Adult zebrafish are a premier vertebrate model to interrogate mechanisms of disease development and tissue regeneration. In this issue, Shaw and Mokalled used high efficiency genome editing tools to introduce 28 mutations in 17 genes with efficiencies exceeding 85%. A quantifiable swim endurance test identified 7 mutations with reduced functional recovery after spinal cord injury. This study provides an experimental pipeline to perform medium- to large-scale neurobehavioral screens in adult zebrafish.
Keywords: spinal cord injury, zebrafish, regeneration, CRISPR/Cas9, genetic screen
Abstract
Adult zebrafish are widely used to interrogate mechanisms of disease development and tissue regeneration. Yet, the prospect of large-scale genetics in adult zebrafish has traditionally faced a host of biological and technical challenges, including inaccessibility of adult tissues to high-throughput phenotyping and the spatial and technical demands of adult husbandry. Here, we describe an experimental pipeline that combines high-efficiency CRISPR/Cas9 mutagenesis with functional phenotypic screening to identify genes required for spinal cord repair in adult zebrafish. Using CRISPR/Cas9 dual-guide ribonucleic proteins, we show selective and combinatorial mutagenesis of 17 genes at 28 target sites with efficiencies exceeding 85% in adult F0 “crispants”. We find that capillary electrophoresis is a reliable method to measure indel frequencies. Using a quantifiable behavioral assay, we identify seven single- or duplicate-gene crispants with reduced functional recovery after spinal cord injury. To rule out off-target effects, we generate germline mutations that recapitulate the crispant regeneration phenotypes. This study provides a platform that combines high-efficiency somatic mutagenesis with a functional phenotypic readout to perform medium- to large-scale genetic studies in adult zebrafish.
Introduction
Zebrafish are a premier model to interrogate mechanisms of vertebrate biology. Embryonic and larval zebrafish are traditionally employed to probe vertebrate embryogenesis (Streisinger et al. 1981; Eisen 1996; Schier and Talbot 2005). But more recently, adult zebrafish have been eminently used to model tissue physiology and disease mechanisms, including arthritis, scoliosis, cancer, blood disorders, and undiagnosed diseases (Langenau et al. 2003; Askary et al. 2016; Kaufman et al. 2016; Wangler et al. 2017; Van Gennip et al. 2018; Gray et al. 2021). Due to their renowned regenerative capacity, adult zebrafish are widely used to uncover injury responses and repair mechanisms in multiple tissues such as fin, heart, and pancreas (Vihtelic and Hyde 2000; Poss et al. 2002b; Yurco and Cameron 2005; Moss et al. 2009; Kroehne et al. 2011; Tu and Johnson 2011). The remarkable capacity to regenerate neural tissues, including brain and spinal cord, is attracting a growing community of scientists into adult neurobehavioral studies (Becker and Becker 2015; Orger and de Polavieja 2017; Mokalled and Poss 2018). Thus, there is a pressing need to refine and expand the genetic and molecular toolkit for adult zebrafish research.
Developing zebrafish embryos are ideal for large-scale genetic screening. Zebrafish generate large clutches of transparent embryos, undergo rapid external development, and allow for direct visualization of phenotypes at embryonic and larval stages. These experimental advantages empowered forward genetic screening in nervous system development and behavior (Brockerhoff et al. 1995; Granato et al. 1996; Moens et al. 1996; Wolman et al. 2015). Conversely, studies of the much larger, nontransparent tissues of adult zebrafish have historically presented a number of technical and practical challenges. (1) Genetic screening requires the ability to grow large numbers of animals, preferably within a small footprint and at a relatively reduced cost. (2) Genetic screening involves multiple generations of breeding, with only a fraction of the resulting F3 animals displaying homozygosity. (3) The success of a genetic screen relies on obtaining highly penetrant and accessible phenotypic readouts. (4) Importantly, in the absence of conditional targeting approaches, genetic screening in adult zebrafish is restricted to genes that are dispensable for embryonic development. The limited ability to identify adult phenotypes, coupled with the spatial and technical demands of adult husbandry, has curtailed the use of adult zebrafish for high-throughput genetic studies.
A number of genetic screens have defied the challenges of large-scale genetics in adult zebrafish. These screens preferentially targeted accessible tissues such as skin pigmentation or fin regeneration, or sought easily discernable phenotypes in less accessible tissues such as sterility or scoliosis (Haffter et al. 1996; Maderspacher and Nusslein-Volhard 2003; Dosch et al. 2004; Wagner et al. 2004; Henke et al. 2017; Gray et al. 2021). Notably, temperature-sensitive adult genetic screens provided an additional advantage, by screening for mutations that bypass early development at permissive temperature but impair fin regeneration at nonpermissive temperature (Johnson and Weston 1995; Poss et al. 2002a; Oppedal and Goldsmith 2010). While many of these adult genetic screens are nonsaturating, they remain unfeasible in less accessible tissues such as the adult spinal cord, precluding large-scale genetic studies in adult zebrafish.
Over the past decade, increased accessibility to large-scale transcriptomics and the advent of CRISPR/Cas9 genome editing revolutionized reverse genetic studies in zebrafish. Standard mutagenesis protocols showed inconsistent efficiencies and variable phenotype penetrance, limiting their application to small-scale studies of single or double germline mutant lines (Hwang et al. 2013; Jao et al. 2013; Gagnon et al. 2014; Kotani et al. 2015; Shah et al. 2015; Varshney et al. 2015; Burger et al. 2016; Wu et al. 2018). Recent development of high-efficiency targeting protocols reinvigorated the prospect of larger-scale somatic mutagenesis in adult zebrafish (Hoshijima et al. 2019; Xu et al. 2020). Chemically synthesized CRISPR/Cas9 dual-guide ribonucleic protein (dgRNP) complexes were shown to reliably produce somatic mutations with >90% efficiency, and to mimic germline mutant phenotypes in transiently targeted, developing zebrafish embryos (Hoshijima et al. 2019). Yet, the efficiency and applicability of CRISPR/Cas9 dgRNP induced mutagenesis in adult zebrafish remains to be determined.
Here, we combined high-efficiency CRISPR/Cas9 mutagenesis with a functional neurobehavioral readout to identify genes necessary for spinal cord regeneration in adult zebrafish. A total of 17 genes, including three sets of duplicate paralogs and one pair of functionally redundant genes, were individually and combinatorially targeted. CRISPR/Cas9 dgRNPs eliminated more than 85% of wild-type gene copies in larval and adult crispants. Ten genes showed comparable mutagenesis rates between larvae and adults. For seven genes, wild-type alleles were recovered at higher frequency in adult animals, suggesting a subset of mutant alleles are subject to negative selection in juvenile zebrafish. Using next-generation sequencing (NGS), we found capillary electrophoresis effectively measures indel frequency in individual fish. Using swim capacity as a readout of motor function recovery, we identified five single genes, one gene duplicate pair, and one pair of functionally redundant genes that are required for functional spinal cord repair after injury. Finally, we generated five germline mutations that recapitulated the histological and functional regeneration phenotypes of targeted crispants. Taken together, this study provides an experimental framework that combines high-efficiency somatic mutagenesis with a functional phenotypic readout to perform medium- to large-scale genetic studies in adult zebrafish.
Methods
Zebrafish
Adult zebrafish of the Tubingen strain were maintained at the Washington University Zebrafish Core Facility. All experiments were performed in compliance with institutional animal protocols. Male and female animals between 3 and 6 months of ∼2 cm length were used. Experimental fish and control clutchmate siblings of similar size and equal sex distribution were used for all experiments. Spinal cord transection surgeries and regeneration analyses were completed in a blinded manner, and two independent experiments were repeated using different clutches of animals.
