Abstract
Plant diseases caused by the Cercospora genus of ascomycete fungi are a major concern for commercial agricultural practices. Several Cercospora species can affect soybeans, such as Cercospora kikuchii which causes soybean leaf blight. Speciation in Cercospora on soybean has not been adequately studied. Some cryptic groups of Cercospora also cause diseases on soybean. Moreover, it has been known C. kikuchii population genetic structure is different between countries. Consequently, further genomic information could help to elucidate the covert differentiation of Cercospora diseases in soybean. Here, we report for the first time, a chromosome-level genome assembly for C. kikuchii. The genome assembly of 9 contigs was 34.44 Mb and the N50 was 4.19 Mb. Based on ab initio gene prediction, several candidates for pathogenicity-related genes, including 242 genes for putative effectors, 55 secondary metabolite gene clusters, and 399 carbohydrate-active enzyme genes were identified. The genome sequence and the features described in this study provide a solid foundation for comparative and evolutionary genomic analysis for Cercospora species that cause soybean diseases worldwide.
Keywords: Cercospora kikuchii, soybean, Cercospora leaf blight, purple seed stain
Introduction
Cercospora is one of the major groups of plant pathogenic fungi and Cercospora spp. can cause necrotic leaf spots on many different species of plants (Groenewald et al. 2013). Cercospora kikuchii (Tak. Matsumoto & Tomoy.) M. W. Gardner is a soybean pathogen, identified in eastern Asia in the early 20th century (Suzuki 1921; Matsumoto and Tomoyasu 1925; Walters 1980). It causes two types of pathogenic symptoms in soybean: purple seed stain (PSS) on seed pods and seeds, and Cercospora leaf blight (CLB) on leaves and petioles. Both symptoms present a typical dark-purple-colored lesion. This is caused by cercosporin, a pigment produced by the pathogen. The pigment induces cell death of the host plant in conditions with light (Kuyama and Tamura 1957; Yamazaki et al. 1975; Daub and Hangarter 1983).
Symptoms caused by C. kikuchii are frequently observed in soybean fields. Currently, CLB is one of the biggest problems for soybean production in many areas of South America, such as Argentina (Wrather et al. 2010). It has been suggested that the C. kikuchii populations in South America and Japan are different, based on phylogenetic analysis (Imazaki et al. 2006). It has also been reported that other Cercospora species such as C. cf. flagellaris, C. cf. sigesbeckiae (Albu et al. 2016), and C. cf. nicotianae (Sautua et al. 2020) can also cause soybean leaf blight. Moreover, some cryptic species of Cercospora infect soybean (Soares et al. 2015). Phylogenetic studies of Cercospora species, including CLB pathogens, have been reported (Groenewald et al. 2013; Soares et al. 2015). Such investigations will significantly benefit from additional high-quality genomic resources.
The genomes of the Cercospora spp. such as C. zeae-maydis (causal agent of gray leaf spot on corn, Haridas et al. 2020) and C. beticola (causal agent of leaf spot on sugar beet and other plant species, Vaghefi et al. 2017) have been analyzed and deposited in public databases. Additionally, genome sequences of the species infecting soybean have been published, such as Cercospora sojina (frogeye leaf spot, Luo et al. 2018; Gu et al. 2020) and Cercospora cf. sigesbeckiae (leaf blight, Albu et al. 2017). For C. kikuchii, two genomes have recently been deposited. These genomes were obtained from isolates originating from the United States (isolate A3, NCBI GenBank accession: GCA_005356855.1, contig N50: 364,948 bp) and Argentina (isolate ARG_18_001, NCBI GenBank accession: GCA_009193115.1, Sautua et al. 2019, contig N50: 675,846 bp), but have low contiguity.
The aim of this study was to obtain a reference genome sequence for C. kikuchii for further comparative genomic studies of the Cercospora species. The Japanese C. kikuchii isolate MAFF 305040 was sequenced. The isolate was subjected to species-wide phylogenetic study of the genus Cercospora in the previous study (Groenewald et al. 2013). Sufficient depth of sequencing coverage by long-read sequences generated a chromosome-level assembly. This high-quality genome sequence data provide fundamental genomic information and deepen our understanding of the pathogen.
Materials and methods
Fungal strain and culture conditions
Cercospora kikuchii MAFF 305040 was provided by the National Agriculture and Food Research Organization (NARO) Genebank (Tsukuba, Ibaraki, Japan). The isolate was maintained and cultured by following the method (Kashiwa et al. 2021).
