Abstract
The prevalence of penicillin-resistant Streptococcus pneumoniae in Thailand has dramatically increased over the last decade. During a national survey, which was conducted from 1992 to 1994, 37.2% of the pneumococci isolated from the nasopharynges of children with acute respiratory tract infections were penicillin resistant (MIC, ≥0.1 μg/ml). In order to investigate the prevalence and clonal relatedness of nasopharyngeal carriage of penicillin-resistant S. pneumoniae in Thailand, a molecular epidemiological survey was undertaken. To this end, 53 penicillin-resistant pneumococcal isolates from children who suffered from acute respiratory tract infections and who originated from five distinct regions of the country were characterized in detail. DNA fingerprint analysis demonstrated 13 clusters, i.e., genotypes shared by two or more strains, and 14 unique genotypes. The cluster size varied from 2 (nine clusters) to 11 strains (one cluster). Six of the 13 restriction fragment end labeling clusters consisted of two or more distinct serotypes, indicating frequent horizontal transfer of capsular genes. Geographical distribution of the genotypes among the five regions of Thailand demonstrated that only four genetic clusters were restricted to single areas of the country, whereas the other nine clusters represented isolates collected in two or more districts. These observations demonstrate that the majority of the genetic clusters are spread throughout the country. The most predominant genetic cluster, representing 21% of the isolates, was identical to the Spanish pandemic clone 23F. In addition, the second largest cluster matched the Spanish-French international clone 9V. These data indicate that the genetic clones 23F and 9V, which are widely spread throughout the world, are the most predominant multidrug-resistant pneumococcal clones in Thailand. Therefore, we conclude that these pandemic clones are primarily responsible for the increase in the prevalence of pneumococcal penicillin resistance in Thailand.
Streptococcus pneumoniae is one of the leading bacterial pathogens causing illness and death among young children, the elderly, and persons with underlying medical conditions, such as immunocompromised patients (5). Pneumococcal infection is usually preceded by colonization of the human nasopharynx. This is an important step toward infection. Consequently, pneumococcal colonization is an important risk factor for developing disease. For instance, young children who are frequently colonized with pneumococci more often develop acute otitis media than do children who are not or less frequently colonized (12, 19, 33, 43).
The prevalence of penicillin resistance among pneumococci, both high-level resistance (MIC of >1 μg/ml) and intermediate-level resistance (MIC of 0.1 to 1 μg/ml), is alarmingly increasing worldwide (4, 8, 14, 21–24, 42). International spread of a restricted number of multiresistant pneumococcal clones has significantly contributed to this increase. Soares and coworkers have documented the spread of a multiresistant clone of serotype 6B from Spain to Iceland in the late 1980s (35). This has resulted in an epidemic of this clone, which was isolated with a frequency of up to 12% as early as 1992 (22). In 1991, Munoz and colleagues reported evidence for the intercontinental spread of a multiresistant clone of S. pneumoniae serotype 23F from Spain to the United States (26). This clone has subsequently disseminated throughout the United States (25). Finally, Gasc and colleagues in 1995 reported the spread of a multiresistant pneumococcal clone of serogroup 9 from Spain to France (13). The international clones 23F and 9V have been identified in many countries all over the world (16, 30). Besides the international spread of the clones 6B, 23F, and 9V, novel penicillin-resistant and multiresistant clones in the former Czechoslovakia, Spain, Japan, and South Africa that tend to spread in an epidemic manner within these countries have been reported (6, 30, 31, 34, 41).
In Thailand, a national survey conducted from 1992 to 1994 demonstrated a prevalence of penicillin-resistant pneumococci (MIC of ≥0.1 μg/ml) as high as 37.2% (1, 29, 37). This figure was much higher than those in the surveys in 1978 (6.7%) (36) and in 1987 (10.6%) (20). In order to identify the nature of the increase in the prevalence of penicillin-resistant pneumococci in Thailand, a molecular epidemiological study was undertaken. To this end, strains isolated from the nasopharynges of 53 pediatric patients, who suffered from acute respiratory tract infections and who originated from different regions across the country, were examined. The pneumococcal isolates were characterized by restriction fragment end labeling (RFEL) analysis, BOX PCR typing, penicillin-binding protein (PBP) genotyping, serotyping, and drug susceptibility testing.
MATERIALS AND METHODS
Bacterial strains.
In the survey conducted in 1978, susceptibility testing was performed with 446 isolates. One hundred seventeen isolates were collected in Bangkok from patients who were admitted to three university hospitals (Siriraj Hospital, Chulalongkorn Hospital, and Ramathibodi Hospital) and a private hospital (Seventh Day Adventist Hospital). These pneumococci were collected mainly from blood (26%), throat swab (23%), sputum (17%), cerebrospinal fluid (10%), and pus (7%) samples. In addition, 329 isolates were obtained from healthy carriers (ranging from 3 to 45 years of age) in Bangkok (80%) and Samutprakarn (20%), a province near Bangkok. The survey conducted in 1986 comprised 95 pneumococcal isolates from patients who were admitted to the Siriraj University Hospital in Bangkok. The isolates were collected mainly from blood (57%), sputum (20%), pus (12%), cerebrospinal fluid (4%), and pleural fluid (4%) samples.