CRISPR/Cas9 mutagenesis
CRISPR/Cas9 design and mutagenesis were performed as previously described (Hoshijima et al. 2019). Briefly, crRNA guide RNA sequences were selected using CHOPCHOP (https://chopchop.cbu.uib.no/). Sequences with no predicted off-target sites, including three or fewer mismatches in the genome, were selected. To maximize the effect of small indels, target sites were chosen within essential domains when possible. For genes that were targeted twice, the second target site was selected in an early exon. Target sequences used for this study are outlined in Supplementary Table S1.
Lyophilized Alt-R tracrRNA and crRNA gRNAs (IDT, Cat# 1072534) were reconstituted using manufacturer’s specifications at 100 µM stocks and stored at −20°. Prior to the day of injection, crRNA and tracrRNA were mixed at a final concentration of 50 µM and annealed by heating to 95° and then gradual cooling to 25° (−0.1°/second). The resulting dgRNA duplexes were stored at −20° until use. Alt-R S.p. Cas9 nuclease V3 (IDT, Cat# 1081059, supplied at 61.7 uM in 50% glycerol) was diluted in Cas9 dilution buffer [1 M HEPES (pH 7.5), 2 M KCl] to a working concentration of 25 uM and stored in single-use aliquots at −80°. On the day of injection, annealed dgRNA duplexes were diluted 1:1 in duplex buffer (IDT, Cat# 11-05-01-03) to a working concentration of 25 µM. Equal volumes of dgRNA were added to Cas9 protein and incubated at 37° for 5 minutes. For samples where two (aste1, bach1a, bach1b, dlb, junba, junbb, nr1d1, m17, pfkfb1, rab44, spi1a/spi1b, stab1, and taz/yap1) or four (bach1a/bach1b and junba/junbb) sites were simultaneously targeted, Cas9 protein was added in equal molar amounts to the total concentration of dgRNA. CRISPR/Cas9 solutions were maintained at room temperature during injections. Tubingen wild-type embryos were injected with 1 nL of CRISPR/Cas9 solution at the one-cell stage and grown to adulthood for spinal cord surgeries and functional analysis.
Capillary electrophoresis
Capillary electrophoresis (fragment analysis) was used to calculate the indel frequency for each CRISPR/Cas9 target site. For DNA extraction, whole 2 dpf larvae or ∼3 mm of excised adult tail fins were added to 50 mM NaOH in 50 µl (larvae) or 100 µl (adult fin). DNA samples were incubated at 95° for 20 minutes and then rapidly cooled to 4°. DNA extractions were neutralized by adding 5 µl (larvae) or 10 µl (adult fins) of 1 M Tris-HCl (pH 8.0). Small 100–200 bp PCR products were amplified using NEB Taq Polymerase (Cat# M0273) with gene-specific primers (Supplementary Table S2) in a volume of 10 µl in Pryme PCR semi-skirted PCR plates (MidSci, Cat# AVRT1). Samples were diluted to 24 µl with TE dilution buffer (Agilent) and loaded into the 5200 Fragment Analyzer System (Agilent, Cat# M5310AA). Capillary electrophoresis was carried out using the Agilent Fragment Analyzer Qualitative DNA Kit (Cat# DNF-905-K1000) according to the manufacturer’s specifications.
To calculate indel frequency, three wild-type siblings were genotyped to determine the size of the wild-type product at each targeting site. Because of significant noise due to primers (<70 bp) and nonspecific products (>200 bp) in wild-type samples, only amplicons between 70 and 200 bp were used to calculate indel frequency. Peaks within 1 bp of wild-type amplicons were considered wild-types (nonindels). All other peaks between 70 and 200 bp were considered indels. Indel frequency was calculated as the proportion of indel peaks relative to total (indel and nonindel) peaks. The same capillary electrophoresis primers were used to genotype stable mutant lines.
Next-generation sequencing
DNA extracts from larvae or adult fins were submitted to the Washington University Genome Engineering & iPSC Center (GEiC) for NGS using the 2 × 250 Illumina MiSeq platform (Sentmanat et al. 2018). The same capillary electrophoresis primers were used for NGS.
Spinal cord transection
Complete spinal cord transections were performed on adult zebrafish as previously described (Mokalled et al. 2016). Zebrafish were anesthetized using MS-222. Fine scissors were used to make a small incision that transects the SC 4 mm caudal to the brainstem region. Complete transection was visually confirmed at the time of surgery. Injured animals were also assessed at 2 or 3 dpi to confirm loss of swim capacity post-surgery.
Immunohistochemistry
Sixteen-micrometer cross cryosections of paraformaldehyde-fixed SC tissues were used. Tissue sections were imaged using a Zeiss LSM 800 confocal microscope. For Gfap staining, mouse anti-Gfap (ZIRC, Zrf1, AB_10013806, 1:1000) and Alexa Fluor-488 anti-mouse secondary antibody (Invitrogen, 1:200) were used. For anterograde axon labeling, zebrafish were anesthetized using MS-222 and fine scissors were used to transect the spinal cord 4 mm rostral to the lesion site. Biocytin-soaked Gelfoam Gelatin Sponge was applied at the new injury site (Gelfoam, Pfizer, cat# 09-0315-08; Biocytin, saturated solution, Sigma, cat# B4261). Fish were euthanized 4 hours post-treatment and Biocytin was histologically detected using Alexa Fluor 594-conjugated Streptavidin (Molecular Probes, cat# S-11227).
Quantification and statistical analysis
All procedures and quantifications were performed blind to condition. For functional recovery, (Figures 4B, C, and 5B), the swim time for individual fish was normalized to the average swim time of its uninjected siblings. For calculation of glial bridging (Figures 4H and 5D), the cross-sectional Gfap+ area at the lesion core was normalized to the cross-sectional area of the intact spinal cord rostral to the lesion. For calculation of axon growth (Figure 5G), biocytin labeled axons were quantified using the “threshold” and “particle analysis” tools in Fiji. Four sections per fish at 0.5 mm (proximal) and 2 mm (distal) caudal to the lesion core, and two sections 1 mm rostral to the lesion, were analyzed. Axon growth was normalized to the efficiency of Biocytin labeling rostral to the lesion for each fish. Axon growth was then normalized to the control group for each experiment.
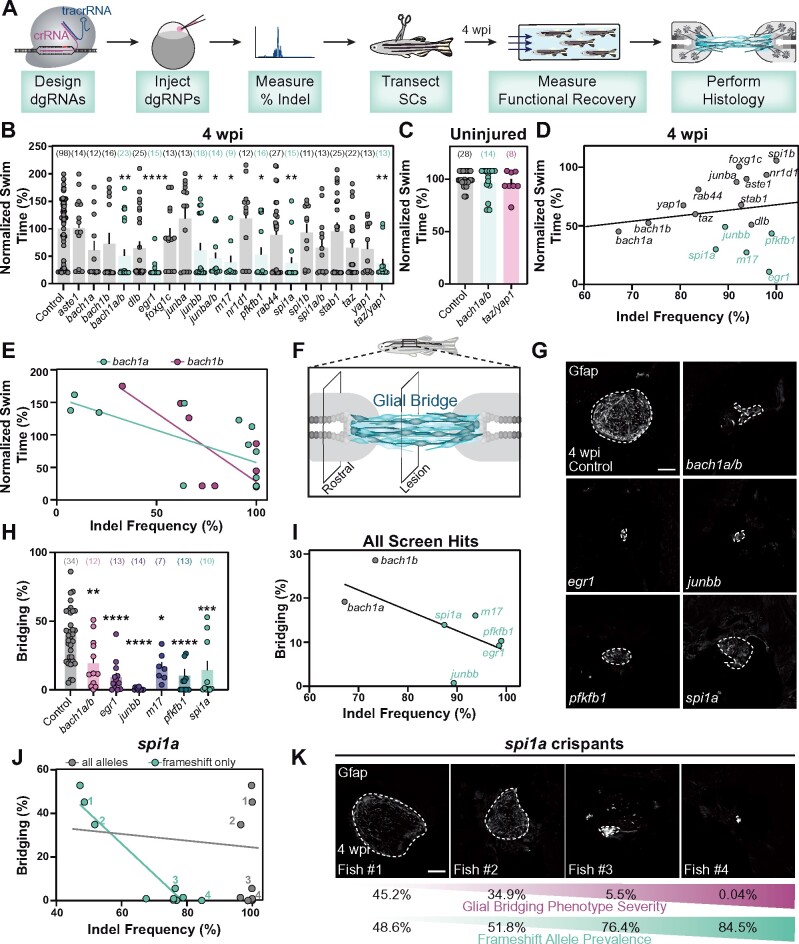
Figure 4.