DNA extraction from the fungus
Mycelia were collected from 3- to 4-day-old liquid cultures and dehydrated on paper towels. Collected mycelia (approximately 1 g) was ground in liquid nitrogen and extracted with 10 mL of CTAB lysis buffer (100 mM Tris-HCl, 2% CTAB, 1.4 M NaCl, 10 mM EDTA) at 65°C for 1 h. DNA was purified using a double extraction by mixing for 15 min with an equal volume of chloroform: iso-amylalcohol mixture (24:1) and then centrifuged at 8,370 × g for 10 min. The DNA was precipitated with ethanol and then centrifuged at 20,400 × g for 5 min at 4°C. The pellet was washed with 70% ethanol and dried. The DNA was resuspended in 10 mM Tris-HCl solution and stored at −30°C prior to genome sequencing.
Genome sequencing and data manipulation
The genome DNA library was prepared and subjected to sequencing by PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA). RNA removal from the DNA solution, DNA library preparation, and sequencing was performed by Macrogen Japan (Tokyo, Japan). Statistics for the sequence data was calculated by SeqKit 0.12.1 (Shen et al. 2016). The obtained sequence data were assembled by Canu 1.9 (Koren et al. 2017). Default values for Canu were kept for the assembly step. The estimated genome size was set to 34 Mb. Assembled sequences were polished by Arrow 2.3.3 (Pacific Biosciences). The polished assembly was checked by Tapestry 1.0.0 (Davey et al. 2021) with default values. The telomeric repeat TTAGGG (Cohn et al. 2005) was for the detection of telomeric repeats. The software Tapestry is also used for the evaluation of read depth and GC content of each contigs.
The Dfam TE Tools 1.4 for Docker was used for soft masking the genome based on de novo repeat finding by RepeatModeler (Flynn et al. 2020). The container incorporating the programs obtained from http://www.repeatmasker.org/ (RepeatModeler, RepeatMasker, coseg, RMBlast, RepeatScout, and RECON) and other tools such as HMMER (http://hmmer.org/), CD-HIT (Fu et al. 2012; Li and Godzik 2006), GenomeTools (Gremme et al. 2013), LTR_retriever (Ou and Jiang 2018), MAFFT (Katoh and Standley 2013), NINJA (Wheeler 2009), and Tandem Repeats Finder (Benson 1999). Gene prediction was performed using BRAKER2 (braker.pl version 2.1.6; Hoff et al. 2016; Hoff et al. 2019; Brůna et al. 2021) incorporating Augustus 3.4.0 (Stanke et al. 2006; Stanke et al. 2008). Genes were predicted with proteins of any evolutionary distance (--epmode). The fungal protein database was obtained from OrthoDB (https://v100.orthodb.org/download/odb10_fungi_fasta.tar.gz, accessed on August 24, 2020) and subjected to gene prediction using the ProtHint pipeline (Lomsadze et al. 2005; Iwata and Gotoh 2012; Gotoh et al. 2014; Buchfink et al. 2015; Brůna et al. 2020) in BRAKER2. Genes predicted by Augustus with hints were used for downstream analysis. Completeness of the genome assembly was assessed by BUSCO v4.1.2 (Seppey et al. 2019) for the predicted proteins, using the dataset dothideomycetes_odb10 (2020-08-05) obtained from the database.
Secreted proteins were predicted using the deep learning-based program DeepSig (docker image: bolognabiocomp/deepsig:latest, accessed on June 9, 2021; Savojardo et al. 2018), then candidate effector proteins were predicted using EffectorP 2.0 (Sperschneider et al. 2018) from the proteins predicted to have signal sequence at the N-terminus. Prediction of the carbohydrate-active enzymes (CAZymes) was performed using the dbCAN meta server (Yin et al. 2012; Zhang et al. 2018; http://bcb.unl.edu/dbCAN2/, accessed on June 9, 2021). The result of HMMER was used when other tools (DIAMOND and/or Hotpep) also predicted the protein as CAZymes. Secondary metabolite (SM) gene clusters were predicted using antiSMASH 5.1.1 (Blin et al. 2019). Genome information was visualized using Circos v0.69-8 (Krzywinski et al. 2009).
Comparative analysis with the C. sojina genome
The genome assembly of the C. sojina Race 15 (Gu et al. 2020, NCBI GenBank accession: GCA_004299825.1, accessed on September 3, 2020) was obtained from the database. The genome assembly was subjected to gene prediction and annotation following the aforementioned scheme that was used for the MAFF 305040 genome.