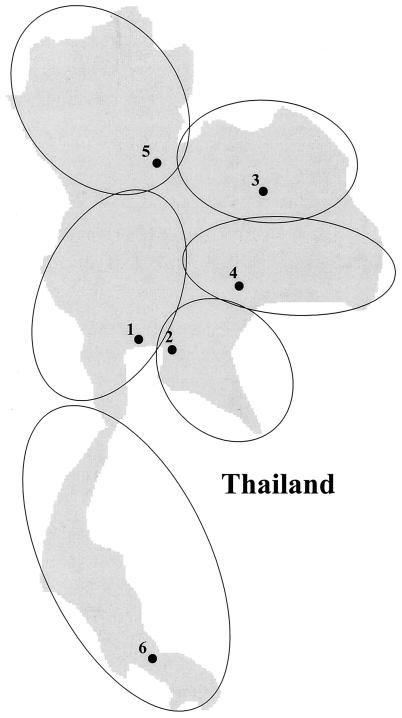
During a national survey of antimicrobial resistance in Thailand from 1992 to 1994 (1, 29, 37), nasopharyngeal swabs were taken from 1,783 children under the age of 5 years with acute respiratory infections. The children were hospitalized in six hospitals representing five regions of Thailand (Fig. 1): Children’s Hospital Bangkok (central region), Cholburi Hospital (eastern region), Khonkaen Hospital and Maharaj Nakorn Ratchasima Hospital (Northeastern region), Phra Buddha Chinaraj Hospital (northern region), and Hatyai Hospital (southern region). S. pneumoniae species identification was determined by optochin sensitivity and bile solubility testing (27). In this survey, 615 pneumococci were isolated (isolation rate, 34.5%). All 615 strains were referred to our laboratory, and 530 strains were viable at the time of arrival. The antibiotic resistance profile of these 530 strains was determined (see below). The collection represented 197 penicillin-resistant pneumococcal isolates (MIC of ≥0.1 μg/ml). Twenty-six of 36 high-level penicillin-resistant pneumococcal isolates (MIC of >1.0 μg/ml) and 27 of 161 intermediate-level-resistant pneumococci (MIC of 0.1 to 1 μg/ml) were randomly selected for this study (Table 1).
FIG. 1.
Geographical map of Thailand. The medical centers collaborating in this study are indicated by numbers: 1, Children’s Hospital Bangkok (central region); 2, Cholburi Hospital (eastern region); 3, Khonkaen Hospital (northeastern region); 4, Maharaj Nakorn Ratchasima Hospital (northeastern region); 5, Phra Buddha Chinaraj Hospital (northern region); 6, Hatyai Hospital (southern region). The geographical regions serviced by the medical centers are depicted by open circles.
TABLE 1.
Prevalence of penicillin-resistant pneumococci among 1,783 children with acute respiratory tract infectionsa
| Hospital | No. of pneumo-coccal isolates | HPRP
|
IPRP
|
% Penicillin-resistant pneumococci (HPRP plus IPRP) | ||||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. randomly selected for this study | No. | % | No. randomly selected for this study | |||
| B | 216 | 15 | 6.9 | 7 | 82 | 38.0 | 16 | 44.9 |
| C | 44 | 2 | 4.5 | 2 | 16 | 36.4 | 4 | 40.9 |
| K | 53 | 1 | 1.9 | 1 | 10 | 18.9 | 3 | 20.8 |
| N | 86 | 3 | 3.5 | 2 | 25 | 29.1 | 2 | 32.6 |
| P | 49 | 14 | 28.6 | 13 | 15 | 30.6 | 2 | 59.2 |
| S | 82 | 1 | 1.2 | 1 | 13 | 15.9 | 0 | 17.1 |
| Total | 530 | 36 | 6.8 | 26 | 161 | 30.4 | 27 | 37.2 |
Data were collected during the National Pneumococcal Antimicrobial Resistance Surveillance between 1992 and 1994. From this collection, 26 high-level and 27 intermediate-level penicillin-resistant isolates were randomly selected for detailed molecular epidemiological analysis. Abbreviations: B, Children’s Hospital Bangkok; C, Cholburi Hospital; K, Khonkaen Hospital; N, Maharaj Nakorn Ratchasima Hospital; P, Phra Buddha Chinaraj Hospital; S, Hatyai Hospital; HPRP, high-level penicillin-resistant pneumococci; IPRP, intermediate-level penicillin-resistant pneumococci.
The MICs for the pneumococcal strains were determined by agar dilution. The MIC was defined as the lowest concentration of the antimicrobial agent preventing visible growth. To this end, serial log2 concentrations of antibiotics were prepared in IsoSensitest agar (Oxoid, Unipath Ltd., Basingstoke, United Kingdom), supplemented with 5% horse blood. The pneumococcal isolates were removed from storage at −70°C and subcultured at 37°C on Columbia agar (Oxoid) supplemented with 5% sheep blood with 5% CO2. Bacterial suspensions were prepared in 0.9% NaCl from 24-h agar cultures and adjusted to a McFarland turbidity standard of 0.5. Suspensions were further diluted (1:10) in saline. The inocula were applied on the test plates with a multipoint inoculator, resulting in about 104 CFU per spot. MICs were read after 24 h of incubation at 37°C with 5% CO2.
The antimicrobial agents tested were penicillin G (Sigma Chemical Co., St. Louis, Mo.), erythromycin (Abbott Laboratories, Ltd., Queenborough, Kent, United Kingdom), doxycycline (Pfizer S. A., Brussels, Belgium), vancomycin (Eli Lilly & Co., Indianapolis, Ind.), rifampin (Sigma), cotrimoxazole, the combination (1:19) of trimethoprim (Sigma) and sulfamethoxazole (Sigma), and ciprofloxacin (Bayer, Wuppertal, Germany). Breakpoints of the antibiotics to discriminate between susceptible and nonsusceptible strains were used according to the National Committee for Clinical Laboratory Standards guidelines for susceptibility testing (28).