Spinal cord regeneration defects in dgRNP targeted zebrafish. (A) Schematic pipeline to screen for spinal cord regeneration phenotypes in dgRNP targeted zebrafish crispants. (B) Functional recovery in dgRNP targeted animals 4 wpi. Data points represent individual animals and sample sizes are indicated. For each group of targeted animals, uninjected siblings were subjected to injury and swim assays. Swim times were normalized to their respective wild-type siblings. Groups with significantly diminished swim function are shown in teal. (C) Quantification of swim function in uninjured bach1a/b and taz/yap1 targeted crispants. Data points represent individual animals and sample sizes are indicated. (D) Functional recovery relative to indel frequency (as measured by capillary electrophoresis) for each targeted gene at 4 wpi. Crispants with significant functional recovery defects are shown in teal. For genes that were targeted at more than one site, indel frequency is averaged across both sites. (E) Functional recovery relative to indel frequency for bach1a (teal) and bach1b (magenta) zebrafish at 4 wpi. Data points represent individual animals. The indel frequency is averaged for target sites #1 and #2, since bach1a and bach1b were targeted at two sites each. (F) Schematic of regenerating zebrafish spinal cord. Bridging glia are shown in teal. Percent bridging was calculated as the ratio of the cross-sectional areas of the glial bridge (lesion) and the area of the intact spinal cord (rostral). (G, H) Glial bridging at 4 wpi. Representative immunohistochemistry shows Gfap+ bridges at the lesion site in bach1a/b, egr1, junbb, pfkfb1, and spi1a animals. Percent bridging was quantified for seven to 14 animals per group. (I) Glial Bridging relative to average indel frequency for each gene (as measured by capillary electrophoresis). Data points represent individual genes. Crispants with significantly diminished bridging are shown in teal. Percent bridging was only measured for crispants that displayed a swim phenotype (B), with the exception of bach1a and bach1b single crispants. For genes that were targeted at more than one site, indel frequency was averaged across both sites. (J) Glial bridging relative to indel frequency (as measured by NGS) for spi1a animals. Data points represent individual animals. Gray dots represent indel frequency of all nonwild-type alleles. Teal dots represent indel frequency of nonwild-type alleles predicted to generate frameshift mutations. Linear regression lines are statistically different (P < 0.0001). (K) Representative immunohistochemistry shows Gfap+ glial bridges in spi1a crispants at 4 wpi. Fish 1 through 4 are indicated in panel J. Percent Bridging (magenta) is indicated for each fish along with the frameshift-only indel frequency (teal). For bar plots (B, C, and H), the bar indicates the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Scale bars, 50 μm.
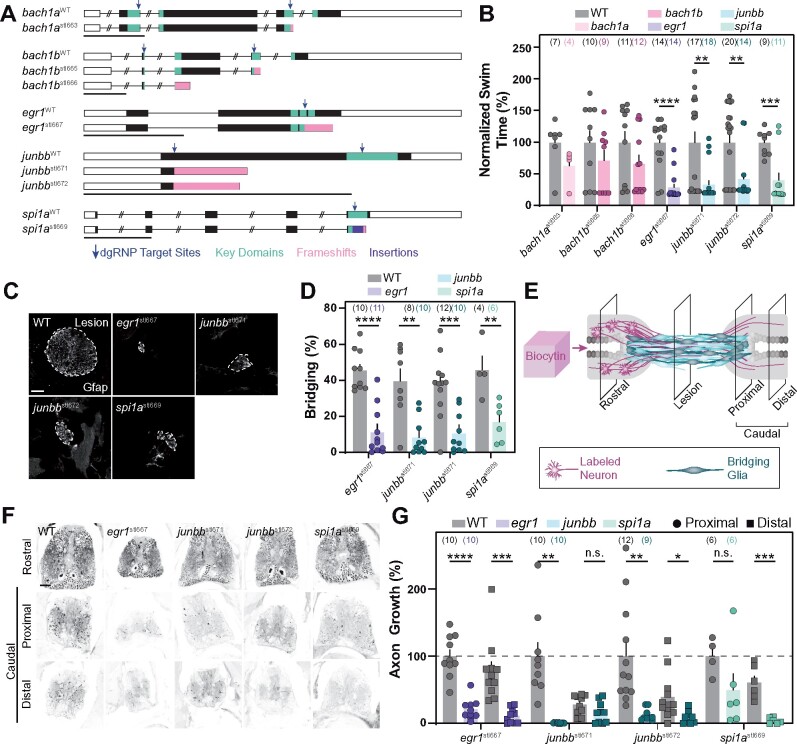
Figure 5.
Stable homozygous mutants recapitulate the spinal cord regeneration defects observed in dgRNP crispants. (A) Germline mutations generated in bach1a, bach1b, egr1, junbb, and spi1a. Black boxes are exons, white boxes are UTRs, and lines are introns. Key domains are indicated in teal. dgRNP target sites are indicated by blue arrows. Predicted frameshift mutations are indicated in pink. A 23 bp insertion in spi1a is indicated in violet. (B) Functional recovery in stable homozygous mutant lines at 4 wpi. Data points represent individual animals. Sample sizes are indicated. For each clutch of siblings, mutant swim times were normalized to their wild-type siblings. (C, D) Glial bridging at 4 wpi. Representative immunohistochemistry shows Gfap+ bridges at the lesion site in egr1, junbb, and spi1a mutants. Percent bridging was quantified for 4–12 animals per group. Data points represent individual animals in (D). Sample sizes are indicated. (E) Anterograde axon biocytin labeling paradigm. Biocytin-labeled neurons (magenta) and bridging glia (teal) are schematized. Axon growth caudal to lesion (proximal and distal) was normalized to the efficiency of biocytin labeling rostral to the lesion (rostral) for each fish. (F, G) Anterograde axon labeling of wild-type control siblings and homozygous mutants proximal (circles) and distal (squares) to the lesion site. n.s. indicates P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Scale bars, 50 μm.
Statistics were performed in GraphPad Prism. In cases where two groups were compared (Figures 3H and 5, B, D, G), t-tests with Welch’s correction were used. When three or more groups were compared (Figure 4, B, C, and H), One-way ANOVA with Dunnett’s correction was used. For survival analysis (Figure 3G), a Log Rank Mantel-Cox test was used. To compare linear regressions in Figure 4J, an Ancova test was used.
