Abstract
A major challenge in regenerative medicine is replacing cells lost through injury or disease. While significant progress has been made, much remains unknown about the accuracy of native regenerative programs in cell replacement. Here, we capitalized on the regenerative capacity and stereotypic retinal organization of zebrafish to determine the specificity with which retinal Müller glial cells replace lost neuronal cell types. By utilizing a targeted genetic ablation technique, we restricted death to all or to distinct cone photoreceptor types (red, blue, or UV-sensitive cones), enabling us to compare the composition of cones that are regenerated. We found that Müller glia produce cones of all types upon non-discriminate ablation of these photoreceptors, or upon selective ablation of red or UV cones. Pan-ablation of cones led to regeneration of the various cone types in relative abundances that resembled those of nonablated controls, that is, red > green > UV blue cones. Moreover, selective loss of red or UV cones biased production toward the cone type that was ablated. In contrast, ablation of blue cones alone largely failed to induce cone production at all, although it did induce cell division in Müller glia. The failure to produce cones upon selective elimination of blue cones may be due to their low abundance compared to other cone types. Alternatively, it may be that blue cone death alone does not trigger a change in progenitor competency to support cone genesis. Our findings add to the growing notion that cell replacement during regeneration does not perfectly mimic programs of cell generation during development.
Keywords: cone genesis, neuronal repair, nitro-reductase cell ablation, photoreceptor cell proliferation, retinal regeneration, zebrafish photoreceptors
1 ∣. INTRODUCTION
Like elsewhere in the nervous system, the vertebrate retina is highly susceptible to damage by injury or disease. The loss of neurons in the retina often results in visual impairment or blindness, and in mammals, these outcomes are largely irreversible because the lost neurons are not replaced spontaneously. In contrast, some nonmammalian vertebrates, such as chick and teleost fish, are able to restore diverse neuronal populations after retinal injury via regeneration (Fischer & Reh, 2003; Lenkowski & Raymond, 2014). The retinas of teleost fish and mammals share a common anatomical plan, and both possess Müller glia, cells that are activated by injury and dedifferentiate to support neuronal regeneration (Karl & Reh, 2010; Wilken & Reh, 2016). As such, many studies have sought to understand the cellular and molecular mechanisms that underlie native retinal regeneration (Goldman, 2014; Gorsuch & Hyde, 2014; Lenkowski & Raymond, 2014), in order to develop strategies to stimulate the same process in mammals (Karl & Reh, 2010; Wilken & Reh, 2016). Further strategies are needed to accurately match neuronal replacement to the injury, especially because many degenerative diseases of the retina primarily impact specific neuronal cell types (D'Orazi, Suzuki, & Wong, 2014; Hoon, Okawa, Della Santina, & Wong, 2014). Indeed, retinal circuitry comprises numerous specialized neuronal cell types (Demb & Singer, 2015; Masland, 2012), which form stereotypic circuit patterns that are fundamental to proper visual processing. However, recent evidence suggests that retinal regeneration can result in genesis of neuronal cell types that were not lost in the original injury, potentially disrupting the normal complement of cell type proportions and connectivity (Powell, Cornblath, Elsaeidi, Wan, & Goldman, 2016; Yoshimatsu et al., 2016). With recent success in stimulating endogenous cell replacement in the adult mammalian retina (Jorstad et al., 2017), it is now increasingly important to further our understanding of the factors that dictate the accuracy by which endogenous cell regeneration re-establishes neuronal populations of the appropriate cell type or types after damage.
Here, we investigated the regeneration of cone photoreceptor populations after their ablation in larval zebrafish to directly assess the specificity of regeneration. Like in humans, disparate cone types in zebrafish express distinct opsins, each with maximal sensitivity to a specific wavelength of light. Zebrafish possess four cone types, including red, green, ultraviolet (UV), or blue cones (Fadool & Dowling, 2008). These cone populations are arranged in an organized mosaic across the retina, and are present in stereotypic ratios (Allison et al., 2010), approximately 1.8 red:1.3 green:1.3 UV:1 blue cone in larvae. We took advantage of the uniquely stereotypic organization of the zebrafish cone populations to (a) determine whether regeneration is conditional, that is, whether regeneration is only stimulated after the death of only specific or all cone photoreceptor types, and to (b) investigate the accuracy of endogenous neuronal replacement in re-establishing the stereotypic proportions of the cone types.
To achieve these aims, we fate-mapped the regenerated cone population after ablating either the entire cone population or select cone types in larval zebrafish. The loss of all cones, red cones, or UV cones resulted in cone regeneration. In contrast, ablation of blue cones failed to trigger substantial cone genesis, indicating that regeneration responds to specific cell death conditions. Analysis of the composition of regenerated populations after red or UV cone ablation demonstrated that although cone regeneration is nonselective, it is biased toward the cone type that was ablated. Global cone ablation induced generation of all cone types, and regenerated cones were present in relative densities that approximate the hierarchy observed in intact larvae. However, across cone ablation paradigms, the ablated cone types failed to repopulate the retina completely, demonstrating that regenerative neurogenesis may be limited. Collectively, these results suggest that regeneration does not completely recapitulate the steps that lead to the generation of the appropriate numbers and proportions of neuronal cell types during early retinal development.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Transgenic zebrafish
All procedures were conducted in accordance with University of Washington Institutional Animal Care and Use Committee guidelines. Embryonic and larval fish were raised at 28°C in a room with a normal light cycle, lights on from 9:00 to 23:00. Embryos were maintained in system water until 12–24 hr post fertilization (hpf), at which point embryos were placed in system water containing 0.2 mM N-Phenylthiorurea (PTU) (Sigma P7629) to prevent pigmentation. Transgenic larvae were screened for fluorescent protein (FP) expression after hatching, typically at 4 days postfertilization (dpf). Screened larvae were removed from PTU-containing system water, and transferred to a University of Washington zebrafish facility where they were fed regularly. Zebrafish larvae were euthanized by MS-222-(Sigma A5040) overdose (200–500 mg/L). See Table 1 for a list of all the transgenic lines used. The Tg(gnat2:nfsBmCherry) transgenic line was generated by injecting pTol2pA-gnat2-nfsBmCherry plasmid into fertilized eggs at the one-cell stage, and progeny were screened by mCherry expression. The pTol2pA-gnat2-nfsBmCherry plasmid was generated in a Gateway recombination reaction: p5E:gnat2 (Suzuki et al., 2013), pME:nfsBmCherry (Yoshimatsu et al., 2016), p3EpA, and pDestTol2pA (Kwan et al., 2007).
TABLE 1.
List of transgenic lines
| Transgenic | Source | Retinal cells labeled | Shorthand |
|---|---|---|---|
| Tg(gnat2:nfsBmCherry) | Wong lab | All cone types | gnat2 |
| Tg(thrb:G4VP16; UAS:nfsBmCherry) | Q31 (Yoshimatsu et al., 2016); c264 (Davison et al., 2007) | Red cones | trβ2 |
| Tg(thrb:Tomato) | Q22 (Suzuki et al., 2013) | Red cones | trβ2 |
| Tg(thrb:MA-YFP) | Q23 (Suzuki et al., 2013) | Red cones | trβ2 |
| Tg(opn1sw1:nfsBmCherry) | Q28 (Yoshimatsu et al., 2016) | UV cones | sws1 |
| Tg(−5.5opn1sw1:EGFP) | kj9 (Takechi, Hamaoka, & Kawamura, 2003) | UV cones | sws1 |
| Tg(opn1sw2:nfsBmCherry) | Q30 (Yoshimatsu et al., 2016) | Blue cones | sws2 |
| Tg(−3.5opn1sw2:EGFP | kj11 (Takechi, Seno, & Kawamura, 2008) | Blue cones | sws2 |
| Tg(Xla.Rho:EGFP) | fl1 (Fadool, 2003) | Rods | Xops |
| Tg(gfap:GFP) | mi2001 (Bernardos & Raymond, 2006) | Müller glia | Gfap |
2.2 ∣. Selective cell ablation
To ablate specific cone populations, NTR-expressing larvae were immersed in Metronidazole (Met) solution (10 mM Met in system water) at 7 dpf for 1 or 6 hr, according to the experimental paradigm. Larvae were fed regularly, washed in clean system water at the end of treatment, and raised normally.
2.3 ∣. EdU labeling
Mitotic cells were labeled in live larvae by immersion in a solution containing 0.5 mM F-ara-EdU (Yoshimatsu et al., 2016) (Sigma T511293) in system water. The duration of treatment was timed according to the experimental paradigm. Half the solution volume was replaced every other day. For visualization of EdU labeling, fixed whole retinas were permeabilized in 0.3–0.5% TritonX-100 (Sigma T8787) in 0.1 M PBS for 30 min at room temperature, and then washed three times in PBS. Click reactions were carried out in PBS solution with 10 μM Cy5-azide (Lumiprobe A2020), 2 mM copper(II) sulfate (Sigma 45,167), and 20 mM sodium ascorbate (Sigma A7631) for 1 hr at room temperature. Samples were processed for immunohistochemistry after three PBS washes.
