Abstract
X inactivation in female mammals is controlled by a key locus on the X chromosome, the X-inactivation center (Xic). The Xic controls the initiation and propagation of inactivation in cis. It also ensures that the correct number of X chromosomes undergo inactivation (counting) and determines which X chromosome becomes inactivated (choice). The Xist gene maps to the Xic region and is essential for the initiation of X inactivation in cis. Regulatory elements of X inactivation have been proposed to lie 3′ to Xist. One such element, lying 15 kb downstream of Xist, is the DXPas34 locus, which was first identified as a result of its hypermethylation on the active X chromosome and the correlation of its methylation level with allelism at the X-controlling element (Xce), a locus known to affect choice. In this study, we have tested the potential function of the DXPas34 locus in Xist regulation and X-inactivation initiation by deleting it in the context of large Xist-containing yeast artificial chromosome transgenes. Deletion of DXPas34 eliminates both Xist expression and antisense transcription present in this region in undifferentiated ES cells. It also leads to nonrandom inactivation of the deleted transgene upon differentiation. DXPas34 thus appears to be a critical regulator of Xist activity and X inactivation. The expression pattern of DXPas34 during early embryonic development, which we report here, further suggests that it could be implicated in the regulation of imprinted Xist expression.
In female mammals, one of the two X chromosomes in each cell becomes transcriptionally inactive early in embryonic development. This process is known as X inactivation and is closely associated with cellular differentiation (see reference 22 for a review). The initiation and spread of X inactivation are dependent on a unique region of the X chromosome, the X-inactivation center (Xic). The Xic is also thought to be involved in determining how many (counting) and which (choice) X chromosomes to inactivate (see reference 2 for a review). The Xist gene, which lies within the Xic region, encodes a large nuclear, untranslated RNA that coats the inactive X chromosome at interphase (5–8). The developmental expression pattern of Xist suggests a causal role in X inactivation: prior to inactivation, Xist is expressed at low levels from every X chromosome in a cell (27), and at the onset of inactivation, steady-state levels of Xist RNA increase and the transcript coats the X chromosome that will be inactivated. This transition is associated with stabilization of Xist RNA on the X chromosome that will become inactive (36, 42). The regulated changes in stability of the Xist transcript and the coating of the X chromosome are thought to be concomitant with a switch in promoter use (26). The elements that regulate all of these changes remain to be defined functionally, however.
Targeted deletions of the Xist gene have demonstrated that it is essential for inactivation in cis (33, 37), but since counting is not abolished in these mutants, it has been suggested that elements outside Xist itself may also be involved in overall Xic function. Indeed, when a 65-kb region 3′ to Xist exon 6 is deleted, cis inactivation is not affected but, instead, the deleted X is systematically inactivated upon differentiation, even in the absence of a second X chromosome (12). Elements lying outside Xist that have been proposed to be implicated in the X-inactivation process include the Xce locus (10), which is known to affect choice and has been shown to be genetically separable from Xist (37a, 43). Another is the DXPas34 locus, which lies 15 kb 3′ to Xist (13).
The DXPas34 locus is a CpG-rich region consisting of a SalI site and a neighboring cluster of HpaII sites lying within a 34-mer minisatellite repeat, which was originally identified as a result of its unusual methylation profile. Methylation of DXPas34 is specifically associated with the active X chromosome and the transcriptionally silent Xist gene in somatic tissues and differentiated embryonic stem (ES) cells (3, 13). Analysis of this methylation profile in strains carrying different Xce alleles suggested a correlation between the Xce allele present in cis and the methylation status of the region, although the Xce locus actually maps distal to DXPas34 (37a, 43). Given its proximity to the Xist gene and its unusual methylation profile, it has been proposed that the DXPas34 region might play a role in regulating Xist expression and the initiation of X inactivation. The DXPas34 locus also displays a number of features that are strongly reminiscent of the differentially methylated regions (DMRs) frequently found in the vicinity of imprinted genes (38). Like DXPas34, DMRs show differential methylation between alleles, occur in or near CpG-rich regions, and are often associated with blocks of short tandem repeats. Elements involved in the establishment of imprinted gene expression lie within such DMRs in several cases. There are also an increasing number of examples where DMRs overlap with the transcribed region of unusual noncoding RNAs. In this context, Lee et al. (28) recently described the presence of an antisense transcript initiating close to the DXPas34 region and stretching as far as the 5′ end of Xist. As part of the study we present here, we show that the antisense transcription in this region is actually more widespread and complex than was originally thought.
To investigate the potential role of the DXPas34 locus in Xist regulation and the initiation of X inactivation, we have deleted it in the context of yeast artificial chromosome (YAC) transgenes in ES cells. We have chosen a YAC transgene-based approach for the ease and rapidity with which transgene mutations can be generated in yeast. Transgenes containing Xist are known to function as ectopic Xics when introduced into ES cells (29–31), provided that they are present as multicopy arrays (20). Both inactivation of autosomal sequences in cis around the transgene and inactivation of the single X chromosome (counting) can be observed in such transgenic male ES cell lines. This parallels the process seen in XX ES cells, where in vitro differentiation is accompanied by inactivation of one of the two X chromosomes. Although single-copy Xist YAC transgenes do not seem to be able to induce inactivation, they correctly express Xist prior to differentiation in ES cells (20). Thus, even single-copy Xist transgenes can be used to study the effects of a DXPas34 deletion on Xist regulation prior to inactivation.
Based on the analysis of several different ES cell lines carrying YAC transgenes with DXPas34 deleted, we demonstrate that this locus is essential for Xist expression in undifferentiated ES cells and may affect the normally random nature of X inactivation in ES cells. We also show that the presence of this locus is associated with widespread antisense transcription. Finally, we show that transcription at the DXPas34 locus during early development is imprinted, suggesting that this locus may play a regulatory role in the early steps of Xist expression and X inactivation.
MATERIALS AND METHODS
RT-PCR analysis.
Reverse transcription (RT) was performed on total RNA isolated with RNAzol B (Bioprobe) and treated with RNase-free DNase I (Pharmacia; 10 U/μg of RNA) for 30 min at 37°C to destroy genomic DNA. Genomic DNA contamination artifacts were controlled for in every RT reaction by including a RNA sample where reverse transcriptase was omitted. Random-primed RT was performed on 10 μg of RNA by using SuperScriptII reverse transcriptase as recommended by the manufacturer (Gibco-BRL) with random hexamers (Pharmacia) to prime first-strand cDNA synthesis in a 50-μl reaction mixture. The first round of PCR was performed on 2 μl of the RT reaction products and involved 40 cycles with forward and reverse primers under standard PCR conditions with an annealing temperature of 55°C unless otherwise stated (see Table 1). The second round of PCR was performed on 1 μl of the first-round reaction product by using nested primers (see Table 1) and a further 30 cycles of PCR. For strand-specific RT, 20 μg of DNase I-treated total RNA was divided into five aliquots and reactions were performed in a 50-μl volume for 1 h at 42°C with primers (26pmol) with or without reverse transcriptase as follows (see Fig. 1C also). The five aliquots were as follows: 1, forward primer, reverse transcriptase present, to detect the antisense transcript (with respect to Xist); 2, forward primer, reverse transcriptase absent; 3, reverse primer, reverse transcriptase present, to detect the sense transcript; 4, reverse primer, reverse transcriptase absent; 5, no-primer control, reverse transcriptase present. A single round of 40 cycles of PCR was performed on each reaction with nested forward and reverse primers adjacent to the primer used for cDNA synthesis. Sites further downstream were also tested to detect the extent of the cDNAs generated. In each case 15 μl of the 50-μl reaction mixture was loaded onto an ethidium bromide-stained agarose gel.
