ABSTRACT
Listeria monocytogenes is a saprophytic gram-positive bacterium, and an opportunistic foodborne pathogen that can produce listeriosis in humans and animals. It has evolved an exceptional ability to adapt to stress conditions encountered in different environments, resulting in a ubiquitous distribution. Because some food preservation methods and disinfection protocols in food-processing environments cannot efficiently prevent contaminations, L. monocytogenes constitutes a threat to human health and a challenge to food safety. In the host, Listeria colonizes the gastrointestinal tract, crosses the intestinal barrier, and disseminates through the blood to target organs. In immunocompromised individuals, the elderly, and pregnant women, the pathogen can cross the blood-brain and placental barriers, leading to neurolisteriosis and materno-fetal listeriosis. Molecular and cell biology studies of infection have proven L. monocytogenes to be a versatile pathogen that deploys unique strategies to invade different cell types, survive and move inside the eukaryotic host cell, and spread from cell to cell. Here, we present the multifaceted Listeria life cycle from a comprehensive perspective. We discuss genetic features of pathogenic Listeria species, analyze factors involved in food contamination, and review bacterial strategies to tolerate stresses encountered both during food processing and along the host’s gastrointestinal tract. Then we dissect host–pathogen interactions underlying listerial pathogenesis in mammals from a cell biology and systemic point of view. Finally, we summarize the epidemiology, pathophysiology, and clinical features of listeriosis in humans and animals. This work aims to gather information from different fields crucial for a comprehensive understanding of the pathogenesis of L. monocytogenes.
KEYWORDS: Listeriosis, stress response, intracellular pathogen, food contamination, pathogenesis
Graphical abstract
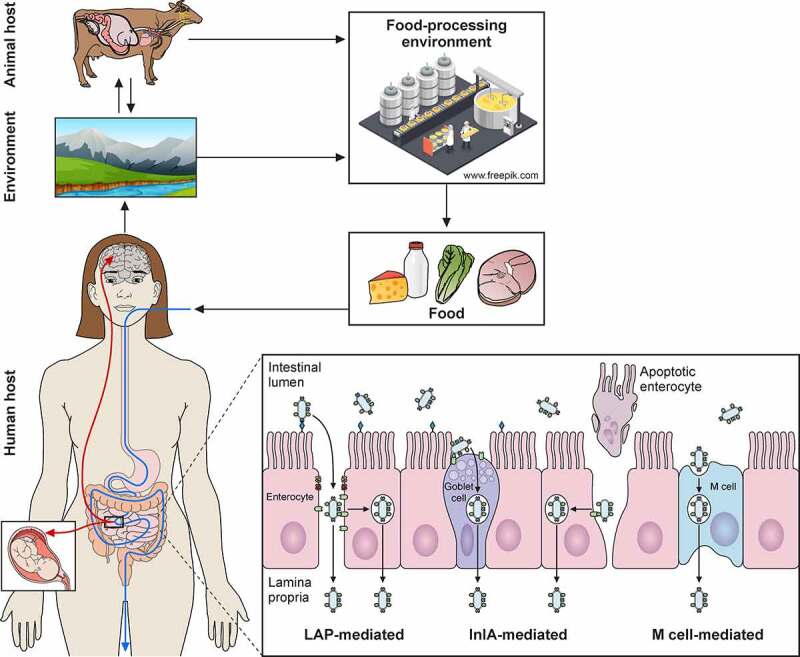
Introduction
L. monocytogenes is the causative agent of listeriosis, a sporadic disease in humans and animals with very high hospitalization and case-fatality rates, and considered one of the most serious foodborne diseases [1,2]. The genus Listeria currently includes 21 recognized species of ubiquitous small rod-shaped gram-positive bacteria, of which only Listeria ivanovii and L. monocytogenes are mammalian pathogens [3]. Pathogenic Listeria species emerged as a major foodborne pathogen in western countries in the second half of the twentieth century, and human and animal listeriosis outbreaks have had a significant economic impact on society and the food industry. Importantly in Europe, the incidence of listeriosis has increased since 2008 [2].
Although Listeria genus was a saprophytic bacterium in origin, some species have successfully adapted to different environmental niches associated with human activity, including farms (mammalian and avian feces), food, and food-processing environments, thanks to an unparalleled capacity to sense and respond to environmental stress. This stress tolerance also allows Listeria to pass from contaminated food into the gastrointestinal tract of mammalian hosts [4].
Once inside the host, L. monocytogenes evolves varied and sophisticated mechanisms to invade different eukaryotic cell types, survive intracellularly, evade the immune system, and disseminate through the body [5]. Moreover, this pathogen can cross the blood-brain and placental barriers, with tragic consequences in disease progression (meningitis, abortion), and a fatal outcome in immunocompromised individuals and pregnant women, respectively [6,7].
L. monocytogenes is a multifaceted pathogen that represents a serious threat to human and farm animals and a challenge to food safety [1,2]. It has become a high priority for molecular and cellular pathogenesis studies due to the urgent need to develop targeted therapies to help to reduce mortality. In this review, we approach Listeria from an interdisciplinary and comprehensive perspective. We discuss its contamination routes and risk factors, as well as its epidemiology, pathophysiology, and clinical signs in humans and animals. We also consider, from a molecular and clinical angle, the genetic features of pathogenic Listeria species that allow them to survive environmental and host-associated stresses and bacterial regulation mechanisms, as well as the host-pathogen interactions underlying listerial pathogenesis in mammals.
The genus Listeria encompasses 21 species
Not all Listeria species are pathogenic
The genus Listeria currently includes 21 species of ubiquitous gram-positive rods found in different environmental niches [3]. Two Listeria species, L. monocytogenes and L. ivanovii, have been historically considered pathogenic [8,9]. L. monocytogenes infects animals and humans, and is in its genus the zoonotic species of greatest importance for global public health and economics. L. ivanovii has been considered to infect mainly ruminants [10], since human cases are rare, involving mainly immunocompromised, debilitated patients [11–13]. Two explanations may support the low occurrence of L. ivanovii infections in humans: 1) this species could have low pathogenicity for humans, or 2) the sporadic occurrence and limited distribution in nature (including food) of L. ivanovii would limit the exposure of humans to this bacterium [9,11,14].
Although L. innocua was initially considered a non-hemolytic and nonpathogenic Listeria species, natural atypical hemolytic isolates have been isolated from different food products [15–17], and rare cases of L. innocua septicemia and meningitis infections have been reported in both ruminants and humans [18–21]. These atypical hemolytic L. innocua challenge the concept that this species is innocuous. Moreover, these L. innocua isolates possess a functional Listeria pathogenicity island 1 (LIPI-1) and internalin A (inlA) genes, which encode important virulence factors for the intestinal infection stage, the entry into the host cells, and the adaptation to an intracellular lifestyle [22]. Importantly, atypical hemolytic L. innocua can actively cross the intestinal barrier and invade deeper organs like the liver and spleen [22]. Moreover, some L. innocua strains bear LIPI-3, a pathogenicity island specific to L. monocytogenes lineage I, which is overrepresented in epidemic listeriosis outbreaks [23–25]. LIPI-3 encodes a bacteriocin highly expressed in the intestine to alter host intestinal microbiota, allowing L. monocytogenes colonization of the intestine (Table 1) [23–25]. Remarkably, one of these L. innocua strains possessing LIPI-3 was isolated from a human patient with meningitis [23]. Human exposure to hemolytic L. innocua is rare, since no isolate exhibiting this phenotype has been detected during Listeria surveillance in the context of food and clinical samples performed by the World Health Organization Collaborating Center for Listeria (Institut Pasteur, France) after analyzing more than 7236 L. innocua isolates between 1987 and 2018 [22].
Table 1.
Characteristics of species of genus Listeria related to human and animal cases [8,9,22,26,27,29,32,35,46,335]. PI-PLC is an important Listeria enzymatic marker used routinely in selective chromogenic culture media for discrimination and enumeration of L. monocytogenes-L. ivanovii and other Listeria spp. in human specimens, food products and environmental samples (e.g. ALOA and RAPID’L.mono Agar Plates)
| L. monocytogenes Lineage I | L. monocytogenes Lineage II | L. monocytogenes Lineage III and IV | L. ivanovii | L. innocua | L. seeligeri | |
|---|---|---|---|---|---|---|
| LIPI −1 | + | + | + | + | -(rare isolates are +) | Some isolates are + (with some gene rearrangements) |
| InlA/B locus | + | + (>30% isolates present truncated InlA) |
+ | + | -(Rare isolates possess InlA) | - |
| LIPI −3 | Present in ≈ 50% Lineage I isolates | - | - | - | Present in ≈ 20% isolates | - |
| LIPI −4 | Mostly present in CC4 | - | Some isolates are + | - | + | - |
| LIPI −2 | - | A highly virulent hybrid sub-lineage harbors a truncated LIPI-2 | - | + | - | - |
| Virulence | +++ (specially CC1, CC2, CC4, and CC6) Overrepresented among human isolates |
+ Overrepresented among food and food-related isolates |
+ Most isolates obtained from animals |
Mainly animal cases Rare human cases |
-Rare human cases | -Rare human cases |
| Hemolysis | + | + | + | +++ (due to the sphingomyelinase SmcL) |
-(Rare isolates are +) | LIPI −1 + strains are hemolytic |
| Phosphatidylinositol-specific phospholipase C (PI-PLC coded by plcA) | + | + | + | + | -(Rare LIPI −1 + strains have PI-PLC activity) | LIPI −1 + strains have PI-PLC activity |
Some L. seeligeri isolates possess LIPI-1 and are hemolytic [26,27]. Rare human cases caused by L. seeligeri have been documented, including a previously healthy adult presenting acute purulent meningitis [28] (Table 1). However, despite atypical isolation from human clinical cases, there is no evidence that L. seeligeri should be considered a pathogen or presents a human health risk comparable to L. monocytogenes [8].
Characterization and Subtyping of L. monocytogenes: Lineage differences
L. monocytogenes was initially classified into 13 serotypes, based on agglutination of somatic (O) and flagellar (H) antigens, with only three (1/2a, 1/2b, and 4b) causing more than 90% of invasive human infections [29,30]. Further characterization and differentiation on the strain level was achieved by the development of molecular methods including pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) [30–32]. In L. monocytogenes, MLST was carried out by sequencing internal portions of seven housekeeping genes [33]. MLST showed that: 1) L. monocytogenes forms a structured population consisting of four divergent lineages (I–IV); and, 2) the isolates belong to groups of genetically highly similar strains, called clonal complexes (CCs) [33]. Each lineage includes specific serotypes: lineage I comprises serotypes 1/2b, 3b, 4b, 4e and 7; lineage II, serotypes 1/2a, 1/2 c, 3a, and 3 c; lineage III, serotypes 4b, 1/2a, 4a, and 4 c; and lineage IV, 4a and 4 c [31,33]. Although most of the clinically relevant strains belong to lineages I and II, major listeriosis epidemics are associated with lineage I isolates and, more specifically, with serotype 4b [34–36]. Lineage II or serotypes 1/2a, 1/2b, and 1/2 c are more frequent in foods [35,37]. Lineages III and IV are rarely isolated, predominantly from animal sources [29,32]. Although MLST provides highly standardized genotypes and nomenclature, it lacks the discriminatory power required for epidemiological surveillance [32]. Nowadays, whole-genome sequencing (WGS) is the most powerful epidemiological typing tool for investigating outbreaks and performing biological population studies [32,38,39]. WGS techniques use a combination of core genome multilocus sequence typing (cgMLST, which covers 1748 genes) and single-nucleotide variant methods [32].
To gain experimental reproducibility among laboratories, the majority of studies focused on L. monocytogenes pathogenicity have been carried out with laboratory reference strains: EGDe, EGD, or 10403S. These strains belong to lineage II (EGD and 10403S to CC7, and EGDe to CC9), which is underrepresented in patients with clinical signs [35,40,41]. The recurrent use of these CC7 and CC9 reference strains has led to an underestimation of overall L. monocytogenes biodiversity and, as a result, of the heterogeneity that may exist in the virulence mechanisms used by strains of lineages I and II (Table 1) [42]. CC1, CC2, CC4, and CC6 all belonging to lineage I (Table 1) are associated with human clinical cases (some of them even without immunosuppressive comorbidities). This evidence suggests that these CCs are hypervirulent, while other CCs, such as CC9 and CC121 belonging to lineage II, are strongly associated with food environments (Table 1) [35]. Hypervirulent L. monocytogenes clones, particularly CC1, associate strongly with dairy products, while hypovirulent clones, CC9 and CC121, are frequently isolated in meat products [36,43]. Hypervirulent clones colonize better in the intestine and display a higher invasion rate of the intestinal mucosa than hypovirulent clones. Conversely, hypovirulent clones adapt well to food-processing environments, with higher prevalence of genes involved in stress resistance and tolerance to disinfectants [36]. In domestic animals, listeriosis is most common in ruminants. Interestingly, CC1 (which associates strongly with clinical origin in humans) also associates with cases of ruminant rhombencephalitis and is highly overrepresented in milk-derived products. These findings suggest that L. monocytogenes virulence in humans could relate to its ability to associate with dairy ruminants [35,36,44,45].
Virulence factors of L. monocytogenes are either scattered across the genome (e.g., the inlA-inlB locus, bsh, inlC, lap, among others) or clustered in pathogenicity islands LIPI-1, LIPI-3, and LIPI-4. The inlA-inlB locus encodes two surface proteins, internalins InlA and InlB, involved in pathogen internalization by non-phagocytic cells after binding of the host cell receptors E-cadherin and Met, respectively [44]. The inlA-inlB locus, together with LIPI-1, is necessary for key steps of intracellular parasitism (e.g. adhesion, internalization, intracellular survival and dissemination). Importantly, the inlA-inlB locus and LIPI-1 are conserved in almost all L. monocytogenes isolates, highlighting their crucial role for pathogenicity (Table 1) [5]. Importantly, more than 30% of lineage II isolates (overrepresented in food) harbor premature stop codons in the inlA gene, leading to virulence attenuation [29,46]. Comparative genomics between hypo- and hypervirulent clones led to the discovery of new virulence factors such as LIPI-3 (present only in a subset of lineage I strains) and LIPI-4. LIPI-4 is present in L. monocytogenes lineage I CC4, (a CC significantly associated with materno-fetal listeriosis and neurolisteriosis in humans), in L. monocytogenes lineages III and IV, and lately, in L. innocua (Table 1) [22,25,32,35,47,48]. The virulence factors encoded in LIPI-1 hly and actA are indispensable for virulence [49–51]. Recent studies of human epidemiological and clinical data integrated with bacterial population genomics identified full-length InlA, LIPI-3 and LIPI-4 as being strongly associated with infectious potential at the population level [35]. The functions of these LIPIs and the inlAB locus will be further discussed below (Table 1). LIPI-2, which encodes several internalins and the hemolytic enzyme sphingomyelinase, is specific for L. ivanovii (Table 1) [9]. Other important L. monocytogenes virulence genes like bsh or inlC are not clustered in pathogenic islands but instead scattered across the genome [52,53]. Instead of providing an exhaustive list of the virulence factors of L. monocytogenes, we will in each section of the manuscript describe the most important factors that play key roles in the different steps of the infection process (e.g., gut colonization and intestinal barrier traversal, brain and placental invasion or cell entry, vacuolar scape and cell-to-cell spread, among others). For comprehensive reviews of Listeria virulence factors see [9,54–56].
Recent studies characterizing L. monocytogenes isolates from severe ovine listeriosis outbreaks identified a highly virulent hybrid sub-lineage of the major lineage II. These L. monocytogenes isolates harbored LIPI-1, the inlA-inlB locus, and a truncated LIPI-2 locus (encoding sphingomyelinase) from L. ivanovii. Importantly, LIPI-3 and LIPI-4 were absent in these isolates [57] (Table 1).
Acquisition and loss of genetic elements have provided the properties necessary for specialization of L. monocytogenes to an environment or to a host. Three distinct patterns among major L. monocytogenes clones have been proposed: (i) clones that persist efficiently in food-production environments owing to efficient tolerance to disinfectants and biofilm formation, but with low adaptation to the host (e.g. CC9 and CC121); (ii) clones that are host-associated, exhibiting a low adaptation to food-production environments and rarely harboring disinfectant resistance genes (e.g. CC1 and CC4); and (iii) intermediary clones that may be in the process of transitioning from host-associated to saprophytic lifestyles. These intermediary clones would combine vertically transmitted features of host-adapted clones and horizontally transferred disinfectant resistance genes [36,58]. Genome-wide association studies have pointed to potential genes involved in adaptation to the host or to a saprophytic lifestyle [36]. Future experiments testing the contribution of these genes to L. monocytogenes lifestyle preferences will be crucial to understand its saprophyte-to-pathogen transition.
Listeria is a foodborne pathogen
Food associated with Listeria outbreaks
The majority of large listeriosis outbreaks are associated with the consumption of ready-to-eat (RTE) food products, including meat and seafood, as well as milk and dairy products. Outbreaks have also been reported due to consumption of produce, including fresh fruit and frozen vegetables, in the home environment (Figure 1) [2,59].
Figure 1.
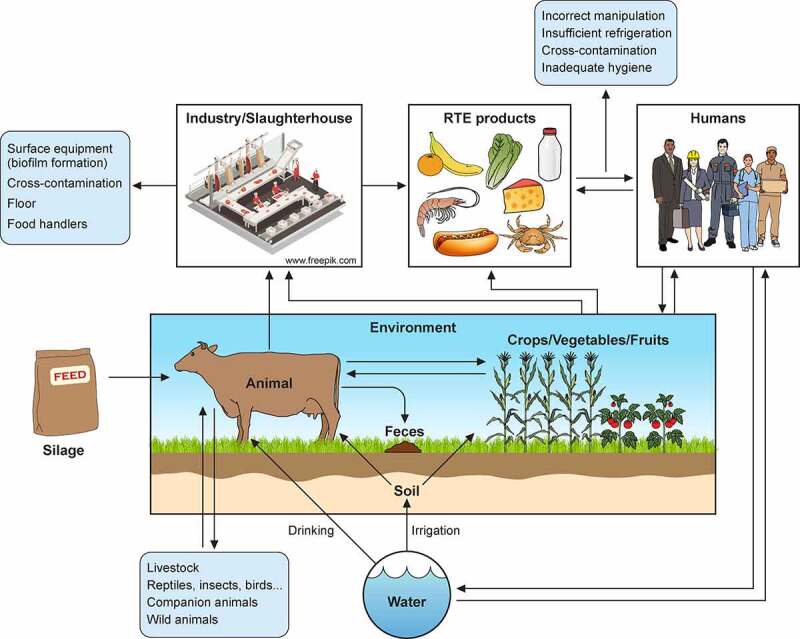
L. monocytogenes contamination sources. Transmission scenarios for L. monocytogenes between soil/water, animals, crop/vegetables/fruits, industries, food products, humans, and environment. Potential transmission pathways indicated by arrows
Foods mostly associated with foodborne listeriosis include RTE products that: (i) support growth of L. monocytogenes; (ii) are consumed without receiving any antibacterial treatment (e.g., thermic treatment); and (iii) have long refrigerated shelf-life [60]. Despite frozen foods do not support the growth of L. monocytogenes, consumption of this type of food may contribute to the risk of listeriosis, mainly when it is added directly (e.g. to salads or smoothies) [61]. L. monocytogenes adaptation mechanisms to cold and their consequences are described in the next sections. The most severe worldwide listeriosis outbreak reported in history, with 937 cases and 216 deaths, occurred in 2018 in South Africa, and was due to the consumption of an RTE processed meat product (bologna-style sausage) [62]. Other reported foodborne outbreaks associated with meat and poultry products were linked to hot dogs [63], RTE pork meats [64], turkey meat products [65], or rillettes and jellied pork tongue [66]. Dairy products, such as pasteurized and unpasteurized cheeses [67–69], pasteurized milk [70], ice cream [71], and butter [72], have also been frequently associated with listeriosis outbreaks, as well as seafood such as rainbow trout [73], crabmeat [74], and smoked salmon (Table 2) [75]. Although most contaminations occur during processing, for instance, while cutting or packaging, outbreaks are also associated with unprocessed raw or fresh fruits and vegetables, such as coleslaw [76] or cantaloupe [59]. During 2011, a cantaloupe-associated listeriosis outbreak impacted 28 US states with 147 cases, 33 deaths, and 1 miscarriage [59], highlighting the significance of produce contamination within farm and processing environments. The preparation process of raw and fresh products entails an additional risk since a contamination on the surface during peeling and cutting can be transferred to the inner part of the food (e.g. pulp) [59,77]. The 2018 report of the European Food Safety Authority (EFSA) and the European Center for Disease Prevention and Control (ECDC) [2] points out that RTE food represents one of the most important routes of Listeria transmission, with an occurrence of 2.7% for fish and fishery products, 1.4% for meat and meat products, 1.8% for RTE fruit and vegetables, and <0.8% for soft and semi-soft cheeses and <0.5% for hard cheeses [2].
Table 2.