2.4 ∣. Immunohistochemistry
After humane killing, larvae were fixed in a solution of 4% paraformaldehyde and 3% sucrose in 0.1 M phosphate-buffered saline (PBS), pH 7.4 at room temperature, and retinas were dissected out within 1–3 days. Fixed, whole retinas were blocked in PBS containing 5% normal donkey serum and 0.5% TritonX-100 for 1–4 hr at room temperature. Tissue was incubated in primary antibody in blocking solution for 1–5 days at 4°C. After three washes in 0.5% TritonX-100 in PBS, samples were incubated in secondary antibody solution for 1 day at 4°C. Samples were washed three times in PBS, mounted in 0.7% agarose, and coverslipped in Vectashield (Vector Labs). See Tables 2 and 3 for lists of all primary and secondary antibodies used.
TABLE 2.
List of primary antibodies
| Antibody | Host | Concentration | Source | RRID |
|---|---|---|---|---|
| Anti-arrestin3a | Mouse | 1:100 | ZIRC (Larison & Bremiller, 1990) | AB_10013803 |
| Anti-DsRed | Rabbit | 1:500 | Clontech 632496 | AB_10013483 |
| Anti-GFP | Chicken | 1:500 | Abcam ab13970 | AB_300798 |
| Anti-GFP | Mouse | 1:200 | Neuromab 75-132 | Not available |
| Anti-GFP | Rabbit | 1:500 | Abcam ab13970 | AB_371416 |
| Anti-UV opsin | Rabbit | 1:1,000 | Gift of Jeremy Nathans (Luo et al., 2004) | Not available |
TABLE 3.
List of secondary antibodies
| Antibody | Host | Concentration | Source | RRID |
|---|---|---|---|---|
| Anti-chicken IgY DyLight488 | Goat | 1:500 | Jackson ImmunoResearch | AB_2336973 |
| Anti-mouse IgG DyLight405 | Goat | 1:500 | Jackson ImmunoResearch | AB_2338986 |
| Anti-mouse IgG DyLight488 | Donkey | 1:500 | Jackson ImmunoResearch | AB_2572300 |
| Anti-mouse IgG DyLight649 | Goat | 1:500 | Jackson ImmunoResearch | AB_2338811 |
| Anti-rabbit IgG DyLight488 | Donkey | 1:500 | Jackson ImmunoResearch | AB_2492289 |
| Anti-rabbit IgG Alexa Fluor 568 | Donkey | 1:500 | Life technologies | AB_2534017 |
| Anti-rabbit IgG Alexa Fluor 647 | Donkey | 1:500 | Jackson ImmunoResearch | AB_2340625 |
2.5 ∣. Combinations of transgenic lines and immunolabeling for quantifying cone type regeneration
It was not always possible to visualize all four cone types in a single retina. The cone labeling methods (transgenic lines and immunolabeling) used for quantifying cone density under different experimental conditions (Figure 7) are detailed here.
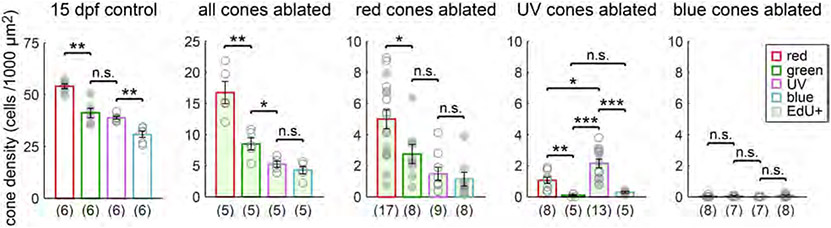
FIGURE 7.
Selective cone ablation induces biased and incomplete regeneration of specific cone types. Quantification of the densities of each cone type in control retinas, and of each cone type in the regenerated population (EdU-positive) after selective cone ablation. Plots show the mean population density across control and cone ablation conditions in 15 dpf (8 dpa) fish. Each circle represents one retina, with the numbers of retinas analyzed in parentheses. Because it was not technically feasible in most selective ablation conditions to visualize all cone populations together within a single retina, cone density measurements were pooled across retinas in which different cone populations were labeled. For each experimental condition, filled and open circles indicate measurements from transgenic lines in which different combinations of cone types were fluorescently labeled (see Section 2). Error bars are ± SEM. *P < .05, **P < .01, ***P < .001; Wilcoxon–Mann–Whitney rank sum test
15 dpf controls:
UV and blue cones visualized together in Tg(sws1:nfsBmCherry; sws2:GFP). Red and green cones visualized together in Tg(tr/β2:G4VP16; UAS:nfsBmCherry) with arrestin 3a immunostaining.
All cones ablated:
Red, green, UV, and blue cones visualized together using Tg(gnat2:nfsBmCherry; trβ2:MYFP) with immunostaining for arrestin 3a and UV-opsin.
Red cones ablated:
Red, green, and blue cones visualized together using Tg(trβ2:G4VP16; UAS:nfsBmCherry; sws2:GFP) with arrestin 3a immunostaining.
Red and UV cones visualized together using Tg(trβ2:G4VP16;UAS:nfsBmCherry, sws1:GFP).
UV cones ablated:
Red, green, and UV cones visualized together using Tg(sws1:nfsBmCherry; trβ2:tdTomato) with arrestin 3 a immunostaining.
2.6 ∣. Confocal image acquisition
Image stacks were acquired on a confocal microscope (Olympus FV1000 or Leica TCS SP8) using a 1.35 numerical aperture (NA) 60× oil (Olympus), 63× oil (1.4 NA) (Leica), 20× oil (0.85 NA) (Olympus), or a 20× oil (0.75 NA) (Leica) objective lens. Images were acquired at the following resolutions: high magnification images for orthogonal rotations, 0.18 μm per pixel XY and 0.3 μm Z step; identifying regenerated cone cell types, 0.18 μm per pixel XY and 0.5 μm Z step; whole retinas, between 0.1 and 0.4 μm per pixel XY, and 1 μm Z steps.
2.7 ∣. Image analysis
Image stacks were median filtered in Fiji (NIH) (Schindelin et al., 2012). Maximum intensity projections were generated in Amira (FEI). Three-dimensional (3D) image reconstructions were digitally sliced using the Amira slice functions. All measurements were made in Fiji. Image brightness, contrast, and hue were further adjusted in Photoshop (Adobe) or GIMP (GNU Image Manipulation Platform).
Cell densities across the retina were assessed by counting the number of labeled cells within an area in central, dorsal retina. In 10 dpf larvae, cones were counted within the area of a rectangle, 5,000 μm2. In 15 dpf larvae, cones were counted within the area of an oval, about 15,000 μm2.
2.8 ∣. Statistical analysis
Because it was not feasible in most experimental conditions to visualize all cone populations together in a single retina, quantitative data was pooled for statistical comparisons. In Figure 2b, a one-way ANOVA was used to test for differences in cone densities across cone ablation conditions. Pair-wise comparisons between cone ablation conditions were made using the Wilcoxon–Mann–Whitney rank sum test. In Figure 7, a one-way ANOVA was used to test for differences in cone densities across cone types in each ablation condition. Pair-wise comparisons between cone types were made using the Wilcoxon–Mann–Whitney rank sum test. In Table 4 (see Section 3), pair-wise comparisons between cone types in control populations and regenerated populations were made using the Wilcoxon–Mann–Whitney rank sum test. All statistical tests were performed using a significance level of 0.05. All quantitative measures are reported as the mean ± standard error (SEM).
FIGURE 2.
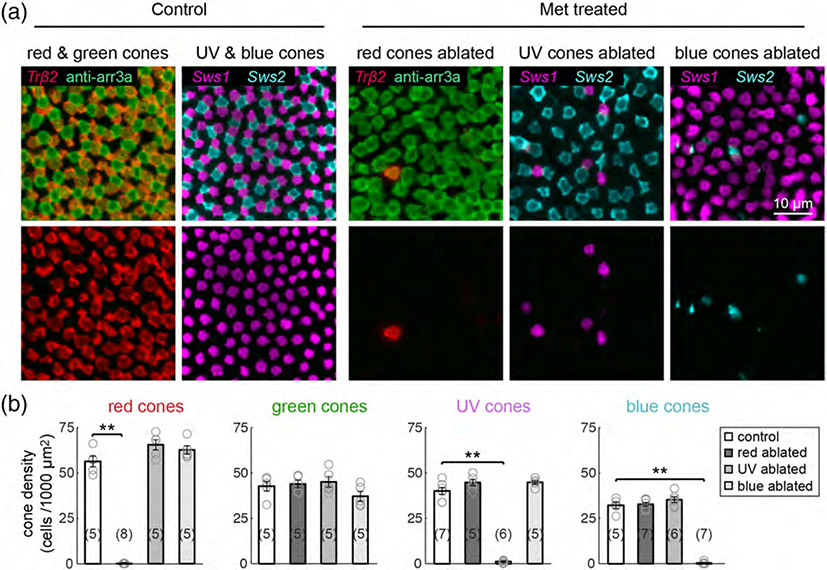
Selective cone ablation does not damage neighboring cones. Ablation of red, UV, or blue cone populations in the background of transgenically-labeled or immunostained cones. nfsB-expressing fish were crossed with the Tg(trβ2:tdTomato) line to visualize nontargeted red cones, with Tg(sws1:GFP) fish to visualize nontargeted UV cones, or with Tg(sws2:GFP) to visualize nontargeted blue cones. Anti-arrrestin3a immunostaining labels both red and green cones. (a) Maximum intensity projections of confocal image stacks from 10 dpf (3 dpa) control or Met-treated retinas. Met was applied for 1 hr at 7 dpf. (b) Plots show the mean cell density of each cone type from Met-treated larvae at 3 dpa, and from age-matched control fish. Each open circle represents a single retina, with the numbers of retinas analyzed shown in parentheses. Error bars are ± SEM. **p < .01; Wilcoxon–Mann–Whitney rank sum test
TABLE 4.
