Abstract
Nutrient sensing is important for cell growth, aging, and longevity. In Saccharomyces cerevisiae, Sch9, an AGC-family protein kinase, is a major nutrient sensing kinase homologous to mammalian Akt and S6 kinase. Sch9 integrates environmental cues with cell growth by functioning downstream of TORC1 and in parallel with the Ras/PKA pathway. Mutations in SCH9 lead to reduced cell growth in dextrose medium; however, reports on the ability of sch9Δ mutants to utilize non-fermentable carbon sources are inconsistent. Here, we show that sch9Δ mutant strains cannot grow on non-fermentable carbon sources and rapidly accumulate suppressor mutations, which reverse growth defects of sch9Δ mutants. sch9Δ induces gene expression of three transcription factors required for utilization of non-fermentable carbon sources, Cat8, Adr1, and Hap4, while sch9Δ suppressor mutations, termed sns1 and sns2, strongly decrease the gene expression of those transcription factors. Despite the genetic suppression interactions, both sch9Δ and sns1 (or sns2) homozygous mutants have severe defects in meiosis. By screening mutants defective in sporulation, we identified additional sch9Δ suppressor mutants with mutations in GPB1, GPB2, and MCK1. Using library complementation and genetic analysis, we identified SNS1 and SNS2 to be IRA2 and IRA1, respectively. Furthermore, we discovered that lifespan extension in sch9Δ mutants is dependent on IRA2 and that PKA inactivation greatly increases basal expression of CAT8, ADR1, and HAP4. Our results demonstrate that sch9Δ leads to complete loss of growth on non-fermentable carbon sources and mutations in MCK1 or genes encoding negative regulators of the Ras/PKA pathway reverse sch9Δ mutant phenotypes.
Keywords: Sch9, Ira1/2, Mck1, Gpb1/2, respiratory metabolism
Introduction
In eukaryotic cells, physiological adaptations to changes in the internal and external environments are regulated by dynamic signal transduction networks that are generally conserved throughout evolution. For Saccharomyces cerevisiae, nutrient availability is the major environmental factor driving cell growth and survival (reviewed in Schuller 2003; Zaman et al. 2008; Turcotte et al. 2010; Loewith and Hall 2011; Broach 2012). Yeast cells utilize interconnected signaling pathways and transcriptional regulatory networks to respond to the external supply of nutrients and adjust their growth and metabolism. Carbon and nitrogen nutrients in particular serve as both substrates to support cell growth and as signaling molecules to guide cellular processes necessary for survival. An important mediator of this coordinated response is the protein kinase Sch9, which functions in both glucose and nitrogen sensing pathways.
Sch9 is a member of the AGC family of serine/threonine protein kinases and is a homolog of mammalian Akt/Protein Kinase B and ribosomal protein (RP) S6 kinase (S6K). Several AGC-family protein kinases are phosphorylated and activated by the evolutionarily conserved Target of Rapamycin (TOR) kinase, a major regulator of cell growth and metabolism in response to environmental signals (Wullschleger et al. 2006). Akt is directly phosphorylated in its hydrophobic motifs by mammalian TOR complex 2 (mTORC2) in response to hormones and other growth factors. Another well-characterized AGC-family kinase is S6K, which is phosphorylated in its hydrophobic motifs and activated by mTORC1 (Burnett et al. 1998) to stimulate ribosomal biogenesis (Ribi) and protein synthesis. In yeast, Sch9 was identified as a major downstream effector of TORC1 signaling and functional ortholog of S6K or PKB/Akt1 (Geyskens et al. 2001; Sobko 2006; Urban et al. 2007). Sch9 is directly phosphorylated in its C-terminal hydrophobic motifs and an adjacent site by TORC1 and in its activation loop by the yeast PDK1 orthologs Pkh1 and Pkh2. When nitrogen is abundant or preferred nitrogen sources are not limiting, TORC1 is activated and promotes mass accumulation and cell growth by inducing ribosome biosynthesis, which is achieved partly through the activation of Sch9 (reviewed in Loewith and Hall 2011). Sch9 positively regulates the expression of genes in Ribi and RP regulons by phosphorylating repressors Dot6, Tod6, and Stb3 and thereby inhibiting the recruitment of the RPD3L histone deacetylase complex to target gene promoters (Urban et al. 2007; Huber et al. 2009; Lee et al. 2009; Lippman and Broach 2009; Huber et al. 2011). Sch9 also positively regulates RNA polymerase III-dependent transcription of 5S ribosomal RNA and tRNA genes by directly phosphorylating and inhibiting Maf1, as well as RNA polymerase I-dependent expression of 35S pre-ribosomal RNA (Huber et al. 2009; Lee et al. 2009; Wei and Zheng 2009).
In addition to its functions in the TORC1-mediated nitrogen sensing branch of nutrient signaling, Sch9 may also play a role in the regulation of cell growth in response to the quality of carbon sources. Glucose-grown cells exhibit an increase in both protein level and phosphorylation of Sch9 (Jorgensen et al. 2004; Urban et al. 2007). There has been much evidence to suggest that Sch9 functions in a pathway parallel to cAMP-dependent protein kinase PKA and that the two kinases exhibit some functional redundancy (reviewed in Loewith and Hall 2011; Broach 2012). SCH9 was originally identified as a multicopy suppressor of growth defects associated with loss of Ras or PKA activity and, conversely, hyperactivation of PKA due to TPK1 overexpression, bcy1Δ, or expression of a constitutive activating mutant of RAS2, RAS2Val19, suppresses the slow growth defects of sch9Δ strain (Toda et al. 1988; Prusty and Keil 2004). The activity of PKA is regulated by cAMP, whose synthesis depends on the activity of adenylate cyclase (reviewed in Thevelein and de Winde 1999; Broach 2012). PKA is a tetramer of two identical regulatory subunits encoded by BCY1 and two catalytic subunits encoded by the three functionally redundant genes, TPK1, TPK2, and TPK3. cAMP activates PKA by binding to Bcy1, relieving its inhibition of the catalytic subunits. Activation of PKA is positively regulated by the G protein-coupled receptor Gpr1 and its associated Gα protein, Gpa2 (Kraakman et al. 1999; Lorenz et al. 2000). Adenylate cyclase activity is also mediated by Ras proteins Ras1 and Ras2, which are regulated positively by Cdc25, the guanine nucleotide exchange factor of Ras, and negatively by Ira1 and Ira2, the GTPase-activating proteins of Ras. Two other related proteins, Gpb1 and Gpb2, also negatively regulate the activity of the Ras/PKA signaling pathway (Harashima and Heitman 2002; Harashima et al. 2006; Budhwar et al. 2011). Gpb1 promotes ubiquitin-dependent proteolysis of Ira2 and Gpb2 inhibits Ras activity through direct interactions with Ira1 and Ira2 (Harashima et al. 2006; Phan et al. 2010). PKA stimulates cell growth in part by inhibiting transcription factors Msn2/4 and Gis1, which regulate the expression of stress-responsive element- and post-diauxic shift-driven genes, respectively, and Rim15, a protein kinase that induces progression into stationary phase and meiosis (reviewed in Roosen et al. 2005; Broach 2012). Despite the suggested functional redundancy with PKA, the role of Sch9 in glucose sensing is still unclear. On one hand, 90% of the glucose-induced changes in gene expression can be recapitulated by PKA activation or Sch9 overexpression in glycerol-grown cells. On the other hand, Sch9 inactivation does not interfere with the glucose response, while PKA inactivation does (Zaman et al. 2009). The common substrates of Sch9 and PKA include Dot6, Tod6, and Maf1, which are important factors for the regulation of ribosome synthesis and the expression of tRNA genes (Huber et al. 2009; Lee et al. 2009; Lippman and Broach 2009; Soulard et al. 2010; Huber et al. 2011). Sch9 and PKA have similar phosphorylation site motifs and share overlapping target substrates (Plank et al. 2020), which helps to explain their close relationship in mediating cell growth and survival in response to nutrient signals.
Consistent with its role as an important kinase sensing nutrient and stress conditions, Sch9 has been implicated in many cellular processes. Sch9 is involved in the regulation of stress-responsive genes (Pedruzzi et al. 2003; Roosen et al. 2005; Lavoie and Whiteway 2008). Inhibition of both Sch9 and PKA is known to induce autophagy (Yorimitsu et al. 2007). Sch9 negatively regulates the expression of genes in the nitrogen discrimination pathway (Smets et al. 2008), which are controlled by several GATA transcription factors including Gln3 and Gat1 (Cooper 2002; Smets et al. 2008). Interestingly, a gln3 gat1 double mutation confers resistance to rapamycin treatment and partially suppresses the growth defects of sch9Δ mutant cells (Schmelzle et al. 2004; Urban et al. 2007). Sch9 and PKA converge on Rim15 to regulate transcription of genes involved in the stress response and during the post diauxic shift (Pedruzzi et al. 2003; Roosen et al. 2005). Sch9 regulates both chronological and replicative life span (Morano and Thiele 1999; Fabrizio et al. 2001, 2003; Kaeberlein et al. 2005), which is partly mediated by Rim15 (Fabrizio et al. 2001; Wei et al. 2008; Burtner et al. 2009). Sch9 has also been shown to regulate cell size, osmotic stress response, sphingolipid metabolism, ceramide biosynthesis, V-ATPase assembly, and pH homeostasis (Jorgensen et al. 2002; Pascual-Ahuir and Proft 2007; Swinnen et al. 2014; Wilms et al. 2017). Together, the evidence suggests that Sch9 participates in a dynamic signaling network to regulate cell growth and survival in response to nutrient and stress conditions.
Sch9 and its eukaryotic homologs are broadly implicated in integrating extracellular and intracellular signals to promote cell growth and survival. It is compelling to understand the functions and regulatory mechanisms of Sch9. There are currently discrepancies in the literature with regards to how sch9 mutations impact cells’ ability to utilize non-fermentable carbon sources. Sch9 mutations have been reported to cause a range of severe to no growth defects on non-fermentable carbon sources (Crauwels et al. 1997; Jorgensen et al. 2004; Roosen et al. 2005; Lavoie and Whiteway 2008; Zhang et al. 2011; Teixeira et al. 2014). There are also conflicting reports about the effects that sch9 mutations have on the expression of genes encoding mitochondrial proteins, the HAP4 gene, which encodes the regulatory subunit of the Hap2/3/4/5 transcription factor complex, and stress-responsive genes such as HSP12 and HSP26 (Fabrizio et al. 2003; Roosen et al. 2005; Lavoie and Whiteway 2008; Smets et al. 2008; Pan and Shadel 2009; Pan et al. 2011; Teixeira et al. 2014). These discrepancies could be due to fast accumulation of sch9Δ suppressor mutations, originally reported by Huber et al. (2009) who used analog-sensitive sch9 alleles instead of sch9Δ to minimize complications that may be caused by sch9Δ suppressor mutations. Nevertheless, many published results were generated using sch9Δ mutant strains.
Here, we report the isolation and characterization of spontaneous and rapidly accumulating suppressor mutations that occur in slow-growing sch9Δ mutant cells. We found that sch9Δ cells are unable to grow on non-fermentable carbon sources in two commonly used strain backgrounds, W303-1A and W303-1B as well as BY4741 and BY4742. The suppressor mutations rescue the growth defects of sch9Δ mutants on both dextrose medium and non-fermentable carbon sources. We found that mutations in IRA1 and IRA2 account for all of the spontaneous, recessive sch9Δ suppressor mutations that were isolated. We further found that mutations in GPB1, GPB2, and MCK1 can suppress the growth defects of sch9Δ mutant cells. We also show that PKA negatively regulates the expression of CAT8, ADR1, and HAP4. Altogether, we have identified genes responsible for spontaneous sch9Δ suppressor mutations that may complicate the analysis of the cellular functions of Sch9.