Sources and foods implicated in published reports of foodborne listeriosis
| Source in which L. monocytogenes has been isolated | RTE food in which L. monocytogenes has been isolated |
|---|---|
| Fruits & Vegetables | Cantaloupe [59] Caramel apples [336] Packaged salad [337] Coleslaw [76] Diced celery [338] Stone fruits [339] |
|
Mammals: Cattle [309] Pig [307] Wild Boar [310] Sheep [309] Goat [340] Deer [310] |
Meat: Sausages [62] Hot dogs [63] Pork products [66,341] Beef products [342] Rillettes/Pâté [66] Game meat [343] Dairy: Raw milk [90] Pasteurized milk [70] Cheeses [67,68] Ice cream [71] Butter [72] |
|
Aquatic animals: Fish [1] Crustaceans [1] |
Crab [74] Trout [73] Smoked salmon [75] |
|
Birds: [344–346] |
Egg [347] Poultry products [17] Turkey products [65] |
|
Insects: Ants [348] |
|
|
Nature : Soil [349] Silage [306] Irrigation water [349,350] |
|
|
Farm environment: Soil and silage [87] Milk filter/tanks [91] Equipment [90] Workers [90] |
|
|
Food industry: Floor drains [351] Biofilms [351,352] Cross-contamination [353,354]: Vacuum machine, slicing machine, cutting boards, stainless steel tables, non-food contact surfaces (walls, floor) Workers’ hands [354] |
The infectious dose of L. monocytogenes
The severity of listeriosis in humans depends on the virulence of the bacterial strain. Other key factors are the dose of bacteria ingested, the diversity of a population’s genetic background, the general health and immune status of the host, and any attributes of the food that alter microbial or host status [60].
To date, no conclusive epidemiological data are available to establish the level of contamination involved in most cases of food listeriosis outbreaks. Nonetheless, infective doses of L. monocytogenes have been estimated to be 107 to 109 colony-forming units (CFUs) in healthy hosts, and only 105 to 107 CFUs in high-risk individuals [71,78–81]. Recent outbreaks have shown that even with a widespread distribution of products contaminated with L. monocytogenes, most consumers will not become ill if contamination levels are low and no growth is facilitated [71]. In this regard, documented episodes of human fecal carriage of L. monocytogenes did not coincide with overt illness [82]. These studies, together with available reports on outbreaks in the US and Italy, indicate that even the milder form of the disease, characterized by febrile gastroenteritis in normal hosts, requires the ingestion of high doses of several million bacteria [70,82,83]. On the other hand, highly susceptible populations can develop severe clinical manifestations of listeriosis after consuming products with a low-level of contamination [71].
To control L. monocytogenes food transmission, regulatory agencies have obliged food industries to develop programs for hazard analyses at critical control points and have strictly regulated the L. monocytogenes contamination of food [6,84,85].
Although for regulatory purposes all L. monocytogenes strains are currently treated equally, some strains (CC1, CC2, CC4 and CC6) are hypervirulent, often associated with clinical cases, and moreover associated with patients with few or no immunosuppressive comorbidities, while other strains (CC9 and CC121) are less virulent and infrequently related to clinical infection [35,36]. The severity of the disease, the uncertainty associated with the minimal infectious dose, and the virulence differences observed among strains, would lead to recommending that members of these high-risk groups (immunocompromised individuals, the elderly, and pregnant women) avoid eating food very likely to contain high amounts of L. monocytogenes. For the rest of the population, it would be advisable to handle high-risk foods carefully, and to store them at low temperatures for only short periods.
Contamination sources
The L. monocytogenes ubiquity in the environment determines the high risk of contamination during food manufacturing processes. L. monocytogenes has been isolated from natural environments, farms, soil, water, silage, decaying vegetables, human and animal feces and tissues, food processing industries, and processed food products (Figure 1) [10,60]. Although the environment is the natural reservoir for Listeria spp., where they live as saprophytes, its incidence increases with human and animal activity [86].
Farm environments and animals have a high genetic diversity of Listeria spp [87,88]. Moreover, persistent strains can stay in the farm environment for years, making its control a challenge [89]. In addition, farm animals can behave as silent carriers of L. monocytogenes, resulting in pathogen dissemination by their feces to the environment, farm surfaces, and equipment (e.g. milking equipment) [90], or directly through the milk [91] or meat [92] (Figure 1). These routes result in L. monocytogenes being transmitted as contaminants to foods.
L. monocytogenes can be introduced into food plant industries as a result of cross-contamination by workers (human carriers), transportation of animals (fecal shedders), raw food (e.g., milk), and materials from crops, soil, and silage [82,87,93]. Then, L. monocytogenes can persist in food-processing plants for years, or even decades, facilitating food contamination during processing/handling and packaging, and therefore its foodborne transmission. Biofilm formation, adaptation to stress, especially to cold temperatures and sub-lethal concentrations of disinfectants, together with the existence of niches in facilities and in equipment that are difficult to clean, have been identified as key contributors to persistence of the pathogen [94] (Figure 1).
Gaining insight into the natural reservoirs of L. monocytogenes will help to prevent its entry into the food chain. Surveillance strategies in agricultural settings and food industries will help to understand how L. monocytogenes circulates between the environment, animals, food industries and humans.
Listeria responds to environmental stress
The ubiquity of Listeria results from an outstanding capacity to adapt to stress conditions encountered in different environments. L. monocytogenes proliferates in distinct food matrices, where it can be exposed to high salt concentrations, an acidic pH, refrigeration temperatures or germicidal blue light, among others, and persists in food-processing environments (FPEs) cleaned with disinfectants [4]. In addition, once ingested by mammalian hosts Listeria resists the acidic pH of the stomach, and stressful conditions in the gut lumen, such as a high osmolality, and the presence of bile and the microbiota. The alternative sigma factor (SigB) is partially responsible for this resilience, by inducing hundreds of genes involved in the general stress response (GSR) [95]. However, the response to each specific stress is different, and SigB-independent regulatory mechanisms may be essential for its robustness (Figure 2).
Figure 2.
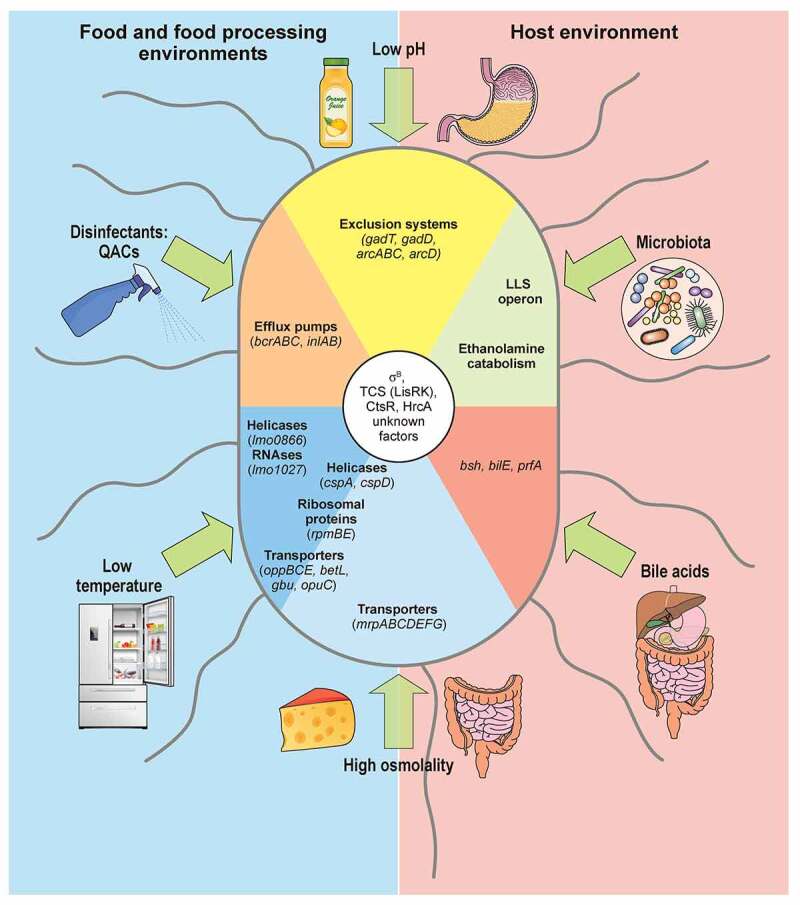
L. monocytogenes response to stress encountered in the environment and within host. Different types of stress encountered in food and food-processing environments (blue) and within host (beige) indicated with green arrows. Low pH and high osmolality are relevant stresses both in host and food. L. monocytogenes responses depicted as sectors. Genes participating in responses to both low temperature and high osmolality (helicases, ribosomal proteins, transporters) written on boundary of respective sectors. L. monocytogenes that tolerates quaternary ammonium compound (QACs) exposure overexpresses efflux pumps, like bcrABC operon. Two representative groups of enzymes induced specifically upon cold exposure: helicases, e.g. lmo0866, an RNA helicase homologue to DEAD-box protein A; and RNases, e.g. lmo1027, protein similar to Ribonuclease J1. Sodium/proton antiporter, encoded by operon mrpABCDEFG, induced exclusively under high osmolality conditions, as in salt-preserved food. Large overlap exists in response to high osmolality and low temperature: helicases, ribosomal proteins, and well-characterized osmolyte transporters like oppBCE, betL, gbu and opuC. cspA and cspD induced in both conditions: cspA predominant in response to cold shock; cspD appears in high osmotic conditions. Acidic environments increase transcription of exclusion systems as GAD and ADI, intending to raise intracellular pH. In gut, L. monocytogenes competes for nutrients with host microbiota by induction of secondary metabolic pathways like ethanolamine catabolism, or production of bacteriocins like listeriolysin S (LLS). Bile acids secreted to intestine promote induction of bsh and bilE that allow L. monocytogenes survival, and prfA which prepares L. monocytogenes for internalization and intracellular lifestyle. Regulation of these responses not yet fully elucidated (unknown factors), but some regulators shown to play role in control of stress responses are: SigB, two-component system LisRK, and transcriptional regulators HrcA and CtsR
L. monocytogenes withstands stresses in food and food-processing environments
Salting is a common and effective method to preserve food. However, Listeria can survive and grow, albeit with difficulty, in high salt concentrations (3 M) mainly by favoring the accumulation of compatible solutes, which are water-soluble organic compounds that increase intracellular osmolality, thus preventing outward water fluxes. Upon osmotic shock, Listeria induces gbu and betL, encoding glycine-betaine transporters and the carnitine ABC transporter opuCA (Figure 2) [96]. L. monocytogenes preferentially uses one of these transporters to adapt under high salt concentrations, depending on the availability of each osmo-protectant in the food matrix; of these, glycine-betaine is more abundant in plants [97] and carnitine in meat [98].
L. monocytogenes is one of the few foodborne pathogens that can grow at temperatures as low as −0.4°C, a condition referred to as psychrotolerance. This capacity renders refrigeration ineffective to restrict its proliferation in RTE. In non-psychrotroph organisms, low temperatures make biological membranes less fluid and stabilize secondary structures of nucleic acids, which impair transport mechanisms and halt gene expression, respectively [99]. Upon cold shock, L. monocytogenes dramatically reduces its growth rate, and induces enzymes participating in the synthesis of precursors of branched-chain fatty acids, and transporters of glycin-betaine (gbu), carnitine (opuC), and oligopeptides (oppA), which may contribute to maintenance of membrane fluidity and increase in the uptake of compatible solutes, respectively [100] (Figure 2). In addition, induction of RNA helicases and the cold shock protein CspA may enable protein synthesis at low temperatures by melting RNA secondary structures [100–103]. Interestingly, the two additional Listerial csp homologues (cspB, and cspD) are downregulated at low temperatures [100,101], suggesting that conserved traits of cold shock proteins may be relevant for physiological functions, other than adaptation to cold, that remain to be described.
Apart from these classical food-preservation methods, L. monocytogenes is exposed to antimicrobial procedures in food or FPEs. L. monocytogenes can also persist in FPEs following routine cleaning and disinfecting procedures [94]. As sanitization does not reach all surfaces homogeneously, concentration gradients of biocides are generated throughout FPEs, enabling microorganisms to colonize niches (or harborage sites) where they may be exposed to sub-inhibitory concentrations of these compounds [104]. This mild selective pressure favors the development of tolerance mechanisms against quaternary ammonium compounds (QAC), which are the most common and effective disinfectants used in FPEs. The mechanisms of tolerance to QACs rely mainly on the expression of efflux pumps encoded by horizontally acquired genetic mobile elements [105–107], and on the formation of biofilms [108]. A widespread dissemination across L. monocytogenes strains of the bcrABC locus, which encodes an efflux pump, renders cells tolerant to benzalkonium chloride – a widely used QAC – regardless of their serotype and the source from where they were isolated [106] (Figure 2). Acquisition of efflux pumps might be an advantage, not only to tolerate QACs and persist in FPEs, but also to tolerate antibiotics commonly used in clinics [109]. For a more detailed overview of Listeria biofilms we refer the reader to recent excellent reviews [110,111]).
Industrial and agricultural activity may facilitate a toxic accumulation of heavy metals. A high tolerance to cadmium (Cd) and arsenic (As) is a frequent trait in L. monocytogenes thanks to efflux pumps present in genetic mobile elements found both in the chromosome and plasmids [112]. The prevalence of Cd resistance is higher in strains associated with food isolates (1/2a and 1/2b) and persistent clones in FPEs that also show resistance to benzalkonium chloride [112,113], although the potential contribution to persistence in FPEs remains to be elucidated. On the other hand, resistance to As is much more prevalent in serotype 4b and, in particular, among clones associated with outbreaks of listeriosis [112,114]. This observation points at a relevant role of As resistance in virulence. However, a mechanistic explanation for increased prevalence of tolerance to Cd and As in persistent and highly virulent strains, respectively, is still elusive.
L. monocytogenes survives the acidic pH of the stomach
Upon ingestion of contaminated food, L. monocytogenes reaches the stomach, where it faces an extremely acidic pH (1–2). This low pH poses the first physicochemical antimicrobial host barrier. Consistently, a higher fecal isolation rate of L. monocytogenes has been found in patients receiving long-term gastric acid suppression with H2-antagonists, and treatment with proton-pump inhibitors is associated with an increased risk of listeriosis [115,116]. L. monocytogenes has different systems in place to regulate intracellular pH (see below), thus allowing it to overcome the acidic pH of the stomach, as well as the mild acidity of particular types of food (Figure 2) [reviewed in [117]].
The glutamate decarboxylase (GAD) system imports a molecule of glutamate by an antiporter (GadT), which is then converted into γ-aminobutyrate (GABA) by the glutamate decarboxylase (GadD). The intracellular glutamate decarboxylation reaction consumes one proton, thereby raising intracellular pH. The GadT antiporter subsequently transports GABA back out of the cell in exchange for glutamate. This system is relevant for withstanding strong acids in synthetic gastric fluid and for oral infection of mice [118,119]. Five gad genes have been identified: two transporters (gadT1-2) and three enzymes (gadD1-3). In EGD-e, SigB plays a crucial role in adaptation to strong acidic stress and regulates the transcription of gadT2, gadD2 and gadD3 upon exposure to pH 4.5 for 1 h [120]. Interestingly, differences in the expression of gad genes have been found among L. monocytogenes isolates from lineage II [119,121]. It would be illuminating to dissect the precise regulation in clinical isolates of listeriosis outbreaks to investigate whether specificities in the functioning of the GAD system may be associated with the virulence of these strains.
The second main mechanism utilized by L. monocytogenes for pH homeostasis is the arginine deiminase system (ADI). In the cytosol, arginine is converted to ornithine by the action of three enzymes encoded by the arcABC operon. Then, a membrane-bound antiporter encoded by arcD exports a molecule of ornithine from the cell while importing a molecule of arginine. Conversion of arginine to ornithine generates ammonia (NH3), which associates with a proton to produce NH4+, thus increasing cytoplasmic pH [122]. arcABC genes are induced upon exposure to acid and in the presence of arginine [123]. Mutants lacking ArcB grow more slowly under mild acidic conditions (pH = 4.8) and show a decreased survival in synthetic gastric fluid [124]. A putative transcriptional regulator, ArgR, has been identified, although the precise regulation of ADI genes as a function of pH and arginine availability, in addition to its interdependence with SigB, is as yet unclear [123,125].
In the intestinal lumen L. monocytogenes competes with endogenous microbiota
The intestine is a complex environment where L. monocytogenes must adapt to harsh conditions and coexist with host intestinal microbiota. The gut microbiota provides so-called colonization resistance, which is a protection against the establishment of foreign (pathogenic) bacteria in the digestive tract. Colonization resistance relies on diverse mechanisms, such as growth inhibition of enteric pathogens by competition, immune system maturation, or enhancing the maintenance of the intestinal barrier by synthesizing short fatty acids. Enteropathogenic bacteria have, however, developed strategies that overcome colonization resistance, such as the usage of alternative metabolic pathways like ethanolamine catabolism, the enhancement of gut inflammatory response, or the production of bacteriocins [126].
To assess the influence of host microbiota on L. monocytogenes infection, germ-free mice were inoculated with two Lactobacillus species prior to oral challenge with this pathogen. Pre-colonization by Lactobacillus induced transcriptomic changes in both host and pathogen, resulting in a limited dissemination of L. monocytogenes [127]. Other beneficial gut microbiota-associated bacteria effective against L. monocytogenes are Clostridia. In germ-free mice, pre-inoculation with clostridial species impaired L. monocytogenes colonization of the intestinal lumen and systemic dissemination [128]. This latter study shows an increased susceptibility to L. monocytogenes infection due to antimicrobial therapy, highlighting the importance of a varied gut microbiota and introducing the possibility of a probiotic preventive treatment in immunocompromised individuals or pregnant women.
L. monocytogenes can respond to trophic competition by utilizing alternative metabolic sources. Upon oral administration of L. monocytogenes in germ-free mice pre-inoculated with Lactobacillus, the pathogen increased levels of enzymes involved in the ethanolamine catabolism pathway, allowing it to exploit an alternative nitrogen source, and thus to bypass the metabolic competition imposed by the commensal microbiota (Figure 2) [127]. The same metabolic adaptation has been reported in Salmonella and enterohaemorrhagic E. coli [129,130]. In addition, L. monocytogenes can produce bacteriocins, such as listeriolysin S (LLS) and Lmo2776, which are proteins that selectively kill or impair growth of neighbor competing bacteria (Figure 2). LLS is encoded in the LIPI-3 island, which is present in some lineage I strains associated with clinical origin, and it is overexpressed in bacteria colonizing the intestine, where it shows its bacteriocin activity [25,47]. Inoculation of L. monocytogenes expressing LLS demonstrated a reduction of Allobaculum and Alloprevotella genera, favoring pathogen persistence at a higher rate than an isogenic strain lacking the lls gene [25]. Lmo2776 is present in lineage I and a few lineage II strains [131]. Lmo2776 targets Prevotella in vitro in a reconstituted human gut environment, as well as in mice. Strikingly, mutants defective in lmo2276 disseminate better to liver and spleen, suggesting that Prevotella could enhance intestinal infection. Selective reduction of Prevotella abundance by L. monocytogenes might prevent excessive inflammation, thus allowing the development of persistent longer-lasting infections [131].
Holding in the gut: Listeria tolerates osmotic stress and bile salts
The intestine poses moderately high osmolality conditions to L. monocytogenes as the pathogen traverses along the gastrointestinal (GI) tract. As in the case of the osmotic stress encountered in food (see above), L. monocytogenes increases the uptake of compatible solutes by overexpressing membrane transporters BetL, Gbu and OpuC (Figure 2) [132]. Other proteins with osmoprotectant activity are: proline synthetase (ProAB), which promotes proline accumulation in high osmolality; guanosine tetra- and pentaphosphate (p)ppGpp synthetase RelA, master regulator of the stringent response; RNA chaperone Hfq; and proteases HtrA and ClpC, which are general stress-induced proteins that contribute to the degradation of misfolded proteins [96]. As most of these proteins are also involved in the response to other stresses, such as acidic or low-temperature conditions, they may have been induced in previous stages of the L. monocytogenes life cycle (e.g., in food) and thus serve as cross-protection against stress within the host (Figure 2) [96,133].
Bile is a complex mixture of bile acids, cholesterol, phospholipids, and biliverdin, a mixture which constitutes a host-natural antimicrobial fluid. Primary bile acids produced in the liver concentrate in the gall bladder and are subsequently released into the duodenum during digestion, increasing oxidation in the bacterial cytosol, protein unfolding and aggregation, and cell membrane damage [134]. L. monocytogenes can colonize the murine gall bladder since the pH of bile (pH = 7–8) is not as low as in the small intestine (pH = 5.5). Therefore, the gall bladder can act as a reservoir for extracellular replication of L. monocytogenes and its further dissemination to the intestine [135]. The bile salt hydrolase (Bsh) is one of the main L. monocytogenes factors contributing to bile tolerance (Figure 2). Bsh hydrolases, present also in intestinal microbiota, deconjugate bile salts, and their maximal expression occurs in the duodenum, coinciding with the release of bile, where it acquires its lowest pH [136]. In addition, the membrane transporter BilE functions as a bile exclusion system (Figure 2) [137]. Interestingly, secondary bile salts produced by commensal microorganisms can modulate Clostridium difficile infections [138], suggesting that host microbiota-derived byproducts of bile may play a role in L. monocytogenes infection. The general stress response regulator SigB, as well as PrfA, the master virulence-regulator of L. monocytogenes, positively regulate bsh and bilE [53,137,139]. Moreover, bile exposure triggers expression of the PrfA regulon but does not induce further SigB-dependent genes. Altogether, these data suggest that bile may be a signal preparing L. monocytogenes for intracellular life, and a critical factor involved in switching from SigB- to PrfA-mediated gene expression [140].
Regulation of the response to stress
The response of Listeria to stress with regard to the gene sets that directly mitigate each specific stress has been well characterized and described in previous sections. However, these stress responses entail an extensive cell reprogramming with pleiotropic effects, which are still poorly understood. For example, recent transcriptomic analyses involving distinct L. monocytogenes strains subjected to acidic conditions (pH = 3.0) showed enhanced expression of genes belonging to both GAD and ADI systems, along with hundreds of differentially expressed genes with no obvious involvement in acid tolerance [141,142]. This is also true of other stress responses, such as those triggered by low temperature [101], high osmolality [143], QACs [144], or bile salts [140]. These analyses reveal a significant overlap in the response, with consistent changes in genes involved in chemotaxis and motility, nutrient uptake and metabolism of alternative sources, and cell wall biosynthesis and modification. As in other microorganisms, disparate stresses can induce in L. monocytogenes a common response with a multifaceted output, known as the general stress response (GSR) [145]. GSR induction allows a prior exposure to a particular stress condition to confer cross-protection to other stresses [143,146–148].
