Abstract
Genetic and environmental factors play a major role in metabolic health. However, they do not act in isolation, as a change in an environmental factor such as diet may exert different effects based on an individual’s genotype. Here, we sought to understand how such gene–diet interactions influenced nutrient storage and utilization, a major determinant of metabolic disease. We subjected 178 inbred strains from the Drosophila genetic reference panel (DGRP) to diets varying in sugar, fat, and protein. We assessed starvation resistance, a holistic phenotype of nutrient storage and utilization that can be robustly measured. Diet influenced the starvation resistance of most strains, but the effect varied markedly between strains such that some displayed better survival on a high carbohydrate diet (HCD) compared to a high-fat diet while others had opposing responses, illustrating a considerable gene × diet interaction. This demonstrates that genetics plays a major role in diet responses. Furthermore, heritability analysis revealed that the greatest genetic variability arose from diets either high in sugar or high in protein. To uncover the genetic variants that contribute to the heterogeneity in starvation resistance, we mapped 566 diet-responsive SNPs in 293 genes, 174 of which have human orthologs. Using whole-body knockdown, we identified two genes that were required for glucose tolerance, storage, and utilization. Strikingly, flies in which the expression of one of these genes, CG4607 a putative homolog of a mammalian glucose transporter, was reduced at the whole-body level, displayed lethality on a HCD. This study provides evidence that there is a strong interplay between diet and genetics in governing survival in response to starvation, a surrogate measure of nutrient storage efficiency and obesity. It is likely that a similar principle applies to higher organisms thus supporting the case for nutrigenomics as an important health strategy.
Keywords: DGRP, diet–gene interactions, starvation, multivariate analysis, glucose transport
Introduction
Diet is a major risk factor for metabolic disease in humans, as indicated by the marked increase in metabolic disease over the past 50 years commensurate with increased consumption of high fructose and processed foods. However, despite being exposed to the same environment not all individuals acquire the disease, and this has given rise to the concept that different diets may affect the health of individuals in discrete ways.
Studies in Indigenous populations have provided some support for this concept. Here, specific populations that subsisted on different diets, whether rich in fats (Greenlandic Innuits) or carbohydrates (American Pima Indians), for thousands of years (Schulz and Chaudhari 2015; Andersen and Hansen 2018) have undergone increased incidence of metabolic diseases like diabetes commensurate with their exposure to westernized foods over the past 30–50 years. This has led to the identification of a handful of genetic variants. However, for the most part, these are not found in people of European ancestry. More recently, studies in mice and humans have demonstrated that individuals display heterogeneous metabolic responses to the same diets (Parks et al. 2015; Zeevi et al. 2015). While genome-wide association studies (GWAS) of body mass index, insulin resistance, and other metabolic traits have identified several causative genetic loci, these loci do not account for the majority of phenotypic variation (Phillips 2013). In fact, studies in mono- and di-zygotic twins revealed that a combination of genetic and environmental factors contributed to variance in body weight (Stunkard et al. 1986; Dubois et al. 2012). Overall, this emphasizes the need to consider not only genetic predisposition or diet alone, but also specifically gene × diet interactions (Heianza et al. 2017). Furthermore, one’s genotype can affect metabolic outcomes in response to a particular diet, and this “diet-responsiveness” can vary markedly between individuals.
The notion that individuals display discrete sensitivity to certain macronutrients, with dietary interventions being based on one’s genotype, is the key premise of the field of personalized nutrition, or nutrigenomics (Sales et al. 2014) and is supported by recent clinical trials (Qi 2014; Tan et al. 2018). However, we do not have a thorough understanding of how genes and diet interact to influence the onset of metabolic diseases (Drabsch and Holzapfel 2019). A major challenge with such studies in humans is that the environment, particularly diet and exercise, are difficult to quantify and control at a scale sufficient to facilitate genetic mapping. Hence, model systems such as mouse or Drosophila have become invaluable. A number of mouse or Drosophila genetic reference panels (DGRPs) have been established comprising a broad range of genetic diversity. These are ideal for studying gene × environment interactions as here it is feasible to carefully control environmental exposures. In this study, we have used the DGRP to compare the effects of different dietary exposures. This is an ideal system because >70% of known human disease genes have fly orthologs (Reiter et al. 2001), and the DGRP comprises an extensive collection of inbred and fully sequenced lines that are freely available, thereby allowing identification of causal genetic variants (Mackay et al. 2012).
We capitalized on the high-throughput nature of the DGRP to study the effect of gene × diet interactions on nutrient storage and utilization. A phenotype that encapsulates these outcomes is survival during starvation, which measures an organism’s ability to metabolize and store dietary nutrients during feeding and utilize them efficiently during fasting. For instance, adult flies fed a high carbohydrate diet (HCD) showed greater resistance to death from starvation, as well as higher triacylglyceride (TAG) content, suggesting a correlation between diets and starvation resistance via energy storage (Lee and Jang 2014; Krycer et al. 2020). However, it is not clear if this principle is broadly applicable across animals of different genetic backgrounds. Notably, previous studies have shown significant variation in starvation resistance between strains across the DGRP when fed one single diet (Mackay et al. 2012).
In this study, starvation resistance varied considerably between flies of distinct genetic backgrounds when fed diets composed of different macronutrient combinations (fat, sugar, and protein). Notably, while the high protein diet (HPD) reduced starvation resistance in most strains, highly variable responses were observed for the high carbohydrate or high-fat diets (HFDs) across the different fly strains. GWAS revealed SNPs that contribute to these gene × diet effects. One such gene CG4607 encoded a putative glucose transporter—knockdown of CG4607 caused lethality in flies fed high carbohydrate but not when fed high protein or HFDs. Our study uncovers a previously under-appreciated influence of diet on the heritability of starvation resistance, providing a rich resource of diet-specific genes for further study.
Materials and methods
Drosophila stocks and procedures
Stocks: RNAi knockdown fly lines were obtained from the VDRC: CGnone, v60000; CG4607 v5450 and v107219; Cip4, v108625. The following lines were obtained from the Bloomington stock center (Bloomington, IN, USA): DGRP; ubiquitous(ubi)-GAL4, BL32551; mef2-GAL4 BL27390; CG-Gal4, 7011 and the midgut NP3084-gal4, 113094 was obtained from the Kyoto Stock center (Kyoto, Japan).
Flies were maintained at 25°C with 12 hours light/dark cycles. DGRP flies were expanded in bottles before collecting adult males for experiments. Five replicates of ten 3–5 days old adult males from each strain were collected and passaged onto each diet. Food was changed every other day, and the mortality rate was monitored for the 10 days of diet treatment. As the HFD has a different consistency compared with the other diets, we assessed whether there was increased lethality during the pre-feeding with this diet, but we observed no lethality on HFD, ensuring that all diets were well tolerated. At the conclusion of the dietary exposure, males were placed into starvation vials with Kim wipes and 1 mL of water and monitored every 12 hours for death. Death from starvation was assessed in two ways: for screening the DGRP, we visually inspected flies every 12 hours; for the RNAi validation screen, we used the Drosophila activity monitoring system (DAMS) and defined activity (i.e., alive) as a beam crossing every 5 minutes. Survival time for each fly was defined as the time-point following the last recording for an activity for that fly. The mean starvation resistance was calculated as the average time of death for 50 flies per line on each diet. Heatmaps to assess qualitative differences between strains on the different diets were created using ggplot2, grid, and ggdendro packages in R. We used default Euclidean distances for hierarchical clustering as part of the hclust function in the ggdendro package.
Experimental diets
The experimental diets that were used throughout this study were made up of agar (Sigma) and torula yeast (H.J Langdon & Co, VIC, Australia), Sucrose (Table Sugar), and extra virgin coconut oil (Absolute Organics, NSW, Australia). The yeast, sucrose, and coconut oil correspond to protein, carbohydrate, and fat, respectively, and were combined as detailed in Supplementary Table S1 to generate a HPD, HCD, and HFD.
Heritability estimates using linear mixed models
We estimated the heritability of log survival, as well as the variance explained by dietary effects, using a series of linear mixed models (LMMs). LMM 1 contained strain as a random effect as well as diet as a fixed effect and was equivalent to:
where yij is the survival time of individual i from strain j, β0 is the survival of a typical strain on the reference diet (diet 0; here diet NF), θ0j is a random effect estimating the deviation of strain j from β0, which is normally distributed with mean 0 and variance σθ02 (i.e., the among-strain variance, which is estimated by the model), βk is a fixed effect giving the average response to diet k (k = 1 … ndiet—1; ndiet is the number of diets), xik is a dummy predictor for individual i on diet k and εi is the residual for individual i, which is normally distributed with mean 0 and variance σε2. In LMM 1 σε2 is the nonstrain variance after accounting the effects of diet. In this model, dietary effects are assumed to be uniform across strains.
LMM 2 was equivalent to:
where βk now gives the effect of diet k in a typical strain, θkj is the deviation in response to diet k by strain j (a “random slope”), which is normally distributed with mean 0 and variance σθk2, ρθ0θk is the correlation between strain-specific response to diet k and diet 0, and all other terms are as above. LMM 2 thus estimates variance between strains in the effect of diet k (σθk2).