Bacterial typing. (i) Serotyping.
Pneumococci were serotyped on the basis of capsular swelling (quellung reaction) observed microscopically after suspension in antisera prepared at Statens Seruminstitut, Copenhagen, Denmark (11).
(ii) RFEL analysis.
Typing of pneumococcal strains by RFEL was performed as described by Van Steenbergen et al. (40) and adapted by Hermans et al. (18). Briefly, purified pneumococcal DNA was digested by the restriction enzyme EcoRI. The DNA restriction fragments were end labeled at 72°C with [α-32P]dATP with Taq DNA polymerase (Goldstar; Eurogentec, Seraing, Belgium). The radiolabeled fragments were denatured and separated electrophoretically on a 6% polyacrylamide sequencing gel containing 8 M urea. Subsequently, the gel was transferred onto filter paper, vacuum dried (Haake Buchler Instruments Inc., Saddlebrook, N.Y.), and exposed for varying periods at room temperature to ECL Hyperfilms (Amersham, Little Chalfont, Buckinghamshire, United Kingdom).
The RFEL types were analyzed with the Windows version of the Gelcompar software version 4 (Applied Maths, Kortrijk, Belgium) after the RFEL autoradiograms were scanned with Image Master DTS (Pharmacia Biotech, Uppsala, Sweden). For this purpose, the DNA fragments in the molecular size range of 160 to 400 bp were examined. The fingerprints were normalized with pneumococcus-specific bands present in the RFEL banding patterns of all strains. Comparison of the fingerprints was performed by the unweighted pair group method using arithmetic averages and by using the Jaccard similarity coefficient applied to peaks. Computer-assisted analysis, methods, and algorithms used in this study were carried out or used according to the instructions of the manufacturer of Gelcompar. A tolerance of 1.5% in band positions was applied during comparison of the fingerprint patterns. Identical DNA types were arbitrarily defined as RFEL homologies higher than 95%. A genetic cluster was defined as a genotype (RFEL or PBP type) that was shared by two or more pneumococcal strains. The degree of genetic clustering was defined as the percentage of strains displaying genotypes (RFEL or PBP types) that were observed twice or more.
(iii) BOX PCR typing.
BOX PCR typing was carried out as described before (39). Briefly, 50 ng of pneumococcal DNA was amplified by PCR (4 min at 94°C [predenaturation]; 40 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 74°C; and 2 min at 74°C [extension]), with primer BOX-A, designed on the primary structure of the BOX repeat motif. The amplified products were separated on a 1.5% agarose gel. Gels were stained with ethidium bromide, after which the banding patterns were evaluated visually. BOX PCR patterns showing a single band difference were defined as nonidentical types. BOX PCR was used as a confirmatory method to verify genetic clustering observed by RFEL analysis.
(iv) PBP genotyping.
Genetic polymorphism of the penicillin resistance genes pbp1a, pbp2b, and pbp2x was investigated by restriction fragment length polymorphism analysis. To this end, we amplified the genes by PCR and analyzed the digested DNA products by agarose gel electrophoresis. PCR amplification of the PBP-encoding genes was performed in a 50-μl PCR buffer system containing 75 mM Tris-HCl (pH 9.0), 20 mM (NH4)2SO4, 0.01% (wt/vol) Tween 20, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, 10 pmol of the individual primers, 0.5 U of Goldstar Taq DNA polymerase (Eurogentec, Liège, Belgium), and 10 ng of purified chromosomal DNA. Cycling was performed in a PTC-100 programmable thermal controller (MJ Research, Watertown, Mass.) and consisted of the following steps: predenaturation at 94°C for 1 min; 30 cycles of 1 min at 94°C, 1 min at 52°C, and 2 min at 72°C; and final extension at 72°C for 3 min. The primers used to amplify the genes pbp1a, pbp2b, and pbp2x were described previously (7, 10, 26). The amplification products (5 μl) were digested by restriction endonuclease HinfI and separated by electrophoresis in 2.5% agarose gels (5 mm thick; Agarose MP; Boehringer Mannheim, Almere, The Netherlands) containing 0.5× Tris-borate-EDTA and 0.1 μg of ethidium bromide per ml. Gels were run in 0.5× Tris-borate-EDTA containing 0.1 μg of ethidium bromide per ml at a constant current of 20 mA for 4 h. Prior to electrophoresis, samples were mixed with a 5× concentrated layer mix consisting of 50% glycerol in water and 0.8 mg of bromophenol blue per ml. Gels were photographed with a Polaroid MP4 Land camera and Polaroid 667 films. The different PBP genotypes are represented by a three-number code (e.g., 1-6-13), referring to the restriction fragment length polymorphism patterns of the genes pbp1a (pattern 1), pbp2b (pattern 6), and pbp2x (pattern 13), respectively.