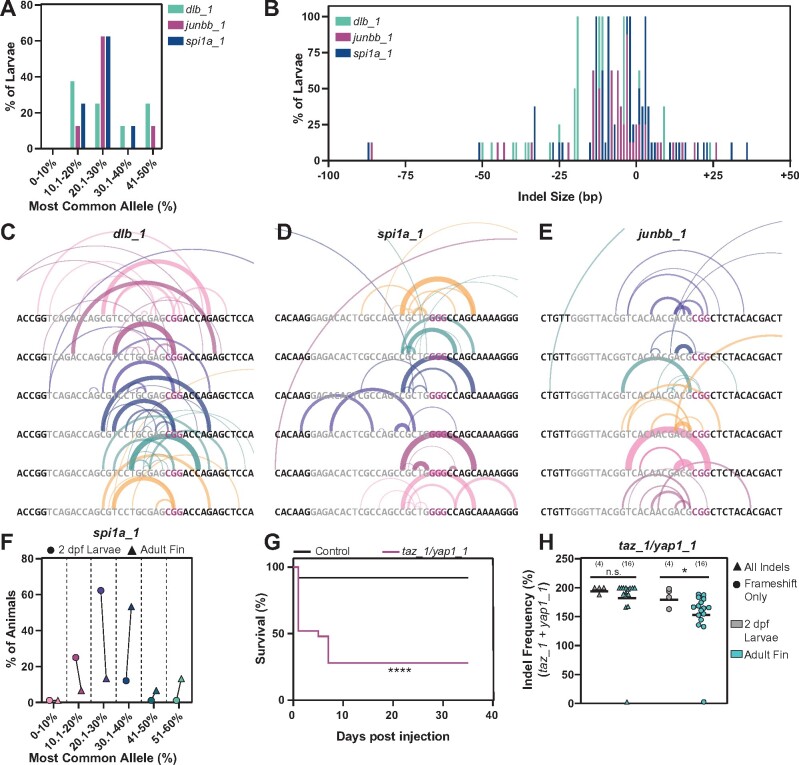
Figure 3.
Characterization of dgRNP generated alleles (A) Frequency of occurrence of dgRNP generated alleles for dlb_1, junbb_1, and spi1a_1. NGS data shown represents eight larvae at each target site. Y-axis indicates percent larvae; X-axis categorizes the most common alleles based on their indel frequency in individual larvae. (B) Indel size distribution of dgRNP generated alleles for dlb_1, junbb_1, and spi1a_1. NGS data shown represents eight larvae at each target site. Y-axis indicates percent larvae; X-axis indicates indel size (ranging from −100 bp deletions to +50 bp insertions) (C–E) Schematics of dgRNP generated deletions in individual dlb_1 (C), spi1a_1 (D), and junbb_1 (E) larvae. Target site protospacers are shown in gray, and PAM sites in magenta. For each allele, semi-circles indicate base deletion. The weights and opacities of the semi-circles are proportional to the indel frequency of each allele. The heights of the semi-circles are proportional to deletion size. (F) The frequency for the most common allele generated by CRISPR/Cas9 dgRNPs at the spi1a_1 target site in whole 2 dpf larvae (circles) and adult caudal fin (triangles). NGS data shown represents eight animals. The percent of animals with common alleles and the prevalence of reads are shown. (G) Survival curves of 25 uninjected wild-type control (black) and 25 taz/yap1 dgRNP injected (magenta) zebrafish. (H) The indel frequency of all alleles (triangles) and frameshift only alleles (circles) at taz_1 and yap_1 target sites in whole 2 dpf larvae (gray) and adult caudal fins (teal) as measured by capillary electrophoresis. Data points represent individual animals. Indel frequency represents the sum of frequencies at the taz_1 and yap_1 target sites. Sample sizes are indicated. n.s. indicates P > 0.05; *P < 0.05; ****P < 0.0001.
Data availability
All zebrafish lines are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplementary Tables S3–S6 contain raw de-barcoded NGS sequences and read counts. Supplemental Material available at figshare: https://doi.org/10.25387/g3.14219252.
Results
Gene targeting by CRISPR/Cas9 dgRNPs
We used high-efficiency CRISPR/Cas9 mutagenesis to identify genes that direct spontaneous spinal cord repair in adult zebrafish (Hoshijima et al. 2019). Using previously generated transcriptomic datasets, we selected 14 genes that are upregulated in the spinal cord after injury (GEO accession # GSE164945) (Klatt Shaw et al. 2021). Genes of interest were filtered based on their biological function and previous characterization during embryonic development. For biological function, we preferentially targeted transcription factors that are enriched in regenerative glial cells after injury. To prevent early developmental lethality, we excluded genes that were previously associated with lethal phenotypes in embryonic or juvenile mutant animals (zfin.org). We also used the EMBL-EBI Danio rerio expression atlas to select genes that were either maternally supplied or not expressed at early zygotic stages (https://www.ebi.ac.uk/gxa/experiments/E-ERAD-475). To reduce the likelihood of genetic compensation between duplicate copies of the same gene (Postlethwait et al. 1998), gene duplicates were targeted alone and in tandem (Figure 1A). For each candidate gene, we designed one or two targeting dgRNAs that do not have any predicted off-target sites in the zebrafish genome (Labun et al. 2019). Overall, we targeted 17 genes at 28 target sites.
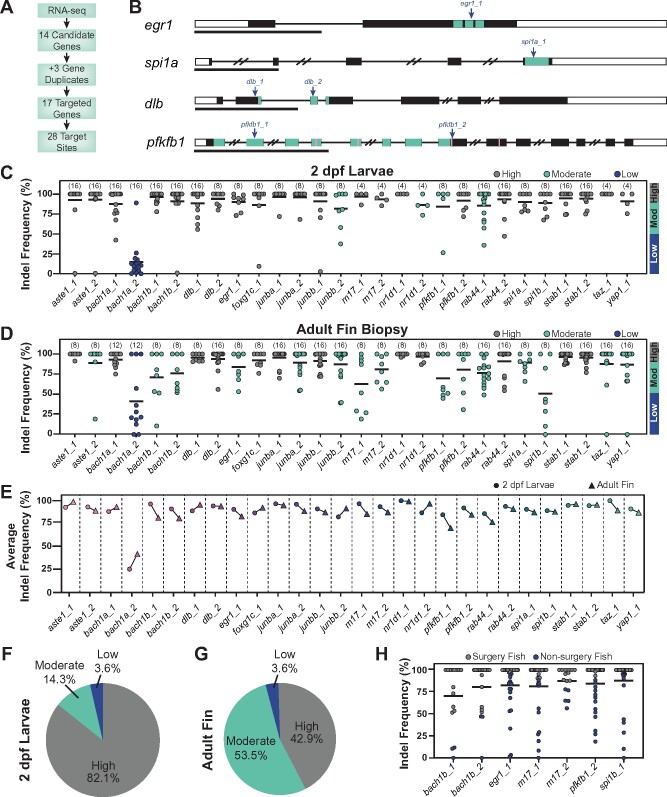
Figure 1.
CRISPR/Cas9 dgRNPs effectively mutate genes in adult zebrafish. (A) Schematic summary of gene targeting. A total of 17 genes were targeted using 28 dgRNPs. (B) Representative schematics for dgRNP target genes. Shown are transcription factors egr1 and spi1a, the dlb ligand, and the enzyme pfkfb1 (black boxes, exons; white boxes, UTRs; and lines, introns). Key domains are indicated in teal: zinc finger (egr1) and ETS (spi1a) DNA binding domains, EGF-like domain 1 (dlb), and 6PF2K domain (pfkfb1). The enzymatic active sites are indicated by pink lines in pfkfb1. dgRNP target sites are indicated by blue arrows. Scale bars, 1 Kilobase (Kb). (C, D) Targeting efficiency in dgRNP injected zebrafish crispants by capillary electrophoresis. Data points represent individual animals . Lines indicate means. Sample sizes are indicated between parentheses. 2 dpf larvae (C) or adult caudal fin (D) were analyzed. Target sites are classified as high-efficiency (indel frequency >90%, gray), moderate-efficiency (indel frequency 50–90%, teal), and low-efficiency (indel frequency <50%, blue). (E) Targeting efficiency in larval (circles) and adult (triangles) zebrafish. Data points represent the average indel frequency for each target site. (F, G) Pie chart representation of dgRNP efficiency. Shown are the fractions of dgRNPs with high (gray), moderate (teal), and low (blue) efficiencies. (H) Capillary electrophoresis genotyping of adult animals in which indel frequency decreased between larvae and adults. Data points represent individual animals. Adult fish with indel frequency >90% (gray) were subjected to spinal cord transection and subsequent phenotyping. Because only 16 taz_1/yap_1 dual-targeted animals survived to adulthood, all were genotyped in D.
