Abstract
We describe in vivo evolution of carbapenem and ceftazidime-avibactam resistance by analyzing four longitudinal Klebsiella pneumoniae clinical isolates from a patient with pneumonia following antimicrobial treatment. The patient had fever, cough associated with expectoration, and new infiltration was found on the chest CT. Antimicrobial susceptibility was determined, and whole genome sequencing (WGS) was performed to investigate its dynamic change of resistance phenotype. Population analysis profile was performed to investigate the population of Klebsiella pneumoniae. The infection started with a KPC-2-producing K. pneumoniae (ZRKP01, ceftazidime-avibactam-S/carbapenem-R). Then, after ceftazidime-avibactam treatment, the strain switched to D179Y mutant that is KPC-33 (ZRKP02, ceftazidime-avibactam-R/carbapenem-S), which restored carbapenem susceptibility. However, the restored carbapenem susceptibility in vivo was not stable and the subsequent use of imipenem against KPC-33-producing K. pneumoniae infection resulted in a reversion of KPC-2 producers (ZRKP03 and ZRKP04, ceftazidime-avibactam-S/carbapenem-R). Genetic analysis demonstrated that all four K. pneumoniae isolates belonged to sequence type 11and had identical capsular polysaccharide (KL47), identical porin genes, and same plasmid replicon types. Phylogenetic analysis indicated that four K. pneumoniae isolates showed a high degree of relatedness. Single nucleotide polymorphisms analysis indicated that the number of mutations observed in the KPC-33 isolate was more than in the wild-type KPC-2 isolates and the four KPC-Kp isolates evolved from a longitudinal evolution of K. pneumoniae harboring blaKPC-2 gene. This is the first report to observe the in vivo evolution of wild-type KPC-2 to KPC-33 and then the reversion to its original wild-type KPC-2. Through WGS, we demonstrated the role of selective pressure of antibiotic in the mutation and reversion of blaKPC genes, which leading to the dynamic change of KPC enzymes and the dynamic emergence of resistance to ceftazidime-avibactam and carbapenems.
Statement: Recently, studies reported the emergence of ceftazidime-avibactam-resistant strains. The KPC mutations mediating ceftazidime-avibactam resistance are generally associated with the restoration of carbapenem susceptibility. However, clinical significance of this observation is unclear. In this manuscript, we demonstrate the role of selective pressure of antibiotic in the mutation and reversion of blaKPC genes, which leading to the dynamic change of KPC enzymes and the dynamic emergence of resistance to ceftazidime-avibactam and carbapenems. To the best of our knowledge, this is the first report to observe the in vivo evolution of wild-type KPC-2 to KPC-33 and then the reversion to its original wild-type KPC-2. It should be noted that understanding the clinical significance of this observation is of critical importance, and reversion to carbapenem susceptibility would not imply a potential role for carbapenems monotherapy. We hope our study will draw attention to clinicians, so that this agent can be used most effectively for the longest period of time.
Keywords: Klebsiella pneumoniae carbapenemase, ceftazidime-avibactam resistance, whole genome sequencing, KPC-33, KPC-producing Klebsiella pneumoniae
Introduction
Ceftazidime-avibactam is a β-lactam/β-lactamase inhibitor combination that was approved for the treatment of complicated intra-abdominal infections, complicated urinary tract infections, hospital-acquired pneumonia, and ventilator-associated pneumonia (Falcone and Paterson, 2016). The agent is highly active against class A β-lactamases including Klebsiella pneumoniae carbapenemases, class C β-lactamases, and OXA-48 carbapenemase, but not metallo-β-lactamases such as NDM, VIM, and IMP (van Duin and Bonomo, 2016). Despite a limited use of ceftazidime-avibactam at a worldwide scale, ceftazidime-avibactam resistance has been reported either in patient with no history of ceftazidime-avibactam therapy (Humphries, 2015) or in patient after short periods of ceftazidime-avibactam exposure (Shields et al., 2016). Resistance to ceftazidime-avibactam has been linked to specific mutations in the blaKPC gene (Shields et al., 2017a), specific mutations in the blaCTX-M gene (Both et al., 2017), porin deficiency combined with high ceftazidime hydrolysis (Shen et al., 2017; Galani et al., 2019), porin inactivation with or without increased expression of the blaKPC gene (Humphries and Hemarajata, 2017; Coppi et al., 2020), or transposition of KPC with porin deficiency (Nelson et al., 2017). The mechanism most often associated with the emergence of ceftazidime-avibactam resistance after treatment has been observed to be mutations in the blaKPC genes encoding for KPC enzymes, such as L169P, A177E, D179Y, D179N, V240G, Y241H, T243M, and H274N mutations in KPC (Shields et al., 2017a; Giddins et al., 2018; Hemarajata and Humphries, 2019; Sun et al., 2020). These mutational changes in KPC that emerged in vivo are often associated with fully or partially reversion to carbapenem susceptibility (Haidar et al., 2017; Shields et al., 2017a; Giddins et al., 2018). However, clinical significance of this observation is unclear, since subsequent exposure to carbapenems can restore resistance to them in vitro passage experiments (Shields et al., 2017b). In the present study, we observed the in vivo evolution of KPC-2 to KPC-33 and then the reversion to KPC-2, which leading to the dynamic emergence of resistance to ceftazidime-avibactam and carbapenems.