RESULTS
From 1992 to 1994, the prevalence of penicillin-resistant pneumococci in Thailand was determined during a national antimicrobial resistance survey. This study showed a country-wide penicillin resistance rate (MIC of ≥0.1 μg/ml) of 37.2%; 6.8% of the pneumococcal isolates showed high-level resistance to penicillin, whereas 30.4% of the isolates were intermediate-level resistant (Table 1). The resistance rates varied from region to region, ranging from 17.1% in Hatyai Hospital (southern region) to 59.2% in Phra Buddha Chinaraj Hospital (northern region). The latter region had the highest prevalence of high-level pneumococcal penicillin resistance (28.6%), followed by the Children’s Hospital Bangkok (central region; 6.9%). Twenty-three of the 26 randomly selected high-level penicillin-resistant isolates and 14 of the 27 intermediate-level penicillin-resistant isolates were resistant to more than three antibiotics with resistance profile penicillin–trimethoprim-sul-famethoxazole–doxycycline–erythromycin–chloramphenicol, penicillin–trimethoprim-sulfamethoxazole–doxycycline–eryth-romycin, or penicillin–trimethoprim-sulfamethoxazole–doxycycline–chloramphenicol. None of the isolates were resistant to vancomycin, rifampin, or ciprofloxacin.
All 53 penicillin-resistant pneumococcal isolates were analyzed by RFEL. This DNA fingerprint method divided the strains into 27 distinct types (Fig. 2 and Table 1). These RFEL types represented 13 clusters, i.e., types shared by two or more strains, and 14 unique types. Thirty-nine strains shared RFEL types with at least one other strain (74%). The cluster size varied from 2 (nine clusters) to 11 strains (one cluster). In addition, two clusters of three strains and one cluster of four strains were observed. For 10 RFEL clusters, genetic identity was confirmed by BOX PCR typing. Within the remaining three RFEL clusters, strong genetic relatedness was observed by BOX PCR typing (data not shown). Although the degree of genetic clustering, i.e., the percentage of strains sharing their RFEL types with one or more other strains, was comparable between the groups of high-level and intermediate-level penicillin-resistant strains (high level, 50%; intermediate level, 48%), a reduced number of RFEL types was observed among the high-level penicillin-resistant isolates (15 high-level penicillin resistance RFEL types and 20 intermediate-level penicillin resistance RFEL types [Fig. 2]).
FIG. 2.
Genetic relatedness of 53 penicillin-resistant pneumococcal isolates from Thailand. The RFEL fingerprint patterns of the strains (designated by numbers) and their genetic relatedness (dendrograph) are depicted (for details, see text). High-level penicillin-resistant isolates are marked in boldface.
Comparison of the 27 Thai RFEL types with 107 additional types present in the international RFEL data library and representing 15 other countries (15, 16) revealed that the most predominant cluster matched the Spanish pandemic clone 23F (RFEL type 15), whereas RFEL cluster 23, representing four 9V strains, was identical to the Spanish-French international clone 9V, respectively. Strain P390, displaying RFEL type 17, was genetically identical to a penicillin-resistant pneumococcal strain isolated in The Netherlands. In addition, the remaining 24 Thai RFEL types did not match any of the 107 types present in the international library.
The 53 pneumococcal isolates comprised eight distinct serotypes. In addition, one strain displayed a nontypeable capsular type. The capsular types of the high-level-resistant isolates were restricted to 6A, 6B, 9V, 19F, and 23F. The intermediate-level penicillin-resistant pneumococci harbored the serotypes 6A, 6B, 9V, 14, 15B, 15C, 19F, and 23F (Table 2). Six of the 13 RFEL clusters consisted of two or more distinct serotypes. The most predominant RFEL cluster, 15, consisting of 11 strains, harbored three distinct serotypes, 23F, 19F, and 14 (Table 2). Based on the serotype distribution of the 53 penicillin-resistant pneumococci, 89% of the strains display capsular types that are covered by the 7-valent conjugate vaccine in which the serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F are represented.
TABLE 2.
Microbiological parameters of 53 penicillin-resistant pneumococcal isolates and demographic data of the patients
| Strain | Hospitala | RFEL type | PBP genotype
|
Sero-type | Penicillin MICd | Resistance profileb | ||
|---|---|---|---|---|---|---|---|---|
| 1a | 2b | 2x | ||||||
| P410 | P | 39 | 01 | 06 | 13 | 6B | 2.000 | PTDEC |
| P550 | C | 39 | 02 | 04 | 41 | 15B | 0.500 | P |
| P320 | B | 38 | 16 | 01 | 13 | 6B | 1.000 | PTDE |
| P386 | P | 38 | 01 | 01 | 01 | 23F | 2.000 | PTDEC |
| P042 | P | 42 | 02 | 02 | 34 | 19F | 0.125 | PTDEC |
| P519 | B | 42 | 11 | 02 | 30 | 19F | 0.250 | PDE |
| P390 | B | 17 | 01 | 10 | 13 | 6B | 1.000 | PTDEC |
