Abstract
INTRODUCTION:
The liver’s remarkable capacity to maintain proper tissue mass after injury has been known since ancient times. However, there has been considerable debate about the source of new liver cells that contribute to tissue growth, maintenance, and regeneration. Multiple studies have reported that disparate cell populations in the liver serve as rare stem cells, whereas others have proposed that most hepatocytes are similar in their regenerative activity regardless of position or function. Although hepatocytes appear histologically homogeneous, the liver lobule is actually organized into concentric zones, or rings, in which hepatocytes express different metabolic enzymes across the portal vein–to–central vein axis through which blood flows. Recently, single-cell profiling has enriched our understanding of the extraordinary diversity of hepatocytes, but this “zonal” heterogeneity has not been functionally interrogated in the context of tissue homeostasis, because the critical genetic labeling tools have not been available.
RATIONALE:
Previous efforts to identify the most-regenerative hepatocytes have not definitively resolved fundamental questions about whether regenerative activity is spatially restricted within particular zones or whether rare or common subsets of hepatocytes are responsible. This uncertainty was in part because fate mapping had only been performed on a few hepatocyte subsets and without side-by-side comparisons. We sought to systematically address fundamental questions about the source of new liver cells by generating a panel of 11 new CreER knock-in mouse models that label zonal subpopulations across the liver lobule. By using these tools in tandem with three existing CreER lines, tissue maintenance and regeneration as a function of zonal position were assessed.
RESULTS:
In contrast to the idea that all hepatocytes across the lobule contribute equally to regeneration, we identified major differences between hepatocytes from different locations. During steady-state homeostasis, zone 1 cells near the portal vein decreased in number over time, as did zone 3 cells near the central vein on the opposite end of the lobule. However, midlobular zone 2 hepatocytes marked by the hepcidin antimicrobial peptide 2 (Hamp2) gene were in large part responsible for homeostatic repopulation. Zone 2 cells were also shelteredfrom toxic injuries affecting either end of the lobule and thus were well positioned to contribute to regeneration after these insults. To define the mechanistic basis of these lineage-tracing results, single-cell and bulk RNA sequencing transcriptomics were used to define genes that were specifically up- or down-regulated in zone 2. We then used in vivo CRISPR knock-out and activation screening to identify functionally important pathways that regulate zone 2 proliferation. These methods revealed that zone 2 repopulation is driven by theinsulin-like growth factor binding protein 2–mechanistic target of rapamycin–cyclin D1 (IGFBP2-mTOR-CCND1) axis.
CONCLUSION:
Different regions of the liver lobule exhibit differences in their contributions to hepatocyte turnover, and zone 2 is an important source of new hepatocytes during homeostasis and regeneration. These results challenge the idea that stem cells near the portal or central veins have the highest rates of liver repopulation, but they also support the principle that there are important zonal differences in hepatocyte biology. This study reconciles findings from multiple groups and offers a more unified view of hepatocyte repopulation. The identification of zone 2 hepatocytes as a regenerative population has far-reaching implications for the cellular basis of chronic disease pathogenesis, cancer development, and regenerative medicine strategies.
Graphical abstract
Fate-mapping strains label different zones across the liver lobule. Cross-sectional images of a liver lobe from a glutamines 2 (Gls2)–CreER reporter mouse in which hepatocytes from different metabolic zones are labeled. Panels from left to right: (i) 4′,6-diamidino-2-phenylindole (DAPI) staining (blue) of cell nuclei, (ii) glutamine synthetase staining (green) of hepatocytes adjacent to central veins, (iii) Tomato fluorescence (red) that labels Gls2-expressing hepatocytes in zone 1, and (iv) a merged composite image of all three channels. This reporter strain is one of 14 used to track metabolically heterogeneous hepatocytes in the liver. Collectively, these strains were used to understand the regenerative capacity of different liver cell subtypes under homeostatic and injury conditions.
The liver is organized into zones in which hepatocytes express different metabolic enzymes. The cells most responsible for liver repopulation and regeneration remain undefined, because fate mapping has only been performed on a few hepatocyte subsets. Here, 14 murine fate-mapping strains were used to systematically compare distinct subsets of hepatocytes. During homeostasis, cells from both periportal zone 1 and pericentral zone 3 contracted in number, whereas cells from midlobular zone 2 expanded in number. Cells within zone 2, which are sheltered from common injuries, also contributed to regeneration after pericentral and periportal injuries. Repopulation from zone 2 was driven by the insulin-like growth factor binding protein 2–mechanistic target of rapamycin–cyclin D1 (IGFBP2-mTOR-CCND1) axis. Therefore, different regions of the lobule exhibit differences in their contribution to hepatocyte turnover, and zone 2 is an important source of new hepatocytes during homeostasis and regeneration.
The growing burden of liver disease caused by viral infections, medications, and over-nutrition has become a widespread clinical problem (1). Tissue damage from these etiologies leads to chronic liver inflammation and increased cell turnover. However, the identity of the cells responsible for repopulation under steady-state conditions and for regeneration after injury continues to be debated. Fate mapping has only been performed in a few subsets of hepatocytes (2–6), leaving the activities of other subpopulations untested. Even among the subsets of hepatocytes that have been shown to contribute to liver homeostasis or regeneration, there has been no side-by-side comparison of their activities. Moreover, different subsets of hepatocytes have often been studied under varying experimental conditions (e.g., homeostasis versus different injury models), contributing to the challenge of comparing results. Thus, it remains possible that different subsets of hepatocytes contribute to regeneration under different physiological circumstances or that the magnitudeof their contributions differ. Hereafter, werefer to tissue mass recovery during homeostasis as “repopulation”and recovery after injury as “regeneration.”
Lineage-tracing studies have shown that hepatocytes are the major source of de novo hepatocytes during homeostasis and regeneration after many types of liver injury (7, 8). More recently, several rare hepatocyte subpopulations have been shown to contribute to homeostasis and regeneration, suggesting the possibility of stem cells in certain regions of the lobule. Zone 3 hepatocytes, near the central vein, labeled with Axin2-CreER have been reported to give rise to substantial numbers of hepatocytes under homeostatic conditions in the adult liver (3); however, this observation was most recently challenged by an alternative Axin2 reporter allele using a bacterial artificial chromosome CreER transgene that did not disrupt endogenous Axin2 expression (9). This second allele showed that Axin2-positive cells did not contribute substantially to hepatocyte repopulation. On the opposite end of the lobule, zone 1 hepatocytes near the portal vein labeled with Sox9-CreER become highly proliferative after injury but not during homeostasis (2). Finally, hepatocytes labeled with Tert-CreER are sparsely distributed throughout all zones of the lobule and contribute to regeneration both during homeostasis and after injury (4). Another recent report challenged the stem cell paradigm in the liver by showing that proliferation was similar between randomly labeled hepatocytes across the lobule (10). These conflicting studies continue to raise fundamental questions about whether repopulating activity is spatially restricted within the liver, whether specialized and/or rare subsets of hepatocytes are primarily responsible, or whether most hepatocytes contribute.
The lobule is the histologic unit iterated throughout the liver (11, 12). Historically, crosssectioned lobules have been divided into three arbitrarily defined concentric rings, or zones, extending from the portal vein (PV) to the central vein (CV), although zonal transitions are gradual (fig. S1A). Venous blood from the gut mixes with oxygenated arterial blood in the portal triads (zone 1) then travels through the sinusoids in zone 2 (midlobule) before draining into the CV in zone 3 and returning to the heart. Zones 1 and 3 are associated with specific metabolic functions: lipid b-oxidation and gluconeogenesis are prominent on the portal side, whereas lipogenesis, ketogenesis, and glycolysis are increased near the CV (12). Zone 2 is a transitional zone with no known specialized functions. Although proliferative hepatocytes likely exist in all zones (4, 10), other potential differences in location-dependent events (death, senescence, or cell size changes) make it possible for there to be differences in how much particular zones contribute to liver homeostasis or regeneration. The clearest way to define the summation of these events would be to lineage trace distinct zones, but this has not been performed because the required tools were not available.
