Abstract
Genetic diversity is the raw material for germplasm enhancement. Landraces and wild species relatives of potato, which contain a rich gene pool of valuable agronomic traits, can provide insights into the genetic diversity behind the adaptability of the common potato. The diploid plant, Solanum stenotomum (Sst), is believed to have an ancestral relationship with modern potato cultivars and be a potential source of resistance against disease. Sequencing of the Sst genome generated an assembly of 852.85 Mb (N50 scaffold size, 3.7 Mb). Pseudomolecule construction anchored 788.75 Mb of the assembly onto 12 pseudochromosomes, with an anchor rate of 92.4%. Genome annotation yielded 41,914 high-confidence protein-coding gene models and comparative analyses with closely related Solanaceae species identified 358 Sst-specific gene families, 885 gene families with expansion along the Sst lineage, and 149 genes experiencing accelerated rates of protein sequence evolution in Sst, the functions of which were mainly associated with defense responses, particularly against bacterial and fungal infection. Insights into the Sst genome and the genomic variation of cultivated potato taxa are valuable in elaborating the impact of potato evolution in early landrace diploid and facilitate modern potato breeding.
Keywords: Solanum stenotomum, primitive cultivated potato, genome assembly, comparative genome, genomic variations
Introduction
The cultivated potato, Solanum tuberosum L., was domesticated nearly 8000 years ago, from its origin and domestication in the southern Andes of Peru, where it was essential for feeding a growing population. Nowadays, it is widely recognized as the most important nongrain food crop and is central to global food security (Spooner et al. 2005; Pearsall 2008). It has been declared as the “Food for the Future” (FAO 2019), and it was estimated that more than two billion people worldwide will depend on the potato for food, feed, or income by 2020 (Ortiz and Mares 2017). Over the past 10 years, potato production has increased at an annual rate of 4.5%, exceeding the rate of new varieties generation. Scientists have begun to study potato genetics, with the aim of variety improvement in terms of yield, quality traits, disease resistance, and adaption to the changing climate and agro-ecosystems (G’alvez et al. 2017; Ortiz and Mares 2017); Potato improvement efforts have been hindered by a relative lack of genetic resources, mainly refer to the alleles lost to domestication bottlenecks or never introgressed into cultivated germplasm from landraces and tuber-bearing wild potato relatives. Fortunately, tuber-bearing Solanum species, are widely distributed in the Americas, represent an extraordinary resource of genes and allelic diversity that are lacking in modern cultivars (Spooner and Hijmans 2001; Machida-Hirano and Niino 2017). Hence, these plants are potentially valuable sources of variation for genetic enhancement and improvement of potato. As concluded by several studies, mining and use of genetic diversity within the gene pools of wild relatives and landraces can be used to optimize new cultivar breeding (Harlan and de Wet 1971; Bradeen et al. 2011; Spooner et al. 2014).
The diploid plant, Solanum stenotomum (Sst), is believed to be the most primitive form of the current cultivated potato, and has an ancestral relationship with modern cultivars (Hawkes et al. 1990; Spooner et al. 2005). As an early landrace diploid, Sst is a potential source of resistance against disease in potato breeding programs (Fock et al. 2001), as it contains important agronomic traits, such as resistance to soft rot erwinias, bacterial wilt, potato virus X and Y, early blight, and late blight and, most importantly, it can be crossed with S. tuberosum (Herriott et al. 1986; Vallejo et al. 1994; Wolters and Collins 1995; Christ and Haynes 2001). Furthermore, in addition to potentially contributing resistance to biotic and abiotic stress, Sst also represents good source of germplasm for high-quality breeding, because of the high Ca and K concentrations in its tubers, as well as its high carotenoid and crude protein content (Lu et al.2001, 2012; Subramanian et al. 2017). Attempts have also been made to broaden the genetic base of European and North American potatoes through creation of populations of long-day adapted Solanum phureja (Sph) × Sst, which are reported to exhibit resistance against early blight and late blight (Bradshaw et al. 2006). Genomic efforts in Sst may help to identify important genes or molecular pathways required for specific traits, which could be further applied to improve cultivated potatoes.
A significant amount of baseline work has previously been conducted to aid advances in potato genomics and genome-based breeding strategies. The further development of high quality, well-annotated genomes could radically enhance our ability to explore intraspecific allelic variants of genes and link them with important agronomic traits (Patil et al. 2017). A reference genome sequence was generated from a doubled monoploid clone of the original cultivated potato, Sph (The Potato Genome Sequencing Consortium 2011). In recent years, genome sequences of two diploid wild relatives of potato, Solanum commersonii (Sco) and Solanum chacoense (Sch), have been reported (Aversano et al. 2015; Leisner et al. 2018), reaffirming the importance of potato genomic resources within wild relatives and triggering extensive post-genomic work on gene discovery, marker development, evolution and diversity, and engineering of new phenotypes in potato. Crop genomics have gone beyond generation of single reference genomes, as large amounts of both sequences and genes from single reference genomes are not detected in the genomes of other individuals in the species (Tao et al. 2019a). For example, a comparison of two maize inbred lines (B73 and Mo17) revealed that over 10% of genes did not have homologs in the alternative genome (Sun et al. 2018). Therefore, the development of multiple reference-quality genomes is required to capture the full landscape of genetic diversity within a species and explore structural variation to aid crop improvement. In addition to maize, reference-quality genomes have recently been assembled for rice, soybean, sorghum, and other crops (Zhang et al. 2016; Deschamps et al. 2018; Yang et al. 2019).