We calculated heritability as the percentage of variance in log starvation response due to strain. Using LMM 1 this was calculated as σθ02/(σθ02 + σε2) × 100, which is the genetic variance after accounting for the effects of diet. For LMM 2, the variance among strains is diet-specific. The heritability of diet 0 was calculated as σθ02/(σθ02 + σε2) × 100 and that on diet k was calculated as calculated as (σθ02 + σθk2 + 2 × COVθ0θk)/(σθ02 + σθk2 + 2 × COVθ0θk + σε2) × 100. Note the covariance between responses to diet 0 and effect of diet k was found as ρθ0θk× σθ0× σθ1. Models were implemented using the “lme” function in the package nlme in R. Code is available in Figshare Supplementary Figures.
Statistical SNP analysis—overview
Prior to subjecting the starvation resistance data to SNP analysis, the data were processed. To adjust for the starvation resistance on normal food (NF), the starvation resistance time for each diet was divided by the line’s starvation resistance time on NF to calculate the fold change, which was subsequently log2-transformed and z-scored (per diet), a process termed “standardization” in this study. This standardized starvation resistance was fed into the analysis pipeline. DGRP SNP genotypes and gene annotations (FB5.57 annotation file) were downloaded from the DGRP Freeze 2 online resource http://dgrp2.gnets.ncsu.edu/data.html for all lines used in this study. SNPs were filtered to only contain SNPs with a minor allele count of at least 10, resulting in testing across 1,883,157 SNPs. Lines with missing allele information for a given variant were not considered. In other words, statistical tests for association of each SNP included only lines with allele information but discarded those with missing allele information. Statistical testing included multivariate analysis of variance (MANOVA) testing accounting for outliers, with Wolbachia status, large genomic inversions and the population structure relationship matrix (with first 10 principal components associated with the lines’ population structure relationship matrix, downloaded from DGRP, Huang et al. 2014) as covariates per variant, with an unadjusted P-value <1×10-3 selected for post-hoc analysis (see [a] below for details). The overall enrichment of the multivariate response was assessed by comparison to the null P-value distribution (Supplementary Figure S3). The post-hoc analysis was performed on selected SNPs for each diet separately using a Wilcoxon Rank Sum Test (see [b] below for details). Here, we included a minimum log2 fold change cut-off in starvation resistance comparing lines containing the “variant” allele with those containing the “reference” allele. We selected diet-responsive SNPs for further consideration if they were significant in the Wilcoxon Rank Sum test (P < 0.005) and had an absolute difference in mean phenotypes of at least 0.3 (log2 fold change).
[a] MANOVA model
For the diet survival outcomes Yij, we standardized by the control diet NF, (i.e., for a line i and diet j)
Y*ij= log10((Yij+a)/(YiD+a)), where YiD corresponds to the survival outcome for line i in the NF diet, and a is an offset value to avoid division by zero. Thus, deviation from zero in Y* corresponds to a relative change of survival outcome between each treatment diet and the control diet NF. To account for outliers, we reset extremely negative “relative diet” survival outcomes (i.e., with a value < −4), to the minimum value of −4.
For a given SNP, we fit the following multivariate ANOVA model:
where Y is the multivariate diet outcome (three diets, standardized to the control diet NF), x is the SNP allele status, w is wolbachia status, z1, z2,…, zi corresponds to status of the i inversion events, p1, p2,…, p10 correspond to the first 10 principal components associated with the lines’ population structure relationship matrix, and E corresponds to multivariate normally distributed noise. Only SNPs with a nonmissing allele value were considered for the MANOVA test, and inversion covariates were filtered to include at least 10 lines to ensure stability in model estimation.
To assess the MANOVA model, we generated a classic quantile–quantile plot (Q–Q plot) of the P-values (Supplementary Figure S3). The distribution of the Q–Q plot shows substantial deviation from the expected uniform distribution throughout the range of P-values.
[b] Wilcoxon Rank Sum test per diet
For the Wilcoxon rank-sum test, we first used linear regression to remove the effect of Wolbachia and inversion status from the data as well as accounting for the population structure (with first 10 principal components associated with the lines’ population structure relationship matrix), and then performed the Wilcoxon test on the residuals from that model. It is important to note that this resulted in a “corrected fold-change”, which in theory is technically different from the classical fold-change calculation. However, in practice, these two values are extremely similar (Pearson correlation coefficient = 0.98).
Validation/automated starvation resistance (DAMS assay)
We used the GAL4-UAS gene expression system to validate the two candidate diet-responsive genes. RNAi knockdown fly lines were mated with 20 ubi-Gal4 females. Sixteen 3–5-day old males were placed on four different diets for 10 days. The food was changed every other day for 10 days until males were placed into the DAMS apparatus (Trikinetics, inc., USA). The flies were loaded into DAMS tubes (one fly per tube) containing 2% agar and monitored every 5 minutes for death. Candidate genes were considered validated when the fold change in survival on each respective diet (relative to NF) corroborated with the SNP analysis (Table 2). The gene × diet interaction P-values were generated using the Cox proportional hazards regression model (survival package, R).
Table 2.
Validated candidate genes
| Gene/orthologa | SNP class | Manova P-value b | Dietc | vHCDd | vHPDe | vHFDf | G × D P-valueg | Functionh |
|---|---|---|---|---|---|---|---|---|
| CG4607/SLC2A6 | UPSTREAM | 7.97E-05 | HPD | DOA | −1.29 | −0.11 | 8.77E-09 | Glucose transmembrane transporter activity |
| Cip4/TRIP10 | SYNONYMOUS _CODING | 3.94E-04 | HFD | 0.42 | −0.51 | −0.67 | 0.0013 | Lipid binding |
a Gene symbol/human ortholog.
b The significance from the multivariate ANOVA of the diet responsive SNP.
c The diet upon which the gene is predicted to affect starvation resistance.
d–f The log fold change in starvation resistance (Log 2 (SRcontrol Diet/NF/SRKD Diet/NF)).
g The significance of the gene-by-diet interaction for each gene and it’s fold change on the predicted diet (from d).
h The functional annotation of the gene from www.flybase.org.
Table 1.
Analysis of the mean starvation resistance data on all diets
| Diet | Mean | SEM | Broad sense heritability (%) |
|---|---|---|---|
| NF | 51.46 | 1.15 | 61.6 |
| HCD | 68.88 | 1.64 | 62.7 |
| HFD | 55.10 | 1.09 | 59.1 |
| HPD | 36.77 | 0.97 | 68.9 |
A summary table of the mean starvation data, SEM, genetic and environmental variance from the log transformed starvation data across all the DGRP lines on each diet.
Estimates of genetic variance and heritability are derived from LMMs of log survival (see Supplementary Table S2).
Activity measures
To measure fly activity while eating NF or HCD, we placed 3–5-day old male flies into DAMs tubes with either NF or HCD and monitored activity for 10 days. Because CG4607 KD flies died within the first 24 hours after being placed on HCD, we analyzed the activity data from the beginning of the monitoring. DAMS data were analyzed using the R survminer and rethomics packages (Geissmann et al. 2019).
Capillary feeding assay
We used the CAFE assay as previously described (Ja et al. 2007) to determine the amount of the experimental diet eaten. Ten replicates of five 3–5-day old adult males were placed into vials with water soaked Kim wipes and sealed with a rubber stopper with two holes. Capillary tubes (5 µl) containing food were placed into the vials to allow flies to feed for 24 hours after which food intake was assessed by measuring loss of capillary volume. The diets were composed of yeast (MP Biomedicals cat # 2232731) and sucrose.
Gas trap assay to measure CO2 and triglycerides
We used a Gas Trap Assay to assess the capability of CG4607 KD flies to utilize glucose. The gas trap protocol has been previously described (Francis et al. 2019). Briefly, four replicates of 10 male adult 3–5-day old flies were starved overnight in vials with a Kim wipe and 1 mL of water. Flies were then placed into 12 well plates containing glucose radiolabeled food and blue dye. We measured glucose oxidation and processed the flies for measurement of TAGs and blue dye content as described.
Blue dye extraction and measurements
To determine the amount of food ingested during the gas-trap assay, we extracted and measured blue dye that was mixed with the radiolabeled food. In short-term experiments, the blue dye serves as a proxy for food intake (Krycer et al. 2019). Four replicates of 10 flies were collected and homogenized (Reche MM400) in 100 µl of water. Samples were briefly centrifuged, and the supernatant was dried in a Genevac evaporator. Dried samples were reconstituted in 50 µl of water, vortexed, and placed into a 96 well plate for absorbance measurement at 628 nm in a spectrophotometer. Dilutions of blue dye (Queenie Brand, Coles Supermarket, Australia) were used as a standard. Data were analyzed in Excel (Microsoft) and plotted in Prism (GraphPad). Statistical significance was calculated between genotypes using Student’s t-test.
