prediction of miRNA targets for miR-263b using the PITA and TargetScan algorithms (Kertesz et al., 2007; Agarwal et al., 2018) indicate a considerable number (n = 186) of potential targets of this miRNA in the Drosophila transcriptome highlighted simultaneously by both algorithms; approximately one-third of these targets (28%) show expression in the nervous system (Fig. 4A). Within the predicted set of neural targets, 33% have demonstrated expression in the PNS. Among the top-ten predicted miR-263b molecular targets with PNS expression, most genes encode factors with known functions in eye morphogenesis and differentiation, including: WASp (Wiskott–Aldrich syndrome protein; Ben-Yaacov et al., 2001), tup (tailup, also known as islet; Thor and Thomas, 1997), Kr-h1 (Krüppel homolog 1; Fichelson et al., 2012), arr (arrow; Wehrli et al., 2000), sca (scabrous; Mlodzik et al., 1990), repo (reverse polarity; Xiong et al., 1994), Vav (Vav guanine nucleotide exchange factor; Malartre et al., 2010), and the bHLH transcription factor Atonal encoded by the atonal (ato) gene. The latter was of particular interest to us because of its role as a proneural gene during the specification and development of chordotonal organs (Jarman et al., 1993; Jarman and Ahmed, 1998), among other sensory organs (Jarman et al., 1995; Gupta and Rodrigues, 1997). Chordotonal organs are internal stretch receptors that detect cuticle movement and vibration, and have been shown to be important for detecting sound (Zhang et al., 2013). As we expect chordotonal organs to play roles in all three behaviors tested (i.e., self-righting, sound-response and touch-response), we selected atonal as an entry point for detailed functional studies.
Figure 4.
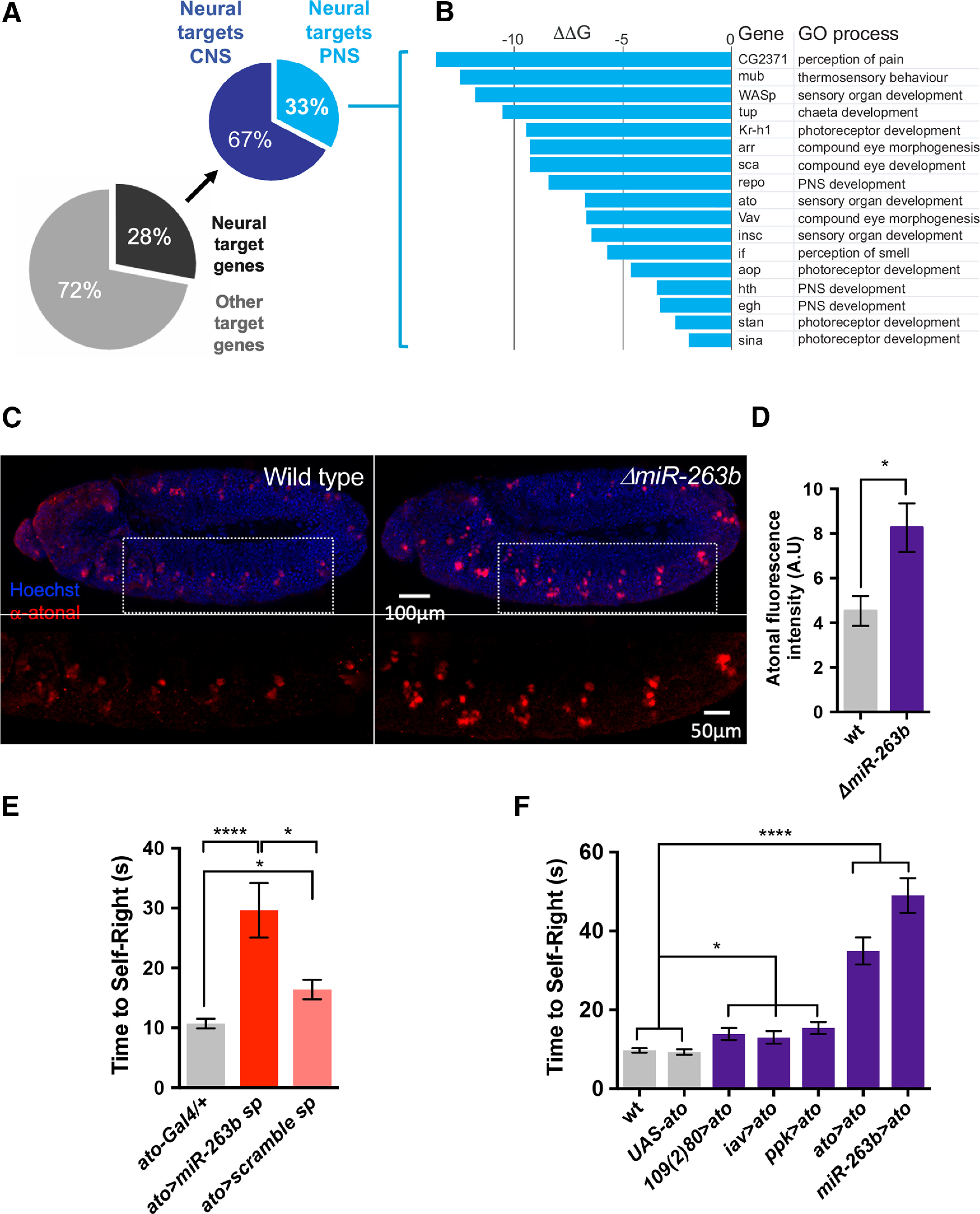
Potential molecular targets of miR-263b and their roles in self-righting. A, Pie charts representing shared predicted target genes of miR-263b through the combined output of the PITA and TargetScan algorithms. Out of 186 total miRNA targets highlighted by both computational approaches, 52 (28%) can be classified (GO terms) as “neuronal targets”; within this group, 17 relate to the PNS. B, Details on the 17 predicted target genes involved in the PNS, note the proneural gene atonal (ato) within the top-ten predicted target genes of miR-263b [NB: expression/process associations are according to Gene Ontology (GO) analysis]. C, Atonal protein expression analysis in wild-type and miR-263b mutant specimens reveals that Atonal expression is elevated in miR-263b mutants. D, Quantification of antibody labeling