With further reference-quality potato genome sequences, comparative genome studies among related wild and cultivated species will increase the ability to identify genes of interests shared with the tuber-bearing Solanum species and shed light on the phylogenetic relationships and taxonomy of potatoes. Here, we report the assembly of a high-quality Sst reference genome by BioNano optical-mapping and high-throughput chromosome conformation capture (Hi-C) technologies. Through genomic comparison, sets of genes with potential for introgression of important agronomic traits into modern cultivated potato were identified and played important roles in defending against herbivores and pathogens. This study will help capture the full landscape of potato genetic diversity and facilitate mining for novel genetic variation that can be applied for potato improvement.
Materials and methods
Plant material
We sequenced the genome of Sst, derived from a single tuber seedling with accession F172 from the China National Potato Improvement Center, Keshan, Heilongjiang, China, which was introduced from the Center of International Potato with accession CIP719909. This wild relative of potato has been widely characterized in our breeding program and made important contributions to potato breeding programs as a source of resistance genes to biotic and abiotic stresses. Tissue culture plantlets were grown in MS media on light racks set to a 16 h/8 h day/night photoperiod at 25°C and high molecular weight DNA extracted and purified using a DNeasy Plant Maxi Kit (Qiagen). DNA concentration was measured using a NanoDrop spectrophotometer (NanoDrop Technologies, USA).
Illumina sequencing and de novo assembly
Illumina-compatible paired-end libraries with insert sizes of 250 and 450 bp, as well as mate-pair libraries with insert sizes of 2, 5, 10, and 20 kb, were constructed and sequenced on an Illumina HiSeq 2500 instrument. Finally, a total of 198.03 Gb raw Illumina reads were generated. All Illumina data reads were corrected using the BFC package (https://github.com/lh3/bfc) and the filtered read-pairs de novo assembled using SOAP-denovo2 with the parameter, K = 8 (Luo et al. 2012). SSPACE software was employed for additional scaffolding, with the following parameters: -x 0 -m 46 -k 10 -a 0.4 -p 1 (Boetzer et al. 2011). Gap closing of all corrected Illumina reads was conducted using Platanus GapCloser with default parameters (Kajitani et al. 2014). To remove redundancy in the preliminary assembly scaffolds, a self-to-self Blast strategy was employed to identify redundancy, which summarized all hits with identity values >85 and >85% covered by a longer scaffold. Several long candidates in the Blast results were manually checked. The gene space of the assembled genome was assessed using CEGMA (Core Eukaryotic GenesMapping Approach, http://korflab.ucdavis.edu/dataseda/cegma/). BUSCO was used to further evaluate genome-assembly completeness (Simão et al. 2015). To estimate the assembly consensus quality (QV) accuracy, Illumina reads were aligned to the assemblies using BWA. Base pair errors were called using Free-Bayes –skip-coverage 600.
BioNano map-assisted gap filling
High molecular weight DNA was extracted and assessed as described in the “Plant material” section, then digested using the nicking endonuclease, Nt.BspQI, and labeled using an IrysPrep Reagent Kit, according to standard BioNano protocols. Labeled DNA was linearized in nanochannel arrays and imaged automatically using the Nt. BspQI BioNano Irys system (Jiao et al. 2017). BioNano raw BNX files were de novo assembled into genome maps using IrysSolve (https://bionanogenomics.com/support/software-downloads/), and the sequence maps were compared with Irys genome maps to identify all molecule overlaps using BioNano’s proprietary alignment tool, RefAligner. Then, filtered sequence and Irys maps were merged using RefAligner with a P-value of 10−10, to create super scaffolds. A cumulated length of 358.9 Gb, with an average label density of 12.19 labels/100 kb, was generated and de novo assembled using a layout-overlap-consensus method. The de novo map assembly yielded 1312.5 Mb, with a map N50 value of 1.2 Mb.