Materials and Methods
Bacterial Strains and Susceptibility Testing
The patient underwent a routine culture of sputum or bronchoalveolar lavage fluid (BALF) over a 3-month hospitalization. A total of 19 respiratory tract specimens from the patient were collected, including 12 sputum and 7 bronchoalveolar lavage. Four of these specimens yield K. pneumoniae. Isolates were identified as K. pneumoniae by MALDI-TOF MS (Bruker Daltonics, Billerica, MA, United States), and antimicrobial susceptibility testing (AST) was performed using the VITEK-2 compact system (bioMerieux, Marcy-l’Etoile, France). AST was further performed by means of broth microdilution, which was performed and interpreted according to the guidelines established by the Clinical and Laboratory Standards Institute, United States (CLSI, 2019). Avibactam was tested at a fixed concentration of 4mg/l in combination with increasing concentrations of ceftazidime.
DNA Sequencing, de novo Assembly, and Annotation
For whole genome sequencing (WGS), the genomic DNA of four isolates was subjected to both short- and long-read massively parallel sequencing. Short-read sequencing was performed on the Illumina HiSeq 2,500 sequencing platform (Illumina, San Diego, CA), and long-read sequencing was performed using the Oxford Nanopore MinION platform (Oxford Nanopore, Oxford, United Kingdom). Raw reads were filtered to remove low-quality sequences and adaptors. De novo assembly was conducted using SPAdes Genome Assembler v3.13.1 (Bankevich et al., 2012; Nurk et al., 2013) and Unicycler (Wick et al., 2013). Gene prediction was performed using Prokka 1.12 (Seemann, 2014). Antimicrobial resistance genes and plasmid replicon analysis were performed using ResFinder and PlasmidFinder tools via the CGE server.1 Prophages and insertion sequences in the genome were identified with PHASTER (Amdt et al., 2016) and ISsaga (Varani et al., 2011), respectively. Mutations present in isolates were analyzed as follows: Sequence reads were mapped to the reference genome of strain ZRKP01 using Bowtie 2 (Langmead and Salzberg, 2012), and single nucleotide polymorphisms (SNPs) were identified with SAMtools (Li et al., 2009) and Genome Analysis Toolkit (Mckenna et al., 2010). All SNPs were manually checked. Subsequently, we constructed a maximum likelihood phylogenic tree by using RAxML (Stamatakis, 2014) with general time reversible model of nucleotide substitution and a Gamma distribution of rate heterogeneity.
Population Analysis Profile (PAP) of Four KPC-Producing Klebsiella pneumoniae (KPC-Kp) Isolates
To investigate the presence of meropenem and ceftazidime/avibactam heteroresistance, population analysis profiles were determined by spiral plating 50-μl aliquots of the starting bacterial cell suspension on Mueller-Hinton agar plates without or with various concentrations of meropenem and ceftazidime-avibactam (0, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, 256, and 512mg/l) as previously described (Li et al., 2006; Gaibani et al., 2018). Avibactam was tested at a fixed concentration of 4μg/ml in combination with increasing concentrations of ceftazidime. The analysis was conducted in three replicates, and carbapenem-susceptible K. pneumoniae ATCC 25922 was used as the control strain. After 48h of incubation at 35°C, the number of colonies was counted.
Results
Patient-Clinical Context
The patient was a 75-year-old man who had been diagnosed with Mycobacterium avium-intracellulare complex lung disease 2years earlier and had been treated with antimicrobial therapies since then. Antimicrobial therapy included ethambutol, clarithromycin, moxifloxacin, and linezolid. The patient had been admitted to the hospital several times over the past 2years for the lung disease. However, the infective process and radiological findings persisted despite the repeated courses of antimicrobial therapies. M. avium-intracellulare in sputum samples also persisted. He was admitted to the department of pulmonary and critical care medicine, China-Japan Friendship Hospital due to a fever. During hospitalization, patient was treated with clarithromycin (0.5g every day; 0.25g qn) and ethambutol (750mg every day) for M. avium-intracellulare complex lung disease. On day 32 post-admission, under therapy with ceftazidime-avibactam plus amikacin for 3days, a carbapenem-resistant KPC-producing K. pneumoniae (isolate ZRKP01) was recovered from a sputum. On hospital day 47, under therapy with ceftazidime-avibactam plus amikacin for 12days, a ceftazidime-avibactam-resistant but imipenem-susceptible isolate was recovered from sputum (isolate ZRKP02, exhibited a phenotype compatible with an ESBL producer). The antibacterial therapy was changed to imipenem plus polymyxin B. 15days after imipenem therapy, bronchoalveolar cultures yielded a carbapenem-resistant K. pneumoniae (isolate ZRKP03 and isolate ZRKP04) on hospital day 61 and again on day 67, respectively, which prompted reinitiation of amikacin plus ceftazidime-avibactam with the continued addition of imipenem. After imipenem plus ceftazidime-avibactam plus amikacin was started, five respiratory tract specimens were collected between days 72 and 90 that did not yield microorganisms. However, the patient developed respiratory failure and died. The patient’s antibiotic treatment course and the timeline of the K. pneumoniae isolates acquired are summarized in Figure 1.
