Abstract
Studying genetic variation of gene expression provides a powerful way to unravel the molecular components underlying complex traits. Expression quantitative trait locus (eQTL) studies have been performed in several different model species, yet most of these linkage studies have been based on the genetic segregation of two parental alleles. Recently, we developed a multiparental segregating population of 200 recombinant inbred lines (mpRILs) derived from four wild isolates (JU1511, JU1926, JU1931, and JU1941) in the nematode Caenorhabditis elegans. We used RNA-seq to investigate how multiple alleles affect gene expression in these mpRILs. We found 1789 genes differentially expressed between the parental lines. Transgression, expression beyond any of the parental lines in the mpRILs, was found for 7896 genes. For expression QTL mapping almost 9000 SNPs were available. By combining these SNPs and the RNA-seq profiles of the mpRILs, we detected almost 6800 eQTLs. Most trans-eQTLs (63%) co-locate in six newly identified trans-bands. The trans-eQTLs found in previous two-parental allele eQTL experiments and this study showed some overlap (17.5–46.8%), highlighting on the one hand that a large group of genes is affected by polymorphic regulators across populations and conditions, on the other hand, it shows that the mpRIL population allows identification of novel gene expression regulatory loci. Taken together, the analysis of our mpRIL population provides a more refined insight into C. elegans complex trait genetics and eQTLs in general, as well as a starting point to further test and develop advanced statistical models for detection of multiallelic eQTLs and systems genetics studying the genotype–phenotype relationship.
Keywords: multiparental RILs, expression QTL, eQTL, SNPs, C. elegans, MPP, Multiparental Populations, Multiparent Advanced Generation Inter-Cross (MAGIC)
Introduction
Investigation of the genotype–phenotype relationship is at the heart of genetic research. The detection and description of allelic variants and genetic mechanisms have been a demanding task due to the quantitative nature of most phenotypic variation. Quantitative trait locus (QTL) mapping has been one of the methods of choice for finding the loci on which these allelic variants can be found. Many functional polymorphisms in plants and animals, including many model species such as model nematode Caenorhabditis elegans, have been discovered using QTL mapping (Tijsterman et al. 2002; Rogers et al. 2006; Kammenga et al. 2007; Gloria-Soria and Azevedo 2008; Palopoli et al. 2008; Reiner et al. 2008; Seidel et al. 2008; McGrath et al. 2009; Reddy et al. 2009; Bendesky et al. 2011; Seidel et al. 2011; Bendesky et al. 2012; Ghosh et al. 2012; Andersen et al. 2014; Noble et al. 2015; Schmid et al. 2015; Cook et al. 2016; Greene et al. 2016; Large et al. 2016; Ben-David et al. 2017; Zdraljevic et al. 2017; Hahnel et al. 2018; O'Donnell et al. 2018; Brady et al. 2019; Zdraljevic et al. 2019). Over the last decade, molecular phenotypes such as transcript levels, protein levels, and metabolites have also been used in QTL mapping (Li et al. 2010; Vinuela et al. 2010; Singh et al. 2016; Snoek et al. 2017; Sterken et al. 2017; Gao et al. 2018; Sterken et al. 2019). Heritable variation in these molecular phenotypes often plays a role in heritable phenotypic variation (Jimenez-Gomez et al. 2010; Schmid et al. 2015; Sterken et al. 2017). Mapping expression QTLs (eQTLs) can provide insight into the transcriptional architecture of complex traits and have been conducted in model species such as Arabidopsis thaliana and C. elegans as well as several other taxa (Li et al. 2006; Keurentjes et al. 2007; West et al. 2007; Li et al. 2010; Rockman et al. 2010; Vinuela et al. 2010; Snoek et al. 2012, 2017; Cubillos et al. 2014; Ranjan et al. 2016; Sterken et al. 2017, 2019; Hartanto et al. 2020).
Most eQTL studies have been done on populations of recombinant inbred lines (RILs) originating from a cross between two different parental genotypes (Li et al. 2006; Keurentjes et al. 2007; West et al. 2007; Li et al. 2010; Rockman et al. 2010; Vinuela et al. 2010; Snoek et al. 2012, 2017; Cubillos et al. 2014; Sterken et al. 2017, 2019; Hartanto et al. 2020). The inclusion of more than two parents can capture more genetic variation, increasing the number of detected QTLs, potentially allowing more precise mapping and therefore reducing the number of potential candidate causal genes to be verified (King et al. 2012). Such a strategy was first used for Arabidopsis by developing a Multiparent Advanced Generation Inter-Cross (MAGIC) lines population consisting of 527 RILs developed from 19 different parental accessions (Kover et al. 2009). Several other MAGIC populations have been developed since then for a range of species, including C. elegans (de Koning and McIntyre 2017; Noble et al. 2017; Snoek et al. 2019).
Recently multiparental RIL (mpRILs) populations have been developed in C. elegans (Noble et al. 2017; Snoek et al. 2019). These populations have been created using other strains than the most frequently used N2 strain and the Hawaiian CB4856 strain (Li et al. 2006; Doroszuk et al. 2009; Rockman and Kruglyak 2009; Li et al. 2010; Vinuela et al. 2010; Rodriguez et al. 2012; Vinuela et al. 2012; Snoek et al. 2013, 2014, 2014, 2017, 2020; Stastna et al. 2015; Sterken et al. 2015, 2017, 2019, 2021; Thompson et al. 2015; Kamkina et al. 2016; Nakad et al. 2016; Singh et al. 2016; Jovic et al. 2017; Jovic et al. 2019; Evans et al. 2021). In this study, we used the population of 200 mpRILs, derived from an advanced cross between four wild types: JU1511 and JU1941 isolated from Orsay (France) and JU1926 and JU1931 isolated from Santeuil (France) (kindly provided by MA Félix, Paris, France; Volkers et al. 2013; Snoek et al. 2019). In a previous study, the RNA-seq data of these mpRILs were used to obtain almost 9000 SNPs variable between the four parental genotypes and used to identify QTLs for life-history traits (Snoek et al. 2019). The RNA was sampled from the mpRILs grown under standardized conditions (24°C, OP50, 48 h after bleaching) and obtained from animals from two 6-cm dishes, with one RNA-seq replicate per mpRIL and two per parental isolate. To investigate the effect of multiple genetic backgrounds on gene expression, we used the RNA-seq data to associate gene expression levels to genetic variants present in the population. We compared the gene expression level differences between the parental wild isolates, calculated transgression, as well as heritability and mapped eQTLs. We identified six trans-bands (TBs), hotspots at which many trans-eQTLs colocate, which we further studied by gene ontology enrichment. Lastly, we compared the eQTLs found in this study to the eQTLs found in previous eQTL studies in C. elegans (Li et al. 2006, 2010; Rockman et al. 2010; Vinuela et al. 2010; Snoek et al. 2017; Sterken et al. 2017). Together these results present the first insights into the genetic architecture of gene expression in a C. elegans multiparental RIL population.