TABLE 1.
Primers used for RT-PCR analysis
| Site | Size of first-round PCR product (bp) | First-round PCR primers
|
Second-round nested PCR primers
|
||
|---|---|---|---|---|---|
| Forward | Reverse | Forward | Reverse | ||
| MX6a | 520 | tcctcctgactctgcctactg | cactccttctgttattaatatgcc | ggaaggagcatcaggagtt | atgcacagtcaagcaggag |
| MX3a | 1,210 | tgtgaagagcccttccttg | gttgcacgcctttaactgat | gtaacctttacttctcaattcag | cagaagaggagtggccacaaag |
| X1b | 541 | gagatacatttatttgctca | gacttagtttggtttcttta | agtcttttctcaaggtcctaa | tatgtttacattacaggtggc |
| X2b | 582 | gagtttctttctgtgccacccd | ccgtccagtggtctgttctgd | ctgtgccacccactgaaa | gtcgtctggaccaacaagg |
| X3b | 561 | ggggtttctatctgatctcca | ctccatccaagttctttctgg | ggcaagtcaataaagcactcc | cactggcaaggtgaatagcat |
| X4b | 722 | aactgggtcttcagcgtgat | catgcaactccagggcatatt | aaatgacccgaggatcaacat | taagcagcaaacaccataagc |
| 1c | 597 | 3003–3023 | 3581–3600 | 3267–3286 | 3445–3465 |
| 2c | 221 | 5928–5947 | 6129–6149 | 5941–5958 | 6084–6103 |
| 3c | 628 | 10867–10886d | 11478–11495d | 10920–10938 | 11419–11439 |
| 4c | 964 | 11365–11384d | 12310–12329d | 11384–11404d | 12288–12308d |
| 5c | 830 | 12310–12329 | 13120–13140 | 12348–12366d | 13079–13096d |
| 6c | 809 | 13120–13140d | 13907–13929d | 13141–13162 | 13892–13913 |
| 7c | 516 | 13907–13929d | 14403–14423d | 13941–13961 | 14367–14387 |
| 8c | 521 | 14403–14423d | 14904–14924d | 14412–14433 | 14880–14901 |
| 9c | 1,284 | 14810–14827 | 16074–16094 | 14822–14842 | 15989–16009 |
| 10c | 520 | 16304–16323 | 16807–16824 | 16327–16344d | 16783–16800d |
| 11c | 982 | 16831–16848 | 17794–17813 | 16895–16915 | 17674–17694 |
| 12c | 878 | 17794–17813d | 18653–18672d | 17874–17894d | 18615–18635d |
| 13c | 1,022 | 18653–18672d | 19656–19675d | 18724–18744 | 19605–19625 |
| 14c | 1,116 | 19656–19675 | 20753–20772 | 19778–19798 | 20657–20677 |
| 15c | 962 | 25554–25574 | 26496–26516 | 25749–25769 | 26451–26471 |
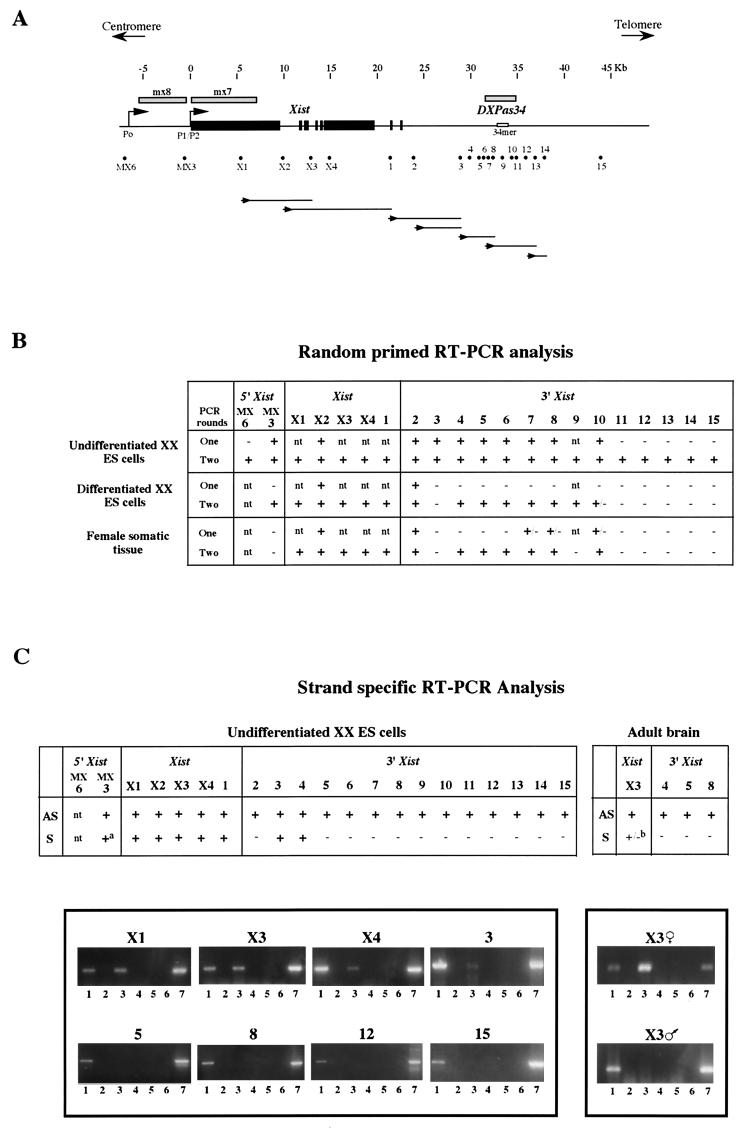
FIG. 1.