Comparison of cone population densities in control fish and fish with cone regeneration
| all ablated |
R ablated |
UV ablated |
B ablated |
|||||
|---|---|---|---|---|---|---|---|---|
| regen. R | regen. G | regen. UV | regen. B | regen. R | regen. UV | regen. B | ||
| Control | R | 0.004 | 0.004 | |||||
| G | 0.004 | |||||||
| UV | 0.004 | 0.00007 | ||||||
| B | 0.004 | 0.0007 | ||||||
Note: p-values from pair-wise comparisons of cone densities in 15 dpf unablated, control fish vs. regenerated (EdU-positive) cone densities in Met-treated fish, Wilcoxon–Mann–Whitney rank sum test. (“Regen.” regenerated, “R” red cones, “G” green cones, “UV” UV cones, and “B” blue cones).
3 ∣. RESULTS
3.1 ∣. Selective and pan in vivo ablation of cone photoreceptors in larval zebrafish retina
To gain a deeper understanding of the limits of cell type replacement during regeneration, we targeted specific photoreceptor populations for cell death using the Nitroreductase/Metrondazole technique. Expression of Nitroreductase (NTR; nfsB) can be genetically targeted to specific cell types, such that application of its prodrug Metronidazole (Met) only induces cytotoxicity in NTR-expressing cells (Curado et al., 2007). We investigated the efficacy of this approach to ablate the three cone photoreceptor populations for which transgenic tools are currently available: Tg(trβ2:G4VP16; UAS:nfsBmCherry) (red cones), Tg(sws1:nfsBmCherry) (UV cones), or Tg(sws2:nfsBmCherry) (blue cones) (Yoshimatsu et al., 2016). We treated red, UV, and blue cone ablation larvae with 10 mM Met for 1 hr at 7 days postfertilization (dpf) (Figure 1a), a stage by which retinal circuitry is functionally mature (Easter Jr & Nicola, 1996). The fusion of mCherry fluorescent protein (FP) to NTR facilitated visualization of each of the targeted populations. Examination of whole retinas from Met-treated larvae at 10 dpf, or 3 days postablation (dpa) (Figure 1a) revealed that only sparse, punctate mCherry signal remained in central regions (Figure 1b). The remaining FP-expressing cones in the retinal peripheral margin are likely cells that were generated after Met treatment, as the periphery hosts a stem cells niche that supports ongoing cell genesis (Lenkowski & Raymond, 2014). To confirm the specificity of NTR-induced ablation, we visualized neighboring cone populations in fixed tissue at 3 dpa (Figure 2a). The mosaic arrangement of nontargeted cone types was preserved after the ablation of red, UV, or blue cones (Figure 2a). Whereas the population densities of targeted cone types were almost completely diminished, the densities of each nontargeted cone population remained unchanged at 3 dpa (Figure 2b). Thus, brief Met treatment was effective in eliminating the majority of cells of each cone type, without killing neighboring cones via bystander effects.
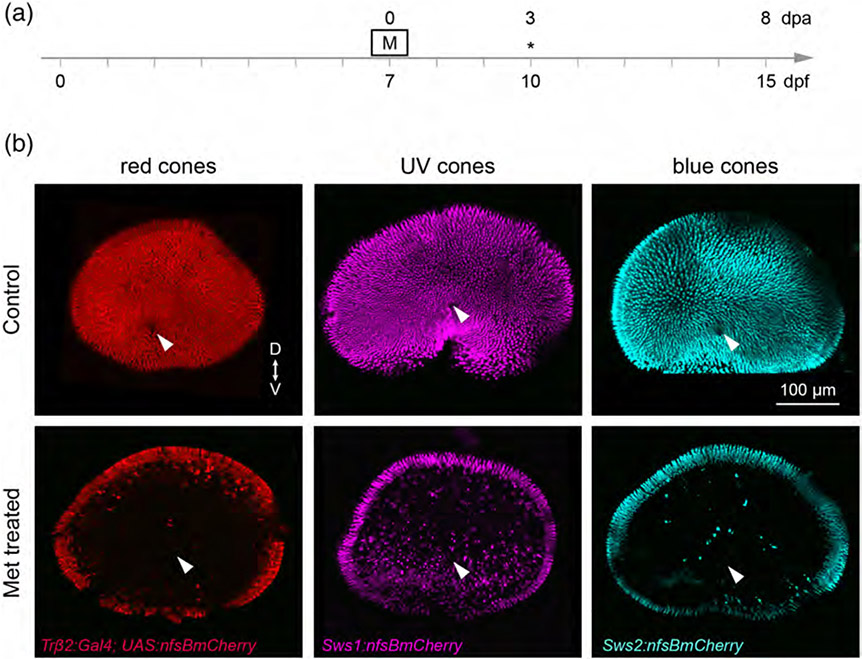
FIGURE 1.
Selective ablation of specific cone populations in larval zebrafish. (a) Timeline demonstrates timing of Met (M) treatment. Asterisks denotes the age at which larvae were fixed for analysis. (b) En face views of wholemount, fixed retinas from 10 dpf (3 dpa) Met-treated and control fish. Met was applied for 1 hr at 7 dpf. Specific cone populations were targeted for ablation by selective expression of nfsB. Tg(trβ2:G4VP16; UAS:nfsBmCherry) fish were used to ablate red cones, Tg(sws1:nfsBmCherry) fish were used to ablate UV cones, and the Tg(sws2:nfsBmCherry) line was used to ablate blue cones. Arrowheads denote the optic nerve head. (“D” dorsal, “V” ventral)
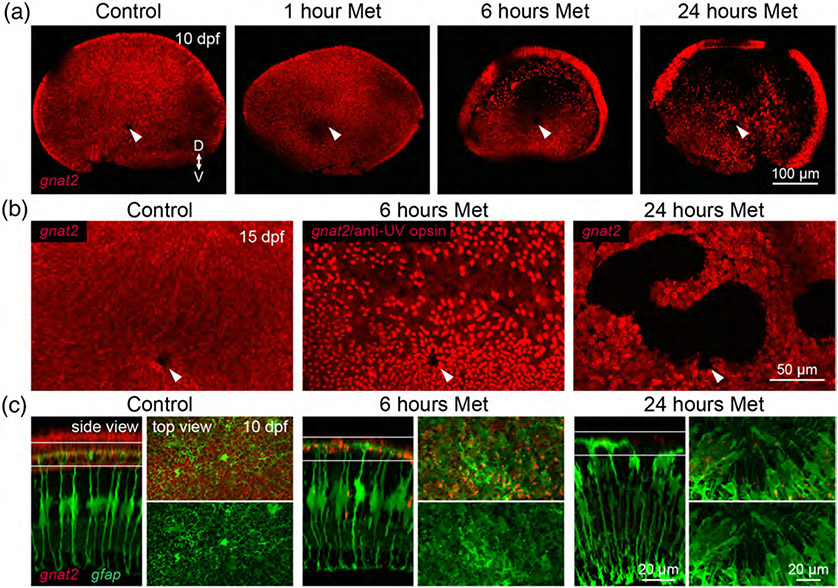
To gain further insight into the specificity and robustness of cone regeneration, we targeted the entire cone population for ablation. We generated the Tg(gnat2:nfsBmCherry) line, in which the guanine nucleotide binding protein, alpha transducing activity polypeptide 2 (gnat2) promoter (Kennedy et al., 2007) drives nfsB-mCherry expression in all cone types, and treated these fish with Met for 1 hr at 7 dpf as before (Figure 2a). Examination of whole retinas from Met-treated fish at 3 dpa revealed that brief Met treatment was insufficient to induce death of cone photoreceptors (Figure 3a). We therefore extended Met treatments to 6 or 24 hr. Whereas cone ablation was primarily restricted to dorsal retinal regions in fish treated with Met for 6 hr at 3 dpa, ablation spanned almost the entire retina in fish treated for 24 hr (Figure 3a). Moreover, analysis at a later stage (8 dpa) revealed that the ablation persisted in gnat2 fish that had been treated with Met for 24 hr. Cones repopulated dorsal regions of the retina by 8 dpa in fish treated for 6 hr, but in 24 hr-treated fish, large regions still completely lacked cones (Figure 3b). We hypothesized that the lack of cone repopulation may have resulted from damage to the cells that produce regenerative progenitors, Müller glia. Indeed, visualization of Müller cells at 3 dpa in 6 or 24 hr-treated larvae demonstrated that the Müller glia became disorganized after extended Met treatment (Figure 3c). There were apparent breaks in the outer limiting membrane, indicating that Müller glia had retracted their apical processes. This may be a consequence of the fact that Müller glia respond to photoreceptor death by phagocytosing the dead or dying cells (Bailey, Fossum, Fimbel, Montgomery, & Hyde, 2010; Morris, Scholz, Brockerhoff, & Fadool, 2008), which would potentially render Müller cells vulnerable to cytotoxic agents, and especially so when prodrug treatment is long-lasting. Thus, in order to ablate cone photoreceptors without inducing secondary damage that might inhibit cone repopulation, we proceeded with a 6-hr Met treatment paradigm in gnat2 fish.