Methods
Nematode strains and culturing, RNA-sequencing, construction of the genetic map
The C. elegans strains and culturing condition, RNA-sequencing, and construction of the genetic map can be found in Snoek et al. (2019). In short, the mpRILs used were grown in five separate batches with two 6-cm dishes per strain (24°C, OP50, 48 h after bleaching; the plates were randomized within incubators) and per strain, the two samples were pooled for RNA isolation, with one RNA-seq replicate per mpRIL and two replicates per parental isolate (Supplementary Table S4). Collecting and freezing the samples for one batch took approximately 30 min. The genetic map and eQTL profiles can found on WormQTL2 (Li et al. 2009) (http://www.bioinformatics.nl/EleQTL; Snoek et al. 2020).
SNP calling and gene expression levels
The paired-end reads were mapped against the N2 reference genome (WS220) using Tophat (Trapnell et al. 2009), allowing for four read mismatches, and a read edit distance of 4. SNPs were called using samtools (Li et al. 2009), mpileup with bcftools and vcfutils as described in Snoek et al. (2019). Expression levels were determined using the tuxedo pipeline, giving length normalized fragments per kilobase per million (fpkm) values (Trapnell et al. 2012). Transcripts were assembled from the mapped reads using cufflinks (Trapnell et al. 2012). Raw RNA-seq data can be found in the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) with ID PRJNA495983. Normalized read counts can be found on WormQTL2 (http://www.bioinformatics.nl/EleQTL; Snoek et al. 2020). Normalization was done after the selection of the consistently detected transcripts (see QTL mapping and FPR) by taking the fpkm per gene per million fpkm per sample.
Heritability and transgression
Heritability of gene expression levels was calculated using the heritability package in “R.” A narrow-sense heritability was calculated using the function marker_h2 (Kruijer et al. 2015). The required kinship matrix was calculated using the emma.kinship function from the EMMA package (Kang et al. 2008). To determine a per-gene significance, we used a permutation approach where we shuffled the expression levels per transcript. After 100 permutations, the 95th highest value was taken as the 0.05 false discovery rate (Speed et al. 2012; Kruijer et al. 2015; Gilmour 2019). Transgression was determined by counting the number of mpRILs with an expression level beyond the mean + 2 SD of the most extreme parental lines. SD was calculated on the within-line variation of the parental samples. False-positive rate (FPR) was determined by permutations, randomly assigning the parental labels to gene-expression values. The threshold for transgression was set at an arbitrary 50 mpRILs (25% of all lines; FPR = 0.08) beyond the most extreme parental line(s).
eQTL mapping and FPR
For eQTL mapping, we first selected the genes with consistently detected transcripts, meaning those expressed with a mean log2 expression (fpkm) >−5, which resulted in a set of 12,029 genes with transcripts that were detected in all samples. eQTLs were mapped by a linear model using a single marker model explaining gene expression (as log2 ratio with the mean) by one SNP-marker at the time for the whole genome. FPR was determined by one round of permutations where for each transcript, the counts were randomly distributed over the RILs before eQTL mapping. The -log10(p) value when number of false positives divided by the number of true positives was <0.01 [-log10(p) > 5.35]. Transbands (or eQTL hotspots) were determined for those loci that harbor more than 100 eQTLs in a 1-Mbp window to both sides of the marker under consideration. Genome-wide eQTL significance profiles [-log10(p)] can be found on WormQTL2 (http://www.bioinformatics.nl/EleQTL; Snoek et al. 2020).
Enrichment analysis and figures
Enrichment of GO terms was done using the hypergeometric test in “R” (R Core Team 2017). GO term genes associations were download from Wormbase (http://www.wormbase.org) version WS276. Only genes that passed the filtering step for eQTL mapping were used as background genes. For significant enrichment, a P-value < 1e−5 was used and a geneset size per GO term >3. Most figures were made using the R package ggplot2 (Wickham 2009) except Figure 1 which was made using the UpSetR library.
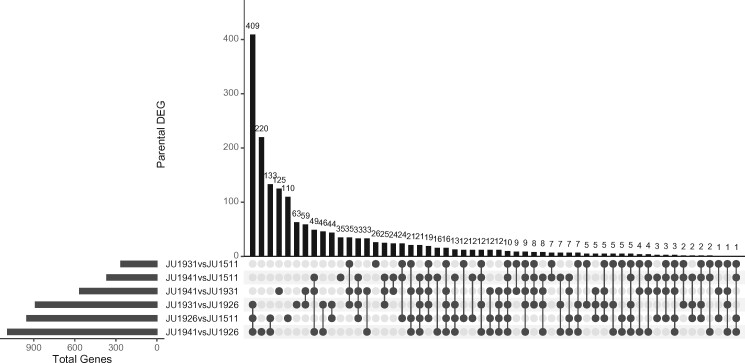
Figure 1.
Gene expression differences between the four mpRIL parental lines. Upset plot shows the pairwise comparisons and the overlap between the pairs (Tukey’s HSD; P < 0.001; FPR = 0.05). The horizontal bar plot shows the number of DEG per parental pair, while the vertical bar plot indicates the number of shared DEG per comparison. For example, an overlap of 409 genes was found between the three comparisons that include the JU1926 parental line, which shows that JU1926 differed most from all other lines.
eQTL comparison between experiments/studies
To compare how many genes with an eQTL overlapped between the different studies (Li et al. 2006, 2010; Rockman et al. 2010; Vinuela et al. 2010; Snoek et al. 2017, 2020; Sterken et al. 2017) available in WormQTL2 (Snoek et al. 2020), we downloaded the eQTL profiles and markers used per experiment and listed the genes with a cis- or a trans-eQTL. For eQTL determination, the most significant marker per gene was taken as the peak. A -log10(p) > 3.5 was used as threshold for calling the eQTL. An eQTL was determined cis when the peak position was within 1 Mbp of the start position of the gene. These lists were compared with the genes having an eQTL in this study. The percentage overlap was calculated against the original study.