(A) Map of the 50-kb region containing the Xist gene (exons shown as solid boxes) and the DXPas34 locus (which includes the 34-mer minisatellite repeat shown). Shaded bars above the map represent probes mx8, mx7, and DXPas34, used for RNA FISH. The positions of all the sites tested by RT-PCR are shown beneath the map as dots. Each dot represents the primer pairs used for RT-PCR analysis (for sequences of both outer and inner nested primer pairs, see Table 1). Lines below the map represent antisense transcript continuity: first-strand antisense cDNA was synthesized by using a sense primer (shown as an arrowhead below the appropriate site) and then amplified with primer pairs at sites downstream. Each line represents the extent of positive amplification obtained for the cDNA in question. (B) Summary of the RT-PCR data obtained by using random hexamers to prime cDNA synthesis. Control samples where reverse transcriptase was omitted were always included to rule out possible genomic DNA contamination. The results were assessed by using ethidium bromide-stained gels. Undifferentiated male and female ES cells gave identical results. Differentiated female ES cells represent 10-day EBs. Differentiated male ES cells gave identical results, apart from sites within Xist and site 2, which were negative after a single round of PCR. Somatic tissues tested were adult brain and in some cases liver. Male and female adult tissues gave identical results apart from site 2 (as above) and site MX3, which was positive in male but not in female cells. (C) Strand-specific RT-PCR analysis. (Top) Summary of the data; (bottom) representative sample of the data is shown. Antisense transcription was detected at all 20 sites from MX3 through to site 15 in undifferentiated ES cells. Sites X1 through to 1 within Xist also showed sense transcription. Sense transcripts were also detected at sites 3 and 4. (a) At MX3, sense transcript was only faintly, and not systematically, detected. In adult brain, sites X3, 4, 5, and 8 were tested and all showed antisense transcription. (b) Sense transcript was detected within Xist at X3 in females but not in males. cDNA synthesized from antisense transcript (AS) at each site was primed by using the sense primer, and cDNA synthesized from sense transcript (S) was primed by using an antisense primer. Lanes: 1, AS plus reverse transcriptase; 2, AS minus reverse transcriptase; 3, S plus reverse transcriptase; 4, S minus reverse transcriptase; 5, reverse transcriptase without any primer; 6, H2O control; 7, genomic DNA control. The specificity of the RT reactions was controlled by including an RNA sample where no primer was present (lane 5).
YAC manipulation and mutagenesis.
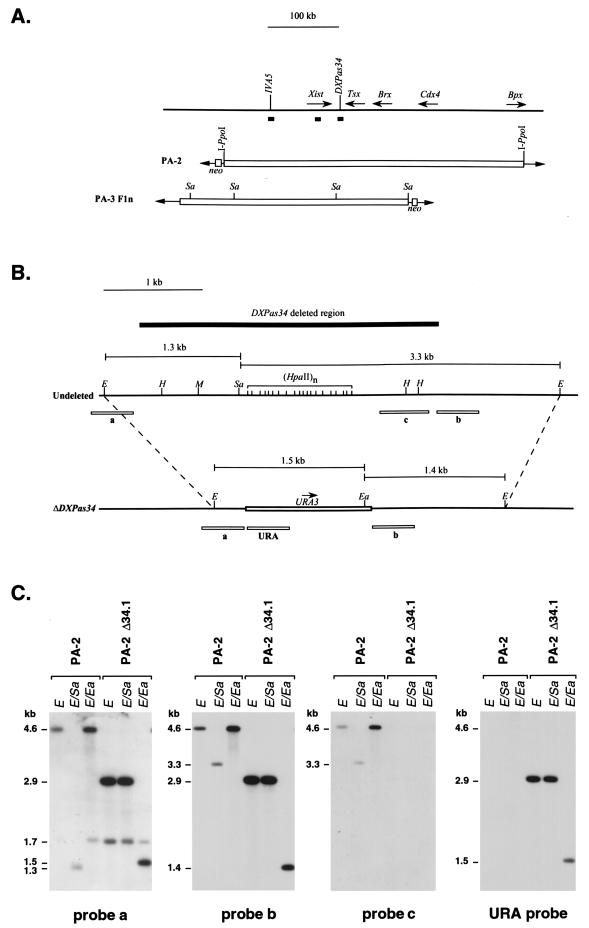
The YACs PA-2 and PA-3 F1n contain extensive sequences 5′ (130 kb for PA-2 and 150 kb for PA-3 F1n) and 3′ (310 kb for PA-2 and 100 kb for PA-3 F1n) to Xist and have previously been described in detail (18–21). Retrofitting of PA-2 and PA-3 F1n with the Pgk1-neo cassettes and I-PpoI sites (for PA-2 only) has been described previously (15, 20). DXPas34 was deleted by homologous recombination in yeast. The pUR and pRA vectors can be used to delete the target sequence by replacement with a selectable marker (URA3) (16). Regions of homology for targeting of the DXPas34 region were generated with the following primers, designed from published sequence (accession no. X99946) (44): primer 34-1, 5′ cgggatccCTTGGTGGCTTAAAGCAACA 3′ (positions 13421 to 13440); primer 34-2, 5′ ggggtaccTTCGGAAGAGAAGAAAGATT 3′ (positions 13861 to 13842); primer 34-3, 5′ cgggatccGGGGCAGAGCTTAGGGGAAC 3′ (positions 16879 to 16898); and primer 34-4, 5′ ggggtaccTTTGCATTGATACTGACTGT 3′ (positions 17320 to 17301). The primers contain a BamHI (34-1 and 34-3) or KpnI (34-2 and 34-3) 5′ extension to enable the subsequent cloning of PCR products into the pUR and pRA plasmids. PCR was performed with primer pair 34-1 and 34-2, which amplifies a 441-bp 5′ fragment (34-1/2), and with pair 34-3 and 34-4, which give rise to a 442-bp 3′ fragment (34-3/4). PCR was performed on 100 ng of C3H/He genomic DNA under standard conditions (16), with an annealing temperature of 55°C. The PCR products were digested with BamHI and KpnI, and 50 fmol was ligated to equimolar amounts of BamHI-KpnI pUR (5′ PCR product 34-1/2)- or BamHI-KpnI pRA (3′ PCR product 34-3/4)-digested plasmids. Recombinant plasmids were sequenced to verify the absence of mutations prior to their introduction into the YACs. The pUR-34-1/2 (homology 5′ to DXPas34) and pRA 34-3/4 (homology 3′ to DXPas34) plasmids were linearized with NotI, and 5 μg of each fragment was cotransformed into YAC PA-2 or PA-3 F1n-containing AB1380 yeast spheroplasts (40). URA+ transformants were selected for in complete synthetic medium lacking uracil (SD COM-Ura) and then replica plated onto SD COM-Trp-Lys-Ura (PA-2) or SD COM-Trp-Lys-His-Ura (PA-3 F1n) to eliminate clones which had lost only one of the selectable markers present in the YAC arms and were unlikely to represent true recombinants. Agarose plugs of yeast genomic DNA were prepared, and pulsed-field gel electrophoresis (PFGE) and Southern blot analysis of YAC clones were performed as previously described (19). Seven PA-2 clones (35%) and four PA-3 F1n clones (22%) were obtained that had retained an intact YAC (460 and 320 kb, respectively) which was positive for the URA3 gene (URA probe generated by HindIII/NotI digestion of pUR vector). The use of primers URA34f (5′ GTTTCTTCCTCCAATGTAAT 3′) and URA34r (5′ CCATTCTTTTCTGTGACTCC 3′), which should amplify a 1.5-kb fragment from the correctly deleted and URA3-replaced region, allowed initial characterization of deletion clones. Southern analysis of recombinant clones, following EcoRI, EcoRI-SalI, and EcoRI-EagI DNA digestion, enabled a more detailed assessment of the structure of the deletion. The overall structure of the YACs was verified by hybridization of various probes lying in and around the Xist gene to EcoRI- and HindIII-digested DNA (see Fig. 3).
FIG. 3.