Hi-C sequencing
To anchor the scaffold to chromosomes, leaf nuclear DNA samples were cross-linked and then cut with the restriction enzyme, DpnII, according to the Hi-C procedure. The sticky ends of the resulting fragments were biotinylated and then ligated to each other to form chimeric circles. Through standard library construction and sequencing on an Illumina HiSeq 2500 instrument, a total of 102 Gb raw data were obtained. After read filtering, 659.61 million clean paired-end reads and 122.99 million valid interaction pairs were obtained for chromosome-level assembly. Scaffolds within the assemblies were placed into groups and merged together by agglomerative hierarchical clustering using LACHESIS with parameters CLUSTER MIN RESITES = 300, CLUSTER MAX LINK DENSITY = 3, CLUSTER NONINFORMATIVE RATIO = 1.4. Scaffolds were ordered following construction of the trunk tree and modification of the spanning tree, which kept the total edge weight heuristically low and the conversion in full order within chromosome groups. Finally, 1034 scaffolds, with a total length of 788.75 Mb were anchored and oriented to their respective 12 chromosomes, with an anchor rate of 92.9%.
PacBio full-length cDNA sequencing
Total RNA was isolated from two different tissues (seedling and tuber) using Trizol reagent (Invitrogen) followed by treatment with RNase-free DNase I (Promega, USA), according to the manufacturer’s protocols. The quantity and quality of RNA was checked using a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, USA) and an Agilent 2100 Bioanalyzer. cDNA was synthesized using SMARTer PCR cDNA Synthesis Kits, optimized for preparing full-length cDNA (Takara, Japan). Size fractionation and selection (1–2, 2–3, and >3 kb) were performed using the BluePippinTM Size Selection System (Sage Science, USA). SMRT bell libraries were constructed using a Pacific Biosciences DNA Template Prep Kitsv.2.0. SMRT sequencing was then performed on the Pacific Bioscience Sequel platform using the provided protocol.
Genome annotation
Repetitive elements in the Sst genome were identified through a combination of de novo and homolog-based approaches. Repeat components were first estimated by building a de novo transposable element library employing the programs LTR-FINDER (Xu and Wang, 2007), RepeatScout (Price et al. 2005) and RepeatModeler (http://www.repeatmasker.org/RepeatModeler/). The outputs were merged and classified using PASTEClassifier (Wicker et al. 2007). The RepeatMasker program was then used to discover and identify repeats in the Sst genome, based on a combination of the de novo constructed database and Repbase (Tarailo-Graovac and Chen 2009). Repetitive elements in the Sph and Sch genomes were identified using the same method. For prediction of protein-coding genes, a combination of de novo, protein homology, and Iso-Seq consensus isoforms alignment approaches were used. In detail, Augustus (Stanke and Waack 2003), Genscan (Burge and Karlin 1997), GlimmerHMM (Majoros et al. 2004), and SNAP (Blanco et al. 2007) were used for de novo gene prediction; GeMoMa (Keilwagen et al. 2016) was used to align the homologous protein sequences collected from Arabidopsis thaliana, Vitis vinifera, Solanum lycopersicum, Capsicum annuum, and Coffea canephora. Transcript evidence referred to high-quality, full-length whole seedling and tuber tissue transcripts from Iso-Seq aligned to the repeat-masked assemblies using BLAT (Kent 2002). Furthermore, the de novo predictions, homologous protein alignments, and full-length transcript data were integrated to generate consensus gene models using EVidenceModeler (Haas et al. 2008) and further refined using PASA (Haas et al. 2003). For annotation of predicted genes, Blast alignments against a series of nucleotide and protein databases, including NBCI-NR, SwissProt, InterPro, and KEGG, were conducted with an e-value cutoff of 1e-5.
Lineage-specific evolutionary rate analysis
The CodeML utility in the PAML package was used to calculate synonymous and nonsynonymous mutation rates for syntenic gene pairs (Yang 2007). Two different classes of branch models were implemented with the null model (model = 0) assuming that all branches have been evolving at the same rate and the alternative model (model = 2), allowing the foreground branch to evolve at a different rate. These models were used to generate maximum likelihood (ML) estimates of ratios and attached a log likelihood (lnL) value to each examined alignment and tree topology. An LRT was conducted to discriminate between alternative models for each ortholog in the gene set by examining the significance of differences between their lnL values (calculated as 2ΔLnL—twice the difference between their lnL values). LRT values asymptotically follow a χ2 distribution, with the number of degrees of freedom equal to the differences in the number of parameters between the models being compared. Genes with bonferroni adjust P-values < 0.05 and a higher value for the foreground than the background branches were considered to be evolving at a significantly faster rate in the foreground branch (Denoeud et al. 2014).