Results and discussion
Assembly and completeness of the genome sequence
Genome sequencing generated more than 3.5 Gb from 3 SMRT cells of the PacBio RS II. Read length N50 was approximately 14 kb. The polished genome assembly for MAFF 305040 consisted of 15 contigs comprising approximately 34.55 Mb. GC content of the polished assembly was 53.0%. Based on the size of contigs, we obtained nine large contigs (2.36–5.86 Mb) and six small contigs (1.2–54.3 kb). To filter contigs, we used two criteria based on Tapestry results: contig GC content ranges 53 ± 10%; and median of read depth for contig is above 20. After filtering, six small contigs were excluded. Telomeric repeats were detected on both ends for eight contigs out of the nine selected contigs. This suggested that the eight contigs were close-to-complete chromosomes. The nine contigs comprising approximately 34.44 Mb were subjected to downstream analysis (Table 1). GC content of the nine contigs was 53.0% (Table 1).
Table 1.
Summary of the genome assembly
| Features | Value |
|---|---|
| Isolate name | MAFF 305040 |
| Culture collection | NARO Genebank, Japan |
| Origin of isolatea | Kagoshima, Japan |
| Assembly size (bp) | 34,440,063 |
| Number of contigs | 9 |
| Maximum contig length (bp) | 5,855,908 |
| N50 of contigs | 4,190,438 |
| GC content (%) | 53.0 |
| BUSCO completeness (%) | 99.4 |
| Predicted genes | 13,001 |
| Genome accessionb | BOLY00000000 |
Data obtained from NARO Genebank (https://www.gene.affrc.go.jp, accessed on August 31, 2020).
DDBJ/EMBL/GenBank accession number for the assembled contigs.
As a result of repeat masking, approximately 1.45 Mb (4.21% of total sequence) was masked. Among the masked regions, 119 LINEs, 426 LTR elements, and 92 DNA transposons were detected.
Recently, genome sequences of soybean pathogens were deposited in public databases, and some of their assemblies produced chromosome-level contigs. Compared to C. sojina (12 chromosomes comprising approximately 40.12 Mb, NCBI GenBank accession: GCA_004299825.1, Gu et al. 2020), the genome of C. kikuchii MAFF 305040 was found to be somewhat smaller, while that of C. cf. sigesbeckiae (approximately 34.94 Mb, NCBI GenBank accession: GCA_002217505.1, Albu et al. 2017) was of a similar size. However, consideration should be given to the fragmentation of the C. cf. sigesbeckiae genome (1,945 contigs) for size comparison.
A total of 13,001 genes were predicted by Augustus that was trained by the BRAKER2 pipeline. The pipeline used a fungal gene ortholog database, and Augustus was trained by the proteins from any evolutionary distance for gene prediction. Based on the predicted coding protein sequences, the completeness of the genome assembly was estimated to be 99.4% by BUSCO (Table 1). In the 3,786 groups, there were only 21 missing and 4 fragmented BUSCOs for MAFF 305040.
Pathogenicity-related genomic features and comparisons to C. sojina, one of the soybean pathogen of genus Cercospora
To obtain information about the pathogenicity-related genes in the genome, several programs were used for functional annotation. Effectors are an important feature of plant pathogenic fungi, as they suppress host immunity. Effector proteins possess several characteristics, such as being small, secreted, and they have a high proportion of cysteine residues (Sperschneider et al. 2018). The DeepSig program estimated that a total of 1,393 proteins harbored an N-terminus signal peptide sequence for secretion. From the predicted secretome proteins, the EffectorP 2.0 program identified 245 proteins as effector candidates. The number of the genes for effector candidates was 242 (Table 2) after removing duplicated predictions for two transcripts from one gene.
Table 2.
Candidates for pathogenicity-related genes and gene clusters
| Features | Number |
|---|---|
| Effector candidatesa | 242 |
| SM gene clustersb | 55 |
| Polyketide | 15 |
| Nonribosomal peptide | 22 |
| Terpene | 6 |
| Beta-lactone | 1 |
| Siderophore | 2 |
| Fungal-RiPP | 1 |
| Others | 8 |
| CAZymesc | 399 |
|
GH |
220 |
| GT | 86 |
| CBM | 10 |
| AA | 66 |
| CE | 19 |
| PL | 7 |
Genes encoding effector candidates predicted by EffectorP.
Numbers of SM gene clusters for each metabolite type predicted by antiSMASH. Fungal-RiPP, fungal ribosomally synthesized and post-translationally modified peptide.