Figure 1.
History of the K. pneumoniae isolation and clinical antimicrobial treatment course of a patient with KPC-Kp infection. Blue bar represents the imipenem, purple bars represent the amikacin, red bars represent ceftazidime-avibactam, and green bars represent polymyxin B. The red arrows show the isolation times of K. pneumoniae (isolate ZRKP01, ZRKP02, ZRKP03, and ZRKP04). CZA, Ceftazidime-avibactam; IMP, Imipenem; MEM, Meropenem; BALF, bronchoalveolar lavage fluid.
Emergence of an Apparent Phenotypic Change of K. pneumoniae Isolates During Infection
During the course of illness, four serial K. pneumoniae isolates were cultured from respiratory tract specimens on hospital 32, 47, 61, and 67days, respectively. Testing of susceptibility to ceftazidime-avibactam and carbapenems showed three distinct susceptibility phenotypes as shown in Table 1. Phenotype 1 (isolate ZRKP01), collected at the baseline (under therapy with ceftazidime-avibactam for 3days), was susceptible to ceftazidime-avibactam (MIC, 0.5mg/l) and resistant to carbapenem (IMP, MIC 32mg/l; MEM, MIC 128mg/l). Phenotype 2 (isolate ZRKP02), collected following 12days ceftazidime-avibactam, displayed ceftazidime-avibactam resistance (MIC, 64mg/l) and restored susceptibility to carbapenem (IMP, MIC 0.06mg/l; MEM, MIC 2mg/l). Of note, phenotype 3 (isolate ZRKP03), collected after 15days imipenem plus 6days polymyxin B, reverted to carbapenem resistance (IMP, MIC 128mg/l; MEM, MIC 512mg/l) and restored susceptibility to ceftazidime-avibactam (MIC, 2mg/l; although with elevated MIC compared with baseline isolate ZRKP01). Phenotype 3 (isolate ZRKP04), collected during imipenem, ceftazidime-avibactam plus amikacin combination therapy, was susceptible to ceftazidime-avibactam (MIC, 2mg/l) and resistant to carbapenem (IMP, MIC 64mg/l; MEM, MIC 256mg/l; a lower MIC compared with ZRKP03). These isolates displayed in an apparent phenotypic change from carbapenem-resistant to susceptible and then reverted to resistant for carbapenem, while the phenotypic change from ceftazidime-avibactam-susceptible to resistant then reverted to susceptible for ceftazidime-avibactam.
Table 1.
Summary of antimicrobial susceptibility testing for four K. pneumoniae isolates recovered from a patient with pneumonia.
| Isolates No. | MIC (mg/L) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CZA | IMP | MEM | CAZ | ATM | FEP | TZP | TGC | CST | |
| ZRKP01 (KPC-2) | 0.5 | 32 | 128 | >=64 | >=64 | >=64 | >=128 | 1 | 0.5 |
| ZRKP02 (KPC-33) | 64 | 0.06 | 2 | >=64 | >=64 | >=64 | >=128 | 1 | 0.5 |
| ZRKP03 (KPC-2) | 2 | 128 | 512 | >=64 | >=64 | >=64 | >=128 | 1 | 1 |
| ZRKP04 (KPC-2) | 2 | 64 | 256 | >=64 | >=64 | >=64 | >=128 | 1 | 1 |
CZA, Ceftazidime-avibactam; CAZ, Ceftazidime; IMP, Imipenem; MEM, Meropenem; ATM, Aztreonam; FEP, Cefepime; TZP, Piperacillin/tazobactam; TGC, Tigecycline; CST, Colistin.