Results
Genome sequencing and assembly
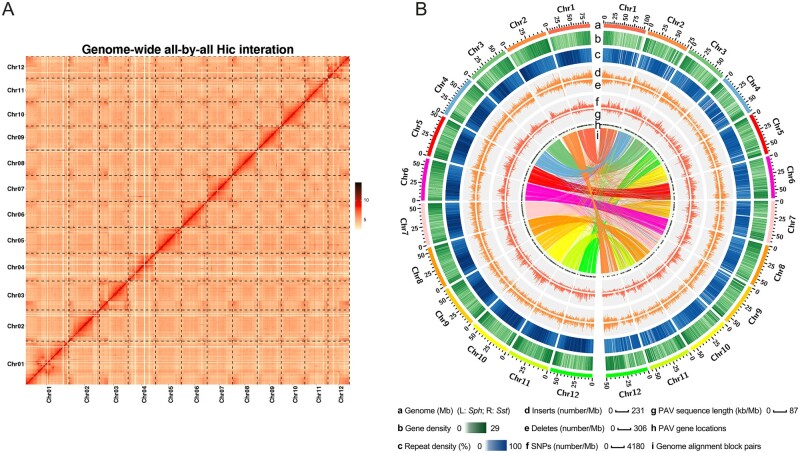
To perform a de novo assembly of the Sst genome, we integrated three sequencing and assembly technologies. First, Illumina deep sequencing (198.03 Gb, depth 237.76×) was used to generate short-read sequences from paired-end and mate pair libraries with a variety of insert sizes (Supplementary Table S1). The genome size was estimated to be 847 Mb, based on k-mer analysis. The Illumina reads were initially assembled using SOAP-denovo2 with contig N50 of 41 kb. Following scaffolds construction and gap filling using SSPACE and GapCloser, an assembly of 769.59 Mb in 39,308 scaffolds with an N50 scaffold size of 1.49 Mb, was obtained (Supplementary Table S2). The short reads were further mapped onto the assemblies, with a mapping rate of 99.4% and coverage of 99.66%, indicating high identity between reads and assemblies. The assembly consensus quality value (QV) was estimated to be 34.1, which represented a log-scaled probability of error for the consensus base calls. Approximately 97.18 and 95%, respectively, of conserved Embryophyta and orthologous single-copy genes were detected in our assembly, according to CEGMA and BUSCO analyses. Second, a hybrid scaffolding approach was applied to improve the assembly of super-scaffold sequences. An approximately 424-fold-coverage BioNano optical map, with 358.9 Gb BioNano molecules, was generated and the second assembly was 852.85 Mb and contained 39,554 scaffolds, with an N50 scaffold size of 3.7 Mb (Supplementary Table S3). To aid the assembly of scaffolds into chromosomes, we constructed Hi-C libraries and generated 122.99 million valid interaction pairs. With the aid of Hi-C data, we finally anchored and oriented 788.75 Mb of the assembly onto 12 pseudochromosomes, with an anchor rate of 92.4% (Figure 1A, Supplementary Table S4).
Figure 1.
Features of the Sst genome. (A) Sst Hi-C contact data mapped on the Sst genome. Strong signals were observed on diagonal regions, indicating that the scaffolds were accurately oriented on the pseudochromosomes. (B) Comparison of the genomic landscapes of Sst and Sph. (a) Distribution of chromosomes in the Sph (left) and Sst (right) genomes. (b and c) Gene and repetitive element density in 1 Mb sliding windows. (d and e) Distribution of insertion and deleted regions in the Sst genome in 1 Mb sliding windows. (f) Numbers of single nucleotide polymorphisms between the two genomes in 1 Mb sliding windows. (g) Distribution of presence-absence variation sequences in 1 Mb sliding windows. (h) Locations of PAV sequences on chromosomes. (i) Gene pairs between Sst and Sph genomes, identified using the best-hit method.
Genome annotation
Repetitive elements are the major components driving genome divergence in most genomes and are widely dispersed throughout the Sst genome, accounting for approximately 61.91% of assembly sequences. In addition to tandem repeats (4.48%), 59.10% of repetitive sequences were transposable elements, according to RepeatMasker, based on a combination of data from Repbase and the outputs from multiple de novo prediction software (see Materials and Methods); 17.54% of transposable elements were predicted using RepeatProteinMask (Supplementary Table S5). Among these transposable elements, retrotransposons represented 52.2% of all sequences and DNA transposons 6.78% of assembly sequences (Supplementary Table S6 and Figure S1). The proportion and composition of different classes of repetitive elements in the Sst genome were highly similar to those in the Sph and Sch genomes (Supplementary Table S7).
To annotate protein-coding genes in the Sst genome, we used a comprehensive strategy, combining general annotation and accurate homolog annotation pipelines. For the general gene set, we combined results obtained from ab initio, protein-homology-based, and IsoSeq-based prediction (see Materials and Methods for details) and a total of 30,041 gene models were predicted (Supplementary Table S8). Furthermore, we aligned the homologous protein sequences of the Sph reference genome to our assemblies and identified regions containing optimal homologs. Using Genewise, a total of 34,544 gene models were annotated in the accurate homolog gene set. After integration of the two gene sets, 41,914 high-confidence protein-coding gene models were predicted (Supplementary Table S9) and 60.22% were supported by full-length transcripts. Of these, 37,559 gene models (89.61%) were functionally annotated in six public databases (Supplementary Table S10). Furthermore, 39,125 (93.35%) of predicted genes were allocated to the 12 pseudochromosomes. Protein-coding genes were primarily within chromosome arms and were inversely correlated with transposable-element density (Figure 1B). Comparative analysis with the reference Sph genome revealed that predicted gene number and average transcript and CDS lengths were larger in the Sst genome than those in the Sph genome (Supplementary Table S11).