Structural analysis of the DXPas34 deletion in YACs PA-2 and PA-3 F1n. (A) Structures of the two YACs used for the deletion of DXPas34, PA-2 (460 kb), and PA-3F1n (320 kb). The DNA probes, in and around the Xist gene, used to characterize the YACs and transgenic clones are shown as solid boxes along with the I-PpoI (PA-2 only) and SalI (Sa) sites that were informative in the analysis of the transgenes. (B) Structure of the DXPas34 locus before and after deletion. The 4.6-kb EcoRI fragment containing DXPas34 in the undeleted YACs and the structure of the same region after the deletion of DXPas34 and replacement with the URA3 yeast gene are indicated. A solid box above the map represents the extent of the 3-kb DXPas34 deletion, and dashed lines indicate the correspondence between the EcoRI sites of the intact and the deleted (ΔDXPas34) sequences. EcoRI (E), HpaII (H), MluI (M), EagI (Ea), and SalI (Sa) indicate restriction sites used for the structural analysis of this region. The sizes of informative EcoRI-SalI and EcoRI-EagI fragments are shown. Probes (a, b, c, and URA) used to assess the structure of the region, following homologous recombination, are shown as open boxes below each map. The region containing probe c disappears following the replacement of DXPas34 by the yeast URA3 gene (open box). Transcriptional orientation of URA3 is shown by an arrow (5′-3′). (C) Example of Southern blot analysis with probes a, b, c, and URA on YAC PA-2 Δ34.1 DNA compared to the parental undeleted YAC PA-2 DNA. EcoRI (E), EcoRI-SalI (E/Sa), and EcoRI-EagI (E/Ea) informative digests were used to demonstrate the disappearance of a SalI site and the region containing probe c, concomitant with the replacement by the URA3 and a new EagI site (B). Probe a detects 4.6-kb (undeleted), 2.9-kb (ΔDXPas34), and 1.7-kb EcoRI fragments, the last of these corresponding to the additional restriction fragment recognized by this probe which overlaps an EcoRI site (B). Similar results were obtained when this analysis was performed on YAC PA-3 F1n Δ34.3 and parental YAC PA-3 F1n DNAs (data not shown).
Generation and characterization of transgenic ES cell lines.
YAC DNA was purified and lipofected into CK35 ES cells (9), as previously described (20). Neor clones were selected 24 h after YAC transfer by G418 treatment (0.25 mg/ml). Agarose plugs of genomic DNA from transgenic and control ES cells were prepared as described previously (20). I-Ppo-I digestion of agarose-embedded DNA from YAC PA-2 Δ34.1-derived lines was performed as specified by the manufacturer (Promega). Southern analysis of EcoRI- or HindIII-digested DNA from YAC PA-2 Δ34.1- and YAC PA-3 F1n Δ34.3-derived lines was carried out by standard procedures. The transgene copy number was quantitated by PhosphorImager analysis (Molecular Dynamics) of blots hybridized with a Xist probe (nucleotides 5379 to 5547) together with an X chromosome probe Xpct (probe 128E2) which does not lie within the YACs (14). Normalization of Xist versus Xpct signals was performed with male and female controls and other YACs containing both Xist and Xpct genes (14).
Culture and differentiation of ES cells.
Male CK35 and female HP310 ES cells (20) and transgenic ES cell derivatives were maintained in the undifferentiated state by culture in ES cell medium (Dulbecco’s modified Eagle’s medium [DMEM], 4.5 g of glucose per liter, 2 mM glutaMAX I [Gibco-BRL], 0.1 mM 2-mercaptoethanol, 15% fetal calf serum (Gibco-BRL), 103 U of recombinant leukemia inhibitory factor (LIF) [Gibco-BRL] per ml, 0.05 mg of streptomycin per ml, 50 U of penicillin per ml) on mitomycin C-treated mouse fibroblast feeder cells with appropriate G418 selection for transgenic cells. For differentiation into embryoid bodies (EBs), feeders were first removed by successive adsorptions on gelatinized dishes and then cultivated for 3 days under adherent conditions in ES cell medium (39). Aggregates were formed following mild trypsinization and transferred to suspension culture (day 0) in EB medium (DMEM, 10% newborn calf serum, 0.1 mM 2-mercaptoethanol, 2 mM glutaMAX I, antibiotics [as above]) without G418 selection. Four-day-old EBs were attached to LabTek chamber slides (Nunc) for monolayer outgrowth for 3 to 8 days in DMEM–2mM glutaMAX I–antibiotics (as above)–10% fetal calf serum.
Embryo preparation for RNA fluorescent in situ hybridization (FISH).
Preimplantation embryos were flushed from the oviducts or uterus and the zona pellucida was removed as described previously (25). Postimplantation embryos were dissected from maternal tissue and Richert’s membrane and the ectoplacental cone were removed as described previously (4). Embryo collection was performed in M2 medium (25). Embryos of 3.5, 6.5, and 7.5 days postcoitum (p.c.) were disaggregated in trypsin-EDTA. Whole (≤ 3.5 days p.c.) or disaggregated embryos were transferred in 100 μl of DMEM–10% fetal calf serum to siliconized cytofunnels and collected on Superfrost Plus glass slides (BDH) by cytocentrifugation at 600 to 800 rpm (Shandon Cytospin 3) for 4 min. Embryo slides were fixed and stored as described below for ES cells.
DNA and RNA FISH analysis on ES cells and EBs.
Cells grown on chamber slides or cytocentrifuged (4 min at 400 rpm) onto baked glass slides were permeabilized with Triton X-100 in ice-cold cytoskeletal buffer for 7 min and fixed with 4% paraformaldehyde for 10 min on ice; they were then stored in 70% ethanol at 4°C. ES cells and EBs were prepared for RNA and DNA FISH as described previously (20). For RNA FISH, the slides were dehydrated and used directly for hybridization. For DNA FISH, nuclei were denatured for 2 min in 70% formamide–2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 75°C, while for simultaneous RNA and DNA FISH, nuclei were denatured for just 1.5 min. The hybridization and washing conditions were as described previously (11). Nuclei were mounted in Vectashield (Vector) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). A Quantix charge-coupled device camera and IPLab and Photoshop software were used for image acquisition and treatment. Probes were labelled either by nick translation (Vysis) with Spectrum Green-dUTP or Spectrum Red-dUTP (Vysis) or by biotin incorporation via random priming (Gibco-BRL). Biotinylated probes were detected with fluorescein avidin (Vector) or Texas red avidin (Vector). Amplification of signal, when used (at sites 11 to 15), involved one layer of biotinylated anti-avidin (Vector) followed by another layer of fluorescein avidin or Texas red avidin. The Xist probes were λ 510 (41), which covers 20 kb of the Xist gene, mx8 (26), which covers 5 kb between the Xist P0 and P1 promoters, and mx7 (26), which covers 7 kb within Xist exon I. The DXPas34 probe was a plasmid obtained by cloning of the 3.2-kb PCR product amplified from C3H/He mouse genomic DNA with primers URA34 for and URA 34 rev (see “YAC manipulation and mutagenesis” above) in pGEM-T easy (Promega). Probes corresponding to sites 1 to 15 were generated by pGEM-T-cloning of PCR products amplified from genomic DNA with the primers listed in Table 1. Strand-specific probes were either single-stranded phage DNA corresponding to mx8, isolated from pBluescript as recommended by Stratagene and verified for template specificity by Southern hybridization with radioactively end-labelled oligonucleotides, or pools of 50-mer fluorescein-labelled oligonucleotides, corresponding to sequences within Xist exon I, synthesized by Eurogentec. The YAC-specific probe was pYAC4 vector DNA. The X chromosome-specific probe (BAC X) was a BAC isolated by using the microsatellite marker DXMit158; it lies outside the X chromosome region covered by YACs PA-2 and PA-3 F1n.