Development of CreER alleles for lineage tracing
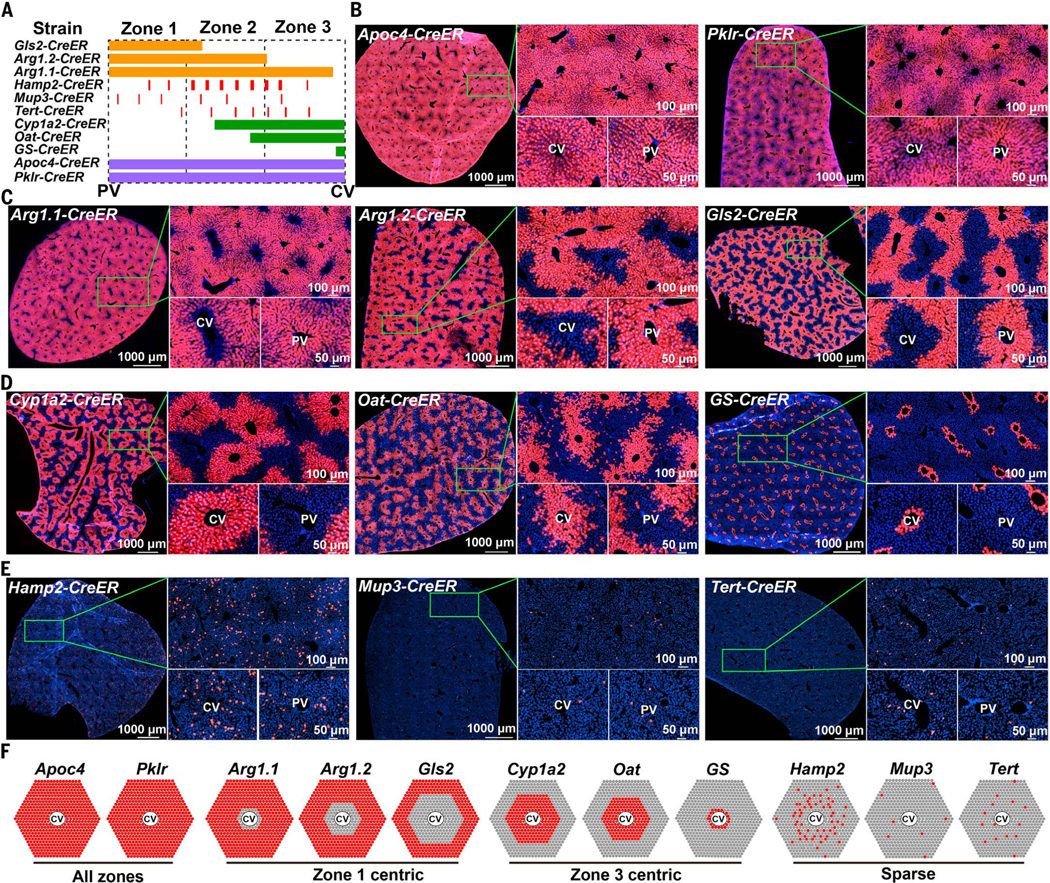
Single-cell RNA sequencing (scRNA-seq) has provided a high-resolution picture of spatial heterogeneity among hepatocytes (13–15). However, the experimental tools to assess the fates of these cell types have not been available. To systematically ascertain the locations and identities of the hepatocytes that contribute the most to mouse hepatocyte repopulation, we created a panel of 11 CreER alleles that mark hepatocytes in different regions of the liver lobule. Mouse lines with these alleles were mated with mice expressing a conditional Tomato reporter and analyzed in tandem with the three existing CreER lines that mark hepatocytes or bile duct cells with regenerative activity in the liver: Axin2-CreER (3), Sox9-CreER (4), and Krt19-CreER (16). For the 11 lines we generated (Fig. 1A), CRISPR was used to integrate an IRES cassette followed by CreERT2 recombinase (hereafter called CreER) into the endogenous 3′ untranslated regions (3′UTRs) of genes with defined zonal expression patterns (see fig. S1, B to K, for design and genotyping and table S1 for methods and primers). Each line was crossed to Rosa-LSL-tdTomato (abbreviated tdTomato) reporter mice to allow permanent Tomato protein expression in cells that express CreER, and in their progeny, after tamoxifen administration (Fig. 1, B to E, and fig. S2A). One to five doses of tamoxifen (100 mg per kilogram of mouse, intraperitoneally) were given at 6 to 8 weeks of age, and livers were analyzed 1 week later (fig. S2B). Only the glutamine synthetase (GS)–CreER transgene was active in the absence of tamoxifen, but this strain did faithfully induce Tomato labeling only in a small subset of pericentral hepatocytes (Fig. 1D).
Fig. 1. Development of CreER knock-in strains using CRISPR mouse engineering.
(A) Eleven CreER knock-in strains used to label different zones across the liver lobule. (B) Each CreER strain was crossed to Rosa-LSL-tdTomato reporter mice, enabling permanent labeling by Tomato protein expression. Shown here are the pan-zonal, all-hepatocyte strains. For all strains, 100 mg of tamoxifen per kilogram of mouse was injected intraperitoneally once, unless otherwise indicated in fig. S2B. For all experiments in this figure, livers were harvested and analyzed ~1 week after the last dose of tamoxifen. (C) The zone 1 strains. (D) The zone 3 strains. For GS-CreER, no tamoxifen was needed. (E) The zone 2 strains. For Tert-CreER, 100 mg of tamoxifen per kilogram of mouse was injected intraperitoneally for 5 days. (F) Diagram of Tomato labeling in each CreER strain. At the corners are PVs and in the centers are CVs. Red spots reflect reporter expression in hepatocytes 1 week after tamoxifen administration.
Tomato expression patterns agreed with reported immunohistochemistry or RNA in situ zonation patterns [Fig. 1, B to F, and (5, 17)]. To inducibly label all hepatocytes (but not bile ducts) from all zones, we generated apolipoprotein C4 (Apoc4)–CreER and pyruvate kinase L/R (Pklr)–CreER mice (Fig. 1B). To label hepatocytes occupying different-sized rings surrounding the PV in zone 1, we generated glutaminase 2 (Gls2)–CreER and two independent alleles of arginase 1 (Arg1)–CreER mice (Fig. 1C). To label hepatocytes occupying different-sized rings surrounding the CV in zone 3, we generated cytochrome P450 family 1 subfamily A member 2 (Cyp1a2)–CreER, ornithine aminotransferase (Oat)–CreER, and GS-CreER mice (Fig. 1D). To label midlobular zone 2 hepatocytes, we generated hepcidin antimicrobial peptide 2 (Hamp2)–CreER and major urinary protein 3 (Mup3)–CreER mice (Fig. 1E). The zone 2 CreER alleles labeled more sparsely than the zones 1 and 3 alleles. We also generated a new Tert-CreER line in which CreER was knocked into the 3′UTR rather than into exon 1 (4). Nonetheless, this Tert-CreER allele marked a similarly rare population as the previously described allele (4) (Fig. 1E). For each CreER allele, labeling patterns were consistent among individual mice 7 days after tamoxifen administration (fig. S2A). Labeling also did not appreciably change between 7 and 21 days, suggesting that there was no ectopic labeling after tamoxifen clearance (fig. S3). When oil without tamoxifen was administered, there was no reporter activation at 7 days or 1 year for any of the tested lines other than GS-CreER, indicating a lack of ectopic labeling by CreER (fig. S4).
Hepatocyte homeostasis receives no contributions from biliary or other cellular compartments
To identify the hepatocyte subpopulations that repopulate liver tissues during homeostasis, all of the above CreER mice were traced for up to 52 weeks after a single injection of tamoxifen at 6 to7 weeks of age. CreER;tdTomato mice were sacrificed 1 week, 6 months, or 12 months after tamoxifen administration. Glutamine synthetase (GS) immunofluorescence, which exclusively marks hepatocytes around CVs, was used to distinguish hepatocytes associated with CVs from those associated with PVs. We quantified the percentage of the lobular area marked by Tomato, which assesses the tissue mass contributed by the originally labeled cells and their offspring. Any change in this percent must be caused by a change in cell size or cell number of either labeled or unlabeled populations. Within zones, hepatocyte size does not change over time (fig. S5), so a change in Tomato area is a reasonable proxy for changes in cell number. For example, if the Tomato percent decreases, this could be due to a decrease in the number of Tomato-positive cells or an increase in the number of negative cells.
Because biliary populations are capable of replenishing both biliary and hepatocyte pools after severe injuries (18–20), we wanted to first confirm that transdifferentiation did not occur during normal homeostasis. We traced Krt19CreER;tdTomato mice, which labeled only biliary epithelial cells but not hepatocytes. The Krt19-positive compartment showed no expansion into the lobule nor was there any Tomato labeling of hepatocytes over a 6-month period after tamoxifen administration (fig. S6A). This experiment showed that bile duct epithelial cells did not contribute to the hepatocyte pool. To perform the opposite type of lineage labeling, we traced PklrCreER;tdTomato and Apoc4-CreER;tdTomato mice, both of which labeled 100% of hepatocytes but not biliary epithelial cells. In these mice, Tomato-positive cells did not contribute to the biliary compartment and Tomatonegative clones did not appear among hepatocytes over a 6-month period (fig. S6, B and C). The percent area occupied by Tomato-positive cells was unchanged for each of these tracing experiments (fig. S6, A to C), confirming no contribution by hepatocytes to biliary compartments or vice versa during homeostasis.