experiments for Atonal demonstrate that the levels of this proneural protein are significantly increased in miR-263b mutants (n = 10, Mann–Whitney test). E, Reduction of miR-263b function within the atonal expression domain increases self-righting time (n > 60, Mann–Whitney test). F, Artificial increase of atonal expression (i) within different sensory fields (activated via 109(2)80, ppk, and iav drivers); (ii) within the endogenous atonal expression domain; or (iii) within the miR-263b transcriptional domain all result in significantly increased self-righting times (n > 60, ANOVA and Kruskal–Wallis test). These data suggest that atonal might be one of the molecular targets that mediates the roles of miR-263b in the sensory system.
Three independent series of experiments support the model that atonal is one of the factors that mediates the actions of miR-263b in the sensory system with effects on self-righting control. First, atonal is expressed in sensory neurons, and its expression is increased in miRNA null mutants, in line with the expected de-repression effect caused by removal of a repressive regulatory miRNA (Fig. 4C,D). Second, expression of a miR-263b-sponge within the atonal domain is sufficient to trigger a self-righting phenotype (Fig. 4E). Third, artificial upregulation of atonal within its natural transcriptional domain in wild-type larvae, so that its expression level emulates those observed in miR-263b mutants, is sufficient to phenocopy the self-righting defect observed in miRNA mutants (Fig. 4F); similar effects are observed when ato is specifically delivered within a sub-field of the sensory system or when driven within the transcriptional domain of miR-263b (Fig. 4F).
We next decided to further probe the relation between miR-263b and atonal. For this, we conducted a series of experiments in which we artificially increased (UAS-miR-263b) or decreased (UAS-miR-263b sponge) the function of miR-263b within the natural atonal expression domain: should there be a direct regulatory link between miR-263b and atonal genes, an increment of miRNA repression is predicted to reduce Atonal protein signal, while a reduction of miRNA function should lead to Atonal upregulation (Fig. 5A–C). The data shown in Figure 5D–J provide experimental validation of both these predictions. Expression of miR-263b leads to a significant decrease in atonal expression (Fig. 5E,H,H'), while expression of miR-263b-sp induces an increase of Atonal signal (Fig. 5F,I,I'). These data support a regulatory interaction between miR-263b and atonal within neurons.