Materials and methods
Strains, plasmids, growth media, growth conditions, and yeast transformation
Yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Yeast strains were grown at 30° in YPD (1% Bacto-yeast extract, 2% Bacto-peptone; and 2% glucose), YPL [1% Bacto-yeast extract, 2% Bacto-peptone and 3.7% DL-lactic acid (w/w, 85%), adjusted to pH 5.3 using NaOH], YPEG (1% Bacto-yeast extract, 2% Bacto-peptone, 2% ethanol, and 3% glycerol), minimal dextrose medium (SD) (0.67% yeast nitrogen base and 2% glucose), YPR (1% Bacto-yeast extract, 2% Bacto-peptone, and 2% raffinose), sporulation media (0.1% Bacto-yeast extract, 1% potassium acetate, and 0.02% raffinose), YNBcasD or YNBcas5D (0.67% yeast nitrogen base, 0.25% casamino acids, 2% or 5% dextrose), YNBcasR (0.67% yeast nitrogen base, 0.25% casamino acids, and 2% raffinose), or YNBcasL [0.67% yeast nitrogen base, 1% casamino acids, and 3.7% DL-lactic acid, adjusted to pH 5.3] as indicated. Amino acids, adenine, and uracil were added to the growth medium at standard concentrations to cover auxotrophic requirements if needed (Amberg et al. 2005). Agar was added to a final concentration of 2% to solidify growth medium. For transformation, yeast cells were freshly grown in YPD media and transformed with plasmids using the high-efficiency method (Gietz et al. 1992).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source | Application |
|---|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met17Δ0 ura3Δ0 | Lab. stock | Figures 2B, 3, 4, 5, 6C, and 7 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Lab. stock | Figure 2A |
| W303-1A | MATa ura3-1 ade2-1 leu2-3,112 trp1-1 his3-11,15 | Lab. stock | Figures 1 and 3D |
| W303-1B | MATα ade2-1, his3-11, 15, ura3-1, leu2-3, 112, trp1-1, can1-100 | Lab. stock | Figure 3D |
| PPY566 | BY4741 sch9Δ::kanMX4 sns1 | This study | Figures 2A and 3B |
| ZLY6649 | BY4741 sch9Δ::kanMX4 | This study | Figures 2B, 4D, 5B, and 5C |
| ZLY6650 | BY4741 sch9Δ::kanMX4 | This study | Figures 4C, 4E, 5C, and 8 |
| PPY730 | BY4741 sns1 | This study | Figures 2B and 3B |
| PPY733 | BY4741 sch9Δ::kanMX4 sns1 | This study | Figures 2B and 5D |
| PPY632 | BY4741 sch9Δ::kanMX4 | This study | Figures 3B, 4F, and 5E |
| PPY633 | BY4742 sch9Δ::kanMX4 | This study | For constructing PPY745 |
| PPY731 | BY4742 sns1 | This study | For constructing PPY737 |
| PPY734 | BY4742 sch9Δ::kanMX4 sns1 | This study | For constructing PPY741 |
| BY4743 | BY4741 × BY4742 | This study | Figures 2C, 3, 4A, 5, and 8 |
| PPY745 | PPY632 × PPY633 | This study | Figures 2C, 3, 4A, 5A, 5D, and 8 |
| PPY737 | PPY730 × PPY731 | This study | Figures 2C, 3, 4A, and 8 |
| PPY741 | PPY733 × PPY734 | This study | Figures 2C, 3, 4A, 5A, and 8 |
| PPY624 | W303-1B MATα sch9Δ::kanMX4 | This study | Figures 1 and 3D |
| PPY673 | W303-1A MATa sch9Δ::kanMX4 sns1 | This study | Figure 1 |
| PPY622 | W303-1A MATa sch9Δ::kanMX4 | This study | Figure 3D |
| ZLY6614 | BY4741 sns2 | This study | Figures 2B, 3C, and 4A |
| ZLY6619 | BY4741 sch9Δ::kanMX4 sns2 | This study | Figures 2, A, B, 3C, 4A, and 5D |
| ZLY6626 | BY4742 sch9Δ::kanMX4 sns2 | This study | Figures 4A and 5D |
| ZLY6581 | BY4742 sns2 | This study | Figure 4A |
| ZLY6519 | BY4741 isc1Δ::kanMX4 | This study | Figure 4B |
| ZLY6606 | BY4741 mck1Δ::kanMX4 | This study | Figure 4, B–F |
| ZLY6609 | BY4741 ira2Δ::kanMX4 | This study | Figure 5, B and E |
| ZLY6629 | BY4741 get2Δ::kanMX4 | This study | Figure 4B |
| ZLY6634 | BY4742 vma6Δ::kanMX4 | This study | Figure 4B |
| ZLY6633 | BY4742 paf1Δ::kanMX4 | This study | Figure 4B |
| ZLY6637 | BY4741 atg15Δ::kanMX4 | This study | Figure 4B |
| ZLY6640 | MATα his3Δ1 leu2Δ0 ura3Δ0 met17Δ0 def1Δ::kanMX4 | This study | Figure 4B |
| ZLY6642 | BY4742 tps2Δ::kanMX4 | This study | Figure 4B |
| ZLY6644 | BY4741 dep1Δ::kanMX4 | This study | Figure 4B |
| ZLY6645 | BY4742 sem1Δ::kanMX4 | This study | Figure 4B |
| ZLY6647 | BY4741 ira1Δ::kanMX4 | This study | Figure 5E |
| ZLY6662 | BY4741 vac8Δ::kanMX4 | This study | Figure 4B |
| ZLY6665 | BY4742 erv14Δ::kanMX4 | This study | Figure 4B |
| ZLY6669 | BY4741 vma2Δ::kanMX4 | This study | Figure 4B |
| ZLY6604 | BY4741 gpb2Δ::kanMX4 | This study | Figures 4, B, C, E, and F |
| ZLY6660 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 met17Δ0 sch9Δ::kanMX4 ira2Δ::kanMX4 | This study | Figure 5E |
| ZLY6661 | BY4741 sch9Δ::kanMX4 ira2Δ::kanMX4 | This study | Figure 5, B and D |
| ZLY6601 | BY4742 ira2Δ::kanMX4 | This study | Figure 5B |
| ZLY6648 | BY4742 ira1Δ::kanMX4 | This study | Figure 5C |
| ZLY6651 | BY4742 sch9Δ::kanMX4 | This study | Figure 4C |
| ZLY6602 | BY4742 gpb2::kanMX4 | This study | Figure 4C |
| ZLY6652 | BY4741 sch9Δ::kanMX4 mck1Δ::kanMX4 | This study | Figure 4, D and F |
| PPY754 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 met17Δ0 sch9Δ::kanMX4 ira1Δ::kanMX4 | This study | Figure 5C |
| PPY755 | BY4742 sch9Δ::kanMX4 ira1Δ::kanMX4 | This study | Figure 5, D and E |
| PPY734 | BY4742 sch9Δ::kanMX4 sns1 | This study | Figure 5D |
| PPY777 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 met17Δ0 sch9Δ::kanMX4 gpb1Δ::kanMX4 | This study | Figure 4F |
| PPY757 | BY4741 gpb1Δ::kanMX4 | This study | Figure 4, C, E, and F |
| ZLY6677 | BY4742 sch9Δ::kanMX4 gpb2Δ::kanMX4 | This study | Figure 4, C, E, and F |
| PPY763 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 gpb1Δ::kanMX4 gpb2Δ::kanMX4 | This study | Figure 4, E and F |
| PPY767 | BY4741 sch9Δ::kanMX4 gpb1Δ::kanMX4 gpb2Δ::kanMX4 | This study | Figure 4, E and F |
| ZLY4313 | BY4741 yak1Δ::kanMX4 | This study | Figures 6 A, B, and 7A |
| ZLY5023 | BY4741 yak1Δ::HIS3 tpk1Δ::kanMX4 tpk2Δ::kanMX4 tpk3Δ::kanMX4 | Figure 6, A and B | |
| ZLY6715 | BY4741 bcy1Δ::kanMX4 | This study | Figure 6C |
| PPY776 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pde1Δ::kanMX4 pde2Δ::kanMX4 | This study | Figures 6C and 7B |
| ZLY6578 | BY4743 ira2Δ/IRA2 | Yeast genome deletion project | Figure 5B |
| ZLY6656 | BY4743 ira1Δ/IRA1 | Figure 5C | |
| ZLY6707 | BY4741 sch9Δ::kanMX4 yak1Δ::kanMX4 | This study | Figure 7A |
| ZLY4368 | BY4741 pde2Δ::kanMX4 | This study | Figure 7B |
| PPY758 | BY4741 sch9Δ::kanMX4 pde2Δ::kanMX4 | This study | Figure 7B |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference | Application |
|---|---|---|---|
| pDC115 | pHAP4-lacZ, expressing lacZ under the control of a 1.8kbp HAP4 promoter. | This study | Figures 3, A–D, 4, B, F, 5E, and 6 |
| pZL2140 | pCAT8-lacZ, expressing lacZ under the control of an 866 bp CAT8 promoter. | This study | Figures 3, A–D, and 6 |
| pZL2108 | pADR1-lacZ, expressing lacZ under the control of a 1249 bp ADR1 promoter. | This study | Figures 3, A–D, and 6 |
| pZL3929 | pRS416-CAT8-HA, encoding Cat8 with a C-terminal 3xHA tag under the control of the endogenous promoter. | This study | Figure 3, E and G |
| pZL1324 | pRS416-HAP4-HA, encoding Hap4 with a C-terminal 3xHA tag under the control of the endogenous promoter. | This study | Figure 3, F and H |
| pZL2416 | YCp50 | Lab. stock | Figure 5A |
| pPP322 | YCp50-lib., a sns1-complementing library plasmid encoding IRA2 and REX4. | This study | Figure 5A |
Yeast cell extract preparation, Ponceau S staining, and Western blotting
Total cellular proteins were prepared by treating yeast cells with a freshly made solution of 7.5% β-mercaptoethanol and 1.85 N NaOH and precipitated with trichloroacetic acid as described previously (Yaffe and Schatz 1984). Protein samples were resuspended in SDS-PAGE sample buffer with 100 mM dithiothreitol and boiled for three minutes before being separated by SDS-PAGE. Pre-stained protein ladder (broad range, 10–230 kDa, P7710S, New England Biolabs) was used in protein gels. Proteins were transferred to a nitrocellulose membrane for immunoblotting with monoclonal anti-HA antibody, clone 3F10. HRP-conjugated secondary antibody from Jackson ImmunoResearch Laboratories (West Grove, PA) was used to probe the primary antibody. Membranes were stained in 0.1% Ponceau S in 0.1% acetic acid for 10 minutes and images of Ponceau-stained membranes were taken before immunoblotting. Chemiluminescence images of Western blots were captured using the Bio-Rad ChemiDoc MP imaging system and processed using Bio-Rad Image Lab software. Protein bands on Western blots were quantified using the same software.
β-Galactosidase assays
Yeast strains were grown at 30° to mid-log phase for at least six generations to reach an optical density at 600 nm (OD600) of 0.6–0.8. Cells were collected by centrifugation and β-galactosidase activity assays were conducted as described previously (Amberg et al. 2005). Assays were carried out in duplicate for each sample from three independent cultures. β-galactosidase activities were given as the mean ± SD, n = 3.
Chronological lifespan assay
Yeast cultures (two replicates per sample) were grown in YPR media with a starting OD600 of 0.01 and allowed to grow until cells reached stationary phase (two days for wild type, sns1, and sch9Δ sns1 strains; four days for sch9Δ mutant strain), which was set to be day 0 for the lifespan assay. Cultures were removed from the flask every five days starting from day 0 and live cells were quantified by colony forming assay. For colony forming assay, 500 cells from each culture were plated on YPD plates and colony forming units (CFUs) were counted and averaged after 3 days’ growth at 30°. The percentage of viable cells relative to day 0 (100% survival) was calculated and plotted. The values are the mean ± SD of two independent experiments.
Sporulation efficiency assay
Diploid strains were grown on SD plate for 2 days and then transferred into 1 ml sporulation media with starting OD600 of 0.3. After eight days of incubation with shaking at 220 rpm at 30°, 400 total cells were counted for each independent strain and the percentage of cells that had formed dyads and tetrads was recorded. The data were generated from the results of three replicate samples per strain.
Data availability
The authors affirm that all data necessary to draw conclusions in this article are present within the texts, figures, and tables. Strains, plasmids, and other noncommercial research reagents are available upon request.
