Abstract
The ability to introduce mutations, or transgenes, of choice to precise genomic locations has revolutionized our ability to understand how genes and organisms work. In many mosquito species that are vectors of various human diseases, the advent of CRISPR genome editing tools has shed light on basic aspects of their biology that are relevant to their efficiency as disease vectors. This allows a better understanding of how current control tools work and opens up the possibility of novel genetic control approaches, such as gene drives, that deliberately introduce genetic traits into populations. Yet for the Anopheles funestus mosquito, a significant vector of malaria in sub-Saharan Africa and indeed the dominant vector species in many countries, transgenesis has yet to be achieved. We describe herein an optimized transformation system based on the germline delivery of CRISPR components that allows efficient cleavage of a previously validated genomic site and preferential repair of these cut sites via homology-directed repair (HDR), which allows the introduction of exogenous template sequence, rather than end-joining repair. The rates of transformation achieved are sufficiently high that it should be able to introduce alleles of choice to a target locus, and recover these, without the need to include additional dominant marker genes. Moreover, the high rates of HDR observed suggest that gene drives, which employ an HDR-type mechanism to ensure their proliferation in the genome, may be well suited to work in A. funestus.
Keywords: mosquitoes, funestus, anopheles, CRISPR, HDR, transgenesis, genetic control, vector biology, gene drive
Introduction
Mosquitoes belonging to the genus Anopheles are vectors of a range of agents of human disease, including viruses, parasitic worms, and, most notably, malaria parasites. A way to introduce defined genetic changes precisely into these insects can drastically improve our understanding of the genes responsible for key traits that determine its capacity as a disease vector, such as parasite susceptibility, female reproductive output, human-biting behavior, as well as helping in elucidating mechanisms of resistance to insecticides. Moreover, the ability to introduce genes of choice opens up the possibility of genetic control—the deliberate introduction of traits into a mosquito population to reduce its capacity to transmit disease.
To date, most attention has focused on two members of the Anopheles gambiae species complex, A. gambiae (s.s.) and Anopheles coluzzi, which are the dominant vectors of human malaria across most of sub-Saharan Africa, where malaria burden is highest. However, Anopheles funestus, not a member of this species complex, is the dominant vector in many places, notably in southerly regions of sub-Saharan Africa, and insecticide resistance in this species has been associated with local malaria resurgence there and loss of efficacy of control interventions (Cohuet et al. 2004; Riveron et al. 2019). Because the first demonstrations of a working, transposon-based transgenic technology for mosquitoes over 20 years ago, most of the major mosquito vectors have now been transformed (Coates et al. 1998; Jasinskiene et al. 1998; Catteruccia et al. 2000; Allen et al. 2001; Grossman et al. 2001), yet A. funestus still remains the exception. This may in part simply reflect the logistical difficulty associated with rearing this species in the laboratory (Hunt et al. 2005; Morgan et al. 2010; Ngowo et al. 2021), but may also reflect a quirk in its biology that make it more refractory to the process of germline transformation.
The prospects for transforming a mosquito species of choice have been greatly improved by the general applicability of CRISPR-Cas9 genome engineering tools to a wide range of species (Gantz et al. 2015; Hammond et al. 2016; Purusothaman et al. 2021). CRISPR-mediated cleavage can be repaired via one of two pathways: nonhomologous end-joining (NHEJ) repair and homology-directed repair (HDR). Recently, CRISPR-mediated cleavage was used to introduce random, heritable mutations via NHEJ at specific sites in the genome of A. funestus (Li et al. 2018). However, to date there still has been no documented report of HDR introduction (often referred to as “knock-in”) of either transgene or specified pre-determined mutations, into this species. This method relies on the provision of a DNA “donor” template containing regions of homology on either side of the cut site, designed in such a way that the cell’s HDR machinery uses it as a template for repair, resulting in the incorporation of an allele or transgene of choice. A method of this type is crucial for two reasons: it would allow confirmation of genetic factors/signatures identified in field populations that putatively confer important phenotypes, such as insecticide resistance, with strong relevance to control programs; it opens the door to novel genetic control approaches, such as gene drive, which result in the specific introduction and spread of transgenic traits into mosquito populations as a form of vector control.
The relative propensity for CRISPR-induced DNA breaks to be repaired by HDR or NHEJ can show strong variation according to whether the tissue is of somatic or germline origin, and even within the germline it can be dependent on the level and timing of Cas9 expression (Li et al. 2017; Kandul et al. 2020; Hammond et al. 2021). For the purposes of generating stable transformed mosquito strains it is essential that the CRISPR-induced modifications occur in the germline, so that they are transmitted to their offspring from which a modified line can be established. We have previously demonstrated high rates of CRISPR-mediated HDR in generating transgenic A. gambiae by microinjecting embryos with a plasmid-based source of Cas9 under a germline-specific promoter together with a donor template designed to incorporate a dominant marker gene. Given this success in A. gambiae, we investigated whether a similar approach could work in A. funestus, using components likely to work broadly across both species.
