Abstract
Aspergillus flavus is an opportunistic pathogen of crops, including peanuts and maize, and is the second leading cause of aspergillosis in immunocompromised patients. A. flavus is also a major producer of the mycotoxin, aflatoxin, a potent carcinogen, which results in significant crop losses annually. The A. flavus isolate NRRL 3357 was originally isolated from peanut and has been used as a model organism for understanding the regulation and production of secondary metabolites, such as aflatoxin. A draft genome of NRRL 3357 was previously constructed, enabling the development of molecular tools and for understanding population biology of this particular species. Here, we describe an updated, near complete, telomere-to-telomere assembly and re-annotation of the eight chromosomes of A. flavus NRRL 3357 genome, accomplished via long-read PacBio and Oxford Nanopore technologies combined with Illumina short-read sequencing. A total of 13,715 protein-coding genes were predicted. Using RNA-seq data, a significant improvement was achieved in predicted 5’ and 3’ untranslated regions, which were incorporated into the new gene models.
Keywords: Aspergillus flavus, NRRL 3357, PacBio, Nanopore, genome sequence
Introduction
Aspergillus flavus is an opportunistic plant pathogen, human pathogen, and a saprophyte. Agriculturally, A. flavus colonizes crops such as maize, peanuts, and cotton, both pre- and post-harvest (Klich 2007). While colonizing these crops, A. flavus produces aflatoxin, which is both toxic to mammals and a potent carcinogen (Williams et al. 2004; Liu and Wu 2010; Wild and Gong 2010), chronically impacting an estimated 4.5 billion people (CDC 2016). In the United States, mycotoxin production causes estimated yearly agricultural losses in corn ranging from tens of millions to over $1 billion (Mitchell et al. 2016). In addition to mycotoxin production, A. flavus is also a leading cause of invasive aspergillosis in humans, as well as the leading cause of fungal sinusitis and keratitis in tropical climates (Krishnan et al. 2009; Pasqualotto 2009; Rudramurthy et al. 2019).
The A. flavus isolate NRRL 3357 produces high levels of aflatoxin and has been developed as a model for the development of molecular tools and for dissecting the regulation and production of aflatoxin (Georgianna and Payne 2009; Amaike and Keller 2011). Previously, the genome sequence of A. flavus NRRL 3357 was sequenced using the whole-genome shotgun method (Nierman et al. 2015) and assembled into 958 contigs comprising 331 scaffolds. To enhance genome-wide studies in A. flavus for both functional and population genomics studies, we sought to generate a more complete genome assembly and annotation. Here, we report an updated, near complete, telomere-to-telomere assembly of the A. flavus strain NRRL 3357 genome, with 8 scaffolds corresponding to the 8 chromosomes of this species. Genome annotation, assisted by publicly available RNA-seq data, yielded greater resolution of 5’ and 3’ UTRs in this organism, with nearly half of genes containing annotated UTRs. We manually curated over 200 previously published genes, verifying that new predicted gene models corresponded with prior gene models and RNA-seq datasets.
Materials and methods
Fungal strain culture and DNA extraction
A. flavus NRRL 3357 was originally isolated from peanut, with our sample originating in the Keller lab strain collection (Hesseltine et al. 1966). Conidia from a week-old culture were grown in glucose minimal medium + 0.5% yeast extract at 30°C overnight. Genomic DNA isolation for genome sequencing of A. flavus was previously reported (Drott et al. 2020). Briefly, powdered lyophilized mycelia were resuspended in LETS buffer (20 mM EDTA pH 8.0, 0.5% SDS, 10 mM Tris-HCl pH 8.0, and 0.1 M LiCl). Genomic DNA was extracted using phenol: chloroform: isoamyl alcohol (25:24:1), followed by ethanol precipitation. gDNA was collected either by spooling or centrifugation, then washed with 70% ethanol and allowed to air dry. gDNA was resuspended in 10 mM Tris-HCl (pH 8.0) + 3.33 μg/mL RNase A and heated at 65°C for 30 minutes.