Periportal zone 1 compartment contracts during normal homeostasis
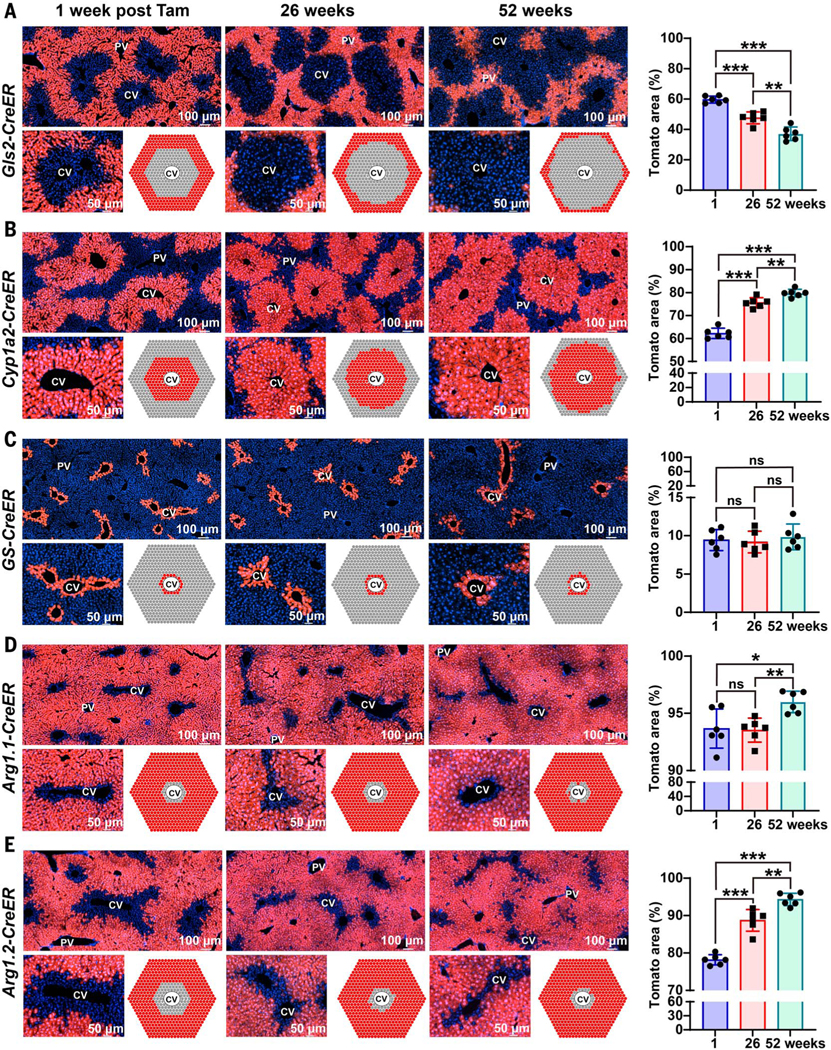
To determine whether zone 1 cells contribute to repopulation, we traced Gls2-CreER; tdTomato mice, which labeled all zone 1 cells surrounding the PV region. The proportion of Tomatopositive cells declined over 6and 12-month periods. Tomato-positive cells went from occupying 60 ± 2.3% to 37 ± 4.4% (mean ± SD) of the total lobular area after 12 months (Fig. 2A). This suggested that Gls2-positive cells in zone 1 make little or no contribution to the generation of new hepatocytes under steady-state conditions. To further test this with an orthogonal approach, we traced Cyp1a2CreER;tdTomato mice, which labeled hepatocytes throughout zones 2 and 3 but not in zone 1 (Fig. 2B). Consistent with the Gls2-CreER results, the proportion of Tomato-positive cells in Cyp1a2-CreER;tdTomato mice increased over 6and 12-month periods, ultimately occupying part of zone 1 in addition to zones 2 and 3. Tomato-positive cells went from occupying 62 ± 2.3% to 80 ± 1.7% (mean ± SD) of the total lobular area after 12 months (Fig. 2B). This increase matched the decrease in labeled area observed in Gls2-CreER;tdTomato mice, further suggesting that zone 1 hepatocytes arise from hepatocytes in zones 2 and/or 3 under steady-state conditions. It is known that repopulating cells that cross zonal boundaries ultimately acquire the differentiation state of the new zone that they are in rather than changing zonal gene expression boundaries (5). To further assess whether zone 1 cells arise from zones 2 and/or 3 cells, we also fate mapped in Oat-CreER;tdTomato mice, which labeled all of zone 3 and a smaller subset of zone 2 hepatocytes (fig. S6D). In a fashion similar to Cyp1a2-positive cells, the proportion of Tomato-positive cells in Oat-CreER;tdTomato mice increased over a 6-month period, coming to occupy more of zones 2 and 1. Tomatopositive cells went from occupying 46 ± 1.9% to 54 ± 1.5% (mean ± SD) of the total lobular area after 6 months (fig. S6D). Altogether, these data showed that the proportion of labeled hepatocytes in zone 1 decreased, whereas those in zones 2 and 3 increased over time, giving rise to hepatocytes in zone 1.
Fig. 2. Zone 1 and zone 3 compartments contract during normal homeostasis.
(A) For each of the lineage tracing experiments in this figure, we measured the proportion of total lobular area covered by Tomato as a proxy measure for hepatocyte repopulation (quantified on the right). Over 6 and 12 months, the percent area labeled by Gls2CreER decreased. (B) Over 6 and 12 months, the percent area labeled by Cyp1a2-CreER increased. (C) Over 6 and 12 months, the percent area labeled by GS-CreER did not change. (D) Over 6 months, the percent area labeled by Arg1.1CreER did not change, but after 12 months, the percent area increased. (E) Over 6 and 12 months, the percent area labeled by Arg1.2-CreER increased. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Error bars indicate standard deviation (SD).
Zone 3 cells near the CV do not repopulate during normal homeostasis
To further dissect populations within zone 3, we performed fate mapping in GS-CreER; tdTomato mice, which labeled a small, oneto three-cell-thick ring of hepatocytes around all CVs (Fig. 2C). This line did not require tamoxifen to induce Tomato expression, possibly because GS is very highly expressed in these cells, leading to leaky CreER activity in the absence of tamoxifen (fig. S4A). Even so, the GS-CreER allele produced a Tomato expression pattern that precisely recapitulated GS immunostaining and Axin2 mRNA in situ hybridization patterns (3). The proportion of Tomato-positive cells in GS-CreER;tdTomato mice did not change over 6and 12-month periods (Fig. 2C and fig. S4B). Tomato-positive cells went from occupying 9.4 ± 1.4% to 9.9 ± 1.7% (mean ± SD) of the total lobular area after 12 months. This result suggested that zone 3 cells that surround CVs are not a significant source of hepatocytes under steady-state conditions. To further test this with a CreER allele that shows the opposite labeling pattern, we traced in Arg1.1-CreER;tdTomato mice, which labeled all hepatocytes in zones 1 and 2 and most hepatocytes in zone 3, except for the most pericentral GS-positive hepatocytes (Fig. 2D). In agreement with the results from the GS-CreER allele, the proportion of Tomato-positive cells in Arg1.1-CreER;tdTomato mice did not change substantially over time. Tomato-positive cells went from occupying 94 ± 1.7% to 96 ± 0.9% of the lobule area, and the unlabeled cells went from occupying 6.3 ± 1.7% to 4.0 ± 0.9% (mean ± SD) after 12 months (Fig. 2D). This observation suggested that pericentral GS-positive hepatocytes are either long-lived or they are maintained independently of other hepatocytes in the lobule.
To further assess the ability of zone 1 and/or 2 hepatocytes to give rise to zone 3 hepatocytes, we traced in Arg1.2-CreER;tdTomato mice, which labeled almost all hepatocytes around the PV in zones 1 and 2 but left most of zone 3 unlabeled (Fig. 2E). The proportion of Tomatopositive cells in Arg1.2-CreER;tdTomato mice increased over time, and the area around the CV occupied by unlabeled cells decreased over 6and 12-month periods. Tomato-positive cells went from occupying 78 ± 1.4% to 94 ± 1.7% of the lobule, and unlabeled cells went from occupying 22 ± 1.4% to 5.7 ± 1.7% (mean ± SD) of the lobule after 12 months (Fig. 2E). This result indicated that zone 1 and/or 2 cells gave rise to zone 3 cells outside of the GSpositive ring over time under homeostatic conditions. Altogether, three independent CreER lines that traced the fates of pericentral zone 3 hepatocytes showed no increase in proportional contribution during normal liver homeostasis, contrasting with the conclusion that Axin2-positive cells, located in the most pericentral region of zone 3, repopulate the liver over time (3).
To better understand the differences between our results and the previously published results with Axin2-CreER (3), we performed fate mapping using the same CreER allele used by Wang et al. (3). Axin2-CreER;tdTomato mice labeled pericentral cells in a sparse fashion, typically within five cell diameters of the CV (fig. S6F). Consistent with the results reported previously by Wang et al. (3), we observed that Tomato-positive cells in Axin2-CreER;tdTomato mice increased in number over time, ultimately contributing to hepatocytes in zones 2 and 3 over a 6-month period. Tomato-positive cells went from occupying 1.2 ± 0.39% to 9.3 ± 0.94% (mean ± SD) of the lobule after 6 months (fig. S6F). Side-by-side comparisons of temporal changes in the labeling patterns in these Axin2-CreER;tdTomato mice and the three CreER alleles that we generated for pericentral zone 3 hepatocytes showed that the disagreement in results did not reflect differ ences in diet, housing conditions, or fatetracing protocols.