Our experimental design allowed us to determine the relative rates of NHEJ compared to HDR following Cas9 activity in injected embryos. We were able to show rates of HDR-mediated germline transformation comparable to those achieved for A. gambiae, suggesting that the wealth of functional genetics tools and options for genetic control that exist in that species may soon be available in A. funestus.
Materials and methods
Generation of transformation constructs for gene targeting by CRISPR
The helper plasmid, containing source of Cas9 and guide RNA, was a derivative of plasmid p165 (accession ID: KU189142) (Hammond et al. 2016), containing a vasa2::SpCas9 construct and a U6::gRNA cassette containing a spacer cloning site based on Hwang et al. (2013). Oligos (forward—TGCTGGTGAGCTCCTTGCGGTGAT; reverse—AAACATCACCGCAAGGAGCTCACC) were designed to make a functional spacer targeting a previously validated target site in the A. funestus white gene (AFUN003538) (Li et al. 2018) were annealed and cloned via BbsI Golden Gate cloning into the spacer cloning site to produce helper plasmid p18.
The HDR “donor” construct contained regions of homology approximately 1 kb long immediately upstream and downstream of the target cut site, amplified from pooled genomic DNA of A. funestus mosquitoes (FANG strain, LSTM) with primer sets designed for Gibson assembly of the two homology arms flanking a dominant marker cassette, designed to express eCFP. Primer sequences (sequences for Gibson cloning in small case) for the homology arms were as follows: 5’ homology, forward—gcgagctcgaattaaccattgtggAACCGTGCCTCTATTCTCAGC; 5’ homology, reverse—tggggtaccggtACCGCAAGGAGCTCACCAC; 3’ homology, forward—atcctgaacgcGTGATGGGTAGTTCCGGTGC; 3’ homology, reverse—tactccacctcacCCATGGGACCCAACATCGGATCCTTCAGAAC. The dominant marker cassette, containing an enhanced cyan fluorescent protein (eCFP) unit under the promotional control of the Drosophila Actin5c promoter (Han et al. 1989) enclosed within two ϕC31 attP recombination sequences, was amplified from plasmid pK104c (gift of K. Kyrou) using primers kk047 (5′-ACCGGTACCCCAATCGTTCA-3′) and kk048 (5′- ACGCGTTCAGGATTATATCT-3′). The homology arms and marker cassette were cloned by Gibson assembly into MluI- and MshTI-digested plasmid K103 (K. Kyrou), to generate the final donor plasmid (p19).
Anopheles funestus rearing—insectary conditions, microinjections, screening of transgenics
A large (>5000 adults) colony of A. funestus mosquitoes, FANG strain (Hunt et al. 2005) is routinely held in insectaries at the Liverpool Insect Testing Establishment (LITE), a dedicated GLP-approved facility, adjoined to our host institution, for the continuous production of mosquitoes and the testing of vector control products. The FANG strain originated from Angola and is fully susceptible to all insecticides (Hunt et al. 2005). For general maintenance, FANG colonies at LITE were provided a blood meal twice prior to oviposition, using a Hemotek Membrane Feeding System (Hemotek Ltd., Blackburn, UK), and with screened human blood procured from NHS bloodbanks. In brief, eggs were floated in purified water with 0.01% w/v yeast hatching solution and reared at larval densities of 0.24 larvae/mL. Larvae were fed increasing amounts of ground TetraMin® Tropical Flakes under the following regime: day 1 (day of hatching) ∼100 µg/larvae; then day 2–7 ∼200 µg/larvae; then day 8–10 ∼233 µg/larvae, with day 10 being the first day of pupation.
Prior to microinjection a cohort of mosquitoes (approximately 1500 adults) was maintained in dedicated insectary facilities adjacent to the microinjection facility, at a temperature of 26°C ± 2°C, a relative humidity of 70 ± 10%, and under an 11-hours light/dark cycle with 1-hour dawn/dusk transitions. Female mosquitoes were provided a blood meal from a human volunteer in accordance with LSTM’s code of practice for arm-feeding Anopheles mosquitoes. Gravid females were encouraged to oviposit 60–96 hours after blood feeding by introducing them into oviposition “chambers,” comprising of a 50 ml falcon tube with the conical base removed, covered at one end with dual-layered netting to create a secure way of entry, and plugged at the other end with a shallow, concave plastic cap. Mosquitoes were introduced inside the chambers in small groups (of approximately 10–15 individuals) and left to acclimatize in the dark for at least 30 minutes before initiating oviposition by adding 2 ml distilled H2O to the cap.