Figure 5.

Regulatory effects of miR-263b on the expression of the bHLH transcription factor Atonal. A–C, Predicted regulatory effects of miR-263b on atonal expression. A, According to our regulatory model, in control conditions, miR-263b is expected to repress the expression of atonal, contributing to the maintenance of concentration values of Atonal protein within a physiological range compatible with normal sensory neuron function. B, Reduction of miR-263b function is expected to de-repress atonal expression, leading to an increase in Atonal protein concentration. C, In contrast, upregulation of miR-263b expression is predicted to reduce the levels of expression of Atonal below normal concentration. D–F, Experimental validation of the predicted regulatory effects of miR-263b on Atonal expression within the natural atonal transcriptional domain (Ato-Gal4). D, G, G', Normal expression of Atonal (red) as detected by immunohistochemistry using anti-Atonal antibodies. E, H, H', Functional reduction of miR-263b within the atonal expression domain by means of expressing a miR-263b-sponge leads to an increase in Atonal signal (red), in line with predicted de-repression effects (see B). F, I, I', Increase of miR-263b expression leads to downregulation of Atonal expression (red), as predicted by the miR-263b-dependent Atonal expression control model, see C (blue signal: DAPI). J, Quantification of the regulatory interactions between miR-263b and Atonal shows a statistically significant increase of Atonal expression when miR-263b function is reduced (ato>miR-263b-sp), and reduction of Atonal signal when miR-263b is overexpressed (ato>miR-263b). These observations validate the proposed miR-263b-dependent regulatory model for atonal expression control in sensory neurons (n ≥ 13, Mann–Whitney test).
Together, these results suggest a model by which miR-263b-mediated repression of atonal in sensory neurons is essential for a normal self-righting response.
Discussion
We have employed Drosophila larvae to investigate the sensory elements required for triggering an evolutionarily-conserved 3-D postural behavior (self-righting) and established that normal expression of a miRNA gene, miR-263b, in the larval sensory system is necessary for a normal self-righting response. Our experiments also explore the causal links between the absence of miR-263b and the resulting behavioral effects, and show that absence of miR-263b does not seem to affect the complex morphologic organisation of sensory neurons in the fruit fly larvae. Instead, we observe an impairment in several sensory functions (touch-response, sound detection) as well as a reduced level of spontaneous neural activity in the sensory neurons of miR-263b mutants. Based on these results we suggest that miR-263b is required for the normal physiological control of sensory function in Drosophila larvae, with no detectable effects in the formation of the sensory system. At the mechanistic level, the combination of bioinformatic, gene expression and behavioral analyses indicate that miR-263b might exert its actions, at least in part, through repression of the bHLH transcription factor atonal, a regulatory proneural gene with known roles in the formation of sensory elements in Drosophila (Jarman et al., 1993, 1995; Bossuyt et al., 2009) and mammals (Bermingham et al., 1999; Zheng and Gao, 2000).
Evidence from behavioral genetic studies across different species indicates that the genetics of behavior is often complex and that the path between individual genes and behavioral traits is frequently intricate and hard to work out (Greenspan, 2008). Here, we see a different picture, in which reduction in the expression of a miRNA gene (caused by either a single loss-of-function mutation or tissue-specific functional attenuation) leads to a consistent behavioral change. Yet, how exactly a reduced level of expression of miR-263b modifies the properties of sensory elements in the fruit fly maggot, causing a change in behavior, remains open. In this respect, our observations suggest that miR-263b absence leads to an increase in the expression of atonal, which, instead of derailing the developmental process, affects the biochemistry and physiology of sensory cells.