Triglyceride assay
To measure TAG content, we used a triglyceride extraction method as previously described (FOLCH et al. 1957). Six to ten replicates of six 3–5-day old flies were collected and washed in four dilutions (1:2,1:4,1:10,1:20) of isopropanol to remove excess food. The aqueous phase was used for glucose assays. The lipids were collected after extraction, evaporated under N2 gas and reconstituted with 95% ethanol. Scintillant was added to samples with radioactive tracer instead of ethanol and analyzed on a beta counter (Beckman Coulter). For nonradioactive samples, samples were spun and placed into 96 well plates (Sigma-Aldrich, # CLS9018BC) and incubated with triglyceride reagent (200 µl; Thermo Fischer Cat #TR22421) at 37°C for 30 minutes. Precimat glycerol reagent (Thermo Fischer # NC0091901) was used as a standard. Total absorbance at 500 nm was measured in a plate reader (Beckman) and subtracted from a blank before determining the amount of triglyceride using the reference standard curve. All calculations were performed in Excel (Microsoft) and graphed in Prism.
Glycogen assay
To determine the amount of glycogen, we collected six replicates of six male flies and washed them in dilutions of isopropanol to remove food. Fasted flies were collected after 24 hours of starvation. Flies were homogenized in 1 M KOH for 30 seconds using steel balls and a tissue lyser (Resche MM400). Samples were heated for 30 minutes at 70°C. Saturated Na2SO4 was added prior to addition of 95% Ethanol for precipitation. The pellet was spun down and then reconstituted in water, heated at 70°C, before adding 95% Ethanol again. The pellet was centrifuged in the water/95% ethanol solution, the supernatant was removed and amyloglucosidase (Merck # A7420) was added overnight at 37°C. Samples were centrifuged and placed into 96-well plates (Sigma-Aldrich, # CLS9018BC) and incubated with glucose oxidase reagent (200 µl; Thermo Fischer, TR15221) at 37°C for 30 minutes. Glucose (1 mg/ml) was used as a standard. Total absorbance at 500 nm was measured in a plate reader (Beckman) and subtracted from a blank before determining the amount of glucose using the reference standard curve. All calculations were performed in Excel (Microsoft) and graphed in Prism.
Glucose assay
Glucose was measured from the aqueous phase of the triglyceride extraction (see above): the aqueous mixture was evaporated in a Genevac evaporator until a dried pellet was visible. The pellet was reconstituted with water and glucose was measured as described for glycogen.
Generation of CG4607-mRuby3 plasmid
CG4607-mRuby3 construct was created through Gibson cloning (Gibson et al. 2009). The CG4607 cDNA (clone RH58543 #11058, Drosophila Genome Resource Center, IN, USA) was PCR amplified using the following primers:
dCG4607GibF1: GGACTCAGATCTCGAGACAAGATGAAGGGCCAGCAGGAGGAG
dCG4607GibR1: CATGCTGCCttCAGCTGAGGACAATTTCTTTAGGAACACTT
The backbone was PCR amplified using GLUT4-mRuby3 plasmid to include overhangs using the following primers:
mRuby3GibF1: TCCTCAGCTGAAGGCAGCATG
mRuby3GibR1: AGCTGAGGATCCCTTGTCTCGAGATCTGAGTCC.
PCR products were combined with Gibson master mix, and the resulting plasmid was sequenced before cell transfection.
Cell culture and immunostaining
HeLa cells were maintained in DMEM with 1% glutamax and 10% FCS at 37°C and 5% CO2. Cells were transfected with lipofectamine 2000 and split onto coverslips at 2 × 105 cells/mL. Coverslips were fixed in 4% paraformaldehyde, washed with PBS, and blocked for 30 minutes with 0.02% saponin (Sigma) and 2% BSA in PBS. Primary antibodies: ms anti-LAMP1 (1:100, Developmental Studies Hybridoma Bank). Secondary antibodies: Gt anti-mouse 488 (1:200, Invitrogen). Coverslips were mounted in Mowiol and imaged using a 60x water objective on the A1R confocal (Nikon). Colocalization analysis was performed using the coloc2 plugin in Fiji (Schindelin et al. 2012, ImageJ, NIH, Bethesda, MD, USA).
qPCR of knockdown
Three replicates of 10 flies were homogenized in TRIzol™ Reagent (Invitrogen, 15596026). RNA isolation and qPCR were performed as previously described (Krycer et al. 2019). Tubulin was used as a housekeeping gene and the following primers were used:
CG4607: F: 5′-ACTCCCACGCGAAGGAGAA-3′ R: 5′-GCTGATTGAGAGTAACTGCCG-3′
Tubulin: F: 5′-TGTCGCGTGTGAAACACTTC-3′ R: 5′- AGCAGGCGTTTCCAATCTG-3′
The samples were run using the ROCHE Lightcycler 480 II (Roche). The knockdown efficiency was calculated using the delta-delta Ct method (Excel and GraphPad, Prism) and the Ct-values were graphed. Significance between the control and knockdown transcript was calculated using a Student’s t-test (P < 0.0001 ****).
Results
Starvation resistance of DGRP across 4 different diets
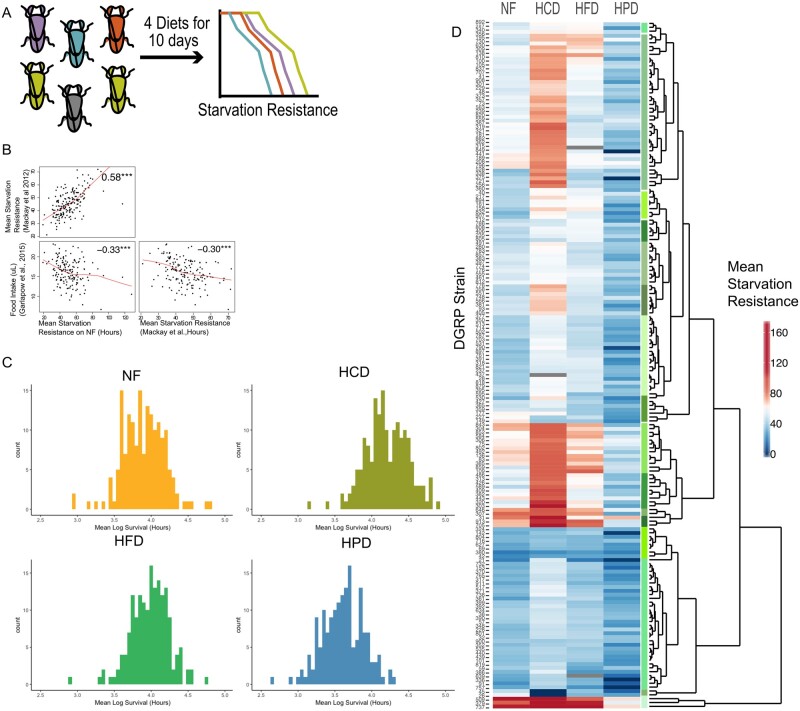
We first sought to identify novel diet-responsive genes that affect nutrient storage and utilization in Drosophila. We used survival during starvation as a surrogate for an obesogenic phenotype to screen for dietary effects. Starvation resistance is the ability of certain fly strains, often with abundant energy stores, to live longer under starvation conditions, compared to others. This is a powerful and sensitive assay as starvation resistant flies are often replete with fat stores immediately prior to starvation and feeding flies a high sugar diet (HCD) increases fat stores and prolongs starvation survival (Djawdan et al. 1998; Harshman et al. 1999; Bjedov et al. 2010). The diets (NF, HCD, HFD, and HPD, Supplementary Table S1) were selected based on previous studies that explored the effect of different sugar and protein concentrations on starvation resistance in a single strain (Skorupa et al. 2008; Lee and Jang 2014). The dietary composition is indicated in Supplementary Table S1: the carbohydrate is sucrose; the protein is yeast, and the fat is coconut oil. We exposed 3–5-day old adult male flies from 178 DGRP strains to the four diets for 10 days and then measured starvation resistance by removing food and assessing survival every 12 hours until all the flies were dead (Figure 1A, Supplementary Table S7). The DGRP has previously been used to examine starvation resistance in flies on a standard diet (Mackay et al. 2012) and there was a strong correlation in mean starvation resistance across the 178 strains between the two studies (males, fed NF, Pearson’s R = 0.58, Figure 1B), demonstrating the robustness of the starvation phenotype and the DGRP resource. Interestingly, we found that previously published food intake data in DGRP (Garlapow et al. 2015) was negatively correlated with starvation resistance across strains (males, fed NF, Pearson’s R = −0.32, Figure 1B) in both our study and previously published starvation resistance data (Mackay et al. 2012). This is intriguing as it suggests that strains that ate the most were the least resistant to starvation. This could be due to differences in metabolic rate, an increase in hunger cues, nutrient storage capacity, or differences in hormonal responses.
Figure 1.
Inter-strain variation in response to diet. (A) DGRP screen schematic outlines the workflow. Each individually colored fly represents an individual DGRP line and survival curves represent the measured starvation resistance (in hours) after exposure of flies to each diet. (B) A pairs plot showing correlations of the mean starvation resistance data fed the NF diet from this study with previously published starvation resistance data (Mackay et al. 2012) and food intake data (Garlapow et al. 2015). The red lines represent local linear regression (locally estimated scatterplot smoothing) of the correlations. (C) Histograms of the mean Log-transformed starvation resistance data for each strain on each diet. (D) Heatmap and Euclidean hierarchical clustering of the raw mean starvation resistance (in hours) of each DGRP strain on each diet.