Global comparison with genomes of closely related species
As a prerequisite to comparing the gene content of Sst with that of other representative organisms at the whole genome scale, we ran OrthoMCL on aligned protein sequences from seven closely related species: a potato relative (Sph), tomato (S. lycopersicum), pepper (C. annuum), eggplant (Solanum melongena), Petunia (Petunia axillaris), glory (Ipomoea nil), and coffee (C. canephora). This analysis generated 33,826 groups of orthologous genes, including 3262 single-copy gene families, shared by all eight species (Supplementary Figure S2). Orthologous gene pairs from the most closely related Solanaceae species (two potato relatives, tomato, pepper, and eggplant) were extracted and 1529 genes clustered into 358 families were identified as unique to Sst. A Venn diagram representing these data is shown in Supplementary Figure S3. Taking into account wild relatives of Solanaceae species, we conducted a further four-way comparison of the potato relatives, Sst and Sph, wild tomato (Solanum pimpinellifolium) and wild pepper (Glabriusculum chiltepin), yielding 29,725 gene families, containing 8880 single-copy gene families (Supplementary Figure S4). Furthermore, 298 gene families comprising 691 genes could not be grouped with any of the genes from the wild species and were annotated as Sst-specific genes (Supplementary Figure S5). Gene ontology (GO) enrichment analysis indicated that these Sst-specific genes were highly enriched in 19 GO terms (P < 0.01, chi-squared test), most of which belonged to the functional categories metal ion binding (mainly Cu2+ and Mg2+), response to biotic stimulus, glycolipid transporter activity, and terpene synthase activity (Supplementary Table S12). The Sst-specific gene sets were further analyzed based on the KEGG orthology pathway database, to provide insights into Sst-specific biology and adaptation. Functional characterization of 16 significantly enriched pathways revealed that Sst-specific genes are primarily involved in sesquiterpenoid and triterpenoid biosynthesis, phenylpropanoid biosynthesis, and monobactam biosynthesis (Supplementary Table S13).
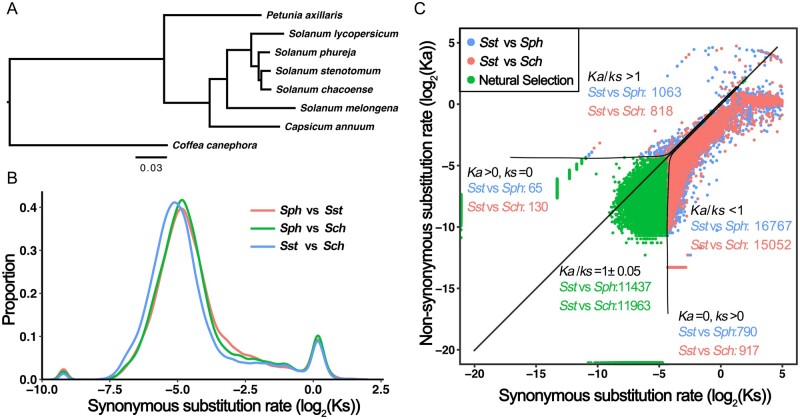
Phylogenetic analysis and divergence time among the eight closely related species were estimated based on the single-copy orthologous genes identified as described above. Divergence times between tomato and coffee (70–120 Mya), tomato and pepper (15–25 Mya), and tomato and potato Sph (7–11 Mya) were used as references for time calibration. Phylogenomic analysis revealed that Sph was most closely related to Sst, with an estimated divergence time of approximately 5.1 Mya (Supplementary Figure S6).
Gene family evolution among the eight closely related species was analyzed using CAFÉ 3. The results suggested that gene family expansions (+885) outnumbered contractions (−664) along the Sst lineage, with the opposite (i.e., families undergoing significant contraction, rather than expansion) in the Sph lineage (+281/−701), relative to the common ancestor between Sst and Sph (Supplementary Figure S7). Of functional categories (generic GO terms), 89 were significantly represented among Sst-specific expanded genes families (P < 0.05) (Supplementary Table S14) and these were involved in two main functional categories: defense response and metabolic process, the former including the synthesis of important compounds, such as catechol oxidase and terpene, and the latter including various catalytic activities. Intriguingly, families undergoing significant expansion in Sst (P < 0.05) were primarily involved in isoquinoline alkaloid (Ko00950); tropane, piperidine and pyridine alkaloid (Ko00960); phenylpropanoid (Ko00400); and diterpenoid (Ko00904) biosynthesis pathways, consistent with the high production of terpene and phenols by some medicinal plants (Supplementary Table S15).