RESULTS
RT-PCR analysis of transcription in the Xist-DXPas34 region in ES cells.
To identify potential transcripts associated with the DXPas34 locus, RT-PCR and Northern analysis of RNA from ES cells and adult tissues were performed. Initial results suggested the presence of multiple, low-level transcripts mainly in the antisense orientation (based on riboprobe analysis) with respect to Xist, which included but extended beyond the DXPas34 locus (data not shown). To examine this low-level transcription pattern further, RT-PCR analysis was carried out at 21 positions (primer positions are given in Fig. 1 and Table 1) in a region spanning just over 50 kb and including Xist and DXPas34. In undifferentiated XX and XY ES cells, when random-primed RT followed by a single round of PCR was performed, RT products were detected from MX3 (a site lying between Xist promoters P0 and P1) through to site 10 (within DXPas34). When a second (nested) round of PCR was performed, widespread transcription was observed across the whole 50-kb region tested. The results are summarized in Fig. 1B. When strand-specific RT-PCR was carried out with undifferentiated XX ES cells, using sense or antisense primers from site MX3 through to site 15, antisense transcription was detected at all positions (Fig. 1C). This technique is apparently more sensitive than random-primed RT, since RT products could be detected after just a single round of PCR. Sense transcription, in addition to antisense transcription, was seen within Xist (site X1 through to site 1 [Fig. 1C]) as expected. Occasional sense transcripts were also found 3′ to Xist (site 3 [Fig. 1C] and site 4 [not shown]).
The continuity of the transcript(s) in this region was also assessed. RT was initiated at single sites on the RNA with strand-specific primers, and the extent of the resulting cDNA was examined by PCR with primers at various sites downstream of the primer used for first-strand synthesis (a summary of these results is given in Fig. 1A). Within the Xist gene itself, antisense transcript continuity appeared to be extensive. For example, when primer X2F was used to prime cDNA synthesis from antisense RNA, sites up to 12 kb away (i.e., as far as site 1 [Fig. 1A]) were positive by PCR. Downstream of Xist, the continuity of antisense transcripts detected was more variable, particularly in the vicinity of the DXPas34 region (Fig. 1A). For example, cDNAs initiated with primers at sites 3 and 6 extended only up to 4 kb (site 8) and 6.5 kb (site 13) away, respectively. This could be a result of the inefficiency of the reverse transcriptase in extending across certain sequences. Alternatively, it could be an indication that multiple transcripts of variable length are present in this region.
We also examined transcription in differentiated ES cells and adult tissues. As expected from previous work (27), RT products were seen within Xist after just a single round of random primed RT-PCR in differentiated female (Fig. 1B) but not male (not shown) ES cells and adult brain. Transcription detected at site 2, just 3′ to Xist exon 8, in females probably represents runthrough transcripts from the 3′ end of Xist. Some low-level transcription was, however, detected at several sites in both male and female differentiated cells following nested, random primed RT-PCR. This was seen in two domains, one across Xist and the other around DXPas34, separated by a gap at site 3 (Fig. 1B). Strand-specific RT-PCR performed at a number of sites revealed this low-level transcription to be antisense (Fig. 1C).
We conclude that widespread, low-level antisense (and occasional sense) transcription occurs in undifferentiated ES cells, across a region covering at least 50 kb and including Xist and DXPas34. Transcripts within the 40-kb region spanning Xist and DXPas34 appear to be more readily detectable than in the flanking regions. In differentiated ES cells and adult tissues, low-level antisense transcription is also present but appears to be more restricted, covering an approximately 23-kb domain centered on Xist and a 5-kb domain over DXPas34.
RNA FISH analysis of transcription in the DXPas34 region in ES cells and early mouse embryos.
Using the complementary approach of FISH, we were able to assess the proportion of ES cells expressing transcripts in the DXPas34 region and also to examine the kinetics of this expression upon differentiation. A 3.2-kb probe covering the DXPas34 locus was used to examine DXPas34 transcription in undifferentiated male (CK35) and female (HP310) ES cells, as well as in male ES cells carrying a two-copy 460-kb Xist YAC transgene (L412) (20). In undifferentiated ES cells, the Xist transcript, present in unstable form at every Xist locus, is detected by RNA FISH as a punctate signal (pinpoint) at its site of synthesis (36, 42). Using dual-color RNA FISH, we found that the DXPas34 RNA signal consistently colocalized with the punctate RNA signal at the Xist locus (λ 510) in undifferentiated ES cells (>90%, n > 500) (Fig. 2A). Probes corresponding to cloned PCR products from sites 2 through 10 (Fig. 1A) gave similar profiles to the 3.2-kb DXPas34 probe. RNA signal could not be detected with probes corresponding to sites 11 through 15, although following amplification (see Materials and Methods), faint signals could occasionally be seen (data not shown). This is consistent with our random primed RT-PCR data suggesting that transcription in the region distal to DXPas34 is present at lower levels or is less readily detectable. Finally, in undifferentiated male, female, and transgenic ES cells, DXPas34 transcripts were found to colocalize with both sense and antisense transcripts at the Xist locus (mx8 and Xist exon I [Fig. 1A]; see Materials and Methods), as expected from our RT-PCR analysis and previously published data (28) (Fig. 2G to L).
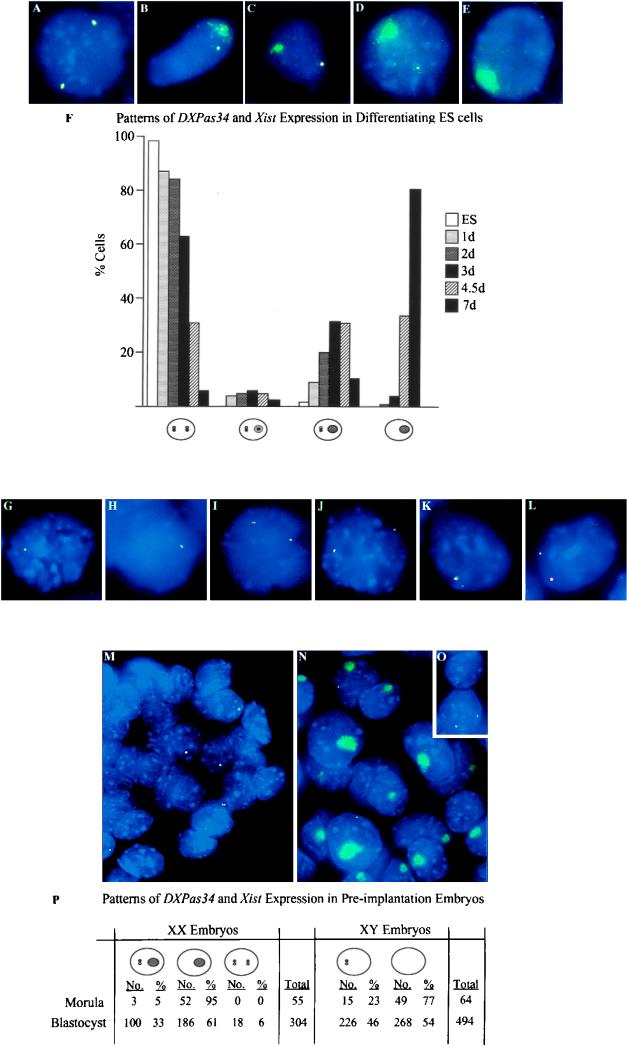
FIG. 2.