Indeed, experiments in rat explants indicate that an excess of atonal's orthologue in the rat (Math1) can promote the formation of hair cells out of a population of postnatal utricular supporting cells (Zheng and Gao, 2000); this shows that going above a particular “set-point” of Atonal protein concentration (as observed in our study) is sufficient to modify the cellular processes in sensory elements. Given that atonal is expressed in Drosophila chordotonal organs (Jarman et al., 1993; Jarman and Ahmed, 1998) as well as in the cochlear system of mammals (Bermingham et al., 1999; Yang et al., 2012; Jarman and Groves, 2013), it might be plausible that atonal-dependent events could impact on behavior in other systems too. Interestingly, in the mouse, expression of Math5 and NeuroD1 (both, murine orthologues of atonal) is detected in postmitotic cells in the retina, consistent with a postdevelopmental role of atonal in mammalian sensory neurons (Yang et al., 2003); in particular, NeuroD expression has been shown to play a role in the terminal differentiation of retinal neurons in mammals (Ahmad et al., 1998). Also, in Caenorhabditis elegans, hlh4, a bHLH transcription factor related to the Atonal/Achaete-Scute complex class, has been shown to be required for the normal postdevelopmental function of nociceptive neurons (Masoudi et al., 2018). More broadly, gene expression analysis of vertebrate and invertebrate proneural proteins, including those encoded by atonal orthologues, shows signal in postmitotic neurons, where these proteins are believed to regulate neuronal migration and axonal/dendritic growth (Guillemot and Hassan, 2017).
A previous investigation in our laboratory (Picao-Osorio et al., 2015) showed that a deficit of another miRNA, miR-iab4, causes a change in the physiology of larval motor neurons involved in self-righting. In this case, miR-iab4 mediates the repression of the Hox protein Ultrabithorax (Ubx), a homeodomain-containing transcription factor (Bridges and Morgan, 1923; Sánchez-Herrero et al., 1985; Mallo and Alonso, 2013); in addition, an increase of Ubx phenocopies the behavioral effects observed in miR-iab4 mutants (Picao-Osorio et al., 2015).
Building on our findings on miR-263b and miR-iab4, we propose the model that miRNA-dependent control of behavior relies on maintaining the expression levels of a small set of transcription factors (ato, Ubx) within a particular concentration range; departures from such range may lead to gene regulatory effects with impact on neuronal biochemistry, which in ultimate instance, modify the physiological properties of the cell. This model, however, may be at odds with current views on the molecular mechanisms of miRNA function, which strongly indicate that miRNAs regulate multiple targets within the cell (Bartel, 2018; McGeary et al., 2019) arguing that regulation via single or few target genes is unlikely. A potential framework that reconciles both scenarios is that for specific cells (i.e., neurons) miRNA-dependent regulation of just a small subset of targets (ato, Ubx) is critical for their biological function, while other regulatory events are simply tolerated, or compensated for, by the regulatory networks of the cell (Alonso, 2012) and/or the neural circuits underlying behavior. Observations of miRNA effects on simple nematode behaviors (CO2 response) seem also to rely on the modulation of a few target genes (Drexel et al., 2016), suggesting that the generality of our proposed model might extend to other animal phyla.
It should also be noted, that self-righting was disrupted in ∼40% of miRNA mutants tested in an earlier study in our lab (Picao-Osorio et al., 2017) suggesting that many defects caused by miRNAs can affect self-righting, in addition to the above-mentioned cases of miR-iab4 and miR-263b. Indeed, while miR-263b mutants often appear to delay efforts to self-right (in line with their sensory deficits), some other miRNA mutants seem simply “sluggish,” or display sequences of unproductive peristaltic waves and head twists (Picao-Osorio et al., 2017); these observations suggest that each miRNA may affect self-righting in a particular way. Investigation of how this comprehensive set of “self-righting miRNAs” affects behavior is currently under way in our laboratory, and, once completed, should allow a more thorough testing of the model suggested above.
Our results show that ablation of either all, or genetically-defined subtypes of sensory elements in the larvae lead to a significant impairment in self-righting, but how sensory information is transformed into the actual self-righting response in the larva is still unknown. A current effort (Picao-Osorio, O'Garro-Priddie, Cardona and Alonso, in preparation) is using neural connectomics and reconstruction at synaptic resolution to map the neural substrates of self-righting behavior and should, when completed, offer a cellular platform for the investigation of how information in the sensory system is transmitted and converted into the motor patterns that underlie the self-righting sequence.