| P276 | C | 40 | 14 | 02 | 38 | 14 | 0.250 | PDC |
| P146 | C | 41 | 16 | 01 | 13 | 6B | 2.000 | PTDE |
| P318 | B | 92 | 15 | 02 | 39 | 14 | 0.125 | PT |
| P218 | N | 35 | 02 | 04 | 13 | 15C | 0.250 | PC |
| P520 | B | 35 | 02 | 04 | 41 | 15B | 0.250 | PTDE |
| P471 | B | 50 | 00 | 12 | 01 | 23F | 1.000 | PTDE |
| P179 | K | 51 | 02 | 05 | 36 | 14 | 0.250 | P |
| P356 | B | 51 | 01 | 10 | 13 | 6B | 1.000 | PTC |
| P246 | P | 51 | 02 | 05 | 36 | 14 | 0.250 | P |
| P034 | C | 58 | 12 | 09 | 01 | NTc | 0.500 | PTDC |
| P349 | N | 15 | 14 | 02 | 38 | 14 | 0.125 | PDC |
| P077 | P | 15 | 01 | 01 | 01 | 23F | 2.000 | PTDEC |
| P053 | P | 15 | 01 | 01 | 01 | 23F | 2.000 | PTDEC |
| P176 | B | 15 | 01 | 01 | 01 | 19F | 2.000 | PTDEC |
| P193 | S | 15 | 01 | 01 | 01 | 23F | 2.000 | PTDEC |
| P462 | B | 15 | 01 | 01 | 01 | 19F | 2.000 | PTDEC |
| P192 | P | 15 | 01 | 01 | 01 | 23F | 2.000 | PTDEC |
| P210 | P | 15 | 01 | 01 | 01 | 23F | 2.000 | PTDEC |
| P469 | B | 15 | 01 | 01 | 01 | 19F | 2.000 | PTDC |
| P477 | B | 15 | 01 | 01 | 01 | 23F | 2.000 | PTDEC |
| P386 | P | 15 | 01 | 01 | 01 | 23F | 2.000 | PTDEC |
| P537 | B | 48 | 01 | 01 | 01 | 9V | 1.000 | PTDE |
| P476 | B | 68 | 01 | 03 | 13 | 6B | 1.000 | PTDEC |
| P499 | B | 68 | 01 | 03 | 13 | 6B | 1.000 | PTDEC |
| P403 | C | 70 | 01 | 10 | 13 | 6A | 1.000 | PTDEC |
| P404 | C | 70 | 01 | 10 | 13 | 6A | 2.000 | PTDEC |
| P329 | B | 69 | 17 | 03 | 40 | 19F | 0.500 | PDE |
| P385 | K | 69 | 02 | 06 | 36 | 14 | 0.125 | PT |
| P441 | B | 67 | 01 | 03 | 13 | 6B | 1.000 | PTDEC |
| P375 | B | 67 | 01 | 10 | 13 | 6B | 2.000 | PTDEC |
| P174 | B | 66 | 01 | 10 | 13 | 6B | 2.000 | PTDEC |
| P458 | B | 66 | 01 | 10 | 13 | 6B | 1.000 | PTDEC |
| P086 | P | 59 | 01 | 03 | 13 | 6B | 2.000 | PTDE |
| P228 | B | 59 | 01 | 10 | 13 | 6B | 1.000 | PTDEC |
| P122 | B | 59 | 01 | 10 | 13 | 6B | 0.060 | PTDEC |
| P216 | N | 62 | 01 | 10 | 13 | 6B | 2.000 | PTDEC |
| P244 | P | 78 | 01 | 06 | 13 | 6B | 2.000 | PTDEC |
| P397 | B | 79 | 15 | 02 | 39 | 14 | 0.250 | PT |
| P411 | P | 74 | 01 | 06 | 13 | 6B | 2.000 | PTDEC |
| P543 | P | 75 | 01 | 06 | 13 | 6B | 2.000 | PTDEC |
| P409 | P | 76 | 01 | 06 | 13 | 6B | 2.000 | PTDEC |
| P304 | B | 77 | 01 | 03 | 13 | 6B | 2.000 | PTDEC |
| P165 | P | 23 | 01 | 01 | 01 | 9V | 2.000 | PT |
| P353 | N | 23 | 01 | 01 | 01 | 9V | 2.000 | PT |
| P117 | K | 23 | 01 | 01 | 01 | 9V | 1.000 | PT |
| P384 | K | 23 | 01 | 01 | 01 | 9V | 2.000 | PT |
B, Children’s Hospital Bangkok; C, Cholburi Hospital; K, Khonkaen Hospital; N, Maharaj Nakorn Ratchasima Hospital; P, Phra Buddha Chinaraj Hospital; S, Hatyai Hospital.
Resistance profile: P, penicillin; T, trimethoprim-sulfamethoxazole; D, doxycycline, E, erythromycin; C, chloramphenicol.
NT, nontypeable.
MICs are in micrograms per milliliter.
The 53 penicillin-resistant pneumococcal strains were collected in six hospitals representing five regions of the country: Children’s Hospital Bangkok (central region; n = 23), Cholburi Hospital (eastern region; n = 6), Khonkaen Hospital and Maharaj Nakorn Ratchasima Hospital (northeastern region; n = 4 and 4, respectively), Phra Buddha Chinaraj Hospital (northern region; n = 15), and Hatyai Hospital (southern region; n = 1). The RFEL clusters 66 (n = 2), 67 (n = 2), 68 (n = 2), and 70 (n = 2) were observed in single regions of the country, whereas the other nine clusters represented isolates collected in two or more regions (Table 2).
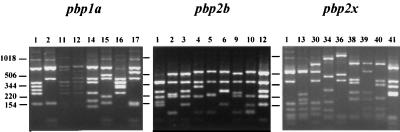
All strains were analyzed by PBP genotyping. Sixteen distinct PBP genotypes were observed. The representative types are summarized in Fig. 3. The PBP types 1-1-1 and 1-10-13 were the most predominant penicillin resistance types and represented 16 and 10 strains, respectively. PBP type 1-1-1 was observed in four distinct RFEL types representing five distinct regions, whereas PBP type 1-10-13 was displayed by seven different RFEL types representing three distinct regions. Fourteen PBP type 1-1-1 strains (88%) and four PBP type 1-10-13 strains (40%) were high-level penicillin resistant (Table 2). The PBP types 1-1-1 and 1-10-13 were observed in 11 and 4 of the 16 countries that are currently represented in the international database (15). Twelve of the 16 PBP genotypes were Thailand specific, as they were not observed so far in any of the other countries representing 80 distinct PBP genotypes (15). The pbp2x type 13 was observed in 23 strains isolated in four distinct regions of the country and represented 17 RFEL types (Table 2). The vast majority of the 23 pbp2x type 13 strains displayed serotype 6B (n = 20). Comparison of the PBP genotypes of the individual genes pbp1a, pbp2b, and pbp2x between the 53 Thai penicillin-resistant strains and 185 Dutch and 10 Thai penicillin-susceptible pneumococcal isolates (17, 32) did not demonstrate any overlap (data not shown).