Results
Gene expression differences between the parental lines
To study the effect of genetic variation on gene expression, we used RNA-seq on a population of 200 multiparental recombinant inbred lines (mpRILs) (Snoek et al. 2019), made from a cross between four parental lines isolated from Orsay, France (JU1511, JU1941) and Santeuil, France (JU1926, JU1931) (Volkers et al. 2013). The animals used were grown on two 6-cm dishes (24°C, OP50, 48 h after bleaching) per sample pooled for RNA isolation, with one RNA-seq replicate per mpRIL and two per parental isolate. First, we determined the expression differences between the parental lines (Supplementary Table S1). Of the 12,029 detected transcripts, we found 1789 genes differently expressed between at least one parental pair (TukeyHSD P < 0.001; FPR < 0.05; Figure 1). Of the four strains, JU1926 was most different when compared to the other lines, with 409 genes being differently expressed between JU1926 and the other three lines. Thereafter, JU1941 was most different from the remaining two lines. These differences in gene expression between the parental lines are likely genotype dependent. To illustrate the reproducibility of the parental lines, we calculated the correlation between the parental samples and found that replicate pairs are formed. The correlation between the parental pairs is JU1511: 0.91, JU1926: 0.91, JU1931: 0.94, and JU1941: 0.82.
Transgression and heritability
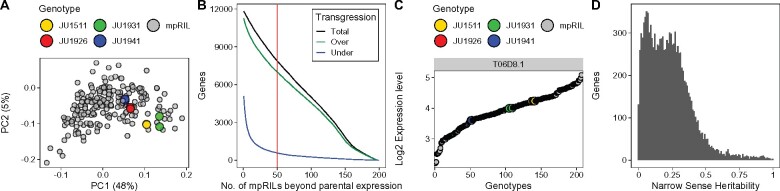
To explore the variation in gene expression between the different parental and mpRIL genotypes, we applied principal component analysis on the log2 gene expression ratios (Figure 2A). From exploration of the PCA axes, we concluded that there were no batch effects. This was also based on mapping (1) growth/sample batch, (2) RNA-isolation batch, and (3) sequencing batch. Neither of these traits mapped to the TBs that we detected. We can see that the expression variation in many of the mpRILs extends beyond the parental expression variation. The extension of variation suggests transgression and/or developmental variation. PC1 most likely corresponds to differences in development as gene families known to be strongly upregulated during L4 progression (Snoek et al. 2014) like vitellogenins (vit), major sperm proteins (msp), and chondroitin proteoglycans (cpg), were highly correlated with PC1. Analysis revealed transgression for 7896 genes (FPR = 0.08; Figure 2, B and C, Supplementary Table S2). Notably, most transgression was one-sided, showing increased expression level beyond the highest expression level found in the parental lines. This suggests that multiple segregated loci, in combination with developmental variation, are involved in regulating the transcription in the mpRILs. The mostly higher than the parental lines type one-sided transgression could also be caused by the developmental differences between the parental lines and the mpRILs, with most of the mpRILs showing a further developed expression profile. As a specific group of genes shows a large and progressing upregulation during L4, this could show up as one-sided transgression. Nevertheless, transgression often indicates that the trait variation, in this case gene expression levels, is heritable. We calculated the narrow-sense heritability (h2) and found significant h2 for expression variation of 9500 genes (per-gene FPR = 0.05; Figure 2D, Supplementary Table S2). Most gene expression variation showed an h2 below 0.5, indicating that part of the variation is caused by other factors than additive genetic effects. These other factors contributing to gene expression variation could be technical, environmental, but also more complex genetic interactions, such as epistasis.
Figure 2.
Gene expression variation in the mpRILs and parental genotypes. (A) Principal component analysis (PCoA) of the log2 ratios, mpRILs shown in gray, parental lines shown in color. (B) Transgression: number of mpRILs beyond the parental expression level (x-axis) against the number of genes (y-axis). The mpRILs below (under) the lowest parental expression level in blue, mpRILs over the highest parental expression level in green, and the sum of these (total) in black. (C) Example of two-sided transgression for expression levels of gene T06D8.1. (D) Genes with significant narrow-sense heritability (h2) and the distribution of heritable variation of gene expression variation at FPR = 0.05.
Expression QTLs
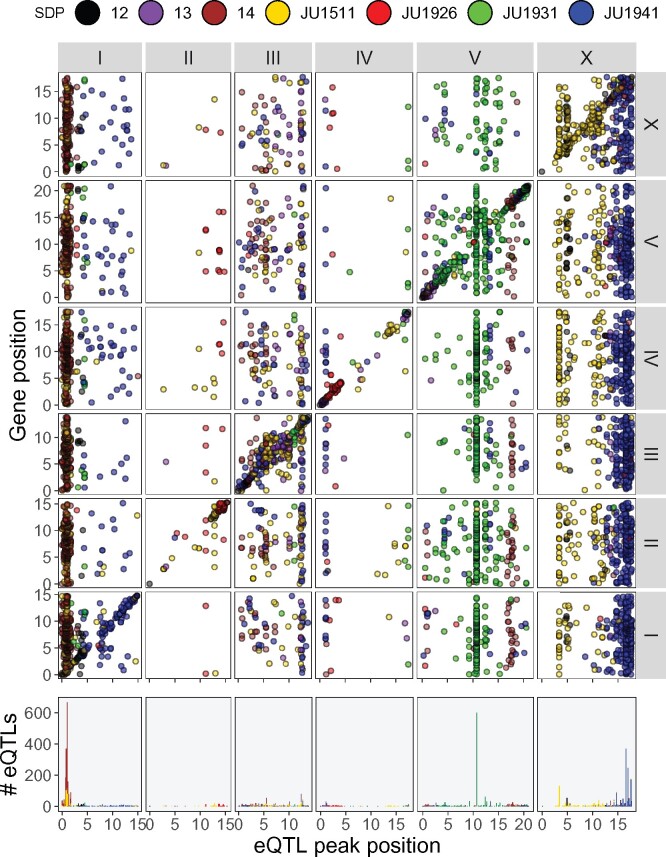
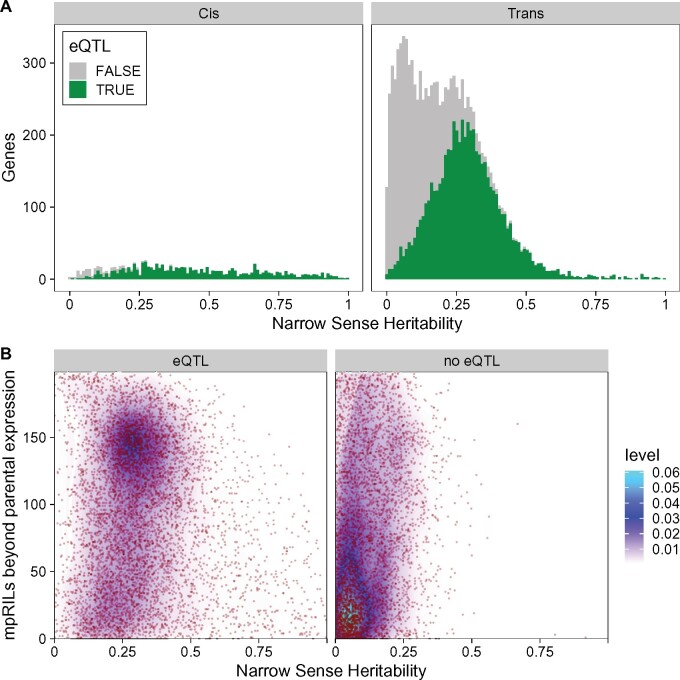