Genes encoding carbohydrate-activate enzymes (CAZymes) predicted by dbCAN. Some genes were annotated with more than one category. GH, glycoside hydrolase; GT, glycosyltransferase; CBM, carbohydrate-binding module; AA, auxiliary activity; CE, carbohydrate esterase; and, PL, polysaccharide lyase
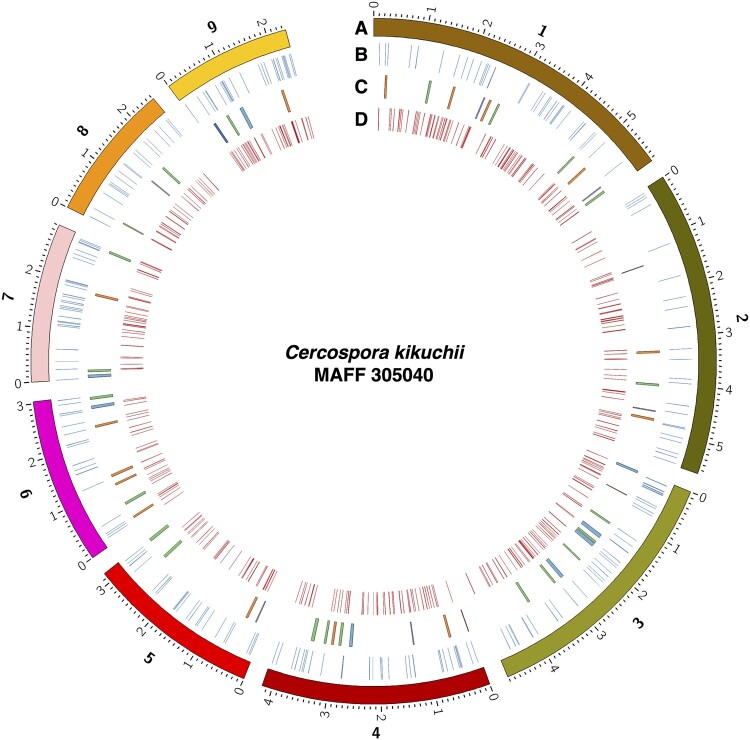
In some plant pathogenic fungi, specific genomic compartments can function as toolboxes for effector genes. For example, in Fusarium oxysporum, effector genes such as SIX (Secreted In Xylem) are enriched in small dispensable chromosomes (approximately 2 Mb). Transmission of the chromosomes between the isolates enables the acquisition of pathogenicity to nonpathogens (Ma et al. 2010). For the genome of C. kikuchii MAFF 305040, the effector candidate genes were distributed among the nine contigs (Figure 1B). However, the frequency of the genes for effector candidates was different among the contigs. For the largest contig_00001, the frequency of the genes for effector candidates in 100 kb of nucleotide sequences was 0.60 (35 genes in 5,855,908 bp). Meanwhile, the frequency was more than double (1.23, 29 genes in 2,357,730 bp) for contig_00009. It is assumed that the benefit of effector-enriched chromosomes is a facile adaptation to environmental changes (Croll and McDonald 2012). Namely, conditionally dispensable small chromosomes fulfill the role of the toolbox for pathogenicity-related factors and accelerate changes in the pathogenicity by altering the content and/or expression profile of the effector genes on the chromosomes (Seidl et al. 2016). It is uncertain whether such genomic regions or chromosomes are in the C. kikuchii genome. Further studies of the gene composition of each chromosome and the virulence functions of the predicted effector candidates are necessary to define the properties of chromosomes.
Figure 1.
Circos plot of MAFF 305040 genome. Tracks indicates: (A) Contigs of MAFF 305040, minor ticks indicate 0.1 Mb. Numbers for tracks indicate identifier for contigs (1, contig_00001; 2, contig_00002; 3, contig_00003; 4, contig_00004; 5, contig_00005; 6, contig_00006; 7, contig_00007; 8, contig_00008; and 9, contig_00009); (B) Positions of the genes encoding effector candidates (blue); (C) Positions of the SM gene clusters. Colors indicate predicted SM type of the cluster: orange, polyketide; green, nonribosomal peptide; red, siderophore; purple, terpene and beta-lactone; and dark blue, ribosomally synthesized and post-translationally modified peptide. Other clusters are colored blue. (D) Positions of the genes encoding CAZymes (red).
SM gene clusters were also predicted in the nine contigs (Figure 1C, Table 2). A total of 55 gene clusters were predicted from the genome (Table 2). Numbers of each metabolite types from the cluster are also listed in Table 2. For instance, 15 polyketides, 22 nonribosomal peptides, and 6 terpenes were predicted. A cluster harboring multiple core biosynthetic genes was grouped into “others” in Table 2.
Carbohydrate-active enzymes (CAZymes) are also important for fungi, for successful infections of their plant hosts. CAZymes are used to degrade plant polysaccharides to obtain nutrients and to enhance the infection in the host (Zhao et al. 2014). A total of 399 genes encoding CAZymes were predicted by dbCAN and classified into six categories (Figures 1D and 2, Table 2). Some of the proteins were matched multiple categories. Among these, 220 genes were categorized as glycoside hydrolase (GH), 86 as glycosyltransferase (GT), 10 as carbohydrate-binding module (CBM), 66 as auxiliary activity (AA), 19 as carbohydrate esterase (CE), and 7 as polysaccharide lyase (PL).