In L. monocytogenes, the GSR is driven by an alternative sigma factor, called SigB [95]. SigB binding to the RNA polymerase core induces a transcriptional reprogramming that comprises up to 300 genes involved in carbohydrate and amino acid metabolism, cell envelope modification, flagellar biosynthesis, pH homeostasis, osmoregulation, antibiotic resistance, and quorum sensing [149]. SigB activity is controlled by a partner-switching mechanism regulated by opposed effects of protein kinase and phosphatase activities on a protein called RsbV (Figure 3). In resting conditions, SigB binding to the RNA polymerase is prevented by the anti-sigma factor RsbW. RsbW availability depends on the phosphorylation status of the anti-anti-sigma factor RsbV. Unphosphorylated RsbV binds RsbW with high affinity, which releases SigB. RsbW in turn phosphorylates RsbV, which dissociates the complex, allowing RsbW to bind and inactivate SigB. This equilibrium is displaced upon stress, a condition in which a proteinaceous macro-complex, the stressosome, activates a protein phosphatase that de-phosphorylates RsbV, thus keeping SigB free to bind to the RNA polymerase (Figure 3) [95].
Figure 3.
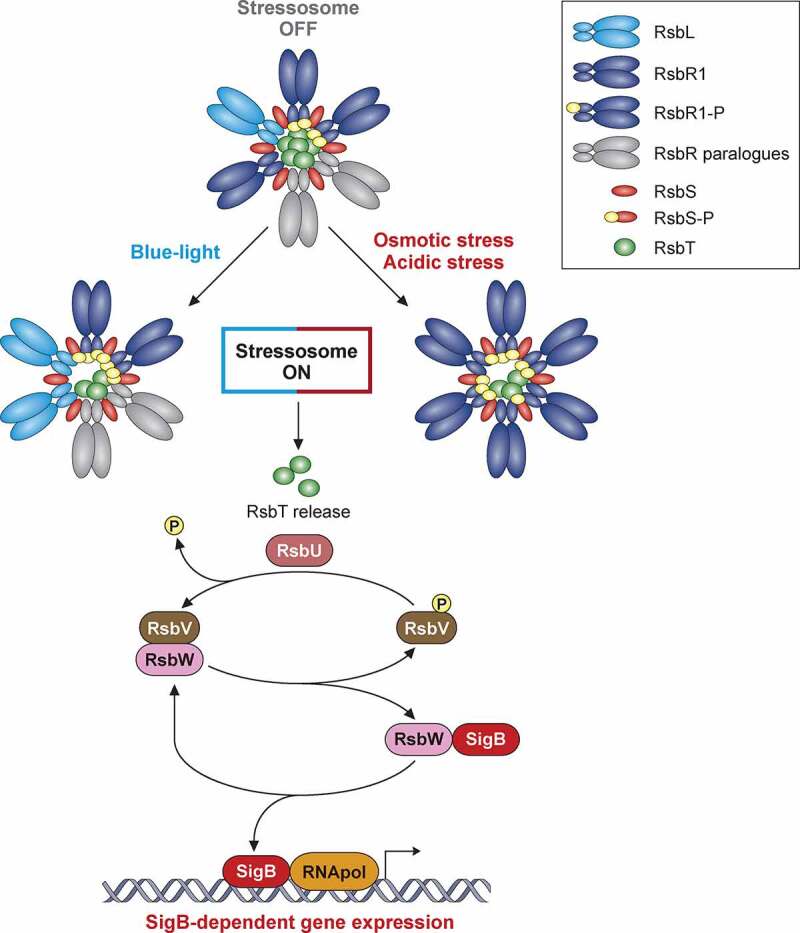
Model of SigB regulation in response to stress in Listeria monocytogenes. The stressosome is composed of RsbR, RsbR paralogues, RsbS and RsbT proteins. In unstressed conditions, RsbT is predicted as sequestrated within core of complex. Different stress types lead to phosphorylation of RsbR and RsbS by kinase RsbT through unknown mechanisms. This phosphorylation event results in release of RsbT from core of stressosome, initiating downstream SigB activation cascade. Stress-induced release of RsbT activates RsbU phosphatase. RsbU dephosphorylates RsbV, which then binds with RsbW, thus facilitating SigB release. L. monocytogenes triggers SigB-dependent general stress response after exposure to diverse environmental conditions. This model highlights the mechanisms of stressosome activation with the most characterized stress conditions reported to date
The stressosome acts as a hub for stress signal integration, as it can transduce varying multiple stresses into SigB activation. Structural studies have shown that the stressosome forms a pseudo-icosahedral core with turrets on its surface. It is in this core where the RsbR1 core domains and RsbS subunits would integrate the signals, while the RsbR1 domains forming the protruding turrets would act as stress sensors [150]. Recent reports point to a critical phosphorylation of RsbR1 core component and the subcellular localization of the kinase RsbT as essential events in the transmission of osmotic and acidic stress signals [151–153]. Nevertheless, a detailed molecular mechanism of signal transduction by the stressosome still warrants elucidation.
Despite the detailed structural analyses of the stressosome, the molecular mechanisms of stress sensing by Listeria in vivo remain mostly obscure [95]. One notable exception is blue-light sensing. Blue light has been used to kill bacteria on fresh produce [154,155]. RsbL (Lmo0799) is a paralogue of the main stressosome component RsbR1, and displays a light-oxygen-voltage (LOV) domain that binds flavin mononucleotide (FMN). Blue-light irradiation induces a transient FMN crosslinking to RsbL, which triggers a conformational change that activates signal transduction from the stressosome (Figure 3) [156–159]. Cycles of dark and blue-light induce an RsbL-dependent adaptive response consisting of the formation of opaque rings; this differentiation in colony formation confers better survival to the excessive production of reactive oxygen species (ROS) induced after exposure to blue light [157,158].
Stress sensing in Listeria may also rely on two-component systems (TCSs). TCSs are sensor and signal transduction modules in bacteria, consisting of a transmembrane sensor histidine kinase (HK) and a cognate cytoplasmic response regulator. A total of 16 TCSs have been described in L. monocytogenes, and their involvement in stress adaptation has been systematically screened [160,161]. One example is LisR-LisK, which contributes to survival under acidic conditions [162], adaptation to increased osmolality [163], growth under cold temperatures [160,164], and tolerance to antibiotics (Figure 2) [165,166] . However, the molecular mechanism(s) by which the bacteria sense these stress signals are still obscure. LisRK activity is enhanced in response to cell envelope stress triggered by treatment with cefuroxime [167]. Given the involvement of LisRK in the adaptation to so many different sources of stress, it is tempting to speculate that these could all affect the cell envelope, which would in turn activate LisRK. For instance, as the plasma membrane becomes more rigid upon temperature downshift, this alteration in membrane fluidity may represent the cold signal that activates LisRK through a yet unknown mechanism.
Although SigB is essential for the ability of L. monocytogenes to adapt to stressful conditions, including osmotic shock and acidic pH (reviewed in [95]), SigB-independent mechanisms are necessary for the specificity and robustness of each particular stress response. However, the relative contribution of SigB-dependent and SigB-independent regulatory mechanisms needs to be clarified for each particular stress response. For instance, deletion of sigB reduces L. monocytogenes survival under osmotic stress in the presence of compatible solutes [168], and opuCA induction in these conditions largely depends on SigB (Figure 2) [169]. In contrast, at 4°C opuCA induction is SigB-independent, and mutants lacking SigB acclimate to cold in the same way as wild-type bacteria [170]. As SigB is present also in other Gram-positive bacteria [171] with limited ability to withstand harsh conditions compared to Listeria, deciphering the precise participation of SigB-independent mechanisms could unveil crucial clues for understanding and preventing Listeria adaptation to stress.
Stress adaptation and virulence
An open question is whether adaptation to stress encountered in food or FPEs influences the pathogenicity of Listeria. Exposure to stress conditions prompts a response leading to phenotypic changes that may remain even after the stress disappears. Indeed, pre-exposure to sub-lethal stresses facilitates adaptation to subsequent challenges [172–174]. Thus, the use of traditional food preservation measures and/or the ineffective inactivation of L. monocytogenes contamination in food and FPEs could introduce a selective pressure on the pathogen.
The selective advantage might not necessarily, or exclusively, be restricted to a protection mechanism for a specific stress, but rather involve a more general effect. For example, stable phenotypic variants isolated after exposure to an acid stress insult (pH = 3.5, 90 min) exhibit a decreased growth rate and an enhanced resistance to other stresses. Whole genome sequencing of these variants identified mutations only in the rpsU gene [175,176], which codes for the S21 ribosomal subunit. As such, this mutation could explain only the slow growth rate in these multiple-stress tolerant variants [177]. However, mounting evidence suggests that a reduced growth rate is a trade-off, allowing for resource allocation to ensure tolerance to any stress [178,179].
Another possibility is that stress pre-conditioning in food and FPEs may result in residual activity of SigB inside the mammalian host, providing cross-protection against the acidic environment of the stomach, and against the presence of bile salts and a moderately high osmolality in the gut. In fact, SigB is the main regulator of stress response genes in the gastro-intestinal (GI) tract [180]. This is consistent with the fact that mutants lacking this sigma factor are attenuated upon oral infection of guinea pigs, but not when administered intravenously [181]. Intriguingly, intragastric inoculation of a SigB mutant in mice did not show bacterial recovery rates in liver and spleen different from those in mice infected with the wild type [182]. These conflicting results may reflect differences in invasion mechanisms in guinea pigs and mice (see below, c.f. Figure 4c), and highlight the importance of SigB-independent regulatory mechanisms of stress adaptation and virulence.
Figure 4.
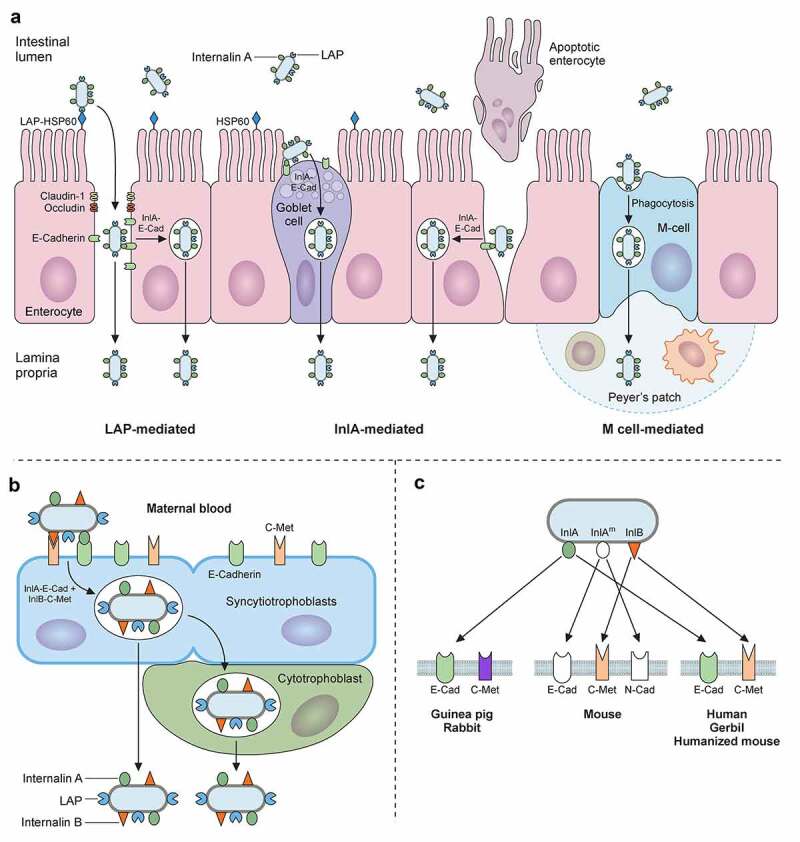
Trespassing the intestinal barrier. A) L. monocytogenes can breach intestinal barrier through three different cellular types: M-cells, goblet cells and enterocytes. L. monocytogenes will reach Peyer’s patches after M-Cell-mediated phagocytosis followed by transcytosis, where it can infect macrophages and dendritic cells, surviving intracellularly. L. monocytogenes invades secretory goblet cells through E-cadherin displayed on its junctions. Trespassing through enterocytes can be mediated in two ways: (i) via E-cadherin exposure during natural cellular extrusion, for instance, as a result of apoptosis of an enterocyte, and (ii) via LAP-HSP60 interaction. This interaction promotes redistribution of claudin-1, occludin and E-cadherin, which perturbs cell junctions, allowing translocation between enterocytes. Also, LAP-HSP60 interaction promotes E-cadherin exposure on lateral face of enterocytes, allowing Listeria cellular invasion and transcytosis. B) Infections of trophoblasts by free-circulating bacteria in blood. C) Species specificities of InlA, InlAm and InlB
Nevertheless, a direct link between an enhanced capacity to adapt to stress conditions in food and FPEs and virulence of L. monocytogenes is not yet clear. Comparison of distinct stress tolerance between subtypes isolated from either food or clinical cases has provided conflicting results [183–186]. Interestingly, genomic comparisons showed that genes involved in tolerance to the QAC benzalkonium chloride, used as surface disinfectant in FPEs, as well as other stress resistance genes, such as the SSI-1 and SSI-2 stress survival islets, are more commonly found in hypo-virulent lineage II strains [42]. This inverse correlation of the presence of stress tolerance genes and virulence suggests a divergent adaption to different environmental niches, where lineage II strains evolved an enhanced resistance to stress encountered in food and FPEs, and lineage I strains adapted to the host. In any case, stress conditions could differently affect the virulence of different strains. For example, serotype 4b (Lineage I) clinical isolates stored at cold temperatures, compared to bacteria maintained at optimal growth conditions, show an enhanced invasion of Caco-2 cells and a higher pathogenicity on chick embryos, while serotype 1/2a (Lineage II) strains, which are more frequently associated with food, do not [187,188]. Overall, while a robust capacity to tolerate a number of stresses does not seem to be a critical trait for hypervirulence, exposure to stress in the environment could protect against host GI-tract conditions and be a signal contributing to deployment of virulent traits.
Transition from stress tolerance in the GI tract into the infection: From Sigma B to PrfA-based regulation
SigB is not only a central regulator in adaptation to stressful conditions, both in host and non-host environments, but also a regulator of virulence genes. The positive regulatory factor A (PrfA) is encoded in LIPI-1 and is the master transcriptional regulator of L. monocytogenes virulence [189]. prfA is transcribed from three promoters, one of them directly and positively regulated by SigB [190,191]. Moreover, SigB regulates transcription of inlA [192] and inlB [193], which are essential for invasion of the intestinal epithelium. Thus, exposure to stress indeed “prepares” the pathogen for invasion and its intracellular life stage. Once inside the host cell, PrfA becomes the predominant regulator of virulence factors that enable intracellular replication and bacterial spread to neighboring cells. At this step, a transition has taken place from SigB to a PrfA-dominated phase of L. monocytogenes infection [180]. Nonetheless, SigB modulates the exacerbated activity of the constitutively active PrfA* mutant by downregulating PrfA regulon through a yet unknown mechanism [194]. Such a strategy, self-limiting the expression of virulence factors, restricts host cell damage, which would prolong the intracellular life of the pathogen, thereby promoting a persistent infection state [195].
PrfA induces transcription of LIPI-1, the main virulence regulon [196]. LIPI-1 includes prfA itself and genes encoding listeriolysin O (hly), phospholipase A (plcA), phospholipase B (plcB), actin assembly inducing-protein (actA), a zinc metalloproteinase (mpl), and OrfX (orfX). PrfA also regulates expression of genes encoding the bile salt hydrolase Bsh and the internalins A, B, and C (InlA, InlB, InlC) [53,196]. PrfA activity is allosterically regulated by reduced glutathione (GSH) [197]. Eukaryotic host cell cytosol is a reducing compartment where glutathione is mainly in its reduced form, and thus a source for GSH uptake by L. monocytogenes during infection. In addition, the activity of L. monocytogenes GSH synthetase, GshF, is highly induced upon mammalian cell infection [197]. Therefore, PrfA activity is enhanced in the intracellular niche, and this stimulation may depend on sensing through the redox status of the cytosol. As an additional layer of regulation, translation of the prfA mRNA is low until L. monocytogenes is inside the host by means of a mechanism that depends on host temperature [198]. In particular, at 30°C the 5’-untranslated region (UTR) of prfA mRNA folds into a secondary structure that maintains the ribosome-binding site (RBS) inaccessible to the ribosome, thus blocking its translation outside the host [198]. At 37°C, this structure melts, releasing the RBS blockade and allowing PrfA translation within the host. The augmented abundance of PrfA at 37°C leads to transcriptional activation of the upstream plcA promoter by PrfA binding, which leads to generation of a bicistronic PlcA-PrfA mRNA. Thus, thermoregulation of prfA transcript translation results in a further increase of PrfA protein levels through a positive feedback loop [198].
Listeria crossing the intestinal barrier
L. monocytogenes deploys several strategies for crossing the intestinal barrier
The next step in the Listeria infectious lifecycle is to trespass the gut epithelial barrier (Figure 4a). To do so, L. monocytogenes utilizes three different main routes [5,199]: (i) transcytosis, mainly through the invasion of goblet cells, and to a lesser extent, of enterocytes located in the tip of villi; (ii) para-cellular translocation, involving Listeria adhesion protein (LAP); and (iii) through M-cells into the Peyer patches.
In the first route, epithelial cell invasion requires binding of the cell wall-anchored protein InlA to its host receptor E-cadherin (Figure 4a). E-cadherin is a transmembrane adherent junction protein which, upon binding to InlA, gets clustered and becomes phosphorylated and ubiquitinated. Then, clathrin endosomal machinery is recruited to that site of the membrane, followed by a cytoskeletal rearrangement that entails actin filament polymerization nucleated by the Arp2/3 complex. This sequence of events eventually results in pathogen internalization [200,201]. InlB can also participate in invasion but is dispensable in intestinal epithelial cells [202]. Due to the species-specific differences in E-cadherin amino acid sequence (Figure 4c), models to study L. monocytogenes adhesion and internalization in vivo had to be adapted to allow the InlA-E-cadherin interaction. This was achieved by using: (i) transgenic mice expressing human E-cadherin, (ii) knock-in mice with humanized E-cadherin, and (iii) L. monocytogenes strains with “murinized InlA” (inlAm; Figure 4c) [203–205].
The second route to cross the intestinal epithelium occurs at the tips of the villi, and is initiated by interaction of LAP with its host receptor, the heat shock protein 60 (Hsp60) (Figure 4a) [206]. LAP is an alcohol acetaldehyde dehydrogenase present in pathogenic and nonpathogenic Listeria species. However, it promotes adhesion only in pathogenic Listeria (L. monocytogenes, L. ivanovii) due to its higher expression and secretion in these species [207]. On the other hand, Hsp60 is a chaperonin expressed in mouse ileal villi enterocytes, where it can localize at the apical domain of plasma membrane. LAP binding to Hsp60 results in NF-κB activation, which triggers the secretion of pro-inflammatory cytokines IL-6 and TNF-α and the activation of Myosin light chain kinase MLCK. The latter promotes cellular redistribution of occludin, claudin-1, and E-cadherin, resulting in distortion of the tight and adherent junctions. These changes weaken the epithelial layer, allowing paracellular translocation of L. monocytogenes from the intestinal lumen to the lamina propria [208]. It has been proposed that the LAP-Hsp60 pathway could be the main mechanism exploited by L. monocytogenes to cross the epithelial barrier at early stages of infection (12–48 h). The InlA-E-cadherin pathway would subsequently become more relevant (48 h), most likely favored by E-cadherin redistribution as a result of LAP-Hsp60 interaction [199,208].
The third route to cross the intestinal epithelium involves the Peyer’s patches (Figure 4a). Peyer’s patches are lymphoid follicles associated with regions of the epithelium highly enriched in M-cells, a specialized epithelial phagocyte. M-cells sample antigens from the intestinal lumen and release them to antigen-presenting cells present in Peyer’s patches. This process helps to trigger a specific intestinal immune response against the invading pathogen. In mice, listerial M-cell invasion depends on InlB but not on InlA [209]. However, L. monocytogenes expressing a murinized InlA (InlAm) exhibit increased M-cell invasion. This can be explained by the fact that InlAm interacts not only with murine E-cadherin, but also with N-cadherin, which is expressed in M-cells (Figure 4c). Therefore, mice infection models using L. monocytogenes expressing InlAm have the drawback of InlA promoting invasion of M-cells in a non-physiological manner. This difference also results in altered immune responses compared to those of humanized mice expressing E-cadherin and infected with wild-type L. monocytogenes [210]. The route through M-cells was thought to proceed through transcytosis. However, recent studies indicate that L. monocytogenes can overturn transcytosis through M-cells and spread to adjacent enterocytes [211].
Listeria entry and proliferation inside eukaryotic cells
L. monocytogenes is able to invade and proliferate within phagocytic and epithelial nonphagocytic cells. Interaction of InlA and InlB with their receptors (Figure 4a) leads to bacterial internalization within a membrane bound compartment. InlA and InlB are the two major surface molecules driving bacterial entry into host cells (Figure 5). Moreover, ActA, LLO and other bacterial surface proteins have been described as supporting bacterial entry [212].
Figure 5.