To maximize the effect of small indels, we selected target sites within key domains (https://www.uniprot.org/), such as DNA or ligand-binding domains (Figure 1B). This strategy has been proved effective at eliminating gene function in larval zebrafish (Shi et al. 2015; Hoshijima et al. 2019). To achieve efficient targeting at specific loci, Alt-R-modified crRNA and tracrRNA were annealed into an RNA duplex and complexed with Cas9 protein (Gagnon et al. 2014; Hoshijima et al. 2019). Assembled dgRNPs were injected into the cytoplasm of one-cell stage embryos. For genes where separate sites were targeted, two dgRNPs were injected in combination. For multigene targeting of gene paralogs or functionally redundant genes, up to four dgRNPs (two dgRNPs for spi1a/b and taz/yap1; four dgRNPS for bach1a/b and junba/b) were injected in combination.
We first used PCR-based capillary electrophoresis to probe indel proportions in dgRNP targeted larvae. PCR primers were designed to amplify 100–200 base pair (bp) amplicons containing each target site and capillary electrophoresis was used to separate PCR amplicons with 1–2 bp resolution (Carrington et al. 2015; Ramlee et al. 2015; Varshney et al. 2015). In 2 days post fertilization (dpf) larvae, the average frequency of nonwild-type sized PCR amplicons (indel frequency) for all target sites was 87.6% (Figure 1C). Out of 28 dgRNPs, bach1a_2 showed poor activity, achieving an average indel frequency of 17.2%. junbb_2 (82.3%), nr1d1_2 (86.7%), pfkfb1_1 (77.9%), and rab44_1 (85.9%) dgRNPs resulted in moderate indel frequencies, ranging between 50 and 90%. The 23 remaining dgRNPs achieved high indel frequencies that exceeded 90%. These studies recapitulated previous findings and revealed that all 17 genes were successfully targeted at high-efficiency (>90%) by at least one dgRNP in zebrafish larvae.
dgRNP-induced mutagenesis efficiency in adult zebrafish
We next examined the efficiency of dgRNP induced mutagenesis in adult zebrafish. To this end, we raised dgRNP injected crispants to adulthood, and measured indel frequency in adult zebrafish fins by capillary electrophoresis. Fin biopsies were shown to accurately represent alleles found in somatic tissues and in the germline (Varshney et al. 2015; McKenna et al. 2016). Indel frequency averaged 85.2% at all targeted sites (Figure 1D). Among 28 dgRNPs, bach1a_2 showed poor activity, 15 dgRNPs showed moderate efficiencies (50–90%), while 12 dgRNPs achieved high efficiencies (>90%). A subset of genes important for adult tissue regeneration is likely required during embryonic development or juvenile growth. We postulated that cells or animals harboring deleterious alleles may be subjected to negative selection, and that wild-type clonal expansion could alter the rates of mutagenesis at adulthood. To test this hypothesis, we compared indel frequencies between larval and adult zebrafish at each of the genomic sites targeted in this study (Figure 1E). For nine target sites in seven of the targeted genes, indel frequency decreased by >10% between larval and adult stages (bach1b, egr1, m17, pfkfb1, spi1b, taz, and yap1). In contrast, bach1a_2 and nr1d1_2 dgRNPs displayed higher indel efficiencies in adult animals (Figure 1E). Overall, we found that 82.1% of dgRNPs achieved high indel frequencies in 2 dpf larvae, while 42.9% of dgRNPs maintained high efficiencies in adult fin biopsies (Figure 1, F and G). For genes that showed moderate mutagenesis efficiency in adult fish, all targeted crispants were genotyped, and 15 crispants with the highest indel frequencies were subjected to spinal cord transection and subsequent phenotyping (Figure 1H). Due to this selection process, indel frequency averaged 91.1% in animals selected for spinal cord transection. Thus, despite evidence that some mutant alleles are subject to negative selection during animal development, dgRNPs achieve efficient mutagenesis in adult zebrafish crispants.
Indel frequency by capillary electrophoresis and next-generation sequencing
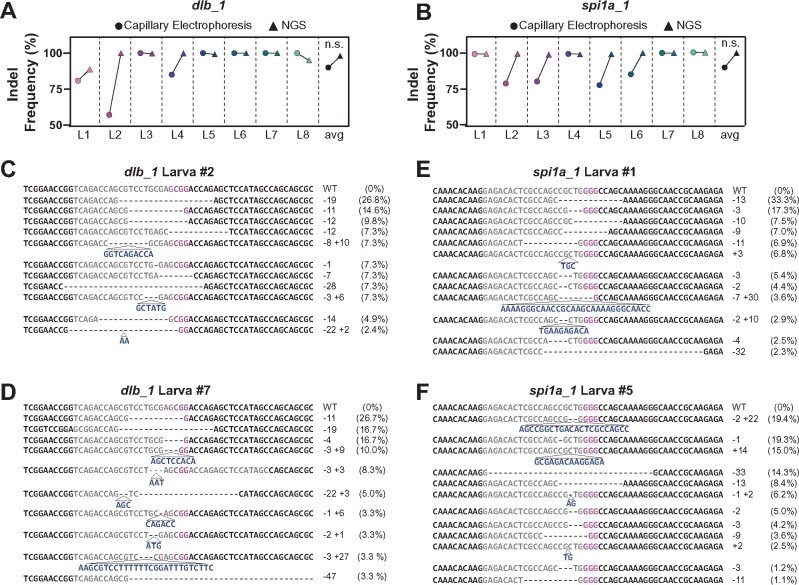
Compared to standard genotyping methods including NGS, restriction enzyme-based genotyping, T7E I-based genotyping, and high-resolution melt analysis, PCR-based capillary electrophoresis offers a cheaper, faster, and less restrictive tool to measure indel frequency (Sentmanat et al. 2018). To confirm the mutagenesis rates derived by capillary electrophoresis, we compared the indel frequencies calculated by capillary electrophoresis to the mutagenesis rates obtained by NGS in individual dgRNP targeted animals at two genes. For dlb_1 and spi1a_1 target sites, capillary electrophoresis slightly underestimated the prevalence of nonwild-type alleles (Figure 2, A and B). The discrepancy between indel calculation methods was largely due to small, 1–2 bp indels that were coupled with insertions or deletions of the same size (Figure 2, C–F). These findings confirmed that capillary electrophoresis measures mutagenesis efficiency with great accuracy.
Figure 2.
Capillary electrophoresis provides accurate measurement of indel frequencies in zebrafish. (A, B) Comparison of indel frequency calculated by capillary electrophoresis (circles) and NGS (triangles) for dlb_1 (A) and spi1a_1 (B) target sites in 2 dpf larvae. Shown are the average indel frequencies for eight individual larvae (L1-L8) . n.s. indicates P-value >0.05. (C–F) Sample NGS results for two larvae at dlb_1 (L2, C and L7, D) and spi1a_1 (L1, E and L5, F) target sites. Sequence reads represent the wild-type (WT) allele and the mutagenized alleles retrieved at each target site. NGS-derived indel frequencies indicate the percentage of reads mapping to each allele. The target site protospacer is in gray, and the PAM sequence is in magenta. Insertions (navy) and deletions (dashed lines) are indicated.