Results
sns1 and sns2 mutations reverse the growth defects of sch9Δ mutants on both dextrose and non-fermentable carbon sources
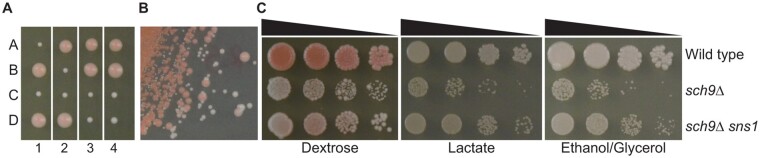
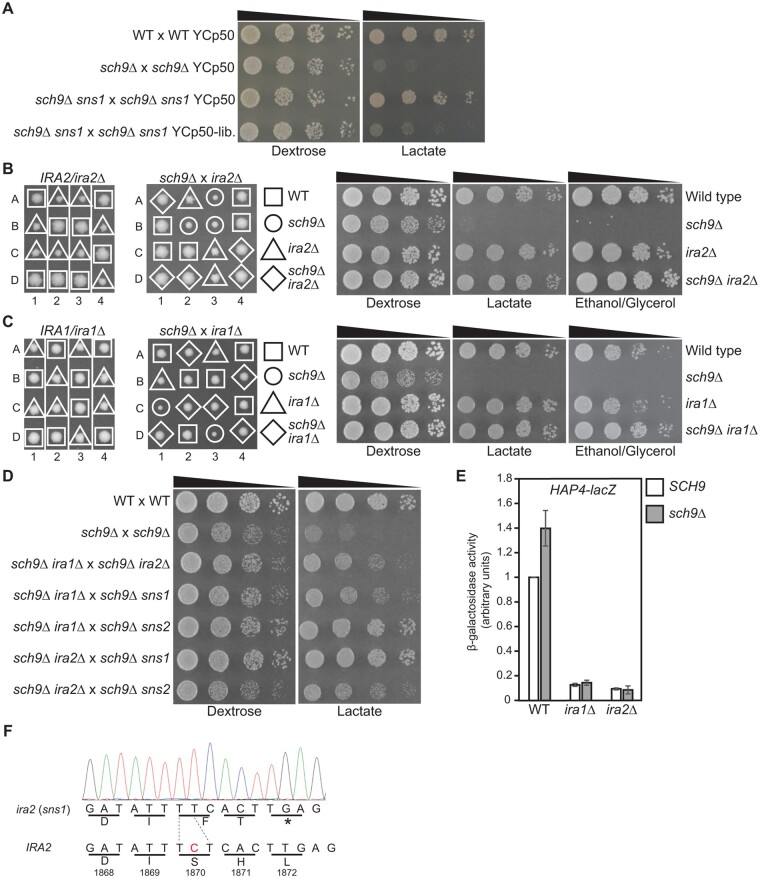
sch9Δ mutations have been reported to cause a range of severe to no growth defects in cells grown on non-fermentable carbon sources. To understand the discrepancies reported in the literature, we generated a heterozygous sch9Δ mutation in the W303 strain background and obtained sch9Δ mutants via tetrad analysis. Figure 1A shows that each tetrad yielded two big wild type and two small sch9Δ mutant colonies on dextrose medium, consistent with published results (Toda et al. 1988; Lorenz et al. 2000). W303 background strains have an ade2 mutation, leading to a red colony phenotype on solid dextrose medium. The smaller sch9Δ mutant colonies were white, suggesting sch9Δ may lead to defects in respiratory metabolism. When sch9Δ mutant cells were streaked onto a dextrose medium plate, fast-growing red colonies appeared among the small white sch9Δ mutant colonies (Figure 1B). These large red sch9Δ mutant colonies were suspected to contain suppressor mutations. Upon further propagation on plates, the number of fast-growing suppressor mutant colonies quickly surpassed that of slow-growing sch9Δ mutant colonies. To quantify the growth phenotypes of sch9Δ mutants and the suppressor mutants, we performed a serial dilution growth assay of a wild type strain, an sch9Δ single mutant, and an sch9Δ sns1 (sch9 suppressor 1) double mutant on both a fermentable carbon source, dextrose, and non-fermentable carbon sources, lactate, and ethanol/glycerol. Figure 1C shows sch9Δ mutants have growth defects on dextrose and show no growth on the lactate and ethanol/glycerol plates except for putative suppressor mutant colonies. However, the sns1 mutation partially suppresses the growth defects of sch9Δ mutant cells on both dextrose and non-fermentable carbon sources. Our data indicate that the growth defects of sch9Δ mutant cells from the W303 background can be reversed by a high frequency of spontaneous suppressor mutations.
Figure 1.
sch9Δ mutant cells in the W303 background have growth defects and rapidly accumulate suppressor mutations. (A) Tetrad analysis of wild type (W303-1A) × sch9Δ (PPY624) on YPD plate medium. Each tetrad, numbered 1-4, was dissected into four spores labeled A-D. (B) sch9Δ mutants quickly accumulate suppressor mutations. An sch9Δ mutant strain (PPY624) was streaked on YPD plate. Small white colonies are sch9Δ mutants, while large red colonies are sch9Δ mutants carrying suppressor mutations. (C) The growth defects of sch9Δ mutants are suppressed by a sns1 mutation. Wild type (W303-1A), sch9Δ (PPY624), and sch9Δ sns1 (PPY673) mutant strains were serially diluted and spotted on YPD (Dextrose), YPL (Lactate), and YPEG (Ethanol/Glycerol) medium.
BY4741 and closely related strains are derived from the S288c background, which were used in the yeast genome deletion project and are commonly utilized in yeast research labs. We wanted to determine whether sch9Δ suppressor mutations would also be easily and spontaneously obtained in this strain background. We generated sch9Δ mutants in S288c background strains by both direct transformation with an sch9Δ::kanMX4 disruption cassette and tetrad analysis. Similar results were obtained: sch9Δ results in a slow growth phenotype on dextrose medium and spontaneous suppressor mutations arise at a high frequency. A high frequency of sch9Δ suppressor mutations could skew some of the reported phenotypes of sch9Δ mutants and mask the cellular functions of Sch9. As such, it was difficult for us to maintain sch9Δ mutant cells free of suppressor mutants in both W303 and S288c background strains. Therefore, it is important to know the identity of genes whose spontaneous mutation suppresses sch9Δ. To that end, we isolated 21 sch9Δ suppressor mutants from both W303 and S288c backgrounds. These sch9Δ sns double mutants were crossed to sch9Δ mutant cells and the resultant diploid strains were analyzed for growth defects on both dextrose and glycerol medium. Based on their growth phenotypes, three of the 21 mutant strains were found to carry dominant sch9Δ suppressor mutations and were not analyzed further. We conducted a complementation group analysis on the remaining 18 mutant strains carrying recessive sch9Δ suppressor mutations. Accordingly, sch9Δ suppressor mutant strains of opposite mating types were crossed to generate diploids and analyzed for their ability to grow on glycerol plates. All resultant diploid strains were homozygous for sch9Δ and would display a sch9Δ suppressor mutant phenotype if the two haploid strains carried mutations in the same gene. From our analysis, we placed the 18 recessive sch9Δ suppressor mutations into two complementation groups, which were designated sns1 and sns2.
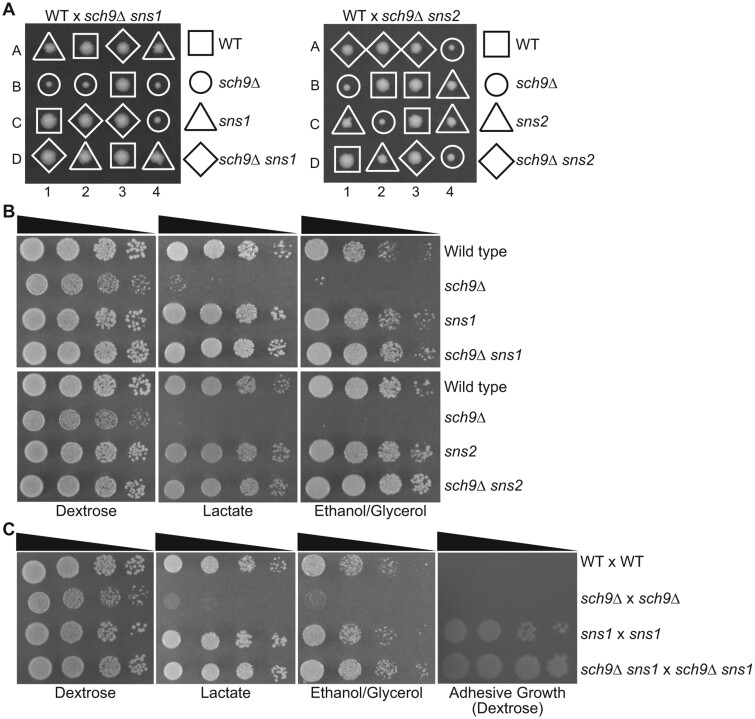
To further characterize the sch9Δ suppressor mutations, we performed tetrad analysis on wild type × sch9Δ sns1 and wild type × sch9Δ sns2 diploid strains (Figure 2A). As in the W303 background strains, sch9Δ mutant colonies from the S288c background were slow-growing on dextrose medium. sns1 and sns2 single mutant colonies on the dissection plate exhibited a slight growth defect compared with the wild type and had a rough edge or “papillae” phenotype. In the sch9Δ sns1 and sch9Δ sns2 double mutant colonies, sns1 and sns2 suppressed the slow growth of sch9Δ and yielded colonies of a similar size to wild type. Interestingly, the double mutant colonies appeared to have a relatively smooth edge like wild type, suggesting that sch9Δ suppresses the papillae phenotype of sns1 and sns2 mutants.
Figure 2.
sns1 and sns2 mutations reverse growth defects of sch9Δ mutants in the S288c background. (A) Tetrad analysis of wild type (BY4742) × sch9Δ sns1 (PPY566) and wild type (BY4742) × sch9Δ sns2 (ZLY6619) on YPD plates. (B) Wild type (BY4741) and isogenic sch9Δ (ZLY6649), sns1 (PPY730), sns2 (ZLY6614), sch9Δ sns1 (PPY733), and sch9Δ sns2 (ZLY6619) mutant strains were serially diluted and spotted on YPD (Dextrose), YPL (Lactate), and YPEG (Ethanol/Glycerol) medium. (C) Wild type (WT × WT) (BY4741 × BY4742) and isogenic sch9Δ × sch9Δ (PPY745), sns1 × sns1 (PPY737), and sch9Δ sns1 × sch9Δ sns1 (PPY741) mutant strains were serially diluted and spotted on YPD, YPL, and YPEG plates. Adhesive growth of strains on dextrose medium was examined by washing the agar surface with a stream of water. The plate was photographed shortly after being washed.
We performed serial dilution growth assays to quantify the suppression of sch9Δ by sns1 and sns2 (Figure 2B). On both dextrose medium and non-fermentable carbon sources, lactate and ethanol/glycerol, sns1 and sns2 single mutants grew like wild type. On dextrose medium, both sns1 and sns2 mutations fully suppressed the slow growth phenotype of sch9Δ mutant cells. On non-fermentable carbon sources, sch9Δ mutant cells did not exhibit any growth, except for a few presumed suppressor mutant colonies, while the sns1 and sns2 mutations completely reversed the growth defect of sch9Δ mutant cells. It often took several trials to get serial dilution growth assay results from sch9Δ mutant strains with no or a small number of suppressor mutants. Since most of the suppressor mutants carried recessive mutations, we decided to test the growth phenotype in diploid strains with the expectation that spontaneous suppressor mutants would be less likely to arise in the diploid setting. To that end, we generated an sch9Δ × sch9Δ homozygous diploid strain and found they showed a lower frequency of suppressors. sch9Δ suppression by sns1 was also observed in serial dilutions of the diploid strains (Figure 2C). When cells were left on dextrose medium for seven days, wild type colonies developed a tan color while sch9Δ mutant cells were white. Yeast cells without mitochondrial DNA, also known as rho0 petites, are unable to grow on non-fermentable carbon sources and form completely white colonies on plate. The white colony phenotype of sch9Δ mutant cells is consistent with their inability to grow on non-fermentable carbon sources. DAPI staining of sch9Δ cells showed that mitochondrial DNA maintenance did not seem to be affected. Furthermore, diploid cells formed between the crossing of sch9Δ mutant cells and a rho0 strain without mitochondrial DNA were able to grow on non-fermentable carbon sources, indicating that sch9Δ mutant cells have functional mitochondrial DNA and thus are not mitochondrial petites. Since the identities of SNS1 and SNS2 were not known at this stage of our study, we looked for other unique phenotypes of sns1 and sns2 mutants. Interestingly, washing of the plate surface with water after seven days of cell growth on dextrose medium showed that the sns1/sns1 and sch9Δ/sch9Δ sns1/sns1 diploid strains displayed increased agar adhesion (Figure 2C). The same phenotype was also observed for haploid sns1, sns2, sch9Δ sns1, and sch9Δ sns2 mutant strains. Together, these results indicate that the growth defects of sch9Δ mutants on both dextrose and non-fermentable carbon sources are reversed by spontaneously arising sns1 and sns2 mutations and that sns1 and sns2 mutants exhibit similar phenotypes.