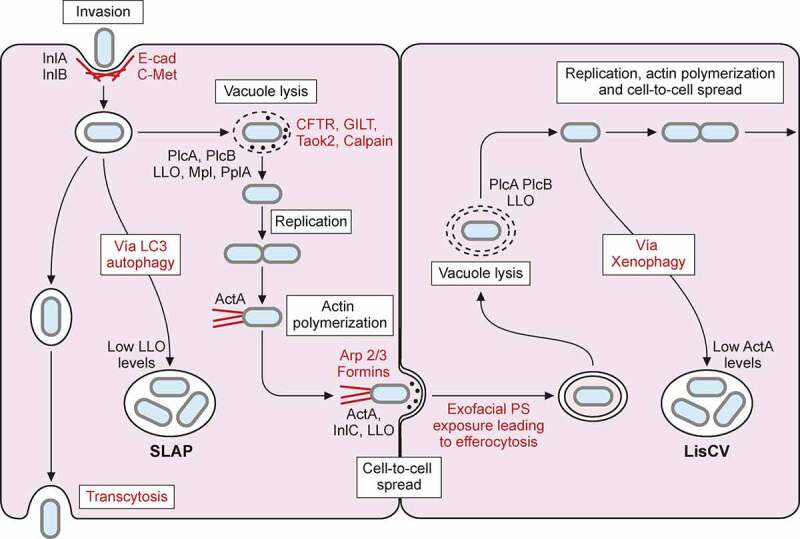
L. monocytogenes intracellular lifecycle. Invasion of non-phagocytic cells mediated by InlA and InlB interaction with host receptors E-cadherin (E-cad) and C-Met respectively, enhances actin polymerization and leads to bacterial internalization. Once inside a primary vacuole in host cytoplasm, L. monocytogenes can follow different pathways. Bacterium remains in vacuole, leading to transcytosis, as in goblet cells. In some macrophages, L. monocytogenes can replicate inside this vacuole, developing spacious Listeria containing phagosomes (SLAPs), whose formation is associated with autophagy and low LLO secretion. The vacuole can be lysed by virulence factors LLO, PlcA, PlcB, Mpl and PplA. Release of LLO into the cytoplasm has different effects on host cell, like histone modifications, mitochondrial fission, etc. In the cytoplasm of trophoblasts and hepatocytes, L. monocytogenes can be engulfed into an acidic vacuole known as Listeria containing vacuole (LisCV). Formation of LisCV’s may be due to xenophagy process in host cell and loss of ActA in L. monocytogenes. Listeria in the cytoplasm induces ActA, which interacts with host Arp 2/3 complex and formins. This promotes actin polymerization, which propels Listeria throughout the cytoplasm and leads to protrusion formation on adjacent cell. Internalin C (InlC) secretion in host cell cytoplasm perturbs apical junctions, facilitating cell-to-cell spread. LLO, also secreted in the protrusion, damages host cell membrane, exposing inner phosphatidylserine in exoplasmic layer of protrusion membrane. Exofacial exposure of phosphatidylserine is recognized as an eat-me signal that promotes Listeria-containing vesicle engulfment by macrophages. Therefore, L. monocytogenes also exploits efferocytosis for cell-to-cell spread. Bacterium will be hosted in new cell within double membrane vacuole that can be lysed again, repeating its infectious lifecycle
Once L. monocytogenes is internalized via receptor-mediated endocytosis or phagocytosis, it produces virulence factors encoded in LIPI-1 (PrfA, PlcA, LLO, Mpl, PlcB, and OrfX), which enable bacteria to escape from the vacuole (endosome or phagosome), proliferate in the cytosol, and spread to adjacent cells (Figure 5) [5,213,214].
Listeriolysin O (LLO) is a cholesterol-dependent cytolysin, encoded by hly, which binds to cholesterol and assembles large pores in the vacuolar membrane. L. monocytogenes utilizes multiple strategies to guarantee that LLO activity is restricted to the endosomal compartments. One strategy relies on the acidic pH of endosomes and phagocytic vacuoles. At low pH, LLO dimerizes and remains active, while at cytosolic pH, it aggregates and gets degraded [215]. In addition, LLO includes a 26-aminoacids segment near its N-terminus, known as PEST-like sequence, that interacts with the AP2 component of the clathrin-dependent endocytic machinery, which could promote rapid removal of LLO from the plasma membrane and targeting to autophagosomes [216]. On the other hand, oxidoreductases located in non-reducing phagosomal compartments prevent an inhibitory cysteine glutathionylation on the PEST-like sequence that keeps LLO activity low in the cytosol [217]. These features allow L. monocytogenes to break down the vacuolar membrane, while surviving in the cytosol without lysing the host cell plasma membrane, hence avoiding host immune system recognition. Indeed, mutants expressing an LLO devoid of its PEST-like sequence show a dramatic increase in cytolytic activity, along with reduced virulence [216].
The relevance of maintaining LLO activity under precise control is supported by the existence of multiple layers of hly gene expression regulation. In addition to the PrfA-mediated transcriptional control of hly, hly mRNA is also regulated post-transcriptionally. Codon restriction in the PEST-encoding RNA region, and its involvement in the formation of a secondary structure that blocks the ribosome binding site, participate in LLO translational repression in the cytosol but not in the phagosome [218]. In addition, hly 3’-UTR binds the 5’UTR of prsA2 mRNA, which protects the latter from degradation. prsA2 encodes a chaperone induced during intracellular life that is necessary for full virulence by stabilizing LLO. Thus, this mechanism, where the mRNA of a critical chaperone is stabilized by the transcript of its substrate, represents a positive feedback loop that guarantees that LLO is fully active only in intracellular bacteria [219,220].
Apart from its role in intracellular vacuolar scape, recent studies have associated LLO with many other functions [56,221]. These include nuclear processes like histone modification and the DNA damage response [222,223], modulation of the immune response [224], alteration of mitochondrial dynamics [225], and lysosomal permeabilization [226], among others [56]. Given the pleiotropic effect of LLO in eukaryotic host cell biology, it would be interesting to assess how LLO-induced alterations of each of these processes impacts disease outcome [56]. Finally, LLO is a dominant antigen for T cells. Based on this notion, LLO devoid of its cholesterol-recognition site, and having lost its pore-forming activity, was developed as a vaccine candidate with capacity to elicit cellular and humoral immune response in mice [227].
Pore formation by LLO activity is complemented by: (i) the action of phospholipases A and B (encoded by plcA and plcB); (ii) the secretion of a PrfA-dependent lipoprotein called peptide pheromone-encoding lipoprotein A (PplA); and (iii) the phage excision from its genome (restoring the activity of the competence apparatus, which promotes phagosomal escape) (Figure 5) [228–231]. Interestingly, phospholipases also inhibit the autophagic flux in infected cells, thus preventing autophagy-mediated clearance of L. monocytogenes [232,233]. During the L. monocytogenes intracellular life stage, host cell factors play a significant role in modulating infection. For example, intracellular sensing of pathogens triggers innate immune signaling, which in turn restricts bacterial infection [5,234,235]. A number of host cell factors have also been shown to modulate vacuolar rupture, including the cystic fibrosis transmembrane conductance regulator (CFTR), calpain and γ-interferon–inducible lysosomal thiol reductase (GILT) in phagocytic cells, as well as the serine threonine kinase Taok2 in epithelial cells [212,236].
L. monocytogenes also resides within vacuoles in a slow/non-growing state. In macrophages of severe combined immunodeficient (SCID) mice, which lack adaptive immunity, L. monocytogenes replicates in large compartments termed spacious Listeria-containing phagosomes (SLAPS) (Figure 5) [237]. SLAPs are nonacidic and non-degradative phagosomes generated in an autophagy dependent manner, which maintain a sub-population of intracellular L. monocytogenes producing low amounts of LLO. More recently, live fluorescent microscopy of Listeria infection of epithelial cells revealed that a subset of bacteria remains within long-term vacuoles, where they can proliferate as quickly as cytosolic ones [238]. Cytoplasmic L. monocytogenes has also been shown to switch from an active motile lifestyle to a stage of persistence within vacuoles in epithelial and trophoblast cells during several days of infection. Upon intercellular spread, L. monocytogenes gradually decreases the production of the actin-nucleating protein ActA. This ceases actin polymerization at the bacterial surface, and intracellular bacteria become trapped in lysosome-like vacuoles termed Listeria-containing vacuoles (LisCVs), where the pathogen enters into a dormant viable but non-culturable state (Figure 5) [239]. The ability of L. monocytogenes to reside inside vacuoles could reduce exposure to antibiotics during listeriosis treatment and contribute to the incubation period of listeriosis and/or the carriage of this pathogen in asymptomatic hosts [240].
Cell-to-cell spread: Importance of ActA for Listeria dissemination
The actA gene is highly upregulated during intracellular growth of the pathogen in the cytoplasm. ActA interacts with the eukaryotic Arp2/3 complex, thereby promoting actin polymerization and cell-to-cell spread [241]. Electron microscopy allowed for observation of this polymerized actin structure, named “actin comet tail” due to its aspect on the images [242]. ActA structurally mimics host nucleation-promoting factor activity – concretely that of the C-terminal domain of WASp family proteins – directly activating the Arp2/3 complex. ActA-mediated recruitment and activation of Arp2/3 promote branching of actin filaments and drive actin polymerization to propel bacteria throughout the cytoplasm (Figure 5) [243]. Other pathogens like Shigella or Burkholderia use similar intracellular motility strategies [244–246]. Formins, which are another type of host actin nucleators, promote protrusion formation [247]. The propulsion generated by actin filament polymerization results in membrane protrusions containing bacteria that penetrate adjacent host cells. Efficient cell-to-cell spread requires the secreted virulence factor Internalin C, which diminishes the cortical tension between cells and recruits the host exocyst complex, thereby facilitating formation and elongation of the protrusion [248,249]. Finally, LLO damages host cell membrane at the protrusion by its pore-forming activity. Another strategy exploits efferocytosis by macrophages. In this process, LLO activity promotes exposure of phosphatidylserine exoplasmic face of the generated Listeria-containing vesicles, which constitutes eat-me signals that promote phagocytosis by macrophages [250].
Once L. monocytogenes has spread from cell to cell, it is located in a two-membrane vacuole originated from the donor and recipient cells. In primary murine macrophages, L. monocytogenes phospholipases are involved in the dissolution of the phagosome’s inner membrane, whereas LLO targets the outer membrane originated from the recipient host cell. In epithelial cells, LLO is dispensable for this process, as phospholipases are sufficient to mediate continued cell-to-cell spread (Figure 5). These data suggest that during infection the spread of L. monocytogenes to distant organs could occur even in the absence of LLO expression [251].
Apart from mediating intracellular motility and cell-to-cell spread, the activity of ActA provides additional advantages to L. monocytogenes. In epithelial cells, ActA-driven actin polymerization prevents autophagic recognition of L. monocytogenes by masking the bacterial surface with host factors. Additionally, actin-based motility allows escape of L. monocytogenes from initial autophagic membranes (phagophore) in the macrophage cytosol [252]. During in vivo infection, ActA is also critical for L. monocytogenes aggregation in the gut lumen. ActA-dependent aggregation facilitates pathogen persistence within the cecum and colon lumen, and shedding in the feces (Figure 6) [253].
Figure 6.
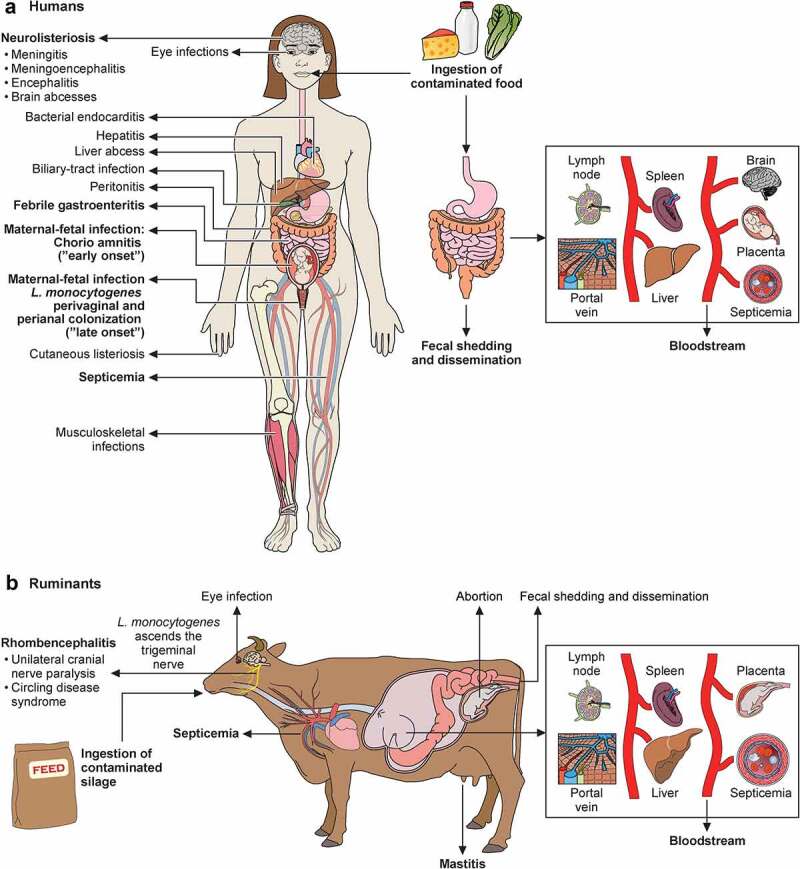
Schematic representation of transmission, pathophysiology (right side) and clinical signs (left side) of listeriosis in humans (a) and ruminants (b). Listeria, via contaminated food products, reaches intestine. In immunocompetent humans, Listeria produces febrile gastroenteritis; in immunocompromised individuals it traverses intestinal barrier, produces septicemia, can cross blood–brain barrier and cause meningoencephalitis. Newborn infection occurs as consequence of maternal chorioamnitis (“early-onset” sepsis) or by contamination from birth canal colonized with Listeria from digestive tract (“late-onset” meningitis). Listeria localized infections occur in multiple organs. In ruminants, Listeria vehiculated through contaminated silage crosses oral epithelium (facilitated by small breaches of the oral mucosa), ascends to brain stem via trigeminal nerve, leading to unilateral cranial nerve paralysis and circling disease syndrome. In ruminants, Listeria also causes septicemia, abortion and, less frequently reported, mastitis and eye infections
As depicted in this section, several host pathways are hijacked by L. monocytogenes to control entry, vacuolar rupture, intracellular motility, and cell-to-cell spread. Future efforts to decipher all the eukaryotic host cells signaling hubs subverted by L. monocytogenes will help to discover new targets to develop anti-Listeria drugs.
Listeriosis as systemic infection
L. monocytogenes in the blood
In immunocompromised individuals, L. monocytogenes traverses the intestinal epithelial barrier into the lamina propria to further disseminate via the lymph and blood to the liver and the spleen (Figure 6) [254]. The main part of the L. monocytogenes burden in the GI tract is extracellular, but the small proportion of intracellular bacteria is crucial for efficient spread to the mesenteric lymph nodes, spleen and liver [255]. In guinea pigs, pathogen dissemination to the liver occurs via two routes. The first one uses a direct pathway from the intestine to the liver via the portal vein as early as 4 h after ingestion. The second wave of dissemination occurs through an indirect pathway from the intestine via the mesenteric lymph nodes into the bloodstream, followed by systemic dissemination. This dissemination leads to the colonization of liver and spleen (Figure 6) [256].
L. monocytogenes circulates in blood either freely or associated with mononuclear phagocytes and polymorphonuclear leukocytes [257]. In vitro studies show that blood leukocytes can kill a portion of the ingested L. monocytogenes [258]. Whilst SigB mediates activation of virulence-associated genes in the host’s intestinal lumen, PrfA controls transcription of virulence genes in the blood [180]. However, in L. monocytogenes exposed to plasma in vitro, a SigB mutant showed reduced survival compared with the wild-type strain [259]. L. monocytogenes remodels its cell surface during the blood stage, selectively altering the number of several surface proteins. At this blood stage, increased levels of Lmo0514 and InlA, two surface proteins covalently bound to peptidoglycan, or LAP, are detected in the pathogen surface, and Lmo0514 is required for survival in this condition [259]. By contrast, other surface proteins, such as Internalin I, are downregulated following blood exposure [259,260]. In the mouse model, overwhelming replication of L. monocytogenes in the liver and spleen leads to a secondary bacteremia composed of a combination of cell-free and intracellular bacteria. It is during this phase that bacteria enter the central nervous system [257,261]. Cell wall remodeling could help L. monocytogenes to survive to the bactericidal activity of blood and plasma and reach target organs such as the brain or the placenta. A complete comprehension of L. monocytogenes physiology during the blood stage could help to discover new bacterial targets to develop anti-Listeria drugs to control bacteriemia.
Listeria evades host immune response
During the early infection stage, accounting for the first few days, L. monocytogenes triggers the innate immunity of the host. Innate immune responses alone can control low bacterial numbers and restrict pathogen growth in resistant mammalian hosts such as C57BL/6 mice. Subsequently, the host triggers a specific T cell response that mediates infection clearance and immunological memory [262]. The reader is referred to excellent recent and classic reviews to get a more comprehensive view related to this topic [262–265].
Although infections by this pathogen elicit strong immune responses, L. monocytogenes has evolved mechanisms to evade and modulate these host defenses. An important strategy of L. monocytogenes is to use actin-based motility to spread from cell to cell, avoiding the extracellular milieu. This explains why cytotoxic CD8+ T cells are crucial for clearance [262,266]. Another strategy is based on InlC. InlC is highly secreted by Listeria in infected cells, where it interacts with IκB kinase (IKKα) preventing NF-κB activation and, consequently, impairing cytokine expression and neutrophil recruitment at the site of infection [52]. Other L. monocytogenes secreted factors termed nucleomodulins induce host cell epigenetic modifications that manipulate host cell transcription to benefit the pathogen [267]. Modification of Listeria cell wall mediated by peptidoglycan deacetylases (PgdA) and acetyltransferases (OatA) confer lysozyme resistance and plays a key role to evade innate host defenses [254,268]. Elucidation of these strategies would help in the identification of Listeria vulnerabilities to prevent systemic dissemination.
L. monocytogenes invades target organs: Liver and spleen
Kupffer cells are professional phagocytes in the liver that experience necroptotic cell death in early L. monocytogenes infection stages. Kupffer cell death leads to a type-1 microbicidal inflammation followed by a type-2-mediated liver repair. The latter consists of massive monocyte recruitment into the liver, where differentiation to macrophages serves to replace dead Kupffer cells [269]. In addition, hepatocytes can promote monocyte recruitment via TLR2-dependent secretion of CCL2 and CXCL1 chemokines, resulting in the formation of micro-abscesses and enhanced phagocytosis of L. monocytogenes, thus limiting its spread [270]. Finally, L. monocytogenes requires InlB for hepatocyte invasion [271]. Consistently, mice infected intravenously with L. monocytogenes lacking a functional InlB show reduced burden in the liver [271,272].
In the spleen, the innate immune system controls L. monocytogenes at early stages of infection through neutrophils, dendritic cells, and macrophages [264]. At later phases, CD8+ T cells primed by CD8α+ dendritic cells (DC) are responsible for killing L. monocytogenes-infected cells [273]. Upon infection, sentinel CD8α+ DCs in the periphery migrate through the lymphatic system to lymphoid organs to participate in adaptative response. DC maturation occurs concomitantly to this migration. However, DCs can also act as a reservoir for L. monocytogenes, where bacteria evade innate immune response and can have access to the reticuloendothelial system. In this regard, Batf3(-/-) mice, which lack CD8α+ DCs, have reduced bacterial burden in the spleen. Under this condition, traffic of L. monocytogenes to the periarteriolar lymphoid sheath from the marginal zone may be impaired. This can keep bacteria longer in the marginal zone and have them more efficiently cleared [274].
Listeria reaches the brain
To date, many aspects of neurolisteriosis remain elusive. Specialized reviews have addressed the invasion of the brain by L. monocytogenes [275,276]. This pathogenic species is the only one from the genus Listeria that leads to central nervous system infection in humans and domestic ruminants [261,276,277]. Mechanisms by which L. monocytogenes accesses the CNS include retrograde axonal transport and crossing of the blood-brain barrier (BBB).
Retrograde axonal transport occurs through two different routes: (i) one that utilizes the cranial nerves – primarily the trigeminal nerve – upon crossing of the oral epithelium; and (ii) one which exploits the olfactory epithelium. In the first case, cranial nerves become infected after L. monocytogenes enters through mucosal injuries in the oropharyngeal cavity. This mechanism is most likely to occur in ruminants, although some rare cases suggest that it could also occur in humans (Figure 6) [276,278]. Consequently, L. monocytogenes induces rhombencephalitis almost exclusively in affected animals [277]. An alternative port of entry and route of infection for neonatal neurolisteriosis occurs through invasion of the olfactory epithelium during birth, when the newborn comes into contact with the vagina of the mother (Figure 6, late-onset) [279].
Blood-borne L. monocytogenes can cross the BBB or the blood-cerebrospinal fluid barrier (BCSFB). Bacteria carried in the blood can be extracellular, where bacteria, both free and/or associated on the surface of circulating cells, may recognize receptors at the surface of the barriers and cross them. Also, bacteria crossing the BBB may be associated with infected leukocytes, utilized as Trojan horses to get access to CNS. This hematogenous route is probably the most frequent in humans, leading to either meningitis or meningoencephalitis (Figure 6) [276,280–282].
Although the role of listerial virulence factors during CNS access is only partially understood, some molecular mechanisms have been elucidated. Three virulence factors from LIPI-1 play a role during brain infection: PlcB, LLO, and ActA [41,236,279,283–286].