To find the loci involved in gene expression variation between the mpRILs, we used a single marker QTL model. We found 6784 eQTLs (one eQTL per gene, -log10(p) > 5.35; FPR = 0.01), of which 929 were cis- and 5855 trans-eQTLs (Table 1; Figure 3; Supplementary Table S2). Most cis-eQTLs were found on chromosome V and most trans-eQTLs on chromosomes I and X. For both cis- and trans-eQTLs, fewest were found on chromosomes II and IV. The SNP Distribution Pattern (SDP) groups SNPs with the same distribution in the parental lines, for example the SNPs found in JU1511 and JU1941, but not in JU1926 and JU1931 share the same SDP. When the SDP is considered, many of the cis-eQTLs were found to have an effect where either the JU1511 or JU1941 allele was different from the three other parental genotypes. For the trans-eQTLs, the largest groups also show this allelic difference or those SNPs that distinguish JU1511/JU1941 from JU1926/JU1931. A substantial group of eQTLs was found for the JU1931 allele, whereas hardly any eQTLs were found for the JU1926 specific SNPs. The lack of JU1926 linked eQTLs is somewhat surprising as it had the most differentially expressed genes (DEG) in the comparison of the parental lines. Yet, we found much more genes with eQTLs than being DEG in the parental comparison. These are much more likely to be caused by new allelic combinations present in the mpRILs. Overall, the majority of the eQTLs are found on a few major effect loci with a specific SDP linkage (Figure 3). Moreover, comparing the h2 to the eQTLs showed that genes with an eQTL have a much higher h2 than those without an eQTL, where genes with an h2 > 0.25 almost all have an eQTL (Figure 4). Comparing cis- and trans-eQTLs showed that genes with a cis-eQTL have a higher h2 on average, yet the h2 distributions of cis- and trans-eQTLs are overlapping.
Table 1.
eQTLs per type (cis/trans) per chromosome per SDP
|
Cis
|
Trans
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SDP | I | II | III | IV | V | X | Tot | I | II | III | IV | V | X | Tot |
|
14 | 2 | 17 | 2 | 23 | 13 | 71 | 35 | 0 | 1 | 1 | 3 | 67 | 107 |
|
6 | 1 | 14 | 39 | 41 | 3 | 104 | 0 | 2 | 106 | 13 | 15 | 27 | 163 |
|
12 | 0 | 19 | 0 | 53 | 11 | 95 | 1373 | 0 | 119 | 5 | 103 | 44 | 1644 |
| JU1511 | 37 | 32 | 61 | 14 | 18 | 81 | 243 | 457 | 28 | 211 | 20 | 9 | 430 | 1155 |
| JU1926 | 0 | 32 | 4 | 59 | 5 | 1 | 101 | 5 | 44 | 5 | 26 | 10 | 5 | 95 |
| JU1931 | 8 | 0 | 15 | 3 | 81 | 1 | 108 | 31 | 0 | 12 | 21 | 919 | 2 | 985 |
| JU1941 | 76 | 0 | 66 | 5 | 38 | 22 | 207 | 150 | 1 | 155 | 35 | 94 | 1,271 | 1,706 |
| Total | 153 | 67 | 196 | 122 | 259 | 132 | 929 | 2,051 | 75 | 609 | 121 | 1,153 | 1,846 | 5,855 |
SDP, Chromosome, Peak position, and left and right borders in Mega-base pairs.
Figure 3.
Cis/trans plot of the identified eQTLs. eQTL position shown on the x-axis, gene position shown on the y-axis (upper plot) or number of eQTLs (bottom plot). SDP shown in color, chromosomes shown in the gray strips on top and on the right of the panels.
Figure 4.
Relation between eQTLs, transgression, and narrow-sense heritability (h2). (A) Narrow-sense heritability (h2; x-axis), distribution in genes (y-axis) with cis- and trans-eQTLs, significance of the eQTLs is TRUE (green) when -log10p >5.35 and FALSE (grey) otherwise. (B) Relation between narrow-sense heritability (h2; x-axis) and transgression (y-axis) for genes with and without a significant eQTL, individual datapoints shown in red, color gradient indicates datapoint density.
Trans-bands
A large majority of the trans-eQTLs (3704; 63% of all trans-eQTLs) were found in six hotspots, so-called TBs (number of trans-eQTLs > 100, window 1 Mbp to both sides; Table 2; Figure 3). Two TBs were found on chromosome I, one on chromosome V, and three on chromosome X. The two TBs on chromosome I colocated but were linked to different SDP: the SDP 14 (JU1511/JU1941 vs JU1926/JU1931) and SDP JU1511 (vs the rest). The TB on chromosome V was linked to SDP JU1931 and the three TBs found on chromosome X were linked to SPD JU1511 and JU1941.
Table 2.
Descriptive overview of the six identified TBs
| SDP | CHR | Peak (Mbp) | Left (Mbp) | Right (Mbp) | eQTLs | GO Enrichment (selection from enrichment table) | Phenotypic QTL [in Snoek et al. ( 2019)] | |
|---|---|---|---|---|---|---|---|---|
| TB1 | 14 (JU1511 and JU1941 vs JU1926 and JU1931) | I | 1.03 | 0.03 | 2.03 | 1,339 | Thermosensory behavior, negative regulation of engulfment of apoptotic cell, DNA replication, embryonic body morphogenesis, establishment or maintenance of actin cytoskeleton polarity, muscle fiber development, epidermis development, response to unfolded protein and, molting cycle, collagen, and cuticulin-based cuticle | Population growth on Erwinia and on B. thuringiensis |
| TB2 | JU1511 | I | 0.83 | 0 | 1.83 | 443 | Regulation of protein stability, regulation of vulval development, DNA replication, anaphase-promoting complex and, microtubule polymerization | NA |
| TB3 | JU1931 | V | 10.74 | 9.74 | 11.74 | 607 | Hemidesmosome assembly, external side of plasma membrane and, negative regulation of response to oxidative stress | NA |
| TB4 | JU1511 | X | 3.40 | 2.40 | 4.40 | 133 | Adenyl-nucleotide exchange factor activity | Heat-shock sensitivity |
| TB5 | JU1941 | X | 14.69 | 13.69 | 15.64 | 225 | Dendrite morphogenesis | Population growth on B. thuringiensis |
| TB6 | JU1941 | X | 16.60 | 15.65 | 17.6 | 957 | Embryonic body morphogenesis, DNA replication, integral component of peroxisomal membrane, anaphase-promoting complex, endosome, phagocytic vesicle membrane, neuronal signal transduction, response to anoxia, cuticle pattern formation, cell fate commitment, hemidesmosome-associated protein complex, and response to lipid | Sensitivity to oxidative stress |
Selection of enriched GO terms from Supplementary Table S3 and overlap with phenotypic QTLs found in Snoek et al. (2019).