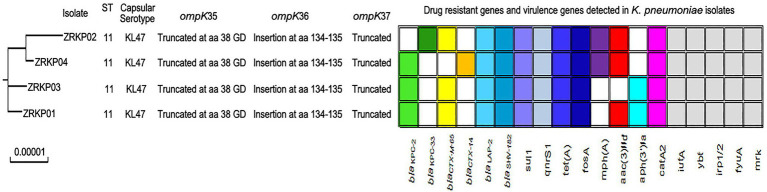
Genome Comparison of Four KPC-Kp Isolates
Genetic analysis demonstrated that all four KPC-Kp isolates belonged to sequence type (ST11) and had identical capsular polysaccharide (KL47; wzc: 47, wzi: 209), a common CRE species in China. All the four isolates had genes encoding KPC, SHV-182, and CTX-M β-lactamases. Other acquired resistance genes justifying the resistance phenotype and virulence genes are depicted in Figure 2. Analysis of outer membrane porin genes demonstrated that all the four isolates had a mutated ompK35 gene encoding truncated porin (premature stop codon at amino acid position 88) and had a mutated ompK36 gene encoding a porin OmpK36 with a GD insertion at amino acid position 134–135.
Figure 2.
Molecular characteristic of four K. pneumoniae isolates.
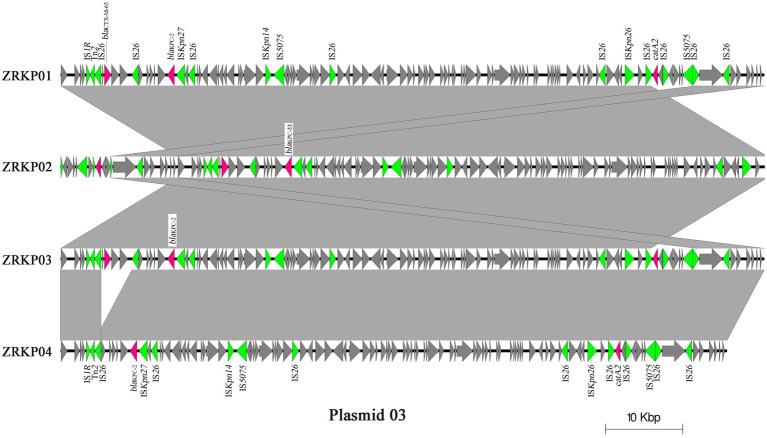
The genomic features of assemblies are presented in Table 2. The chromosome lengths of ZRKP01, ZRKP02, ZRKP03, and ZRKP04 were 5.435 Mbp, 5.438 Mbp, 5.434 Mbp, and 5.436 Mbp, respectively, similar in length to other K. pneumoniae genomes in public databases (range, 5.3–5.6 Mbp). They had an average G + C content of 56.9% and carried 25 ribosomal RNA genes and 85 transfer RNA genes. The four K. pneumoniae isolates harbored five similar plasmids referred as plasmid 01, plasmid 02, plasmid 03, plasmid 04, and plasmid 05. Plasmid content analysis showed that all four KPC-Kp isolates shared the same plasmid replicon types as shown in Table 2. Furthermore, the fusion was observed of IncFII and IncFIB in plasmid 01 and IncFII and IncR in plasmid 03. The plasmid 03 lengths of ZRKP01, ZRKP02, ZRKP03, and ZRKP04 were 91,097, 91,502, 91,097, and 86,253 base pairs, respectively. The blaKPC gene was located in plasmid 03 as shown in Figure 3. Ceftazidime-avibactam-susceptible isolates (ZRKP01, ZRKP03, and ZRKP4) carried blaKPC-2, whereas ceftazidime-avibactam-resistant isolate (ZRKP02) carried mutant blaKPC-2 (blaKPC-33) encoding for KPC enzymes. Deep examination of reads aligning to the blaKPC-2 gene demonstrated that 94% of aligned reads of the ZRKP01 isolate displayed the wild-type (KPC-2) and 6% displayed D179Y mutation (KPC-33). We did not find coexistence of wild-type KPC-2 and mutation D179Y (KPC-33) in the WGS data of the remaining three K. pneumoniae isolates (ZRKP02-ZRKP04). Corresponding to the phenotype change, the K. pneumoniae isolates displayed an genotype change from wild-type KPC-2 to variant KPC-2 (KPC-33) and then reversion to its original wild-type KPC-2.
Table 2.