In an in vitro model of the blood-cerebrospinal fluid barrier based on human choroid plexus papilloma cells (HIBCPP), L. monocytogenes invades these cells (expressing both E-cadherin and Met) in an InlA- and InlB-dependent manner. The deletion of either InlA or InlB leads to a similar decrease in invasion, suggesting an interdependent function of InlA and InlB during the invasion of choroid plexus epithelial cells [282]. Other internalins such as InlF, or InlL, are suggested to be required for optimal colonization of the brain [280,287,288].
L. monocytogenes isolates belonging to clones CC1, CC4 and CC6 (lineage I) are hypervirulent and, most notably, neurotropic in contrast to lineage II reference strains EGDe (CC9) and 10403S (CC7) [35]. A prominent feature among the 19 CC4-specific genes is LIPI-4, a cluster of six genes annotated as a cellobiose-family phosphotransferase system, whose specific mechanism of action is currently unknown. Remarkably, LIPI-4 inactivation reduces brain invasion without affecting bacterial colonization of other tissues [35].
Placental-neonatal listeriosis
In pregnant women, L. monocytogenes crosses the placental barrier and infects the fetus (Figures 4b and 6) [289]. This is achieved through two main pathways: (i) a cell-to-cell spread from maternal infected phagocytes; and (ii) infection of trophoblasts by free-circulating bacteria in the blood. The preference for one of these pathways probably depends on the infectious dose and the bacterial burden in liver and spleen [289]. In vivo infection studies in gerbils, an animal model close to humans in terms of the affinity of E-cadherin and c-Met receptors (Figure 4b), reveal that while InlA and InlB contribute to some extent to placental invasion, mutants lacking these two internalins can still infect the fetus [203]. This result supports the idea of residual placental invasion proceeding through maternal phagocytes, and demonstrates the co-occurrence of both pathways.
In humans, the placental barrier is composed of epithelial cells of fetal origin called cytotrophoblasts, which fuse to originate the syncytiotrophoblasts. Syncytiotrophoblasts are the cells that are in direct contact with maternal blood. Other cellular components, the extravillous cytotrophoblasts, anchor the villous tree in the decidua. L. monocytogenes can invade both cell types [7]. The mechanism of syncytiotrophoblast invasion is different from that of enterocytes [290]. Syncytiotrophoblasts display E-cadherin and c-Met on the surface but, unlike enterocytes, they do not show the intrinsic PI3-K activity required for L. monocytogenes internalization [203]. In syncytiotrophoblasts, InlB binding to c-Met promotes PI3-K activation and, hence, the cytoskeletal rearrangements needed for bacterial internalization. Therefore, the constitutive activity of PI3-K in enterocytes makes InlB dispensable for crossing the intestinal barrier, and it explains why both InlA and InlB are necessary to cross the placental barrier. An internalin-family secreted protein, InlP, is critical for placental invasion [291]. InlP binds to human afadin, a cytosolic protein associated with cell-to-cell junctions. InlP binding to afadin leads to modulation of the basal face of epithelial cells, promoting bacterial transcytosis in epithelial cell monolayers [292].
High rates of materno-fetal listeriosis in humans are associated with particular hypervirulent clonal complexes (CC), identifying InlA and LIPI-4 as relevant virulence factors. Expression of full-length InlA is more significantly associated with maternal-neonatal isolates than with bacteremia ones [203]. LIPI-4 deletion produced a decrease in placental and neuroinvasive listeriosis in mice without affecting other tissue colonization [35].
Future research will shed light into pivotal steps during the infection process, such as understanding the basis of the long latency period in human infection, the molecular and cellular mechanisms that allow intestinal niche colonization, as well as the crossing of the blood–brain barrier and the placental barrier. Additionally, the identification of protective intestinal microbiota species and their potential use as oral probiotics could be paramount for prevention of listeriosis in immunocompromised individuals or pregnant women.
Human and animal listeriosis
Epidemiology and predisposing host factors
Listeriosis is a foodborne sporadic disease with outbreaks occurring after the consumption of contaminated food. The first recognized foodborne outbreak was reported in Canada from 1980 to 1981, and was triggered by the ingestion of contaminated coleslaw [76]. L. monocytogenes is the foodborne pathogen associated with the highest case-fatality rate in the western hemisphere (e.g., 15.6% case-fatality rate in 2018 in the EU), where its incidence is estimated at around two to five cases per 1 million people per year [2,293–295]. A statistically significant increasing trend of confirmed listeriosis cases in the EU/ European Economic Area was observed in 2009–2018, as well as during the last analyzed 5 year- period (2014–2018) [2,296]. Risk factors for bacteremia and neurolisteriosis include the extremes of age (neonates and the elderly), alcoholism, innate and cellular immune defects, HIV infection, malignancy, corticosteroid therapy, immunosuppression, diabetes mellitus, and liver or kidney disease [297]. L. monocytogenes infections are commonly reported in patients over 64 years, and particularly in the age group over 84 years [2].
Regarding pregnancy, the incidence of listeriosis is approximately 13 times higher than in the general population [298,299]. Noteworthy, in France the incidence of pregnancy-related listeriosis decreased by a factor of 12 between 1984 and 2011. This decline was explained by the progressive implementation of specific L. monocytogenes control measures in food production, together with information campaigns on preventive measures to avoid listeriosis [300]. Although maternal-fetal listeriosis may occur during all stages of gestation, it is most frequently diagnosed during the third trimester [301].
A large prospective cohort study called the MONALISA study (4 year period, 818 patients: 107 maternal–neonatal infections, 427 patients with bacteremia, and 252 with neurolisteriosis) was recently performed in France. MONALISA showed that listeriosis is a severe disease with a very poor prognosis: more than 80% of infected mothers experienced major fetal or neonatal complications (fetal loss, very high prematurity, early or late-onset disease); and only 39% of patients with neurolisteriosis survived and fully recovered [302]. This study also showed that for bacteremia and neurolisteriosis, the strongest mortality predictors were aggravation of any preexisting organ dysfunction, monocytopenia, ongoing cancer, and multi-organ failure [302]. Intriguingly, the MONALISA study showed that 10 of 252 patients (4%) with neurolisteriosis did not report specific risk factors: they were younger than 40 years, informed no comorbidity or ongoing pregnancy, and did not state any infection before listeriosis [302]. This suggests that genetic susceptibility, a massive inoculum, or infection with a hypervirulent strain could underlie these rare cases of neurolisteriosis. Indeed, hypervirulent clones CC1, CC2, CC4, and CC6 are more prevalent among patients with few or no immunosuppressive comorbidities than hypovirulent clones CC9 and CC121, which are more often isolated in highly immunocompromised patients [35].
Multiple factors may explain the global increase in L. monocytogenes infections reported during the last decades: (i) the increasing susceptible population: aged and immunosuppressed patients (HIV, cancer or transplant patients); (ii) industrialization of food production and the subsequent risk of large distribution of contaminated food; (iii) the generalization of food preservation methods, such us refrigeration, which allows L. monocytogenes selective growth; (iv) increased consumption of preservative-free RTE foods; (v) use of antacids and gastric-acid-suppressive medications; and (vi) improved diagnostic methods and enhanced public health surveillance [44,93,115].
The average incidence of culture-confirmed fecal carriage in healthy adults is two episodes of L. monocytogenes carriage per person per year [82]. Importantly, the duration of fecal carriage of L. monocytogenes is short, with a maximum of 4 days of shedding [82]. The majority of listeriosis patients develop L. monocytogenes specific antibodies in serum, whilst healthy donors do not [303]. In a screening of healthy volunteers, 12 of 74 blood donors had antibodies that recognized L. monocytogenes [304]. Considering that only a subset of the human population is exposed to a high enough dose to induce circulating antibodies, these studies are consistent with the notion that humans frequently ingest small quantities of L. monocytogenes [262]. These results, together with the data on L. monocytogenes on RTE foods reported by EFSA, reflect the extent of L. monocytogenes contamination in today’s food [2]. Altogether, these data regarding the frequency of subclinical infection due to ingestion of contaminated food, highlight the importance of surveillance to better prevent and manage listeriosis worldwide.
Hospitalized human patients are also at risk of acquiring invasive nosocomial listeriosis; this suggests that non-pregnancy-associated listeriosis could be reduced by making safer food a standard of hospital care for immunocompromised patients [305]. As aging populations will continue to increase in most developed countries in the coming years, it will be important to raise awareness of listeriosis and the risk of certain types of food and consumption patterns/habits.
In animals, L. monocytogenes has been isolated from >40 species of domestic and wild mammals and 22 species of birds, as well as fish, crustaceans, and insects [1]. Most clinical listeriosis cases occur in ruminants, while pigs rarely develop the disease and birds are generally subclinical carriers [1]. Listeriosis in animals is generally transmitted by consumption of spoiled silage. The less acidic pH of spoiled silage favors the multiplication of this bacterium [306,307]. Listeriosis outbreaks in animals normally occur ≥10 days after consuming poor-quality silage [308]. Analyses of fecal samples from 343 herds including sheep, beef cattle, dairy cattle, and swine, revealed that L. monocytogenes was present in 46.3% of dairy cattle, 30.6% of beef cattle, and 14.2% of sheep herds, but not in swine [309]. L. monocytogenes has also been isolated from the feces of wild animals like wild boar and deer [310]. Fecal carriage correlates with shedding in the environment and contamination of raw food materials that are then conveyed to the food processing industries (Figure 1) [311] .
Importantly, most isolates from clinical, food-borne, and environmental sources are still susceptible to a wide range of antibiotics, except cephalosporins and fosfomycin. See seminal papers related to antimicrobial resistance in Listeria spp [312,313].
Clinical features of listeriosis
Mammalian hosts have multiple strategies to limit L. monocytogenes growth and may even reduce bacterial dissemination to secondary infection sites. However, the ingestion of large numbers of this pathogen in hosts with weak or delayed immunity can result in invasive disease [262]. L. monocytogenes causes in both animals and humans a wide variety of clinical syndromes, which are described below (Figure 6).
Human listeriosis: Febrile gastroenteritis
Food-borne transmission of L. monocytogenes causes a self-limited acute febrile gastroenteritis among healthy people. Large outbreaks of L. monocytogenes-associated febrile gastroenteritis syndrome implicated heavily contaminated foods (>109 CFU/ml), an average incubation period of approximately 24 h (variation from 6 to 240 hours), and attack rates up to 72%. L. monocytogenes gastroenteritis is normally self-limited without severe complications in healthy individuals. Primary symptoms include fever, diarrhea, arthromyalgia, and headache (Figure 6) [6,68,70,83,314–317].
Human invasive listeriosis: Septicemia, neurolisteriosis and maternal-fetal infections
Clinical features of invasive listeriosis derive from the ability of L. monocytogenes to cross the intestinal barrier, the blood–brain barrier, and the maternal-fetal barrier (Figure 6). In the MONALISA study, septicemia represented nearly 50% of cases, neurolisteriosis 30%, and maternal-fetal infections 10% of L. monocytogenes infections [302]. The overall median incubation period of invasive listeriosis is 8 days, and differs depending on the clinical form of the disease: pregnancy-associated cases (median: 27.5 days; range: 17–67 days), neurolisteriosis (median: 9 days; range: 1–14 days) and septicemia (median: 2 days; range: 1–12 days) [317].
Septicemia symptoms resemble those of other types of bacterial sepsis and include fever, nausea, vomiting, chills and malaise. Neurolisteriosis manifests in humans as meningitis and meningoencephalitis, which are the most frequent clinical presentations, followed by brain stem infection (rhombencephalitis) and brain abscesses. Clinical features of meningitis include fever, nuchal rigidity, movement disorders such as tremor and/or ataxia, and seizures. The most common non-meningitic form of neurolisteriosis is rhombencephalitis, characterized by fever, headache, nausea, and vomiting. Subsequently, patients develop focal neurological findings in the hindbrain, including ataxia and multiple cranial nerve abnormalities. Fever is not detected in 15% of rhombencephalitis cases. L. monocytogenes may also be present within brain abscesses in about 10% of neurolisteriosis cases [6,301,318,319]. The prospective MONALISA study showed that the prognosis of listeriosis is grim: a 3-month mortality rate of 46% in septicemia cases and 30% in neurolisteriosis cases, and only 39% of patients with neurolisteriosis survived and fully recovered [302].
Symptoms of listeriosis in pregnant women are nonspecific, but it commonly has flu-like or pyelonephritis symptoms. The main risk associated with L. monocytogenes during pregnancy is the infection of the fetus during maternal septicemia and chorioamnitis, or upon delivery, due to perivaginal and perianal listerial colonization in the mother. Transplacental infection of the infant (“early-onset”) can lead to fetal loss, or the preterm birth of a severely ill infant with a very high mortality rate. This infant syndrome is known as “granulomatosis infantiseptica”. If the infant is infected during transition through an L. monocytogenes-colonized birth canal (“late-onset” listeriosis), neonatal meningitis develops 7 to 14 days post-partum (Figure 6) [6,300,301]. Maternal-fetal listeriosis is virtually never associated with maternal neurolisteriosis, and never leads to maternal mortality but, as noted above, more than 80% of infected mothers experience major fetal or neonatal complications [302].
Localized infections
These rare infections represent 10% of listeriosis cases [302]. Cutaneous listeriosis is a rare occupational hazard associated with exposure to animal products of conception removed from the birth canal. Typically, it manifests as nonpainful, nonpruritic, self-limited, localized, papulopustular, or vesiculopustular eruptions from which the organism can be isolated. Cutaneous listeriosis cases are mild and resolve successfully [320,321]. Other rare forms of listeriosis are localized infections involving endocarditis, hepatitis and liver abscesses, peritonitis, biliary-tract, eye and musculoskeletal infections [6,321–326] (Figure 6).
Clinical features in animals
Although most L. monocytogenes infections in animals are subclinical, the invasive disease can occur either sporadically or as an outbreak. Similarly to humans, the clinical features of listeriosis in animals include septicemia, rhombencephalitis, and abortion, especially in domestic ruminants (sheep, goats, and cattle) [1,9]. It is unusual for different clinical manifestations to be diagnosed during the same outbreak. The infrequent septicemic form generally occurs in the neonate. Rhombencephalitis in adult ruminants is the most frequently recognized form. Listeria encephalitis in ruminants is mainly a localized infection of the brain stem that develops when L. monocytogenes ascends the trigeminal nerve, leading to unilateral cranial nerve paralysis, circling disease syndrome, depression, and anorexia. Diffuse meningitis or meningoencephalitis has only exceptionally been reported in these species [277,327]. In ruminants, abortion is usually late term [1,276,306], and although less frequently reported, mastitis has been associated with L. monocytogenes infection [328]. Peri-parturient mice infected intragastrically with L. monocytogenes shed the pathogen in vaginal secretions, facilitating its dissemination to the mammary chain, from which it is shed in the milk of peri-parturient mice [329]. The incubation period for septicemic/abortive listeriosis can be as short as 1 day, but in rhombencephalitis it is usually 2–3 weeks, mainly because L. monocytogenes invades the brainstem via cranial nerves after breaching of the oral mucosa [1]. Gastrointestinal infections can occasionally occur in sheep [330]. Iritis, uveitis, and keratoconjunctivitis have been reported in cattle associated with contact with contaminated silage (Figure 6) [331]. In pigs, the primary manifestation of listeriosis is septicemia, and less frequently, encephalitis and abortions [1,307]. L. monocytogenes has been reported to cause invasive disease in >40 animal species. In contrast, L. ivanovii almost exclusively affects ruminants, causing abortion and neonatal septicemia but not central nervous system infections [8,277]. Due to the L. monocytogenes broad host range, several animal models have been used in pathogenesis research. These animal models include conventional, transgenic, and knock-in mice, gerbils, guinea pigs, rabbits, the zebrafish Danio rerio, chicken embryos, the wax moth Galleria mellonella and the nematode Caenorhabditis elegans [41,203–205,254].
Concluding remarks
Listeria has evolved a plethora of molecular strategies that allow it to tolerate stress very efficiently. This outstanding capacity is crucial to survive and exploit environmental niches under conditions that other microorganisms cannot endure. Part of this success relies on the cross-protection among different types of stress provided by the SigB-induced general stress response, whereby some factors induced to cope with a particular stress turn out to be useful to alleviate a different type of stress. For instance, transporters of compatible solutes expressed at refrigeration temperatures help to adapt Listeria to high salt concentrations [143,146]. Similarly, stress found in food can favor virulence within the host [332]. However, SigB is also present in other pathogenic and nonpathogenic gram-positive bacteria with a significant functional overlap among their SigB-regulated genes. Therefore, in Listeria, SigB-independent mechanisms play a unique role in the versatility of stress adaptation and invasion of host tissues. Interestingly, most regulatory mechanisms that govern SigB-independent responses remain elusive. For example, very few two-component systems, transcriptional regulators, or sRNAs have thus far been linked to stress tolerance. The latter seems intriguing because Listeria can express more than 300 sRNAs and, at least in other microorganisms, sRNAs play a central role in stress responses [333,334]. In addition, besides the blue light sensor RsbL [158], molecular sensors relaying stress signals to activate the stressosome or any other downstream effector are unknown. In the next few years, the identification of stress sensors and stress response regulators could point to potential vulnerabilities, allowing Listeria stress tolerance to be targeted in food and in the GI tract to prevent infections.
An interplay between L. monocytogenes and commensal microbiota has recently been unveiled [5]. Listeria deploys specific strategies such as metabolic adaptations to scavenge ethanolamine and the production of bacteriocins [126]. This suggests that its interaction with bacterial communities in the gut (and in the soil) may be more complex than anticipated. In addition, host factors influence several stages in the infective cycle, including intracellular bacterial growth and dissemination throughout the body. Yet remaining to be determined are the precise advantages obtained by a subset of intracellular bacteria that stay confined within the vacuole [237] in a persistent state, and their resulting impact on disease progression. Would this confinement help to carry bacteria in asymptomatic hosts, or be an adaptation that reduces antibiotic exposure during treatment in clinical listeriosis cases? Future research will shed light on the specific contribution of the interaction both of host factors and of bacterial communities on the evolution of Listeria virulence.
Finally, although most of what we currently know about Listeria molecular and cellular pathogenesis is based on research performed on a handful of strains used as model organisms, mounting evidence highlights the intra-species heterogeneity of L. monocytogenes. We now know that lineage II strains are more prone to be associated with food, while lineage I strains are most commonly found in clinical isolates related to listeriosis outbreaks [34,36,37]. Identification of the relevant virulence factors specific to these epidemic strains [32,35] will help to understand the molecular basis of hypervirulence.
Acknowledgments
We apologize to authors whose relevant work could not be cited owing to space limitations. MGP and ADO research was supported by the Spanish Ministry of Science and Innovation (PGC2018-096364-B-I00). JJQ was supported by the Generalitat Valenciana (GV/2018/A/183) and the Spanish Ministry of Science and Innovation (PID2019-110764RA-I00/AEI/10.13039/501100011033). FG-dP was supported by the European Union’s Horizon 2020 Research-and-Innovation Program under Marie Sklodowska-Curie grant agreement no. 721456. JJQ was a recipient of the “Ramón y Cajal” Programme from the Spanish Ministry of Science, Innovation, and Universities (RYC-2018-024985-I). AM-G was funded by the Youth Employment Initiative from the Comunidad de Madrid. CP-G was funded by the Universidad Cardenal Herrera-CEU. CD was an Early Stage Researcher from the Horizon 2020 Research-and-Innovation Program under Marie Sklodowska-Curie grant agreement no. 721456 as.
Funding Statement
This work has been supported by Ministerio de Ciencia Innovación y Universidades [PGC2018-096364-B-I00; PID2019-110764RA-I00/AEI/10.13039/501100011033]; Conselleria d’Educació, Investigació, Cultura i Esport [GV/2018/A/183]; H2020 Marie Skłodowska-Curie Actions [721456].
References
- [1].World Organisation for Animal Health . Access online: OIE - World Organisation for Animal Health [Internet]. 2018. cited 2020 Jul 31. Available from: https://www.oie.int/en/standard-setting/terrestrial-manual/access-online/.