FIGURE 3.
Optimizing Met treatment to ablate all cones without damaging Müller glia. (a) En face views of wholemount, fixed retinas from 10 dpf (3 dpa) Met-treated and control fish from the Tg(gnat2:nfsBmCherry) line. Fish were treated with Met for 1, 6, or 24 hr at 7 dpf. Arrowheads denote the optic nerve head. (“D” dorsal, “V” ventral). (b) High-magnification, en face views of dorsal retinal regions in 15 dpf control and Met-treated gnat2 larvae. In fish treated with Met for 6 hr, UV opsin immunolabeling was visualized in the same channel as gnat2 transgenic labeling. Arrowheads denote the optic nerve head. (c) Visualization of Müller glia in 10 dpf (3 dpa) larvae after different durations of Met treatment of Tg(gnat2:nfsBmCherry; gfap:GFP) double transgenic fish. (Side view) Orthogonal views of central retinal regions. Shown are images from control animals and from fish treated with Met for 6 or 24 hr. (Top view) En face views of the outer nuclear layer (ONL) together with Müller glia end feet, at the level indicated in the side views
3.2 ∣. Selective cone ablation triggers nonspecific cone genesis
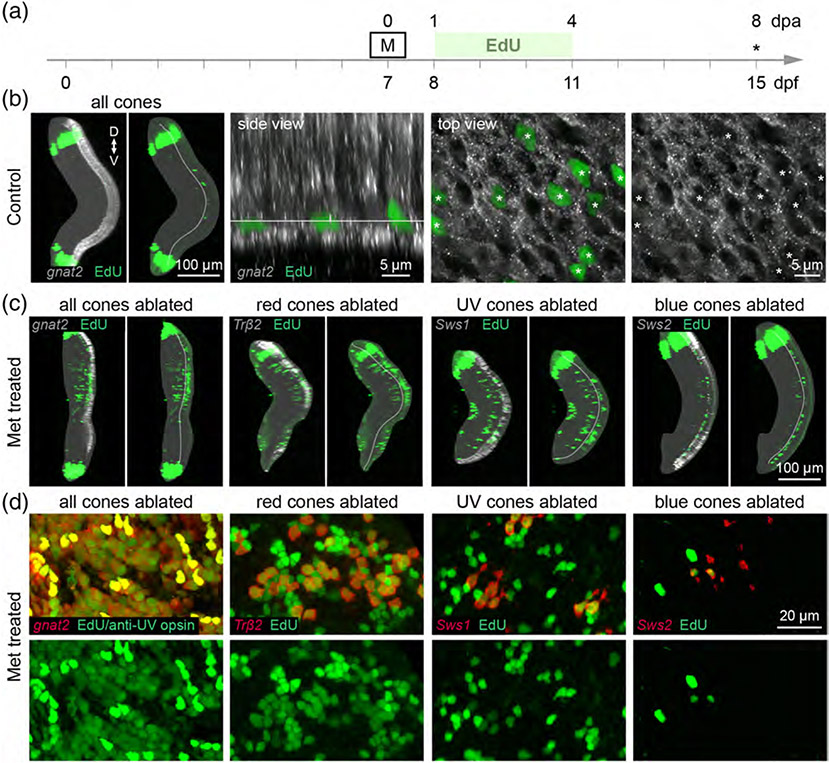
We next evaluated whether selective cone ablation only induced repopulation of the lost cone types. To unequivocally identify newly proliferated cones, we exposed control and cone-ablated larvae to the thymidine analog (2′S)-2′-Deoxy-2′-fluoro-5-ethynyluridine (EdU). Previous studies investigating regeneration after widespread photoreceptor loss in zebrafish reported that progenitor proliferation peaks between 1 and 4 dpa (Bernardos, Barthel, Meyers, & Raymond, 2007; Vihtelic & Hyde, 2000; Yoshimatsu et al., 2016). Further, a recent study examining cone regeneration after selective loss of UV cones in larvae demonstrated that repopulation plateaus between 7 and 10 dpa (Yoshimatsu et al., 2016). As such, we treated cone ablation fish with EdU from 1 to 4 dpa, before analysis of fixed, whole retinas at 8 dpa (Figure 4a). We observed that EdU sparsely labeled nuclei in the outer nuclear layer (ONL) of control larvae (Figure 4b); however, EdU never incorporated into cones, demonstrating that cone genesis is not ongoing in control fish (Figure 4b). Instead, the EdU-positive nuclei likely mark newborn rod progenitors or rods, as these photoreceptors are seeded into central retinal regions from late larval stages through adulthood (Stenkamp, 2011). In contrast to controls, EdU incorporation was robust in the ONL of cone-ablated fish, indicating that new photoreceptors were generated between 1 and 4 dpa in response to the death of all or select types of cone photoreceptors (Figure 4c,d). The level of EdU incorporation in the ONL appeared to occur to different extents, depending on the cone type that was ablated (Figure 4c,d). However, it was apparent that regenerated cone populations did not fully compensate for neuron loss, as they did not reach their original population densities (Figure 4d; Table 4). Regenerated cones often appeared in patches of clusters, such that they did not appear to localize in their typical mosaic organization in the ONL (compare Figure 4d with Figure 2a). We also observed mitotic label in the inner nuclear layer (INL) of cone ablation retinas, but not in control retinas (Figure 4c). The EdU-positive nuclei in undamaged layers may mark newly-generated inner retinal neurons, as observed after photoablation of all photoreceptors in adult zebrafish (Powell et al., 2016).
FIGURE 4.
Cone ablation induces proliferation of photoreceptors within days of cell death. (a) Timeline demonstrates the timing of Met and EdU treatment. Asterisk denotes age at which larvae were fixed for analysis. Met was applied for 1 hr at 7 dpf in select cone type-ablation; Met was applied for 6 hr at 7 dpf in all cones-ablation conditions. (b) Maximum intensity projections of confocal image stacks from 15 dpf control Tg(gnat2:nfsBmCherry) larvae that were treated with EdU. In the orthogonal view of the whole retina, the boundary between the ONL and inner nuclear layer (INL) is outlined in the panel showing EdU labeling alone. The dense bands of EdU-positive cells at the peripheral retina demarcate cells generated in the ciliary marginal zone at the time of EdU treatment. (Side view) Orthogonal rotation showing the photoreceptor layer. (Top view) En face views of photoreceptors together with EdU labeling, at the level indicated in the side view. Arrowheads point to EdU-positive nuclei. (c) Orthogonal views of whole eyes from 15 dpf (8 dpa) fish in which specific cone populations or all cone photoreceptors were ablated. The targeted cone population is visualized by nfsB-mCherry expression (gray scale), and shown together with EdU labeling (green). The boundary between the ONL and INL is outlined in the panels showing EdU labeling alone. (d) En face, high-magnification views showing EdU colabeling in the ONL of 15 dpf cone-ablated fish
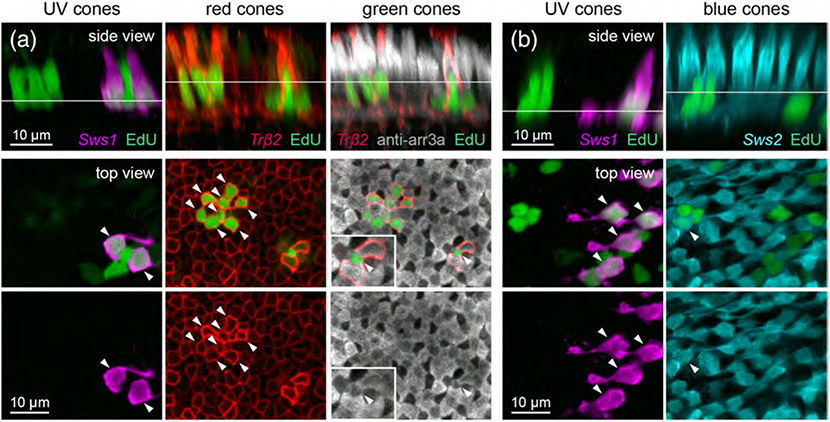
In addition to inducing the regeneration of lost cell type(s), the ablation of specific neurons can provoke the genesis of ectopic neuron types (Powell et al., 2016; Yoshimatsu et al., 2016). Indeed, EdU appeared to incorporate into nontargeted photoreceptors in the ONL, in addition to the ablated cone types (Figure 4d). To address this, we visualized nontargeted cones in retinas in which a specific cone type was eliminated. EdU labeling demonstrated that, in each case in which one cone type was ablated, other cone types were generated together with the ablated type. Figure 5 demonstrates how we determined the identities of cone types that were regenerated. In brief, to visualize nontargeted cone types, NTR-expressing transgenic fish were crossed with other transgenic lines in which specific cone types (red, UV, or blue) express FP (Figure 5a,b), and immunostained for arrestin 3a to label red and green cones (Figure 5a; see Section 2).