Genomic features of four Klebsiella pneumoniae ST11 isolates.
| Feature | ZRKP01 | ZRKP02 | ZRKP03 | ZRKP04 |
|---|---|---|---|---|
| G+C content, % | 56.98 | 56.98 | 56.99 | 56.99 |
| Plasmids, no. | 5 | 5 | 5 | 5 |
| Size, base pairs | ||||
| Chromosome | 5,435,561 | 5,438,841 | 5,434,420 | 5,436,990 |
| Plasmid 01 | 239,469 | 220,761 | 205,940 | 238,333 |
| Plasmid 02 | 110,529 | 110,529 | 110,529 | 110,529 |
| Plasmid 03 | 91,097 | 91,502 | 91,097 | 86,253 |
| Plasmid 04 | 10,060 | 10,060 | 10,060 | 10,060 |
| Plasmid 05 | 5,596 | 5,596 | 5,596 | 5,596 |
| Plasmid (Inc) | ||||
| Plasmid 01 | IncFIB, IncFII | IncFIB, IncFII | IncFIB, IncFII | IncFIB, IncFII |
| Plasmid 02 | IncFIB | IncFIB | IncFIB | IncFIB |
| Plasmid 03 | IncFII, IncR | IncFII, IncR | IncFII, IncR | IncFII, IncR |
| Plasmid 04 | ColRNAI | ColRNAI | ColRNAI | ColRNAI |
| Plasmid 05 | NA | NA | NA | NA |
| Genes, no. | 5,186 | 5,189 | 5,180 | 5,182 |
| CDS, no. | 5,076 | 5,079 | 5,070 | 5,072 |
| Ribosomal RNA genes, no. | 25 | 25 | 25 | 25 |
| Transfer RNA genes, no. | 85 | 85 | 85 | 85 |
| Prophages, no. | 15 | 14 | 14 | 14 |
| IS elements, no. | 50 | 53 | 49 | 51 |
| Chromosome IS family (no.) | IS1 (4), IS3 (9), IS5 (25), IS6 (1), IS481 (1), IS1182 (1), IS1380 (3), ISNCY (6) | IS1 (3), IS3 (9), IS5 (27), IS6 (1), IS110 (2), IS481 (1), IS1182 (1), IS1380 (3), ISNCY (6) | IS1 (3), IS3 (9), IS5 (25), IS6 (1), IS481 (1), IS1182 (1), IS1380 (3), ISNCY (6) | IS1 (3), IS3 (9), IS5 (26), IS6 (1), IS110 (2), IS481 (1), IS1182 (1), IS1380 (3), ISNCY (6) |
CDS, coding DNA sequences; IS, insertion sequence.
Figure 3.
Major structural features of plasmid 03 identified in the four K. pneumoniae isolates. Light grey shading denotes shared regions of homology, and open reading frames (ORFs) are portrayed by arrows. Pink arrows represent antibiotic resistant genes, and green arrows represent transposon-related genes and insertion sequences. The plasmid sequence of sample ZRKP02 was reverse-complemented to get a better presentation.
Using the genome assembly of K. pneumoniae isolate (ZRKP01) as a reference, we identified 17 substitutions among K. pneumoniae isolates comprising 6 synonymous and 11 nonsynonymous substitutions. SNP analysis revealed that the isolate ZRKP02 (KPC-33) evolved by up to 12 SNPs/genome, the ZRKP03 evolved by 6 SNPs/genome, and the ZRKP04 evolved by 7 SNPs/genome over the patient’s hospitalization. Numbers and positions of SNPs for each pairwise comparison of isolates are shown in Figure 4. In our analysis of the evolutionary process in this case, we found that the number of mutations observed in the ZRKP02 (KPC-33) isolate was more than in the wild-type KPC-2 isolates (ZRKP01, ZRKP03, and ZRKP04). These mutations in ZRKP02 isolate accounted for 9 of 11 total nonsynonymous mutations. Of note, 6 of these targets (2 malt, srlB, nifJ, ydjA, and blaKPC-33) reverted to its original wild-type on further imipenem treatment; in particular, wild-type KPC-2 confer resistance to carbapenem. On the other hand, the remaining 3 mutant genes (rhtB, manZ, and a hypothetical protein) showed clonal succession (Figure 4) Therefore, it demonstrated that the four KPC-Kp isolates evolved from a longitudinal evolution of K. pneumoniae harboring blaKPC-2 gene.
Figure 4.
Summary of mutations present in four K. pneumoniae isolates and their effects on annotated coding sequences. The mutations in ZRKP 02 (KPC-33) isolate accounted for 9 of 11 total nonsynonymous mutations. Six of these targets (2 malt, srlB, nifJ, ydjA, and blaKPC-33) reverted to its original wild-type, including wild-type KPC-2 confer resistance to carbapenem in ZRKP 03 and ZRKP04 isolates. The remaining 3 mutant genes (rhtB, manZ, and a hypothetical protein) showed clonal succession in ZRKP 03 and ZRKP04 isolates. Amino acid abbreviations follow the standard one-letter code. Red arrows represent the nonsynonymous substitutions, green arrows represent the synonymous substitutions, and blue arrows represent the reversion of the nonsynonymous substitutions.