Previous work showed that miR-263b plays an important role in the Drosophila PNS, where it forms part of a gene regulatory pathway that represses apoptosis during sense organ development (Hilgers et al., 2010), and other studies report that downregulation or mutation of miR-263b lead to distortions of adult locomotion patterns (Donelson et al., 2020) or affect stereotypical parameters of male–female courtship (Iftikhar et al., 2019), respectively. Further studies in the adult indicate that miR-263b is also involved in the regulation of circadian behaviors (Nian et al., 2019; You et al., 2018). Our work adds to these studies, suggesting that miR-263b underlies a wide-spectrum of neural functions across developmental stages, implying the likely existence of positive selective pressure to retain a functional version of this gene in the Drosophila genome. Indeed, we note that miR-263b is evolutionary conserved within the Drosophilids, but also across long taxonomical distances. Furthermore, regarding the miR-263b gene family, and in addition to its close relative miRNA miR-183 (Fig. 2B), we wish to point out that the evolutionarily conserved miR-96 has an identical “seed” to miR-263b suggesting similar specificity (Pierce et al., 2008); intriguingly, miR-96 is required for normal hearing in mice and humans (Krohs et al., 2021; Lewis et al., 2009).
Building on the extensive phylogenetic preservation of miR-263b and its relative miRNAs, and the similarly broad conservation of atonal (Quan and Hassan, 2005; Cai and Groves, 2015) and that of self-righting itself (Ashe, 1970; Faisal and Matheson, 2001; Jusufi et al., 2011), we speculate that miR-263b and atonal orthologues in other species may play a role in self-righting and other postural control mechanisms in other orders of animals. Although the idea that the same sets of miRNAs and targets may contribute to establish the cellular biochemistry that enables similar behavioral responses in animals with drastically distinct body plans (e.g., insects and mammals), may seem unlikely, a recent observation on miRNA function in our lab does lend support to this possibility: miR-iab4-dependent control of Ubx expression is required for normal self-righting behavior in both Drosophila larvae and adults, two morphs of the fly that bear radically different morphologies, neural constitution and biomechanics (Issa et al., 2019).
Our study shows that the function of sensory neurons may rely on the normal expression of small non-coding RNAs, and that this is relevant for a complex and adaptive behavioral sequence in Drosophila larvae illustrating that behavior emerges from a careful balance in the expression of transcriptional as well as posttranscriptional gene regulators within the nervous system.
Footnotes
This work was supported by a UK Medical Research Council Project Grant MR/S011609/1 (to C.R.A.) and a Wellcome Trust Investigator Award 098410/Z/12/Z (to C.R.A.). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. We thank members of the Alonso lab for helpful discussions and comments; Tom Baden and Virginia Mahou at Sussex Neuroscience for their assistance with functional imaging and behavioral experiments, respectively; Jonathan Wing at the Genome Damage and Stability Centre at Sussex for his help with FACS experiments; Sherry Aw for sharing fly strains; Daniel Marenda for sharing antibodies; and the anonymous reviewers that reviewed our work, for their constructive and helpful comments.
This paper is dedicated to the memory of C. Gustavo Pesce, an exceptional biologist and dear friend, who tragically passed away earlier this year. His wisdom will always guide our science.
The authors declare no competing financial interests.