FIGURE 5.
Example of identification of regenerated cone types after UV cone ablation. Demonstration of how different cone types were distinguished in the population of regenerated photoreceptors after UV cone ablation. (a) Identification of regenerated UV, red, and green cones after UV cone ablation in 15 dpf (8 dpa) Tg(sws1:nfsBmCherry; trβ2:MYFP) double transgenic fish, with arrestin3a immunostaining and EdU labeling. Panels shown are images taken from the same region of tissue. (b) Identification of regenerated UV and blue cones after UV cone ablation in 15 dpf (8 dpa) Tg(sws1:nfsBmCherry; sws2:GFP) double transgenic fish with EdU labeling. Panels shown are images taken from the same region of tissue. In (a) and (b), side views are orthogonal rotations of the ONL from UV cone-ablated retinas. Top views show the nuclei located at the level of the line indicated in the side view. In order to visualize EdU-positive UV cones, a different plane of section was visualized because UV cone cell bodies reside in a lower plane compared to other cone types. Arrowheads point to EdU-positive nuclei
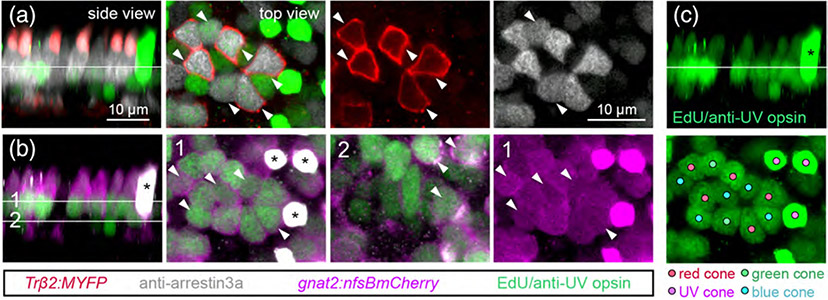
We also evaluated the composition of regenerated populations after ablation of all cone photoreceptor types, as shown in Figure 6. We visualized red cones by crossing Tg(gnat2:nfsBmCherry) fish with the Tg(trβ2:MYFP) line, and distinguished green and UV cones by immunostaining for arrestin 3a and UV-opsin, respectively (Figure 6a, b). Blue cones were identified by the process of elimination; after identifying gnat2-positive cells that colabeled with trβ2, arrestin 3a, or UV-opsin, any remaining gnat2-positive photoreceptors were classified as blue cones (Figure 6b). Thus, in a single larva, we could distinguish which of the EdU-positive nuclei were associated with red, green, UV, or blue cones after cone photoreceptor death and regeneration (Figure 6c).
FIGURE 6.
Identification of regenerated cone types after ablation of all cones. Demonstration of how each cone type was distinguished in retinas from 15 dpf (8 dpa) Met-treated Tg(gnat2:nfsBmCherry; trβ2:MYFP) double transgenic larvae. (a) Identification of regenerated red and green cones. Trβ2 labels red cones alone, and anti-arrestin3a labels both red and green cones (gray). EdU and anti-UV opsin were visualized in the same channel (green). (b) Identification of regenerated UV and blue cones. gnat2 labels all cone types (magenta), anti-UV opsin labels UV cones (green). Line 1 in the side view marks the level at which blue cone nuclei were visualized for top views, line 2 marks the level at which UV cone nuclei were visualized for top views. (c) Representative map of different cone types in the regenerated population. Filled circles denote the identity of each EdU-positive nucleus. In A-C, side views are orthogonal rotations of the ONL; top views show the nuclei located at the level of the line indicated in the side view. Arrowheads point to EdU-positive cones, asterisks mark UV opsin-positive cones
3.3 ∣. Cone type-specific proliferative advantages and biases in cone regeneration
Quantitation of the composition of the regenerated population revealed that specific cone types appear to hold a proliferative advantage, depending on the identity of the population ablated. The robustness of cone regeneration roughly correlated with the density of the population ablated; all > red > UV > blue cone types (Figure 7). Global cone death induced regeneration of all cone types, and the relative abundance of regenerated cells of each type approximates the normal hierarchy: red > green > UV blue cone (Figure 7). However, the stereotypic relationship between the densities of each cone type was not preserved in regeneration; whereas cones in control retinas are present in a ratio of 1.8 red:1.3 green:1.3 UV:1 blue cone, regenerated cones in retinas with general cone death appear in a ratio of 3.9 red:2 green:1.2 UV:1 blue cone on average. Thus, genesis was especially skewed toward red and green cones after ablation of all cones. Like retinas in which all cones were ablated, ablation of red cones induced genesis of all cone types (Figure 7). However, regenerated red cones appeared in the highest density, demonstrating that regeneration was biased toward red cones after their selective ablation. Likewise, UV cone ablation biased regeneration toward UV cones. Other cone types were also generated after UV cone ablation; new red and blue cones were consistently produced, but green cones were only sometimes generated, and in extremely low numbers. Finally, as reported previously, ablation of the blue cone population induced a very weak regenerative response (Yoshimatsu et al., 2016). Although EdU labeling was evident in the ONL of blue-ablated retinas (Figure 4c,d), we observed that ablating blue cones only stimulated cone genesis in half the retinas analyzed (4/8 retinas). No UV cones incorporated EdU, and EdU-positive blue, red, or green cones were present in approximately equal numbers (EdU-positive blue cone density = 0.01 ± 0.03 cells per 1,000 μm2, n = 8 retinas; EdU-positive red cone density = 0.02 ± 0.02 cells per 1,000 μm2, n = 8 retinas; EdU-positive green cone density = 0.02 ± 0.02 cells per 1,000 μm2, n = 7 retinas) (Figure 7). Repeating blue cone ablation in rod-labeled transgenics (Tg(sws2:nfsBmCherry; xops:GFP)) demonstrated that the majority of the EdU-positive cells in the ONL at 8 dpa were rod photoreceptors (EdU-positive cell density in the ONL of blue-ablated retinas = 5.49 ± 1.05 cells per 1,000 μm2, n = 8 retinas; EdU-positive rod density in blue-ablated retinas = 5.37 ± 1.06 cells per 1,000 μm2, n = 8 retinas; p =.75, Wilcoxon–Mann–Whitney rank sum test.) Further, blue cone loss did not increase rod genesis in lieu of replacing lost blue cones (EdU-positive rod density in blue-ablated retinas = 5.37 ± 1.06 cells per 1,000 μm2, n = 8 retinas; EdU-positive rod density in control retinas = 3.76 ± 1.3 cells per 1,000 μm2, n = 7 retinas; p =.28, Wilcoxon–Mann–Whitney rank sum test; Figure S1). Thus, it appears that unlike fish in which red or UV cones were ablated, photoreceptor regeneration was not skewed toward the targeted cone type in blue cone-ablated larvae. Furthermore, blue cone ablation did not increase the level of normal, ongoing rod genesis in lieu of cone regeneration.