Sequencing
DNA quality control, library preparation for PacBio and Illumina, and sequencing were performed at the Vincent Coates Genomics Sequencing Laboratory at the University of California, Berkeley. For the PacBio sequencing, DNA quantification and quality control were performed using a Femto Pulse System (Agilent Technologies), and high molecular weight DNA (∼130 kbp average size) was used to construct BluePippin (Sage Science) size-selected (>30-kbp) SMRTbell libraries (PacBio). PacBio libraries were sequenced on the Sequel Platform using the S/P2-C2 polymerase and version 5.0 chemistry on four Sequel SMRT cells, generating a total of ∼1.8 M reads with average read length of 13,122 bp. For Illumina sequencing, small (∼540 bp) insert libraries were generated using KAPA DNA HyperPrep kit with PCR-free protocol (Roche), generating ∼319 M PE150 reads. Oxford Nanopore libraries were generated using ∼15 μg of high molecular weight DNA and the SQK-LSK308 kit. Oxford Nanopore sequencing was performed in-house using the minION platform and a combination of live base calling using minKNOW App (v1.11.5) and offline base calling using Albacore App (v2.3.3) (Oxford Nanopore). Three FLO-MIN107 flow cells (version 9.5.1 pore chemistry) were used to generate ∼1.4 M reads with average length of 3050 bp. The assembled genome sequence has been used to assess population genomics of A. flavus (Drott et al.2020, 2021).
Genome assembly and annotation
The combined PacBio and Oxford Nanopore long-read datasets were used to generate a hybrid de novo assembly using the CANU assembler (v.1.7.1) (Koren et al. 2017) with default settings, except the genome size was set to 40 Mbp and stopOnReadQuality set to “false.” The final read depth coverage, after filtering for reads <1 kbp, was ∼700X. The CANU scaffolds were polished using the PacBio Sequel data and a combination of pbalign (v.0.3.1), blasr (v.5.3), and arrow (v.2.2.2) from the SMRT Link package (v.5.1.0.26412, PacBio). A final error correction step was performed using the Illumina data (∼650X coverage) with a combination of bwa (v.0.7.17) (Li 2013), samtools (v.1.9) (Li et al. 2009), and pilon (v.1.22) (Walker et al. 2014). For the most accurate final assembly of the A. flavus genome, Illumina data and at least one round of pilon correction were needed. For the discovery of de novo repeats Repeatscout v1.0.5 (Price et al. 2005) was used and for repeat masking, RepBase (Bao et al. 2015) and RepeatMasker (Smit et al. 1996-2010) were used.
Annotation of the genome was performed using the Joint Genome Institute (JGI) Genome Annotation pipeline (Grigoriev et al. 2014) using publicly available RNA-seq data (SRA datasets: SRR2632952, SRR2632961, SRR2632962, SRR2632963, SRR2632966, SRR2633059, SRR2633060, SRR2633061, SRR2633139, SRR5061895, SRR5061899, SRR5061903, SRR5061905, SRR5061908, SRR5061909, SRR544871, SRR544872, SRR544873, SRR8115610, SRR8115611, SRR8115612, SRR8115613, SRR8115614, and SRR8115615). Previously produced gene models available from FungiDB (Stajich et al. 2012) were mapped forward to the new assembly. The final annotation included 13,715 gene models, of which 43.29% represent previously produced models mapped forward from FungiDB (fungidb.org/), while the remainder of gene models were updated/improved based on transcriptomics data and the improved assembly. For predicted short genes (i.e., <200AA), we evaluated predicted annotations including signal peptides, transmembrane domains, InterPro domain annotations, or support through self-clustering. If any of these types of support were detected, short gene models were retained in our final gene set. In addition, although alternative splice forms were not included in our final gene catalog, RNA-seq based models from tools like COMBEST (Zhou et al. 2015) are available as tracks on the Aspfl2_3 genome browser https://mycocosm.jgi.doe.gov/Aspfl2_3 and enable reconstruction of alternative splice forms.