sch9Δ and sns1 mutations have opposing effects on the expression of genes encoding transcription factors Cat8, Adr1, and Hap4
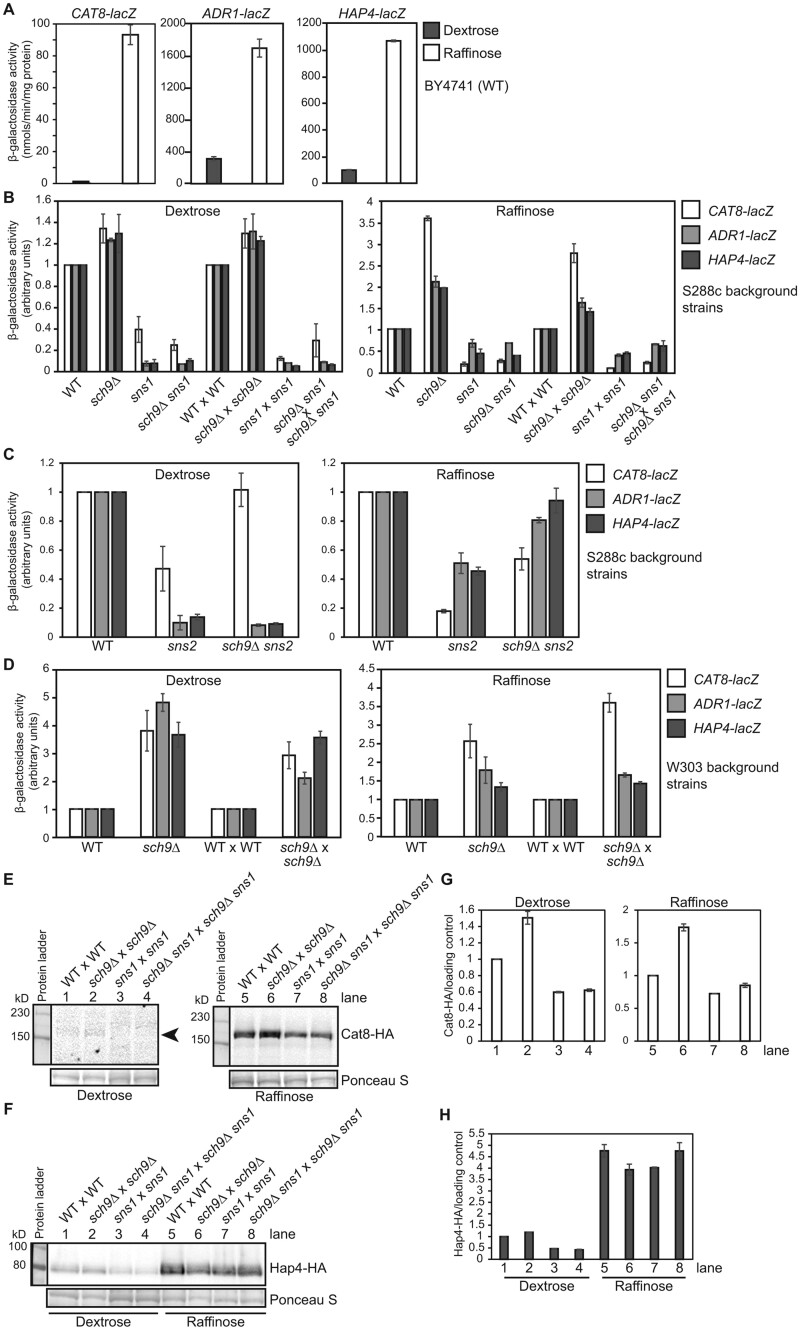
Utilization of non-fermentable carbon sources requires transcriptional activators that regulate the expression of genes involved in both respiratory metabolism and gluconeogenesis. Cat8 and Adr1 are involved in the derepression of genes required for growth on non-fermentative carbon sources, while the Hap2/3/4/5 complex is a master regulator of mitochondrial biogenesis and respiratory metabolism (reviewed in Turcotte et al. 2010). It has been reported that Sch9 is a positive regulator of ADH2 expression, which requires Adr1 (Denis and Audino 1991). Reports on the effects of sch9 mutations on HAP4 expression were inconsistent (Roosen et al. 2005; Lavoie and Whiteway 2008). Discrepancy on the growth phenotype of sch9Δ mutant cells on non-fermentable carbon sources in the published studies and ours prompted us to examine the role of Sch9 in the regulation of Hap4, Cat8, and Adr1. Accordingly, we generated lacZ reporter genes under the control of the CAT8, ADR1, and HAP4 promoters and examined their expression in wild type and sch9Δ mutant cells grown in dextrose as well as in raffinose medium. Raffinose was chosen because it allows sch9Δ mutant cells to grow and is a glucose-limiting carbon source, which is expected to lead to similar changes in gene expression to those in cells grown in non-fermentable carbon source media. Studies on the regulation of the retrograde signaling pathway in which rho0 cells without mitochondrial DNA were grown in raffinose medium to study gene expression under glucose derepression conditions have been successful (Liu and Butow 1999). The expression of HAP4, ADR1, and CAT8 are subject to glucose repression (Forsburg and Guarente 1989; Hedges et al. 1995; Dombek and Young 1997; Randez-Gil et al. 1997). Consistently, Figure 3A shows that all three reporter genes exhibit higher activity in raffinose-grown wild type cells compared with dextrose. In S288c, background strains grown in dextrose and raffinose media, both the haploid and homozygous diploid sch9Δ mutant strains showed an increase in all three reporter genes compared with the wild type haploid and diploid strains, respectively (Figure 3B). The sns1 mutation had an opposite effect on the expression of CAT8, ADR1, and HAP4 and exhibited a significant decrease in β-galactosidase activity compared with the wild type. sch9Δ sns1 double mutant strains, like sns1 single mutants, also showed reduced expression of these three reporter genes compared with the wild type in both dextrose- and raffinose-grown cells (Figure 3B). The effect of sns2 on the expression of these three reporter genes was similar to that of sns1 (Figure 3C). To address whether the difference in reporter gene activity was strain dependent, we also performed β-galactosidase assays on these three reporter genes in wild type and isogenic sch9Δ strains in the W303 background and found that sch9Δ increased the expression of the three reporter genes in cells grown in both dextrose and raffinose media (Figure 3D). In this strain background in dextrose media, sch9Δ leads to an even more significant 3.6 to 4.8-fold increase in CAT8, ADR1, and HAP4 expression in the haploid strains and a 2.1- to 3.6-fold increase in the diploid strains. In raffinose-grown W303 background strains, an increase in reporter gene activity due to sch9Δ was comparable with what was observed for the S288c background strains.
Figure 3.
sch9Δ and sns1 mutations have opposing effects on the expression of CAT8, ADR1, and HAP4. (A) β-galactosidase activities of CAT8-lacZ, ADR1-lacZ, and HAP4-lacZ reporter genes in the BY4741 wild type strain grown in YNBcas5D (Dextrose) and YNBcasR (Raffinose) medium. β-galactosidase activity assays were carried out as described in Materials and Methods. (B) β-galactosidase activities of CAT8-lacZ, ADR1-lacZ, and HAP4-lacZ reporter genes in haploid wild type (WT, BY4741), sch9Δ (PPY632), sns1 (PPY730), and sch9Δ sns1 (PPY566) mutant strains and corresponding diploid strains (WT × WT, PPY745, PPY737, PPY741) grown in YNBcas5D and YNBcasR medium. The reporter gene activity in wild type haploid and diploid strains grown in dextrose and raffinose medium was set as 1, respectively. (C) β-galactosidase activities of CAT8-lacZ, ADR1-lacZ, and HAP4-lacZ reporter genes in haploid wild type (WT, BY4741), sns2 (ZLY6614), and sch9Δ sns2 (ZLY6619) mutant strains grown in YNBcas5D and YNBcasR medium. (D) β-galactosidase activities of CAT8-lacZ, ADR1-lacZ, and HAP4-lacZ in the W303 background strains. Wild type haploid (W303-1B), sch9Δ mutant haploid (PPY622), wild type diploid (W303-1A × W303-1B), and sch9Δ homozygous mutant diploid cells (PPY622 × PPY624) were grown in YNBcas5D or YNBcasR. (E and F) Western blots of Cat8-HA and Hap4-HA in wild type and mutant diploid strains as indicated. Wild type (BY4741 × BY4742), sch9Δ × sch9Δ (PPY745), sns1 × sns1 (PPY737), and sch9Δ sns1 × sch9Δ sns1 (PPY741) mutant strains carrying a plasmid encoding CAT8-HA (pZL3929) (panel E) or HAP4-HA (pZL1324) (panel F) were grown in YNBcas5D and YNBcasR medium. The left and right panels of Cat8-HA were from the same membrane, but the Western blot from dextrose-grown cells required a longer exposure time due to weak signals. The arrowhead indicates the position of Cat8-HA bands. Ponceau S staining was used as the loading control. (G and H) Quantification of the ratios of Cat8-HA (panel G) and Hap4-HA (panel H) protein amount to the loading control by Ponceau S staining. The ratio of HA-tagged protein/loading control was set as 1 in wild type cells. The data are presented as the mean ± standard deviation, n = 2.
To address whether changes in gene expression are reflected in changes in the protein level, plasmids encoding functional C-terminal HA-tagged Hap4 and Cat8 were generated and transformed into wild type, sch9Δ, sns1, and sch9Δ sns1 diploid strains of the S288c background and HA-tagged proteins were analyzed by Western blotting. Figure 3, E and G shows that sch9Δ increased the expression of Cat8-HA in both dextrose- and raffinose-grown cells and that sns1 and sch9Δ sns1 mutants exhibited reduced expression of Cat8-HA compared with wild type. sch9Δ and sns1 increased and decreased the protein level of Hap4-HA in dextrose-grown cells, respectively (Figure 3, F and H). Their effect on Hap4-HA expression is largely absent in raffinose-grown cells, which is somewhat consistent with the smaller effect of sch9Δ and sns1 on HAP4-lacZ expression compared with their effect on CAT8-lacZ expression in diploid cells (Figure 3B, right panel). It is possible that Hap4-HA protein expression was subject to a saturation effect due to its much higher expression level in raffinose-grown cells than CAT8 (Figure 3A).
Mutations in MCK1 and GPB1/GPB2 partially suppress sch9Δ
Since the identities of SNS1 and SNS2 were thus far unknown, we looked for sns1 and sns2 mutant phenotypes that might be useful in cloning SNS1 and SNS2. sns1 and sns2 mutants displayed other phenotypes, including the formation of papillae and tanner-colored colonies, increased adhesive growth, prominent vacuolar structures, and larger cell size (Figure 2, A and C, and data not shown). Phenotypes that are helpful in library complementation such as growth defects at high or low temperatures, or sensitivity to drugs such as caffeine, were not found for sns1 mutants.