GO enrichment
To study the effect of TBs on the biological function, we used GO term enrichment (Table 2, Supplementary Table S3). Each of the TBs was linked to mostly different sets of GO terms, suggesting an effect on different parts of C. elegans biology. The genes mapping to TB1 on chromosome I were enriched for behavior and muscle and epidermis development GO categories. The genes mapping to TB2 on chromosome I were enriched for the GO term “vulval development,” among others. The genes with a trans-eQTL on TB3 on chromosome V were enriched for GO terms associated with oxidative stress. The genes mapping to TB4 and TB5 on chromosome X only showed a few enriched GO terms, among which adenyl-nucleotide exchange factor activity and dendrite morphogenesis. The genes mapping to TB6 on chromosome X were enriched for the GO term “response to anoxia” and many more. This shows that these TBs can be involved in several developmental processes and in the interaction with the environment.
Overlap with other eQTL experiments
To investigate if the genes with eQTLs found in the present mpRIL study also had eQTLs in other studies, we compared them with the studies found in WormQTL2 (Table 3; Li et al. 2006, 2010; Rockman et al. 2010; Vinuela et al. 2010; Snoek et al. 2017, 2020; Sterken et al. 2017). In general, we found that a substantial group of genes with a trans-eQTL in any of the studies had an eQTL in our mpRIL experiment (26.5–36.9%). The groups of genes with trans-eQTLs show much higher overlap than the genes with a cis-eQTL in any of the experiments (10.2–20.0%). Around a third of the genes with a trans-eQTL in Vinuela et al. (2010), Snoek et al. (2010, 2017), and Sterken et al. (2017) also showed a trans-eQTL in the mpRILs, with numbers almost equal between developmental stages and treatments. Slightly fewer overlapping genes with eQTLs were found with Rockman et al. (2010) and Sterken et al. (2017). Comparing the experiments performed with the same N2 × CB4856 in the same lab (Li et al. 2006; Snoek et al. 2010, 2017; Vinuela et al. 2010; Sterken et al. 2017) shows that environmental conditions and developmental stage only have a small effect on the global overlap and difference between cis- and trans-eQTLs. As the genetic backgrounds of the mpRILs are different from the N2 × CB4856 populations used in the other experiments, the low percentage of overlapping cis-eQTLs could be expected. The large group of genes with a trans-eQTL in both experiments shows that the expression levels of a substantial group of genes are more prone to be affected by genetic variation independent of environment or developmental stage, while the loci involved are most likely different in each experiment/condition (Vinuela et al. 2010; Snoek et al. 2017; Sterken et al. 2017).
Table 3.
Overlapping eQTLs between this mpRIL experiment and the RIL experiments available in WormQTL2 (Nijveen et al. 2017)
| eQTL experiment | Total Cis | Cis Overlap (%) | Total Trans | Trans Overlap (%) |
|---|---|---|---|---|
| 16°C (Li et al. 2006) | 240 | 14.6 | 817 | 31.6 |
| 24°C (Li et al. 2006) | 337 | 12.2 | 998 | 30.5 |
| Li et al. (2010) | 752 | 14.5 | 3,544 | 28.7 |
| Rockman et al. (2010) | 1,958 | 12.0 | 2,792 | 28.8 |
| Control (Snoek et al. 2017; Sterken et al. 2017) | 961 | 17.1 | 1,,481 | 36.1 |
| Heat-shock (Snoek et al. 2017; Sterken et al. 2017) | 976 | 20.0 | 2,776 | 36.9 |
| Recovery (Snoek et al. 2017; Sterken et al. 2017) | 992 | 16.1 | 1,519 | 33.4 |
| Sterken et al. (2017) | 719 | 10.2 | 1,116 | 26.5 |
| Juvenile (Snoek et al. 2010; Vinuela et al. 2010) | 303 | 11.9 | 2,206 | 33.4 |
| Old (Snoek et al. 2010; Vinuela et al. 2010) | 220 | 15.0 | 1,790 | 34.9 |
| Reproductive (Snoek et al. 2010; Vinuela et al. 2010) | 348 | 13.2 | 2,010 | 32.7 |
Percentages indicate the percentage of eQTLs found in the indicated experiment that are also found in the mpRILs eQTLs. Threshold used for the eQTL experiments in this table: -log10(p) >3.5; Cis-eQTLs were called if the peak of the eQTL was within 1 Mbp of the gene start, otherwise it was called a trans-eQTL.
Discussion
In this experiment, we used a population of mpRILs and RNA-seq to find 6784 eQTLs, of which 929 were cis-eQTLs and 5855 were trans-eQTLs. A large proportion (63%) of the trans-eQTLs were found in six TBs. The total number of eQTLs found in this mpRIL study (6784) is at the high end of what was previously found in other experiments (mean: 2560; 653–6518) (Li et al. 2006; Rockman et al. 2010; Vinuela et al. 2010; Snoek et al. 2017; Sterken et al. 2017). This number is hard to compare as the number of identified eQTLs depend on many factors, such as population size, number of recombinations, statistical model, and RNA measurement technology used, which are nearly all different between this and the other eQTL studies in C. elegans (Li et al. 2006; Rockman et al. 2010; Vinuela et al. 2010; Snoek et al. 2017; Sterken et al. 2017). Nevertheless, it seems that a combination of RNA-seq and multiple genetic backgrounds increased the number of detected eQTLs. A very clear increase was found for trans-eQTLs (5855) compared to the numbers found in previous studies, even at a much lower significance threshold. For example, the study of Rockman et al. (2010) used a comparable number of recombinant inbred advanced intercross lines (RIAILs) as the number of mpRILs in this study (∼200), yet found fewer trans-eQTLs, however, the different conditions and technologies used prevent any definitive conclusions. With respect to trans-eQTLs, we do know that they depend on environmental conditions or a response to changing conditions. It could be that with a background of four parental genotypes the mpRILs perceive the ambient environment in a broader range than the RIAILs with a background of two parental genotypes used by Rockman et al. (2010), and the RILs in the other studies. For example, the mpRILs could have inherited parts of four different sets of environmental preferences as opposed to two in the RIAILs and RILs, potentially extending the accompanying gene expression patterns and eQTLs. Yet, the most likely reason for the increased number of trans-eQTLs is the use of RNA-seq in this study compared to microarrays in the other studies. Another reason for finding more trans-eQTLs could be due to the generally genome-wide equal allelic distributions in this population (Snoek et al. 2019). Namely, a similar TB as the chromosome I TB at 1 Mb (TB1) related to development has been spotted in other datasets, but has been spurious due to being located near the peel-1 zeel-1 incompatibility locus, therefore, lacking recombinations in the N2 × CB 4856 RIL panel use before (Li et al. 2006; Seidel et al. 2008; Li et al. 2010; Snoek et al. 2017, 2020). Another advantage of using RNA-seq is that the genotype and gene-expression levels can be obtained from the same sample, preventing mislabeling errors and the need for “reGenotyping” in case of microarrays (Zych et al. 2017). In summary, as has been shown for yeast (Albert et al. 2018), the combination of generally smaller effect of trans-eQTLs and higher dynamic range of RNA-seq would at least increase the possibility to pick-up trans-eQTLs in C. elegans and in general.