Data availability
The whole-genome assembly and annotation and the A. flavus mitochondrial DNA sequence are available from the JGI MycoCosm portal (Grigoriev et al. 2014) at https://mycocosm.jgi.doe.gov/Aspfl2_3 and have been deposited at GenBank under accession numbers CP044616-CP044623, Bioproject accession number PRJNA575750. Raw sequencing reads have been deposited under SRA project accession number PRJNA637788. Manually curated genes and references are included in Supplementary Table S1. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14738154.
Results and discussion
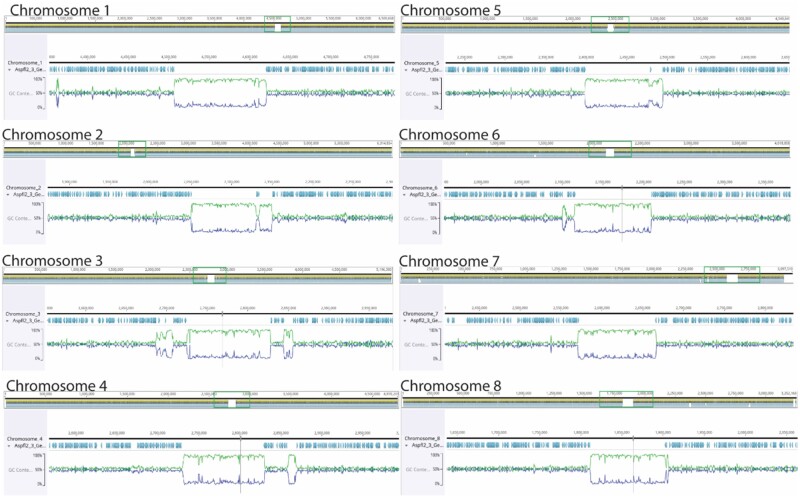
The final genome assembly of NRRL 3357 resulted from a combined long- (PacBio SMRT and Oxford Nanopore) and short-read (Illumina) sequencing methods was 37.75 Mbp in 8 contigs, a significant improvement from the previous assembly which contained 331 scaffolds (Table 1) (Nierman et al. 2015) and a genome size slightly less the 37 Mbp. The industrially relevant species, Aspergillus oryzae, is closely related to A. flavus and A. flavus chromosome names are based on the A. oryzae genome (Machida et al. 2005; Figure 1). The final sequencing read depth was 650X, and the average GC content across the genome was 47.34%. Seven of eight chromosomes are represented by complete telomere-to-telomere assemblies. It was not possible to complete the assembly of the right end of chromosome 7 due to the presence of a large rDNA repeat. The new assembly increased in size relative to the original assembly (Nierman et al. 2015) primarily due to the improved assembly of repetitive regions other than the rDNA region, resulting in an increase from 1.1% of the genome to 3.47% (Table 1), including a significant increase in Mariner Tc1 repeats [43% in previous assembly (Nierman et al. 2015) compared to 83.18% in the current assembly]. We also identified 15 out of 16 telomeres as well as predicted centromeric regions (Figure 1). Each chromosome was flanked by 10–13 telomeric repeats “TTAGGGTCAACA” that were identical to those identified in the A. oryzae (Kusumoto et al. 2003). In filamentous fungi, centromeric regions have high AT content, are ∼100 kb and typically lack coding regions (Smith et al. 2012). In the assembled A. flavus genome, predicted centromeric regions were identified using these criteria and were ∼100 kbp, but some chromosomes had additional AT-rich regions surrounding predicted centromeric regions (Figure 1). Verification of functional centromeric regions of the A. flavus genome would require further experimentation, for example, the identification of the specialized histone H3 variant [called CenH3 in Neurospora crassa) (Smith et al. 2011)], a so-called universal “centromere identifier.” In our new assembly, the mitochondrial DNA was identical to that previously published (Joardar et al. 2012).
Table 1.