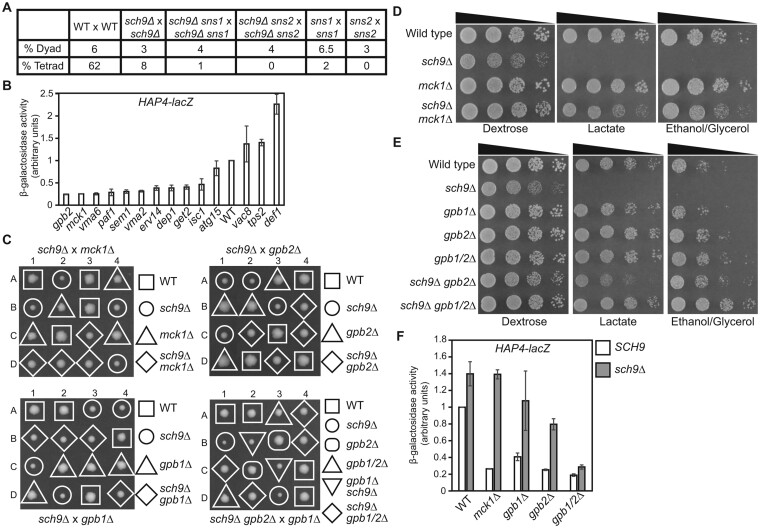
During our genetic analysis of sch9Δ mutants, we found that an sch9Δ homozygous diploid mutant showed reduced tetrad formation by 7.7-fold compared with wild type (Figure 4A). Since sns1 and sns2 mutations reverse the growth defects of sch9Δ mutant cells, we asked whether they would also suppress sporulation defects. Instead, however, the introduction of sns1 and sns2 mutations further reduced efficiency of tetrad formation, suggesting that mutations in SNS1 and SNS2 may have their own sporulation defects. Indeed, both sns1 and sns2 homozygous mutant strains showed almost no tetrad formation. Two large-scale studies have been conducted to identify genes required for sporulation (Deutschbauer et al. 2002; Enyenihi and Saunders 2003). We decided to use the sporulation phenotype to identify SNS1 and SNS2. Accordingly, we chose 85 genes from the study by Enyenihi and Saunders (2003) and seven genes that showed both a genetic or physical interaction with SCH9 and sporulation defects when mutated (curated data from the Saccharomyces genome database). Since HAP4-lacZ expression was greatly reduced in both sns1 and sns2 single mutant cells, we hypothesized that HAP4-lacZ reporter gene expression could be used to narrow down the list of 92 genes by looking for genes that showed reduced expression of HAP4-lacZ when mutated. Accordingly, we transformed a HAP4-lacZ reporter gene into these 92 haploid mutant strains obtained from the yeast genome deletion project and the resulting transformants were analyzed for β-galactosidase activity. We found mutations in ATG15, DEF1, DEP1, ERV14, GPB2, GET2, ISC1, MCK1, PAF1, SEM1, TPS2, VAC8, VMA2, and VMA6 reduced HAP4-lacZ expression by at least threefold. We then generated these 14 mutant strains via tetrad analysis from respective heterozygous mutant strains obtained from the yeast genome deletion project. Re-examination of HAP4-lacZ expression confirmed that HAP4-lacZ expression was reduced by at least twofold in 10 of the 14 mutants (Figure 4B). We crossed each of the 14 mutant strains with an sch9Δ mutant and obtained respective double mutant strains via tetrad analysis. Analysis of colony size on the dissection plates revealed that mutations in MCK1 and GPB2 partially suppressed the growth defects due to sch9Δ, which was confirmed by serial dilution growth assays (Figure 4, C, D, and E). Interestingly, mutations in MCK1 and GPB2 also caused the largest decreases in HAP4-lacZ expression among the 14 mutants (Figure 4B). A large decrease in HAP4-lacZ expression was observed in sns1 and sns2 mutant cells (Figure 3, B and C). Together, our data suggest that reduced HAP4 expression correlates with sch9Δ suppression.
Figure 4.
Mutations in MCK1 and GPB1/2 suppress the growth defects of sch9Δ mutant cells. (A) Mutations in SCH9, SNS1, and SNS2 lead to severe sporulation defects. Frequency of dyads and tetrads in the indicated isogenic diploid strains was calculated after eight days of growth in sporulation media. WT × WT, BY4741 × BY4742; sch9Δ × sch9Δ, PPY745; sch9Δ sns1 × sch9Δ sns1, PPY741; sch9Δ sns2 × sch9Δ sns2, ZLY6619 × ZLY6626; sns1 × sns1, PPY737; sns2 × sns2, ZLY6581 × ZLY6614. (B) β-galactosidase activities of a HAP4-lacZ reporter gene in haploid wild type and 14 deletion mutant strains in genes whose mutation leads to sporulation defects in homozygous mutant cells. Cells were grown to mid-log phase in YNBcas5D medium and assayed for β-galactosidase activities. (C) Tetrad analysis of sch9Δ (ZLY6651) crossed with mck1Δ (ZLY6606), gpb1Δ (PPY757), or gpb2Δ (ZLY6604), and sch9Δ gpb2Δ (ZLY6677) × gpb1Δ (PPY757) on YPD plates. Each dissected tetrad, numbered 1-4, yielded four spores, labeled A-D. (D) Wild type (BY4741) and isogenic sch9Δ (ZLY6649), mck1Δ (ZLY6606), and sch9Δ mck1Δ (ZLY6652) mutant strains were serially diluted and spotted on YPD, YPL, and YPEG plates. (E) Wild type (BY4741) and isogenic sch9Δ (ZLY6650), gpb1Δ (PPY757), gpb2Δ (ZLY6604), gpb1/2Δ (PPY763), sch9Δ gpb2Δ (ZLY6677), and sch9Δ gpb1/2Δ (PPY767) mutant strains were serially diluted and spotted on YPD, YPL, and YPEG plates. (F) β-galactosidase activities of HAP4-lacZ in wild type (BY4741) and isogenic mck1Δ (ZLY6606), gpb1Δ (PPY757), gpb2Δ (ZLY6604), and gpb1/2Δ (PPY763) mutant strains with or without an sch9Δ mutation grown in YNBcas5D medium. The HAP4-lacZ reporter gene activity in the wild type strain is set as 1.
Mck1 is a protein kinase of the glycogen synthase kinase-3 family, which has three other members in budding yeast, Mds1, Ygk3, and Mrk1 (Bianchi et al. 1993; Puziss et al. 1994; Hardy et al. 1995; Hirata et al. 2003). Mck1 and Ygk3 are paralogs, sharing 43% sequence identity and 64% sequence similarity. We generated an sch9Δ ygk3Δ double mutant via tetrad analysis and found that ygk3Δ did not suppress the growth defects of sch9Δ. We then generated sch9Δ mck1Δ ygk3Δ triple mutant strains and compared their growth phenotype with sch9Δ mck1Δ double mutants. We found their growth was indistinguishable. These results suggest that sch9Δ suppression by mck1Δ is not shared by ygk3Δ.
Gpb2 is a negative regulator of the Ras/PKA signaling pathway and has a paralog Gpb1 (Harashima and Heitman 2002; Broach 2012). We crossed a gpb1Δ mutant with an sch9Δ mutant and obtained sch9Δ gpb1Δ double mutant strains via tetrad analysis. We found gpb1Δ marginally suppressed the growth defect of sch9Δ mutant cells (Figure 4C, lower left panel). Nevertheless, tetrad analysis revealed the growth defects of sch9Δ mutant cells on dextrose medium are almost completely suppressed by a gpb1/2Δ double mutation (Figure 4C, lower right panel). We performed a serial dilution growth assay on sch9Δ gpb1Δ double mutant strains and found they accumulated suppressor mutations to a greater extent than sch9Δ mutant cells. This prevented us from drawing a definitive conclusion on whether there was weak suppression of sch9Δ by gpb1Δ. However, Figure 4E shows that a gpb1/2Δ double mutation suppresses the growth defects of sch9Δ mutants better than a gpb2Δ single mutation on both dextrose medium and non-fermentable carbon sources. Together, these results indicate that GPB1 and GPB2 play a redundant role in inhibiting cell growth in sch9Δ mutant cells, with GPB2 playing a bigger role than GPB1.
We examined HAP4-lacZ expression in sch9Δ mck1Δ, gpb1Δ, sch9Δ gpb1Δ, sch9Δ gpb2Δ, and sch9Δ gpb1/2Δ mutant cells grown in dextrose medium. We found that increased expression of HAP4-lacZ due to sch9Δ is not affected by mck1Δ, is partially reduced by gpb2Δ, and is almost completely reversed by a gpb1/2Δ double mutation (Figure 4F). gpb1Δ single and gpb2Δ single mutations reduced HAP4-lacZ expression by 2.5-fold and fourfold, respectively, while a gpb1/2Δ double mutation reduced HAP4-lacZ expression by 5.3-fold. The large error bar of HAP4-lacZ activity in sch9Δ gpb1Δ likely reflects fast accumulation of other suppressor mutations, which could reduce HAP4-lacZ expression as observed in sch9Δ sns1 and sch9Δ sns2 mutant cells. Taken together, there seems to be a correlation between suppression of sch9Δ mutant growth defects and the extent of reduction in HAP4-lacZ expression due to gpb1Δ, mck1Δ, gpb2Δ, gpb1/2Δ, sns1, and sns2 mutations. mck1Δ, gpb1Δ, gpb2Δ, and gpb1/2Δ mutant colonies did not have a clear papillae phenotype like sns1 and sns2 mutants. Therefore, SNS1 and SNS2 are unlikely to be MCK1, GPB1, or GPB2.
IRA1 and IRA2 play a largely non-redundant role in sch9Δ suppression
Alongside our genetic screening of mutant genes that lead to sporulation defects, we sought to clone the SNS1 gene by direct complementation using a yeast genomic DNA library. Our first attempt was to use the color phenotype of W303 background strains, as sch9Δ cells with an ade2 mutation leads to small white colonies, while sch9Δ sns1 double mutant cells form red colonies (Figure 1). We transformed sch9Δ sns1 double mutant cells with a yeast genomic DNA library and looked for transformants that were white, which would indicate complementation of sns1. We conducted several rounds of transformation and recovered many white colonies, none of which contained a sns1 complementing plasmid. We hypothesized that spontaneous mutations might complicate library plasmid complementation of sch9Δ in haploid strains. Accordingly, we used sch9Δ/sch9Δ sns1/sns1 diploid mutant cells in the S288c background and transformed them with a yeast genomic DNA library. In this strain background, we expected that sch9Δ sns1 transformants containing a complementing SNS1 plasmid would display sch9Δ mutant phenotypes of slow growth on dextrose medium and no growth on non-fermentable carbon sources. Of the approximate 12,000 transformants on dextrose medium, dozens of colonies that were slow-growing were chosen for further analysis. Transformant colonies were then examined for severe growth defects on glycerol medium. We found one transformant that showed plasmid-dependent growth defects. We recovered the plasmid from the transformant, re-transformed it into the sch9Δ/sch9Δ sns1/sns1 mutant cells, and found that the library plasmid could complement the sns1 mutation (Figure 5A). Sequencing of the sns1-complementing plasmid found that it carried a ∼15kb chromosomal DNA fragment, which included complete coding sequences of IRA2 and REX4, and partial coding sequences of ATG19 and AVO1.
Figure 5.
SNS1 and SNS2 are IRA2 and IRA1, respectively. (A) A sns1 mutation is complemented by a genomic DNA library plasmid. Wild type (BY4741 × BY4742), sch9Δ × sch9Δ (PPY745), and sch9Δ sns1 × sch9Δ sns1 (PPY741) mutant strains were transformed with control vector YCp50 or the sns1-complementing library plasmid, YCp50-lib. (pPP322). Transformants were serially diluted and spotted on YNBcasD (Dextrose) and YNBcasL (Lactate) plates. (B) Tetrad analysis of IRA2/ira2Δ (ZLY6578, left panel) and ira2Δ (ZLY6601) × sch9Δ (ZLY6649) (middle panel) on YPD plates. Wild type (BY4741) and isogenic sch9Δ (ZLY6649), ira2Δ (ZLY6609), and sch9Δ ira2Δ (ZLY6661) mutant strains were serially diluted and spotted on YPD, YPL, and YPEG plates (right panel). (C) Tetrad analysis of IRA1/ira1Δ (ZLY6656, left panel) and sch9Δ (ZLY6649) × ira1Δ (ZLY6648) (middle panel) on YPD plates. Wild type (BY4741) and isogenic sch9Δ (ZLY6650), ira1Δ (ZLY6648), and sch9Δ ira1Δ (PPY754) mutant strains were serially diluted and spotted on YPD, YPL, and YPEG plates (right panel). (D) Diploid strains obtained from the crossings among sch9Δ ira1Δ (PPY755), sch9Δ ira2Δ (ZLY6661), sch9Δ sns1 (PPY733, PPY734), and sch9Δ sns2 (ZLY6619, ZLY6626) mutant strains as indicated were serially diluted and spotted on YPD and YPL plates. BY4741 × BY4742 and sch9Δ × sch9Δ (PPY745) strains were included as controls. (E) β-galactosidase activities of a HAP4-lacZ reporter gene in wild type (BY4741), ira1Δ (ZLY6647), and ira2Δ (ZLY6609) mutant strains with or without an sch9Δ deletion mutation grown in YNBcas5D medium. The HAP4-lacZ reporter gene activity in the wild type strain is set as 1. (F) IRA2 from sns1 mutant cells has a frameshift mutation. The chromatogram shows the sequence of IRA2 containing the frameshift mutation from a sns1 mutant. The nucleotide C missing in the IRA2 gene from the sns1 mutant was highlighted in red in the wild type IRA2 sequence.
ira2 mutant cells have been reported to exhibit increased adhesive growth and sporulation defects (Tanaka et al. 1990b; Barrett et al. 2012). Our data showed that sns1 mutants also had sporulation defects and inceased agar adhesion (Figures 2C and 4A), suggesting IRA2, not REX4, complements the sns1 mutation. During dissection of tetrads from sporulated IRA2/ira2Δ cells, we observed that ira2Δ mutant colonies grew slower than wild type and formed papillae, rough-edged colonies (Figure 5B, left panel), similar to sns1 colonies (Figure 2A, left panel), suggesting IRA2 is SNS1. We then crossed an ira2Δ mutant with an sch9Δ mutant and obtained sch9Δ ira2Δ double mutant strains via tetrad analysis (Figure 5B, middle panel). A serial dilution growth assay showed that ira2Δ completely suppressed the growth defects of sch9Δ mutant cells on both dextrose and non-fermentable carbon sources (Figure 5B, right panel). IRA2 has a paralog, IRA1. We conducted similar genetic analyses and found that the phenotypes of ira1Δ mutants are similar to those of ira2Δ mutants and that ira1Δ also completely suppressed the growth defects of sch9Δ mutant cells (Figure 5C).