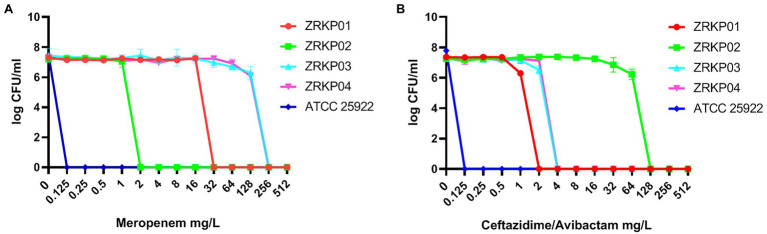
PAP of Four KPC-Kp Isolates
PAP curves of the four KPC-Kp isolates are shown in Figure 5. Population analysis showed that both the meropenem-resistant subpopulation and ceftazidime-avibactam-resistant subpopulation were not found in all four isolates (CRKP01-CRKP04).
Figure 5.
Population analysis profile (PAP) curves of the four KPC-producing K. pneumoniae clinical isolates. PAP at different concentrations of meropenem (A) and ceftazidime-avibactam (B).
Discussion
Here, we report the dynamic emergence of resistance to ceftazidime-avibactam and carbapenems in KPC-Kp during antimicrobial therapy. Using WGS of four longitudinal clinical isolates, we directly documented clonal succession of the blaKPC-2 conferring carbapenems and blaKPC-33 conferring ceftazidime-avibactam resistance.
The patient described in this study was intermittently exposed to 40days of ceftazidime-avibactam. After 12days ceftazidime-avibactam plus amikacin treatment, ceftazidime-avibactam-resistant KPC-33 (KPC-2 D179Y variant) producing K. pneumoniae strain appeared, as indicated from previous study (Shields et al., 2016; Haidar et al., 2017; Shields et al., 2017a; Giddins et al., 2018; Hemarajata and Humphries, 2019; Sun et al., 2020). Of note, KPC-33 producing K. pneumoniae (ceftazidime-avibactam-resistant) isolate was unable to preserve the resistance phenotype without the selective pressure of ceftazidime-avibactam and it reverted to susceptible phenotype on further selective pressure of imipenem. Importantly, the restored carbapenem susceptibility was not stable and subsequently reverted to its original carbapenem resistance phenotype on the selective pressure of imipenem. It should be noted that understanding the clinical significance of this observation is of critical importance, and reversion to carbapenem susceptibility would not imply a potential role for carbapenems monotherapy. In addition, the efficacy of dual ceftazidime-avibactam and carbapenem therapy in these settings is unclear. More studies are needed to establish the precise role of carbapenems in treating such infections and the durability of restored carbapenem susceptibility in vivo.
Similarly, the in vitro reversion of blaKPC-3 mutations by K. pneumoniae isolates was recently described by Shields et al., 2017b; isolates with D179Y substitutions in KPC-3 when exposed to meropenem, blaKPC-3 mutations reverted to wild type, were replaced by new mutations, or were retained (Shields et al., 2017b). This in vitro reversion of blaKPC-3 mutation was also observed by Göttig et al. from Germany (Göttig et al., 2019). They found that isolate with D179Y substitution in KPC-3, blaKPC-3 mutation reverted to wild type, and demonstrated the isolate with mutational change in KPC-3 under selection pressure are associated with ceftazidime-avibactam resistance, while imipenem resistance was solely due to reversion of KPC-3 D179Y to wild-type KPC-3. To the best of our knowledge, this is the first report to observe the in vivo evolution of wild-type KPC-2 to KPC-33 and then the reversion to its original wild-type KPC-2.
Mechanisms of horizontal gene spread among K. pneumoniae often considered to be the main mediators of antibiotic resistance, such as acquisition and loss of antibiotic resistance genes as previously described (Villa et al., 2013; Simner et al., 2018). However, mutational resistance also has primary clinical importance when considering resistance to particular antibiotics, especially to carbapenems and ceftazidime-avibactam (Haidar et al., 2017; Shields et al., 2017a; Giddins et al., 2018; Hemarajata and Humphries, 2019; Sun et al., 2020). In the present study, genetic analysis demonstrated that all four K. pneumoniae isolates belonged to sequence type 11, had identical capsular polysaccharide (KL47), identical porin genes, and same plasmid replicon types. Phylogenetic analysis indicated that four K. pneumoniae isolates showed a high degree of relatedness. PAPs showed no co-mixed infection population. These results demonstrate the four KPC-Kp isolates evolved from a longitudinal evolution of K. pneumoniae harboring blaKPC-2 gene. Our result indicated that the variant KPC can be relatively unstable, which may result in KPC enzymes dynamic change through mutations or reversions to alter their spectra of activity under different selection pressure.