Summary of assembly and annotation statistics of the A. flavus NRRL 3357 genome
| Assembly statistics | Illumina assembly (Nierman 2015 )a | Illumina assembly, reannotated (Hatmaker 2020 )b | Mixed sequencing assembly (this work) |
|---|---|---|---|
| Genome size (Mbp) | 36.89 | 36.89 | 37.75 |
| Coverage | 5X | 5X | 650X |
| Number of scaffolds/contigs | 958/331 | 958/331 | 8 |
| L50 | 6 | 6 | 4 |
| N50, Mbp (Scaffold) | 2.39 | 2.39 | 4.81 |
| Complete chromosomes | — | — | 8 |
| % genes by BUSCO assessmentc: Fungi set | |||
| Single-copy | 92.70 | 96.30 | 98.70 |
| Duplicated | 0.30 | 1.70 | 0.50 |
| Fragmented | 3.00 | 0.90 | 0.00 |
| Annotation statistics | |||
| Number of predicted protein coding genes | 13,485 | 14,313 | 13,715 |
| Percent gene models with annotated UTRs | 9.25% | — | 44.43% |
| Predicted secondary metabolite clusters | 56 | 83d | |
| Predicted CAZymes | 627 | 644 | |
| Total repeat length (bp) | 404,315 (1.1%) | 1,311,342 (3.47%) |
Nierman et al. (2015).
Hatmaker et al. (2020).
Seppey et al. (2019).
Drott et al. (2021).
Figure 1.
Graphic representation of the 8 NRRL 3357 Aspergillus flavus assembled chromosomes. Green traces represent AT content and dark blue traces represent GC content. Teal-colored arrows indicate predicted coding regions1.
1Figure generated using Geneious Prime v.2021.0.3; www.geneiousprime.com.
Our final annotation contained 13,715 predicted protein coding genes, of which 43.29% of the final filtered gene model set represent previously produced models mapped forward from FungiDB (https://fungidb.org/). The remaining gene models were updated and improved based on the improved assembly and on publicly available transcriptomics data (see Materials and Methods). This gene model count was slightly lower than a recently updated, transcriptome-based annotation of the 331 scaffolds of the A. flavus 2015 assembly (Nierman et al. 2015), which relied on RNA isolated from A. flavus in six conditions and used a combination of different ab initio gene predictors (Hatmaker et al. 2020). A BUSCO v5.0 (Seppey et al. 2019) analyses, calculated using the Fungi dataset, revealed that 98.7% of BUSCO genes were captured in single copy in our new assembly and annotation, vs 96.3% in the Hatmaker et al. (2020) and 92.7% from Nierman et al. (2015) versions (Table 1). In addition, fragmented and duplicated genes were less frequent in our version relative to other annotations. The total number of gene models in our version (13,715 genes) and the Nierman et al., annotation (13,485 genes) (Nierman et al. 2015) was slightly lower than gene models predicted by Hatmaker et al., (14,313 genes) (Hatmaker et al. 2020; Table 1). This discrepancy may partially result from our filtering methods, which aimed to remove transposable elements and short, unsupported gene models from the final gene catalog. We also used publicly available transcriptomic data to improve the annotation of 5’ and 3’ UTRs on our filtered gene models, which were absent or rarely predicted in other models. We UTRs for 44.43% of our models (6093 genes), a significant increase from the 9.25% UTRs identified from previous studies.
To validate updated annotation methods using the mapped-forward gene models, we chose a set of 172 genes that had been described in the literature to manually curate and to compare our new JGI models with FungiDB gene models (Supplementary Table S1). Within this set, we used publicly available RNA-seq data to provide 5’ and 3’ untranslated regions (UTRs) information and homology modeling to confirm gene models. In 68/172 genes, the models did not change, while for an additional 47 genes, the gene predictions were 100% identical, but UTRs were added (Supplementary Table S1). In 57 cases, the existing FungiDB gene model was corrected using newly available RNAseq and homology evidence. For a few gene models, it was difficult to define the correct model, primarily due to the absence of RNA-seq data or differences in predicted gene models when orthologs in other species were evaluated. In the Mycocosm portal (https://mycocosm.jgi.doe.gov/Aspfl2_3/Aspfl2_3.home.html), we provide a link to the closest FungiDB AFLA model for users to compare the new models with the previous annotation. Overall, our mapped-forward gene models, using the complete genome assembly and RNA-seq data provided more complete gene models for A. flavus and with significantly more 5’ and 3’ UTR information compared to previous versions.