We next sought to determine whether IRA1 is SNS2. We crossed mutant strains among sch9Δ sns1, sch9Δ sns2, sch9Δ ira1Δ, and sch9Δ ira2Δ mutants and analyzed cell growth of the resultant diploid strains via a serial dilution growth assay. Figure 5D shows that sch9Δ ira1Δ × sch9Δ sns2 and sch9Δ ira2Δ × sch9Δ sns1 diploid strains exhibited similar growth to wild type on both dextrose and lactate medium, suggesting that IRA1 is SNS2 and IRA2 is SNS1. sch9Δ ira1Δ × sch9Δ ira2Δ, sch9Δ ira1Δ × sch9Δ sns1, and sch9Δ ira2Δ × sch9Δ sns2 diploid strains grew slightly better than sch9Δ/sch9Δ homozygous mutant, but still exhibited clear growth defects. This result is somewhat surprising. Ira1 and Ira2 are homologous proteins and play a partially redundant role in inhibiting the activity of Ras1 and Ras2. On the one hand, loss of both copies of IRA1 or IRA2 can restore growth of sch9Δ/sch9Δ mutant cells to wild type level. On the other hand, loss of one copy each of IRA1 and IRA2 only partially suppresses the growth defects of sch9Δ/sch9Δ mutant cells. This result suggests that IRA1 and IRA2 play partially redundant but distinct roles in regulating cell growth of sch9Δ/sch9Δ mutant cells. It has been reported previously that IRA1 and IRA2 have similar but not the same functions (Tanaka et al. 1990b). Our results resemble those of ira1 and ira2 mutations in suppressing the slow growth phenotype of reg1 mutant cells (Barrett et al. 2012). fst1 and fst2 are two of the complementation groups of mutants that suppress the reg1 mutation. fst1 does not complement ira1Δ, and fst2 does not complement ira2Δ for reduced glycogen accumulation and increased agar adhesion. However, fst1 complements fst2 in the said phenotypes. The authors concluded that fst1 is IRA1 and fst2 is IRA2.
We also analyzed the expression of the HAP4-lacZ reporter gene in ira1Δ, ira2Δ, sch9Δ ira1Δ, and sch9Δ ira2Δ mutant cells grown in dextrose medium (Figure 5E). We found that HAP4-lacZ expression was greatly reduced in these mutants, to the same extent as what was observed in sns1, sns2, sch9Δ sns1, and sch9Δ sns2 mutant cells. Taken together, our data suggest that IRA1 and IRA2 play a largely non-redundant role in inhibiting cell growth in sch9Δ mutant cells. To confirm SNS1 is IRA2, we sequenced the IRA2 gene from a sns1 mutant and found a frameshift mutation in the codon for serine residue 1870 (Figure 5F). The deletion of the nucleotide C from codon TCT to TT leads to a stop codon almost right after the mutation. Ira2 is a GTPase-activating protein for Ras1 and Ras2, with its Ras GTPase-activating domain being comprised of amino acid residues 1651–1989. This mutation in IRA2 truncates this domain and helps to explain that sns1 acts like a total-loss-of-function allele of IRA2 (Figures 2, 3, and 5).
Mutations in genes encoding components of the PKA signaling pathway affect the expression of lacZ reporter genes under the control of ADR1, CAT8, and/or HAP4 promoters
Ira1 and Ira2 are GTPase-activating proteins of Ras1 and Ras2 and function as negative regulators of the Ras/PKA signaling pathway (Tanaka et al. 1990a, 1991). Gpb1 and Gpb2 interact with Ira1/2 and indirectly inhibit Ras1/2 by promoting the stability of Ira1/2 (Harashima et al. 2006). Our data show a clear correlation between the degree of suppression of sch9Δ mutant growth defects and reduced expression of HAP4-lacZ reporter genes caused by mutations in GPB1, GPB2, IRA1, IRA2, and MCK1. Activation of PKA signaling is known to suppress the growth defects of sch9Δ mutant cells (Toda et al. 1988). These results raise the possibility that PKA may inhibit the expression of HAP4, CAT8, and ADR1. The effect of sns1 and sns2 mutations on ADR1 expression is consistent with the reported finding that the expression of an ADR1-lacZ reporter gene in dextrose-grown cells is reduced by 9.9-fold due to bcy1Δ, which constitutively activates PKA (Dombek and Young 1997).
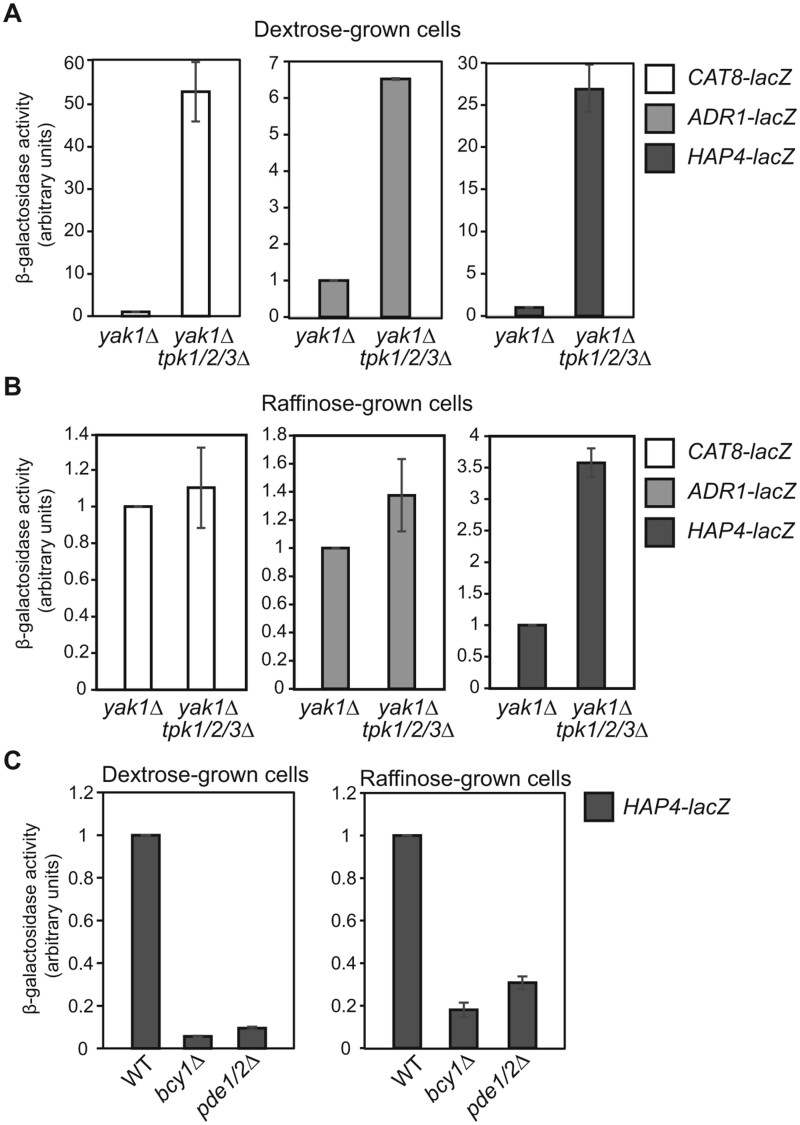
We decided to determine the effect of a PKA mutation on the expression of lacZ reporter genes under the control of these three gene promoters. PKA has three functionally redundant catalytic subunits, Tpk1, Tpk2, and Tpk3 (Toda et al. 1987a). A triple deletion in TPK1/2/3 is lethal, which can be suppressed by a yak1Δ mutation (Garrett and Broach 1989). We generated a tpk1/2/3Δ yak1Δ quadruple mutant and conducted a β-galactosidase assay in cells grown in dextrose as well as raffinose medium. A yak1Δ mutant strain was included as the control and was found to have little to no effect on the expression of these three lacZ reporter genes. In dextrose medium, tpk1/2/3Δ had a dramatic effect on the expression of the CAT8, ADR1, and HAP4 reporter genes and increased their expression by 54-, 6.5-, and 28-fold, respectively (Figure 6). In raffinose medium, tpk1/2/3Δ increased HAP4-lacZ expression by 3.6-fold, while it had little effect on the expression of CAT8-lacZ and ADR1-lacZ reporter genes. In contrast, constitutive activation of PKA activity due to a bcy1Δ mutation or a pde1Δ pde2Δ double mutation resulted in reduced expression of the HAP4-lacZ reporter gene in both dextrose and raffinose-grown cells. bcy1Δ and pde1Δ pde2Δ mutant cells are known to have growth defects on non-fermentable carbon sources (Nikawa et al. 1987; Toda et al. 1987b), which may be explained by a stronger reduction in HAP4 expression, and possibly CAT8 and ADR1 well, than what is observed in ira1 and ira2 single mutant cells. Together, these data indicate that PKA is a strong negative regulator of the expression of ADR1, CAT8, and HAP4 under glucose repression conditions.
Figure 6.
Mutations in genes encoding components of the PKA signaling pathway affect the expression of lacZ reporter genes under the control of ADR1, CAT8, and/or HAP4 promoters. (A-B) β-galactosidase activities of CAT8-lacZ, ADR1-lacZ, and HAP4-lacZ reporter genes in yak1Δ (ZLY4313) and yak1Δ tpk1/2/3Δ (ZLY5023) mutant strains grown in YNBcas5D (A) and YNBcasR (B) medium. β-galactosidase activity from each of the three reporter genes in the yak1Δ strain was set as 1, respectively. (C) β-galactosidase activities of a HAP4-lacZ reporter gene in wild type (BY4741), bcy1Δ (ZLY6715), and pde1Δ pde2Δ (PPY776) mutant cells grown in dextrose and raffinose medium. The activity of HAP4-lacZ in wild type cells was set as 1.