Two-color RNA FISH analysis of ES cells and preimplantation mouse embryos. (A to E) RNA FISH on XX ES cells with a Xist (λ 510) probe (green) and the 3.2-kb DXPas34 probe (red). Overlapping green and red signals are seen as yellow. (A) Undifferentiated XX ES cell showing colocalization of Xist and DXPas34 punctate signals. (B) Differentiating XX ES cell displaying an immature Xist RNA domain containing a DXPas34 signal on one X and a Xist/DXPas34 punctate signal on the other. (C) Differentiating XX ES cell displaying an immature Xist RNA domain without a DXPas34 signal. (D) Differentiating XX ES cell with a mature Xist RNA domain and no DXPas34 signal within it. (E) Fully differentiated XX ES cell with a mature Xist RNA domain and no DXPas34 signal on either X. (F) Frequency of cells exhibiting different patterns of Xist and DXPas34 RNA signals in XX ES cells and EBs at different times of differentiation in days (d). Xist RNA punctate signals (detected by λ 510) are shown as white pinpoints, and DXPas34 RNA signals are shown as black pinpoints. Immature Xist RNA domains are shown as light grey ovals. Mature Xist RNA domains are shown as dark grey ovals. Cell numbers scored were 1 day (n = 56), 2 days (n = 116), 3 days (n = 100), 4.5 days (n = 85), and 7 days (n = 78). A small proportion of cells (<5%), showing either two Xist pinpoints and only one DXPas34 pinpoint or vice versa (and no Xist RNA domain), which were detected at every stage, are not included in the histogram. (G to L) RNA FISH on XY and transgenic XY ES cells by using strand-specific Xist probes mx8 or exon I oligonucleotides (green) and the 3.2-kb DXPas34 probe (red). (G) Undifferentiated male ES cell. The Xist sense transcript colocalizes with the DXPas34 transcript. (H) Undifferentiated male ES cell. The Xist antisense transcript colocalizes with the DXPas34 transcript. (I) Undifferenitated female ES cell. Sense transcripts at both Xist alleles colocalize with DXPas34 transcripts. (J) Undifferentiated female ES cell. Antisense transcripts at both Xist alleles colocalize with DXPas34 transcripts. (K) Undifferentiated male ES cell containing a two-copy YAC PA-2 transgene (L412). The Xist sense transcript colocalizes with the DXPas34 transcript. (L) Undifferentiated L412 ES cell. The Xist antisense transcript colocalizes with the DXPas34 transcript. Note that in panels K and L, one allele (previously shown to be the two-copy transgene [20]) gives a slightly larger signal than the other. (M to P) RNA FISH on male and female blastocysts by using a Xist (λ 510) probe (green) and the 3.2-kb DXPas34 probe (red). Smaller probes from the DXPas34 region (sites 2, 3, 6, and 9) gave similar profiles to the larger DXPas34 probe. (M) Male blastocysts. In a proportion of cells, a Xist/DXPas34 RNA pinpoint signal is detected on the maternal X chromosome. (N) Female blastocysts. Most cells contain a Xist RNA domain corresponding to the paternal X with no sign of DXPas34 RNA within such domains. A proportion of these cells also contain a Xist/DXPas34 RNA pinpoint signal. (O) A small proportion of female blastocyst cells, probably corresponding to the ICM, contains no Xist RNA domain, and either one or two Xist/DXPas34 RNA pinpoint signals. (P) Frequency of the different patterns of Xist/DXPas34 expression observed in morulas and blastocysts.
We next investigated the kinetics of DXPas34 and Xist transcription in differentiating ES cells (data summarized in Fig. 2F). When XX ES cells are differentiated in vitro, Xist transcripts are stabilized and accumulate over the X chromosome to be inactivated, forming a “domain” of Xist RNA. In fully differentiated XX ES cells, DXPas34 transcription was found to be absent from such Xist RNA domains (Fig. 2E). At early stages of differentiation, (Fig. 2A to D), a Xist RNA domain (prospective inactive X) in addition to a punctate Xist signal on the active X was seen in an increasing proportion of cells. DXPas34 RNA was associated only with the punctate Xist signal and not with domains in the majority of such cells (Fig. 2A to D). Occasionally, however, a DXPas34 RNA pinpoint was seen within larger Xist RNA signals that appeared to correspond to “immature” domains (e.g., day 2, 6%, n = 116) (Fig. 2B). Down-regulation of DXPas34 transcription thus seems to be tightly correlated with the onset of Xist RNA coating of the X chromosome to be inactivated but does not actually precede it. On the active X in both male and female ES cells, DXPas34 transcription disappeared as differentiation proceeded, as did the punctate Xist RNA signal (Fig. 2E). Similar results were obtained with transgenic (L412) ES cells.
We also examined DXPas34 transcription in early mouse embryos by RNA FISH. In preimplantation female embryos, the paternally derived X chromosome is coated by Xist RNA from at least the eight-cell stage, while the maternal Xist allele is seen as a punctate signal and is present in only a proportion of cells (42). This pattern may underlie the preferential inactivation of the paternal X, which occurs later, in extraembryonic tissues. In the earliest embryos examined (early morulas, 8 to 16 cells), we detected a DXPas34 RNA signal which colocalized consistently with the Xist punctate signal on the maternal X but never with the Xist RNA domain over the paternal X (Fig. 2M and N). Strand-specific probes at the Xist locus showed that both sense and antisense Xist RNA pinpoint signals colocalized with the DXPas34 signal (data not shown). At a later stage, in female blastocysts, a small proportion of cells which contained one or two Xist/DXPas34 RNA punctate signals and no Xist RNA domains were also observed (Fig. 2O). These presumably corresponded to inner cell mass (ICM) cells destined to form the embryo. The data are summarized in Fig. 2P. In postimplantation (6.5 and 7.5 days p.c.) embryos, DXPas34 transcripts always colocalized with Xist RNA pinpoints on the active X (data not shown). In female embryos, as in differentiating XX ES cells (see above), occasional cells were observed with a DXPas34 RNA signal within an immature Xist RNA domain (as in Fig. 2B) but never within mature Xist RNA domains. In summary, transcription at the DXPas34 locus seems to be imprinted and to correlate with the low-level expression of the maternal Xist gene seen in preimplantation embryos.