Embryos were injected using a modified version of the method of Benedict (2007) as described previously (Fuchs et al. 2013). Briefly, embryos were aligned against semi-moist nitrocellulose paper, wet with distilled water, and injected into the posterior pole at an oblique angle. The injection mix containing helper (100 ng/µl) and donor plasmids (300 ng/µl) was resuspended in 1x injection buffer (0.1 mM sodium phosphate buffer pH6.8; 10 mM KCl). Following injection, eggs were floated in 1 L distilled water and 0.01% w/v yeast solution, in plastic trays line with filter paper around the waterline to prevent embryos adhering to the plastic above the waterline and dessicating. Surviving G0 progeny were screened for transient expression of eCFP or RFP at the 1st larval instar and reared separately, accordingly as “transients” or “nontransients.” We found the injection of A. funestus embryos slightly more challenging in terms of needle entry and survival, compared to A. gambiae and Anopheles stephensi. Because it is certain that transient individuals were injected with a significant amount of injection mix, we prioritized our efforts to maximize the screening of the progeny of these individuals as opposed to the nontransient larvae that may include a significant amount of noninjected, or sub-optimally injected, individuals.
Early-stage larvae of the 1st and 2nd instars were fed 0.015% TetraMin® Baby fish food daily; 3rd and 4th instar larvae were provided with 0.02% ground TetraMin® Tropical Flakes. Once transgenic colonies were established, larval trays were maintained at a density of ≤100–150 larvae per tray of size 30 × 30 cm.
G0 mosquitoes exhibiting transient fluorescent expression as 1st instar larvae were reared in separate groups of males or females and mated to wild-type for ≥7 days before blood-feeding. Subsequent generations were screened for actin5c::eCFP expression as mid-stage larvae; white-eye phenotypes and sex were assessed at pupal stages. For G1, all whiteCFP offspring were female and thus mated to wild-type males for ≥7 days before blood-feeding. For G2, whiteCFP offspring were outcrossed to wild-type in separate groups of males or females. For G3, whiteCFP offspring from whiteCFP G2 males (all female) were outcrossed to wild-type, while whiteCFP offspring from whiteCFP G2 females (males and females) were intercrossed together. Finally at G4, homozygous (white-eye, whiteCFP females) were crossed to whiteCFP males in order to maintain transgene purity in following generations.
T7 endonuclease assay
To assess for nonHDR mutagenic activity, G0 mosquitoes, exhibiting transient fluorescent expression of the helper and donor plasmids, were first assessed for white-eye phenotypes under standard brightfield microscopy. The genomic DNA (gDNA) of individual transient G0 mosquitoes was extracted using the Wizard® Genomic DNA Purification Kit (Promega). In the T7 Endonuclease I (T7EI) assay, a 744 bp region spanning the cut-site was first amplified by PCR (primer sequence forward—GGCTGGTGTATGGTGAGTATG; reverse—GAAGAGCTACGGTTCGGTTAAG). One microliter of T7EI (NEB) was then added to 19 µl of purified (QIAquick PCR Purification Kit, QIAGEN) and hybridized PCR product containing approximately 200 ng of the PCR product, digested for 15 minutes at 37°C, and immediately visualized on a 1% agarose electrophoresis stained with Midori Green Advance (Nippon). T7 endonuclease-mediated cleavage of mismatched base pairs at the target site is expected to yield products of approximately 518 and 226 bp.
