ABSTRACT
Nearly complete genome sequences were obtained for a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant of concern and two variants of interest from nasopharyngeal swab samples obtained during surveillance activities in urban communities, among individuals with no previous travel history, in Santo Domingo, Dominican Republic.
ANNOUNCEMENT
The first severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) case in the Dominican Republic (DR) was reported on 29 February 2020 (1, 2). SARS-CoV-2 variants have been causing global health concerns due to their higher transmissibility, vaccine neutralization, and rapid accumulation of spike (S) protein mutations over time (3). Early in the pandemic, a D614G mutation was associated with high viral loads and increased transmissibility and was responsible for the first identified community transmission in the DR (4). This virus belongs to the Betacoronavirus genus of the family Coronaviridae, the members of which have been emerging as global threats since SARS-CoV was first recognized in 2002 (5).
Here, we report nearly complete genome sequences for three samples of two SARS-CoV-2 variants circulating in the DR. Nasopharyngeal swab samples were obtained from symptomatic individuals suspected to be infected with SARS-CoV-2 during community surveillance interventions in the city of Santo Domingo, DR (Fig. 1A). This study was approved by the Universidad Iberoamericana (UNIBE) institutional review board (approval number CEI-2020-16) and the National Bioethical Committee (CONABIOS) (approval number 020-2021).
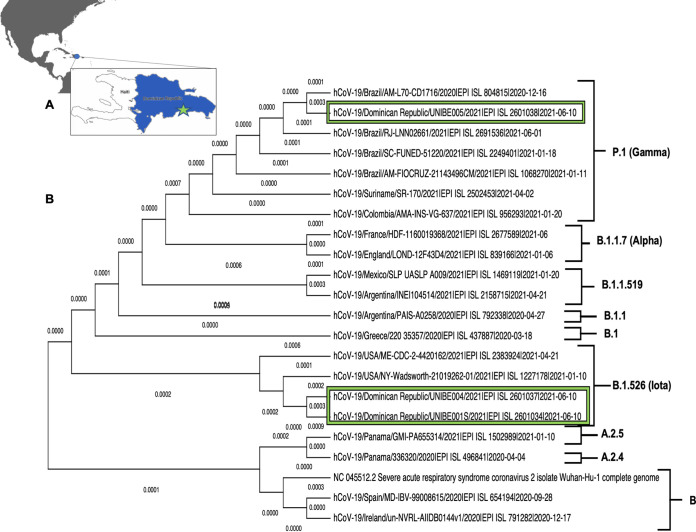
FIG 1.
(A) Sample collection site (green star) in Santo Domingo, DR. Maps were created using ArcGIS and modified using the Sketches app to depict provinces where samples were obtained. (B) Phylogenetic tree of two variants of interest (B.1.526 [Iota]) and one variant of concern (P.1 [Gamma]) detected in DR (green rectangle) with relevant genomes (GISAID accession numbers are indicated). This tree was built with the maximum likelihood algorithm (GTR+G model) using MEGA X.
Samples were processed at the Instituto de Medicina Tropical y Salud Global (IMTSAG)-UNIBE for SARS-CoV-2 RNA amplification. RNA was extracted and purified with a 96-well automated nucleic acid extractor (Biocomma, Shenzhen, China) using the Biocomma viral DNA/RNA purification kit, following the manufacturer’s instructions. For the extraction procedure, 300 μl of sample was utilized, and subsequent amplification of these samples was carried out on an ABI 7500 Fast system (Applied Biosystems, USA). For the real-time quantitative PCR (RT-qPCR) protocol, we utilized the SARS-CoV-2 detection kit (BGI, Beijing, China), following the manufacturer’s instructions for both reagent volumes and the RT-qPCR thermocycling configuration. Samples were later selected based on cycle threshold (CT) values (CT values of <30) to reduce high-concentration inhibition during sequencing. Libraries were prepared according to the third iteration of the Artic Network protocol on SARS-CoV-2 for Oxford Nanopore Sequencing (https://artic.network/ncov-2019). The Artic Network protocol was reproduced completely except for the sequencing run time, for which we chose a value of 24 h so that all regions were represented in our data set. Data processing began with base calling of the raw fast5 data produced by the sequencer. Base calling was performed with high accuracy with Guppy base caller v5.0.7 after the sequencing. The run configuration was -c dna_r9.4.1_450bps_fast.Cfg. Demultiplexing and read filtering were performed so that the barcodes at each end had a read length between 400 and 700 bp. Demultiplexed and filtered reads were then processed through the Artic Network MinION pipeline (https://github.com/artic-network/fieldbioinformatics), ensuring further read filtering, primer trimming, amplicon coverage normalization, variant calling, and consensus building. Genomes obtained were visually curated with the IGV genome browser (6). Consensus genomes obtained from these processes were compared using the Nextstrain clade tool (7), with which the proper clade was identified. To perform phylogenetic analysis, other relevant genomes were downloaded from the GISAID database, the genomes were aligned with MAFFT v7 (https://mafft.cbrc.jp/alignment/server), and a phylogenetic tree was constructed (Fig. 1B) with a maximum likelihood algorithm based on the GTR model using MEGA X v7.0.26 (https://www.megasoftware.net). We used default parameters for all software.