References
- Agarwal V, Subtelny AO, Thiru P, Ulitsky I, Bartel DP (2018) Predicting microRNA targeting efficacy in Drosophila. Genome Biol 19:152. 10.1186/s13059-018-1504-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I, Acharya HR, Rogers JA, Shibata A, Smithgall TE, Dooley CM (1998) The role of NeuroD as a differentiation factor in the mammalian retina. J Mol Neurosci 11:165–178. 10.1385/JMN:11:2:165 [DOI] [PubMed] [Google Scholar]
- Ainsley JA, Pettus JM, Bosenko D, Gerstein CE, Zinkevich N, Anderson MG, Adams CM, Welsh MJ, Johnson WA (2003) Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr Biol 13:1557–1563. 10.1016/S0960-9822(03)00596-7 [DOI] [PubMed] [Google Scholar]
- Alonso CR (2012) A complex 'mRNA degradation code' controls gene expression during animal development. Trends Genet 28:78–88. 10.1016/j.tig.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Ashe MV (1970) The righting reflex in turtles: a description and comparison. Psychon Sci 20:150–152. 10.3758/BF03335647 [DOI] [Google Scholar]
- Bartel DP (2018) Metazoan MicroRNAs. Cell 173:20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaacov S, Le Borgne R, Abramson I, Schweisguth F, Schejter ED (2001) Wasp, the Drosophila Wiskott-Aldrich syndrome gene homologue, is required for cell fate decisions mediated by Notch signaling. J Cell Biol 152:1–13. 10.1083/jcb.152.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S (1967) Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc Natl Acad Sci USA 58:1112–1119. 10.1073/pnas.58.3.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY (1999) Math1: an essential gene for the generation of inner ear hair cells. Science 284:1837–1841. 10.1126/science.284.5421.1837 [DOI] [PubMed] [Google Scholar]
- Bodmer R, Jan YN (1987) Morphological differentiation of the embryonic peripheral neurons in Drosophila. Rouxs Arch Dev Biol 196:69–77. 10.1007/BF00402027 [DOI] [PubMed] [Google Scholar]
- Bossuyt W, De Geest N, Aerts S, Leenaerts I, Marynen P, Hassan BA (2009) The atonal proneural transcription factor links differentiation and tumor formation in Drosophila. PLoS Biol 7:e40. 10.1371/journal.pbio.1000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges CB, Morgan TH (1923) The third-chromosome group of mutant characters of Drosophila melanogaster. Washington, DC: Carnegie Institution of Washington. [Google Scholar]
- Cai T, Groves AK (2015) The role of atonal factors in mechanosensory cell specification and function. Mol Neurobiol 52:1315–1329. 10.1007/s12035-014-8925-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AM, Goldstein LD, Tevlin M, Tavaré S, Shaham S, Miska EA (2010) The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic Acids Res 38:3780–3793. 10.1093/nar/gkq083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson NC, Dixit R, Pichardo-Casas I, Chiu EY, Ohman RT, Slawson JB, Klein M, Fulga TA, Van Vactor D, Griffith LC (2020) MicroRNAs regulate multiple aspects of locomotor behavior in Drosophila. G3 (Bethesda) 10:43–55. 10.1534/g3.119.400793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexel T, Mahofsky K, Latham R, Zimmer M, Cochella L (2016) Neuron type-specific miRNA represses two broadly expressed genes to modulate an avoidance behavior in C. elegans. Genes Dev 30:2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisal AA, Matheson T (2001) Coordinated righting behaviour in locusts. J Exp Biol 204:637–648. 10.1242/jeb.204.4.637 [DOI] [PubMed] [Google Scholar]
- Fichelson P, Brigui A, Pichaud F (2012) Orthodenticle and Kruppel homolog 1 regulate Drosophila photoreceptor maturation. Proc Natl Acad Sci USA 109:7893–7898. 10.1073/pnas.1120276109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field LH, Matheson T (1998) Chordotonal organs of insects. Adv Insect Physiol 28:1–228. [Google Scholar]
- Fulga TA, McNeill EM, Binari R, Yelick J, Blanche A, Booker M, Steinkraus BR, Schnall-Levin M, Zhao Y, DeLuca T, Bejarano F, Han Z, Lai EC, Wall DP, Perrimon N, Van Vactor D (2015) A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs. Nat Commun 6:7279. 10.1038/ncomms8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN (1999) Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev 13:2549–2561. 10.1101/gad.13.19.2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ (2008) Seymour Benzer (1921-2007). Curr Biol 18:R106–R110. 10.1016/j.cub.2007.12.039 [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN (2002) Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129:2867–2878. 10.1242/dev.129.12.2867 [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Yang CH, Younger S, Borden K, Jan LY, Jan YN (2007) Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development 134:55–64. 10.1242/dev.02666 [DOI] [PubMed] [Google Scholar]