In the course of ceftazidime-avibactam treatment, the dynamic change of resistance to ceftazidime-avibactam and carbapenems has been documented in KPC-Kp clinical isolates (Gaibani et al., 2018; Sun et al., 2020). However, little is known regarding the mechanisms of dynamic change. Gaibani et al. found that two different subpopulations harboring wild-type and mutant blaKPC-3 coexisting in the same KPC-Kp clinical isolate and the coexistence of different variants within a single isolate determining a hybrid phenotype resulting in resistance to both carbapenems and ceftazidime/avibactam (Gaibani et al., 2018). Heteroresistance to ceftazidime-avibactam was also observed in our previous study (Sun et al., 2020). We described coexistence of wild-type KPC-2(29.4%) and mutation D179Y (KPC-33, 71.6%) within a single isolate, which may contribute resistance to both carbapenems and ceftazidime-avibactam. In the present study, through WGS, we demonstrated the role of selective pressure of antibiotic in the mutation and reversion of blaKPC genes, which leading to the dynamic change of KPC enzymes and the dynamic emergence of resistance to ceftazidime-avibactam and carbapenems.
It should be noted that K pneumoniae with mutant blaKPC may be identified as ESBL producers (rather than KPC producers), if carbapenemase screening is triggered by elevated carbapenem MICs (Shields et al., 2017a). Targeted sequencing of blaKPC of isolates may prove to be powerful tools for rapidly identifying loss or restored activity of ceftazidime-avibactam and carbapenems, respectively.
Conclusion
This study described the plasticity and speed of evolutionary changes in KPC-Kp strains and stressed the importance of mutations and reversion of blaKPC genes in dynamic change of resistance to ceftazidime-avibactam and carbapenems.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors to any qualified researcher. The assembly genome sequences have been deposited on NCBI with BioProject ‘PRJNA613645’.
Ethics Statement
Permission for using the information in the medical records of the patients and the K. pneumoniae isolates for research purposes was granted by the ethical committee of China-Japan Friendship Hospital (2019-164-K113).
Author Contributions
BC and YL conceived or designed the work. ZL, AS, and BL collected the clinical data. BL and LS collected the laboratory data and performed the tests. CW and JZ analyzed and interpreted the data. CW, JZ, YL, and BC drafted the manuscript. All authors contributed to the article and approved the final version of the manuscript to be published.
Funding
This work was financially supported by the National Key R&D Program of China (2017YFC1309301 and 2017YFC1309300), CAMS Innovation Fund for Medical Sciences (CIFMS 2018-I2M-1-003), and National Science Grant for Distinguished Young Scholars (81425001/H0104) for BC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Lifeng Li, Yanyan Fan, Xiaohui Zuo, Yudi Xia, Haibo Li, Xinmeng Liu, Zhujia Xiong, Yulin Zhang, and Jiajing Han for their assistance.
Footnotes
References
- Amdt D., Grant J. R., Marcu A., Sajied T., Pon A., Liang Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both A., Buttner H., Huang J., Perbandt M., Belmar C. C., Christner M., et al. (2017). Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. J. Antimicrob. Chemother. 72, 2483–2488. doi: 10.1093/jac/dkx179, PMID: [DOI] [PubMed] [Google Scholar]
- CLSI (2019). Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 29th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. 2019. [Google Scholar]
- Coppi M., Pilato V. D., Monaco F., Giani T., Conaldi P. G., Rossolini G. M. (2020). Ceftazidime-avibactam resistance associated with increased blaKPC gene copy number mediated by pKpQIL plasmid derivatives in sequence type 259 Klebsiella pneumoniae. Antimicrob. Agents Chemother. 64, e01816–e01819. doi: 10.1128/AAC.01816-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone M., Paterson D. (2016). Spotlight on ceftazidime-avibactam: a new option for MDR gram-negative infections. J. Antimicrob. Chemother. 71, 2713–2722. doi: 10.1093/jac/dkw239, PMID: [DOI] [PubMed] [Google Scholar]
- Gaibani P., Campoli C., Lewis R. E., Volpe S. L., Scaltriti E., Giannnella M., et al. (2018). In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J. Antimicrob. Chemother. 73, 1525–1529. doi: 10.1093/jac/dky082, PMID: [DOI] [PubMed] [Google Scholar]
- Galani I., Antonisdou A., Karaiskos I., Kontopoulou K., Giamarellou H., Souli M. (2019). Genomic characterization of a KPC-producing Klebsiella pneumoniae ST 258 clinical isolate resistant to ceftazidime-avibactam. Clin. Micro. Infect. 25, 763–e5. doi: 10.1016/j.cmi.2019.03.011, PMID: [DOI] [PubMed] [Google Scholar]