- [2].European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC) . The European Union One Health 2018 Zoonoses Report. EFSA J. 2019;17(12):e05926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Quereda JJ, Leclercq A, Moura A, et al. Listeria valentina sp. nov., isolated from a water trough and the faeces of healthy sheep. Int J Syst Evol Microbiol. 2020;70:5868–5879. [DOI] [PubMed] [Google Scholar]
- [4].NicAogáin K, O’Byrne CP.. The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front Microbiol. 2016;7:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Radoshevich L, Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol. 2018;16:32–46. [DOI] [PubMed] [Google Scholar]
- [6].Schlech WF. Epidemiology and clinical manifestations of Listeria monocytogenes infection. In: Microbiol Spectr. 2019;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Charlier C, Disson O, Lecuit M. Maternal-neonatal listeriosis. Virulence. 2020;11:391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Orsi RH, Wiedmann M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl Microbiol Biotechnol. 2016;100:5273–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vázquez-Boland JA, Kuhn M, Berche P, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Allerberger F, Wagner M. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect. 2010;16:16–23. [DOI] [PubMed] [Google Scholar]
- [11].Guillet C, Join-Lambert O, Le Monnier A, et al. Human listeriosis caused by Listeria ivanovii. Emerg Infect Dis. 2010;16:136–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lessing MPA, Curtis GDW, Bowler ICJ. Listeria ivanovii infection. J Infect. 1994;29:230–231. [DOI] [PubMed] [Google Scholar]
- [13].Cummins AJ, Fielding AK, McLauchlin J. Listeria ivanovii infection in a patient with AIDS. J Infect. 1994;28:89–91. [DOI] [PubMed] [Google Scholar]
- [14].Rocourt J, Seeliger HPR. [Distribution of species of the genus Listeria]. Zentralbl Bakteriol Mikrobiol Hyg A. 1985;259:317–330. [PubMed] [Google Scholar]
- [15].Johnson J, Jinneman K, Stelma G, et al. Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Appl Environ Microbiol. 2004;70:4256–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moreno LZ, Paixão R, Gobbi DD, et al. Characterization of atypical Listeria innocua isolated from swine slaughterhouses and meat markets. Res Microbiol. 2012;163:268–271. [DOI] [PubMed] [Google Scholar]
- [17].Milillo SR, Stout JC, Hanning IB, et al. Listeria monocytogenes and hemolytic Listeria innocua in poultry. Poult Sci. 2012;91:2158–2163. [DOI] [PubMed] [Google Scholar]
- [18].Perrin M, Bemer M, Delamare C. Fatal case of Listeria innocua bacteremia. J Clin Microbiol. 2003;41:5308–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rana F, Shaikh MM, Bowles J. Listeria meningitis and resultant symptomatic hydrocephalus complicating infliximab treatment for ulcerative colitis. JRSM Open. 2014;5:205427041452222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rocha PRD, Dalmasso A, Grattarola C, et al. Atypical cerebral listeriosis associated with Listeria innocua in a beef bull. Res Vet Sci. 2013;94:111–114. [DOI] [PubMed] [Google Scholar]
- [21].Walker JK, Morgan JH, McLauchlin J, et al. Listeria innocua isolated from a case of ovine meningoencephalitis. Vet Microbiol. 1994;42:245–253. [DOI] [PubMed] [Google Scholar]
- [22].Moura A, Disson O, Lavina M, et al. Atypical hemolytic Listeria innocua isolates are virulent, albeit less than Listeria monocytogenes. Freitag NE, editor. Infect Immun. 2019;87(4):e00758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clayton EM, Daly KM, Guinane CM, et al. Atypical Listeria innocua strains possess an intact LIPI-3. BMC Microbiol. 2014;14:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Quereda JJ, Meza-Torres J, Cossart P, et al. Listeriolysin S: a bacteriocin from epidemic Listeria monocytogenes strains that targets the gut microbiota. In: Gut Microbes. 2017;8(4):384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Quereda JJ, Dussurget O, Nahori MA, et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci U S A. 2016;113:5706–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Müller AA, Schmid MW, Meyer O, et al. Listeria seeligeri isolates from food processing environments form two phylogenetic lineages. Appl Environ Microbiol. 2010;76:3044–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].den Bakker HC, Cummings CA, Ferreira V, et al. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2010;11:688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rocourt J, Hof H, Schrettenbrunner A, et al. [Acute purulent Listeria seelingeri meningitis in an immunocompetent adult]. Schweiz Med Wochenschr. 1986;116:248–251. [PubMed] [Google Scholar]
- [29].Orsi RH, den Bakker HC, Wiedmann M. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol. 2011;301:79–96. [DOI] [PubMed] [Google Scholar]
- [30].Burall LS, Grim CJ, Mammel MK, et al. A comprehensive evaluation of the genetic relatedness of Listeria monocytogenes serotype 4b variant strains. Front Public Health. 2017;5:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Painset A, Björkman JT, Kiil K, et al. LiSEQ - whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. In: Microb genomics. 2019;5(2):e000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Moura A, Criscuolo A, Pouseele H, et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol. 2016;2:16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ragon M, Wirth T, Hollandt F, et al. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008;4:1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jeffers GT, Bruce JL, McDonough PL, et al. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology. 2001;147:1095–1104. [DOI] [PubMed] [Google Scholar]
- [35].Maury MM, Tsai YH, Charlier C, et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet. 2016;48:308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Maury MM, Bracq-Dieye H, Huang L, et al. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat Commun. 2019;10:2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gray MJ, Zadoks RN, Fortes ED, et al. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl Environ Microbiol. 2004;70:5833–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Van WI, Björkman JT, Cormican M, et al. Retrospective validation of whole genome sequencing enhanced surveillance of listeriosis in Europe, 2010 to 2015. Eurosurveillance. 2018;23:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jackson BR, Tarr C, Strain E, et al. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis. 2016;63:380–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bécavin C, Bouchier C, Lechat P, et al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio. 2014;5:e00969–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Quereda JJ, Andersson C, Cossart P, et al. Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet Res. 2018;49:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Disson O, Moura A, Lecuit M. Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol. 2021. DOI: 10.1016/j.tim.2021.01.008 [DOI] [PubMed] [Google Scholar]
- [43].Moura A, Lefrancq N, Leclercq A, et al. Emergence and global spread of Listeria monocytogenes main clinical clonal complex. bioRxiv. 2020. DOI: 10.1101/2020.12.18.423387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lecuit M. Listeria monocytogenes, a model in infection biology. Cell Microbiol. 2020;22:e13186. [DOI] [PubMed] [Google Scholar]
- [45].Dreyer M, Aguilar-Bultet L, Rupp S, et al. Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci Rep. 2016;6:36419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jacquet C, Doumith M, Gordon JI, et al. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J Infect Dis. 2004;189:2094–2100. [DOI] [PubMed] [Google Scholar]
- [47].Cotter PD, Draper LA, Lawton EM, et al. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4:e1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Quereda JJ, Nahori MA, Meza-Torres J, et al. Listeriolysin S is a streptolysin S-like virulence factor that targets exclusively prokaryotic cells in vivo. MBio. 2017;8:e00259–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cossart P, Vicente MF, Mengaud J, et al. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect Immun. 1989;57:3629–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Barry RA, Bouwer HGA, Portnoy DA, et al. Pathogenicity and immunogenicity of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect Immun. 1992;60:1625–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Angelakopoulos H, Loock K, Sisul DM, et al. Safety and shedding of an attenuated strain of Listeria monocytogenes with a deletion of actA/plcB in adult volunteers: a dose escalation study of oral inoculation. Infect Immun. 2002;70:3592–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gouin E, Adib-Conquy M, Balestrino D, et al. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the IκB kinase subunit IKKα. Proc Natl Acad Sci U S A. 2010;107:17333–17338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dussurget O, Cabanes D, Dehoux P, et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;45:1095–1106. [DOI] [PubMed] [Google Scholar]
- [54].Camejo A, Carvalho F, Reis O, et al. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence. 2011;2:379–394. [DOI] [PubMed] [Google Scholar]
- [55].Carvalho F, Sousa S, Cabanes D. How Listeria monocytogenes organizes its surface for virulence. Front Cell Infect Microbiol. 2014;4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nguyen BN, Peterson BN, Portnoy DA, et al. A phagosome-specific cytolysin revisited. Cell Microbiol. 2019;21:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yin Y, Yao H, Doijad S, et al. A hybrid sub-lineage of Listeria monocytogenes comprising hypervirulent isolates. Nat Commun. 2019;10:4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Maury MM, Chenal-Francisque V, Bracq-Dieye H, et al. Spontaneous Loss of Virulence in Natural Populations of Listeria monocytogenes. Infect Immun. 2017;85(11):e00541-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].McCollum JT, Cronquist AB, Silk BJ, et al. Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med. 2013;369:944–953. [DOI] [PubMed] [Google Scholar]
- [60].FAO/WHO . Risk assessment of Listeria monocytogenes in ready-to-eat foods. FAO/WHO Microbiol Risk Assess Ser. 2004. ;4:1–48. [Google Scholar]
- [61].Zoellner C, Wiedmann M, Ivanek R. An assessment of listeriosis risk associated with a contaminated production lot of frozen vegetables consumed under alternative consumer handling scenarios. J Food Prot. 2019;82:2174–2193. [DOI] [PubMed] [Google Scholar]
- [62].Thomas J, Govender N, McCarthy KM, et al. Outbreak of listeriosis in South Africa associated with processed meat. N Engl J Med. 2020;382:632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mazzotta AS, Gombas DE. Heat resistance of an outbreak strain of Listeria monocytogenes in hot dog batter. J Food Prot. 2001;64:321–324. [DOI] [PubMed] [Google Scholar]
- [64].WHO | Listeriosis– Spain [Internet] . cited 2020 Aug 7. Available from: https://www.who.int/csr/don/16-september-2019-listeriosis-spain/en/.
- [65].Gelbíčová T, Zobaníková M, Tomáštíková Z, et al. An outbreak of listeriosis linked to Turkey meat products in the Czech Republic, 2012-2016. Epidemiol Infect. 2018;146:1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].De Valk H, Vaillant V, Jacquet C, et al. Two consecutive nationwide outbreaks of listeriosis in France, October 1999-February 2000. Am J Epidemiol. 2001;154:944–950. [DOI] [PubMed] [Google Scholar]
- [67].Linnan MJ, Mascola L, Lou XD, et al. Epidemic listeriosis associated with Mexican-style cheese. N Engl J Med. 1988;319:823–828. [DOI] [PubMed] [Google Scholar]
- [68].Carrique-Mas JJ, Hökeberg I, Andersson Y, et al. Febrile gastroenteritis after eating on-farm manufactured fresh cheese - An outbreak of listeriosis? Epidemiol Infect. 2003;130:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fretz R, Pichler J, Sagel U, et al. Update: multinational listeriosis outbreak due to “quargel”, a sour milk curd cheese, caused by two different L. monocytogenes serotype 1/2a strains, 2009-2010. Eurosurveillance. 2010;15:2–3. [PubMed] [Google Scholar]
- [70].Dalton CB, Austin CC, Sobel J, et al. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N Engl J Med. 1997;336:100–105. [DOI] [PubMed] [Google Scholar]
- [71].Pouillot R, Klontz KC, Chen Y, et al. Infectious dose of Listeria monocytogenes in outbreak linked to ice cream, United States, 2015. Emerg Infect Dis. 2016;22:2113–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lyytikäinen O, Autio T, Maijala R, et al. An outbreak of Listeria monocytogenes serotype 3a infections from butter in Finland. J Infect Dis. 2000;181:1838–1841. [DOI] [PubMed] [Google Scholar]
- [73].Miettinen MK, Siitonen A, Heiskanen P, et al. Molecular epidemiology of an outbreak of febrile gastroenteritis caused by Listeria monocytogenes in cold-smoked rainbow trout. J Clin Microbiol. 1999;37:2358–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Elson R, Awofisayo-Okuyelu A, Greener T, et al. Utility of whole genome sequencing to describe the persistence and evolution of Listeria monocytogenes strains within crabmeat processing environments linked to two outbreaks of listeriosis. J Food Prot. 2019;82:30–38. [DOI] [PubMed] [Google Scholar]
- [75].Schjørring S, Gillesberg Lassen S, Jensen T, et al. Cross-border outbreak of listeriosis caused by cold-smoked salmon, revealed by integrated surveillance and whole genome sequencing (WGS), Denmark and France, 2015 to 2017. Euro Surveill. 2017;22:17–00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Schlech WF, Lavigne PM, Bortolussi RA, et al. Epidemic Listeriosis — evidence for Transmission by Food. N Engl J Med. 1983;308:203–206. [DOI] [PubMed] [Google Scholar]
- [77].Ukuku DO, Fett W. Behavior of Listeria monocytogenes inoculated on cantaloupe surfaces and efficacy of washing treatments to reduce transfer from rind to fresh-cut pieces. J Food Prot. 2002;65:924–930. [DOI] [PubMed] [Google Scholar]
- [78].Farber JM, Ross WH, Harwig J. Health risk assessment of Listeria monocytogenes in Canada. Int J Food Microbiol. 1996;30:145–156. [DOI] [PubMed] [Google Scholar]
- [79].Pérez-Trallero E, Zigorraga C, Artieda J, et al. Two outbreaks of Listeria monocytogenes infection, Northern Spain. Emerg Infect Dis. 2014;20:2155–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Heiman KE, Garalde VB, Gronostaj M, et al. Multistate outbreak of listeriosis caused by imported cheese and evidence of cross-contamination of other cheeses, USA, 2012. Epidemiol Infect. 2016;144:2698–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Johnsen BO, Lingaas E, Torfoss D, et al. A large outbreak of Listeria monocytogenes infection with short incubation period in a tertiary care hospital. J Infect. 2010;61:465–470. [DOI] [PubMed] [Google Scholar]
- [82].Grif K, Patscheider G, Dierich MP, et al. Incidence of fecal carriage of Listeria monocytogenes in three healthy volunteers: a one-year prospective stool survey. Eur J Clin Microbiol Infect Dis. 2003;22:16–20. [DOI] [PubMed] [Google Scholar]
- [83].Aureli P, Fiorucci GC, Caroli D, et al. An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N Engl J Med. 2000;342:1236–1241. [DOI] [PubMed] [Google Scholar]
- [84].Archer DL. The evolution of FDA’s policy on Listeria monocytogenes in ready-to-eat foods in the United States. Curr Opin Food Sci. 2018;20:64–68. [Google Scholar]
- [85].The Commission of the European Communities . Commission Regulation (EC) No 2073/2005. Off J Eur Union. 2005;48:1-26. [Google Scholar]
- [86].Velge P, Roche SM. Variability of Listeria monocytogenes virulence: a result of the evolution between saprophytism and virulence? Future Microbiol. 2010;5:1799–1821. [DOI] [PubMed] [Google Scholar]
- [87].Castro H, Jaakkonen A, Hakkinen M, et al. Occurrence, persistence, and contamination routes of Listeria monocytogenes genotypes on three Finnish dairy cattle farms: a longitudinal study. Appl Environ Microbiol. 2018;84:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gómez-Laguna J, Cardoso-Toset F, Meza-Torres J, et al. Virulence potential of Listeria monocytogenes strains recovered from pigs in Spain. Vet Rec. 2020;187:e101–e101. vetrec-2020-105945 [DOI] [PubMed] [Google Scholar]
- [89].Haley BJ, Sonnier J, Schukken YH, et al. Diversity of Listeria monocytogenes within a U.S. dairy herd, 2004-2010. Foodborne Pathog Dis. 2015;12:844–850. [DOI] [PubMed] [Google Scholar]
- [90].Tahoun ABMB, Rmm AE, Abdelfatah EN, et al. Listeria monocytogenes in raw milk, milking equipment and dairy workers: molecular characterization and antimicrobial resistance patterns. J Glob Antimicrob Resist. 2017;10:264–270. [DOI] [PubMed] [Google Scholar]
- [91].Kim SW, Haendiges J, Keller EN, et al. Genetic diversity and virulence profiles of Listeria monocytogenes recovered from bulk tank milk, milk filters, and milking equipment from dairies in the United States (2002 to 2014). PLoS One. 2018;13:e0197053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Farber J, Pagotto F, Scherf C. Incidence and behavior of Listeria monocytogenes in meat products. In: List List Food Safety. Third Ed ed. 2007:503–570. [Google Scholar]
- [93].Edwards D, Dunlop DJ. Food Microbiology. In: editors, Mp D, Diez-Gonzalez F, Hill C. Food Microbiol. 5th ed Washington, DC, USA: ASM Press; 2019:1093. [Google Scholar]
- [94].Ferreira V, Wiedmann M, Teixeira P, et al. Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot. 2014;77:150–170. [DOI] [PubMed] [Google Scholar]
- [95].Guerreiro DN, Arcari T, O’Byrne CP. The σB-Mediated general stress response of Listeria monocytogenes: life and death decision making in a pathogen. Front Microbiol. 2020;11:1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Burgess CM, Gianotti A, Gruzdev N, et al. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int J Food Microbiol, Elsevier B.V. 2016;37–53. [DOI] [PubMed] [Google Scholar]
- [97].Sleator RD, Gahan CGM, Abee T, et al. Identification and disruption of BetL, a secondary glycine betaine transport system linked to the salt tolerance of Listeria monocytogenes LO28. Appl Environ Microbiol. 1999;65:2078–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Demarquoy J, Georges B, Rigault C, et al. Radioisotopic determination of l-carnitine content in foods commonly eaten in Western countries. Food Chem. 2004;86:137–142. [Google Scholar]
- [99].Tasara T, Stephan R. Cold stress tolerance of Listeria monocytogenes: a review of molecular adaptive mechanisms and food safety implications. J Food Prot. 2006;69:1473–1484. [DOI] [PubMed] [Google Scholar]
- [100].Hingston P, Chen J, Allen K, et al. Strand specific RNA-sequencing and membrane lipid profiling reveals growth phase-dependent cold stress response mechanisms in Listeria monocytogenes. PLoS One. 2017;12:e0180123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chan YC, Raengpradub S, Boor KJ, et al. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase Cells. Appl Environ Microbiol. 2007;73:6484–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Markkula A, Mattila M, Lindström M, et al. Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3°C. Environ Microbiol. 2012;14:2223–2232. [DOI] [PubMed] [Google Scholar]
- [103].Zhang Y, Burkhardt DH, Rouskin S, et al. A stress response that monitors and regulates mRNA structure is central to cold shock adaptation. Mol Cell. 2018;70:274–286.e7. DOI: 10.1016/j.molcel.2018.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Martínez-Suárez JV, Ortiz S, López-Alonso V. Potential impact of the resistance to quaternary ammonium disinfectants on the persistence of Listeria monocytogenes in food processing environments. Front Microbiol. 2016;7:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kovacevic J, Ziegler J, Walecka-Zacharska E, et al. Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl Environ Microbiol. 2016;82:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Dutta V, Elhanaf D, Kathariou S. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl Environ Microbiol. 2013;79:6067–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kropac AC, Eshwar AK, Stephan R, et al. New insights on the role of the pLMST6 Plasmid in Listeria monocytogenes biocide tolerance and virulence. Front Microbiol. 2019;10:1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Rodríguez-López P, Rodríguez-Herrera J, Vázquez-Sánchez D, et al. Current knowledge on Listeria monocytogenes biofilms in food-related environments: incidence, resistance to biocides, ecology and biocontrol. Foods. 2018;7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Yu T, Jiang X, Zhang Y, et al. Effect of benzalkonium chloride adaptation on sensitivity to antimicrobial agents and tolerance to environmental etresses in Listeria monocytogenes. Front Microbiol. 2018;9:2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Valderrama WB, Cutter CN. An ecological perspective of Listeria monocytogenes biofilms in food processing facilities. Crit Rev Food Sci Nutr. 2013;53:801–817. [DOI] [PubMed] [Google Scholar]
- [111].Kannan S, Balakrishnan J, Govindasamy A. Listeria monocytogenes - Amended understanding of its pathogenesis with a complete picture of its membrane vesicles, quorum sensing, biofilm and invasion. In: Microb. Pathog. Academic Press; 2020;149:104575. [DOI] [PubMed] [Google Scholar]
- [112].Parsons C, Lee S, Kathariou S. Heavy metal resistance determinants of the foodborne pathogen Listeria monocytogenes. In: Genes (Basel). 2019;10(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ratani SS, Siletzky RM, Dutta V, et al. Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl Environ Microbiol. 2012;78:6938–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lee S, Rakic-Martinez M, Graves LM, et al. Genetic determinants for cadmium and arsenic resistance among Listeria monocytogenes serotype 4B isolates from sporadic human listeriosis patients. Appl Environ Microbiol. 2013;79:2471–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Cobb CA, Curtis GDW, Bansi DS, et al. Increased prevalence of Listeria monocytogenes in the faeces of patients receiving long-term H2-antagonists. Eur J Gastroenterol Hepatol. 1996;8:1071–1074. [DOI] [PubMed] [Google Scholar]
- [116].Kvistholm Jensen A, Simonsen J, Ethelberg S. Use of proton pump inhibitors and the risk of listeriosis. A nationwide registry-based case-control study. Clin Infect Dis. 2016;64:ciw860. [DOI] [PubMed] [Google Scholar]
- [117].Gahan CGM, Hill C. Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front Cell Infect Microbiol. 2014;5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Cotter PD, Gahan CGM, Hill C. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol. 2001;40:465–475. [DOI] [PubMed] [Google Scholar]
- [119].Feehily C, Finnerty A, Casey PG, et al. Divergent evolution of the activity and regulation of the glutamate decarboxylase systems in Listeria monocytogenes EGD-e and 10403S: roles in virulence and acid tolerance. PLoS One. 2014;9:e112649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Wemekamp-Kamphuis HH, Wouters JA, De Leeuw PPLA, et al. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl Environ Microbiol. 2004;70:3457–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Karatzas K-AG, Suur L, O’Byrne CP. Characterization of the intracellular glutamate decarboxylase system: analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl Environ Microbiol. 2012;78:3571–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Cunin R, Glansdorff N, Pierard A, et al. Microbiol. Rev. In: Biosynthesis and metabolism of arginine in bacteria. American Society for Microbiology (ASM);, 1986;50:314–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Ryan S, Begley M, Gahan CGM, et al. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ Microbiol. 2009;11:432–445. [DOI] [PubMed] [Google Scholar]