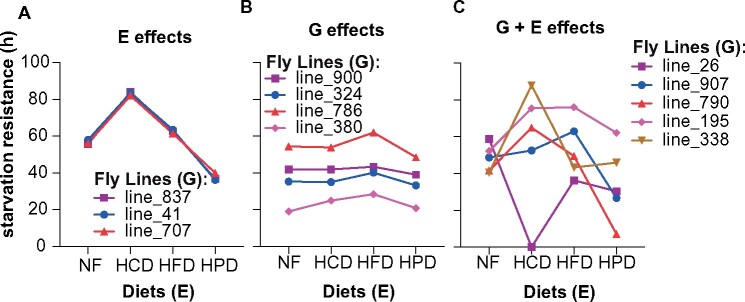
We observed variation in starvation resistance across all DGRP strains within each diet (Figure 1C). Unsupervised hierarchical clustering of the raw starvation resistance data revealed considerable variability in how strains responded to different diets with fly strains clustering into groups with distinct diet-specific starvation responses (Figure 1D). Starvation resistance on NF was not a predictor for starvation resistance on other diets. Some strains showed improved starvation resistance on both HCD and HFD, while others only displayed extended starvation resistance on HCD, and some did not show any difference between NF, HCD, or HFD. Notably, most strains showed reduced starvation resistance on HPD. Clustering was used to segregate flies into discrete starvation response groups. As illustrated in Figure 2, three different classes of response were observed: the “E” responders were those strains that responded only to the different diets in the same way (Figure 2A); the “G” responders were those that did not elicit any difference in their starvation response regardless of type of diet but showed differences between strains (Figure 2B); and the “G + E” responders were those that showed differences amongst strains and between diets as illustrated by the cross-over patterns (Figure 2C). Notably, the latter class was the most prevalent. To specifically identify diet-specific effects on starvation resistance, we next standardized each diet response to that observed in response to NF. This analysis clearly revealed clusters of strains that exhibited vastly different diet-responses (Figures 1D, 2, Supplementary Figure S1). While many strains exhibited high starvation responses on high fat and HCDs and low responses on HPD, there were many strains that showed quite different patterns. For example, some strains showed high responses to carbohydrate but low responses to HFD while others showed converse responses. Notably, these data included strains that did not tolerate a particular diet, where all flies died during the 10-day feeding period, prior to commencement of the starvation resistance analysis. These strains were assigned the lowest observed log2 fold change (−5.9) and include two strains that died on HCD (#26, #45) and HPD (#336, #849) (Supplementary Figure S1).
Figure 2.
Gene-by-diet interactions. Mean starvation resistance is shown for selected DGRP lines on the four diets. Selected DGRP lines to illustrate either only environmental (E) effects (A), only (or mainly) genetic (G) effects (B) or a gene by environment (G + E) interaction (C) on starvation resistance.
Gene × diet interactions and heritability
To accurately determine gene-by-diet interactions would require comparison of the effects of different diets in the same individuals. Obviously, this is not possible in this case. However, because we are using inbred lines we reasoned that it would be feasible to obtain gene-by-diet estimates using more qualitative approaches. We quantified the differential contributions of diet and gene-by-diet interactions to the variation in starvation resistance among DGRP strains. We determined the broad-sense heritability (H2) of the starvation response using two linear-mixed models (LMMs). LMM1, which assumed common effects of diets across all strains, and LMM2 (random-regression), which assumed strain-specific effects of diet. LMM 1 estimated the H2 of starvation resistance, after accounting for dietary effects at 48.9% (see Supplementary Table S2 for LMM coefficients). LMM2 had a significantly better fit than LMM1 (likelihood ratio test, L = 85543, d.f. = 15, P < 0.001) indicating the presence of gene by environment (G×E) interactions. Within-diet, the H2 of starvation resistance on NF, HCD, HFD, and HPD was 61.6, 62.7, 59.1, and 68.9%, respectively (Supplementary Table S2), thus illustrating the importance of genetics in governing phenotypic outcomes such as starvation resistance in response to diets of different macronutrient compositions.
Mapping “diet-responsive genes”
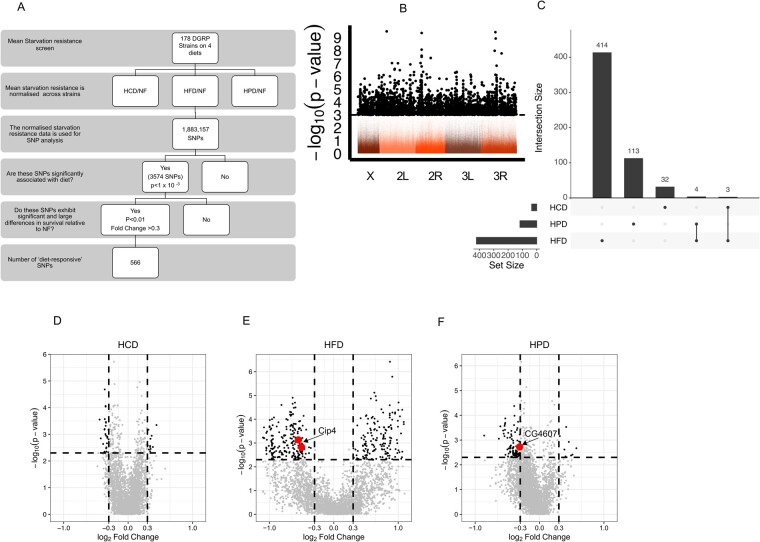
We aimed to identify gene–diet interactions by uncovering SNPs associated with diet-responsive starvation resistance using a multi-step analysis (Figure 3A). For this analysis, the starvation resistance data (log2 fold change) was z-scored for each diet. This resulted in relatively normally distributed data with the exception of several outliers, which comprised the strains that were intolerant to particular diets, including the strains (#26 and #45) that displayed lethality on HCD (Supplementary Figure S2). As we were interested in mapping the genetic contribution to starvation resistance in flies exposed to three separate diets compared to NF, we used a multivariate approach to identify SNPs rather than three separate univariate analyses. With more than one trait, multivariate approaches have proven to be more powerful in identifying biologically meaningful associations (Pitchers et al. 2019). We included 1,883,157 SNPs with a minor allele count of ≥10 in the multivariate ANOVA (MANOVA) (Garlapow et al. 2015). Large genomic inversions, Wolbachia status, and the population structure relationship matrix were included as covariates in the analysis. We observed no significant association of the starvation resistance responses to Wolbachia, inversion status, or population structure (Supplementary Table S5). A quantile–quantile (Q–Q) plot was generated to examine the distribution of the P-values, which is expected to have uniform distribution under the null assumption. This Q–Q plot showed an enrichment of significant SNPs compared to the expected P-value distribution (Supplementary Figure S3), notably persistent after accounting for possible confounders such as population structure, large genomic inversions, or Wolbachia infection status. Overall conventional SNP analysis resulted in an enrichment of SNPs associated with starvation resistance across all diets.
Figure 3.
Mapping the SNPs in diet-responsive genes (A) Schematic of SNP analysis used in this study. (B) Manhattan plot of the MANOVA SNP analysis shows all SNPs with 3574 significant SNPs (P-value of < 1 × 10−3, dashed line) in black. The x-axis represents the fly chromosomes and y-axis displays -Log (P-value) of the MANOVA. (C) An upset plot indicating the degree of overlap of the numbers of highly significant SNPs between each diet and the number of SNPs per diet. (D–F) Volcano plots of Wilcoxon Test of the 566 significant SNPs in (C) for HCD (D), HFD (E), and HPD (F). The y-axis shows the -Log (P-value) of the Wilcoxon Test and the x-axis displays the Log2 fold change in starvation resistance of the indicated diet compared to NF. Highly significant SNPs with a P-value <0.01 and a fold change >±0.3 are in black with validated genes labeled and indicated in red.
Using the MANOVA, accounting for outliers, we selected 3574 SNPs across all diets (P-value < 10−3) for further analysis. These SNPs were distributed across the entire fly genome (Figure 3B, black dots). This approach is a departure from traditional DGRP screens which tend to use a more stringent statistical threshold and these studies often fail to identify significant SNPs. Such sub-threshold approaches are valid provided they are accompanied by rigorous secondary screens. Similar approaches are now being used in human GWAS to identify biologically relevant sub-threshold loci (Wang et al. 2016; Nelson et al. 2017). In our case, we performed a post-hoc analysis on the 3574 SNPs using a Wilcoxon test. We next performed a post-hoc analysis on the 3574 SNPs using a Wilcoxon Test. This enabled us to identify diet-responsive SNPs that showed a significant difference between a particular diet and NF (P < 0.005). In addition, we applied a more stringent filter with a minimum log2 fold change of ±0.3 in starvation resistance. This yielded 566 variants that showed a significant change in starvation resistance on at least one diet (Supplementary Table S9). Of these, 7 were found to have significant variance in response to two diets (4 in HPD & HFD and 3 in HCD & HFD, Figure 3C). Notably, most of the 566 SNPs were associated with reduced starvation resistance in HFD and HPD, with fewer SNPs identified in HCD, as illustrated in volcano plots in Figure 3, D–F.