Characterization of dgRNP generated alleles
We next investigated the range and diversity of allelic mutations generated by dgRNPs. NGS revealed more than eight different alleles in individual larval animals (Figure 2, C–E and data not shown). However, only two to four alleles were present in >10% of sequenced reads in each animal (data not shown). In 2 dpf larvae, the most common allele was typically present in 20–30% of NGS reads for junbb_1 and spi1a_1, and in 10–20% for dlb_1 (Figure 3A). These values were consistent with allele prevalence calculated by capillary electrophoresis at all sites (data not shown). Consistent with previous studies, we concluded that dgRNPs were most active at the two- to four-cell stage (Burger et al. 2016). In 2 dpf larvae, most alleles were small indels of <10 bp, although larger insertions and deletions were present (Figure 3B). At three separate target sites (dlb_1, junbb_1, and spi1a_1), the same indel was generated in multiple, independent animals (Figure 3, C–E). In dlb_1 and spi1a_1 targeted animals, the most common indel shared between independent animals was a frameshift-causing deletion (an 11 bp deletion in dlb_1 [TCCTGCGAGCG] found in seven out of eight animals and a 13 bp deletion in spi1a [CGCTGGGGCCAGC] found in six out of eight animals, respectively) (Figure 3, C and D). In junbb_1 targeted animals, the most common allele was a 3 bp deletion of ACG found in six out of eight independent animals (Figure 3E). Consistent with previous studies, these findings suggested a bias in the double-strand break repair process (Gagnon et al. 2014; Burger et al. 2016; Ata et al. 2018) and emphasized the need to target key protein domains to maximize the effect of small indels on gene function.
To investigate how allelic prevalence changes from larval to adult stages, we compared indel frequency in spi1a_1 dgRNP animals at 2 dpf and in adult fin biopsies. While the most common allele in spi1a_1 crispant larvae was present in 20–30% of reads, the most common allele in adults could be found at much higher frequencies (30–60%, Figure 3F), suggesting clonal cell expansion occurs throughout development (McKenna et al. 2016). In spi1a_1 targeted animals, the expanded alleles did not appear to have a bias for in-frame mutations, suggesting clonal expansion may be random for spi1a. However, a bias for in-frame mutations was observed for genes expected to cause early embryonic lethality. Taz and Yap1 are functionally redundant downstream effectors of Hippo signaling (Plouffe et al. 2018). Double homozygous taz/yap1 zebrafish mutants die during gastrulation (Miesfeld et al. 2015). Due to their functional redundancy, we targeted taz and yap1 alone and in combination. Indeed, 50% of animals injected with both taz_1 and yap1_1 dgRNPs arrested prior to 24 hpf and only 30% survived to adulthood (Figure 3G). Among taz/yap1 targeted adults, significant proportions of wild-type or presumptive in-frame indels (as measured by capillary electrophoresis) were present at one or both taz_1/yap1_1 target sites (Figure 3H). Taken together, these data support a model in which small indels may minimize the effect of dgRNP induced mutagenesis on certain phenotypes (Gagnon et al. 2014). This may be exacerbated in cases where genes are necessary for development, and mutant cells are removed upon growth to adulthood. Notably, we did not observe this trend in other target sites that were specifically chosen to disrupt key domains. Together, these findings suggested hypomorphic or loss-of-function alleles are considerably prevalent in the majority of genes disrupted by in-frame mutations.
Screening F0 crispants for spinal cord regeneration defects
To identify genes that direct spinal cord regeneration, we assessed functional recovery in CRISPR/Cas9 dgRNP injected F0 crispants (Mokalled et al. 2016). Mutagenized 3–4 months-old animals were subjected to complete spinal cord transection. At 4 weeks post injury (wpi) mutagenized animals and control siblings were subjected to an increasing water current inside an enclosed swim tunnel (Figure 4A). Swim capacity was significantly decreased in egr1, junbb, pfkfb1, m17, and spi1a crispants relative to their respective control siblings (Figure 4B). Additionally, functional recovery was more defective in bach1a/b double crispants compared to single bach1a or bach1b crispants, suggesting bach1 paralogs are additively required for functional spinal cord repair (Figure 4B). Furthermore, taz/yap1 crispants exhibited a more pronounced phenotype than either taz or yap1 alone (Figure 4B), despite taz/yap1 adult crispants possessing a bias for in-frame mutations. To rule out the effects of developmental or gross morphological defects in targeted crispants, we assessed swim capacity in uninjured bach1a/b and taz/yap1 crispants. In this assay, bach1a/b and taz/yap1 crispants showed comparable swim function to their control siblings, indicating their functional regeneration defects were injury-induced (Figure 4C). These studies revealed dgRNP induced spinal cord regeneration defects in F0 crispants.
To evaluate the impact of mutagenesis efficiencies on neurobehavioral phenotyping, we compared the extent of functional regeneration relative to indel frequency for different genes and individual animals per gene. At the gene level, average indel frequency did not correlate with swim capacity after injury (R2 = 0.02) (Figure 4D). We next performed similar comparisons for individual bach1a and bach1b targeted animals and observed a negative correlation between indel frequency and normalized swim time (R2 = 0.45 for bach1a and 0.65 for bach1b) (Figure 4E). These findings suggested differences in mutagenesis efficiencies among individual animals may underlie increased variability during neurobehavioral phenotyping.
We next assessed anatomical regeneration in dgRNP targeted crispants. After spinal cord transection, regenerative glial cells form a bridge and axons regrow across the lesion site (Goldshmit et al. 2012). To assess glial bridging, we measured the cross-sectional area of Gfap+ bridges at the lesion core relative to the intact spinal cord (Figure 4F). Crispants in bach1a/b, egr1, junbb, m17, pfkfb1, and spi1a displayed less glial bridging (Figure 4, G and H). To test if indel frequency correlated with glial bridging defects, we compared indel frequencies to percent bridging for different genes and individual spi1a targeted animals. At the gene level, we observed minor negative correlation between indel frequencies and glial bridging (R2 = 0.4) (Figure 4I), suggesting mutagenesis efficiency may influence phenotypic severity. To specifically explore how frameshift-causing alleles may influence phenotypic readout, we performed NGS on spinal cords of individual spi1a crispants and did not observe a correlation between indel frequency and glial bridging (R2 = 0.001) (Figure 4J, gray). However, frameshift-causing indels showed a strong negative correlation with percent bridging (R2 = 0.90) (Figure 4J, teal). The three spi1a crispants with the lowest proportion of frameshift alleles displayed comparable glial bridging levels as their control uninjected siblings (Figure 4, J and K). spi1a crispants alleles were remarkably similar between 2 dpf larvae and adult spinal cord tissues (Figure 3D). The most common spi1a allele in adult fish was a 13 bp deletion that was present in 10 out of 10 animals at 5.2–29.0% (averaging 19.3%) of sequence reads per animal. The second most common allele in spi1a adults was a 3 bp deletion present in nine out of 10 adult spinal cord samples at 2.6–29.8% (averaging 10.6%) per animal. The three spi1a crispants with mild glial bridging defects (Figure 4, J and K) had three of the four highest percentages of reads of the 3 bp CTG deletion. Because the other seven spi1a crispants analyzed possessed a smaller fraction of in-frame alleles and a strong glial bridging phenotype, the weak glial bridging phenotype in nonframeshift animals did not affect our interpretation. We propose genes that cause mild phenotypes may be overlooked due to an occasional lack of frameshift mutagenesis or repetitive generation of nonframeshift alleles. These findings emphasized the importance of using a reliable, sensitive assay to phenotype dgRNP derived crispants.