It is noted that genetic variation in development across the mpRILs could be an important driver of trans-eQTL hotspots (Francesconi and Lehner 2014). We found two eQTL hotspots that are enriched in GO terms related to development. As the mpRILs differ in developmental speed, it would be interesting to include it as a covariate and assess its weight in the mapping as has been described in Francesconi and Lehner (2014). To allow for comparison with previous eQTL studies, we decided not to include development as cofactor in our analysis since this was also not done in the earlier studies (Rockman et al. 2010; Francesconi and Lehner 2014).
We previously found QTLs for several different phenotypes, such as population growth on different bacteria, sensitivity to heat shock and oxidative stress (Snoek et al. 2019). Four TBs were found to colocate with the previously found phenotypic QTLs (Table 2). Population growth on Erwinia and on Bacillus thuringiensis DSM was found to colocate with TB1, which was enriched for GO terms related to muscle, epidermis, and molting. This could indicate a difference in these structures that can affect the interaction with different types of bacteria or could indicate that there is a difference in developmental speed through which differences in the expression, and subsequent eQTLs, of molting-related genes are picked up. A QTL for heat-shock sensitivity was inferred to colocate with TB4, however, no indication for a link with this phenotype was found in the annotation of the genes with an eQTL at this position. The same was observed for TB5 and the overlap with population growth on B. thuringiensis, where GO enrichment also did not provide any leads to a potential mechanistic link. The overlap between the QTL for sensitivity to oxidative stress and TB6, however, did show some clues from GO enrichment as genes involved in the peroxisome as well as DNA replication and cuticle formation could be involved in dealing with oxidative stress.
We expect to have only found a fraction of the eQTLs, as we only used a simple additive mapping model, a conservative score of one eQTL per gene, and standard lab conditions with only one time point for RNA isolation. Both the number of eQTLs and genes with one or more eQTLs are expected to increase when more complex models are applied to these data and/or different experimental conditions and time points are considered (Vinuela et al. 2010; Francesconi and Lehner 2014; Snoek et al. 2017). Moreover, we use an SNP-based method for eQTL mapping, which has a binary option for each marker and therefore does not consider the genetic origin (parent) of the SNP. Using the genetic origin of the SNPs could reveal the more complex genetic interactions that could underlie the differences in transcript levels between the mpRILs. These complex genetic interactions are suggested to be present in this mpRIL population, by the heritability and transgression found. A model in which each marker has the four parental options might indicate loci with more than two alleles affecting gene expression. Furthermore, some (relatively small) genetic loci might have been missed all together as our investigations are based on the N2 reference genome and wild isolates can have vastly divergent regions of which sequences reads fail to align to the N2 reference genome with conventional methods (Thompson et al. 2015; Lee et al. 2021).
This study provides a more detailed insight into the genetic architecture of heritable gene expression variation in a multiparent recombinant inbred population. The use of RNA-seq data in combination with more than two alleles allows for a more precise detection of QTLs and incorporates a wider band of standing genetic variation, resulting in a substantial increase in eQTLs especially trans-eQTLs. Comparison to bi-allelic studies supports the position of eQTLs and may be used to detect a more detailed pattern of associated loci. We expect this study, data, and results to provide new insights into C. elegans genetics and eQTLs in general as well as to be a starting point to further test and develop advanced statistical models for detection of eQTLs and systems genetics studying the genotype–phenotype relationship.
Data availability
The data underlying this article available in Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) and can be accessed with ID PRJNA495983 and in WormQTL2 (http://www.bioinformatics.nl/EleQTL; Snoek et al. 2020).
Supplementary material is available at G3 online.
Supplementary Material
Acknowledgments
The authors acknowledge financial support from the Deutsche Forschungsgemeinschaft to H.S. (grant number SCHU 1415/11 and project A1 within the CRC 1182) and to PCR (Competence Centre for Genomic Analysis (CCGA) No. 07495230). J.E.K. was funded by NIH grant 1R01AA 026658. Furthermore, financial support from the NWO-ALW (project 855.01.151) to R.J.M.V. was given. M.G.S. was supported by NWO domain Applied and Engineering Sciences VENI grant (17282). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
B.L.S., H.S., and J.E.K. conceived the study; R.J.M.V. and J.R. performed the experiments; P.C.R. coordinated and supervised transcriptome sequencing; B.L.S., M.G.S., and H.N. analyzed the data, B.L.S. wrote the paper with contributions from all authors.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) nr.1415/11 to H.S., the Competence Centre for genomic analysis nr. 07495230 to P.C.R., the National Institutes of Health (NIH) nr. 1R01AA 026658 to J.E.K., the Netherlands Organisation for Scientific Research nr. 855.01.151 to B.L.S., and the Netherlands Organisation for Scientific Research nr. 17282 to M.G.S.
Conflicts of interest
The authors declare that there is no conflict of interest.
Contributor Information
Basten L Snoek, Laboratory of Nematology, Wageningen University, NL-6708 PB Wageningen, The Netherlands; Theoretical Biology and Bioinformatics, Utrecht University, 3584 CH Utrecht, The Netherlands.
Mark G Sterken, Laboratory of Nematology, Wageningen University, NL-6708 PB Wageningen, The Netherlands.
Harm Nijveen, Bioinformatics Group, Wageningen University, NL-6708 PB Wageningen, The Netherlands.
Rita J M Volkers, Laboratory of Nematology, Wageningen University, NL-6708 PB Wageningen, The Netherlands.