The 566 significant diet-responsive SNPs were located within 293 genes (Supplementary Table S9), 174 of which had human orthologs, with a high proportion of the SNPs (>60%) found in noncoding regions (Supplementary Tables S3 and S4), consistent with previous studies using DGRP (Mackay et al. 2012). These data provide an abundant and valuable resource of diet-responsive genes.
Candidate gene validation
Aside from the post-hoc Wilcoxon cut-off P-value of <0.005 and >±0.3 log2 fold change, we employed additional selection criteria to select genes for validation. These criteria included: the presence of a human ortholog with an ortholog score greater than five; an annotated gene function; and an annotated role in “transcription factor binding, enhancer or regulatory region”; excluding any gene with multiple SNPs with opposing phenotype directions (Supplementary Table S10). This resulted in 91 diet-responsive SNPs in 72 genes. In view of starvation resistance phenotype involving nutrient storage and/or metabolism, these 72 genes were screened for a potential role in nutrient sensing. Only two genes matched this criterion, one encoded for a glucose transporter (CG4607) and the other has been implicated in insulin-regulated glucose transport (Cip4), thus these two genes were selected for further validation. CG4607 is an ortholog of the glucose transporter GLUT8/GLUT6 and Cdc42-interacting protein 4 (Cip4) is involved in the trafficking of the insulin-responsive glucose transporter GLUT4 to the cell surface to facilitate glucose uptake in mice and is also implicated in phospholipid binding in Drosophila (Feng et al. 2010; Zobel et al. 2015). We used RNAi knockdown with the GAL4-UAS system (Brand and Perrimon 1993) to generate whole-body knockdown flies for these genes in the W1118 strain and assessed starvation resistance on all four diets using a more precise (low-throughput) method for monitoring starvation with the DAMS (Pfeiffenberger et al. 2010). Based on the Wilcoxon test, the SNPs in CG4607 (upstream SNP) and Cip4 (synonymous-coding SNPs) were associated with reduced starvation resistance on HPD or HFD, respectively. Notably, whole body knockdown of both genes confirmed the reduction in starvation resistance on the respective diet compared to NF (Figure 4 and Supplementary Figure S4, Table 2). We used the significance test from a cox hazard multivariate regression analysis, which investigates the effect of multiple variables on the time it takes for a specific event to happen, to determine if the relationship between diet and starvation survival was significant (Table 2). The results using RNAi knockdown suggest that the SNPs identified here likely result in a change in expression of the associated gene.
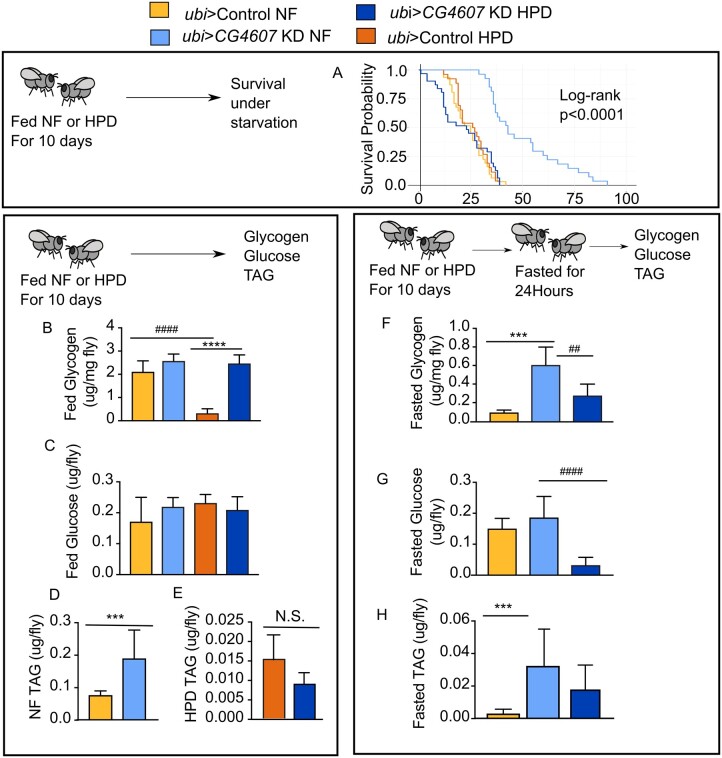
Figure 4.
CG4607 interacts with HPD to regulate starvation resistance. Assessment of energy storage in control and ubi-GAL4>CG4607KD male flies on NF or HPD. (A) Starvation survival curves of control and ubi-GAL4>CG4607KD male flies fed NF or HPD for 10 days followed by food removal and monitoring until flies died (data from two independent experiments, n = 32 flies/genotype). (B–E) Fed nutrient levels were measured in control and ubi-GAL4>CG4607KD flies after feeding NF or HPD for 10 days, including glycogen (B), glucose (C), triglycerides (TAG) (D, E). (F–H) Fasted nutrient levels were measured in control and ubi-GAL4>CG4607KD flies fed NF or HPD for 10 days and then starved for 24 hours including glycogen (F), glucose (G), TAGs (H). (B–H): n = 36 flies/genotype and representative of two experiments. Significance between genotypes (*) and diets (#) was calculated using a one-way ANOVA, **/## P < 0.01, ***/### P < 0.001 (Prism).
Analysis of CG4607, an example of a diet responsive gene
In view of the striking starvation resistance phenotype observed in flies bearing the CG4607 variant allele in response to HPD and because CG4607 encodes a putative glucose transporter, which is likely involved in diet responses, we decided to focus on this gene. CG4607 is homologous to the human glucose transporters SLC2A6 (GLUT6) and SLC2A8 (GLUT8). GLUT6 (formerly GLUT9) is overexpressed in endometrial cancer and is highly expressed in the brain, spleen, and leukocytes in mice (Doege et al. 2000). GLUT8 is highly expressed in the testis, heart, brain, liver, fat, and kidneys in mouse and has been shown to respond to insulin and transports trehalose in hepatocytes (Carayannopoulos et al. 2000; Mueckler and Thorens 2013; Mayer et al. 2016). In flies, CG4607 is highly expressed in the adult midgut and is modestly expressed in other tissues such as the heart, fat body, salivary gland, and fly heads (Robinson et al. 2013). Two RNAi hairpins were available to knockdown CG4607, CG4607 KK104152, and CG4607 GD3268. Whole-body expression of the latter was lethal while whole-body expression of the former was viable presumably because it led to an incomplete, 79% reduction in CG4607 expression (Supplementary Figure S5).
We next investigated the mechanism by which CG4607 controlled nutrient storage or utilization in response to HPD compared to NF. Starvation resistance was not significantly different between NF or HPD pre-fed control flies (Figure 4A). However, NF pre-fed flies expressing UAS-CG4607 KK104152 RNAi in the whole body (CG4607 KD) were starvation resistant but showed starvation sensitivity on HPD, suggesting they were low carbohydrate stressed (Figure 4A, Table 2). We posited that the starvation phenotype was reflected in the levels and handling of energy stores. To determine whether the change in starvation resistance arose from differences in nutrient storage during feeding or utilization during starvation, we measured energy stores during feeding and after 24 hours of fasting. Fed nutrient levels were similar between CG4607KD and control flies on both NF and HPD (Figure 4, B–E), with the exception of TAGs on NF and glycogen on HPD, both of which were higher in CG4607KD animals (Figure 4, B and D). However, major differences in nutrient storage levels between control and CG4607KD flies were observed upon starvation on both diets. Fasted glycogen and TAG levels were significantly higher in CG4607KD flies compared to control flies when pre-fed NF (Figure 4, F and H). While fasting of HPD-fed flies for 24 hours resulted in a marked depletion of nutrients in both genotypes, the CG4607KD flies showed detectable levels of energy storage, in contrast to control flies, where none of the nutrients were detected (Figure 4, F–H).