Mutations in YAK1 and PDE2 partially suppress the growth defects of sch9Δ mutant cells
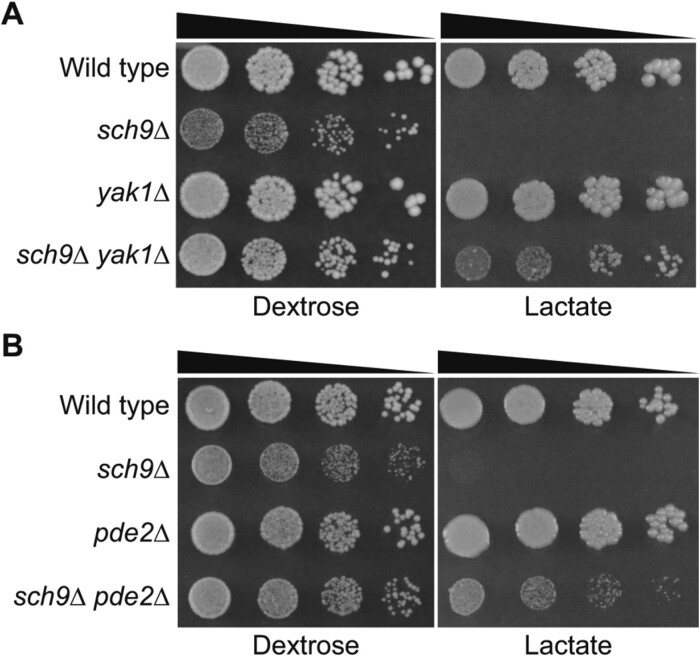
IRA1, IRA2, GPB1, and GPB2 are negative regulators of Ras1 and Ras2. Mutations in RAS2 lead to growth defects on nonfermentable carbon sources (Tatchell et al. 1985; Fasano et al. 1988). It has been suggested that Ras2 can regulate mitochondrial biogenesis and function via both PKA-dependent and independent mechanisms (Hlavata et al. 2003; Hlavata and Nystrom 2003). We sought to determine whether the suppression of an sch9Δ mutant’s growth defects on non-fermentable carbon source is mediated through PKA-dependent or independent Ras2 functions. To that end, we analyzed the effect of mutations in genes functioning in the PKA pathway downstream of Ras1 and Ras2. Since bcy1Δ and pde1Δ pde2Δ lead to growth defects on non-fermentable carbon sources and may complicate the analysis of sch9Δ suppression, we determined the effect of single gene mutations of YAK1, PDE1, and PDE2 on sch9Δ suppression by generating double mutant strains via tetrad analysis. We found that yak1Δ and pde2Δ, but not pde1Δ, partially suppressed the growth defect of sch9Δ mutant cells on dextrose dissection plate. A cell growth assay via serial dilution confirmed that yak1Δ and pde2Δ single mutations partially suppressed the growth defects of sch9Δ mutant cells on both dextrose and lactate medium (Figure 7, A and B). Our results suggest that the suppression of growth defects of sch9Δ mutant cells on dextrose medium and non-fermentable carbon sources is mediated at least through the activation of the PKA pathway. We cannot exclude the possibility that activation of PKA-independent Ras2 function(s) may also contribute to sch9Δ suppression in some of the suppressor mutants.
Figure 7.
Mutations in YAK1 and PDE2 partially suppress the growth defects of sch9Δ mutant cells. (A) sch9Δ suppression by yak1Δ. Wild type (BY4741) and isogenic sch9Δ (ZLY6650), yak1Δ (ZLY4313), and sch9Δ yak1Δ (ZLY6707) mutant strains were serially diluted and spotted on dextrose (YPD) and lactate (YPL) medium. (B) sch9Δ suppression by pde2Δ. Wild type (BY4741) and isogenic sch9Δ (ZLY6650), pde2Δ (ZLY4368), and sch9Δ pde2Δ (PPY758) mutant cells were serially diluted and spotted on YPD and YPL plates.
Chronological lifespan extension due to sch9Δ is reversed by an ira2 mutation
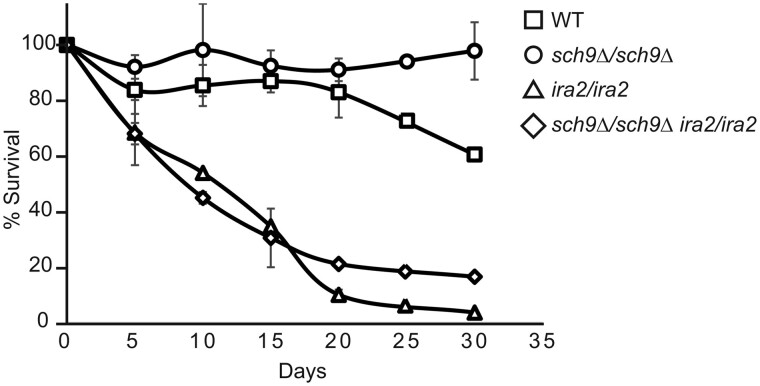
sch9Δ mutant cells have been previously shown to extend chronological lifespan (Fabrizio et al. 2001; Longo 2003; Wei et al. 2008). Mutations in IRA2 reverse the growth defects of sch9Δ mutant cells. We wanted to determine the effect of an ira2 mutation on chronological lifespan in sch9Δ mutant cells. To that end, we generated a survival curve of wild type, sch9Δ, ira2, and sch9Δ ira2 diploid strains grown in raffinose media (Figure 8). Cell growth in raffinose medium mimics glucose limitation conditions, which accounts for the relatively long chronological life span in wild type cells. As expected, sch9Δ mutant cells had better survival than wild type. Both ira2 and sch9Δ ira2 mutant strains showed a significant decrease in cell survival. Mutations in genes in the PKA signaling pathway that inhibit or activate PKA lead to a longer or shorter chronological lifespan, respectively (Fabrizio et al. 2001, 2003; Longo 2003). Reductions in Sch9 activity and PKA pathway signaling have been reported to extend life span in an additive manner (Wei et al. 2008). ira2 mutants are known to have increased Ras/PKA pathway activity (Tanaka et al. 1990b). Together, these data suggest that activation of PKA signaling due to an ira2 mutation antagonizes lifespan extension in sch9Δ mutant cells.
Figure 8.
Chronological lifespan extension due to an sch9Δ mutation is reversed by an ira2 mutation. Wild type (BY4741 × BY4742) and isogenic sch9Δ × sch9Δ (PPY745), ira2 × ira2 (PPY737), and sch9Δ ira2 × sch9Δ ira2 (PPY741) mutant strains were grown in YPR media for 30 days after cultures reached saturation. Percent survival was calculated using colony forming assay at the indicated time points. Errors bars represent the standard deviation of the results from two independent experiments.
Discussion
Deletion of SCH9 has been previously reported to cause a slow growth phenotype on dextrose medium, but the growth phenotypes demonstrated on non-fermentable carbon sources are inconsistent. In this report, we propose that the rapid accumulation of suppressor mutations in sch9Δ mutant cells could account for the discrepancies in the published results. All 18 of the isolated spontaneously arising, recessive mutations that suppressed the growth defects of sch9Δ mutant cells were proposed to be in either IRA1 or IRA2. Using genetic techniques, we also found that mutations in GPB1, GPB2, and MCK1 suppress the growth defects of sch9Δ mutant cells in both dextrose medium and non-fermentable carbon sources. Many studies on the cellular functions of Sch9 employed strains of the W303 and S288c backgrounds, which we also used in this report. The discrepancies in the published literature regarding changes in the expression of HAP4, genes encoding mitochondrial proteins, stress-responsive genes, and oxygen consumption due to sch9 mutations may be attributed to the presence or absence of sch9 suppressor mutations (Crauwels et al. 1997; Fabrizio et al. 2003; Pedruzzi et al. 2003; Jorgensen et al. 2004; Roosen et al. 2005; Lavoie and Whiteway 2008; Smets et al. 2008; Pan and Shadel 2009; Pan et al. 2011; Teixeira et al. 2014). It is also possible that the use of other strain backgrounds, growth conditions, and growth phase could account for the discrepancies. By removing the complication of sch9Δ suppressor mutations, future studies will help reconcile the reported conflicting functions of Sch9 and may uncover novel functions for Sch9.
Fast accumulation of suppressor mutations in sch9Δ mutant cells was so prevalent that it was difficult for us to maintain sch9Δ mutant strains. It is not clear why there are frequent spontaneous mutations in IRA1 and IRA2 loci, which were calculated to arise at a frequency of 1.1 × 10−3 (Halme et al. 2004). Halme et al. further showed that missense mutations and single base-pair deletions or insertions are behind the high-frequency genetic events in IRA1 and IRA2 genes. During the review of this manuscript, we were brought to the attention of a report by van Leeuwen et al., showing that IRA2 is a candidate suppressor gene in a spontaneous sch9Δ suppressor mutant (2016). sch9Δ cells are slow-growing on dextrose media, and the suppressor mutants grow considerably faster and quickly take over the population. We found it was necessary to purify sch9Δ strains right before they were used in each experiment. When purified sch9Δ mutant strains are grown in liquid cultures, it is also important to save small aliquots of cultured cells and examine their growth on dextrose plate medium to determine the extent of “contamination” by spontaneous suppressor mutants. Since most suppressor mutations we isolated were recessive, we propose that sch9Δ homozygous diploid mutant strains should be used for studies on the cellular functions of Sch9. The other alternative is to use analog-sensitive sch9 alleles, but the accumulation of suppressor mutations is still expected to take place. Given that all 18 recessive suppressor mutations we isolated were in either IRA1 or IRA2, another option is to use sch9Δ haploid mutant strains carrying an extra copy of IRA1 and IRA2. However, extra copies of IRA1 and IRA2 may reduce the activity of the Ras/PKA signaling pathway, which could skew conclusions on the cellular functions for Sch9.
Suppression of the growth defects of sch9Δ mutants by IRA1, IRA2, GPB1, and GPB2 is consistent with the notion that PKA and Sch9 play overlapping roles in regulating cell growth in response to nutrient conditions. It has been reported that activation of PKA can suppress the mutant phenotype of sch9Δ and vice versa. IRA1 and IRA2 are GTPase-activating proteins of Ras1/2 and function as negative regulators of the Ras/PKA signaling pathway. Gpb1/2 are negative regulators of PKA signaling by inhibiting Ras, directly regulating PKA, and/or by other means (reviewed in Broach 2012). Therefore, the suppression of growth defects of sch9Δ mutant cells due to mutations in IRA1/2 and GPB1/2 can be explained by increased activity of Ras/PKA signaling. The mechanism behind mck1Δ suppression of sch9Δ is less clear. Mck1 was originally characterized for its role in meiosis and chromosome segregation (Neigeborn and Mitchell 1991; Shero and Hieter 1991). It has also been shown that Mck1 may be required for the activity of Msn2/4 (Hirata et al. 2003; Sadeh et al. 2011; Gutin et al. 2015, 2019). Mutations in IRA2, MCK1, MSN2/4, YAK1, and RIM15 lead to a similar transcriptional response of a HSP12 reporter, while sch9Δ has an opposite effect (Gutin et al. 2015, 2019). Mutations in YAK1, RIM15, and MSN2/4 have also been reported to suppress cell lethality associated with a tpk1/2/3 triple mutation (reviewed in Broach 2012). It is conceivable that mck1Δ may suppress the growth defects of sch9Δ through the Ras/PKA pathway. A potential role of Mck1 in PKA signaling or in another pathway will be examined in future studies.
The effect of sch9Δ, ira1Δ, ira2Δ, gpb1/2Δ, and mck1Δ on the expression of HAP4, ADR1, and CAT8 presents an interesting paradox between cell growth and gene expression. Hap4, Adr1, and Cat8 are global transcriptional activators that are important for the expression of genes involved in gluconeogenesis, the glyoxylate cycle, mitochondrial biogenesis, and/or oxidative metabolism. sch9Δ mutants are unable to utilize non-fermentable carbon sources for growth, which suggests a decrease in the expression of genes involved in respiratory metabolism and gluconeogenesis. Instead, however, sch9Δ increases the expression of genes encoding key transcriptional activators that are required for the utilization of non-fermentable carbon sources. On the other hand, sch9Δ ira1 and sch9Δ ira2 mutant strains can grow robustly on non-fermentable carbon sources, but they exhibit reduced promoter activity of these three transcriptional activators. One possibility may have to do with greatly increased expression of HAP4, CAT8, and ADR1 genes under glucose limitation conditions (Figure 3A). Due to high levels of transcripts, a large reduction in the promoter activity in sch9Δ ira1 and sch9Δ ira2 mutant cells may not translate into reduced protein levels to the same degree. Functional redundancy may also help explain the lack of growth defects in sch9Δ ira1 and sch9Δ ira2 strains on non-fermentable carbon sources. For example, in raffinose-grown cells, CAT8 expression is reduced in these two mutant cells compared with wild type. However, the function of Cat8 is redundant with other transcriptional activators and its downregulation in these mutants may not prevent cells from utilizing non-fermentable carbon sources (reviewed in Schuller 2003; Turcotte et al. 2010). It is also possible that increased expression of HAP4, CAT8, and ADR1 due to sch9Δ may be an adaptative response to cell growth defects. It is important to note that a large difference in transcriptional response determined by the lacZ reporter genes is not translated to the same degree of change in HA-tagged Cat8 or Hap4 determined by Western blotting (Figure 3). This may suggest that translation of CAT8 and HAP4 transcripts may be different from that of lacZ. It is also possible that Cat8 and Hap4 may be subjected to post-translational controls that are not available to the lacZ protein.