Joost Riksen, Laboratory of Nematology, Wageningen University, NL-6708 PB Wageningen, The Netherlands.
Philip C Rosenstiel, Institute for Clinical Molecular Biology, University of Kiel, 24098 Kiel, Germany; Competence Centre for Genomic Analysis (CCGA) Kiel, University of Kiel, 24098 Kiel, Germany.
Hinrich Schulenburg, Zoological Institute, University of Kiel, 24098 Kiel, Germany; Max Planck Institute for Evolutionary Biology, 24306 Ploen, Germany.
Jan E Kammenga, Laboratory of Nematology, Wageningen University, NL-6708 PB Wageningen, The Netherlands.
Literature cited
- Albert FW, Bloom JS, Siegel J, Day L, Kruglyak L. 2018. Genetics of trans-regulatory variation in gene expression. Elife. 7:e35471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Bloom JS, Gerke JP, Kruglyak L. 2014. A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLoS Genet. 10:e1004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E, Burga A, Kruglyak L. 2017. A maternal-effect selfish genetic element in Caenorhabditis elegans. Science. 356:1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A, Pitts J, Rockman MV, Chen WC, Tan MW, et al. 2012. Long-range regulatory polymorphisms affecting a GABA receptor constitute a quantitative trait locus (QTL) for social behavior in Caenorhabditis elegans. PLoS Genet. 8:e1003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A, Tsunozaki M, Rockman MV, Kruglyak L, Bargmann CI.. 2011. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature. 472:313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SC, Zdraljevic S, Bisaga KW, Tanny RE, Cook DE, et al. 2019. A novel gene underlies bleomycin-response variation in Caenorhabditis elegans. Genetics. 212:1453–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Zdraljevic S, Tanny RE, Seo B, Riccardi DD, et al. 2016. The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics. 204:371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, Stegle O, Grondin C, Canut M, Tisne S, et al. 2014. Extensive cis-regulatory variation robust to environmental perturbation in Arabidopsis. Plant Cell. 26:4298–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning DJ, McIntyre LM. 2017. Back to the future: multiparent populations provide the key to unlocking the genetic basis of complex traits. Genetics. 206:527–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroszuk A, Snoek LB, Fradin E, Riksen J, Kammenga J. 2009. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res. 37:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KS, van Wijk MH, McGrath PT, Andersen EC, Sterken MG. 2021. From QTL to gene: C. elegans facilitates discoveries of the genetic mechanisms underlying natural variation. Trends Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi M, Lehner B. 2014. The effects of genetic variation on gene expression dynamics during development. Nature. 505:208–211. [DOI] [PubMed] [Google Scholar]
- Gao AW, Sterken MG, Uit de Bos J, van Creij J, Kamble R, et al. 2018. Natural genetic variation in C. elegans identified genomic loci controlling metabolite levels. Genome Res. 28:1296–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Andersen EC, Shapiro JA, Gerke JP, Kruglyak L. 2012. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science. 335:574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour AR. 2019. Average InformationREML: an efficient algorithm for variance parameter estimation in linear mixed models. J Anim Breed Genet. 136: 262–272.31247685 [Google Scholar]
- Gloria-Soria A, Azevedo RB. 2008. npr-1 regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr Biol. 18:1694–1699. [DOI] [PubMed] [Google Scholar]
- Greene JS, Brown M, Dobosiewicz M, Ishida IG, Macosko EZ, et al. 2016. Balancing selection shapes density-dependent foraging behaviour. Nature. 539:254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnel SR, Zdraljevic S, Rodriguez BC, Zhao Y, McGrath PT, et al. 2018. Extreme allelic heterogeneity at a Caenorhabditis elegans beta-tubulin locus explains natural resistance to benzimidazoles. PLoS Pathog. 14:e1007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartanto M, Joosen RVL, Snoek BL, Willems LAJ, Sterken MG, et al. 2020. Network analysis prioritizes DEWAX and ICE1 as the candidate genes for two major eQTL hotspots in seed germination. BioRxiv. [DOI] [PMC free article] [PubMed]
- Jimenez-Gomez JM, Wallace AD, Maloof JN. 2010. Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genet. 6:e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic K, Grilli J, Sterken MG, Snoek BL, Riksen JAG, et al. 2019. Transcriptome resilience predicts thermotolerance in Caenorhabditis elegans. BMC Biol. 17:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic K, Sterken MG, Grilli J, Bevers RPJ, Rodriguez M, et al. 2017. Temporal dynamics of gene expression in heat-stressed Caenorhabditis elegans. PLoS One. 12:e0189445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamkina P, Snoek LB, Grossmann J, Volkers RJ, Sterken MG, et al. 2016. Natural genetic variation differentially affects the proteome and transcriptome in Caenorhabditis elegans. Mol Cell Proteomics. 15:1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga JE, Doroszuk A, Riksen JA, Hazendonk E, Spiridon L, et al. 2007. A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 3:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, et al. 2008. Efficient control of population structure in model organism association mapping. Genetics. 178:1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, Terpstra IR, Garcia JM, van den Ackerveken G, et al. 2007. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci U S A. 104:1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EG, Merkes CM, McNeil CL, Hoofer SR, Sen S, et al. 2012. Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22:1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, et al. 2009. A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5:e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, et al. 2015. Marker-based estimation of heritability in immortal populations. Genetics. 199:379–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Large EE, Xu W, Zhao Y, Brady SC, Long L, et al. 2016. Selection on a subunit of the NURF chromatin remodeler modifies life history traits in a domesticated strain of Caenorhabditis elegans. PLoS Genet. 12:e1006219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Zdraljevic S, Stevens L, Wang Y, Tanny RE, et al. 2021. Balancing selection maintains hyper-divergent haplotypes in Caenorhabditis elegans. Nat Ecol Evol. 5:794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. ; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, et al. 2006. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2:e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Breitling R, Snoek LB, van der Velde KJ, Swertz MA, et al. 2010. Global genetic robustness of the alternative splicing machinery in Caenorhabditis elegans. Genetics. 186:405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, et al. 2009. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 61:692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakad R, Snoek LB, Yang W, Ellendt S, Schneider F, et al. 2016. Contrasting invertebrate immune defense behaviors caused by a single gene, the Caenorhabditis elegans neuropeptide receptor gene npr-1. BMC Genomics. 17:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijveen H, Ligterink W, Keurentjes JJ, Loudet O, Long J, et al. 2017. AraQTL—workbench and archive for systems genetics in Arabidopsis thaliana. Plant J. 89:1225–1235. [DOI] [PubMed] [Google Scholar]