Knockdown of CG4607 results in HCD-intolerance
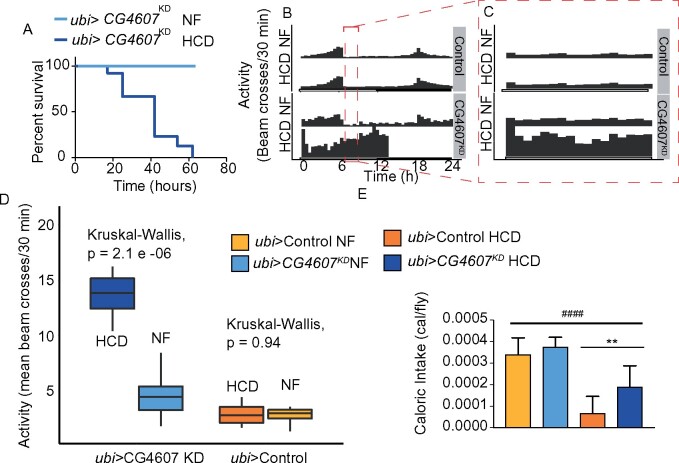
As part of the validation process, CG4607KD and control flies were subjected to the other diets. Remarkably, CG4607KD flies died after only 3 days exposure to the HCD (Figure 5A), while surviving on the other diets. It is worth emphasizing that these flies died prior to any food removal, indicating that they were intolerant to HCD. Notably, one of the two DGRP strains (#26) that died only while feeding on the HCD but not on any other diets, contained the CG4607 variant allele (Supplementary Table S9), suggesting that the HCD-intolerance of strain 26 may in part be due to the presence of the variant in CG4607. However, other strains containing the CG4607 variant allele survived on HCD suggesting that additional genetic modifiers are involved in the HCD intolerance. To further assess this HCD-intolerance phenotype, we sought to monitor the behavior of single-housed control and CG4607KD flies while they were eating NF or HCD. We found a striking hyperactivity in CG4607KD flies on HCD that abruptly stopped after 12 hours when flies died, as indicated by a total lack of activity (Figure 5, B and C). For the first 6 hours, control flies fed NF displayed a mean activity of 2.85 (n = 16) beam crosses, and this did not change upon feeding HCD (mean = 2.85, n = 14) (Figure 5D). Knockdown of CG4607 increased activity, with a mean of 4.22 (n = 14) for CG4607 KD flies fed NF, and this was exacerbated with the HCD diet (mean = 14.24, n = 13) (Figure 5D). This hyperactivity was reminiscent of flies that experience starvation (Keene et al. 2010) and we speculate that the lethality of the CG4607KD flies was due to their inability to consume and/or metabolize HCD, thereby experiencing starvation. However, upon measuring caloric intake in control and CG4607KD flies we observed that although both ate less calories when fed HCD compared to NF, the CG4607KD flies ate 38% more calories when fed HCD compared to controls on HCD (Figure 5E). This indicates a genotype × diet interaction, albeit not significant by two-way ANOVA (Supplementary Table S6). Thus, the observed hyperactivity of CG4607KD flies on HCD may be due to a perception of hunger brought about by the loss of glucose sensitivity, or a defect in the ability to utilize stored nutrients that ultimately lead to lethality.
Figure 5.
Whole-body knockdown of CG4607 is lethal upon HCD feeding. Survival of ubi-GAL4>CG4607KD flies on NF and HCD. (A) Adult male ubi-GAL4>CG4607KD flies (n = 60) were placed on either NF or HCD and manually monitored for lethality every 24 hours. (B) Actogram of activity (n = 16 flies) wrapped over 24 hours of single housed individual control and ubi-GAL4>CG4607KD flies in DAMS monitors and beam crosses were monitored every 5 minutes while feeding on NF or HCD (C) The same actogram as in (B), but the activity between 6 and 9 hours is shown. (D) A box plot of the mean activity (beam crosses from 0 to 6 hours) of control and ubi-GAL4>CG4607KD flies fed NF or HCD. Kruskal-Wallis test was used to calculate significance. (E) Caloric intake of adult male flies (n = 50/genotype) using the CAFE assay. Data are mean ± SD and significance between genotypes (*) and diets (#) were calculated using a t-test, **P < 0.01, #### P < 0.0001 (Prism).
We next attempted to determine which tissue is responsible for these CG4607 knockdown phenotypes. While depletion of CG4607 with both midgut and fat body drivers was viable, they did not reproduce the phenotype observed with whole-body knockdown of CG4607 as described above. Intriguingly, the expression of two hairpins using the muscle-specific driver mef2-GAL4 was lethal (data not shown). Hence, while we were unable to pursue the starvation sensitivity of these flies, these data raise the possibility that the major site of CG4607 starvation sensitivity is the heart, where both mef2 and CG4607 are expressed.
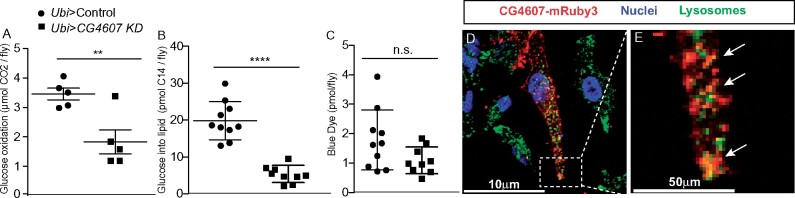
To further probe the role that CG4607 plays in mobilizing energy stores during starvation, we wanted to explore if CG4607 functions like a glucose transporter. To address this, we measured glucose utilization by feeding flies a NF diet containing 14C-radiolabelled glucose and monitoring glucose incorporation into CO2 and lipids (Francis et al. 2019; Krycer et al. 2019). We observed that CG4607KD flies exhibited lower levels of glucose oxidation and incorporation into lipids compared to controls (Figure 6, A and B), indicating reduced glucose utilization. This was not due to lower food intake in the CG4607KD flies (Figure 6C). Given the effect of CG4607 knockdown on glucose utilization, we wanted to determine its subcellular localization and posited that its localization would be similar to its mammalian homologs. The mammalian GLUT6 and GLUT8 both localize to lysosomes (Lisinski et al. 2001; Diril et al. 2009; Maedera et al. 2019). To determine if CG4607 is also localized to lysosomes, we expressed mRUBY3-tagged CG4607 in HeLa cells, a commonly-used mammalian cell-line that is adherent and easily genetically manipulatable. We used immunofluorescence microscopy to observe that, like GLUT6 and GLUT8, CG4607 partially co-localized (Avg. Pearson’s r = 0.40, n = 15 cells) with lysosomal markers (Figure 6, D and E). Taken together, our data show that CG4607, a GLUT6/8 ortholog, is an example of a diet-responsive gene and that responds to dietary sugar possibly by regulating lysosomal glucose metabolism.
Figure 6.
CG4607 controls glucose utilization and uptake. (A) Glucose oxidation was measured after 4 hours of administering 14C labeled glucose. (B) 14C incorporation into TAGs was measured in control and ubi-GAL4>CG4607KD flies. (C) The accumulation of blue dye, which indicates food intake was measured in control and ubi-GAL4>CG4607KD flies during the assay. (A–C) The data are from two independent experiments with n = 40 flies per genotype. Significance between genotypes was calculated using a Student’s t-test, **P < 0.01, ****P < 0.0001 (Prism). (D, E) Immunofluorescence images of fixed HeLa cells transfected with pCMV-CG4607-mRuby3 (red), immunostained with the lysosome marker anti-LAMP1 antibody (green) and counterstained with the nuclear dye, Hoechst (blue). (D inset [E]).
Discussion
The DGRP is a powerful tool for understanding the genetics driving variation of metabolic phenotypes (Mackay et al. 2012; Garlapow et al. 2015; Unckless et al. 2015; Nelson et al. 2016; Jin et al. 2020). Our approach measured starvation resistance in the adult male population after exposure to diets that vary in sugar, protein, and fat content. Using this method, the DGRP supported the notion that nutrition responses vary considerably between individuals of different genetic backgrounds. Furthermore, gender has been shown to have a significant impact on diet phenotypes (Chandegra et al. 2017) and thus should be explored in future diet–gene interaction studies. Although diet had a major influence on starvation resistance, the diets that enhanced or reduced starvation resistance differed markedly between Drosophila strains. In particular, exposure to a diet high in protein (HPD) elicited a greater genetic contribution to phenotypic variation. These data highlight a heterogeneity in the response to diet that underlies the fundamental principles of personalized nutrition. The SNPs we identified provide a resource dataset for further study and we validated 2 genes as bona fide diet responsive genes. Knock down of CG4607, was linked to increased caloric intake and nutrient storage combined with reduced glucose utilization.
In relation to our study, since the natural food sources of Drosophila are rotting fruits that contain sugar and yeast (Markow 2015), we postulate that genetic variants involved in the processing, storage, and utilization of sugar or protein (from yeast) contribute to differences in starvation resistance. Indeed, changes in sugar and lipid handling enzymes have previously been linked to starvation resistance (Harshman et al. 1999). Alternatively, sugar and protein (from yeast) are dominant drivers of food intake with major sensory systems to control intake of these macronutrients, and hence these are more likely to be under genetic control (Chng et al. 2017; May et al. 2019). Overall, this demonstrates that the mechanisms that underpin diet sensitivity are complex and involve differences in food preference and thus food intake as well as differences in nutrient metabolism (energy storage and utilization), all of which are regulated at least in part by one’s genotype.
This study provides a rich resource of diet-responsive genes and pathways. The majority of the SNPs in candidate genes were in noncoding regions (>60%), a finding that is consistent with previous DGRP (Mackay et al. 2012) and human GWAS (Gallagher and Chen-Plotkin 2018), and the functional consequences of such SNPs remain to be identified. As a proof of principle, we were able to validate candidate genes with synonymous coding SNPs using RNAi knockdown indicating that these types of mutations may control expression of these genes. However, further studies examining transcriptional control are required to validate this conclusion.