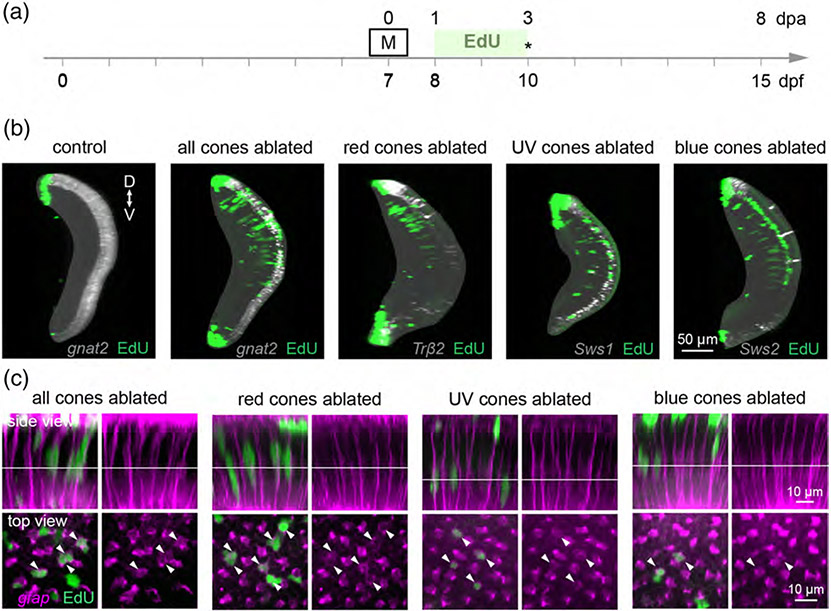
3.4 ∣. Müller glia proliferate in response to global and selective cone death
Several studies in fish retinal regeneration have suggested that a minimum number of cells must die in order to stimulate Müller glia-mediated regeneration (Braisted & Raymond, 1992; Montgomery, Parsons, & Hyde, 2010). In a recent investigation, UV cones were selectively ablated by light lesioning in adult zebrafish, and although Müller glia upregulated markers of dedifferentiation, they did not incorporate mitotic markers (Nagashima, Barthel, & Raymond, 2013). We thus wondered whether the loss of blue cones, the least abundant cone type in the larval retina, was insufficient to induce proliferation of Müller glia. To test this hypothesis, we treated control and cone-ablated larvae with EdU from 1–3 dpa, and fixed at 3 dpa in order to detect any EdU-positive Müller glia before the end of the proliferation period (Figure 8a). Visualizing EdU labeling across the whole retina in control larvae showed that as expected, there was little to no EdU incorporation in the inner retina (Figure 8b). In contrast, there was robust EdU incorporation in the INL of retinas from larvae in which all cones, or only red, UV, or blue cones were ablated. Across ablation paradigms, EdU-positive nuclei appeared in columns, reminiscent of the neurogenic columns commonly observed during Müller glia-mediated regeneration (Lenkowski & Raymond, 2014; Figure 8b). To confirm that these columns were associated with proliferating Müller glia, we visualized Müller glia by crossing cone ablation fish with the Tg(gfap:GFP) line. Indeed, EdU colabeled the nuclei of gfap-positive nuclei after ablation of all or only red, UV, or blue cones (Figure 8c). We therefore conclude that the loss of any cone type population in larval zebrafish is sufficient to provoke Müller glia proliferation. However, the death of a single cone population is not always sufficient to induce production of photoreceptors.
FIGURE 8.
Müller glia undergo cell division in response to death of cone populations, regardless of type. (a) Timeline demonstrates the timing of EdU application after Met treatment in cone-ablated fish. Asterisk denotes the age at which larvae were fixed for analysis. Met was applied for 1 hr at 7 dpf in select cone type-ablation; Met was applied for 6 hr at 7 dpf in all cones-ablation conditions. (b) Orthogonal rotations of whole retinas from 10 dpf control and cone-ablated fish treated with EdU: Tg(gnat2:nfsBmCherry) (all cones), Tg(trβ2:G4VP16; UAS:nfsBmCherry) (red cones), Tg(sws1:nfsBmCherry) (UV cones), and Tg(sws2:nfsBmCherry) (blue cones). (“D” dorsal, “V” ventral). (c) Cone-ablated fish were crossed with Tg(gfap:GFP) to visualize Müller glia for EdU colabeling. (Side view) Orthogonal rotations showing Müller glia together with EdU labeling. (Top view) En face views of EdU-positive nuclei, taken at the level indicated in the side views. Arrowheads mark EdU-positive nuclei
4 ∣. DISCUSSION
4.1 ∣. Determinants of the composition of regenerated neuronal populations
It is clear that even in a system with the native capacity to regenerate diverse neuronal cell types, regeneration is not selective. After ablating single cone types, we found that in addition to the ablated cohort, other cone types were also regenerated. Our observation is in line with previous work in which specific neuron types (D'Orazi, Zhao, Wong, & Yoshimatsu, 2016; Kei, Currie, & Jusuf, 2017; Yoshimatsu et al., 2016) or select retinal layers (Powell et al., 2016; Raymond, Barthel, Bernardos, & Perkowski, 2006) in zebrafish were ablated, which resulted in genesis of nonablated neuronal cell types. The “non-ablated” cell types can persist until at least 30 days postablation (Powell et al., 2016), suggesting that excess neurons are not later culled back. As such, even in an animal capable of endogenous regeneration, neuron replacement is imprecise. Such imprecision may introduce a negative impact on retinal function and organization, and thus underscores the need to understand the determinants of cell fate and proliferation in regeneration, to better tailor repair to specifically replace the lost cell type or types.
Our quantitative analysis of cone regeneration in larval zebrafish after eliminating a single cone type (red, UV or blue) suggests that cell replacement is biased toward the ablated cell type (see also [Fraser, DuVal, Wang, & Allison, 2013; Kei et al., 2017]). Similar conclusions were made by studies of adult zebrafish and larval frogs. Ablation of select retinal layers in adult zebrafish biased regenerative proliferation toward the affected layer(s) (Powell et al., 2016). Also, ablation of specific amacrine cells or inner retinal neuronal subsets in the developing frog retina biased progenitors toward producing the ablated neuronal cell type over others (Reh, 1987; Reh & Tully, 1986). Recent work in zebrafish larvae further suggests that this bias can change as regeneration progresses over time (Kei et al., 2017). Our observations here add to the current notion that neuronal progenitors “detect” which cell type is lost after retinal injury in a dynamic manner, as cells are replaced. Even though the mechanisms by which regeneration is biased toward the lost neuronal population are still unknown, clues come from studies of neurogenesis and cell fate specification during retinal development (Agathocleous & Harris, 2009; Bassett & Wallace, 2012; Brzezinski & Reh, 2015; Cayouette, Poggi, & Harris, 2006).
There are several potential mechanisms by which biased regeneration may occur. First, cell fate specification of neuronal progenitors may be biased toward the ablated neuronal population. Although cell fate specification is largely controlled by a hierarchy of cell-intrinsic transcription factors (Boije, MacDonald, & Harris, 2014), extrinsic cues impinge upon intrinsic programs, often by regulating the timing of cell cycle exit and differentiation in neuroepithelial-derived progenitors. Several of these signaling pathways are activated during regeneration, such as Notch signaling (Lenkowski & Raymond, 2014). Further, extrinsic signals that specify cell fate during vertebrate retinal development, including retinoic acid and thyroid hormone, bind ligand-regulated transcription factors to promote the differentiation of rods and cones, respectively (Sernagor, Eglen, Harris, & Wong, 2006; Swaroop, Kim, & Forrest, 2010). Such extrinsic signals may be redeployed in regeneration to influence cell fate specification, but as yet remain largely unexplored.
Another potential mechanism that could bias neuronal regeneration toward specific cell types is the selective proliferation of fate-restricted progenitors or precursors. Indeed, differentiated neurons in the developing retina can limit their population size via negative feedback signaling, which inhibits the proliferation of particular fate-restricted progenitors (44). For example, RGCs secrete SHH to inhibit the proliferation of progenitor cells during the early retinal stages when RGCs are typically produced in mice (Wang, Dakubo, Thurig, Mazerolle, & Wallace, 2005). Depletion of RGCs from the developing mouse retina results in an increased proportion of the progenitor pool expressing transcription factors necessary for RGC fate, indicating that negative feedback is cell type-specific (Mu et al., 2005). It has been hypothesized that a similar mechanism maintains homeostasis in the mature zebrafish retina, such that ablation of particular populations disinhibits the proliferation of specific progenitors (Powell et al., 2016). Surviving neurons from nontargeted populations may continue to provide negative feedback, and thus act to reduce the generation of ectopic cell types during regeneration. Direct evidence for the proliferation of select progenitor types during regeneration comes from studies of salamander midbrain in which dopaminergic neuron populations are normally fully restored after selective chemical ablation, but their regeneration is blocked when pharmacological agents are applied to compensate for the loss of dopaminergic signaling after neuronal cell death (Berg et al., 2011). There is also recent evidence showing that there are dedicated precursors for each cone type during zebrafish retinal development (Suzuki et al., 2013), providing a potential foundation for the selective regulation of progenitor proliferation during cone regeneration. However, unlike salamander dopaminergic neurons, it is unlikely that each cone type regulates proliferation of its specific precursor via neurotransmitter signaling because all cone types release the same neurotransmitter, glutamate.
Finally, it is possible that selective cell death plays a role in sculpting the composition of the regenerated cone population. Our observations do not exclude the potential scenario in which all cone types are generated after ablation of any distinct cone population, but only a subset of regenerated neuronal types survive. It is evident that there is upregulation of cell death signaling pathways upon cell loss (Gorsuch & Hyde, 2014; Lenkowski & Raymond, 2014). These pathways are often sufficient to initiate regeneration, but all so far appear to be “general” death signals that are unlikely to convey signals to specify select cell fates (Nelson et al., 2013). Differential expression of Fgf signaling components in disparate retinal layers in larval and adult zebrafish retinas, however, has been suggested to provide a framework for layer-specific death signaling (Gorsuch & Hyde, 2014). How such diffusible cues could mediate the specification of cells intermingling within a single layer, like the various cone photoreceptors, is unclear. In the developing chick retina, differentiated, stratified RGCs secrete the neurotrophin NGF, which induces the death of RGCs still migrating into the ganglion cell layer via activation of the p75NTR receptor and downstream pro-apoptotic pathways (Frade, Rodríguez-Tébar, & Barde, 1996; González-Hoyuela, Barbas, & Rodríguez-Tébar, 2001). Neuronal populations can therefore regulate their numbers via targeted death of homotypic, postmitotic cells. In the future, assessing whether or not cell death is elevated after regeneration is complete, and whether ectopic, nonablated cones persist, will provide insight into the extent to which cell death dictates the ultimate composition of regenerated neuronal populations. Further studies identifying cues that control the generation of the various cone types during normal development will also be highly useful in understanding the regeneration program.