FIG. 3.
DNA fingerprint patterns of the pbp1a (n = 8), pbp2b (n = 9), and pbp2x (n = 9) genotypes represented by the 53 Thai penicillin-resistant pneumococcal strains. Lane numbers indicate PBP genotype codes. Numbers at left indicate the sizes of standard DNA fragments in base pairs.
DISCUSSION
In Thailand, a national survey conducted from 1992 to 1994 demonstrated a prevalence of penicillin-resistant pneumococci of 37.2% (1, 29, 37). This figure was much higher than that demonstrated in the surveys in 1978 (6.7%) (36) and in 1987 (10.6%) (20) (details of the 1978 and 1987 surveys are available in Materials and Methods). In order to identify the nature of the increase in the prevalence of penicillin-resistant pneumococci in Thailand, a molecular epidemiological study in which 53 penicillin-resistant pneumococcal isolates collected from 1992 to 1994 were analyzed in detail was conducted.
RFEL analysis of 53 penicillin-resistant pneumococcal isolates that were collected from children with acute respiratory tract infections in five distinct regions divided the strains into 27 distinct types. These RFEL types represented 13 clusters, i.e., types shared by two or more strains, and 14 unique types. Thirty-nine strains shared RFEL types with at least one other strain (74%). The degree of clustering among the penicillin-resistant isolates was comparable with that in data obtained in The Netherlands (17), a country with a very low prevalence of penicillin resistance (<1%). These data clearly demonstrate that the transmission behavior of these strains is comparable in both countries. The cluster size varied from 2 (nine clusters) to 11 strains (one cluster). The most predominant RFEL cluster, representing 21% of the isolates, was identical to the Spanish pandemic clone 23F (RFEL type 15) (26). In addition, the second largest RFEL cluster (RFEL type 23; four isolates) matched the Spanish-French international clone 9V (13). These data indicate that the pandemic clones 23F and 9V, which are widely spread throughout the world (15, 16, 38), are the most predominant multidrug-resistant pneumococcal clones in Thailand.
Although the five regions were not equally represented by pneumococcal isolates, geographical distribution of the genotypes among the five Thai regions demonstrated that only the RFEL clusters 66 (n = 2), 67 (n = 2), 68 (n = 2), and 70 (n = 2) were observed in single areas of the country, whereas the other nine clusters represented isolates collected in two or more districts. These data clearly suggest the national spread of the majority of the RFEL clusters. The majority of the RFEL types (24 of 27 types) were Thailand specific, as they did not match with any of the 107 RFEL types representing 15 other countries. In addition, the PBP genotypes also suggest a Thailand-specific origin: the majority of the PBP types (12 of 16 types) were not observed in the international collection. Nevertheless, since the pandemic clones 23F and 9V are most predominantly present in Thailand, we conclude that these imported clones are primarily responsible for the high prevalence of pneumococcal penicillin resistance in this country.
The increasing rate of antibiotic resistance in S. pneumoniae complicates the elimination of pneumococci by therapy and strongly supports the application of new vaccine strategies. Conjugate capsular vaccines contain a limited number of capsular serotypes, linked to a carrier protein (3, 9). Although the results of early trials with these vaccines look promising, care should be taken since several investigators have observed horizontal transfer of capsular genes (2, 15–17). Horizontal transfer of capsular genes may interfere with vaccination programs in the long run if (antibiotic-resistant) strains with a vaccine-type capsule switch to nonvaccine capsular types. In Thailand, horizontal transfer of capsular genes appears to occur frequently. Six of the 13 RFEL clusters consisted of two or more distinct serotypes. The pandemic clone 23F (RFEL cluster 15; 11 strains), harbored three distinct serotypes, 23F, 19F, and 14. Based on the serotype distribution of the 53 penicillin-resistant pneumococci, 89% of the strains display capsular types that are covered by the 7-valent conjugate vaccine. However, it is important to notice that in the collection investigated in this study the number of high-level penicillin-resistant isolates is overrepresented. Since the number of nonvaccine serotypes is higher in the group of intermediate-level-resistant isolates, vaccine coverage of penicillin-resistant pneumococci (MIC of ≥0.1) is therefore expected to be lower than calculated.
The emergence of resistant strains and the rapid spread of resistant clones raise the need for an effective global surveillance system. Detailed studies on the epidemiology and epidemic behavior of (multi)resistant pneumococci will assist in identifying emerging clones. To this respect, close collaboration between the laboratories sharing interests in pneumococcal molecular epidemiology is of utmost importance. Extensive collaboration can be facilitated by the establishment of a freely accessible electronic network. Such a network can be used to exchange epidemiological information. Consequently, this network can be used to construct and distribute an international data library containing DNA fingerprints of (multi)resistant pneumococcal strains. This approach will facilitate adequate worldwide monitoring of the epidemiology of emerging (multi)resistant pneumococcal strains.