- [124].Chen J, Cheng C, Xia Y, et al. Lmo0036, an ornithine and putrescine carbamoyltransferase in Listeria monocytogenes, participates in arginine deiminase and agmatine deiminase pathways and mediates acid tolerance. Microbiology. 2011;157:3150–3161. [DOI] [PubMed] [Google Scholar]
- [125].Cheng C, Dong Z, Han X, et al. Listeria monocytogenes 10403s arginine repressor ArgR finely tunes arginine metabolism regulation under acidic conditions. Front Microbiol. 2017;8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Rolhion N, Chassaing B. When pathogenic bacteria meet the intestinal microbiota. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Archambaud C, Nahori MA, Soubigou G, et al. Impact of lactobacilli on orally acquired listeriosis. Proc Natl Acad Sci U S A. 2012;109:16684–16689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Becattini S, Littmann ER, Carter RA, et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J Exp Med. 2017;214:1973–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Bertin Y, Girardeau JP, Chaucheyras-Durand F, et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011;13:365–377. [DOI] [PubMed] [Google Scholar]
- [130].Thiennimitr P, Winter SE, Winter MG, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011;108:17480–17485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Rolhion N, Chassaing B, Nahori MA, et al. A Listeria monocytogenes bacteriocin can target the commensal Prevotella copri and modulate intestinal Infection. Cell Host Microbe. 2019;26:691–701.e5. DOI: 10.1016/j.chom.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Gahan CGM, Hill C. Gastrointestinal phase of Listeria monocytogenes infection. J Appl Microbiol. 2005;98:1345–1353. [DOI] [PubMed] [Google Scholar]
- [133].Okada Y, Makino S, Okada N, et al. Identification and analysis of the osmotolerance associated genes in Listeria monocytogenes. Food Addit Contam - Part A Chem Anal Control Expo Risk Assess. 2008;25:1089–1094. [DOI] [PubMed] [Google Scholar]
- [134].Cremers CM, Knoefler D, Vitvitsky V, et al. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo. Proc Natl Acad Sci U S A. 2014;111:E1610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Dowd GC, Joyce SA, Hill C, et al. Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect Immun. 2011;79:369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–651. [DOI] [PubMed] [Google Scholar]
- [137].Sleator RD, Wemekamp-Kamphuis HH, Gahan CGM, et al. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol Microbiol. 2005;55:1183–1195. [DOI] [PubMed] [Google Scholar]
- [138].Ridlon JM, Harris SC, Bhowmik S, et al. Gut Microbes. In: Consequences of bile salt biotransformations by intestinal bacteria. Taylor and Francis Inc, 2016;7:22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Sue D, Boor KJ, Wiedmann M. σB-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology. 2003;149:3247–3256. [DOI] [PubMed] [Google Scholar]
- [140].Guariglia-Oropeza V, Orsi RH, Guldimann C, et al. The Listeria monocytogenes bile stimulon under acidic conditions is characterized by strain-specific patterns and the upregulation of motility, cell wall modification functions, and the PrfA Regulon. Front Microbiol. 2018;9:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Horlbog JA, Stevens MJA, Stephan R, et al. Global transcriptional response of three highly acid-tolerant field strains of Listeria monocytogenes to HCl stress. Microorganisms. 2019;7:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Cortes BW, Naditz AL, Anast JM, et al. Transcriptome sequencing of Listeria monocytogenes reveals major gene expression changes in response to lactic acid stress exposure but a less pronounced response to oxidative stress. Front Microbiol. 2020;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Bergholz TM, Bowen B, Wiedmann M, et al. Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl Environ Microbiol. 2012;78:2602–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Casey A, Fox EM, Schmitz-Esser S, et al. Transcriptome analysis of Listeria monocytogenes exposed to biocide stress reveals a multi-system response involving cell wall synthesis, sugar uptake, and motility. Front Microbiol. 2014;5:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Gottesman S. Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J Biol Chem. 2019;294:11685–11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Pittman JR, Buntyn JO, Posadas G, et al. Proteomic analysis of cross protection provided between cold and osmotic stress in Listeria monocytogenes. J Proteome Res. 2014;13:1896–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Schmid B, Klumpp J, Raimann E, et al. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl Environ Microbiol. 2009;75:1621–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Begley M, Gahan CGM, Hill C. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl Environ Microbiol. 2002;68:6005–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Liu Y, Orsi RH, Gaballa A, et al. Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism. Future Microbiol. 2019;14:801–828. [DOI] [PubMed] [Google Scholar]
- [150].Marles-Wright J, Grant T, Delumeau O, et al. Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science. 2008;322(80–):92–96. [DOI] [PubMed] [Google Scholar]
- [151].Williams AH, Redzej A, Rolhion N, et al. The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation. Nat Commun. 2019;10:3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Dessaux C, Guerreiro DN, Pucciarelli MG, et al. Impact of osmotic stress on the phosphorylation and subcellular location of Listeria monocytogenes stressosome proteins. Sci Rep. 2020;10:20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].He K, Xin Y-P, Shan Y, et al. Phosphorylation residue T175 in RsbR protein is required for efficient induction of sigma B factor and survival of Listeria monocytogenes under acidic stress. J Zhejiang Univ Sci B. 2019;20:660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Josewin SW, Kim MJ, Yuk HG. Inactivation of Listeria monocytogenes and Salmonella spp. on cantaloupe rinds by blue light emitting diodes (LEDs). Food Microbiol. 2018;76:219–225. [DOI] [PubMed] [Google Scholar]
- [155].Kim MJ, Tang CH, Bang WS, et al. Antibacterial effect of 405 ± 5 nm light emitting diode illumination against Escherichia coli O157: H7,Listeria monocytogenes, and Salmonella on the surface of fresh-cut mango and its influence on fruit quality. Int J Food Microbiol. 2017;244:82–89. [DOI] [PubMed] [Google Scholar]
- [156].Ondrusch N, Kreft J. Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS One. 2011;6:e16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Tiensuu T, Andersson C, Rydén P, et al. Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol Microbiol. 2013;87:909–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].O’Donoghue B, NicAogáin K, Bennett C, et al. Blue-light inhibition of Listeria monocytogenes growth is mediated by reactive oxygen species and is influenced by σB and the blue-light sensor Lmo0799. Appl Environ Microbiol. 2016;82:4017–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Dorey AL, Lee B-H, Rotter B, et al. Blue light sensing in Listeria monocytogenes is temperature-dependent and the transcriptional response to it is predominantly SigB-dependent. Front Microbiol. 2019;10:2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Pöntinen A, Markkula A, Lindström M, et al. Two-component-system histidine kinases involved in growth of Listeria monocytogenes EGD-e at low temperatures. Appl Environ Microbiol. 2015;81:3994–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Pöntinen A, Lindström M, Skurnik M, et al. Screening of the two-component-system histidine kinases of Listeria monocytogenes EGD-e. LiaS is needed for growth under heat, acid, alkali, osmotic, ethanol and oxidative stresses. Food Microbiol. 2017;65:36–43. [DOI] [PubMed] [Google Scholar]
- [162].Cotter PD, Emerson N, Gahan CGM, et al. Identification and disruption of lisRK, a genetic locus encoding a two- component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J Bacteriol. 1999;181:6840–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Sleator RD, Hill C. A novel role for the LisRK two-component regulatory system in listerial osmotolerance. Clin Microbiol Infect. 2005;11:599–601. [DOI] [PubMed] [Google Scholar]
- [164].Chan YC, Hu Y, Chaturongakul S, et al. Contributions of two-component regulatory systems, alternative sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth. J Food Prot. 2008;71:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Cotter PD, Guinane CM, Hill C. The LisRK Signal Transduction System Determines the Sensitivity of Listeria monocytogenes to Nisin and Cephalosporins. Antimicrob Agents Chemother. 2002;46(9):2784–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Aslan H, Petersen ME, De Berardinis A, et al. Activation of the two-component system LisRK promotes cell adhesion and high ampicillin tolerance in Listeria monocytogenes. Front Microbiol. 2021;12:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Nielsen PK, Andersen AZ, Mols M, et al. Genome-wide transcriptional profiling of the cell envelope stress response and the role of LisRK and CesRK in Listeria monocytogenes. Microbiology. 2012;158:963–974. [DOI] [PubMed] [Google Scholar]
- [168].Fraser KR, Sue D, Wiedmann M, et al. Role of σB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC Is σB dependent. Appl Environ Microbiol. 2003;69:2015–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Utratna M, Shaw I, Starr E, et al. Rapid, transient, and proportional activation of σB in response to osmotic stress in Listeria monocytogenes. Appl Environ Microbiol. 2011;77:7841–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Chan YC, Boor KJ, Wiedmann M. σB-dependent and σB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl Environ Microbiol. 2007;73:6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Hecker M, Pané-Farré J, Völker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–236. [DOI] [PubMed] [Google Scholar]
- [172].Faleiro ML, Andrew PW, Power D. Stress response of Listeria monocytogenes isolated from cheese and other foods. Int J Food Microbiol. 2003;84:207–216. [DOI] [PubMed] [Google Scholar]
- [173].Davis MJ, Coote PJ, O’Byrne CP. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology. 1996;142:2975–2982. [DOI] [PubMed] [Google Scholar]
- [174].Giotis ES, Julotok M, Wilkinson BJ, et al. Role of sigma B factor in the alkaline tolerance response of Listeria monocytogenes 10403S and cross-protection against subsequent ethanol and osmotic stress. J Food Prot. 2008;71:1481–1485. [DOI] [PubMed] [Google Scholar]
- [175].Metselaar KI, Hmw DB, Boekhorst J, et al. Diversity of acid stress resistant variants of Listeria monocytogenes and the potential role of ribosomal protein S21 encoded by rpsU. Front Microbiol. 2015;6:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Metselaar KI, Hmw DB, Abee T, et al. Isolation and quantification of highly acid resistant variants of Listeria monocytogenes. Int J Food Microbiol. 2013;166:508–514. [DOI] [PubMed] [Google Scholar]
- [177].Akanuma G, Nanamiya H, Natori Y, et al. Inactivation of ribosomal protein genes in bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J Bacteriol. 2012;194:6282–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Radzikowski JL, Schramke H, Heinemann M. Bacterial persistence from a system-level perspective. Curr Opin Biotechnol. 2017;46:98–105. [DOI] [PubMed] [Google Scholar]
- [179].Gollan B, Grabe G, Michaux C, et al. Bacterial persisters and infection: past, present, and progressing. Annu Rev Microbiol. 2019;73:359–385. [DOI] [PubMed] [Google Scholar]
- [180].Toledo-Arana A, Dussurget O, Nikitas G, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. [DOI] [PubMed] [Google Scholar]
- [181].Garner MR, Njaa BL, Wiedmann M, et al. σB contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun. 2006;74:876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Wiedmann M, Arvik TJ, Hurley RJ, et al. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180:3650–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Bergholz TM, den Bakker HC, Fortes ED, et al. Salt Stress Phenotypes in Listeria monocytogenes Vary by Genetic Lineage and Temperature. Foodborne Pathog Dis. 2010;7:1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Ribeiro VB, Destro MT. Listeria monocytogenes serotype 1/2b and 4b isolates from human clinical cases and foods show differences in tolerance to refrigeration and salt stress. J Food Prot. 2014;77:1519–1526. [DOI] [PubMed] [Google Scholar]
- [185].Horlbog JA, Kent D, Stephan R, et al. Surviving host - and food relevant stresses: phenotype of L. monocytogenes strains isolated from food and clinical sources. Sci Rep. 2018;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Zoz F, Grandvalet C, Lang E, et al. Listeria monocytogenes ability to survive desiccation: influence of serotype, origin, virulence, and genotype. Int J Food Microbiol. 2017;248:82–89. [DOI] [PubMed] [Google Scholar]
- [187].Buncic S, Avery SM, Rocourt J, et al. Can food-related environmental factors induce different behaviour in two key serovars, 4b and 1/2a, of Listeria monocytogenes? Int J Food Microbiol. 2001;65:201–212. [DOI] [PubMed] [Google Scholar]
- [188]. A, Magalhães R, TRS B, et al. Impact of exposure to cold and cold-osmotic stresses on virulence-associated characteristics of Listeria monocytogenes strains. Food Microbiol. 2020;87:103351. [DOI] [PubMed] [Google Scholar]
- [189].Tiensuu T, Guerreiro DN, Oliveira AH, et al. Flick of a switch: regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer. Microbiol (United Kingdom). 2019;165:819–833. [DOI] [PubMed] [Google Scholar]
- [190].Nadon CA, Bowen BM, Wiedmann M, et al. σB contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect Immun. 2002;70:3948–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Rauch M, Luo Q, Müller-Altrock S, et al. SigB-dependent in vitro transcription of prfA and some newly identified genes of Listeria monocytogenes whose expression is affected by PrfA in vivo. J Bacteriol. 2005;187:800–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Sue D, Fink D, Wiedmann M, et al. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology. 2004;150:3843–3855. [DOI] [PubMed] [Google Scholar]
- [193].Ollinger J, Wiedmann M, Boor KJ. σB- and PrfA-dependent transcription of genes previously classified as putative constituents of the Listeria monocytogenes PrfA regulon. Foodborne Pathog Dis. 2008;5:281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Ollinger J, Bowen B, Wiedmann M, et al. Listeria monocytogenes σB modulates PrfA-mediated virulence factor expression. Infect Immun. 2009;77:2113–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Guldimann C, Boor KJ, Wiedmann M, et al. Resilience in the face of uncertainty: sigma factor B fine-tunes gene expression to support homeostasis in gram-positive bacteria. Appl Environ Microbiol. 2016;82:4456–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].de las Heras A, RJ C, Mk B, et al. Regulation of Listeria virulence: prfA master and commander. Curr Opin Microbiol. 2011;14:118–127. [DOI] [PubMed] [Google Scholar]
- [197].Reniere ML, Whiteley AT, Hamilton KL, et al. Glutathione activates virulence gene expression of an intracellular pathogen. Nature. 2015;517:170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Johansson J, Mandin P, Renzoni A, et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. [DOI] [PubMed] [Google Scholar]
- [199].Drolia R, Bhunia AK. Crossing the Intestinal Barrier via Listeria Adhesion Protein and Internalin A. In: Trends Microbiol. Elsevier Ltd; 2019;27(5): 408–425. [DOI] [PubMed] [Google Scholar]
- [200].Kühbacher A, Emmenlauer M, Rämo P, et al. Genome-Wide siRNA screen identifies complementary signaling pathways involved in Listeria infection and reveals different actin nucleation mechanisms during Listeria cell invasion and actin comet tail formation. MBio. 2015;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Nikitas G, Deschamps C, Disson O, et al. Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J Exp Med. 2011;208:2263–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Lecuit M, Ohayon H, Braun L, et al. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect Immun. 1997;65:5309–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Disson O, Grayo S, Huillet E, et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455:1114–1118. [DOI] [PubMed] [Google Scholar]
- [204].Wollert T, Pasche B, Rochon M, et al. Extending the host range of Listeria monocytogenes by rational protein design. Cell. 2007;129:891–902. [DOI] [PubMed] [Google Scholar]
- [205].Lecuit M, Vandormael-Pournin S, Lefort J, et al. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. [DOI] [PubMed] [Google Scholar]
- [206].Burkholder KM, Bhunia AK. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation and induces expression of LAP receptor Hsp60. Infect Immun. 2010;78:5062–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Jagadeesan B, Koo OK, Kim KP, et al. LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria, promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology. 2010;156:2782–2795. [DOI] [PubMed] [Google Scholar]
- [208].Drolia R, Tenguria S, Durkes AC, et al. Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe. 2018;23:470–484.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Chiba S, Nagai T, Hayashi T, et al. Listerial invasion protein internalin B promotes entry into ileal Peyer’s patches in vivo. Microbiol Immunol. 2011;55:123–129. [DOI] [PubMed] [Google Scholar]
- [210].Tsai Y-H, Disson O, Bierne H, et al. Murinization of internalin extends its receptor repertoire, altering Listeria monocytogenes cell tropism and host responses. PLoS Pathog. 2013;9:e1003381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Rey C, Chang -Y-Y, Latour-Lambert P, et al. Transcytosis subversion by M cell-to-enterocyte spread promotes Shigella flexneri and Listeria monocytogenes intracellular bacterial dissemination. PLoS Pathog. 2020;16:e1008446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Pizarro-Cerdá J, Cossart P. Listeria monocytogenes: cell biology of invasion and intracellular growth. In: Microbiol Spectr. 2018; 6(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Prokop A, Gouin E, Villiers V, et al. OrfX, a Nucleomodulin required for Listeria monocytogenes virulence. MBio. 2017;8:e01550–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Pizarro-Cerdá J, Cossart P. Microbe profile: listeriamonocytogenes: a paradigm among intracellular bacterial pathogens. Microbiol (United Kingdom). 2019;165:719–721. [DOI] [PubMed] [Google Scholar]
- [215].Bavdek A, Kostanjšek R, Antonini V, et al. pH dependence of listeriolysin O aggregation and pore-forming ability. FEBS J. 2012;279:126–141. [DOI] [PubMed] [Google Scholar]
- [216].Chen C, Nguyen BN, Mitchell G, et al. The listeriolysin O PEST-like sequence Co-opts AP-2-mediated endocytosis to prevent plasma membrane damage during Listeria infection. Cell Host Microbe. 2018;23:786–795.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Portman JL, Huang Q, Reniere ML, et al. Activity of the pore-forming virulence factor listeriolysin O is reversibly inhibited by naturally occurring Sglutathionylation. Infect Immun. 2017;85(4):e00959-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Peterson BN, Portman JL, Feng Y, et al. Secondary structure of the mRNA encoding listeriolysin O is essential to establish the replicative niche of Listeria monocytogenes. Proc Natl Acad Sci U S A. 2020;117:23774–23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Alonzo F, Port GC, Cao M, et al. The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect Immun. 2009;77:2612–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Ignatov D, Vaitkevicius K, Durand S, et al. An mRNA-mRNA interaction couples expression of a virulence factor and its chaperone in Listeria monocytogenes. Cell Rep. 2020;30:4027–4040.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Hamon MA, Ribet D, Stavru F, et al. Listeriolysin O: the Swiss army knife of Listeria. In: Trends Microbiol. 2012;20:360–368. [DOI] [PubMed] [Google Scholar]
- [222].Hamon MA, Batsché E, Régnault B, et al. Histone modifications induced by a family of bacterial toxins. Proc Natl Acad Sci U S A. 2007;104:13467–13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Samba-Louaka A, Pereira JM, Nahori M-A, et al. Listeria monocytogenes dampens the DNA damage response. PLoS Pathog. 2014;10:e1004470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Witte CE, Archer KA, Rae CS, et al. Innate immune pathways triggered by Listeria monocytogenes and their role in the induction of cell-mediated immunity. Adv Immunol. 1st ed. 2012;113:135–156. [DOI] [PubMed] [Google Scholar]
- [225].Stavru F, Palmer AE, Wang C, et al. Atypical mitochondrial fission upon bacterial infection. Proc Natl Acad Sci U S A. 2013;110:16003–16008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Malet JK, Cossart P, Ribet D. Alteration of epithelial cell lysosomal integrity induced by bacterial cholesterol-dependent cytolysins. Cell Microbiol. 2017;19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Phelps CC, Vadia S, Boyaka PN, et al. A listeriolysin O subunit vaccine is protective against Listeria monocytogenes. Vaccine. 2020;38:5803–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Portnoy DA, Chakraborty T, Goebel W, et al. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 1992;60:1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Quereda JJ, Cossart P, Pizarro-Cerdá J. Role of Listeria monocytogenes exotoxins in virulence. In: Microb Toxins. Springer Netherlands: Dordrecht; 2016:1–20. [Google Scholar]
- [230].Xayarath B, Alonzo F, Freitag NE. Identification of a peptide-pheromone that enhances Listeria monocytogenes escape from host cell vacuoles. PLoS Pathog. 2015;11:e1004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Rabinovich L, Sigal N, Borovok I, et al. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell. 2012;150:792–802. [DOI] [PubMed] [Google Scholar]
- [232].Tattoli I, Sorbara MT, Yang C, et al. Listeria phospholipases subvert host autophagic defenses by stalling pre-autophagosomal structures. EMBO J. 2013;32:3066–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Mitchell G, Ge L, Huang Q, et al. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect Immun. 2015;83:2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Radoshevich L, Impens F, Ribet D, et al. ISG15 counteracts Listeria monocytogenes infection. Elife. 2015;4:e06848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Radoshevich L, Dussurget O. Cytosolic innate immune sensing and signaling upon infection. Front Microbiol. 2016;7:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Quereda JJ, Morel C, Lopez-Montero N, et al. A role for Taok2 in Listeria monocytogenes vacuolar escape. J Infect Dis. 2020. DOI: 10.1093/infdis/jiaa367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Birmingham CL, Canadien V, Kaniuk NA, et al. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2008;451:350–354. [DOI] [PubMed] [Google Scholar]