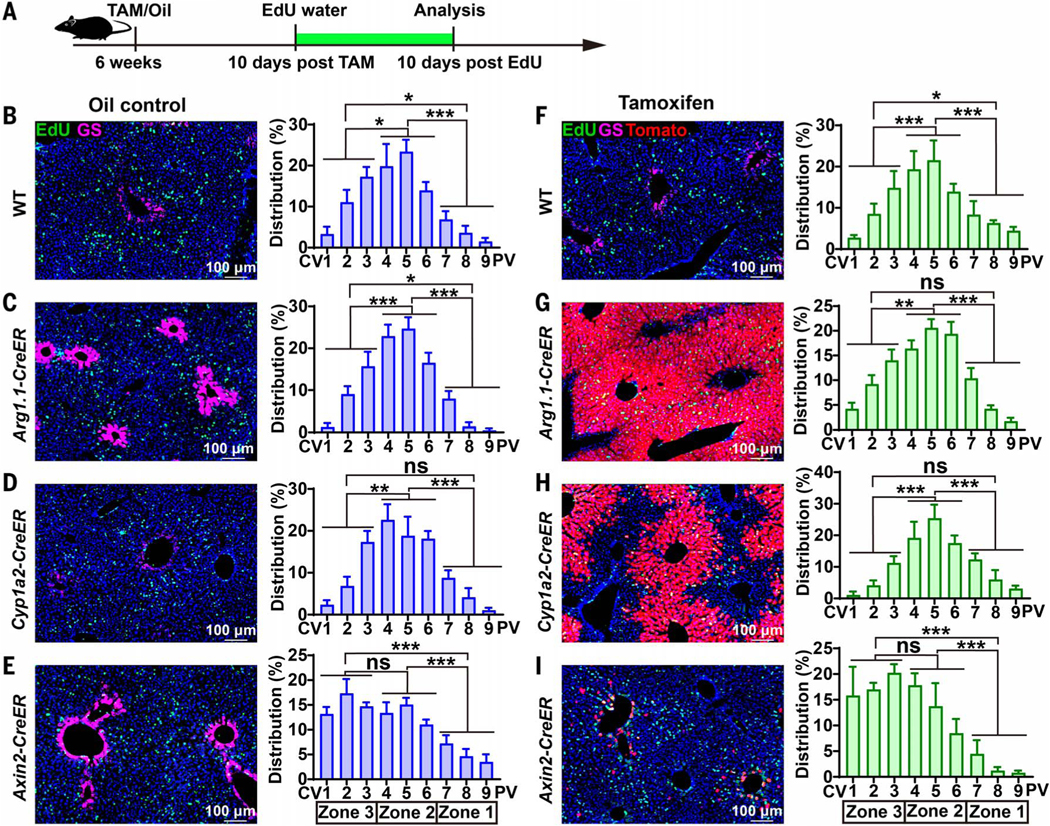
To test whether Axin2-CreER;tdTomato mice exhibited altered hepatocyte turnover relative to wild-type mice or other alleles, we quantified the frequency and spatial distribution of dividing cells by administering a 10-day pulse of 5-ethynyl-2′-deoxyuridine (EdU) (Fig. 3A). All strains (wild-type, Arg1.1CreER, and Cyp1a2-CreER) except for Axin2CreER showed that EdU-positive hepatocytes were enriched in midlobular zone 2 (Fig. 3, B to D). In contrast, EdU-positive hepatocytes in Axin2-CreER livers were more likely to be found in pericentral zone 3 (Fig. 3E). These data showed that the Arg1.1-CreER and Cyp1a2-CreER mice had the same quantity and spatial distribution of proliferating cells as wild-type mice, mainly in zone 2, whereas Axin2-CreER mice had the highest relative rate of proliferation in zone 3. The same analysis in the presence of tamoxifen showed the same results, ruling out potential proliferative effects related to tamoxifen (Fig. 3, F to I). Normalization of EdU distribution on the basis of the area occupied by each zone still revealed that zone 2 had the highest frequency of proliferative cells (fig. S7). Increased zone 3 versus zone 2 proliferation may account for the clonal expansion of Axin2-positive cells that is specifically observed in Axin2-CreER; tdTomato mice, but this is not observed in multiple other CreER mice.
Fig. 3. EdU labeling of wild-type and CreER strains during homeostasis.
(A) Schema for tamoxifen (TAM) administration followed by 10 days of EdU incorporation. (B) EdU florescence images for the detection of proliferating cells (left) and the quantification of spatial distribution of EdU-positive cells (right) from wild-type mice without CreER (n = 6 mice, three images from each were quantified). Distribution (%) of EdU represents the number of EdU-positive cells within a zonal layer divided by the total number of EdU-positive cells in all zones. See methods for positional quantification methods. All images are 7 × 105 μm2. (C) The same EdU analysis in Arg1.1-CreER;tdTomato mice (n = 7 mice, three images from each).(D) EdU analysis in Cyp1a2-CreER;tdTomato mice (n = 5 mice, three images from each). (E) EdU analysis in Axin2-CreER;tdTomato mice (n = 5 mice, three images from each). (F to I) The analysis from (B) to (E) was repeated after tamoxifen administration (n = 8, 9, 8, and 9 mice, respectively). Three images from each mouse were analyzed. *P < 0.05; **P < 0.01; ***P < 0.001. Error bars indicate standard error (SEM).
Midlobular zone 2 cells marked by Hamp2 expand in number during normal homeostasis
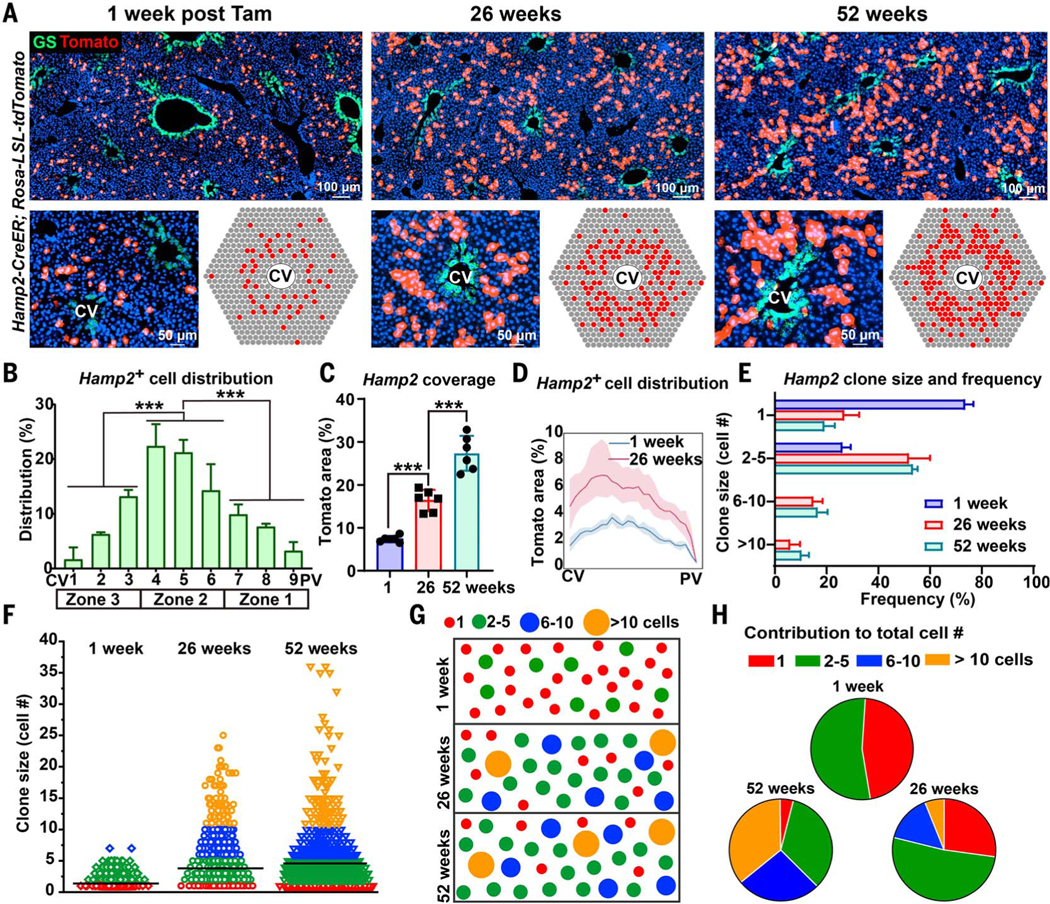
The results above showed that the proportion of labeled hepatocytes from zone 1 (Gls2 positive) and unlabeled hepatocytes from zone 3 (Arg1.2 negative) declined over time. Moreover, zone 2 hepatocytes were more proliferative than hepatocytes in other zones (Fig. 3). Collectively, these data suggest that midlobular zone 2 cells may be the major source of hepatocyte repopulation under steady-state conditions. To further test this hypothesis, we traced hepatocytes in Hamp2-CreER;tdTomato mice (Fig. 4A), since Hamp2 is expressed by hepatocytes in zone 2 according to scRNA-seq results (14). Indeed, Hamp2-CreER recombined mainly in midlobular zone 2 hepatocytes, although some labeled cells were found in other zones (Fig. 4B). The proportion of Tomatopositive cells in Hamp2-CreER;tdTomato mice increased over 6 and 12 months (Fig. 4, A and C), particularly in the midlobular zone 2 region (Fig. 4D). Tomato-positive cells went from occupying 7.4 ± 0.7% to 27.4 ± 4.1% of the lobule after 12 months (mean ± SD) (Fig. 4, C and D). These results further suggest that the main source of new hepatocytes under steadystate conditions is in zone 2.