Molecular genotyping
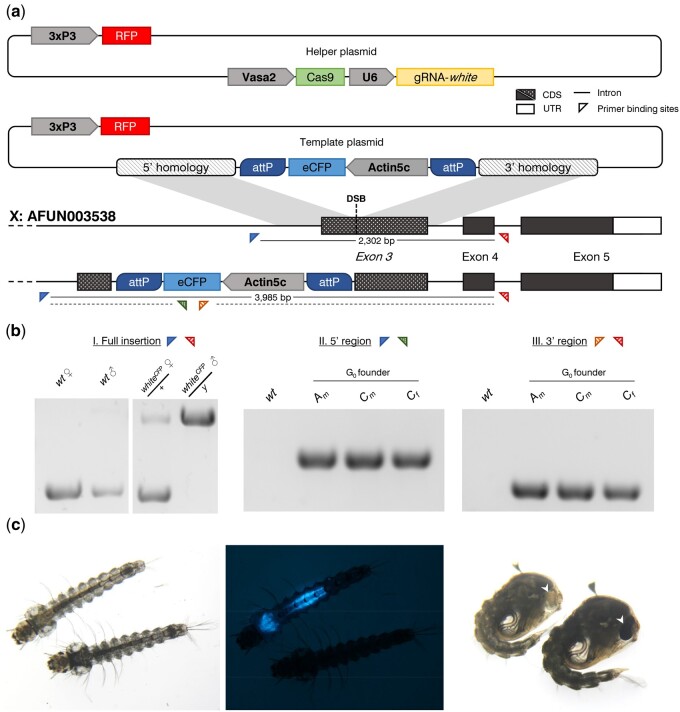
To molecularly characterize HDR knock-in events, the genomic DNA (gDNA) of G1 and G2 mosquitoes fluorescently expressing the actin5c::eCFP construct was extracted as individual samples using the Wizard® Genomic DNA Purification Kit (Promega). Regions of gDNA were then amplified according to the three primer pairs as indicated in Figure 1. Briefly, the primer pairs amplified three regions localized to the insertion site within the white gene: primer pair I (forward—GGTTAACGTATGCGGCAAACAC; reverse—GTGTCGCCACCATCTGTGGTAAG) flanked the full insertion region including the 5’ and 3’ homology arms, amplifying a product of 3985 bp in individuals containing the whiteCFP allele or 2302 bp in wild-type individuals; primer pair II [forward—GGTTAACGTATGCGGCAAACAC (as in primer pair I); reverse—CGACAACCACTACCTGAGC], which spanned the 5’ region of the insertion and amplified a product of 1625 bp in whiteCFP individuals, and primer pair III [forward—GCAGATGAACTTCAGGGTCAGC; reverse—GTGTCGCCACCATCTGTGGTAAG (as in primer pair II)] which spanned the 3’ region of the insertion and amplified a product of 1917 bp.
Figure 1.
Targeted disruption of the white gene (AFUN003538) by CRISPR-mediated HDR. (A) Schematic representation of the HDR knock-in process. DNA repair is mediated through the concurrent microinjection of two plasmid assemblies: a “helper” plasmid, designed to induce a double-stranded break (DSB) at the target locus upon expression, containing a source of Cas9 and gRNA under the control of the vasa2 and U6 promoters respectively; and a “template” plasmid containing the insert region (an actin5c::eCFP cassette enclosed within two reversible ϕC31 attP recombination sequences) flanked both 5’ and 3’ by regions of homology approximately 1 kbp upstream and downstream of the cut site. (B) Diagnostic PCRs of transgenic offspring (G2) deriving from different founders. PCR primer binding sites are represented by triangles. “External” primers flanking the full insertion and binding outside the regions of homology included in the donor construct (red and blue triangles, I) are used to discriminate between wild-type individuals and females heterozygous or males hemizygous for the whiteCFP allele. “Internal” primers complementary to the knocked-in eCFP sequence (green and orange triangles) are used with external primers to amplify the 5’ region upstream (II) or 3’ region downstream (III) of the eCFP cassette. Three separate transgenic lines were produced and assigned according to injection set (A–D) and sex of founder individual or group (m—males or f—females). wt—wild-type control. (C) Bright-field (left and right) or standard fluorescent (middle) microphotographs of representative individuals demonstrating either the wild-type or whiteCFP mutant phenotype. Under bright-field illumination, mutant larvae (left) and pupae (right) exhibit lighter global pigmentation compared to wild-type, and are white-eyed (arrows). Transgenic whiteCFP individuals additionally express cyan fluorescent protein from the actin5C promoter, in a pattern (lower midgut and gastric caecae) consistent with the activity of this promoter in other Anopheles species (middle).
Data availability
The data generated as part of this study are available within the Supplementary material available at G3 online.
Results
As a proof of principle, we determined to target the A. funestus X-linked gene white, which when mutated produces a visible phenotype in the eye (Li et al. 2017), with an HDR template designed to introduce a dominant fluorescent marker gene expressing cyan fluorescent protein (eCFP) (Figure 1A). We produced a “helper” plasmid containing the Cas9 gene under the control of the germline-specific promoter of the A. gambiae vasa gene (Papathanos et al. 2009). Given that this promoter sequence from A. gambiae manages to confer germline expression even in the distantly related mosquito Aedes aegypti (Akbari et al. 2014), we reasoned that it should work similarly in A. funestus. The helper plasmid also contained a construct producing a white-specific guide RNA under transcriptional control of the U6 promoter (Hammond et al. 2016).