We focused on CG4607, a validated diet-responsive gene. The SNP in CG4607 was noncoding and 506 bp upstream of the start site, suggesting that it could affect transcription, splicing, or post-transcriptional processing. Additionally, the SNP was within a regulatory transcription factor binding site for the invective transcriptional repressor. While it may be of interest to determine if the SNP alters the expression of CG4607, our validation using RNAi knock down clearly showed that CG4607 is a diet-responsive gene. SNPs in CG4607 were identified in a GWAS of sleep/activity (Harbison et al. 2013). Consistent with this, CG4607 KD flies showed significantly elevated activity when fed a HCD. Although CG4607 is highly expressed in the midgut, CG4607 knockdown in this tissue did not reproduce the starvation resistance phenotype on HPD, nor lethality on HCD. We found a similar result when CG4607 was selectively knocked down in the fat body. These results suggest that the function of CG4607 in the midgut or fat body alone are not required to regulate starvation survival in response to these diets and it may be that CG4607 mediates its metabolic effects through multiple tissues. Notably, we observed lethality when using the muscle-specific promoter mef2 to knock down CG4607, and so we cannot exclude the possibility that the muscle or cardiac tissue is the site of CG4607 starvation resistance sensitivity. Further studies using alternate muscle-specific promoters are required to resolve this.
Our study provides several lines of evidence that CG4607 mediates its effects via regulating glucose metabolism. First, whole body depletion of CG4607 resulted in lethality after only 3 days on a HCD (Figure 5A). The HCD lethality was accompanied by hyperactivity and an increase in caloric intake on HCD compared to control flies. This is symptomatic of a starvation phenotype (Yang et al. 2015; Yu et al. 2016). Second, CG4607 KD animals have higher energy stores on NF compared to control flies. Third, our metabolic labeling experiments show that CG4607 KD animals exhibit reduced glucose utilization. Finally, the closest mammalian orthologs of CG4607 are the human glucose transporters GLUT6 and GLUT8. Recently, GLUT8 has been shown to transport trehalose in mammalian cells (Mayer et al. 2016) suggesting that CG4607 may play a similar role in flies given that trehalose is the major circulating sugar in flies. Furthermore, GLUT8 was shown to regulate AMPK phosphorylation and signaling via trehalose transport (Mayer et al. 2016; Narita et al. 2019) and AMPK deficient flies are starvation sensitive (Johnson et al. 2010). It would be of interest to determine if CG4607 regulates AMPK in a similar manner as its mammalian ortholog. Given that AMPK is an energy sensor perhaps energy-sensing pathways play a part in diet-dependent differences in starvation resistance.
CG4607 is targeted to lysosomes instead of the plasma membrane, unlike other facilitative sugar transporters (Lisinski et al. 2001; Maedera et al. 2019). This is intriguing as mTORC1 (Lee et al. 2009; Efeyan and Sabatini 2013), which regulates numerous metabolic processes including glycogen breakdown in autophagic vesicles also localizes to lysosomes (Mony et al. 2016; Zhao et al. 2018). Given that compared to control flies, CG4607 KD flies exhibited higher glycogen levels after starvation, and broke down more TAGs during starvation, the metabolic phenotype of CG4607 KD flies resembles a lysosomal glycogen storage disease, where lysosomal glycogen cannot be accessed for energy utilization in the cytosol. Together, this provides strong evidence that CG4607 mediates its effects on starvation resistance by regulating glucose metabolism. Hence this highlights the importance of glucose metabolism as a potential diet responsive pathway.
Overall, our findings demonstrate that gene–diet interactions are an important factor to consider in metabolic homeostasis. This has substantial ramifications for human health because it means that the concept of a “healthy” diet varies between individuals, thus questioning population-wide nutritional recommendations. While our study provides the basis for a nutrigenomics initiative such an endeavor is likely to require a substantial future investment at the clinical level.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials. Supplementary Figure S1 contains a heatmap of NF-standardised mean starvation survival data. Supplementary Figure S2 contains a histogram of NF-standardized data. Supplementary Figure S3 contains a Q–Q plot of the MANOVA P-values. Supplementary Figure S4 contains the starvation survival plot for control and knockdown of Cip4. Supplementary Figure S5 contains plot validating CG4607 RNAi knockdown with qPCR. Supplementary Table S1 contains the composition of diets used in the study. Supplementary Table S2 contains the Linear-Mixed Model Estimates, and Derived Heritability. Supplementary Table S3 contains the numbers of SNPS associated with decreased starvation resistance on 1 diet and Supplementary Table S4 contains the numbers of SNPs associated with increased starvation resistance on 1 diet. Supplementary Table S5 contains the ANOVAs for covariates for HCD, HFD, and HPD. Supplementary Table S6 contains ANOVA tests to determine the significance of gene–diet interaction on caloric intake of NF or HCD. Supplementary Table S7 contains the raw mean starvation survival data for each DGRP line on each diet. Supplementary Table S8 is a dataset containing 3574 SNPs above the P < 1 × 10−3 MANOVA threshold. Supplementary Table S9 is a dataset containing 566 SNPs filtered from the 3574 (Supplementary Table S8) by Wilcoxon P-value <0.005 and a dietary fold change greater than ±0.3. Supplementary Table S10 is a dataset containing the 91 SNPs (out of the 566 from Supplementary Table S9) that meet criteria outlined in the “candidate gene validation” section of the Results. Supplementary File S1 is the code used to generate the heritability data, which includes two accompanying files: Francis_Long_Data.csv and ln_data_long.csv. These supplementary files, Figures, and datasets are found in Figshare DOI: https://doi.org/10.6084/m9.figshare.9740801.
Acknowledgement
This work was supported in part by National Health and Medical Research Council (NHMRC) Project Grant GNT1086851 (to D.E.J.) and Diabetes Australia Research Program Project Grant (to D.F., J.R.K and D.E.J.). D.E.J. is an Australian Research Council (ARC) Laureate Fellow. S.G. is supported by the Royal Society Newton International Fellowship (NIF\R1\181950) and funding from the Judith and Coffey Life Laboratory at the Charles Perkins Centre. J.R.K. is supported by the Australian Diabetes Society Skip Martin Early-Career Fellowship. A.S is supported by the ARC Discovery Early Career Research Award (DECRA\DE180101520). The authors acknowledge the facilities, and the scientific and technical assistance, of the Australian Microscopy and Microanalysis Research Facility and the Fly Facility at the Charles Perkins Centre, University of Sydney.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of NHMRC or ARC.
Literature cited
- Andersen MK, Hansen T.. 2018. Genetics of metabolic traits in Greenlanders: lessons from an isolated population. J Intern Med. 284:464–477. [DOI] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, et al. 2010. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N.. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Dev Camb Engl. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Carayannopoulos MO, Chi MM-Y, Cui Y, Pingsterhaus JM, McKnight RA, et al. 2000. GLUT8 is a glucose transporter responsible for insulin-stimulated glucose uptake in the blastocyst. Proc Natl Acad Sci USA. 97:7313–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandegra B, Tang JLY, Chi H, Alic N.. 2017. Sexually dimorphic effects of dietary sugar on lifespan, feeding and starvation resistance in Drosophila. Aging (Albany NY). 9:2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng WA, Hietakangas V, Lemaitre B.. 2017. Physiological adaptations to sugar intake: new paradigms from Drosophila melanogaster. Trends Endocrinol Metab. 28:131–142. [DOI] [PubMed] [Google Scholar]
- ΡDiril MK, Schmidt S, Krauss M, Gawlik V, Joost H-G, et al. 2009. Lysosomal localization of GLUT8 in the testis – the EXXXLL motif of GLUT8 is sufficient for its intracellular sorting via AP1‐ and AP2‐mediated interaction. FEBS J. 276:3729–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djawdan M, Chippindale AK, Rose MR, Bradley TJ.. 1998. Metabolic reserves and evolved stress resistance in Drosophila melanogaster. Physiol Zool. 71:584–594. [DOI] [PubMed] [Google Scholar]
- Doege H, Bocianski A, Joost H-G, Schürmann A.. 2000. Activity and genomic organization of human glucose transporter 9 (GLUT9), a novel member of the family of sugar-transport facilitators predominantly expressed in brain and leucocytes. Biochem J. 350:771–776. [PMC free article] [PubMed] [Google Scholar]
- Drabsch T, Holzapfel C.. 2019. A scientific perspective of personalised gene-based dietary recommendations for weight management. Nutrients. 11:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois L, Kyvik KO, Girard M, Tatone-Tokuda F, Perusse D, et al. 2012. Genetic and environmental contributions to weight, height, and BMI from birth to 19 years of age: an international study of over 12,000 twin pairs. PLoS One. 7:e30153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Sabatini DM.. 2013. Nutrients and growth factors in mTORC1 activation. Biochem Soc T. 41:902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Hartig SM, Bechill JE, Blanchard EG, Caudell E, et al. 2010. The Cdc42-interacting Protein-4 (CIP4) Gene knock-out mouse reveals delayed and decreased endocytosis. J Biol Chem. 285:4348–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Stanley GHS.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 226:497–509. [PubMed] [Google Scholar]
- Francis D, Krycer JR, Cooney GJ, James DE.. 2019. A modified gas-trapping method for high-throughput metabolic experiments in Drosophila melanogaster. Biotechniques. 67:123–125. [DOI] [PubMed] [Google Scholar]