Fig. 4. Zone 2 hepatocytes repopulate the liver during normal homeostasis.
(A) Hamp2CreER Tomato labeling at 1, 26, and 52 weeks. Endogenous GS immunofluorescence shown in green marks the CV region. Cartoon depicting the pattern of Tomato labeling in Hamp2-CreER livers over time (bottom right).(B) Manual measurements of Hamp2-CreER–labeled Tomatopositive cells as a function of position along the CV-to-PV axis. (C) The percent area labeled by Hamp2-CreER at 1, 26, and 52 weeks after tamoxifen administration. (D) Image analysis of Hamp2-labeled area at 1 and 26 weeks after tamoxifen administration. (E) Labeled clone size frequency in Hamp2-CreER livers at 1, 26, and 52 weeks after tamoxifen administration. (F) Individual clone size distribution in Hamp2-CreER livers at 1, 26, and 52 weeks after tamoxifen administration. The mean clone size (horizontal black line) increased from 1.38 ± 0.37 to 3.81 ± 0.35 to 4.74 ± 0.48 cells at 1, 26, and 52 weeks, respectively (mean ± SD). (G) Statistically representative cartoon of clone size changes over time. (H) Pie chart showing how different-sized clones contributed to the total number of labeled cells. ***P < 0.001. Error bars indicate SD for all panels except (B), where SEM is used.
Expansion occurs in a large fraction of Hamp2-positive clones
We performed clonal analysis of the fates of sparsely labeled zone 2 hepatocytes in Hamp2CreER;tdTomato mice. Labeling was sparse at the time of initial labeling (1 week after tamoxifen administration), because only 7.4 ± 0.7% of the lobular area was Tomato positive (Fig. 4, A and C), and 74 ± 3.0% of these labeled clones were single cells, 26 ± 3.0% were 2 to 5 cells, and no clones were >5 cells (mean ± SD; Fig. 4E). At 6 months, 27 ± 5.7% were single cells, 52 ± 7.3% were 2 to 5 cells, 15 ± 3.1% were 6 to 10 cells, and 6.1 ± 3.4% were >10 cells (mean ± SD; Fig. 4E), indicating that a large proportion of clones had expanded and suggesting that proliferation is common among Hamp2-positive cells. Between 0, 6, and 12 months, we observed that clone size increased modestly for most of the clones, as opposed to large increases for rare clones (from 1.38 ± 0.37 to 3.81 ± 0.35 to 4.74 ± 0.48 cells per clone) (mean ± SD; Fig. 4, F and G). Furthermore, the contribution to the total number of labeled cells after 6 and 12 months came predominantly from smaller clones (Fig. 4H). Thus, many Hamp2-positive hepatocytes divide and form additional hepatocytes during steadystate conditions in the adult liver.
Midlobular zone 2 hepatocytes marked in other ways also proliferate during homeostasis
To independently assess the repopulating activity of midlobular zone 2 hepatocytes, we traced more sparsely labeled hepatocytes in Mup3-CreER;tdTomato mice (fig. S8A). Mup3 mRNA is most abundantly expressed by hepatocytes located in zones 1 and 2 (14), and the Mup3-CreER allele sparsely recombined in hepatocytes across all zones, although predominantly in zones 1 and 2 (fig. S8B). The recombination pattern was sparse enough that we were better able to determine the locations of clones that enlarged over time. Labeling was very sparse at the time of initial labeling and after 6 months, because Tomatopositive cells went from occupying only 0.31 ± 0.06% to 0.85 ± 0.10% (mean ± SD) of the total lobular area (fig. S8, A and C). Seven days after tamoxifen administration, 92.8 ± 5.4% of labeled clones were single cells, 7.2 ± 5.4% were two cells, and no clones were more than two cells (mean ± SD; fig. S8, D and E). Thus, any clones of three or more cells at the 6-month time point represented clones that proliferated. At 6 months, 52.0 ± 4.3% of labeled clones were single cells, 32.5 ± 7.3% were two cells, and 15.6 ± 4.0% were more than two cells (mean ± SD; fig. S8, D and E), again indicating modest proliferation of a large percentage of clones. Most importantly, the expansion of clones from single cells to three or more cells was much more frequently observed in zone 2 than in zones 1 or3 (fig. S8F).
We also sought to test repopulation from zone 2 using a non-CreER or tamoxifen–based method. Tomato reporter mice were injected with low-dose adeno-associated virus–thyroxine binding globulin–Cre (AAV-TBG-Cre), which turns on Tomato in a very small fraction of hepatocytes (fig. S8G). At 1 week, all clones were one to two cells (fig. S8H) and distributed among all three zones (fig. S8I). After 6 months, clones that had expanded to three or more cells were predominantly found in zone 2 (fig. S8, G and J).
Finally, we quantified the spatial distribution of Tert-positive cells, which were reported to be randomly distributed throughout the lobule (4). Tomato-positive cells in the TertCreER;tdTomato mice we generated, which contain a different Tert-CreER knock-in allele, were most likely to appear at the junction of zones 2 and 3 (fig. S9). This result shows that Tert-positive cells are not randomly distributed and may contribute to hepatocyte repopulation from a midlobular position near zone 2. It is possible that these cells overlap with the Hamp2 and Mup3 populations that contributed to homeostasis in the above experiments.
While the compartments originally labeled in zones 1 and 3 appear to contract and zone 2 appears to expand, it is possible that the absolute number of cells in zones 1 and 3 do not change and that, instead, lobular size increases over time as a mechanism of organ growth. This could give the false appearance that cells derived from zone 1 or 3 are lost at the expense of cells from zone 2. Indeed, liver size increases with age in mice (21), but this growth was associated with minimal lobular size increases over a 52-week period (fig. S10). This increase in lobular diameter could, in principle, be caused by zone 2 proliferation without a concomitant decrease in the number of cells from zone 1 or 3. However, we observed a clear reduction in the absolute number of labeled cells in the zone 1 or 3 region over time (Fig. 2,A and E), indicating that zone 2 expansion is accompanied by a decrease in cells originally from zones 1 and 3.
Midlobular cells contribute to regeneration after chronic biliary and centrilobular injuries
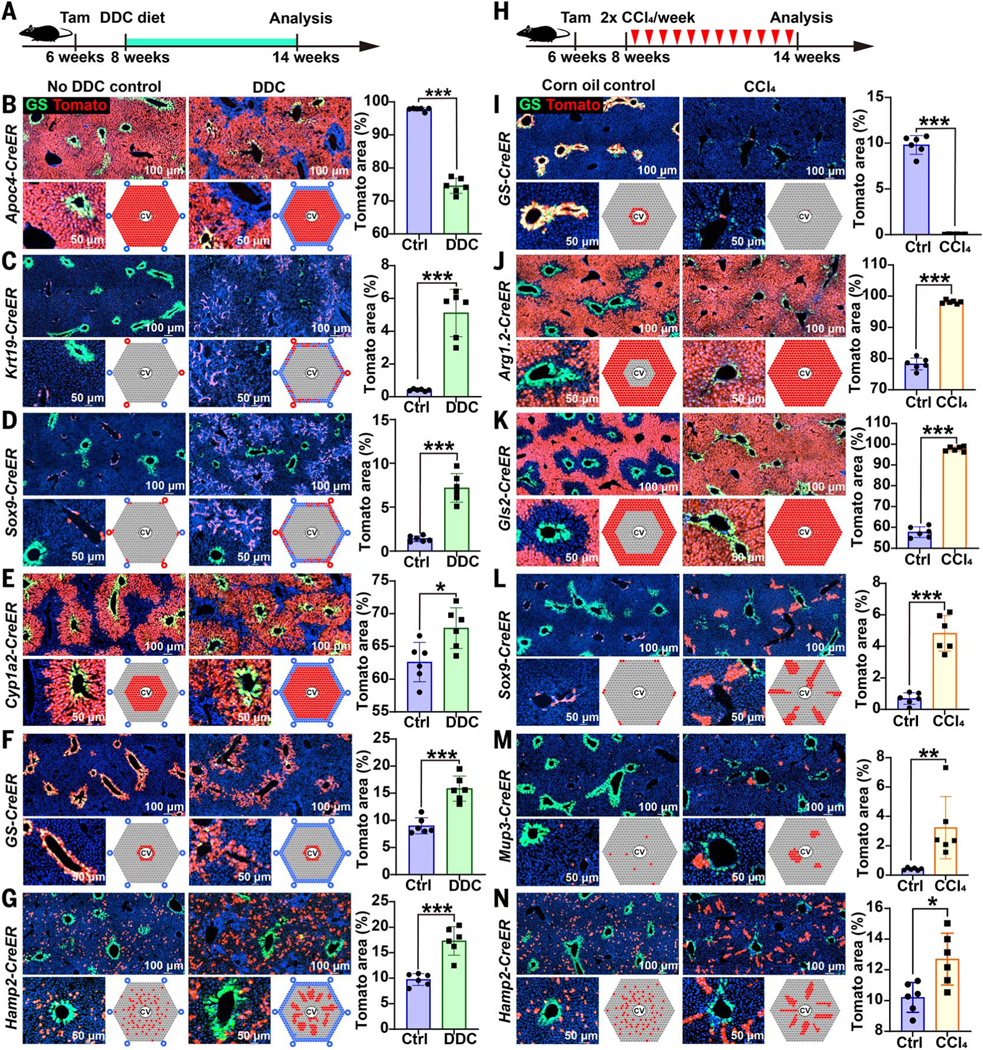
We sought to determine whether zone 2 cells also contributed to regeneration after common liver injuries, which often have the most damaging effects on hepatocytes in zones 1 and 3. We first fed mice with 0.1% 3,5diethoxycarbonyl-1,4-dihydrocollidine (DDC) for 6 weeks (Fig. 5A), which models human cholangiopathies, such as primary sclerosing cholangitis, primary biliary cirrhosis, and drug-induced bile duct damage (22). Tracing of hepatocytes (Apoc4-CreER) and bile duct cells (Krt19-CreER) during DDC treatment revealed expansion in the numbers of biliary epithelial and stromal cells (Fig. 5, B and C). This ductular reaction, which is commonly observed in human cirrhosis, is mostly derived from nonhepatocyte cell populations such as biliary cells. Tracing of periportal Sox9-CreER–expressing cells showed that these cells also contributed to the ductular reaction (Fig. 5D), as reported previously (2, 23). Tracing of Gls2-CreER–, Arg1.1-CreER–, Arg1.2-CreER–, and Mup3-CreER–expressing cells showed that each of these periportal zone 1 cell populations contracted in number significantly, potentially owing to cell death in zone 1 caused by DDC (fig. S11, A to D). Tracing of Cyp1a2-CreER–, GS-CreER–, and Hamp2-CreER–expressing cells revealed that both zone 2 and 3 cell populations expanded in number considerably (Fig. 5, E to G). This shows that zone 2 hepatocytes also contribute to hepatocyte regeneration after DDC injury.