- Guillemot F, Hassan BA (2017) Beyond proneural: emerging functions and regulations of proneural proteins. Curr Opin Neurobiol 42:93–101. 10.1016/j.conb.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Gupta BP, Rodrigues V (1997) Atonal is a proneural gene for a subset of olfactory sense organs in Drosophila. Genes Cells 2:225–233. 10.1046/j.1365-2443.1997.d01-312.x [DOI] [PubMed] [Google Scholar]
- Hartenstein V (1988) Development of Drosophila larval sensory organs: spatiotemporal pattern of sensory neurones, peripheral axonal pathways and sensilla differentiation. Development 102:869–886. 10.1242/dev.102.4.869 [DOI] [Google Scholar]
- Harzer H, Berger C, Conder R, Schmauss G, Knoblich JA (2013) FACS purification of Drosophila larval neuroblasts for next-generation sequencing. Nat Protoc 8:1088–1099. 10.1038/nprot.2013.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Bushati N, Cohen SM (2010) Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol 8:e1000396. 10.1371/journal.pbio.1000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Benzer S (1972) Mapping of behaviour in Drosophila mosaics. Nature 240:527–535. 10.1038/240527a0 [DOI] [PubMed] [Google Scholar]
- Iftikhar H, Johnson NL, Marlatt ML, Carney GE (2019) The role of miRNAs in Drosophila melanogaster. Male Courtship Behavior. Genetics 211:925–942. 10.1534/genetics.118.301901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa AR, Picao-Osorio J, Rito N, Chiappe ME, Alonso CR (2019) A single microRNA-Hox gene module controls equivalent movements in biomechanically distinct forms of Drosophila. Curr Biol 29:2665–2675.e4. 10.1016/j.cub.2019.06.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Ahmed I (1998) The specificity of proneural genes in determining Drosophila sense organ identity. Mech Dev 76:117–125. 10.1016/S0925-4773(98)00116-6 [DOI] [PubMed] [Google Scholar]
- Jarman AP, Groves AK (2013) The role of Atonal transcription factors in the development of mechanosensitive cells. Semin Cell Dev Biol 24:438–447. 10.1016/j.semcdb.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN (1993) atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73:1307–1321. 10.1016/0092-8674(93)90358-W [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN (1995) Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 121:2019–2030. 10.1242/dev.121.7.2019 [DOI] [PubMed] [Google Scholar]
- Jusufi A, Zeng Y, Full RJ, Dudley R (2011) Aerial righting reflexes in flightless animals. Integr Comp Biol 51:937–943. 10.1093/icb/icr114 [DOI] [PubMed] [Google Scholar]
- Kernan M, Cowan D, Zuker C (1994) Genetic dissection of mechanosensory transduction: mechanoreception-defective mutations of Drosophila. Neuron 12:1195–1206. 10.1016/0896-6273(94)90437-5 [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39:1278–1284. 10.1038/ng2135 [DOI] [PubMed] [Google Scholar]
- Krohs C, Körber C, Ebbers L, Altaf F, Hollje G, Hoppe S, Dörflinger Y, Prosser HM, Nothwang HG (2021) Loss of miR-183/96 alters synaptic strength via presynaptic and postsynaptic mechanisms at a central synapse. J Neurosci 41:6796–6811. 10.1523/JNEUROSCI.0139-20.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y, Shen WL, Shim HS, Montell C (2010) Fine thermotactic discrimination between the optimal and slightly cooler temperatures via a TRPV channel in chordotonal neurons. J Neurosci 30:10465–10471. 10.1523/JNEUROSCI.1631-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Ye L, Deisseroth K (2016) Communication in neural circuits: tools, opportunities, and challenges. Cell 164:1136–1150. 10.1016/j.cell.2016.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP (2009) An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet 41:614–618. 10.1038/ng.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malartre M, Ayaz D, Amador FF, Martín-Bermudo MD (2010) The guanine exchange factor vav controls axon growth and guidance during Drosophila development. J Neurosci 30:2257–2267. 10.1523/JNEUROSCI.1820-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M, Alonso CR (2013) The regulation of Hox gene expression during animal development. Development 140:3951–3963. 10.1242/dev.068346 [DOI] [PubMed] [Google Scholar]
- Masoudi N, Tavazoie S, Glenwinkel L, Ryu L, Kim K, Hobert O (2018) Unconventional function of an Achaete-Scute homolog as a terminal selector of nociceptive neuron identity. PLoS Biol 16:e2004979. 10.1371/journal.pbio.2004979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, Bartel DP (2019) The biochemical basis of microRNA targeting efficacy. Science 366:eaav1741. 10.1126/science.aav1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M, Baker NE, Rubin GM (1990) Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev 4:1848–1861. 10.1101/gad.4.11.1848 [DOI] [PubMed] [Google Scholar]