Generation and neurobehavioral phenotyping of stable germline mutants
To confirm that the phenotypes exhibited by somatic F0 crispants were not due to off-target effects, we outcrossed dgRNP injected animals into wild-type Tubingen fish to establish germline mutations. Stable homozygous mutants were generated for bach1a, bach1b, egr1, junbb, and spi1a (Figure 5A) (Klatt Shaw et al. 2021). Adult homozygous mutants were subjected to spinal cord transection followed by functional and anatomical phenotyping. At 4 wpi, bach1a, bach1b, egr1, junbb, and spi1a exhibited functional swim phenotypes with similar severities to their somatic crispants (Figures 4B and 5B). Mutants that displayed reduced functional recovery after injury (egr1, junbb, and spi1a) also displayed defective glial bridging at 4 wpi (Figure 5, C and D). To measure the effect of egr1, junbb, and spi1a mutagenesis on axon regeneration, we performed anterograde axon tracing and quantified the extent of axon regrowth across the lesion site (Figure 5E; Mokalled et al. 2016). At 4 wpi, axon regrowth was significantly diminished in egr1, junbb, and spi1a mutants relative to their wild-type siblings (Figure 5, F and G). These results confirmed dgRNP targeted crispants phenocopy stable germline mutations, suggesting high-efficiency somatic mutagenesis could be used to prescreen for neurobehavioral defects in adult zebrafish.
Discussion
This study presents a pipeline to achieve efficient CRISPR/Cas9 mutagenesis and to screen for neurobehavioral defects at medium- or high-throughput in adult zebrafish crispants (Figure 6). To this end, we targeted 17 genes with 28 dgRNPs and achieved somatic mutagenesis efficiencies exceeding 85% in adult animals. Using a quantifiable swim assay as a neurobehavioral readout, we identified seven genes or gene duplicate pairs that failed to recover swim function after injury. We show that germline mutations displayed functional and anatomical defects that phenocopied somatic crispants following injury.
Figure 6.
Efficient CRISPR/Cas9 mutagenesis for neurobehavioral screening in adult zebrafish. dgRNPs were used to achieve high-efficiency mutagenesis in targeted crispants. Swim function was used to identify spinal cord regeneration defects after injury.
To date, performing large-scale genetics in adult zebrafish has proven bulky and challenging. Classical reverse screening techniques rely on easily accessible tissues and highly penetrant phenotypes. We here demonstrate that CRISPR/Cas9 dgRNP mutagenesis is highly efficient and reproducible. Thus, parsing through long lists of candidate genes for adult phenotypes is now more feasible. CRISPR/Cas9 dgRNP mutagenesis is cost- and space- effective, making large-scale genetics accessible to even the smallest zebrafish lab. For instance, in our hands, raising two tanks of 40 crispants was sufficient to phenotype each gene in two independent experimental replicates. Our study used a specialized pipeline to screen for regenerative factors after spinal cord injury. However, our CRISPR/Cas9-based mutagenesis methods can be combined with a host of phenotypic readouts to determine the biological importance of candidate genes in various settings.
Although dgRNPs are an effective tool, there are current limitations to their utility. We found small in-frame alleles were generated at various frequencies at all the target sites we sequenced. The phenotypic noise generated by such fully or partially functional alleles may cloud the interpretation of certain phenotypes. Consistent with this notion, we found that stable homozygous frameshift mutants in junbb displayed a more severe functional phenotype compared to junbb crispants (Figures 4B and 5B). The most common allele present in junbb crispants was a 3 bp deletion (Figure 3E) that was also recovered in multiple, independent germlines (data not shown). These findings are consistent with the 3 bp deletion being common in adult somatic tissues and likely underlying the difference in phenotypic penetrance between junbb crispants and mutants. Targeting essential, functional domains will increase the likelihood of inhibiting or minimizing gene function, but this does not exclude the possibility that mild phenotypes may be missed during screening. Targeting a gene at two distal sites increases the likelihood of achieving a frameshift-causing mutation. However, this may not be as effective for junbb or other single-exon genes, as proximal editing events are likely to re-establish the reading frame prior to the introduction of a stop codon in the resulting transcript. Another method for preventing small in-frame mutations is to utilize Microhomology-Mediated End Joining (MMEJ) to achieve precise and homogenous mutations. Recent work has established determinants for MMEJ-mediated repair of Cas9-induced double-strand breaks (Shen et al. 2018). Although not used in this study, publicly available MMEJ-predictive software can suggest target sites where the prediction of resulting indels is possible, and MMEJ-mediated repair often results in a reduction of the heterogeneity of edits within and across animals at certain sites (Ata et al. 2018). Combining the use of MMEJ-predictive software with targeting key protein domains offers a two-pronged approach to maximize the effect of dgRNPs on gene function. Furthermore, the possibility of off-target effects caused by CRISPR/Cas9 dgRNPs cannot be ruled out. Therefore, as we present somatic mutagenesis as an efficient platform to pre-screen for adult phenotypes, confirming F0 crispant phenotypes with multiple, independent germline mutations is indispensable for subsequent phenotypic analysis.
Another limitation to dgRNP-based mutagenesis is the inability to achieve temporal or spatial specificity in adult tissues. To avoid developmental lethality, we pre-screened candidate genes that may cause developmental phenotypes. Our approach was to exclude genes that are known to cause developmental phenotypes, and to favor genes that encode maternally supplied transcripts to avoid early developmental defects. In previous genetic screens for adult fin regeneration, and assuming many genes important for development are also necessary for regeneration, temperature-sensitive alleles were generated (Johnson and Weston 1995; Poss et al. 2002a). Similar temporal or spatial control could be applied for large-scale reverse genetics by optimizing transgenesis of temporally or tissue-specific reagents. Driving Cas9 expression using specific promoters could achieve a more targeted approach, and the repertoire of neuronal and glial promoters is continually expanding (Ablain et al. 2015; Chen et al. 2020; Hans et al. 2021). Further work is needed to combine tissue-specific with high-efficiency CRIPSR/Cas9 mutagenesis tools.
Adult zebrafish are a leading vertebrate system to model human diseases and dissect tissue regeneration mechanisms. However, the anatomy of the adult spinal cord is complex and requires prolonged histological processing to acquire and analyze tissue architecture. Using a functional swim assay to pre-screen for phenotypes prior to histology, we identified seven genes or gene-duplicate pairs that direct functional spinal cord repair. Combining transcriptomics, reliable CRISPR/Cas9 dgRNP mutagenesis, and robust functional phenotyping proved to be a powerful approach for neurobehavioral phenotyping in adult zebrafish.
Acknowledgments
We thank A. Johnson, H. McNeill, and L. Solnica-Krezel for discussion; D. Grunwald and A. Stratman for sharing protocols and reagents and the Washington University zebrafish Shared Resource for animal care.
Funding
This research was supported by the W.M. Keck Post-doctoral Fellowship (to D.K.S), the Washington University Center of Regenerative Medicine T32 (T32 EB028092 to D.K.S), grants from the National Institute of Health (R01 NS113915 to M.H.M.), the Curators of the University of Missouri (Spinal Cord Injury and Disease Training Program to M.H.M.), and the McDonnell Center for Cellular Neuroscience (to M.H.M.).
Conflicts of interests: The authors declare no competing interests.