- Gallagher MD, Chen-Plotkin AS.. 2018. The post-GWAS era: from association to function. Am J Hum Genet. 102:717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlapow ME, Huang W, Yarboro MT, Peterson KR, Mackay TFC.. 2015. Quantitative genetics of food intake in Drosophila melanogaster. PLoS One. 10:e0138129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann Q, Rodriguez LG, Beckwith EJ, Gilestro GF.. 2019. Rethomics: an R framework to analyse high-throughput behavioural data. PLoS One. 14:e0209331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, et al. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6:343–345. [DOI] [PubMed] [Google Scholar]
- Harbison ST, McCoy LJ, Mackay TF.. 2013. Genome-wide association study of sleep in Drosophila melanogaster. Bmc Genomics. 14:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman Hoffmann Clark 1999. Selection for starvation resistance in Drosophila melanogaster: physiological correlates, enzyme activities and multiple stress responses. J Evol Biol. 12:370–379. [Google Scholar]
- Heianza Y, Qi L, Heianza Y, Qi L.. 2017. Gene-diet interaction and precision nutrition in obesity. Int J Mol Sci. 18:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Ramia M, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster genetic reference panel lines. Genome Res. 24:1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, et al. 2007. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci USA. 104:8253–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Wilson KA, Beck JN, Nelson CS, Brownridge GW, et al. 2020. Genetic and metabolomic architecture of variation in diet restriction-mediated lifespan extension in Drosophila. PLoS Genet. 16:e1008835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Kazgan N, Bretz CA, Forsberg LJ, Hector CE, et al. 2010. Altered metabolism and persistent starvation behaviors caused by reduced AMPK function in Drosophila. PLoS One. 5:e12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, , DubouéER, , McDonaldDM, , DusM, , Suh GS, et al. . 2010. Clock and cycle Limit Starvation-Induced Sleep Loss in Drosophila. Current Biology. 20:1209–1215. 10.1016/j.cub.2010.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krycer JR, Quek L-E, Francis D, Fazakerley DJ, Elkington SD, et al. 2019. Lactate production is a prioritised feature of adipocyte metabolism. J Biol Chem. 295:83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krycer JR, Quek L-E, Francis D, Zadoorian A, Weiss FC, et al. 2020. Insulin signalling requires glucose to promote lipid anabolism in adipocytes. J Biol Chem. 295:13250–13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MN, Ha SH, Kim J, Koh A, Lee CS, et al. 2009. Glycolytic flux signals to mTOR through Glyceraldehyde-3-Phosphate Dehydrogenase-Mediated Regulation of Rheb▿. Mol Cell Biol. 29:3991–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Jang T.. 2014. Exploring the nutritional basis of starvation resistance in Drosophila melanogaster. Funct Ecol. 28:1144–1155. [Google Scholar]
- Lisinski I, Schürmann A, Joost H-G, Cushman SW, Al-Hasani H.. 2001. Targeting of GLUT6 (formerly GLUT9) and GLUT8 in rat adipose cells. Biochem J. 358:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature. 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maedera S, Mizuno T, Ishiguro H, Ito T, Soga T, et al. 2019. GLUT6 is a lysosomal transporter that is regulated by inflammatory stimuli and modulates glycolysis in macrophages. FEBS Lett. 593:195–208. [DOI] [PubMed] [Google Scholar]
- Markow TA. 2015. The secret lives of Drosophila flies. eLife. 4:e06793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May CE, Vaziri A, Lin YQ, Grushko O, Khabiri M, et al. 2019. High Dietary sugar reshapes sweet taste to promote feeding behavior in Drosophila melanogaster. Cell Rep. 27:1675–1685.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AL, Higgins CB, Heitmeier MR, Kraft TE, Qian X, et al. 2016. SLC2A8 (GLUT8) is a mammalian trehalose transporter required for trehalose-induced autophagy. Sci Rep. 6:38586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mony VK, Benjamin S, O'Rourke EJ.. 2016. A lysosome-centered view of nutrient homeostasis. Autophagy. 12:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M, Thorens B.. 2013. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 34:121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita H, Tanji K, Miki Y, Mori F, Wakabayashi K.. 2019. Trehalose intake and exercise upregulate a glucose transporter, GLUT8, in the brain. Biochem Biophys Res Commun. 514:672–677. [DOI] [PubMed] [Google Scholar]
- Nelson CS, Beck JN, Wilson KA, Pilcher ER, Kapahi P, et al. 2016. Cross-phenotype association tests uncover genes mediating nutrient response in Drosophila. BMC Genomics. 17:867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, et al. 2017. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 49:1385–1391. [DOI] [PubMed] [Google Scholar]
- Parks BW, Sallam T, Mehrabian M, Psychogios N, Hui ST, et al. 2015. Genetic architecture of insulin resistance in the mouse. Cell Metab. 21:334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C, Lear BC, Keegan KP, Allada R.. 2010. Locomotor activity level monitoring using the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010:pdb.prot5518. [DOI] [PubMed] [Google Scholar]
- Phillips CM. 2013. Nutrigenetics and metabolic disease: current status and implications for personalised nutrition. Nutrients. 5:32–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchers W, Nye J, Márquez EJ, Kowalski A, Dworkin I, et al. 2019. A multivariate genome-wide association study of wing shape in Drosophila melanogaster. Genetics. 211:1429–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. 2014. Gene-diet interaction and weight loss. Curr Opin Lipidol. 25:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E.. 2001. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 11:1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SW, Herzyk P, Dow JAT, Leader DP.. 2013. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic Acids Res. 41:D744–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales NMR, Pelegrini PB, Goersch MC.. 2014. Nutrigenomics: Definitions and Advances of This New Science. J Nutrition Metabolism. 2014:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz LO, Chaudhari LS.. 2015. High-risk populations: the pimas of Arizona and Mexico. Curr Obes Rep. 4:92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupa DA, Dervisefendic A, Zwiener J, Pletcher SD.. 2008. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell. 7:478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkard AJ, Foch TT, Hrubec Z.. 1986. A twin study of human obesity. JAMA. 256:51–54. [PubMed] [Google Scholar]
- Tan PY, Mitra SR, Amini F.. 2018. Lifestyle interventions for weight control modified by genetic variation: a review of the evidence. Public Health Genomics. 21:169–185. [DOI] [PubMed] [Google Scholar]
- Unckless RL, Rottschaefer SM, Lazzaro BP.. 2015. A genome-wide association study for nutritional indices in Drosophila. G3 (Bethesda). 5:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tucker NR, Rizki G, Mills R, Krijger PH, et al. 2016. Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. eLife. 5:e10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yu Y, Zhang V, Tian Y, Qi W, et al. 2015. Octopamine mediates starvation-induced hyperactivity in adult Drosophila. Proc Natl Acad Sci USA. 112:5219–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Huang R, Ye J, Zhang V, Wu C, et al. 2016. Regulation of starvation-induced hyperactivity by insulin and glucagon signaling in adult Drosophila. eLife. 5:e15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, et al. 2015. Personalized nutrition by prediction of glycemic responses. Cell. 163:1079–1094. [DOI] [PubMed] [Google Scholar]
- Zhao H, Tang M, Liu M, Chen L.. 2018. Glycophagy: an emerging target in pathology. Clin Chim Acta. 484:298–303. [DOI] [PubMed] [Google Scholar]
- Zobel T, Brinkmann K, Koch N, Schneider K, Seemann E, et al. 2015. Cooperative functions of the two F-BAR proteins Cip4 and Nostrin in the regulation of E-cadherin in epithelial morphogenesis. J Cell Sci. 128:499–515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials. Supplementary Figure S1 contains a heatmap of NF-standardised mean starvation survival data. Supplementary Figure S2 contains a histogram of NF-standardized data. Supplementary Figure S3 contains a Q–Q plot of the MANOVA P-values. Supplementary Figure S4 contains the starvation survival plot for control and knockdown of Cip4. Supplementary Figure S5 contains plot validating CG4607 RNAi knockdown with qPCR. Supplementary Table S1 contains the composition of diets used in the study. Supplementary Table S2 contains the Linear-Mixed Model Estimates, and Derived Heritability. Supplementary Table S3 contains the numbers of SNPS associated with decreased starvation resistance on 1 diet and Supplementary Table S4 contains the numbers of SNPs associated with increased starvation resistance on 1 diet. Supplementary Table S5 contains the ANOVAs for covariates for HCD, HFD, and HPD. Supplementary Table S6 contains ANOVA tests to determine the significance of gene–diet interaction on caloric intake of NF or HCD. Supplementary Table S7 contains the raw mean starvation survival data for each DGRP line on each diet. Supplementary Table S8 is a dataset containing 3574 SNPs above the P < 1 × 10−3 MANOVA threshold. Supplementary Table S9 is a dataset containing 566 SNPs filtered from the 3574 (Supplementary Table S8) by Wilcoxon P-value <0.005 and a dietary fold change greater than ±0.3. Supplementary Table S10 is a dataset containing the 91 SNPs (out of the 566 from Supplementary Table S9) that meet criteria outlined in the “candidate gene validation” section of the Results. Supplementary File S1 is the code used to generate the heritability data, which includes two accompanying files: Francis_Long_Data.csv and ln_data_long.csv. These supplementary files, Figures, and datasets are found in Figshare DOI: https://doi.org/10.6084/m9.figshare.9740801.