- [238].Peron-Cane C, Fernandez J-C, Leblanc J, et al. Fluorescent secreted bacterial effectors reveal active intravacuolar proliferation of Listeria monocytogenes in epithelial cells. PLOS Pathog. 2020;16:e1009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Kortebi M, Milohanic E, Mitchell G, et al. Listeria monocytogenes switches from dissemination to persistence by adopting a vacuolar lifestyle in epithelial cells. PLoS Pathog. 2017;13:e1006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Bierne H, Milohanic E, Kortebi M. To be cytosolic or vacuolar: the double life of Listeria monocytogenes. Front Cell Infect Microbiol. 2018;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Kühn S, Enninga J. The actin comet guides the way: how Listeria actin subversion has impacted cell biology, infection biology and structural biology. Cell Microbiol. 2020;22:e13190. [DOI] [PubMed] [Google Scholar]
- [242].Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Boujemaa-Paterski R, Gouin E, Hansen G, et al. Listeria protein ActA mimics WASP family proteins: it activates filament barbed end branching by Arp2/3 complex. Biochemistry. 2001;40:11390–11404. [DOI] [PubMed] [Google Scholar]
- [244].Sitthidet C, Korbsrisate S, Layton AN, et al. Identification of motifs of Burkholderia pseudomallei BimA required for intracellular motility, actin binding, and actin polymerization. J Bacteriol. 2011;193:1901–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Egile C, Loisel TP, Laurent V, et al. Activation of the Cdc42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol. 1999;146:1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Gouin E, Quereda -J-J, Torres -J-JQ, et al. Intracellular bacteria find the right motion. Cell. 2015;161:199–200. [DOI] [PubMed] [Google Scholar]
- [247].Fattouh R, Kwon H, Czuczman MA, et al. The diaphanous-related formins promote protrusion formation and cell-to-cell spread of Listeria monocytogenes. J Infect Dis. 2015;211:1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Rajabian T, Gavicherla B, Heisig M, et al. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat Cell Biol. 2009;11:1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Dowd GC, Mortuza R, Bhalla M, et al. Listeria monocytogenes exploits host exocytosis to promote cell-to-cell spread. PNAS. 2020;117:3789–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Czuczman MA, Fattouh R, van Rijn JM, et al. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature. 2014;509:230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Alberti-Segui C, Goeden KR, Higgins DE. Differential function of Listeria monocytogenes listeriolysin O and phospholipases C in vacuolar dissolution following cell-to-cell spread. Cell Microbiol. 2007;9:179–195. [DOI] [PubMed] [Google Scholar]
- [252].Cheng MI, Chen C, Engström P, et al. Actin-based motility allows Listeria monocytogenes to avoid autophagy in the macrophage cytosol. Cell Microbiol. 2018;20:e12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Travier L, Guadagnini S, Gouin E, et al. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 2013;9:e1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA. 2011;108:19484–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Jones GS, Bussell KM, Myers-Morales T, et al. Intracellular Listeria monocytogenes comprises a minimal but vital fraction of the intestinal burden following foodborne infection. Infect Immun. 2015;83:3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Melton-Witt JA, Rafelski SM, Portnoy DA, et al. Oral infection with signature-tagged Listeria monocytogenes reveals organ-specific growth and dissemination routes in guinea pigs. Infect Immun. 2012;80:720–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Drevets DA, Tuomanen EI. Dissemination of Listeria monocytogenes by Infected Phagocytes. Infect Immun. 1999;67:3512–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Czuprynski CJ, Noel EJ, Doyle MP, et al. Ingestion and killing of Listeria monocytogenes by blood and milk phagocytes from mastitic and normal cattle. J Clin Microbiol. 1989;27:812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Quereda JJ, García-del Portillo F, Pucciarelli MG. Listeria monocytogenes remodels the cell surface in the blood-stage. Environ Microbiol Rep. 2016;8:641–648. [DOI] [PubMed] [Google Scholar]
- [260].Quereda JJ, Ortega AD, Pucciarelli MG, et al. The Listeria small RNA Rli27 regulates a cell wall protein inside eukaryotic cells by targeting a long 5’-UTR Variant. PLoS Genet. 2014;10:e1004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Drevets DA, Leenen PJM, Greenfield RA. Invasion of the central nervous system by intracellular bacteria.Clin Microbiol Rev. 2004;323–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].D’Orazio SEF. Innate and adaptive immune responses during Listeria monocytogenes infection. In: Microbiol Spectr. 2019:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Maudet C, Levallois S, Disson O, et al. Curr. Opin. Microbiol. In: Innate immune responses to Listeria in vivo. Elsevier Ltd, 2021;59:95–101. [DOI] [PubMed] [Google Scholar]
- [264].Witter AR, Okunnu BM, Berg RE. The essential role of neutrophils during infection with the intracellular bacterial pathogen Listeria monocytogenes. J Immunol. 2016;197:1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Stavru F, Archambaud C, Cossart P. Cell biology and immunology of Listeria monocytogenes infections: novel insights. Immunol Rev Immunol Rev. 2011;160–184. [DOI] [PubMed] [Google Scholar]
- [266].Ladel CH, Flesch IE, Arnoldi J, et al. Studies with MHC-deficient knock-out mice reveal impact of both MHC I- and MHC II-dependent T cell responses on Listeria monocytogenes infection. J Immunol. 1994;153:3116–3122. [PubMed] [Google Scholar]
- [267].Lebreton A, Lakisic G, Job V, et al. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science. 2011;331:1319–1321. [DOI] [PubMed] [Google Scholar]
- [268].Boneca IG, Dussurget O, Cabanes D, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104:997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Blériot C, Dupuis T, Jouvion G, et al. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity. 2015;42:145–158. [DOI] [PubMed] [Google Scholar]
- [270].Wang G, Zhao H, Zheng B, et al. TLR2 promotes monocyte/macrophage recruitment into the liver and microabscess formation to limit the spread of Listeria monocytogenes. Front Immunol. 2019;10:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Dramsi S, Biswas I, Maguin E, et al. Entry of Listeria monocytogenes into hepatocytes requires expression of InIB, a surface protein of the internalin multigene family. Mol Microbiol. 1995;16:251–261. [DOI] [PubMed] [Google Scholar]
- [272].Quereda JJ, Rodríguez-Gómez IM, Meza-Torres J, et al. Reassessing the role of internalin B in Listeria monocytogenes virulence using the epidemic strain F2365. Clin Microbiol Infect. 2019;25:252.e1–252.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Qiu Z, Khairallah C, Sheridan BS. Listeria monocytogenes: a model pathogen continues to refine our knowledge of the CD8 T cell response. Pathog (Basel, Switzerland). 2018;7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Edelson BT, Bradstreet TR, Hildner K, et al. CD8α+ dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity. 2011;35:236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Banović F, Schroten H, Schwerk C. Potential roles and functions of listerial virulence factors during brain entry. Toxins (Basel). 2020;12:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Disson O, Lecuit M. Targeting of the central nervous system by Listeria monocytogenes. Virulence. 2012;3:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Oevermann A, Zurbriggen A, Vandevelde M. Rhombencephalitis caused by Listeria monocytogenes in humans and ruminants: a zoonosis on the rise? Interdiscip Perspect Infect Dis. 2010;2010:632513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Karlsson WK, Harboe ZB, Roed C, et al. Early trigeminal nerve involvement in Listeria monocytogenes rhombencephalitis: case series and systematic review. J Neurol. 2017;264:1875–1884. [DOI] [PubMed] [Google Scholar]
- [279].Pägelow D, Chhatbar C, Beineke A, et al. The olfactory epithelium as a port of entry in neonatal neurolisteriosis. Nat Commun. 2018;9:4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Ghosh P, Halvorsen EM, Ammendolia DA, et al. Invasion of the brain by Listeria monocytogenes is mediated by InlF and host cell vimentin. MBio. 2018;9:e00160–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Greiffenberg L, Goebel W, Kim KS, et al. Interaction of Listeria monocytogenes with human brain microvascular endothelial cells: inlB-dependent invasion, long-term intracellular growth, and spread from macrophages to endothelial cells. Infect Immun. 1998;66:5260–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Gründler T, Quednau N, Stump C, et al. The surface proteins InlA and InlB are interdependently required for polar basolateral invasion by Listeria monocytogenes in a human model of the blood-cerebrospinal fluid barrier. Microbes Infect. 2013;15:291–301. [DOI] [PubMed] [Google Scholar]
- [283].Schlüter D, Domann E, Buck C, et al. Phosphatidylcholine-specific phospholipase C from Listeria monocytogenes is an important virulence factor in murine cerebral listeriosis. Infect Immun. 1998;66:5930–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Zhang T, Bae D, Listeriolysin WC. O mediates cytotoxicity against human brain microvascular endothelial cells. FEMS Microbiol Lett. 2015;362(12). [DOI] [PubMed] [Google Scholar]
- [285].Kayal S, Listeriolysin. O: a key protein of Listeria monocytogenes with multiple functions. FEMS Microbiol Rev. 2006;30:514–529. [DOI] [PubMed] [Google Scholar]
- [286].Kayal S, Lilienbaum A, Poyart C, et al. Listeriolysin O-dependent activation of endothelial cells during infection with Listeria monocytogenes: activation of NF-κB and upregulation of adhesion molecules and chemokines. Mol Microbiol. 1999;31:1709–1722. [DOI] [PubMed] [Google Scholar]
- [287].Popowska M, Krawczyk-Balska A, Ostrowski R, et al. InlL from Listeria monocytogenes is involved in biofilm formation and adhesion to mucin. Front Microbiol. 2017;8:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Autret N, Dubail I, Trieu-Cuot P, et al. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect Immun. 2001;69:2054–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Vázquez-Boland JA, Krypotou E, Scortti M. Listeria placental infection. MBio. 2017;8:e00949–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Gessain G, Tsai YH, Travier L, et al. PI3-kinase activation is critical for host barrier permissiveness to Listeria monocytogenes. J Exp Med. 2015;212:165–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Faralla C, Rizzuto GA, Lowe DE, et al. InlP, a new virulence factor with strong placental tropism. Infect Immun. 2016;84:3584–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Faralla C, Bastounis EE, Ortega FE, et al. Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing. PLoS Pathog. 2018;14:e1007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis. 2011;17:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].de Noordhout CM, Devleesschauwer B, Angulo FJ, et al. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Pohl AM, Pouillot R, Bazaco MC, et al. Differences among incidence rates of invasive listeriosis in the U.S. FoodNet Population by age, sex, race/ ethnicity,and pregnancy status, 2008-2016. Foodborne Pathog Dis. 2019;16:290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Herrador Z, Gherasim A, López-Vélez R, et al. Listeriosis in Spain based on hospitalisation records, 1997 to 2015: need for greater awareness. In: Eurosurveillance. 2019:24(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Goulet V, Hebert M, Hedberg C, et al. Incidence of listeriosis and related mortality among groups at risk of acquiring listeriosis. Clin Infect Dis. 2012;54:652–660. [DOI] [PubMed] [Google Scholar]
- [298].Committee Opinion . Management of pregnant women with presumptive exposure to Listeria monocytogenes. Obstet Gynecol. 2014;124:1241–1244. [DOI] [PubMed] [Google Scholar]
- [299].Silk BJ, Date KA, Jackson KA, et al. Invasive listeriosis in the foodborne diseases active surveillance network (FoodNet), 2004–2009: further targeted prevention needed for higher-risk groups. Clin Infect Dis. 2012;54:S396–S404. [DOI] [PubMed] [Google Scholar]
- [300].Girard D, Leclercq A, Laurent E, et al. Pregnancy-related listeriosis in France, 1984 to 2011, with a focus on 606 cases from 1999 to 2011. Eurosurveillance. 2014;19:20909. [DOI] [PubMed] [Google Scholar]
- [301].Doganay M. Listeriosis: clinical presentation. FEMS Immunol Med Microbiol. 2003;35:173–175. [DOI] [PubMed] [Google Scholar]
- [302].Charlier C, É P, Leclercq A, et al. Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect Dis. 2017;17:510–519. [DOI] [PubMed] [Google Scholar]
- [303].Gentschev I, Sokolovic Z, Kohler S, et al. Identification of p60 antibodies in human sera and presentation of this listerial antigen on the surface of attenuated salmonellae by the HlyB-HlyD secretion system. Infect Immun. 1992;60:5091–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Leong ML, Hampl J, Liu W, et al. Impact of preexisting vector-specific immunity on vaccine potency: characterization of Listeria monocytogenes-specific humoral and cellular immunity in humans and modeling studies using recombinant vaccines in mice. Infect Immun. 2009;77:3958–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Silk BJ, McCoy MH, Iwamoto M, et al. Foodborne listeriosis acquired in hospitals. Clin Infect Dis. 2014;59:532–540. [DOI] [PubMed] [Google Scholar]
- [306].Vázquez-Boland JA, Dominguez L, Blanco M, et al. Epidemiologic investigation of a silage-associated epizootic of ovine listeric encephalitis, using a new Listeria-selective enumeration medium and phage typing. Am J Vet Res. 1992;53:368–371. [PubMed] [Google Scholar]
- [307].Stein H, Stessl B, Brunthaler R, et al. Listeriosis in fattening pigs caused by poor quality silage - a case report. BMC Vet Res. 2018;14:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Scott PR Overview of Listeriosis - Generalized Conditions - Merck Veterinary Manual [Internet]. 2014. cited 2020 Jul 31. Available from: https://www.merckvetmanual.com/generalized-conditions/listeriosis/overview-of-listeriosis.
- [309].Esteban JI, Oporto B, Aduriz G, et al. Faecal shedding and strain diversity of Listeria monocytogenes in healthy ruminants and swine in Northern Spain. BMC Vet Res. 2009;5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Weindl L, Frank E, Ullrich U, et al. Listeria monocytogenes in different specimens from healthy red deer and wild boars. Foodborne Pathog Dis. 2016;13:391–397. [DOI] [PubMed] [Google Scholar]
- [311].Walland J, Lauper J, Frey J, et al. Listeria monocytogenes infection in ruminants: is there a link to the environment, food and human health? A review. Schweiz Arch Tierheilkd. 2015;157:319–328. [DOI] [PubMed] [Google Scholar]
- [312].Luque-Sastre L, Arroyo C, Fox EM, et al. Antimicrobial resistance in Listeria species. Microbiol Spectr. 2018;6:237–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Charpentier E, Courvalin P. Antibiotic resistance in Listeria spp. Antimicrob Agents Chemother. 1999;43:2103–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Maurella C, Gallina S, Ru G, et al. Outbreak of febrile gastroenteritis caused by Listeria monocytogenes 1/2A in sliced cold beef ham, Italy, may 2016. In: Eurosurveillance. 2018;23(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Sim J, Hood D, Finnie L, et al. Series of incidents of Listeria monocytogenes non-invasive febrile gastroenteritis involving ready-to-eat meats. Lett Appl Microbiol. 2002;35:409–413. [DOI] [PubMed] [Google Scholar]
- [316].Ooi ST, Lorber B. Gastroenteritis due to Listeria monocytogenes. Clin Infect Dis. 2005;40:1327–1332. [DOI] [PubMed] [Google Scholar]
- [317].Goulet V, La K, Vaillant V, et al. What is the incubation period for listeriosis? BMC Infect Dis. 2013;13:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Mylonakis E, Hohmann EL, Calderwood SB. Central nervous system infection with Listeria monocytogenes: 33 Years’ experience at a general hospital and review of 776 episodes from the literature. Medicine (Baltimore). 1998;77:313–336. [DOI] [PubMed] [Google Scholar]
- [319].Armstrong RW, Fung PC. Brainstem encephalitis (rhombencephalitis) due to Listeria monocytogenes: case report and review. Clin Infect Dis. 1993;16:689–702. [DOI] [PubMed] [Google Scholar]
- [320].Zelenik K, Avberšek J, Pate M, et al. Cutaneous listeriosis in a veterinarian with the evidence of zoonotic transmission - A case report. Zoonoses Public Health. 2014;61:238–241. [DOI] [PubMed] [Google Scholar]
- [321].McLauchlin J, Low JC. Primary cutaneous listeriosis in adults: an occupational disease of veterinarians and farmers. Vet Rec. 1994;135:615–617. [PubMed] [Google Scholar]
- [322].García-Granja PE, López J, Vilacosta I, et al. Infective Endocarditis due to Listeria monocytogenes: a Report of 4 Patients. Rev Española Cardiol English Ed. 2016;69:700–702. [DOI] [PubMed] [Google Scholar]
- [323].Cardoso C, Cremers I, Oliveira AP. Spontaneous bacterial peritonitis caused by Listeria monocytogenes: a case report and literature review. Ann Hepatol. 2012;11:955–957. [PubMed] [Google Scholar]
- [324].Charlier C, Fevre C, Travier L, et al. Listeria monocytogenes-associated biliary tract infections: a study of 12 consecutive cases and review. Medicine (Baltimore). 2014;93:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [325].Charlier C, Leclercq A, Cazenave B, et al. Listeria monocytogenes-associated joint and bone infections: a study of 43 consecutive cases. Clin Infect Dis. 2012;54:240–248. [DOI] [PubMed] [Google Scholar]
- [326].Hof H. Listeria infections of the eye. Eur J Ophthalmol. 2017;27:115–121. [DOI] [PubMed] [Google Scholar]
- [327].Henke D, Rupp S, Gaschen V, et al. Listeria monocytogenes spreads within the brain by actin-based intra-axonal migration. Infect Immun. 2016;84:866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Winter P, Schilcher F, Bagò Z, et al. Clinical and histopathological aspects of naturally occurring mastitis caused by Listeria monocytogenes in cattle and ewes. J Vet Med Ser B Infect Dis Vet Public Heal. 2004;51:176–179. [DOI] [PubMed] [Google Scholar]
- [329].Poulsen KP, Pillers DM, Conway JH, et al. Post-parturient shedding of Listeria monocytogenes in breast milk of infected mice Listeria monocytogenes shed in murine milk. J Neonatal Perinatal Med. 2013;6:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Clark RG, Gill JM, Swanney S. Listeria monocytogenes gastroenteritis in sheep. N Z Vet J. 2004;52:46–47. [DOI] [PubMed] [Google Scholar]
- [331].Laven RA, Lawrence KR. An outbreak of iritis and uveitis in dairy cattle at pasture associated with the supplementary feeding of baleage. N Z Vet J. 2006;54:151–152. [DOI] [PubMed] [Google Scholar]
- [332].Neuhaus K, Satorhelyi P, Schauer K, et al. Acid shock of Listeria monocytogenes at low environmental temperatures induces prfA, epithelial cell invasion, and lethality towards Caenorhabditis elegans. BMC Genomics. 2013;14:285. [DOI] [PMC free article] [PubMed]
- [333].Fröhlich KS, Gottesman S, Storz G. Small Regulatory RNAs in the enterobacterial response to envelope damage and oxidative stress. Microbiol Spectr. 2018;6:213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Holmqvist E, Wagner EGH. Impact of bacterial sRNAs in stress responses. Biochem Soc Trans. 2017;45:1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [335].Becker B, Schuler S, Lohneis M, et al. Comparison of two chromogenic media for the detection of Listeria monocytogenes with the plating media recommended by EN/DIN 11290-1. Int J Food Microbiol. 2006;109:127–131. [DOI] [PubMed] [Google Scholar]
- [336].Angelo KM, Conrad AR, Saupe A, et al. Multistate outbreak of Listeria monocytogenes infections linked to whole apples used in commercially produced, prepackaged caramel apples: united States, 2014-2015. Epidemiol Infect. 2017;145:848–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Self JL, Conrad A, Stroika S, et al. Notes from the field : outbreak of listeriosis associated with consumption of packaged salad — united States and Canada, 2015–2016. MMWR Morb Mortal Wkly Rep. 2016;65:879–881. [DOI] [PubMed] [Google Scholar]
- [338].Gaul LK, Farag NH, Shim T, et al. Hospital-acquired listeriosis outbreak caused by contaminated diced celery-texas, 2010. Clin Infect Dis. 2013;56:20–26. [DOI] [PubMed] [Google Scholar]
- [339].Jackson BR, Salter M, Tarr C, et al. Notes from the field: listeriosis associated with stone fruit–United States, 2014. MMWR Morb Mortal Wkly Rep. 2015;64:282–283. [PMC free article] [PubMed] [Google Scholar]
- [340].Addis MF, Cubeddu T, Pilicchi Y, et al. Chronic intramammary infection by Listeria monocytogenes in a clinically healthy goat - a case report. BMC Vet Res. 2019;15:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Hächler H, Marti G, Giannini P, et al. Outbreak of listerosis due to imported cooked ham, Switzerland 2011. Euro Surveill. 2013;18:20469. [PubMed] [Google Scholar]
- [342].Smith B, Larsson JT, Lisby M, et al. Outbreak of listeriosis caused by infected beef meat from a meals-on-wheels delivery in Denmark 2009. Clin Microbiol Infect. 2011;17:50–52. [DOI] [PubMed] [Google Scholar]
- [343].Filipello V, Mughini-Gras L, Gallina S, et al. Attribution of Listeria monocytogenes human infections to food and animal sources in Northern Italy. Food Microbiol. 2020;89:103433. [DOI] [PubMed] [Google Scholar]
- [344].Gu Y, Liang X, Huang Z, et al. Outbreak of Listeria monocytogenes in pheasants. Poult Sci. 2015;94:2905–2908. [DOI] [PubMed] [Google Scholar]
- [345].Cao X, Wang Y, Wang Y, et al. Isolation and characterization of Listeria monocytogenes from the black-headed gull feces in Kunming, China. J Infect Public Health. 2018;11:59–63. [DOI] [PubMed] [Google Scholar]
- [346].Hellström S, Kiviniemi K, Autio T, et al. Listeria monocytogenes is common in wild birds in Helsinki region and genotypes are frequently similar with those found along the food chain. J Appl Microbiol. 2008;104:883–888. [DOI] [PubMed] [Google Scholar]
- [347].Ohkochi M, Nakazawa M, Sashihara N. Detection of Listeria monocytogenes in commercially broken unpasteurized liquid egg in Japan. J Food Prot. 2009;72:178–181. [DOI] [PubMed] [Google Scholar]
- [348].Simothy L, Mahomoodally F, Neetoo H. A study on the potential of ants to act as vectors of foodborne pathogens. AIMS Microbiol. 2018;4:319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Iwu CD, Okoh AI, Sekaran SD. Characterization of antibiogram fingerprints in Listeria monocytogenes recovered from irrigation water and agricultural soil samples. PLoS One. 2020;15:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Gúevremont E, Lamoureux L, Genereux M, et al. Irrigation water sources and time intervals as variables on the presence of Campylobacter spp. and Listeria monocytogenes on romaine lettuce grown in muck soil. J Food Prot. 2017;80:1182–1187. [DOI] [PubMed] [Google Scholar]
- [351].Zhao T, Podtburg TC, Zhao P, et al. Control of Listeria spp. by competitive-exclusion bacteria in floor drains of a poultry processing plant. Appl Environ Microbiol. 2006;72:3314–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Galié S, García-Gutiérrez C, Miguélez EM, et al. Biofilms in the food industry: health aspects and control methods. Front Microbiol. 2018;9:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Bolocan AS, Oniciuc EA, Alvarez-Ordóñez A, et al. Putative cross-contamination routes of Listeria monocytogenes in a meat processing facility in Romania. J Food Prot. 2015;78(9):1664–1674. [DOI] [PubMed] [Google Scholar]
- [354].Chiarini E, Tyler K, Farber JM, et al. Listeria monocytogenes in two different poultry facilities: manual and automatic evisceration. Poult Sci. 2009;88:791–797. [DOI] [PubMed] [Google Scholar]