Literature cited
- Ablain J, Durand E M, Yang S, Zhou Y, Zon LI.. 2015. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell. 32:756–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary A, Smeeton J, Paul S, Schindler S, Braasch I, et al. 2016. Ancient origin of lubricated joints in bony vertebrates. Elife. 5:e16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ata H, Ekstrom TL, Martinez-Galvez G, Mann CM, Dvornikov AV, et al. 2018. Robust activation of microhomology-mediated end joining for precision gene editing applications. PLoS Genet. 14:e1007652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker CG, Becker T.. 2015. Neuronal regeneration from ependymo-radial glial cells: cook, little pot, cook!. Dev Cell. 32:516–527. [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE, Hurley JB, Janssen-Bienhold U, Neuhauss SC, Driever W, et al. 1995. A behavioral screen for isolating zebrafish mutants with visual system defects. Proc Natl Acad Sci USA. 92:10545–10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A, Lindsay H, Felker A, Hess C, Anders C, et al. 2016. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143:2025–2037. [DOI] [PubMed] [Google Scholar]
- Carrington B, Varshney GK, Burgess SM, Sood R.. 2015. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 43:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Poskanzer KE, Freeman MR, Monk KR.. 2020. Live-imaging of astrocyte morphogenesis and function in zebrafish neural circuits. Nat Neurosci. 23:1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, et al. 2004. Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell. 6:771–780. [DOI] [PubMed] [Google Scholar]
- Eisen JS. 1996. Zebrafish make a big splash. Cell. 87:969–977. [DOI] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, et al. 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9:e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmit Y, Sztal TE, Jusuf PR, Hall TE, Nguyen-Chi M, et al. 2012. Fgf-dependent glial cell bridges facilitate spinal cord regeneration in zebrafish. J Neurosci. 32:7477–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, et al. 1996. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 123:399–413. [DOI] [PubMed] [Google Scholar]
- Gray RS, Gonzalez R, Ackerman SD, Minowa R, Griest JF, et al. 2021. Postembryonic screen for mutations affecting spine development in zebrafish. Dev Biol. 471:18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Odenthal J, Mullins MC, Lin S, Farrell MJ, et al. 1996. Mutations affecting pigmentation and shape of the adult zebrafish. Dev Genes Evol. 206:260–276. [DOI] [PubMed] [Google Scholar]
- Hans S, Zöller D, Hammer J, Stucke J, Spieß S, et al. 2021. Cre-Controlled CRISPR mutagenesis provides fast and easy conditional gene inactivation in zebrafish. Nat Commun. 12:1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke K, Daane JM, Hawkins MB, Dooley CM, Busch-Nentwich EM, et al. 2017. Genetic screen for postembryonic development in the zebrafish (Danio rerio): dominant mutations affecting adult form. Genetics 207:609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima K, Jurynec MJ, Klatt Shaw D, Jacobi AM, Behlke MA, et al. 2019. Highly efficient CRISPR-Cas9-based methods for generating deletion mutations and F0 embryos that lack gene function in zebrafish. Dev Cell 51:645–657.e644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, et al. 2013. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS ONE 8:e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W.. 2013. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 110:13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Weston JA.. 1995. Temperature-sensitive mutations that cause stage-specific defects in zebrafish fin regeneration. Genetics 141:1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, et al. 2016. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351:aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt Shaw D, Saraswathy VM, Zhou L, McAdow AR, Burris B, et al. 2021. Localized EMT reprograms glial progenitors to promote spinal cord repair. Dev Cell 56:613–626.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani H, Taimatsu K, Ohga R, Ota S, Kawahara A.. 2015. Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 protein complex in zebrafish. PLoS ONE 10:e0128319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M.. 2011. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development 138:4831–4841. [DOI] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, et al. 2019. Chopchop v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47:W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, et al. 2003. Myc-induced T cell leukemia in transgenic zebrafish. Science 299:887–890. [DOI] [PubMed] [Google Scholar]
- Maderspacher F, Nusslein-Volhard C.. 2003. Formation of the adult pigment pattern in zebrafish requires leopard and obelix dependent cell interactions. Development 130:3447–3457. [DOI] [PubMed] [Google Scholar]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, et al. 2016. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353:aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld JB, Gestri G, Clark BS, Flinn MA, Poole RJ, et al. 2015. Yap and Taz regulate retinal pigment epithelial cell fate. Development 142:3021–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Yan YL, Appel B, Force AG, Kimmel CB.. 1996. Valentino: a zebrafish gene required for normal hindbrain segmentation. Development 122:3981–3990. [DOI] [PubMed] [Google Scholar]
- Mokalled MH, Patra C, Dickson AL, Endo T, Stainier DY, et al. 2016. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354:630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokalled MH, Poss KD.. 2018. A regeneration toolkit. Dev Cell 47:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss JB, Koustubhan P, Greenman M, Parsons MJ, Walter I, et al. 2009. Regeneration of the pancreas in adult zebrafish. Diabetes 58:1844–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppedal D, Goldsmith MI.. 2010. A chemical screen to identify novel inhibitors of fin regeneration in zebrafish. Zebrafish 7:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orger MB, de Polavieja GG.. 2017. Zebrafish behavior: opportunities and challenges. Annu Rev Neurosci. 40:125–147. [DOI] [PubMed] [Google Scholar]
- Plouffe SW, Lin KC, Moore JL, 3rd Tan FE, Ma S, et al. 2018. The hippo pathway effector proteins Yap and Taz have both distinct and overlapping functions in the cell. J Biol Chem. 293:11230–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT.. 2002a. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development 129:5141–5149. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT.. 2002b. Heart regeneration in zebrafish. Science 298:2188–2190. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, et al. 1998. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 18:345–349. [DOI] [PubMed] [Google Scholar]
- Ramlee MK, Yan T, Cheung AM, Chuah CT, Li S.. 2015. High-throughput genotyping of CRISPR/cas9-mediated mutants using fluorescent PCR-capillary gel electrophoresis. Sci Rep. 5:15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF, Talbot WS.. 2005. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 39:561–613. [DOI] [PubMed] [Google Scholar]
- Sentmanat MF, Peters ST, Florian CP, Connelly JP, Pruett-Miller SM.. 2018. A survey of validation strategies for crispr-cas9 editing. Sci Rep. 8:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB.. 2015. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods 12:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, et al. 2018. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563:646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, et al. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 33:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G, Walker C, Dower N, Knauber D, Singer F.. 1981. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291:293–296. [DOI] [PubMed] [Google Scholar]
- Tu S, Johnson SL.. 2011. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell 20:725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gennip JLM, Boswell CW, Ciruna B.. 2018. Neuroinflammatory signals drive spinal curve formation in zebrafish models of idiopathic scoliosis. Sci Adv. 4:eaav1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Pei W, LaFave MC, Idol J, Xu L, et al. 2015. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25:1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR.. 2000. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 44:289–307. [DOI] [PubMed] [Google Scholar]
- Wagner DS, Dosch R, Mintzer KA, Wiemelt AP, Mullins MC.. 2004. Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev Cell. 6:781–790. [DOI] [PubMed] [Google Scholar]
- Wangler MF, Yamamoto S, Chao HT, Posey JE, Westerfield M, et al. 2017. Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 207:9–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolman MA, Jain RA, Marsden KC, Bell H, Skinner J, et al. 2015. A genome-wide screen identifies PAPP-AA-mediated IGFR signaling as a novel regulator of habituation learning. Neuron 85:1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Lam II, Clay H, Duong DN, Deo RC, et al. 2018. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev Cell 46:112–125.e114. [DOI] [PubMed] [Google Scholar]
- Xu B, Tang X, Jin M, Zhang H, Du L, et al. 2020. Unifying developmental programs for embryonic and postembryonic neurogenesis in the zebrafish retina. Development 147:dev185660. [DOI] [PubMed] [Google Scholar]
- Yurco P, Cameron DA.. 2005. Responses of muller glia to retinal injury in adult zebrafish. Vision Res. 45:991–1002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All zebrafish lines are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplementary Tables S3–S6 contain raw de-barcoded NGS sequences and read counts. Supplemental Material available at figshare: https://doi.org/10.25387/g3.14219252.