Figure 2.
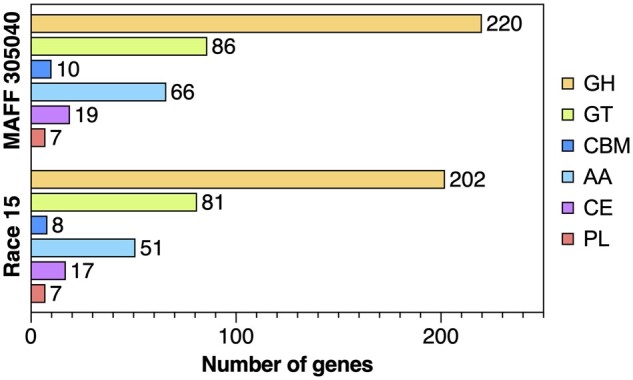
Number of genes in the C. kikuchii MAFF 305040 genome and C. sojina Race 15 genome for each CAZyme category. GH, glycoside hydrolase; GT, glycosyltransferase; CBM, carbohydrate-binding module; AA, auxiliary activity; CE, carbohydrate esterase; PL, polysaccharide lyase.
The genomic composition of CAZymes is key to understanding the infection strategy of plant pathogenic fungi (Zhao et al. 2014; Hane et al. 2020). As described, different species of the genus Cercospora can utilize soybeans as their hosts. To understand their difference, their CAZyme profiles were compared. The CAZymes in the C. sojina Race 15 genome were also checked for this purpose. A total of 13,253 genes were predicted from 12 chromosomes of Race 15 by BRAKER2-trained Augustus. Then, the dbCAN meta server selected 359 genes as having CAZyme encoding sequences. This number is higher than in previous C. sojina genome analyses (number of CAZyme genes was 340, Gu et al. 2020). Interestingly, the number of genes encoding CAZymes in the C. kikuchii genome was greater than in C. sojina (Figure 2). As described, C. kikuchii has a smaller genome (34.44 Mb) than C. sojina (40.12 Mb, Gu et al. 2020). For instance, the number of genes coding for the GH family enzyme was different between the two genomes. The genome of C. kikuchii has some additional genes cording GH (Figure 2 and Table 3). It is known that some enzymes belonging to the GH family are critical for the infection of the host plant. For example, PsGH7a (GH7) contributes for virulence in soybean oomycete pathogen Phytophthora sojae (Tan et al. 2020). Based on the previous report (Zhao et al. 2014), 61 genes from C. kikuchii MAFF 305040 were grouped into plant cell wall (PCW in Table 3) as substrate of the encoding enzyme. For C. sojina Race 15, 53 genes were grouped into PCW (Table 3). Functional analysis of their pathogenicity-related factors, such as CAZymes, is needed for further study.
Table 3.
Number of genes related to GH families identified from C. kikuchii MAFF 305040 and C. sojina Race 15
| Family a | MAFF 305040 | Race 15 | Substrate b |
|---|---|---|---|
| GH64 | 4 | 4 | CW (β-1,3-glucan) |
| GH1 | 3 | 3 | CW (β-glycans) |
| GH2 | 6 | 6 | CW (β-glycans) |
| GH3* | 17 | 16 | CW (β-glycans) |
| GH5* | 14 | 13 | CW (β-glycans) |
| GH32 | 3 | 3 | ESR (sucrose/inulin) |
| GH37 | 2 | 2 | ESR (trehalose) |
| GH65 | 1 | 1 | ESR (trehalose) |
| GH15* | 2 | 1 | ESR (α-glucans) |
| GH30 | 2 | 2 | FCW |
| GH85 | 1 | 1 | FCW |
| GH18* | 6 | 5 | FCW (chitin) |
| GH20 | 1 | 1 | FCW (chitin) |
| GH76 | 9 | 9 | FCW (chitin) |
| GH17 | 4 | 5 | FCW (β-1,3-glucan) |
| GH55 | 5 | 5 | FCW (β-1,3-glucan) |
| GH71 | 2 | 2 | FCW (β-1,3-glucan) |
| GH72 | 6 | 6 | FCW (β-1,3-glucan) |
| GH81 | 1 | 1 | FCW (β-1,3-glucan) |
| GH16* | 11 | 9 | FCW (β-glycans) |
| GH13 | 15 | 15 | FCW + ESR (α-glucans) |
| GH7 | 1 | 1 | PCW (cellulose) |
| GH12 | 1 | 1 | PCW (cellulose) |
| GH10* | 4 | 3 | PCW (hemicellulose) |
| GH11 | 3 | 3 | PCW (hemicellulose) |
| GH27 | 2 | 3 | PCW (hemicellulose) |
| GH29* | 3 | 2 | PCW (hemicellulose) |
| GH35* | 2 | 1 | PCW (hemicellulose) |
| GH36* | 2 | 1 | PCW (hemicellulose) |
| GH39 | 0 | 1 | PCW (hemicellulose) |