ACKNOWLEDGMENTS
We are grateful to our colleagues responsible for the national survey of antimicrobial resistance of S. pneumoniae and Haemophilus influenzae in Thailand for providing the pneumococcal isolates: in particular, P. Sunakorn (chairman), A. Vejabhuti, S. Lochindarat, A. Teeraraatkul, V. Veerawerapong, S. Darnswang, P. Ratanakasetsin, P. Kittikhun, Y. Prompunjai, R. Pinyosmosorn, P. Yutayong, S. Chup-Uppakarn, Y. Sutivichit, S. Dusadeepituk, S. Wasanawat, and P. Wongveerakhan. T. Wattana, P. Rattanapiriyakul, and S. Poosup are greatly acknowledged for their technical assistance. We are also grateful to M. Kusum and P. Warachit for encouraging this study.
Part of this study was supported by the World Health Organization.
REFERENCES
- 1.Bamrungtrakul T, Sunakorn P, Sareebutara W, Vejabhuti A, Lochindarat S, Teeraratkul A, Kittikhun P, Pinyosmosorn R, Wasanawat S, Darnswang S, Chup-Uppakarn S, Kusum M, Dejsirilert S. Surveillance of antimicrobial resistance of Streptococcus pneumoniae and Haemophilus influenzae in Thailand. J Med Assoc Thail. 1994;77:572–579. [PubMed] [Google Scholar]
- 2.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 3.Butler J C, Breiman R F, Lipman H B, Hofmann J, Facklam R R. Serotype distribution of Streptococcus pneumoniae infections among preschool children in the United States, 1978–1994: implications for development of a conjugate vaccine. J Infect Dis. 1995;171:885–889. doi: 10.1093/infdis/171.4.885. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Emergence of penicillin-resistant Streptococcus pneumoniae—Southern Ontario, Canada. Morbid Mortal Weekly Rep. 1995;44:207–209. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Prevention of pneumococcal disease—recommendation of the Advisory Committee on Immunization Practices (ACIP) Morbid Mortal Weekly Rep. 1997;46:1–24. [PubMed] [Google Scholar]
- 6.Coffey T J, Berron S, Daniels M, Garcia-Leoni E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 8.Dagan R, Melamed R, Muallem M, Piglansky L, Yagupsky P. Nasopharyngeal colonization in Southern Israel with antibiotic-resistant pneumococci during the first 2 years of life: relation to serotypes likely to be included in pneumococcal conjugate vaccines. J Infect Dis. 1996;174:1352–1355. doi: 10.1093/infdis/174.6.1352. [DOI] [PubMed] [Google Scholar]
- 9.Dagan R, Yagupsky P, Goldbart A, Wasas A, Klugman K P. Increasing prevalence of penicillin-resistance pneumococcal infections in children in Southern Israel: implication for future immunization policies. Pediatr Infect Dis J. 1994;13:782–786. doi: 10.1097/00006454-199409000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Dowson C G, Hutchison A, Spratt B G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 11.Facklam R R, Washington J A., II . Streptococcus and related catalase-negative gram-positive cocci. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991. pp. 238–257. [Google Scholar]
- 12.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y. Relationship between nasopharyngeal colonization and the development of otitis media in children. J Infect Dis. 1997;175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 13.Gasc A M, Geslin P, Sicard A M. Relatedness of penicillin-resistant Streptococcus pneumoniae serogroup 9 strains from France and Spain. Microbiology. 1995;141:623–627. doi: 10.1099/13500872-141-3-623. [DOI] [PubMed] [Google Scholar]
- 14.Geslin P, Buu-Hoi A, Fremaux A, Acar J F. Antimicrobial resistance in Streptococcus pneumoniae: an epidemiology survey in France, 1970–1990. Clin Infect Dis. 1992;15:95–98. doi: 10.1093/clinids/15.1.95. [DOI] [PubMed] [Google Scholar]
- 15.Hermans P W M, Overweg K, Sluijter M, de Groot R. Penicillin-resistant Streptococcus pneumoniae: an international molecular epidemiological study. In: Tomasz A, editor. Streptococcus pneumoniae: molecular biology and mechanisms of disease—update for the 1990s. New York, N.Y: Mary Ann Liebert, Inc.; 1999. [Google Scholar]
- 16.Hermans P W M, Sluijter M, Dejsirilert S, Lemmens N, Elzenaar K, van Veen A, Goessens W H F, de Groot R. Molecular epidemiology of drug-resistant pneumococci: toward an international approach. Microb Drug Resist. 1997;3:243–251. doi: 10.1089/mdr.1997.3.243. [DOI] [PubMed] [Google Scholar]
- 17.Hermans P W M, Sluijter M, Elzenaar K, van Veen A, Schonkeren J J M, Nooren F M, van Leeuwen W J, de Neeling A J, van Klingeren B, Verbrugh H A, de Groot R. Penicillin-resistant Streptococcus pneumoniae in The Netherlands: results of a 1-year molecular epidemiologic survey. J Infect Dis. 1997;175:1413–1422. doi: 10.1086/516474. [DOI] [PubMed] [Google Scholar]
- 18.Hermans P W M, Sluijter M, Hoogenboezem T, Heersma H, van Belkum A, de Groot R. Comparative study of five different DNA fingerprint techniques for molecular typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1995;33:1606–1612. doi: 10.1128/jcm.33.6.1606-1612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homoe P, Prag J, Farholt S, Henrichsen J, Hornsleth A, Kilian M, Jensen J S. High rate of nasopharyngeal carriage of potential pathogens among children in Greenland: results of a clinical survey of middle-ear disease. Clin Infect Dis. 1996;23:1081–1090. doi: 10.1093/clinids/23.5.1081. [DOI] [PubMed] [Google Scholar]