Two genomes of these three viral samples correspond to the B.1.526 (Iota, 20C) lineage, which was first described in the New York region (8), and one to P.1 (Gamma, 20J/501Y.V3), which was detected first in Japan and later in Brazil, where it has become the dominant circulating variant (9). Genomic characteristics are presented in Table 1. The development of variants of SARS-CoV-2 is and will be quintessential as a result of natural selection; therefore, variant surveillance is of utmost importance to strengthen nonpharmaceutical interventions and to envision the negative impact on vaccines campaigns in low- and middle-income countries.
TABLE 1.
Bioinformatic details for each sequence
| Parameter | Data for variant with genome identification no.: |
||
|---|---|---|---|
| 202106001S | 202106004S | 202106005S | |
| SRA accession no. | SRR15697814 | SRR15697813 | SRR15697812 |
| GISAID clade | GH/253G.V1 | GR/501Y.V3 | GH/253G.V1 |
| PANGO lineage | B.1.526 | P.1 | B.1.526 |
| WHO classificationa | VOI (Iota) | VOC (Gamma) | VOI (Iota) |
| No. of raw reads | 80,557 | 33,553 | 37,553 |
| Genome size (bp) | 29,882 | 29,894 | 29,898 |
| No. of spots | 41,725 | 34,007 | 88,091 |
| Mutations in S proteinb | 4 mutations: A701V, D253G, D614G, E484K; 3 deletions: NSP6 F108del, NSP6 G107del, NSP6 S106del | 2 mutations: T1027I, V1176F; 3 deletions: NSP6 F108del, NSP6 G107del, NSP6 S106del | 7 mutations: A701V, D253G, D614G, E484K, H69Y, L5F, T95I; 3 deletions: NSP6 F108del, NSP6 G107del, NSP6 S106del |
| Read length (range [avg]) (bp) | 480–550 (550) | 460–550 (550) | 460–550 (550) |
| Coverage (%) | 96.49 | 80.61 | 95.31 |
| GC content (%) | 40 | 41.43 | 39.8 |
WHO SARS-CoV-2 variant classification: VOI, variant of interest; VOC, variant of concern.
NSP, nonstructural protein.
Data availability.
Sequences of the SARS-CoV-2 variants were deposited in the NCBI database (GenBank accession numbers MZ645039, MZ576192, and MZ621913 and BioSample accession numbers SAMN21197472, SAMN21197473, and SAMN21197474) and the GISAID database (https://www.gisaid.org) (accession numbers EPI_ISL_2601034, EPI_ISL_2601037, and EPI_ISL_2601038). The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database (SRA accession numbers SRR15697814, SRR15697813, and SRR15697812).
ACKNOWLEDGMENTS
This work is part of the Observatory of Pandemics and Emerging Pathogen initiative at the IMTSAG-UNIBE in Santo Domingo, DR.
We thank the IMTSAG-UNIBE molecular laboratory team for their dedication and constant support throughout the COVID-19 pandemic response.
Contributor Information
Robert Paulino-Ramirez, Email: r.paulino1@unibe.edu.do.
Simon Roux, DOE Joint Genome Institute.
REFERENCES
- 1.Paulino-Ramirez R, Báez AA, Vallejo Degaudenzi A, Tapia L. 2020. Seroprevalence of specific antibodies against SARS-CoV-2 from hotspot communities in the Dominican Republic. Am J Trop Med Hyg 103:2343–2346. doi: 10.4269/ajtmh.20-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Epidemiological Directorate. 2021. COVID-19 weekly epidemiological bulletin 20. http://www.digepisalud.gob.do/documentos/?drawer=Vigilancia%20Epidemiologica*Alertas%20epidemiologicas*Coronavirus*Nacional*Boletin%20Especial%20COVID-19. Accessed 18 July 2021.
- 3.Freitas ARR, Giovanetti M, Alcantara LCJ. 2021. Emerging variants of SARS-CoV-2 and its public health implications. InterAm J Med Health 4:181. doi: 10.31005/iajmh.v4i.181. [DOI] [Google Scholar]
- 4.Paulino-Ramirez R, Riego E, Vallejo-Degaudenzi A, Calderon VV, Tapia L, León P, Licastro D, Dal Monego S, Rajasekharan S, Marcello A, ARGO Open Lab Platform for Genome Sequencing. 2021. Whole genome sequence of a SARS-CoV-2 isolate from Dominican Republic. GenBank accession number PRJNA691021. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA691021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med 26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson J, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West AP, Jr, Barnes CO, Yang Z, Bjorkman PJ. 2021. SARS-CoV-2 lineage B.1.526 emerging in the New York region detected by software utility created to query the spike mutational landscape. bioRxiv 2021.02.14.431043. doi: 10.1101/2021.02.14.431043. [DOI]
- 9.Faria NR, Morales Claro I, Candido D, Moyses Franco LA, Andrade PS, Coletti TM, Silva CAM, Sales FC, Manuli ER, Aguiar RS, Gaburo N, Camilo CdC, Fraiji NA, Esashika Crispim MA, Carvalho MdPS, Rambaut A, Loman N, Pybus OG, Ester C, Sabino EC. 2021. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586. Accessed 18 July 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences of the SARS-CoV-2 variants were deposited in the NCBI database (GenBank accession numbers MZ645039, MZ576192, and MZ621913 and BioSample accession numbers SAMN21197472, SAMN21197473, and SAMN21197474) and the GISAID database (https://www.gisaid.org) (accession numbers EPI_ISL_2601034, EPI_ISL_2601037, and EPI_ISL_2601038). The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database (SRA accession numbers SRR15697814, SRR15697813, and SRR15697812).