Fig. 5. Cellular contributions to regeneration after chronic periportal and centrilobular injuries.
(A) Periportal zone 1 injury was induced by feeding lineage-tracing mice with 0.1% DDC for 6 weeks. DDC was initiated 2 weeks after using tamoxifen to label populations, then we harvested livers and examined the distribution of Tomato-labeled cells. Endogenous GS immunofluorescence is shown in green for all of the images in this figure. (B) Apoc4CreER tracing after 6 weeks of DDC. (C) Krt19-CreER tracing after 6 weeks of DDC. (D) Sox9CreER tracing after 6 weeks of DDC. (E) Cyp1a2-CreER tracing after 6 weeks of DDC. (F) GSCreER tracing after 6 weeks of DDC. (G) Hamp2-CreER tracing after 6 weeks of DDC. (H) For all of the following experiments, mice were injected with 12 biweekly doses of CCl4 (1 ml per gram of mouse) to induce chronic centrilobular liver injury 2 weeks after using tamoxifen to label different populations. One week after the last dose, we harvested livers and examined the distribution of Tomatolabeled cells. (I) GS-CreER tracing after 6 weeks of CCl4. (J) Arg1.2-CreER tracing after 6 weeks of CCl4. (K) Gls2-CreER tracing after 6 weeks of CCl4. (L) Sox9-CreER tracing after 6 weeks of CCl4. (M) Mup3 tracing after 6 weeks of CCl4. (N) Hamp2-CreER tracing after 6 weeks of CCl4. *P < 0.05; **P < 0.01; ***P < 0.001. Error bars indicate SD. ctrl, control.
To probe injuries occurring at the opposite end of the lobule in zone 3, we injected mice with 12 doses of biweekly CCl4 to induce chronic centrilobular necrosis, which models drug (i.e., acetaminophen) toxicity or ischemic injury (17) (Fig. 5H). Pericentral zone 3 cells labeled by GS-CreER, Cyp1a2-CreER, and Axin2-CreER expression almost completely disappeared after injury (Fig. 5I and fig. S11, E and F). Hepatocytes from zones 1 and/or 2 labeled by Arg1.2-CreER and Gls2-CreER expression expanded in number, regenerating the entire lobule after zone 3 ablation (Fig. 5, J and K). However, there was no evidence of a contribution to hepatocyte regeneration from Apoc4-negative or Krt19-positive biliary cells (fig. S11, G and H). To determine which hepatocytes in zone 1 or 2 expanded after centrilobular injury, we traced multiple periportal and midlobular strains. Hepatocytes marked by Sox9-CreER expression proliferated and appeared to stream from their original periportal sites toward the CV (Fig. 5L), as was reported previously (2). We also traced Mup3and Hamp2expressing hepatocytes and found significant increases in these populations after chronic CCl4 administration (Fig. 5, M and N). Overall, these data suggested that most zone 3 cells were eliminated by CCl4 treatment, but that multiple populations of remaining hepatocytes in zones 1 (Sox9 positive) and 2 (Hamp2 positive) were able to regenerate to replace lost cells. These data do not support the model of a singular stem or progenitor cell source, but it is still likely that there are heterogeneous rates of regeneration between different hepatocytes. Because most liver injuries occur in zone 1 or 3, zone 2 hepatocytes are sheltered from damage and are positioned to regenerate the liver after these insults.
Zone 2 proliferation is driven by the IGFBP2-mTOR-CCND1 axis
To examine zone-specific gene expression patterns and to begin to understand the mechanistic underpinnings of zone 2 hepatocyte expansion, we performed scRNA-seq on unsorted cells from Gls2-CreER;tdTomato, Cyp1a2CreER;tdTomato, Hamp2-CreER;tdTomato, and dual Gls2-CreER;Oat-CreER;tdTomato livers. These four strains were chosen because they marked cells in the periportal, pericentral, and midlobular zones (fig. S12, A and B). We detected expected patterns of Tomato coexpression with endogenous Gls2, Cyp1a2, Hamp2, or Gls2/Oat, information that provided additional spatial landmarking confirmation (fig. S12, C to F). Pooling of these four scRNA-seq datasets allowed us to more effectively cluster zone 1, 2, and 3 hepatocytes in an unbiased fashion (fig. S12B). We used this clustering to define differentially expressed genes between zones and within specific genetically marked subpopulations (fig. S12G and table S2). The expected mRNA expression patterns were found: Gls2, Hal, Cyp2f2, Sds, Cdh1, and Hsd17b13 were among the most highly expressed genes in zone 1, while Cyp1a2, Cyp2e1, Gulo, Oat, and Lect2 were among the most highly expressed genes in zone 3. Igfbp2, Hamp2, Hamp, and Mug2 expression was specifically enriched in zone 2 (fig. S12G and table S2). The four scRNA-seq datasets also allowed us to identify differentially expressed genes in cell populations expressing GS, Sox9, or Hamp2, and this analysis was included as reference data (table S2).