- 20.Komolpis P, Leelaporn A, Gherunpong V, Visuthiserewong W. Antimicrobial susceptibility of Streptococcus pneumoniae isolated from patients at Siriraj Hospital. J Infect Dis Antimicrob Agents. 1991;8:209–213. [Google Scholar]
- 21.Koornof H J, Wasas A, Klugman K P. Antimicrobial resistance in Streptococcus pneumoniae: a South African perspective. Clin Infect Dis. 1992;15:84–97. doi: 10.1093/clinids/15.1.84. [DOI] [PubMed] [Google Scholar]
- 22.Kristinsson K G, Hjalmardottir M A, Steingrimsson O S. Increasing penicillin resistance in pneumococci in Iceland. Lancet. 1992;339:1606–1607. doi: 10.1016/0140-6736(92)91868-9. [DOI] [PubMed] [Google Scholar]
- 23.Linares J, Alonso T, Perez J L, Ayats J, Dominguez M A, Pallares R, Martin R. Trends in antimicrobial resistance of clinical isolates of Streptococcus pneumoniae in Bellvitge Hospital, Barcelona, Spain (1979–1990) Clin Infect Dis. 1992;15:99–105. doi: 10.1093/clinids/15.1.99. [DOI] [PubMed] [Google Scholar]
- 24.Marton A. Pneumococcal antimicrobial resistance: the problem in Hungary. Clin Infect Dis. 1992;15:106–111. doi: 10.1093/clinids/15.1.106. [DOI] [PubMed] [Google Scholar]
- 25.McDougal L K, Facklam R R, Reeves M, Hunter S, Swenson J M, Hill B C, Tenover F C. Analysis of multiply antimicrobial-resistant isolates of Streptococcus pneumoniae from the United States. Antimicrob Agents Chemother. 1992;36:2176–2184. doi: 10.1128/aac.36.10.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz R, Coffey T R, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 27.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed., vol. 17, p. 1–29. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 29.Pinyosmosorn R, Yutawong P, Dejsirilert S. Acute respiratory tract infection in children: antimicrobial resistance of Streptococcus pneumoniae and Haemophilus influenzae. Nakorn Ratchasima Hosp Med J. 1996;20:365–373. [Google Scholar]
- 30.Reichmann P, Varon E, Günther E, Reinert R R, Lüttiken R, Marton A, Geslin P, Wagner J, Hakenbeck R. Penicillin-resistant Streptococcus pneumoniae in Germany: genetic relationship to clones from other European countries. J Med Microbiol. 1995;43:377–385. doi: 10.1099/00222615-43-5-377. [DOI] [PubMed] [Google Scholar]
- 31.Sa Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and in Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 32.Sluijter, M. Unpublished observations.
- 33.Sluijter M, Faden H, de Groot R, Lemmens N, Goessens W H F, van Belkum A, Hermans P W M. Molecular characterization of pneumococcal nasopharynx isolates collected from children during their first 2 years of life. J Clin Microbiol. 1998;36:2248–2253. doi: 10.1128/jcm.36.8.2248-2253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South-African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 35.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 36.Sornchai C. Serotype distribution and antibiotic-resistance pattern of Streptococcus pneumoniae in Bangkok, Thailand. Thesis. Bangkok, Thailand: Mahidol University; 1978. pp. 67–76. [Google Scholar]
- 37.Sunakorn P, Bamrungtrakul T, Kusum M, Dejsirilert S, Sareebutara W. Antimicrobial resistance patterns of Streptococcus pneumoniae isolated from ARI patients. Thai J Epidemiol. 1996;4:61–67. [Google Scholar]
- 38.Tomasz A, Corso A Members of the PAHO/Rockefeller University Workshop. Molecular epidemiologic characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six Latin-American countries: an overview. Microb Drug Resist. 1998;4:195–207. doi: 10.1089/mdr.1998.4.195. [DOI] [PubMed] [Google Scholar]
- 39.Van Belkum A, Sluijter M, de Groot R, Verbrugh H A, Hermans P W M. A novel BOX-repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1996;34:1176–1179. doi: 10.1128/jcm.34.5.1176-1179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Steenbergen T J M, Colloms S D, Hermans P W M, de Graaff J, Plasterk R H A. Genomic DNA fingerprinting by restriction fragment end labeling (RFEL) Proc Natl Acad Sci USA. 1995;92:5572–5576. doi: 10.1073/pnas.92.12.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida R, Hirakata Y, Kaku M, Takemura H, Tanaka H, Tomono K, Koga H, Kohno S, Kamahira S. Genetic relationship of penicillin resistant Streptococcus pneumoniae serotype 19B strains in Japan. Epidemiol Infect. 1997;118:105–110. doi: 10.1017/s0950268896007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida R, Kaku M, Kohno S, Ishida K, Mizukane R, Takemura H, Tanaka H, Usui T, Tomono K, Koga H, Hara K. Trends in antimicrobial resistance of Streptococcus pneumoniae in Japan. Antimicrob Agents Chemother. 1995;39:1196–1198. doi: 10.1128/aac.39.5.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zenni M K, Cheatham S H, Thompson J M, Reed G W, Batson A B, Palmer P S, Holland K L, Edwards K M. Streptococcus pneumoniae colonization in the young child: association with otitis media and resistance to penicillin. J Pediatr. 1995;127:533–537. doi: 10.1016/s0022-3476(95)70108-7. [DOI] [PubMed] [Google Scholar]