- Nian X, Chen W, Bai W, Zhao Z, Zhang Y (2019) miR-263b controls circadian behavior and the structural plasticity of pacemaker neurons by regulating the LIM-only protein Beadex. Cells 8:923. 10.3390/cells8080923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picao-Osorio J, Johnston J, Landgraf M, Berni J, Alonso CR (2015) MicroRNA-encoded behavior in Drosophila. Science 350:815–820. 10.1126/science.aad0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picao-Osorio J, Lago-Baldaia I, Patraquim P, Alonso CR (2017) Pervasive behavioral effects of microRNA regulation in Drosophila. Genetics 206:1535–1548. 10.1534/genetics.116.195776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA (2008) MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev 10:106–113. 10.1111/j.1525-142X.2007.00217.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan XJ, Hassan BA (2005) From skin to nerve: flies, vertebrates and the first helix. Cell Mol Life Sci 62:2036–2049. 10.1007/s00018-005-5124-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Herrero E, Vernós I, Marco R, Morata G (1985) Genetic organization of Drosophila bithorax complex. Nature 313:108–113. 10.1038/313108a0 [DOI] [PubMed] [Google Scholar]
- Sun K, Westholm JO, Tsurudome K, Hagen JW, Lu Y, Kohwi M, Betel D, Gao F-B, Haghighi AP, Doe CQ, Lai EC (2012) Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet 8:e1002515. 10.1371/journal.pgen.1002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney ST, Broadie K, Keane J, Niemann H, O'Kane CJ (1995) Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 14:341–351. 10.1016/0896-6273(95)90290-2 [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas JB (1997) The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron 18:397–409. 10.1016/S0896-6273(00)81241-6 [DOI] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S (2000) arrow encodes an LDL-receptor-related protein essential for wingless signalling. Nature 407:527–530. 10.1038/35035110 [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C (1994) repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev 8:981–994. 10.1101/gad.8.8.981 [DOI] [PubMed] [Google Scholar]
- Yang SM, Chen W, Guo WW, Jia S, Sun JH, Liu HZ, Young WY, He DZ (2012) Regeneration of stereocilia of hair cells by forced Atoh1 expression in the adult mammalian cochlea. PLoS One 7:e46355. 10.1371/journal.pone.0046355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L (2003) Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol 264:240–254. 10.1016/j.ydbio.2003.08.005 [DOI] [PubMed] [Google Scholar]
- You S, Fulga TA, Van Vactor D, Jackson FR (2018) Regulation of circadian behavior by astroglial microRNAs in Drosophila. Genetics 208:1195–1207. 10.1534/genetics.117.300342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawarzin A (1912) Histologische Studien uber Insekten. II Das sensible Nervensystem der Aeschnalarven. Z Wess Zool 100:245–286. [Google Scholar]
- Zhang W, Yan Z, Jan LY, Jan YN (2013) Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae. Proc Natl Acad Sci USA 110:13612–13617. 10.1073/pnas.1312477110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ (2000) Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci 3:580–586. 10.1038/75753 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Substrate exploration of a Drosophila first instar larva under normal gravitational conditions. The movie shows substrate exploratory behavior of a wild-type Drosophila first instar larva (∼30 min old) when the gravitational field is in a normal orientation (right-side up). Note that during the sequence, the specimen displays normal forward locomotion and turning behavior, but does not engage in self-righting behavior (Fig. 1).
Substrate exploration of a Drosophila first instar larva under inverted gravitational conditions. The movie shows substrate exploratory behavior of a wild-type Drosophila first instar larva (∼30 min old) when the gravitational field is in an inverted orientation (wrong-side up). Note that, just as in when the experiment is conducted under a normal orientation of the gravitational field, the specimen displays normal forward locomotion and turning behavior but does not engage in self-righting behavior (Fig. 1). These observations demonstrate that an inversion of the polarity of the gravitational field is insufficient to trigger self-righting behavior, indicating that Drosophila larvae must utilize other sensory information than gravity, to trigger the self-righting sequence.