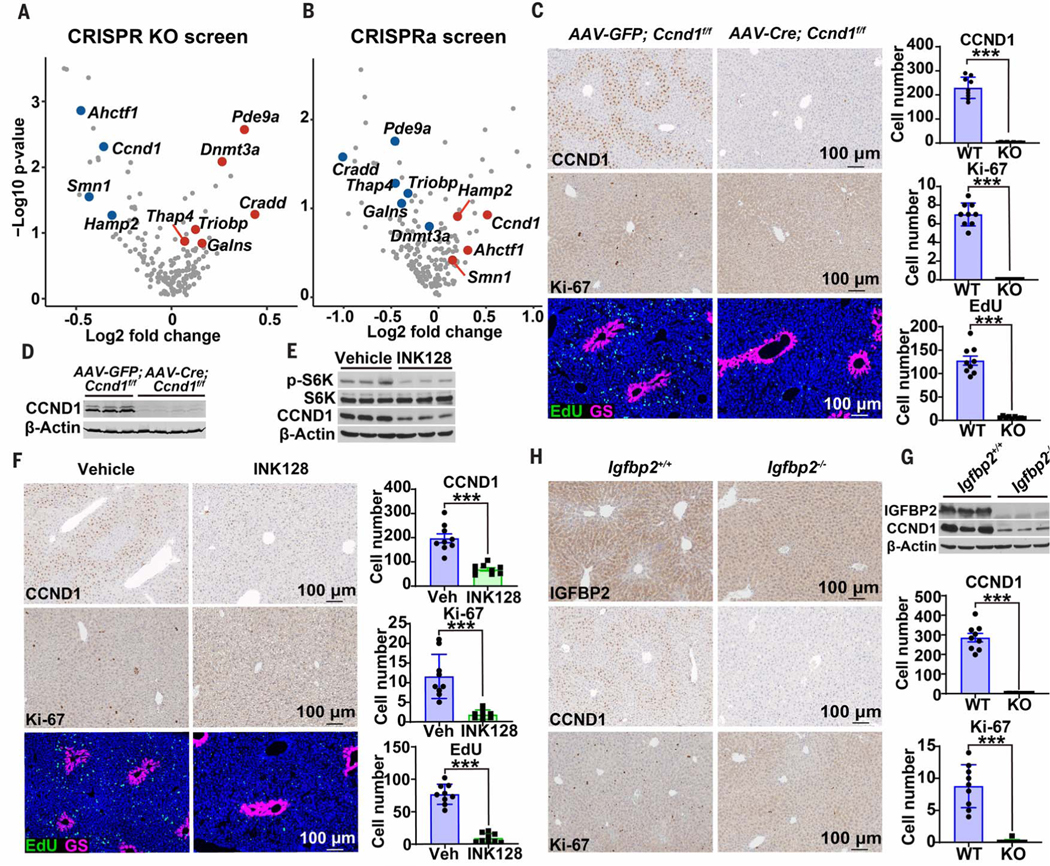
To identify in an unbiased fashion the zone 2 genes and pathways that could account for increased repopulation, we performed pooled in vivo CRISPR screens to identify genes that regulate the proliferative capacity of hepatocytes. We used our scRNA-seq data in addition to previously published bulk RNA-seq datasets from zonal populations (14, 24) to select 176 genes that are upor down-regulated specifically in zone 2 versus zones 1 or 3 (fig. S13 and table S3). We performed in vivo CRISPR knockout (KO) and activation (CRISPRa) screening to test the functional impact of these genes on hepatocyte regeneration as previously described (25) (see also methods and table S4). After CRISPR KO or CRISPRa single guide RNA (sgRNA) transposon libraries were delivered into Fah KO livers via hydrodynamic transfection, hepatocyte clones carrying fumarylacetoacetate hydrolase (FAH), Cas9, and sgRNAs regenerated the Fah-deficient liver. After 4 weeks, the relative abundance of sgRNAs in repopulated livers was measured by deep sequencing. Three genes normally up-regulated in zone 2 (Ahctf1, Ccnd1, and Hamp2) had associated sgRNAs that were simultaneously depleted in the CRISPR KO screen and enriched in the CRISPRa screen, suggesting that these genes were potentially necessary and sufficient for promoting proliferation from zone 2 (Fig. 6, A and B). Similarly, two genes normally down-regulated in zone 2 (Dnmt3a and Triobp) had sgRNAs that were simultaneously enriched in the CRISPR KO screen and depleted in the CRISPRa screen, suggesting that these could be involved in suppressing hepatocyte proliferation outside of zone 2 (Fig. 6, A and B). Altogether, in vivo screening was able to nominate genes, the expression of which was specifically changed in zone 2, that could functionally influence hepatocyte proliferation under homeostatic conditions.
Fig. 6. Zone 2 proliferation is driven by the IGFBP2-mTOR-CCND1 axis.
(A) Pooled transposon plasmids were used for gene deletion and hepatocyte repopulation of Fah KO mouse livers. Plasmids containing Cas9, mouse Fah, and sgRNAs targeting zone 2 genes were injected with Sleeping Beauty transposase (SB100) intravenously using hydrodynamic transfection. After 4 weeks, sgRNAs were deep sequenced and the results are shown here in the form of volcano plots. Only genes scoring as significant (P < 0.15) in both screens are highlighted as red or blue dots. (B) Pooled transposon plasmids were used for gene activation and repopulation of Fah KO mouse livers. Plasmids containing dCas9-VP64 (catalytically dead Cas9 fused with transcriptional activator VP64), mouse Fah cDNA, and sgRNAs targeting the promoters of zone 2 genes were injected with SB100. (C) Sevenweek-old Ccnd1f/f mice were given control AAV-GFP (n = 3 mice) or liver-specific AAV-TBG-Cre (n = 4 mice). After 48 hours, EdU water was provided for 15 days. Immunohistochemistry (IHC) showing CCND1 and Ki-67 along with immunofluorescence (IF) showing EdU (green) and GS (red) in livers of control and Ccnd1-deleted mice. Cell numbers for three image fields per mouse are quantified on the right. All quantified images in this figure are 7 × 105 mm2. WT, wild type; KO, knockout. (D) Western blot analysis from harvested Ccnd1 livers. (E) Eight-week-old wild-type mice were treated with INK128, an mTORC1/2 specific inhibitor (n = 3, 3 mice). Two days after INK128 initiation, EdU water was provided for 10 days. Western blot analysis from harvested livers is shown. (F) IHC showing CCND1 and Ki-67 along with IF showing EdU (green) and GS (red) in livers of control and INK128-treated mice. Cell numbers for three image fields per mouse are quantified on the right. Veh, vehicle. (G) Eight-week-old wild-type and Igfbp2 whole-body KO mice were analyzed by Western blot analysis. (H) IHC showing IGFBP2, CCND1, and Ki-67 in livers of wild-type and Igfbp2 KO mice (n = 3, 3 mice). Cell numbers for three image fields per mouse are quantified on the right. ***P < 0.001. Error bars indicate SD.
We focused on understanding the mechanisms associated with zone 2–specific expression and activity of CCND1. We found that CCND1 was exquisitely expressed in a midlobular pattern even in the absence of injury (Fig. 6C). To define the functional impact of CCND1 under homeostatic conditions, we inhibited the cyclin-dependent kinase 4 and 6 (CDK4/6)–CCND1 complex with palbociclib in wild-type mice, which led to a complete ablation of zone 2 proliferation as measured by Ki-67 and a 10-day EdU trace (fig. S14, A and B). To ensure that this was a CCND1specific effect, we deleted Ccnd1 by delivering the hepatocyte-specific AAV-TBG-Cre into floxed mice. This also led to the disappearance of zone 2 proliferation (Fig. 6, C and D). Because CCND1 is a known target of mTOR and S6K1 in the liver (26), we asked whether zone-specific CCND1 expression and activity was mTOR dependent. Treatment with INK128, a potent and selective mTORC1/2 inhibitor, resulted in a complete ablation of CCND1 expression and almost no proliferation (Fig. 6, E and F). To identify upstream regulators of mTOR, we sought zone 2 genes that might regulate phosphatidylinositol 3-kinase–mTOR signaling. Because the zone 2 secreted factor IGFBP2 is known to act in part through the IGF signaling system, this represented a promising candidate. Upon IGFBP2 overexpression, we observed increased mTOR signaling in the form of increased p-Akt, p-S6K, and p-4E-BP1 (fig. S14, C and D). To determine whether IGFBP2 is necessary for preferential zone 2 proliferation, we showed that whole-body Igfbp2 deletion resulted in ablation of CCND1 and Ki-67 in zone 2 (Fig. 6G, H). Our data shows that the IGFBP2-mTOR-CCND1 axis is one mechanism responsible for the preferential repopulation of zone 2 hepatocytes.
Discussion
Whether there are locationor subtypedependent differences in liver homeostasis or regeneration has been controversial. Currently, debates between two general models for liver repopulation have not been resolved. One is that hepatocytes on either end of the lobular axis preferentially repopulate either during homeostasis or upon injury (2, 3). The other suggests that hepatocytes across the lobule repopulate equivalently in a locationagnostic fashion (4, 10). Until now, it has not been possible to definitively distinguish between these models, because the strains to fate trace location-specific hepatocytes were not available. By comparative lineage tracing of a large panel of zonal CreER mice, we reconciled disparate conclusions from the field and showed that there are significant spatial differences in the repopulation activities from different zones. Even within zone 3 there is evidence of heterogeneity: there are differences between the GS/Lgr5-positive domain that does not change (27, 28), and the Arg1.2negative domain that contracts over time. Most importantly, we discovered that hepatocytes from zone 2 preferentially repopulate the lobule during normal homeostasis and represent a major contributor to regeneration after liver injuries (fig. S15). Zone 2 likely repopulates most but not all hepatocytes. Similar to the way the GS domain is likely maintained by a self-renewing population, the hepatocytes closest to the PV may also be maintained by a self-renewing population, given that we did not observe complete disappearance of Tomatolabeled hepatocytes in zone 1.
Our data challenge the notion that cells near the portal or central veins have the highest rates of repopulation and also disagree with the model that all hepatocytes across the lobule are equivalent in their repopulating activity. The observation that proliferative rates of hepatocytes are highest in zone 2 already suggested the existence of spatial heterogeneity (4, 10, 29), but without zonal lineage tracing tools, it was not possible to definitively demonstrate the expansion of the midlobular compartment and the contraction of other populations. Parallel tracing of CreER lines for all zones served to show that zone 2 expands, and at least one cause of this expansion is the increased rate of cell division found in zone 2. While the concentrated expression of IGFBP2 and CCND1 in zone 2 is important for preferential midlobular proliferation, changes in compartment size are unlikely to be solely caused by proliferative differences. In human livers, cell death rates are increased in pericentral zone 3 (30, 31), suggesting that spatial disequilibrium of cell death could also be important. The possibility that there are differences in cellular disappearance makes room for the simultaneous observations that (i) hepatocytes with high rates of proliferation can exist in all zones and (ii) there are zonal differences in repopulation. For example, Tert-positive cells exist within the GS-positive compartment (Fig. 1E and fig. S9), but the compartment as a whole does not expand. Ultimately, longterm persistence of cells during aging and chronic injury are dependent on competing factors beyond proliferation rates alone, and can best be measured using the lineage tracing approaches used here to follow zonal compartments over time.