The transcriptional response mediated by PKA has been proposed to be conducted mainly through Msn2/4, Gis1, and Rim15 (Roosen et al. 2005; Broach 2012; Pfanzagl et al. 2018). Our data show that PKA is a negative regulator of HAP4, CAT8, and ADR1 (Figure 6). The expression of ADR1 is under negative control by PKA signaling (Dombek and Young 1997). Downregulation of the expression of these three genes in ira1, ira2, and gpb1/2 mutants is likely to be caused by the activation of PKA signaling (Figures 3–5). How does PKA regulate the expression of HAP4, ADR1, and CAT8? Both HAP4 and CAT8 are subject to Mig1-dependent downregulation under glucose repression conditions (reviewed in Schuller 2003; Turcotte et al. 2010; Broach 2012). However, it is unknown whether the effect is direct or indirect. Expression of HAP4 is dependent on Hap1 and is subject to auto-regulatory control (Zhang et al. 2017). Increased expression of CAT8 is dependent on Hap2/3/4/5 under glucose limitation conditions (Rahner et al. 1996). Apart from the findings from this study and others that PKA negatively regulates ADR1 expression, transcriptional regulation of ADR1 is largely unknown. Future studies will be needed to understand the molecular mechanisms underlying the regulation of HAP4, ADR1, and CAT8 by PKA.
Acknowledgments
The authors thank Manika Bhondeley and Savannah Rappold for technical support and Tammy Pracheil Bavaret for editing this manuscript.
Funding
This study was supported by a grant from the U.S. National Institutes of Health (1R15GM121998-01).
Conflicts of interest
Authors have no conflict of interest to declare.
Literature cited
- Amberg DC, Burke DJ, Strathern JN.. 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. New York, NY: Cold Spring Harbor Laboratory, Cold Spring Harbor. [Google Scholar]
- Barrett L, Orlova M, Maziarz M, Kuchin S.. 2012. Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot Cell. 11:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi MW, Plyte SE, Kreis M, Woodgett JR.. 1993. A Saccharomyces cerevisiae protein-serine kinase related to mammalian glycogen synthase kinase-3 and the Drosophila melanogaster gene shaggy product. Gene. 134:51–56. [DOI] [PubMed] [Google Scholar]
- Broach JR. 2012. Nutritional control of growth and development in yeast. Genetics. 192:73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhwar R, Fang G, Hirsch JP.. 2011. Kelch repeat proteins control yeast PKA activity in response to nutrient availability. Cell Cycle. 10:767–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM.. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 95:1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M.. 2009. A molecular mechanism of chronological aging in yeast. Cell Cycle. 8:1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev. 26:223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crauwels M, Donaton MC, Pernambuco MB, Winderickx J, de Winde JH, et al. 1997. The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable-growth-medium-induced (FGM) pathway. Microbiology. 143:2627–2637. [DOI] [PubMed] [Google Scholar]
- Denis CL, Audino DC.. 1991. The CCR1 (SNF1) and SCH9 protein kinases act independently of cAMP-dependent protein kinase and the transcriptional activator ADR1 in controlling yeast ADH2 expression. Mol Gen Genet. 229:395–399. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Williams RM, Chu AM, Davis RW.. 2002. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 99:15530–15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek KM, Young ET.. 1997. Cyclic AMP-dependent protein kinase inhibits ADH2 expression in part by decreasing expression of the transcription factor gene ADR1. Mol Cell Biol. 17:1450–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi AH, Saunders WS.. 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 163:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Liou LL, Moy VN, Diaspro A, Valentine JS, et al. 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 163:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD.. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 292:288–290. [DOI] [PubMed] [Google Scholar]
- Fasano O, Crechet JB, De Vendittis E, Zahn R, Feger G, et al. 1988. Yeast mutants temperature-sensitive for growth after random mutagenesis of the chromosomal RAS2 gene and deletion of the RAS1 gene. Embo J. 7:3375–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L.. 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3:1166–1178. [DOI] [PubMed] [Google Scholar]
- Garrett S, Broach J.. 1989. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3:1336–1348. [DOI] [PubMed] [Google Scholar]
- Geyskens I, Kumara S, Donaton M, Bergsma J, Thevelein J, et al. 2001. Expression of mammalian PKB partially complements deletion of the yeast protein kinase Sch9. In: Proceedings of NATO Conference. Amsterdam, Netherlands: IOS Press. p. 117–126. [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH.. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutin J, Joseph-Strauss D, Sadeh A, Shalom E, Friedman N.. 2019. Genetic screen of the yeast environmental stress response dynamics uncovers distinct regulatory phases. Mol Syst Biol. 15:e8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutin J, Sadeh A, Rahat A, Aharoni A, Friedman N.. 2015. Condition-specific genetic interaction maps reveal crosstalk between the cAMP/PKA and the HOG MAPK pathways in the activation of the general stress response. Mol Syst Biol. 11:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR.. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 116:405–415. [DOI] [PubMed] [Google Scholar]
- Harashima T, Anderson S, Yates JR 3rd, Heitman J.. 2006. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol Cell. 22:819–830. [DOI] [PubMed] [Google Scholar]
- Harashima T, Heitman J.. 2002. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol Cell. 10:163–173. [DOI] [PubMed] [Google Scholar]
- Hardy TA, Wu D, Roach PJ.. 1995. Novel Saccharomyces cerevisiae gene, MRK1, encoding a putative protein kinase with similarity to mammalian glycogen synthase kinase-3 and Drosophila Zeste-White3/Shaggy. Biochem Biophys Res Commun. 208:728–734. [DOI] [PubMed] [Google Scholar]
- Hedges D, Proft M, Entian KD.. 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 15:1915–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavata L, Aguilaniu H, Pichova A, Nystrom T.. 2003. The oncogenic RAS2(val19) mutation locks respiration, independently of PKA, in a mode prone to generate ROS. Embo J. 22:3337–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavata L, Nystrom T.. 2003. Ras proteins control mitochondrial biogenesis and function in Saccharomyces cerevisiae. Folia Microbiol (Praha). 48:725–730. [DOI] [PubMed] [Google Scholar]
- Hirata Y, Andoh T, Asahara T, Kikuchi A.. 2003. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol Biol Cell. 14:302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, et al. 2009. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23:1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, et al. 2011. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 30:3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M.. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science. 297:395–400. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, et al. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, Steffen KK, Westman EA, Hu D, et al. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 310:1193–1196. [DOI] [PubMed] [Google Scholar]
- Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, et al. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 32:1002–1012. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Whiteway M.. 2008. Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot Cell. 7:1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Moir RD, Willis IM.. 2009. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J Biol Chem. 284:12604–12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SI, Broach JR.. 2009. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc Natl Acad Sci U S A. 106:19928–19933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA.. 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 19:6720–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN.. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 189:1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD. 2003. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp Gerontol. 38:807–811. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Pan X, Harashima T, Cardenas ME, Xue Y, et al. 2000. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 154:609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Thiele DJ.. 1999. The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. Embo J. 18:5953–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Mitchell AP.. 1991. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 5:533–548. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Sass P, Wigler M.. 1987. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol Cell Biol. 7:3629–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS.. 2011. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 13:668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Shadel GS.. 2009. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging (Albany NY). 1:131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Ahuir A, Proft M.. 2007. The Sch9 kinase is a chromatin-associated transcriptional activator of osmostress-responsive genes. Embo J. 26:3098–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, et al. 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 12:1607–1613. [DOI] [PubMed] [Google Scholar]
- Pfanzagl V, Gorner W, Radolf M, Parich A, Schuhmacher R, et al. 2018. A constitutive active allele of the transcription factor Msn2 mimicking low PKA activity dictates metabolic remodeling in yeast. Mol Biol Cell. 29:2848–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan VT, Ding VW, Li F, Chalkley RJ, Burlingame A, et al. 2010. The RasGAP proteins Ira2 and neurofibromin are negatively regulated by Gpb1 in yeast and ETEA in humans. Mol Cell Biol. 30:2264–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank M, Perepelkina M, Muller M, Vaga S, Zou X, et al. 2020. Chemical genetics of AGC-kinases reveals shared targets of Ypk1, Protein Kinase A and Sch9. Mol Cell Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusty R, Keil RL.. 2004. SCH9, a putative protein kinase from Saccharomyces cerevisiae, affects HOT1-stimulated recombination. Mol Genet Genomics. 272:264–274. [DOI] [PubMed] [Google Scholar]
- Puziss JW, Hardy TA, Johnson RB, Roach PJ, Hieter P.. 1994. MDS1, a dosage suppressor of an mck1 mutant, encodes a putative yeast homolog of glycogen synthase kinase 3. Mol Cell Biol. 14:831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahner A, Scholer A, Martens E, Gollwitzer B, Schuller HJ.. 1996. Dual influence of the yeast Cat1p (Snf1p) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene CAT8. Nucleic Acids Res. 24:2331–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randez-Gil F, Bojunga N, Proft M, Entian KD.. 1997. Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol Cell Biol. 17:2502–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, et al. 2005. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol. 55:862–880. [DOI] [PubMed] [Google Scholar]
- Sadeh A, Movshovich N, Volokh M, Gheber L, Aharoni A.. 2011. Fine-tuning of the Msn2/4-mediated yeast stress responses as revealed by systematic deletion of Msn2/4 partners. Mol Biol Cell. 22:3127–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE, Hall MN.. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 24:338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller HJ. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet. 43:139–160. [DOI] [PubMed] [Google Scholar]
- Shero JH, Hieter P.. 1991. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1). Genes Dev. 5:549–560. [DOI] [PubMed] [Google Scholar]
- Smets B, De Snijder P, Engelen K, Joossens E, Ghillebert R, et al. 2008. Genome-wide expression analysis reveals TORC1-dependent and -independent functions of Sch9. FEMS Yeast Res. 8:1276–1288. [DOI] [PubMed] [Google Scholar]
- Sobko A. 2006. Systems biology of AGC kinases in fungi. Sci Stke. 2006:re9. [DOI] [PubMed] [Google Scholar]
- Soulard A, Cremonesi A, Moes S, Schutz F, Jeno P, et al. 2010. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell. 21:3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen E, Wilms T, Idkowiak-Baldys J, Smets B, Snijder PD, et al. 2014. The protein kinase Sch9 is a key regulator of sphingolipid metabolism in Saccharomyces cerevisiae. Mol Biol Cell. 25:196–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Lin BK, Wood DR, Tamanoi F.. 1991. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc Natl Acad Sci U S A. 88:468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Satoh T, Marshall MS, Gibbs JB, et al. 1990a. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 60:803–807. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Tamanoi F, Kaziro Y, Matsumoto K, et al. 1990b. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 10:4303–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K, Robinson LC, Breitenbach M.. 1985. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc Natl Acad Sci U S A. 82:3785–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira V, Medeiros TC, Vilaca R, Moradas-Ferreira P, Costa V.. 2014. Reduced TORC1 signaling abolishes mitochondrial dysfunctions and shortened chronological lifespan of Isc1p-deficient cells. Microb Cell. 1:21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH.. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 33:904–918. [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Wigler M.. 1988. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 2:517–527. [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Wigler M.. 1987a. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 50:277–287. [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Scott JD, et al. 1987b. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 7:1371–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B, Liang XB, Robert F, Soontorngun N.. 2010. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 10:2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 26:663–674. [DOI] [PubMed] [Google Scholar]
- van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, et al. 2016. Exploring genetic suppression interactions on a global scale. Science. 354:aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Hu J, Ge H, Cheng C, et al. 2008. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 4:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zheng XF.. 2009. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle. 8:4085–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilms T, Swinnen E, Eskes E, Dolz-Edo L, Uwineza A, et al. 2017. The yeast protein kinase Sch9 adjusts V-ATPase assembly/disassembly to control pH homeostasis and longevity in response to glucose availability. PLoS Genet. 13:e1006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN.. 2006. TOR signaling in growth and metabolism. Cell. 124:471–484. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G.. 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 81:4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Zaman S, Broach JR, Klionsky DJ.. 2007. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 18:4180–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR.. 2009. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol. 5:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR.. 2008. How Saccharomyces responds to nutrients. Annu Rev Genet. 42:27–81. [DOI] [PubMed] [Google Scholar]
- Zhang A, Shen Y, Gao W, Dong J.. 2011. Role of Sch9 in regulating Ras-cAMP signal pathway in Saccharomyces cerevisiae. FEBS Lett. 585:3026–3032. [DOI] [PubMed] [Google Scholar]
- Zhang T, Bu P, Zeng J, Vancura A.. 2017. Increased heme synthesis in yeast induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J Biol Chem. 292:16942–16954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary to draw conclusions in this article are present within the texts, figures, and tables. Strains, plasmids, and other noncommercial research reagents are available upon request.