The synonymous (Ks) and nonsynonymous (Ka) substitution rates and selection pressure (Ka/Ks) of the gene set were used to identify rapidly evolving genes in the Sst genome. A total of 2324 groups of syntenic orthologous genes present in seven Solanaceae species, plus C. canephora as the most closely related outgroup, were identified, with high quality published reference genome sequences (Figure 2A). The overall distribution of synonymous substitution (Ks) values for branches leading to individual species scaled with branch length are presented in Figure 2B. Based on the results of the likelihood ratio test (LRT) (see Materials and Methods for details), we obtained 341 genes with signatures of rapid evolution in potato species, where the branch leading to all three species within a potato clade exhibited the highest Ka/Ks ratio among the other five species examined. Taking Sst as the foreground branch only, in 149 cases the branch leading to Sst had the highest Ka/Ks ratio of all branches examined, and in 46 the branch leading to background branches had the highest Ka/Ks ratio (Figure 2C). The set of genes experiencing accelerated rates of protein sequence evolution in Sst tended to be associated with the functional annotations: “ion binding,” “ATP binding,” and “process of isoprenoid biosynthetic and metabolic.” Furthermore, we generated a dataset of syntenic orthologous genes across wild relative species in the Solanaceae family, including wild tomatoes (S. pennellii and S. lycopersicoides) and pepper (C. annuum var. glabriusculum and C. annuum var. mexicon), with P. axillaris used as an outgroup. Among these wild relative species, 176 genes showed the highest Ka/Ks values in Sst rather than in background species, with a significant 2ΔLnL value in χ2 distribution, and were also identified as genes with signatures of rapid evolution.
Figure 2.
Signatures of rapid evolution among potato species Sst, Sph, and Sch. (A) Phylogenetic tree of eight closely related Solanum species constructed from a concatenated alignment of single-copy genes. (B) Synonymous substitution rate (Ks) distribution of orthologous gene sets among Sst, Sph, and Sch. Horizontal and vertical axes represent the log2 values of the synonymous substitution rate and proportion, respectively. (C) Chart showing the synonymous (Ks) and nonsynonymous (Ka) substitution rates, and selection pressure (Ka/Ks) between Sst and Sph and between Sst and Sch. Ka/Ks = 1 indicates genes with neutral selection, Ka/Ks > 1 indicates positive selection, and Ka/Ks < 1 indicates negative selection.
Discussion
Here, we report de novo genome assemblies for an original potato cultivar of central importance, the cultivated diploid Sst (S. tuberosum, Andigenum group), illustrating the advantages of whole-genome sequencing for detecting genetic variation that would not have been found by resequencing alone. As one of the richest genetic resources of any cultivated plant, with 107 wild and 4 cultivated species, combining both molecular and morphological evidence, a single potato reference genome does not adequately represent the diversity of these plants. As much within-species genetic variation is case-dependent, it is essential to sequence additional original cultivated potato species to capture a representative proportion of the potato’s gene pool and resolve the allelic structure and gene variation within the species, thereby facilitating investigation of domestication and environmental adaptation. Several factors can contribute to the extensive genome diversity among phylogenetically related species, among which, the pressure of artificial selection on DNA rearrangement is key. As reported here and previously hypothesized, Sph exhibits a relatively strong impact of artificial selection on traits, including rapid maturation and lack of tuber dormancy, during a quick domestication process, resulting in abundant genomic structural variation, with a high level of genome diversity and gene sets with accelerated rates of protein sequence evolution. Nevertheless, Sph had relatively greater differences in speciation compared with Sst and the wild relative Sch (Figure 2, A and B). The domestication process of the modern tetraploid potato S. tuberosum Andigenum Group is believed to have started as a single domestication event of wild progenitors from the Solanum brevicaule complex in southern Peru. The result of domestication was diploid Sst, from which other cultivated species were derived. The hypothesis of extensive gene flow between Sst and wild species has been tested by other researchers, using isozyme markers specific to these populations. Indeed, cultivated species were genetically enriched by incorporating wild species germplasm, resulting in gene flow from wild species. These functional genes, identified from expanded gene families, specific gene sets, and rapidly evolving genes encoding proteins in the Sst genome, could be used to design crosses to determine whether they are associated with phenotypic variation of agronomic traits and then develop targeted molecular inbreeding. As reported in our analysis, for example, the copper-containing enzyme, catechol oxidase, which contributes to production of the natural antiseptic, ortho-quinone, could help protect plants from damage by both bacterial and fungal infection. Furthermore, phenylpropanoids have important roles in defending against herbivores and pathogens, mediating plant-pollinator interactions, and providing protection from ultraviolet light. Therefore, our data should enable the crop breeding community to make more effective use of molecular approaches, such as genomic selection, to increase resistance to biotic stress, which is often associated with introgression from wild species.