The mechanisms discussed above are not mutually exclusive; retinal regeneration likely involves to some extent the orchestrated regulation of progenitor proliferation, fate specification, and neuron survival. However, it is clear that the spatially and temporally coordinated mechanisms that generate retinal neurons in the correct numbers and proportions during development are not perfectly recapitulated after selective ablation in mature circuits. The hierarchy of relative cone type abundances is approximately recaptured after pan cone ablation and regeneration. However, none of the cone types recover their original population densities, as previously observed in adult zebrafish (Raymond & Barthel, 2004; Stenkamp & Cameron, 2002). Likewise, the targeted cone populations do not fully repopulate after single cone population elimination, and as a result, the regenerated cone populations fail to organize into their typical mosaic arrangements. Understanding the relative contributions of selective repopulation versus selective maintenance of new cells during regeneration will be critical to specifying regeneration to appropriately replace lost cell types.
4.2 ∣. Activation of Müller glia-mediated cone regeneration
By selectively ablating specific cone types, we found that photoreceptor regeneration is “conditional.” Whereas ablating red or UV cone populations alone induces robust to moderate cone regeneration, blue cone death only sometimes induces cone regeneration, and only to an extremely weak degree. It is unlikely that these differences arise from a limitation in the competency of Müller glia-derived progenitors, because previous work has clearly demonstrated that Müller glia-derived progenitors are multipotent, that is, capable of differentiating into each of the major retinal neuron classes (Fausett & Goldman, 2006; Goldman, 2014; Ramachandran, Reifler, Parent, & Goldman, 2010). In our current and past work, we find that new blue cones are certainly produced after the ablation of all cones, or of red or UV cones (Yoshimatsu et al., 2016). Thus, the failure to reproduce cones after the ablation of blue cones raises the question of what factors dictate whether or not Müller glia-mediated regeneration to replace specific lost neuronal cell types is initiated.
Some of the earliest investigators of teleost retinal regeneration posited that substantial cell death is a major factor in triggering a robust regeneration response from Müller glia (Braisted & Raymond, 1992). Ablation of different proportions of a single neuron type, rod photoreceptors, suggested that Müller glia are sensitive to the extent of cell death (Montgomery et al., 2010). Comparisons of retinas in which the cone and rod photoreceptor populations were ablated to different degrees by light lesioning also revealed that the proportion of Müller glial cells that undergo cell division, as well as the extent of progenitor proliferation, corresponds to the magnitude of photoreceptor death (Thomas, Nelson, Luo, Hyde, & Thummel, 2012). Similarly, we found that the extent of cone regeneration roughly correlated with the normal density of the ablated population, that is, all cone types > red cones > UV cones > blue cones. However, the normal densities of UV versus blue cones only differ slightly: 1.3 UV cones: 1 blue cone. Thus, it may be challenging to pinpoint if a simple threshold of cell loss determines whether cone regeneration is triggered or not.
We found that although Müller glia underwent mitosis in response to the death of blue cones, cones were not produced. Why? Müller glia act as stem cells under two different circumstances; in addition to producing neuronal progenitors in response to cell loss, they support ongoing rod genesis in intact circuits by undergoing slow asymmetric divisions that generate new rod precursors (Bernardos et al., 2007). Thus, Müller glia-derived progenitors must undergo a fate switch in order to support cone regeneration (Fraser et al., 2013). As such, we hypothesized that blue cone death may be insufficient to drive the rod-to-cone competency change in Müller glia-derived progenitors. However, despite the fact that Müller glia increase their rate of division after blue cone loss, we observed blue cone death did not increase the normal production of rods at the expense of cone regeneration. It remains to be seen whether or not neuronal progenitors are instead biased toward nonphotoreceptor fates after selective loss of a single photoreceptor population. Further, it remains possible that cone regeneration only occurs after the death of nonblue cone types because of differences in the origin, not the extent, of cell death.
In conclusion, retinal regeneration appears to be inherently limited after the loss of select neuronal populations. These limitations are especially pertinent to retinal diseases that affect specific retinal neuron types, such as retinitis pigmentosa, an inherited retinal degeneration disorder that primarily affects photoreceptors (Hamel, 2006). Our current findings underscore the need not only to understand the differences between mammalian and teleost systems that dictate whether regeneration is initiated, but also to understand what factors, intrinsic and extrinsic, control the extent and specificity of neuronal regeneration. Of particular interest are the potential intrinsic differences between the neuroepithelial-derived progenitors that support retinal development and the Müller glia-derived progenitors produced in regeneration. Challenges may also arise in recapitulating retinal neurogenesis based on differences in the retinal environment at different stages of maturation.
Supplementary Material
ACKNOWLEDGMENTS
Support from the National Institutes of Health: Vision Training Grant EY07031 and Developmental Biology Training Grant HD07183 to Florence D. D'Orazi, EY14358 to Rachel O. Wong, and Vision Core Grant EY01730 to M. Neitz. We thank Jeremy Nathans for providing the anti-UV opsin antibody; David Raible and Thomas Reh for critical reading of the manuscript in its early stages; Kaori Oda and Mei Zhang for technical assistance, and members of Rachel O. Wong's lab for helpful discussions.
Funding information
Center for Scientific Review, Grant/Award Numbers: EY007031, EY01730, EY14358, HD07183
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
CONFLICT OF INTEREST
The authors declare that there are no potential sources of conflict of interest.
DATA AVAILABILITY STATEMENT
The authors will make available the original data (images) used for generating the results presented in this manuscript upon request to the corresponding authors.
REFERENCES
- Agathocleous M, & Harris WA (2009). From progenitors to differentiated cells in the vertebrate retina. Annual Review of Cell and Developmental Biology, 25, 45–69. 10.1146/annurev.cellbio.042308.113259 [DOI] [PubMed] [Google Scholar]
- Allison WT, Barthel LK, Skebo KM, Takechi M, Kawamura S, & Raymond PA (2010). Ontogeny of cone photoreceptor mosaics in zebrafish. The Journal of Comparative Neurology, 518(20), 4182–4195. 10.1002/cne.22447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TJ, Fossum SL, Fimbel SM, Montgomery JE, & Hyde DR (2010). The inhibitor of phagocytosis, O-phospho-L-serine, suppresses Müller glia proliferation and cone cell regeneration in the light-damaged zebrafish retina. Experimental Eye Research, 91(5), 601–612. 10.1016/j.exer.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, & Wallace VA (2012). Cell fate determination in the vertebrate retina. Trends in Neurosciences, 35(9), 565–573. 10.1016/j.tins.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Berg DA, Kirkham M, Beljajeva A, Knapp D, Habermann B, Ryge J, … Simon A (2011). Efficient regeneration by activation of neurogenesis in homeostatically quiescent regions of the adult vertebrate brain. Development, 138(1), 180–180. 10.1242/dev.061754 [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, & Raymond PA (2007). Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 27(26), 7028–7040. 10.1523/JNEUROSCI.1624-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, & Raymond PA (2006). GFAP transgenic zebrafish. Gene Expression Patterns: GEP, 6(8), 1007–1013. 10.1016/j.modgep.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Boije H, MacDonald RB, & Harris WA (2014). Reconciling competence and transcriptional hierarchies with stochasticity in retinal lineages. Current Opinion in Neurobiology, 27, 68–74. 10.1016/j.conb.2014.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braisted JE, & Raymond PA (1992). Regeneration of dopaminergic neurons in goldfish retina. Development (Cambridge, England), 114(4), 913–919. [DOI] [PubMed] [Google Scholar]
- Brzezinski JA, & Reh TA (2015). Photoreceptor cell fate specification in vertebrates. Development (Cambridge, England), 142(19), 3263–3273. 10.1242/dev.127043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M, Poggi L, & Harris WA (2006). Lineage in the vertebrate retina. Trends in Neurosciences, 29(10), 563–570. 10.1016/j.tins.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, & Stainier DYR (2007). Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Developmental Dynamics, 236(4), 1025–1035. 10.1002/dvdy.21100 [DOI] [PubMed] [Google Scholar]
- D'Orazi FD, Suzuki SC, & Wong RO (2014). Neuronal remodeling in retinal circuit assembly, disassembly, and reassembly. Trends in Neurosciences, 37(10), 594–603. 10.1016/j.tins.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazi FD, Zhao X-F, Wong RO, & Yoshimatsu T (2016). Mismatch of synaptic patterns between neurons produced in regeneration and during development of the vertebrate retina. Current Biology: CB, 26 (17), 2268–2279. 10.1016/j.cub.2016.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, … Parsons MJ (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Developmental Biology, 304(2), 811–824. 10.1016/j.ydbio.2007.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demb JB, & Singer JH (2015). Functional circuitry of the retina. Annual Review of Vision Science, 1(1), 263–289. 10.1146/annurev-vision-082114-035334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter SS Jr., & Nicola GN (1996). The development of vision in the zebrafish (Danio rerio). Developmental Biology, 180(2), 646–663. [DOI] [PubMed] [Google Scholar]