- Giddins M. J., Macesic N., Annavajhala M. K., Stump S., Khan S., McConyville T. H., et al. (2018). Successive emergence of Ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-Harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob. Agents Chemother. 62, e02101–e02117. doi: 10.1128/AAC.02101-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttig S., Frank D., Mungo E., Nolte A., Hogardt M., Besier S., et al. (2019). Emergence of ceftazidime-avibactam resistance in KPC-3-producing Klebsiella pneumoniae in vivo. J. Antimicrob. Chemother. 74, 3211–3216. doi: 10.1093/jac/dkz330, PMID: [DOI] [PubMed] [Google Scholar]
- Haidar G., Clancy C. J., Shields R. K.Hao B., Cheng S., Nguyen M. H.. (2017). Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 61, e02534–e02516. doi: 10.1128/AAC.02534-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemarajata P., Humphries R. M. (2019). Ceftazidime-avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J. Antimicrob. Chemother. 74, 1241–1243. doi: 10.1093/jac/dkz026, PMID: [DOI] [PubMed] [Google Scholar]
- Humphries R. M., Hemarajata P. (2017). Resistance to Ceftazidime-avibactam in Klebsiella pneumoniae due to Porin mutations and the increased expression of KPC-3. Antimicrob. Agents Chemother. 61, e00537–e00517. doi: 10.1128/AAC.00537-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries R. M., Yang S., Hemarajata P., Kevin W. W., Janet A. H., Shelley A. M., et al. (2015). First report of Ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 59, 6605–6607. doi: 10.1128/AAC.01165-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoke R. A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Rayner C. R., Nation R. L., Owen R. J., Spelman D., Tan K. E., et al. (2006). Heteroresistance to Colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50, 2946–2950. doi: 10.1128/AAC.00103-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1298–1303. doi: 10.1101/gr/107524.110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K., Hemarajata P., Sun D., Rubio A. D., Tsivkovski R., Yang S., et al. (2017). Resistance to Ceftazidime-avibactam is due to transposition of KPC in Porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob. Agents Chemother. 61, e00989–e00917. doi: 10.1128/AAC.00989-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S., Bankevich A., Antipov D., Gurevich A. A., Korobeynikov A., Lapidus A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153, PMID: [DOI] [PubMed] [Google Scholar]
- Shen Z., Ding B., Ye M., Wang P., Bi Y., Wu S., et al. (2017). High ceftazidime hydrolysis activity and porin OmpK35 deficiency contribute to the decreased susceptibility to ceftazidime/avibactam in KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 72, 1930–1936. doi: 10.1093/jac/dkx066, PMID: [DOI] [PubMed] [Google Scholar]
- Shields R. K., Chen L., Cheng S., Chavda K. D., Press E. G., Snyder A., et al. (2017a). Emergence of Ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of Carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob. Agents Chemother 61, e02097–e02016. doi: 10.1128/AAC.02097-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. K., Nguyen M. H., Press E. G., Chen L., Kreiswirth B. N., Clancy C. J. (2017b). In vitro selection of meropenem resistance among ceftazidime-avibactam-resistant, meropenem susceptible Klebsiella pneumoniae isolates with variant KPC-3 carbapenemases. Antimicrob. Agents Chemother. 61, e00079–e00017. doi: 10.1128/AAC.00079-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. K., Potoski B. A., Haidar G., Hao B., Doi Y., Chen L., et al. (2016). Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin. Infect. Dis. 63, 1615–1618. doi: 10.1093/cid/ciw636, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simner P. J., Antar A. A. R., Hao S., Gurtowski J., Tamma P. D., Rock C., et al. (2018). Antibiotic pressure on the acquisition and loss of antibiotic resistance genes in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 1796–1803. doi: 10.1093/jac/dky121, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Chen W., Li H., Li L., Zuo X., Zhao J., et al. (2020). Phenotypic and genotypic analysis of KPC-51 and KPC-52, two novel KPC-2 variants conferring resistance to ceftazidime-avibactam in KPC-Kp ST11 clone background. J. Antimicrob. Chemother. 75, 3072–3074. doi: 10.1093/jac/dkaa241, PMID: [DOI] [PubMed] [Google Scholar]
- van Duin D., Bonomo R. A. (2016). Ceftazidime-avibactam and ceftolozane/tazobactam: second-generation beta-lactam/beta-lactamase combinations. Clin. Infect. Dis. 63, 234–241. doi: 10.1093/cid/ciw243, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani A. M., Siguier P., Gourbeyre E., Charneau V., Chandler M. (2011). ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12:R30. doi: 10.1186/gb-2011-12-3-r30, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L., Capone A., Fortini D., Dolejska M., Rodríguez I., Taglietti F., et al. (2013). Reversion to susceptibility of a carbapenem-resistant clinical isolate of Klebsiella pneumoniae producing KPC-3. J. Antimicrob. Chemother. 68, 2482–2486. doi: 10.1093/jac/dkt235, PMID: [DOI] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2013). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors to any qualified researcher. The assembly genome sequences have been deposited on NCBI with BioProject ‘PRJNA613645’.