Deletion of the DXPas34 locus in YACs and generation of ES lines carrying ΔDXPas34 YAC transgenes.
The unusual methylation profile of the DXPas34 locus (13), given its proximity to the Xist gene, provided circumstantial evidence for a role for DXPas34 in the regulation of Xist expression and X inactivation. The antisense transcription detected across the DXPas34 region was also suggestive of some function for this locus. To address this question directly, we deleted the DXPas34 locus in the context of two different YACs, PA-2 (460 kb) and PA-3 F1n (320 kb) (Fig. 3A). Both YACs have previously been shown to carry out aspects of Xic function when introduced at ectopic sites by transgenesis (20). A 3,016-bp region (Fig. 3B) which encompasses the GC-rich HpaII-containing 34-mer repeat array and the SalI site 5′ to it (see Materials and Methods) (13) was targeted by using a combination of two replacement vectors (see Materials and Methods). Homologous recombination resulted in this 3-kb region being replaced by a 1.3-kb fragment containing the yeast URA3 gene (Fig. 3B). Two of the DXPas34 deletion YACs obtained, PA-2 Δ34.1 and PA-3 F1n Δ34.3, were analyzed in detail to verify that their overall structures were correct (Fig. 3C). The expected deletion and replacement of DXPas34 was ascertained in this way. No rearrangements could be detected within or around the Xist gene by Southern blot analysis of these clones (see Materials and Methods).
YACs PA-2 Δ34.1 and PA-3 F1n Δ34.3 were then transferred into CK35 XY ES cells by lipofection of purified DNA and transgenic clones isolated as previously described (20). The structural integrity of the YAC PA-2 Δ34.1 transgenes was verified by I-PpoI digestion of DNA of the PA-2 Δ34.1 transgenic ES cell clones, since sites for the meganuclease I-PpoI had been previously introduced into each arm of this YAC (Fig. 1A) (15, 20). In addition, SalI PFGE Southern blot analysis was performed (data not shown). The integrity of YAC PA-3 F1n Δ34.3 transgenes was assessed by SalI PFGE Southern blot analysis (20). Four PA-2 Δ34.1-containing lines (LP1, LP5, LP6, and LP17) and one PA-3 F1n Δ34.3-containing line (LF1) were chosen for further study based on the integrity of their transgenes as evaluated by such PFGE analysis. The transgene copy number of the lines obtained was assessed by Southern blot analysis of EcoRI-digested ES cell DNA. Simultaneous hybridization of various Xist probes and of another X chromosome probe (Xpct), not contained within the YACs (14), allowed copy number quantification by PhosphorImager analysis. The YAC copy number was estimated to range in these lines from 1 to 3–4 (Table 2).
TABLE 2.
Characteristics of ΔDXPas34 transgenic ES cell lines
| Line | YAC transgene | Copy no.b | Phenotype in ES cellsc
|
|||
|---|---|---|---|---|---|---|
| Undifferentiated, expression in transgene:
|
Differentiated (Xist RNA coating in cis) | |||||
| 5′ Xist | Xist | 3′ Xist | ||||
| L412a | PA-2 (460 kb) | 2 | + | + | + | + |
| L117a | PA3 (320 kb) | 5–8 | + | + | + | + |
| LF1 | PA3 F1n Δ34.3 | 3–4 | − | − | − | + |
| LP5 | PA-2 Δ34.1 | 2–3 | − | − | − | + |
| LP1 | PA-2 Δ34.1 | 1 | − | − | − | − |
| LP6 | PA-2 Δ34.1 | 1 | − | − | − | − |
| LP17 | PA-2 Δ34.1 | 1 | − | − | − | − |
Reference 20 and this study.
Copy number of the transgene determined as described in Materials and Methods.
Assessed by RNA FISH with probes mx8 (5′ Xist), λ510 (Xist) and sites 2 and 3 (3′ Xist; see figure 1A).
ΔDXPas34 transgenes show absence of transcription across the Xist-DXPas34 region in undifferentiated ES cells.
Single-copy and multicopy YAC transgenes have previously been shown to express the Xist transcript in its unstable form in undifferentiated ES cells (20, 31). When RNA FISH is used, two Xist pinpoints corresponding to the transgenic locus and the X chromosome are normally observed in undifferentiated male transgenic ES cell lines. The size of the transgenic Xist transcript signal appears to correlate with copy number (20, 31).
To determine whether the expression profile of Xist was affected by the deletion of DXPas34 in undifferentiated cells, the five transgenic lines (Table 2) were examined by RNA FISH. In all the lines carrying a ΔDXPas34 YAC transgene, only a single RNA pinpoint per cell was observed at the Xist locus (λ 510 probe, 93%, n > 1,000) (Fig. 4B-D). This contrasts with the result for the L412 line, which carries two intact copies of the parental YAC PA-2, where two pinpoints per nucleus were observed in 92% of cells (n > 500), and one of these pinpoints (corresponding to the transgene) was consistently larger than the other (Fig. 4A; see also Fig. 2H to I). DNA FISH with a YAC-specific (pYAC4) probe confirmed that these ΔDXPas34 cells with only one Xist RNA pinpoint were actually transgenic (>95%, n > 500) (data not shown). To determine whether the unique RNA signal at the Xist locus observed in ΔDXPas34 lines was derived from the transgene or the X chromosome, YAC-specific (pYAC4) DNA FISH was performed following Xist RNA FISH detection (shown in Fig. 4E for line LP6). The RNA pinpoint was consistently found to colocalize with the X chromosome rather than with the transgene. Furthermore, dual-color RNA FISH with a Xist (λ 510) probe together with a DXPas34 probe corresponding to the deleted region in the ΔDXPas34 transgene revealed 100% colocalization of the λ 510 and DXPas34 RNA signals (n > 1,000), again demonstrating the X chromosome origin of the unique Xist/DXPas34 RNA pinpoint. Since the Xist RNA signal derived from the undeleted two-copy YAC PA-2 transgene present in L412 cells was actually often more intense than the X chromosome signal, total absence of Xist RNA signal in the ΔDXPas34 lines carrying up to four YAC copies was unlikely to be due to inefficient detection.
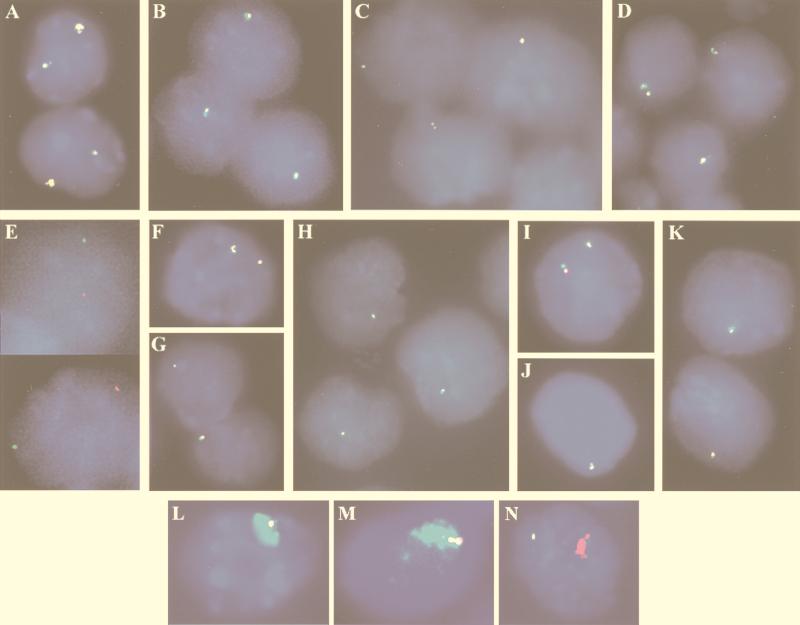
FIG. 4.