Conclusions
In conclusion, we describe identification of valuable germplasm, contributing to understanding of the history of potato evolution, from the diploid primitive species Sst, which is believed to be the primitive form of potato currently cultivated, and a potential source of resistance against disease in potato breeding programs. Genomic efforts in Sst may help to reveal genes important for specific traits, as well as identifying genomic variation among tuber-bearing Solanum species, which could facilitate deeper understanding of the impact of evolution on early landrace diploid and be utilized to improve potato breeding.
Data availability
The genome assembly and gene annotations have been deposited in the NCBI database under BioProject number PRJNA616063 and this whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAERIQ000000000. The version described in this paper is version JAERIQ010000000.
Supplementary Material
Acknowledgments
L.Y., Y.Z., and X.L. designed the research. J.S., C.L., and M.Y. prepared the plant sample. Z.F performed micro-tuber inducement. L.Y., X.L., and Y.Q performed genome sequencing, assembly, and annotation. L.Y., H.W., and X.T performed comparative genomic analysis. Y.Z. and G.C. supervised the experiments and analysis. L.Y. and X.L wrote the initial draft and Y.Z., H.W., and X.T revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by internal funding from Xichang University namely the Five-year funding plan for the Laboratory of Potato Functional Genome and Application (#206805) to L.Y., Y.Q., Z.F., X.L., and G.C., and the Xichang University High-level Talents Introduction Research Project to L.Y. (#50180108) and X.L. (#50190005).
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Aversano R, Contaldi F, Ercolano MR, Grosso V, Iorizzo M, et al. 2015. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell. 27:954–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E, Parra G, Guig ’O R.. 2007. Using geneid to identify genes. Curr Protocols Bioinformatics. 18:Chapter 4:Unit 3. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. 2011. Scaffolding pre-assembled contigs using space. Bioinformatics. 27:578–579. [DOI] [PubMed] [Google Scholar]
- Bradeen JM, Haynes KG, Kole C.. 2011. Introduction to potato. Genetics, Genomics and Breeding of Potatoes. In:Bradeen JM, Haynes KG, editors. Enfield. NH: Scientific Publishing, p. 1–19. [Google Scholar]
- Bradshaw J, Bryan G, Ramsay G.. 2006. Genetic resources (including wild and cultivated solanum species) and progress in their utilisation in potato breeding. Potato Res. 49:49–65. [Google Scholar]
- Burge C, Karlin S.. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 268:78–94. [DOI] [PubMed] [Google Scholar]
- Christ BJ, Haynes K.. 2001. Inheritance of resistance to early blight disease in a diploid potato population. Plant Breed. 120:169–172. [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, et al. 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 345:1181–1184. [DOI] [PubMed] [Google Scholar]
- Deschamps S, Zhang Y, Llaca V, Ye L, Sanyal A, et al. 2018. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat Commun. 9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fock I, Collonnier C, Luisetti J, Purwito A, Souvannavong V, et al. 2001. Use of Solanum stenotomum for introduction of resistance to bacterial wilt in somatic hybrids of potato. Plant Physiol Biochem. 39:899–908. [Google Scholar]
- Héctor Gálvez J, Tai HH, Barkley NA, Gardner K, Ellis D, et al. 2017. Understanding potato with the help of genomics. AIMS Agriculture Food. 2:16–39. [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, et al. 2003. Improving the arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31:5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, et al. 2008. Automated eukaryotic gene structure annotation using evidencemodeler and the program to assemble spliced alignments. Genome Biol. 9:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR, de Wet JMJ.. 1971. Toward a rational classification of cultivated plants. Taxon. 20:509–517. [Google Scholar]
- Hawkes JG. 1990. The Potato: Evolution, Biodiversity and Genetic Resources. London, England: Belhaven Press. [Google Scholar]
- Herriott A, Haynes F, Shoemaker P.. 1986. The heritability of resistance to early blight in diploid potatoes (Solanum tuberosum, subsp. phureja and stenotomum). Am Potato J. 63:229–232. [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, et al. 2017. Improved maize reference genome with single-molecule technologies. Nature. 546:524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, Wenk M, Erickson JL, Schattat MH, Grau J, et al. 2016. Using intron position conser-vation for homology-based gene prediction. Nucleic Acids Res. 44:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. Blat—the blast-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner CP, Hamilton JP, Crisovan E, Manrique-Carpintero NC, Marand AP, et al. 2018. Genome sequence of m6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J. 94:562–570. [DOI] [PubMed] [Google Scholar]