| GH51* | 3 | 2 | PCW (hemicellulose) |
| GH53 | 1 | 1 | PCW (hemicellulose) |
| GH54 | 1 | 1 | PCW (hemicellulose) |
| GH62 | 1 | 1 | PCW (hemicellulose) |
| GH67 | 1 | 1 | PCW (hemicellulose) |
| GH93* | 1 | 0 | PCW (hemicellulose) |
| GH115 | 1 | 2 | PCW (hemicellulose) |
| GH43* | 12 | 10 | PCW (pectin + hemicellulose) |
| GH28 | 5 | 5 | PCW (pectin) |
| GH78* | 3 | 2 | PCW (pectin) |
| GH88* | 2 | 1 | PCW (pectin) |
| GH105 | 3 | 3 | PCW (pectin) |
| GH38 | 1 | 1 | PG (N-/O-glycans) |
| GH47 | 9 | 9 | PG (N-/O-glycans) |
| GH63 | 1 | 1 | PG (N-glycans) |
| GH125 | 3 | 3 | PG (N-glycans) |
| GH31* | 9 | 8 | PG + ESR + PCW (hemicellulose) |
| GH33 | 1 | 1 | NA |
| GH42 | 1 | 1 | NA |
| GH79 | 3 | 4 | NA |
| GH92 | 7 | 7 | NA |
| GH95* | 2 | 0 | NA |
| GH97* | 1 | 0 | NA |
| GH106 | 1 | 1 | NA |
| GH114 | 1 | 1 | NA |
| GH127 | 1 | 1 | NA |
| GH128* | 2 | 1 | NA |
| GH130 | 1 | 1 | NA |
| GH131 | 1 | 1 | NA |
| GH132 | 1 | 1 | NA |
| GH135 | 1 | 2 | NA |
| GH139* | 1 | 0 | NA |
| GH141* | 1 | 0 | NA |
| GH142 | 1 | 1 | NA |
| GH152 | 1 | 1 | NA |
| GH154* | 2 | 1 | NA |
Asterisk indicates that the number of genes identified from the C. kikuchii MAFF 305040 was greater than the number identified from C. sojina Race 15.
Information on the substrate was obtained from Zhao et al. (2014). CW, cell wall; ESR, energy storage and recovery; FCW, fungal cell wall; PCW, plant cell wall; PG, protein glycosylation; NA, not assigned.
As described, several Cercospora species, including cryptic groups, are known to cause diseases in soybeans. The ability to elucidate the relationships between these pathogens provides a strong incentive to define their diversity and classification. This is the first chromosome-level assembly for C. kikuchii, causal agent of CLB and PSS. Our highly contiguous genome assembly of C. kikuchii and the results obtained in this study provide a solid foundation for genome analysis of the genus Cercospora.
Acknowledgments
The authors are grateful to Ms. Kumiko Kitaoka for technical assistance; Drs. Naoki Yamanaka, Kazuo Nakashima, Masayasu Kato, Miguel Angel Lavilla, Antonio Diaz Paleo, and Antonio Juan Gerardo Ivancovich for helpful discussions; and Ms. Yoshii Ishii for useful advice on DNA extraction. The C. kikuchii isolate MAFF 305040 was provided by NARO Genebank in Japan.
Data availability
The data described herein have been deposited at DDBJ/EMBL/GenBank under the BioProject accession PRJDB10872. The BioSample accession number for MAFF 305040 is SAMD00262043. The sequencing reads have been deposited under the accession number DRX296098. The genome assembly has been deposited under accession number BOLY00000000.
Author contributions
T.K. conceived, designed, and performed the experiments. T.K. and T.S. analyzed the genome sequence. T.K. wrote the manuscript in consultation with T.S. All authors have approved the manuscript.
Funding
This study was financially supported by and conducted as part of the JIRCAS research project “Development of technologies for the control of migratory plant pests and transboundary diseases.” This study was also supported by JSPS KAKENHI Grant Numbers JP18K14467 and JP21K14858.
Conflicts of interest
None declared.