- Fadool J, & Dowling J (2008). Zebrafish: A model system for the study of eye genetics. Progress in Retinal and Eye Research, 27(1), 89–110. 10.1016/j.preteyeres.2007.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadool JM (2003). Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Developmental Biology, 258(2), 277–290. 10.1016/S0012-1606(03)00125-8 [DOI] [PubMed] [Google Scholar]
- Fausett BV, & Goldman D (2006). A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. Journal of Neuroscience, 26(23), 6303–6313. 10.1523/JNEUROSCI.0332-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, & Reh TA (2003). Potential of Müller glia to become neurogenic retinal progenitor cells. Glia, 43(1), 70–76. 10.1002/glia.10218 [DOI] [PubMed] [Google Scholar]
- Frade JM, Rodríguez-Tébar A, & Barde YA (1996). Induction of cell death by endogenous nerve growth factor through its p75 receptor. Nature, 383(6596), 166–168. 10.1038/383166a0 [DOI] [PubMed] [Google Scholar]
- Fraser B, DuVal MG, Wang H, & Allison WT (2013). Regeneration of cone photoreceptors when cell ablation is primarily restricted to a particular cone subtype. PLoS One, 8(1), e55410. 10.1371/journal.pone.0055410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D (2014). Müller glial cell reprogramming and retina regeneration. Nature Reviews Neuroscience, 15(7), 431–442. 10.1038/nrn3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Hoyuela M, Barbas JA, & Rodríguez-Tébar A (2001). The autoregulation of retinal ganglion cell number. Development (Cambridge, England), 128(1), 117–124. [DOI] [PubMed] [Google Scholar]
- Gorsuch RA, & Hyde DR (2014). Regulation of Müller glial dependent neuronal regeneration in the damaged adult zebrafish retina. Experimental Eye Research, 123, 131–140. 10.1016/j.exer.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel C (2006). Retinitis pigmentosa. Orphanet Journal of Rare Diseases, 1, 40. 10.1186/1750-1172-1-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon M, Okawa H, Della Santina L, & Wong ROL (2014). Functional architecture of the retina: Development and disease. Progress in Retinal and Eye Research, 42, 44–84. 10.1016/j.preteyeres.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, … Reh TA (2017). Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature, 548(7665), 103–107. 10.1038/nature23283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl MO, & Reh TA (2010). Regenerative medicine for retinal diseases: Activating endogenous repair mechanisms. Trends in Molecular Medicine, 16(4), 193–202. 10.1016/j.molmed.2010.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kei JNC, Currie PD, & Jusuf PR (2017). Fate bias during neural regeneration adjusts dynamically without recapitulating developmental fate progression. Neural Development, 12(1), 12. 10.1186/s13064-017-0089-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BN, Alvarez Y, Brockerhoff SE, Stearns GW, Sapetto-Rebow B, Taylor MR, & Hurley JB (2007). Identification of a zebrafish cone photoreceptor-specific promoter and genetic rescue of achromatopsia in the nof mutant. Investigative Ophthalmology & Visual Science, 48(2), 522–529. 10.1167/iovs.06-0975 [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME,Campbell DS, … Chien C-B (2007). The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 236(11), 3088–3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Larison KD, & Bremiller R (1990). Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development, 109(3), 567–576. 10.1016/j.preteyeres.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Lenkowski JR, & Raymond PA (2014). Müller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Progress in Retinal and Eye Research, 40, 94–123. 10.1016/j.preteyeres.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Williams J, Smallwood PM, Touchman JW, Roman LM, & Nathans J (2004). Proximal and distal sequences control UV cone pigment gene expression in transgenic zebrafish. The Journal of Biological Chemistry, 279(18), 19286–19293. 10.1074/jbc.M400161200 [DOI] [PubMed] [Google Scholar]
- Masland RH (2012). The neuronal Organization of the Retina. Neuron, 76(2), 266–280. 10.1016/j.neuron.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery JE, Parsons MJ, & Hyde DR (2010). A novel model of retinal ablation demonstrates that the extent of rod cell death regulates the origin of the regenerated zebrafish rod photoreceptors. The Journal of Comparative Neurology, 518(6), 800–814. 10.1002/cne.22243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AC, Scholz TL, Brockerhoff SE, & Fadool JM (2008). Genetic dissection reveals two separate pathways for rod and cone regeneration in the teleost retina. Developmental Neurobiology, 68(5), 605–619. 10.1002/dneu.20610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Liang S, Maeda H, Frishman LJ, & Klein WH (2005). Ganglion cells are required for normal progenitor- cell proliferation but not cell-fate determination or patterning in the developing mouse retina. Current Biology: CB, 15(6), 525–530. 10.1016/j.cub.2005.01.043 [DOI] [PubMed] [Google Scholar]
- Nagashima M, Barthel LK, & Raymond PA (2013). A self-renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development (Cambridge, England), 140(22), 4510–4521. 10.1242/dev.090738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Ackerman KM, O'Hayer P, Bailey TJ, Gorsuch RA, & Hyde DR (2013). Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Muller glia proliferation during zebrafish retinal regeneration. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 33(15), 6524–6539. 10.1523/JNEUROSCI.3838-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C, Cornblath E, Elsaeidi F, Wan J, & Goldman D (2016). Zebrafish Müller glia-derived progenitors are multipotent, exhibit proliferative biases and regenerate excess neurons. Scientific Reports, 6, 24851. 10.1038/srep24851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Reifler A, Parent JM, & Goldman D (2010). Conditional gene expression and lineage tracing of tuba1a expressing cells during zebrafish development and retina regeneration. The Journal of Comparative Neurology, 518(20), 4196–4212. 10.1002/cne.22448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond PA, & Barthel LK (2004). A moving wave patterns the cone photoreceptor mosaic array in the zebrafish retina. The International Journal of Developmental Biology, 48(8–9), 935–945. 10.1387/ijdb.041873pr [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, & Perkowski JJ (2006). Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Developmental Biology, 6, 36. 10.1186/1471-213X-6-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reh TA (1987). Cell-specific regulation of neuronal production in the larval frog retina. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 7(10), 3317–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reh TA, & Tully T (1986). Regulation of tyrosine hydroxylase-containing amacrine cell number in larval frog retina. Developmental Biology, 114(2), 463–469. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M,Pietzsch T, … Cardona A (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9(7), 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sernagor E, Eglen S, Harris B, & Wong R (2006). Retinal Development (1st ed.). Cambridge: Cambridge University Press. [Google Scholar]
- Stenkamp DL (2011). The rod photoreceptor lineage of teleost fish. Progress in Retinal and Eye Research, 30(6), 395–404. 10.1016/j.preteyeres.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp DL, & Cameron DA (2002). Cellular pattern formation in the retina: Retinal regeneration as a model system. Molecular Vision, 8, 280–293. [PubMed] [Google Scholar]
- Suzuki SC, Bleckert A, Williams PR, Takechi M, Kawamura S, & Wong ROL (2013). Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proceedings of the National Academy of Sciences of the United States of America, 110 (37), 15109–15114. 10.1073/pnas.1303551110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Kim D, & Forrest D (2010). Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nature Reviews Neuroscience, 11(8), 563–576. 10.1038/nrn2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi M, Hamaoka T, & Kawamura S (2003). Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish. FEBS Letters, 553(1–2), 90–94. 10.1016/S0014-5793(03)00977-3 [DOI] [PubMed] [Google Scholar]
- Takechi M, Seno S, & Kawamura S (2008). Identification of cis-acting elements repressing blue opsin expression in zebrafish UV cones and pineal cells. Journal of Biological Chemistry, 283(46), 31625–31632. 10.1074/jbc.M806226200 [DOI] [PubMed] [Google Scholar]
- Thomas JL, Nelson CM, Luo X, Hyde DR, & Thummel R (2012). Characterization of multiple light damage paradigms reveals regional differences in photoreceptor loss. Experimental Eye Research, 97(1), 105–116. 10.1016/j.exer.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, & Hyde DR (2000). Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. Journal of Neurobiology, 44(3), 289–307. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, & Wallace VA (2005). Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development (Cambridge, England), 132(22), 5103–5113. 10.1242/dev.02096 [DOI] [PubMed] [Google Scholar]
- Wilken MS, & Reh TA (2016). Retinal regeneration in birds and mice. Current Opinion in Genetics & Development, 40, 57–64. 10.1016/j.gde.2016.05.028 [DOI] [PubMed] [Google Scholar]
- Yoshimatsu T, D'Orazi FD, Gamlin CR, Suzuki SC, Suli A, Kimelman D, … Wong RO (2016). Presynaptic partner selection during retinal circuit reassembly varies with timing of neuronal regeneration in vivo. Nature Communications, 7, 10590. 10.1038/ncomms10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors will make available the original data (images) used for generating the results presented in this manuscript upon request to the corresponding authors.