- Lu W, Haynes K, Wiley E, Clevidence B.. 2001. Carotenoid content and color in diploid potatoes. J Am Soc Horticul Sci. 126:722–726. [Google Scholar]
- Lu W, Yu M, Bai Y, Li W, Xu X.. 2012. Crude protein content in diploid hybrid potato clones of Solanum phureja–S.stenotomum. Potato Res. 55:315–322. [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, et al. 2012. Soapdenovo2: anempirically improved memory-efficient short-read de novo assembler. Gigascience. 1:2047–217X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida-Hirano R, Niino T.. 2017. Potato genetic resources. In: Kumar Chakrabarti S, Xie C, Kumar Tiwari J. (Editors) The Potato Genome. Springer, Cham, Chapter 2: 11–30. [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL.. 2004. Tigrscan and glimmerhmm: two open source ab initio eukaryotic gene-finders. Bioinformatics. 20:2878–2879. [DOI] [PubMed] [Google Scholar]
- Ortiz O, Mares V.. 2017. The historical, social, and economic importance of the potato crop. In: Kumar Chakrabarti S, Xie C, Kumar Tiwari J. (Editors). Cham: The Potato Genome. Springer, Chapter 1:1–10. [Google Scholar]
- Patil VU, Sharma NN, Chakrabarti SK.. 2017. High-throughput sequencing of the potato genome. In: Kumar Chakrabarti S, Xie C, Kumar Tiwari J. (Editors). Cham: The Potato Genome. Springer, Chapter 6: 95–107. [Google Scholar]
- Pearsall DM. 2008. Plant domestication and the shift to agriculture in the andes. In: Silverman H, Isbell WH. (Editors)The Handbook of South American archaeology. New York, NY: Springer, Chapter 7: 105–120. [Google Scholar]
- Price AL, Jones NC, Pevzner PA.. 2005. De novo identification of repeat families in large genomes. Bioinformatics. 21:i351–i358. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. Busco: assessing genomeassembly and annotation completeness with single-copy orthologs. Bioinformatics. 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Spooner DM, Ghislain M, Simon R, Jansky SH, Gavrilenko T.. 2014. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot Rev. 80:283–383. [Google Scholar]
- Spooner DM, Hijmans RJ.. 2001. Potato systematics and germplasm collecting, 1989–2000. Am J Potato Res. 78:237–268. [Google Scholar]
- Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ.. 2005. A single domestication for potato basedon multilocus amplified fragment length polymorphism genotyping. Proc Natl Acad Sci USA. 102:14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Waack S.. 2003. Gene prediction with a hidden markov model and a new intron submodel. Bioinformatics. 19:ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Subramanian NK, White PJ, Broadley MR, Ramsay G.. 2017. Variation in tuber mineral concentrations among accessions of solanum species held in the commonwealth potato collection. Genet Resour Crop Evol. 64:1927–1935. [Google Scholar]
- Sun S, Zhou Y, Chen J, Shi J, Zhao H, et al. 2018. Extensive intraspecific gene order and gene structural variations between mo17 and other maize genomes. Nat Genet. 50:1289–1295. [DOI] [PubMed] [Google Scholar]
- Tao Y, Jordan DR, Mace ES.. 2019a. Crop genomics goes beyond a single reference genome. Trends Plant Sci. 24:1072–1074. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N.. 2009. Using repeatmasker to identify repetitive elements in genomic sequences. Curr Protocols Bioinformatics. 25:4–10. [DOI] [PubMed] [Google Scholar]
- The Potato Genome Sequencing Consortium 2011. Genome sequence and analysis of the tuber crop potato. Nature. 475: 189–195. [DOI] [PubMed] [Google Scholar]
- Vallejo RL, Collins WW, Schiavone RD, Lommel SA, Young J.. 1994. Extreme resistance to infection bypotato virus y and potato virus x in an advanced hybridsolanum phureja—S. stenotomum diploid potato population. Am Potato J. 71:617–628. [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8:973–982. [DOI] [PubMed] [Google Scholar]
- Wolters PJ, Collins WW.. 1995. Estimation of genetic parameters for resistance to erwinia soft rot, specific gravity, and calcium concentration in diploid potatoes. Crop Sci. 35:1346–1352. [Google Scholar]
- Xu Z, Wang H.. 2007. Ltrfinder: an efficient tool for the prediction of full-length ltr retrotransposons. Nucleic Acids Res. 35:W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. Paml 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ge X, Yang Z, Qin W, Sun G, et al. 2019. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat Commun. 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen LL, Xing F, Kudrna DA, Yao W, et al. 2016. Extensive sequence divergence between the reference genomes of two elite indica rice varieties zhenshan 97 and minghui 63. Proc Natl Acad Sci USA. 113:E5163–E5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assembly and gene annotations have been deposited in the NCBI database under BioProject number PRJNA616063 and this whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAERIQ000000000. The version described in this paper is version JAERIQ010000000.