Literature cited
- Albu S, Schneider RW, Price PP, Doyle VP.. 2016. Cercospora cf. flagellaris and Cercospora cf. sigesbeckiae are associated with Cercospora leaf blight and purple seed stain on soybean in North America. Phytopathology. 106:1376–1385. doi:10.1094/PHYTO-12-15-0332-R. [DOI] [PubMed] [Google Scholar]
- Albu S, Sharma S, Bluhm BH, Price PP, Schneider RW, et al. 2017. Draft genome sequence of Cercospora cf. sigesbeckiae, a causal agent of Cercospora leaf blight on soybean. Genome Announc. 5:e00708–17. doi:10.1128/genomeA.00708-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. doi:10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, et al. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47:W81–W87. doi:10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brůna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M.. 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform. 3:lqaa108. doi:10.1093/nargab/lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brůna T, Lomsadze A, Borodovsky M.. 2020. GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genom Bioinform. 2:lqaa026. doi:10.1093/nargab/lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12:59–60. doi:10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Cohn M, Liti G, Barton DBH.. 2005. Telomeres in fungi. In: Sunnerhagen P, Piskur J, editors. Comparative Genomics. Topics in Current Genetics. Vol. 15. Berlin, Heidelberg: Springer. p. 101–130. doi:10.1007/4735_108. [Google Scholar]
- Croll D, McDonald BA.. 2012. The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog. 8:e1002608. doi:10.1371/journal.ppat.1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub ME, Hangarter RP.. 1983. Light-induced production of singlet oxygen and superoxide by the fungal toxin, cercosporin. Plant Physiol. 73:855–857. doi:10.1104/pp.73.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Catta-Preta CMC, James S, Forrester S, Motta MCM, et al. 2021. Chromosomal assembly of the nuclear genome of the endosymbiont-bearing trypanosomatid Angomonas deanei. G3 (Bethesda). 11:jkaa018. doi:10.1093/g3journal/jkaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, et al. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117:9451–9457. doi:10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 28:3150–3152. doi:10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O, Morita M, Nelson DR.. 2014. Assessment and refinement of eukaryotic gene structure prediction with gene-structure-aware multiple protein sequence alignment. BMC Bioinformatics. 15:189. doi:10.1186/1471-2105-15-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremme G, Steinbiss S, Kurtz S.. 2013. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform. 10:645–656. doi:10.1109/TCBB.2013.68. [DOI] [PubMed] [Google Scholar]
- Groenewald JZ, Nakashima C, Nishikawa J, Shin H-D, Park J-H, et al. 2013. Species concepts in Cercospora: spotting the weeds among the roses. Stud Mycol. 75:115–170. doi:10.3114/sim0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Ding J, Liu W, Yang X, Yao L, et al. 2020. Comparative genomics and association analysis identifies virulence genes of Cercospora sojina in soybean. BMC Genomics. 21:172. doi:10.1186/s12864-020-6581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane JK, Paxman J, Jones D, Oliver RP, de Wit P.. 2020. "CATAStrophy," a genome-informed trophic classification of filamentous plant pathogens—how many different types of filamentous plant pathogens are there? Front Microbiol. 10:3088. doi:10.3389/fmicb.2019.03088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haridas S, Albert R, Binder M, Bloem J, LaButti K, et al. 2020. 101 Dothideomycetes genomes: a test case for predicting lifestyles and emergence of pathogens. Stud Mycol. 96:141–153. doi:10.1016/j.simyco.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M.. 2016. BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 32:767–769. doi:10.1093/bioinformatics/btv661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lomsadze A, Borodovsky M, Stanke M, 2019. Whole-Genome Annotation with BRAKER. In: Kollmar M, editor. Gene Prediction. Methods in Molecular Biology. Vol. 1962. New York: Humana Press. p. 65–95. doi:10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imazaki I, Homma Y, Kato M, Vallone S, Yorinori JT, et al. 2006. Genetic relationships between Cercospora kikuchii populations from South America and Japan. Phytopathology. 96:1000–1008. doi:10.1094/PHYTO-96-1000. [DOI] [PubMed] [Google Scholar]
- Iwata H, Gotoh O.. 2012. Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res. 40:e161. doi:10.1093/nar/gks708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwa T, Lavilla MA, Paleo AD, Ivancovich AJG, Yamanaka N.. 2021. The use of detached leaf inoculation for selecting Cercospora kikuchii resistance in soybean genotypes. PhytoFrontiers. doi:10.1094/PHYTOFR-01-21-0002-TA. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. doi:10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722–736. doi:10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. doi:10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyama S, Tamura T.. 1957. Cercosporin. A pigment of Cercosporina kikuchii Matsumoto et Tomoyasu. I. Cultivation of fungus, isolation and purification of pigment. J Am Chem Soc. 79:5725–5726. doi:10.1021/ja01578a038. [Google Scholar]