Whereas zones 1 and 3 have defined biological and metabolic functions, zone 2 has been thought of as a transitional zone with no singular purpose. It is possible that zone 2 contains hepatocytes with the requisite plasticity and that are the most well positioned to contribute to zone 1 or zone 3 populations. Zone 2 cells not only preferentially repopulate during normal homeostasis, they also contribute to regeneration when either zone 1 or 3 is damaged with hepatotoxins. Although extensive whole-organ injuries such as partial hepatectomy that elicit robust regenerative responses from most hepatocytes were not explored here, these kinds of injuries are somewhat less relevant to common liver diseases, which involve mild-to-moderate chronic insults. Zone 2 is likely to be more important under these modest injury conditions, in part because zone 2 is more proliferative but also because toxic damage to zone 1 or 3 is much more common than damage to zone 2. Thus, it is not surprising for there to be a reserve population of zone 2 cells that is sheltered from death and that can replenish other zones. The identification of such cells that preferentially repopulate over long time scales has implications for understanding the cellular basis of chronic liver pathologies such as fatty liver and the evolution of liver cancers.
Materials and methods summary
The CreER knock-in lines were generated by injecting CRISPR reagents into mouse embryos. IRES and CreER were flanked by homology arms associated with targeted genes, then cloned into plasmids and converted into various CRISPR-mediated genome engineering reagents. Founder mice that resulted in offspring with germline transmission of CreER were maintained as independent lines and bred to tdTomato reporter mice for testing. Lineage tracing was performed in a standard fashion, and ImageJ facilitated the quantification of the results. In vivo CRISPR screens were performed in a similar fashion to what is described in (25). In vivo chemical injury, genetic manipulation using AAV, and hydrodynamic transfection were all performed in the standard fashion as described in (32). Details of the materials and methods, including scRNA-seq and in vivo screening methods, are included in the supplementary materials.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. J. Morrison, M. Agecanotheous, P. Mishra, M. Grompe, B. Tarlow, A. Chung, L. Nguyen, and S. Wang for constructive feedback on the manuscript; C. Lewis and J. Shelton for histopathology; the CRI Sequencing Core (J. Xu) for genomics; and the CRI Mouse Genome Engineering Core for transgenic mouse models.
Funding: J.H.A. was supported by NIH/NIDDK R01DK54921. T.W. was supported by NIH R03ES026397 and CPRIT (RP150596, RP190208). H.Z. was supported by the Pollack Foundation, NIH/NIDDK R01DK111588, NIH/NCI R01CA251928, CPRIT (RP170267, RP180268), a Burroughs Wellcome Career Award for Medical Scientists, and a Stand Up To Cancer Innovative Research Grant (SU2C-AACR-IRG 10–16). Stand Up To Cancer (SU2C) is a program of the Entertainment Industry Foundation, and its research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C.
Footnotes
Competing interests: The authors certify that they have no relevant conflicts of interest.
Data and materials availability: The accession number for all of the scRNA-seq data is GSE162713. All differential gene expression data from this scRNA-seq data are contained in table S1. The bulk RNA-seq data are from (24). All mouse strains reported for the first time here are available from H.Z.’s lab under a material transfer agreement with UT Southwestern.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Kulik L, El-Serag HB, Epidemiology and management of hepatocellular carcinoma. Gastroenterology 156, 477–491.e1 (2019). doi: 10.1053/j.gastro.2018.08.065; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Font-Burgada J. et al. , Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 162, 766–779 (2015). doi: 10.1016/j.cell.2015.07.026; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang B, Zhao L, Fish M, Logan CY, Nusse R, Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature 524, 180–185 (2015). doi: 10.1038/nature14863; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin S. et al. , Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248 (2018). doi: 10.1038/s41586-018-0004-7; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pu W. et al. , Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat. Commun. 7, 13369 (2016). doi: 10.1038/ncomms13369; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuyama K. et al. , Continuous cell supply from a Sox9expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 43, 34–41 (2011). doi: 10.1038/ng.722; [DOI] [PubMed] [Google Scholar]
- 7.Malato Y. et al. , Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Invest. 121, 4850–4860 (2011). doi: 10.1172/JCI59261; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanger K. et al. , Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 15, 340–349 (2014). doi: 10.1016/j.stem.2014.06.003; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun T. et al. , AXIN2+ pericentral hepatocytes have limited contributions to liver homeostasis and regeneration. Cell Stem Cell 26, 97–107.e6 (2020). doi: 10.1016/j.stem.2019.10.011; [DOI] [PubMed] [Google Scholar]
- 10.Chen F. et al. , Broad distribution of hepatocyte proliferation in liver homeostasis and regeneration. Cell Stem Cell 26, 27–33. e4 (2020). doi: 10.1016/j.stem.2019.11.001; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birchmeier W, Orchestrating Wnt signalling for metabolic liver zonation. Nat. Cell Biol. 18, 463–465 (2016). doi: 10.1038/ncb3349; [DOI] [PubMed] [Google Scholar]
- 12.Ben-Moshe S, Itzkovitz S, Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 16, 395–410 (2019). doi: 10.1038/s41575-019-0134-x; [DOI] [PubMed] [Google Scholar]
- 13.MacParland SA et al. , Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018). doi: 10.1038/s41467-018-06318-7; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halpern KB et al. , Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017). doi: 10.1038/nature21065; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aizarani N. et al. , A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019). doi: 10.1038/s41586-019-1373-2; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Means AL, Xu Y, Zhao A, Ray KC, Gu G, A CK19CreERT knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis 46, 318–323 (2008). doi: 10.1002/dvg.20397; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghafoory S. et al. , Zonation of nitrogen and glucose metabolism gene expression upon acute liver damage in mouse. PLOS ONE 8, e78262 (2013). doi: 10.1371/journal.pone.0078262; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raven A. et al. , Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354 (2017). doi: 10.1038/nature23015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng X. et al. , Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell 23, 114–122.e3 (2018). doi: 10.1016/j.stem.2018.05.022; [DOI] [PubMed] [Google Scholar]
- 20.Choi T-Y, Ninov N, Stainier DYR, Shin D, Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 146, 776–788 (2014). doi: 10.1053/j.gastro.2013.10.019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergmann P, Militzer K, Schmidt P, Büttner D, Sex differences in age development of a mouse inbred strain: Body composition, adipocyte size and organ weights of liver, heart and muscles. Lab. Anim. 29, 102–109 (1995). doi: 10.1258/002367795780740447; [DOI] [PubMed] [Google Scholar]
- 22.Fickert P. et al. , A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. Am. J. Pathol. 171, 525–536 (2007). doi: 10.2353/ajpath.2007.061133; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarlow BD, Finegold MJ, Grompe M, Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury. Hepatology 60, 278–289 (2014). doi: 10.1002/hep.27084; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Moshe S. et al. , Spatial sorting enables comprehensive characterization of liver zonation. Nat. Metab. 1, 899–911 (2019). doi: 10.1038/s42255-019-0109-9; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu M. et al. , Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell 177, 608–621.e12 (2019). doi: 10.1016/j.cell.2019.03.026; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Espeillac C. et al. , S6 kinase 1 is required for rapamycin-sensitive liver proliferation after mouse hepatectomy. J. Clin. Invest. 121, 2821–2832 (2011). doi: 10.1172/JCI44203; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planas-Paz L. et al. , The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol. 18, 467–479 (2016). doi: 10.1038/ncb3337; [DOI] [PubMed] [Google Scholar]
- 28.Ang CH et al. , Lgr5+ pericentral hepatocytes are selfmaintained in normal liver regeneration and susceptible to hepatocarcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 116, 19530–19540 (2019). doi: 10.1073/pnas.1908099116; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanami S. et al. , Dynamic zonation of liver polyploidy. Cell Tissue Res. 368, 405–410 (2017). doi: 10.1007/s00441-016-2427-5; [DOI] [PubMed] [Google Scholar]
- 30.Benedetti A, Jézéquel AM, Orlandi F, Preferential distribution of apoptotic bodies in acinar zone 3 of normal human and rat liver. J. Hepatol. 7, 319–324 (1988). doi: 10.1016/S0168-8278(88)80004-7; [DOI] [PubMed] [Google Scholar]
- 31.Benedetti A, Jezequel AM, Orlandi F, A quantitative evaluation of apoptotic bodies in rat liver. Liver 8, 172–177 (1988). doi: 10.1111/j.1600-0676.1988.tb00987.x; [DOI] [PubMed] [Google Scholar]
- 32.Sun X. et al. , Suppression of the SWI/SNF component Arid1a promotes mammalian regeneration. Cell Stem Cell 18, 456–466 (2016). doi: 10.1016/j.stem.2016.03.001; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.