As A. flavus is a plant pathogen and saprophyte, carbohydrate active enzymes (CAZymes) are important for this fungus’s ability to grow on plant material. With our updated annotation, we observed a slight increase in the number of predicted CAZymes, from 627 to 644, in the updated A. flavus genome annotation as predicted by CAZy Database (Lombard et al. 2014). A. flavus also produces a wealth of bioactive secondary metabolites from biosynthetic gene clusters (BGCs); the function and structure of these secondary metabolites is an active area of research with this organism (Greco et al. 2019; Keller 2019). Through prior studies, 56 secondary metabolite clusters have been identified in the A. flavus genome (Georgianna et al. 2010; Marui et al. 2011; Amare and Keller 2014; Nierman et al. 2015). Using this current A. flavus genome assembly for NRRL 3357, coupled with assessing BGC diversity in A. flavus populations, 83 BCGs were identified in the A. flavus NRRL 3357 genome (Drott et al. 2021). The genomic positions for these 83 BCGs in our assembled A. flavus NRRL 3357 genome are available as supplemental data from Drott et al. (2021).
In summary, we provide an improved, near-complete, telomere-to-telomere genome assembly for A. flavus NRRL 3357, resulting from combined Illumina, Oxford Nanopore, and PacBio SMRT sequencing data. This updated assembly has been useful for assessing population genomics of 94 isolates of A. flavus isolated from different geographic locations in the United States (Drott et al. 2020) and BCG diversity (Drott et al. 2021). The genome assembly consists of 8 contigs representing 8 chromosomes, with 15 of 16 telomeric repeats and all centromeric sequences identified and assembled. The total genome assembly size is 37.75 Mbp, and the updated annotation, supported by RNA-seq and homology data, yielded 13,715 predicted protein-coding gene models.
Funding
Funding for this project was provided by a grant to N.L.G., J.M.S., and A.P.A. through the Innovative Genomics Institute at University of California, Berkeley. Genome annotation performed by the Joint Genome Institute, a Department of Energy (DOE) Office of Science User Facility, was supported by the Office of Science of the US DOE under Contract no. DE-AC02-05CH11231.
Conflicts of interest
None declared.
Literature cited
- Amaike S, Keller NP.. 2011. Aspergillus flavus. Annu Rev Phytopathol. 49:107–133. [DOI] [PubMed] [Google Scholar]
- Amare MG, Keller NP.. 2014. Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet Biol. 66:11–18. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC 2016. Health studies. Understanding chemical exposures. Aflatoxin. CDC, Atlanta, GA: http://www.cdc.gov/nceh/hsb/chemicals/aflatoxin.htm.