- Li W, Godzik A.. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22:1658–1659. doi:10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M.. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33:6494–6506. doi:10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Cao J, Huang J, Wang Z, Guo Z, et al. 2018. Genome sequencing and comparative genomics reveal the potential pathogenic mechanism of Cercospora sojina Hara on soybean. DNA Res. 25:25–37. doi:10.1093/dnares/dsx035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi M-J, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 464:367–373. doi:10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Tomoyasu R.. 1925. Studies on purple speck of soybean seed. Jpn J Phytopathol. 1:1–14. doi:10.3186/jjphytopath.1.6_1. [Google Scholar]
- Ou S, Jiang N.. 2018. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176:1410–1422. doi:10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautua FJ, Gonzalez SA, Doyle VP, Berretta MF, Gordó M, et al. 2019. Draft genome sequence data of Cercospora kikuchii, a causal agent of Cercospora leaf blight and purple seed stain of soybeans. Data Brief. 27:104693. doi:10.1016/j.dib.2019.104693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautua FJ, Searight J, Doyle VP, Scandiani MM, Carmona MA.. 2020. Cercospora cf. nicotianae is a causal agent of Cercospora leaf blight of soybean. Eur J Plant Pathol. 156:1227–1231. doi:10.1007/s10658-020-01969-z. [Google Scholar]
- Savojardo C, Martelli PL, Fariselli P, Casadio R.. 2018. DeepSig: deep learning improves signal peptide detection in proteins. Bioinformatics. 34:1690–1696. doi:10.1093/bioinformatics/btx818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl MF, Cook DE, Thomma BPHJ.. 2016. Chromatin biology impacts adaptive evolution of filamentous plant pathogens. PLoS Pathog. 12:e1005920. doi:10.1371/journal.ppat.1005920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM.. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 1962:227–245. doi:10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, Hu F.. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 11:e0163962. doi:10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares APG, Guillin EA, Borges LL, da Silva ACT, de Almeida ÁMR, et al. 2015. More Cercospora species infect soybeans across the Americas than meets the eye. PLoS One. 10:e0133495. doi:10.1371/journal.pone.0133495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM.. 2018. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol. 19:2094–2110. doi:10.1111/mpp. 12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D.. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 24:637–644. doi:10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Stanke M, Schöffmann O, Morgenstern B, Waack S.. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 7:62–11. doi:10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. 1921. Studies on the cause of purple seed stain of soybean. Korean Agric Assoc Rep. 16:24–28. [Google Scholar]
- Tan X, Hu Y, Jia Y, Hou X, Xu Q, et al. 2020. A conserved glycoside hydrolase family 7 cellobiohydrolase PsGH7a of Phytophthora sojae is required for full virulence on soybean. Front Microbiol. 11:1285. doi:10.3389/fmicb.2020.01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaghefi N, Kikkert JR, Bolton MD, Hanson LE, Secor GA, et al. 2017. De novo genome assembly of Cercospora beticola for microsatellite marker development and validation. Fungal Ecol. 26:125–134. doi:10.1016/j.funeco.2017.01.006. [Google Scholar]
- Walters HJ. 1980. Soybean leaf blight caused by Cercospora kikuchii. Plant Dis. 64:961–962. doi:10.1094/PD-64-961. [Google Scholar]
- Wheeler TJ. 2009. Large-scale neighbor-joining with NINJA. In: Salzberg SL, Warnow T, editors. Algorithms in Bioinformatics. WABI 2009. Lecture Notes in Computer Science, Vol. 5724. Berlin, Heidelberg: Springer. p. 375–389. doi:10.1007/978-3-642-04241-6_31. [Google Scholar]
- Wrather A, G, Shannon RB, L, Escobar Carregal R, et al. 2010. Effect of diseases on soybean yield in the top eight producing countries in 2006. Plant Health Prog. 11:1. doi:10.1094/PHP-2010-0102-01-RS. [Google Scholar]
- Yamazaki S, Okubo A, Akiyama Y, Fuwa K.. 1975. Cercosporin, a novel photodynamic pigment isolated from Cercospora kikuchii. Agr Biol Chem. 39:287–288. doi:10.1080/00021369.1975.10861593. [Google Scholar]
- Yin Y, Mao X, Yang J, Chen X, Mao F, et al. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40:W445–W451. doi:10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yohe T, Huang L, Entwistle S, Wu P, et al. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46:W95–W101. doi:10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Liu H, Wang C, Xu J-R.. 2014. Erratum to: comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 15:6. doi:10.1186/1471-2164-15-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data described herein have been deposited at DDBJ/EMBL/GenBank under the BioProject accession PRJDB10872. The BioSample accession number for MAFF 305040 is SAMD00262043. The sequencing reads have been deposited under the accession number DRX296098. The genome assembly has been deposited under accession number BOLY00000000.