Together with the helper plasmid we included an HDR “donor” construct (Figure 1A) that contained, as a dominant marker the gene encoding an enhanced version of the cyan fluorescent protein (CFP) under the transcriptional control of the actin5c promoter from Drosophila melanogaster which has previously been shown to work well in A. stephensi and A. gambiae (Catteruccia et al. 2000; Bernardini et al. 2014). Again, given this demonstration of conservation of the promoter activity across such a large evolutionary distance, coupled with the ubiquitous use of eCFP as a visual reporter in a range of species, we reasoned that these components would function as a dominant marker cassette in A. funestus.
We injected A. funestus eggs of the FANG strain (Hunt et al. 2005) following a standard microinjection protocol (Benedict 2007; Fuchs et al. 2012) containing only minor modifications. Over four separate sets of injections, we aligned approximately 1700 embryos for injection. Among the embryos surviving the procedure, we screened for those larvae showing visual expression of either the CFP or RFP marker genes transiently from either the helper or donor plasmids. Encouragingly, we were able to see either RFP or CFP expression in 44 of 223 surviving larvae (∼19%, Table 1), indicating that these marker genes, and their respective transcriptional units, are suitably expressed in A. funestus cells, at least from an episomal source of the injected plasmid. Since this fraction of larvae (which we term “transient” G0) are sure to have been injected successfully with a considerable quantity of the transformation plasmids, and given that A. funestus can be cumbersome to rear and cross generally, we focused the majority of our efforts on maximizing the screening of their progeny, rather than the “nontransients” that may represent noninjected, or sub-optimally injected, G0 individuals.
Table 1.
Outcome of independent injection experiments (sets A–D)
| Injection set | Embryos aligned | Larvae surviving | G0 Larvae with transient CFP or RFP expression | G0 transient individuals surviving to adulthood | G0 adults with eye color mosaicism | Crossing scheme | Egg batches produced | HDR events/total G1 (%) |
|---|---|---|---|---|---|---|---|---|
| A | 640 | 53 | 11 | 4 | 0 | Transient ♂(3) × wt outcross | 5 | 71/174 (42.5) |
| Transient ♀(1) wt outcross | 0 | 0/0 (0) | ||||||
| NT♂/♀ intercross | 1 | 0/130 (0) | ||||||
| B | 350 | 68 | 7 | 4 | 0 | Transient ♂(2) × wt outcross | 1 | 0/21 (0) |
| Transient ♀(2) × wt outcross | 0 | 0/0 (0) | ||||||
| NT♂/♀ intercross | 1 | 0/55 (0) | ||||||
| C | 420 | 40 | 12 | 7 | 0 | Transient ♂(5) × wt outcross | 4 | 90/564 (16.0) |
| Transient ♀(2) × wt outcross | 4 | 20/140 (14.3) | ||||||
| NT♂/♀ intercross | 1 | 0/61 (0) | ||||||
| D | 300 | 72 | 14 | 11 | 0 | Transient ♂ (5) × wt outcross | 0 | 0/0 (0) |
| Transient ♀(6) × wt outcross | 2 | 0/73 (0) | ||||||
| NT♂/♀ intercross | 1 | 0/75 (0) |
We crossed G0 individuals to wild type and screened for the presence of CFP-positive offspring—suggestive of a HDR-targeting event in the germline of the parent. Across the injection sets, from a total of 26 surviving adult G0 individuals, we recovered 3 separate founders that produced CFP-positive offspring (Figure 1C), representing a transformation rate of ∼11%, considering this class only (or ∼3% considering all G0, transient and nontransient). Among the offspring of these founders, the ratio of transgenic to nontransgenic offspring was relatively high, ranging from 14 to 43% (Table 1), suggesting that the transformation event happens early in germline development when there are relatively few germline stem cells. The fact that most individuals were CFP-positive only (and not RFP-positive also) is consistent with CRISPR-mediated HDR integration of the CFP cassette contained within the regions of homology upstream and downstream of the target site. To confirm this, we performed a diagnostic PCR using primers that bind the genomic target region externally to the regions of homology contained in the donor plasmid (Figure 1A). The presence of the complete CFP cassette at the target locus and the precise nature of the integration at the 5’ and 3’ end of the construct was confirmed (Figure 1B), and we named the allele thus generated as whiteCFP. Our expectation was that the whiteCFP allele would represent a null allele, given that indels at this target site result in a white-eye phenotype (Li et al. 2018). Indeed, since the white gene resides on the X chromosome, hemizygous whiteCFP males had white eyes whereas heterozygous females exhibited a wild-type eye color, confirming that the whiteCFP allele is both recessive and, likely, null. Among the G1 progeny we also recovered a small fraction of offspring (3.8% of all transgenic offspring) that were both RFP+ and CFP+, suggesting that in rare cases semi-legitimate recombination may occur, inserting at least a section of the donor plasmid external to the homology-flanked cassette that includes an RFP cassette in the plasmid backbone (Figure 1A). However, we did not investigate these rare events molecularly. The whiteCFP HDR allele showed normal Mendelian inheritance and, in homozygosity in females, resulted in a white eye phenotype (Supplementary Table S1).