- Noble LM, Chang AS, McNelis D, Kramer M, Yen M, et al. 2015. Natural variation in plep-1 causes male-male copulatory behavior in C. elegans. Curr Biol. 25:2730–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble LM, Chelo I, Guzella T, Afonso B, Riccardi DD, et al. 2017. Polygenicity and epistasis underlie fitness-proximal traits in the Caenorhabditis elegans Multiparental Experimental Evolution (CeMEE) panel. Genetics. 207:1663–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell MP, Chao PH, Kammenga JE, Sengupta P. 2018. Rictor/TORC2 mediates gut-to-brain signaling in the regulation of phenotypic plasticity in C. elegans. PLoS Genet. 14:e1007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, et al. 2008. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 454:1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/. [Google Scholar]
- Ranjan A, Budke JM, Rowland SD, Chitwood DH, Kumar R, et al. 2016. eQTL regulating transcript levels associated with diverse biological processes in tomato. Plant Physiol. 172:328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, Kim DH. 2009. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 323:382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner DJ, Ailion M, Thomas JH, Meyer BJ. 2008. C. elegans anaplastic lymphoma kinase ortholog SCD-2 controls dauer formation by modulating TGF-beta signaling. Curr Biol. 18:1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5:e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Skrovanek SS, Kruglyak L. 2010. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 330:372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Snoek LB, Riksen JA, Bevers RP, Kammenga JE. 2012. Genetic variation for stress-response hormesis in C. elegans lifespan. Exp Gerontol. 47:581–587. [DOI] [PubMed] [Google Scholar]
- Rogers C, Persson A, Cheung B, de Bono M. 2006. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol. 16:649–659. [DOI] [PubMed] [Google Scholar]
- Schmid T, Snoek LB, Frohli E, van der Bent ML, Kammenga J, et al. 2015. Systemic regulation of RAS/MAPK signaling by the serotonin metabolite 5-HIAA. PLoS Genet. 11:e1005236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, et al. 2011. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 9:e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L. 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 319:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KD, Roschitzki B, Snoek LB, Grossmann J, Zheng X, et al. 2016. Natural genetic variation influences protein abundances in C. elegans developmental signalling pathways. PLoS One. 11:e0149418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek BL, Sterken MG, Bevers RPJ, Volkers RJM, Van't Hof A, et al. 2017. Contribution of trans regulatory eQTL to cryptic genetic variation in C. elegans. BMC Genomics. 18:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek BL, Sterken MG, Hartanto M, van Zuilichem AJ, Kammenga JE, et al. 2020. WormQTL2: an interactive platform for systems genetics in Caenorhabditis elegans. Database. 2020:baz1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek BL, Volkers RJM, Nijveen H, Petersen C, Dirksen P, et al. 2019. A multi-parent recombinant inbred line population of C. elegans allows identification of novel QTLs for complex life history traits. BMC Biol. 17:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Joeri van der Velde K, Li Y, Jansen RC, Swertz MA, et al. 2014a. Worm variation made accessible: take your shopping cart to store, link, and investigate! Worm. 3:e28357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Orbidans HE, Stastna JJ, Aartse A, Rodriguez M, et al. 2014b. Widespread genomic incompatibilities in Caenorhabditis elegans. G3 (Bethesda). 4:1813–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Sterken MG, Volkers RJ, Klatter M, Bosman KJ, et al. 2014c. A rapid and massive gene expression shift marking adolescent transition in C. elegans. Sci Rep. 4:3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Terpstra IR, Dekter R, Van den Ackerveken G, Peeters AJ. 2012. Genetical genomics reveals large scale genotype-by-environment interactions in Arabidopsis thaliana. Front Genet. 3:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Van der Velde KJ, Arends D, Li Y, Beyer A, et al. 2013. WormQTL–public archive and analysis web portal for natural variation data in Caenorhabditis spp. Nucleic Acids Res. 41:D738–D743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. 2012. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 91:1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stastna JJ, Snoek LB, Kammenga JE, Harvey SC. 2015. Genotype-dependent lifespan effects in peptone deprived Caenorhabditis elegans. Sci Rep. 5:16259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken MG, Bevers RPJ, Volkers RJM, Riksen JAG, Kammenga JE, et al. 2019. Dissecting the eQTL micro-architecture in Caenorhabditis elegans. BioRxiv. [DOI] [PMC free article] [PubMed]
- Sterken MG, Snoek LB, Kammenga JE, Andersen EC. 2015. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 31:224–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken MG, van Bemmelen van der Plaat L, Riksen JAG, Rodriguez M, Schmid T, et al. 2017. Ras/MAPK modifier loci revealed by eQTL in Caenorhabditis elegans. G3 (Bethesda). 7:3185–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken MG, van Sluijs L, Wang YA, Ritmahan W, Gultom ML, et al. 2021. Punctuated loci on chromosome IV determine natural variation in orsay virus susceptibility of Caenorhabditis elegans strains Bristol N2 and Hawaiian CB4856. J Virol. 95:e02430-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson OA, Snoek LB, Nijveen H, Sterken MG, Volkers RJ, et al. 2015. Remarkably divergent regions punctuate the genome assembly of the Caenorhabditis elegans Hawaiian strain CB4856. Genetics. 200:975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Okihara KL, Thijssen K, Plasterk RH. 2002. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr Biol. 12:1535–1540. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuela A, Snoek LB, Riksen JA, Kammenga JE. 2010. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 20:929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuela A, Snoek LB, Riksen JA, Kammenga JE. 2012. Aging uncouples heritability and expression-QTL in Caenorhabditis elegans. G3 (Bethesda). 2:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkers RJ, Snoek LB, Hubar CJ, Coopman R, Chen W, et al. 2013. Gene-environment and protein-degradation signatures characterize genomic and phenotypic diversity in wild Caenorhabditis elegans populations. BMC Biol. 11:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, et al. 2007. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 175:1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2009. GGplot2: elegant graphics for data analysis.
- Zdraljevic S, Fox BW, Strand C, Panda O, Tenjo FJ, et al. 2019. Natural variation in C. elegans arsenic toxicity is explained by differences in branched chain amino acid metabolism. Elife. 8:e40260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdraljevic S, Strand C, Seidel HS, Cook DE, Doench JG, et al. 2017. Natural variation in a single amino acid substitution underlies physiological responses to topoisomerase II poisons. PLoS Genet. 13:e1006891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zych K, Snoek BL, Elvin M, Rodriguez M, Van der Velde KJ, et al. 2017. reGenotyper: detecting mislabeled samples in genetic data. PLoS One. 12:e0171324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article available in Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) and can be accessed with ID PRJNA495983 and in WormQTL2 (http://www.bioinformatics.nl/EleQTL; Snoek et al. 2020).
Supplementary material is available at G3 online.