- Drott MT, Rush TA, Satterlee TR, Giannone RJ, Abraham PE, et al. 2021. Microevolution in the pan-secondary metabolome of Aspergillus flavus has macroevolutionary implications for filamentous fungi. Proc Natl Acad Sci USA. 118:e2021683118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drott MT, Satterlee TR, Skerker JM, Pfannenstiel BT, Glass NL, et al. 2020. The frequency of sex: population genomics reveals differences in recombination and population structure of the aflatoxin-producing fungus Aspergillus flavus. mBio. 11:e00963-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgianna DR, Fedorova ND, Burroughs JL, Dolezal AL, Bok JW, et al. 2010. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol Plant Pathol. 11:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgianna DR, Payne GA.. 2009. Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet Biol. 46:113–125. [DOI] [PubMed] [Google Scholar]
- Greco C, Keller NP, Rokas A.. 2019. Unearthing fungal chemodiversity and prospects for drug discovery. Curr Opin Microbiol. 51:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, et al. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42:D699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatmaker EA, Zhou X, Mead ME, Moon H, Yu JH, et al. 2020. Revised transcriptome-based gene annotation for Aspergillus flavus Strain NRRL 3357. Microbiol Resour Announc. 9:e01155-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesseltine CW, Shotwell OL, Ellis JJ, Stubblefield RD.. 1966. Aflatoxin formation by Aspergillus flavus. Bacteriol Rev. 30:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joardar V, Abrams NF, Hostetler J, Paukstelis PJ, Pakala S, et al. 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics. 13:698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP. 2019. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol. 17:167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klich MA. 2007. Aspergillus flavus: the major producer of aflatoxin. Mol Plant Pathol. 8:713–722. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Manavathu EK, Chandrasekar PH.. 2009. Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses. 52:206–222. [DOI] [PubMed] [Google Scholar]
- Kusumoto KI, Suzuki S, Kashiwagi Y.. 2003. Telomeric repeat sequence of Aspergillus oryzae consists of dodeca-nucleotides. Appl Microbiol Biotechnol. 61:247–251. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. ; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wu F.. 2010. Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ Health Perspect. 118:818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2.
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B.. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, et al. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature. 438:1157–1161. [DOI] [PubMed] [Google Scholar]
- Marui J, Yamane N, Ohashi-Kunihiro S, Ando T, Terabayashi Y, et al. 2011. Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn(II)(2)Cys(6) transcriptional activator and induced by kojic acid at the transcriptional level. J Biosci Bioeng. 112:40–43. [DOI] [PubMed] [Google Scholar]
- Mitchell NJ, Bowers E, Hurburgh C, Wu F.. 2016. Potential economic losses to the US corn industry from aflatoxin contamination. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 33:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Yu J, Fedorova-Abrams ND, Losada L, Cleveland TE, et al. 2015. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announc. 3:e00168-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualotto AC. 2009. Differences in pathogenicity and clinical syndromes due to Aspergillus fumigatus and Aspergillus flavus. Med Mycol. 47(Suppl.):S261–S270. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA.. 2005. De novo identification of repeat families in large genomes. Bioinformatics. 21(Suppl.):i351–i358. [DOI] [PubMed] [Google Scholar]
- Rudramurthy SM, Paul RA, Chakrabarti A, Mouton JW, Meis JF.. 2019. Invasive Aspergillosis by Aspergillus flavus: epidemiology, diagnosis, antifungal Resistance, and management. J Fungi (Basel). 5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM.. 2019. 2019 BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 1962:227–245. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P.. 1996-2010. RepeatMasker Open-3.0. http://www.repeatmasker.org.
- Smith KM, Galazka JM, Phatale PA, Connolly LR, Freitag M.. 2012. Centromeres of filamentous fungi. Chromosome Res. 20:635–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M.. 2011. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol. 31:2528–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, et al. 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 40:D675–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP, Gong YY.. 2010. Mycotoxins and human disease: a largely ignored global health issue. Carcinogenesis. 31:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH, Phillips TD, Jolly PE, Stiles JK, Jolly CM, et al. 2004. Human aflatoxicosis in developing countries: a breview of toxicology, exposure, potential health consequences, and interventions. Am J Clin Nutr. 80:1106–1122. [DOI] [PubMed] [Google Scholar]
- Zhou K, Salamov A, Kuo A, Aerts AL, Kong X, et al. 2015. Alternative splicing acting as a bridge in evolution. Stem Cell Investig. 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome assembly and annotation and the A. flavus mitochondrial DNA sequence are available from the JGI MycoCosm portal (Grigoriev et al. 2014) at https://mycocosm.jgi.doe.gov/Aspfl2_3 and have been deposited at GenBank under accession numbers CP044616-CP044623, Bioproject accession number PRJNA575750. Raw sequencing reads have been deposited under SRA project accession number PRJNA637788. Manually curated genes and references are included in Supplementary Table S1. Supplementary material is available at figshare: https://doi.org/10.25387/g3.14738154.