One of our motivations for using a plasmid-based source of Cas9 under a germline-specific promoter was to target Cas9 activity to the germline where rates of HDR seem to be very high, in Anopheles species at least, and to have a system that could minimize somatic activity of the Cas9. Previously, it was shown that while injection of a mix of Cas9 protein and gRNA could generate indels at the target site in the germline, it also generated a mosaic of somatic mutations in these individuals, including the mutation of both alleles (bi-allelic mutations) in some cells, resulting in eye color mosaicism in these G0 individuals (Li et al. 2018). On the contrary, in the G0 individuals injected with the vasa-driven source of Cas9 we failed to observe any eye color mosaicism in males or females (Table 1). A T7 assay revealed that rates of end-joining mutations were very low in injected individuals, to the point where they were barely detectable (Supplementary Figure S1). Indeed, in one founder that produced HDR-mediated transgenic offspring we failed to observe any signal of end-joining activity in the T7 assay. Moreover, we also found no evidence of any germline transmission of Cas9-generated indels (i.e., end-joining repair events) from any of the founders—these indels would be expected to result in a nonCFP, white-eyed phenotype in hemizygous males, yet we failed to observe any such events among the siblings of HDR-mediated transgenic offspring. Taken together, these data support the hypothesis that this transformation protocol, using a germline-restricted source of Cas9, strongly favors the generation of HDR-mediated repair over end-joining repair.
Discussion
This is the first report of the generation of transgenic A. funestus. The availability of a CRISPR-based technology to introduce transgenes, or to introduce precise and defined changes, at specific genomic loci, has the potential transform our ability to perform functional genetics in insects, such as exploring the function of genes putatively controlling key phenotypes such as insecticide resistance (Itokawa et al. 2016; Samantsidis et al. 2020), investigating genes that are essential for important insect behaviors (Raji et al. 2019) or identifying genes that are important for key traits such as immunity and female reproductive capacity (Hammond et al. 2016).
For A. funestus, a pressing aspect to investigate is the nature of its resistance to pyrethroid insecticides, a mainstay of insecticide-treated bednets, where the resistance mechanism appears to be different to that observed in A. gambiae and may involve copy number variation, allelic variation of coding regions and/or the insertion of cis-regulatory sequences that lead to upregulation of expression (Ibrahim et al. 2015; Mugenzi et al. 2019; Weedall et al. 2019, 2020). A CRISPR-based HDR tool will be crucial in investigating these hypotheses.
The high frequency with which transgenics were recovered using our approach means that it should be possible, by judicious molecular genotyping of offspring, to recover precisely modified mosquitoes without the need for insertion of a dominant marker gene, that might have confounding effects on phenotype. Of course, the efficiency of targeting may vary according to gRNA: locus combination and additional future experiments will be important to confirm targeting rates across a range of loci. The relative rates of HDR vs end-joining repair following Cas9-induced DNA breaks can vary according to cell type and between germline and soma (Lin et al. 2014; Kandul et al. 2020; Hammond et al. 2021). In addition, there appears to be variation in the rates of CRISPR-based transgenesis between species with the rates for A. aegypti, for example, generally being significantly lower than those observed in A. gambiae (Kistler et al. 2015; Hammond et al. 2016). However, it is difficult to disentangle effects due to differing biology between the species and effects due to different transformation systems/setups. Indeed, the HDR transformation rates were improved several fold in A. aegypti by injecting a line containing a transgenic source of germline Cas9 (Li et al. 2017). Our approach, of injecting a plasmid-based source of Cas9 active in the germline coupled with a gRNA transcription unit has several attractive features: it obviates the need to inject a Cas9-expressing transgenic line; the plasmid-based DNA solution is much less labile than protein/RNA-based mixtures; rates of HDR are high; and the relative ratio of HDR: NHEJ events is very high, as evidenced by the almost complete lack of somatic mosaicism observed after injection. This latter aspect is particularly important if the target gene is essential and extensive mosaicism might reduce the recovery of desired events.
Finally, an important consideration for the feasibility of generating gene drives, which are designed to bias their inheritance among offspring and thereby spread linked traits into a population (Alphey et al. 2020), are the relative rates of HDR vs NHEJ. Homing-based gene drives (Burt 2003; Esvelt et al. 2014) have, to date, shown the most promise in terms of development and proof-of-principle demonstration in the laboratory, in a number of mosquito species (Gantz et al. 2015; Hammond et al. 2016; Li et al. 2020). This type of gene drive encodes a Cas9: gRNA unit that cleaves germline chromosomes at an endogenous target sequence and hijacks the HDR pathway to copy itself into the repaired site, thereby increasing its copy number. Thus, HDR is essential for this type of gene drive to work. On the flip side, NHEJ repair events following gene drive activity not only reduce the probability of the drive copying itself directly but the small indels can generate target sites that are resistant to future gene drive targeting (Champer et al. 2017; Hammond et al. 2017). Therefore, the high rates of HDR that we observed in A. funestus augur well for the prospect of developing gene drives for control of this considerable vector of malaria. This feature, coupled now with a capacity to perform functional genetics, should expand the range of innovative control tools available and augment those that already exist.
Supplementary Material
Acknowledgments
The authors are extremely grateful to Helen Williams and members of the Liverpool Insect Testing Establishment (LITE), in particular James Court and Amy Guy, and their expertise in rearing large populations of A. funestus colonies, which made this project a lot easier than it could have been. Matt Craske provided excellent technical assistance in the breeding of transgenic lines. They are also grateful to Leon Mugenzi, Andrew Hammond, Linta Grigoraki, and Kyros Kyrou for helpful discussions on the initial optimization of injection technique and DNA cloning protocols.
T.N. and C.W.: Conceptualization; C.Q., A.A., and T.N.: Data curation and formal analysis; T.M. and C.W.: Funding acquisition; T.N., A.A., and C.Q.: Investigation; A.A. and T.N.: DNA cloning; C.Q. and T.N.: Transgenesis; C.Q.: Molecular Genotyping; A.A. and T.N.: Methodology; T.N.: Project administration and supervision; C.Q. and T.N.: Visualization; C.Q. and T.N.: Writing—original draft; C.Q., A.A., C.W., and T.N.: Writing—review and editing.
Funding
This work was supported by pump-prime funding from ANTI-VeC (AV-PP20), a BBSRC/GCRF Network Grant.
Conflicts of interest
None declared.
Literature cited
- Akbari OS, Papathanos PA, Sandler JE, Kennedy K, Hay BA.. 2014. Identification of germline transcriptional regulatory elements in Aedes aegypti. Sci Rep. 4:3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen ML, O’Brochta DA, Atkinson PW, Levesque CS.. 2001. Stable, germ-line transformation of Culex quinquefasciatus (Diptera: Culicidae). J Med Entomol. 38:701–710. [DOI] [PubMed] [Google Scholar]
- Alphey LS, Crisanti A, Randazzo F, Akbari OS.. 2020. Opinion: standardizing the definition of gene drive. Proc Natl Acad Sci USA. 117:30864–30867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict MQ. 2007. Microinjection methods for Anopheles embryos. In: Methods in Anopheles Research. Malaria Research and Reference Reagent Centre MR4. (www.beiresources.org) Chapter 3.1.1
- Bernardini F, Galizi R, Menichelli M, Papathanos P-A, Dritsou V, et al. 2014. Site-specific genetic engineering of the Anopheles gambiae Y chromosome. Proc Natl Acad Sci USA. 111:7600–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A. 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 270:921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, et al. 2000. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature. 405:959–962. [DOI] [PubMed] [Google Scholar]
- Champer J, Reeves R, Oh SY, Liu C, Liu J, et al. 2017. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 13:e1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates CJ, Jasinskiene N, Miyashiro L, James AA.. 1998. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci USA. 95:3748–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohuet A, Simard F, Wondji CS, Antonio-Nkondjio C, Awono-Ambene P, et al. 2004. High Malaria transmission intensity due to Anopheles funestus(Diptera: Culicidae) in a Village of Savannah–forest transition area in Cameroon. J Med Entomol. 41:901–905. [DOI] [PubMed] [Google Scholar]
- Esvelt KM, Smidler AL, Catteruccia F, Church GM.. 2014. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 3:e03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Nolan T, Crisanti A.. 2013. Mosquito Transgenic Technologies to Reduce Plasmodium Transmission. Humana Press.923:601–22 [DOI] [PubMed]
- Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, et al. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA. 112:E6736–E6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman GL, Rafferty CS, Clayton JR, Stevens TK, Mukabayire O, et al. 2001. Germline transformation of the malaria vector, Anopheles gambiae, with the piggyBac transposable element. Insect Mol Biol. 10:597–604. [DOI] [PubMed] [Google Scholar]
- Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, et al. 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 34:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond AM, Kyrou K, Bruttini M, North A, Galizi R, et al. 2017. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 13:e1007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A, Karlsson X, Morianou I, Kyrou K, Beaghton A, et al. 2021. Regulating the expression of gene drives is key to increasing their invasive potential and the mitigation of resistance. PLoS Genet. 17:e1009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Levine MS, Manley JL.. 1989. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 56:573–583. [DOI] [PubMed] [Google Scholar]
- Hunt RH, Brooke BD, Pillay C, Koekemoer LL, Coetzee M.. 2005. Laboratory selection for and characteristics of pyrethroid resistance in the malaria vector Anopheles funestus. Med Vet Entomol. 19:271–275. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Yanfang Fu, Reyon D, Maeder Shengdar ML, Shengdar TQ, et al. 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 31:227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim SS, Jacob M, Riveron J, Bibby H, Irving C, Yunta, et al. 2015. Allelic variation of Cytochrome P450s drives resistance to Bednet insecticides in a major Malaria vector. PLoS Genet. 11:e1005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itokawa K, Komagata O, Kasai S, Ogawa K, Tomita T.. 2016. Testing the causality between CYP9M10 and pyrethroid resistance using the TALEN and CRISPR/Cas9 technologies. Sci Rep. 6:24652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinskiene N, Coates CJ, Benedict MQ, Cornel AJ, Rafferty CS, et al. 1998. Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. Proc Natl Acad Sci USA. 95:3743–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandul NP, Liu J, Buchman A, Gantz VM, Bier E, Akbari Omar S., et al. 2020. Assessment of a split homing based gene drive for efficient knockout of multiple genes. G3 (Bethesda). 10:827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler KE, Vosshall LB, Matthews BJ.. 2015. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 11:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Akbari OS, White BJ.. 2018. Highly efficient site-specific mutagenesis in Malaria mosquitoes using CRISPR. G3 (Bethesda). 8:653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Bui M, Yang T, Bowman CS, White BJ, et al. 2017. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc Natl Acad Sci USA. 114:E10540–E10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yang Nikolay T, Kandul P, Bui M, Gamez S, et al. 2020. Development of a confinable gene drive system in the human disease vector Aedes aegypti. eLife. 9:e51701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA.. 2014. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 3:e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JC, Irving H, Okedi LM, Steven A, Wondji CS.. 2010. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS One. 5:e11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugenzi Leon MJ, Benjamin D, Menze M, Tchouakui MJ, Wondji H, Irving, et al. 2019. Cis-regulatory CYP6P9b P450 variants associated with loss of insecticide-treated bed net efficacy against Anopheles funestus. Nat Commun. 10:4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngowo HS, Hape EE, Matthiopoulos J, Ferguson HM, Okumu FO.. 2021. Fitness characteristics of the malaria vector Anopheles funestus during an attempted laboratory colonization. Malar J. 20:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanos PA, Windbichler N, Menichelli M, Burt A, Crisanti A.. 2009. The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: a versatile tool for genetic control strategies. BMC Mol Biol. 10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purusothaman D-K, Shackleford L, Anderson MAE, Harvey-Samuel T, Alphey L.. 2021. CRISPR/Cas-9 Mediated Knock-In by Homology Dependent Repair in the West Nile Virus Vector Culex quinquefasciatus Say. bioRxiv 2021.01.14.426696 [DOI] [PMC free article] [PubMed]
- Raji JIN, Melo JS, Castillo S, Gonzalez V, Saldana MC, et al. 2019. Aedes aegypti mosquitoes detect acidic volatiles found in human odor using the IR8a pathway. Curr Biol. 29:1253–1262.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Huijben S, Tchapga W, Tchouakui M, Wondji MJ, et al. 2019. Escalation of Pyrethroid resistance in the Malaria vector Anopheles funestus induces a loss of efficacy of Piperonyl Butoxide–based Insecticide-treated Nets in Mozambique. J Infect Dis. 220:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samantsidis G-R, Panteleri R, Denecke S, Kounadi S, Christou I, et al. 2020. What I cannot create, I do not understand: functionally validated synergism of metabolic and target site insecticide resistance. Proc Biol Sci. 287:20200838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedall GD, Leon MJ, Mugenzi BD, Menze M, Tchouakui SS, Ibrahim, et al. 2019. A cytochrome P450 allele confers pyrethroid resistance on a major African malaria vector, reducing insecticide-treated bednet efficacy. Sci Transl Med. 11:eaat7386. [DOI] [PubMed] [Google Scholar]
- Weedall GD, Riveron JM, Hearn J, Irving H, Kamdem C, et al. 2020. An Africa-wide genomic evolution of insecticide resistance in the malaria vector Anopheles funestus involves selective sweeps, copy number variations, gene conversion and transposons. PLoS Genet. 16:e1008822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated as part of this study are available within the Supplementary material available at G3 online.