Xist and DXPas34 expression in ES cell lines with intact or ΔDXPas34 YAC transgenes. (A to D) Dual-color RNA FISH, with Xist (λ 510, green) and DXPas34 (red) probes, was performed on undifferentiated ES cells without denaturation of nuclei. Colocalization of green and red signals results in the appearance of yellow signals. (A) Control transgenic male line L412, which carries two copies of YAC PA-2. Xist and DXPas34 probes detect two colocalized signals, the larger one having been previously shown to be associated with the multicopy transgenic locus (20). (B to D) Line LP5 (which contains two or three copies of YAC PA-2 Δ34.1) (B), line LP6 (which contains one copy of YAC PA-2 Δ34.1) (C), and line LF1 (which contains three or four copies of YAC PA-3 F1n Δ34.3) (C) all exhibit a unique Xist/DXPas34 RNA signal. The presence of two closely associated pinpoint signals represents a replicated locus. (E) An example of the X chromosomal origin of the unique Xist RNA pinpoint in the LP6 representative undifferentiated ES cell line. The Xist RNA FISH signals in positioned nuclei were photographed, and the slides were then denatured and DNA FISH with a transgene-specific (pYAC4) probe was performed. This experiment clearly demonstrates that the single Xist RNA signal (green) observed in ΔDXPas34 transgenic cell lines prior to differentiation is not associated with the transgenic locus (red) and must therefore correspond to the X chromosome. (F to K) Study of the transcription in the region 3′ to Xist with probes corresponding to sites 2 (F to H) and 3 (I to K) (see Fig. 1A for probe positions) in control and ΔDXPas34 lines. Double RNA FISH with a Xist probe (green) and probes from site 2 or 3 (red) was performed. Line L412 (F) exhibits two Xist-site 2 colocalized signals per nucleus, whereas two representative ΔDXPas34 transgenic ES cell lines, LP6 (YAC PA-2 Δ34.1) (G) and LF1 (YAC PA-3F1n Δ34.3) (H), exhibit only a single Xist/site 2 signal. Identical results were obtained with the Xist/site 3 probe combination, on the same lines: L412 (I), LP6 (J), and LF1. (K). (L and M) Capacity of ΔDXPas34 multicopy YAC transgenes to trigger Xist RNA coating of an autosome in cis. Simultaneous RNA and DNA FISH was performed on differentiated cells from the LP5 (L) and LF1 (M) lines with a Xist probe (λ 510; green) and a transgene-specific probe (pYAC4). Formation of Xist RNA domains is observed on the autosome carrying the transgene in both lines. (N) Upon differentiation, transition to Xist RNA coating of an autosome in cis appears to be correctly regulated in the multicopy LF1 line: an mx8 probe (green), specific for Xist transcripts initiating upstream of P1/P2 (from the putative P0 promoter), detects only a single punctate signal per cell, while Xist RNA domain formation on the transgene is associated only with an mx7 (Xist exonI) signal (red).
To study whether deletion of DXPas34 affected the expression of the transcript(s) identified in the region 3′ to Xist (see above), dual-color RNA FISH was performed with probes corresponding to cloned products from sites 2 and 3 (Fig. 1A) together with a Xist (λ 510) probe on two representative transgenic lines (LP6 and LF1 [Fig. 4F to K]). In undifferentiated L412 cells (intact YAC PA-2), two RNA signals were clearly detected with each of these probes, and they both colocalized with the two Xist locus RNA pinpoints originating from the X chromosome and from the transgene (n = 44, probe at site 2; n = 34, probe at site 3 [Fig. 4F and I]). In contrast, only a single RNA signal was detectable when these probes were used in ΔDXPas34 transgenic lines. For example, with a probe at site 2, the RNA signal was always associated with the unique Xist pinpoint from the X chromosome and was never associated with the ΔDXPas34 transgene in lines LP6 (n = 69 [Fig. 4G]) and LF1 (n = 87 [Fig. 4H]). Similar results were obtained with sense and antisense probes which detect Xist transcripts upstream of the P1/P2 promoter (mx8 [Fig. 1A and data not shown]).
Taken together, our data suggest that in undifferentiated ES cells the deletion of DXPas34 either eliminates or at least (within the limits of sensitivity of FISH analysis) drastically reduces the transcriptional activity of a large region encompassing Xist itself and a 15-kb region 3′ to it.
Capacity of ΔDXPas34 transgenes to inactivate in cis and to induce counting.
One of the potential effects of the DXPas34 deletion might be to interfere with the capacity of the transgene to initiate cis inactivation. When induced to differentiate in vitro, ES cells carrying multicopy Xist transgenes can undergo X inactivation; the first sign of this is coating of the autosome with Xist RNA. This is detected by RNA FISH as a large nuclear “domain” of Xist RNA (20, 31). When multicopy transgenic lines LP5 (two or three copies) and LF1 (three or four copies) were differentiated in vitro, Xist RNA domains originating from the transgene were readily observed by simultaneous RNA and DNA FISH (see Materials and Methods) with the λ 510 (Xist) and pYAC4 (transgene-specific) probes (Fig. 4L to M). Thus, deletion of DXPas34 does not interfere with the capacity of multicopy transgenes to lead to Xist RNA coating and initiation of inactivation in cis.
Prior to inactivation, Xist transcription has been reported to initiate up to 6.5 kb upstream of the somatic promoters P1 and P2, from a putative P0 promoter. A switch to the P1 and P2 promoters occurs at the Xist allele on the X chromosome to be inactivated at the onset of X inactivation, and this coincides with the production of Xist stable transcripts which accumulate in cis (26). To determine whether this change in promoter use occurs normally on the ΔDXPas34-linked Xist allele upon differentiation, transcription was examined with probes upstream (mx8) and downstream (mx7) of the P1 and P2 promoters (Fig. 1A) (26). In undifferentiated cells of the multicopy ΔDXPas34 transgenic lines LF1 and LP5, no signal was observed from the ΔDXPas34 transgenes when either mx7 or mx8 was used. In differentiating cells, the Xist RNA species coating the autosome was detected only with probe mx7; it was never detected with upstream probe mx8, even in cells where the formation of a domain has just been initiated (n = 62) (Fig. 4N). We conclude that even when the mx8 region upstream of Xist is not transcribed prior to differentiation as a result of the DXPas34 deletion (see above), the transition to P1- and P2-directed expression at the onset of inactivation is correctly regulated. Thus, deletion of DXPas34 does not disrupt Xist expression in a constitutive fashion.
In every multicopy Xist transgenic ES cell line described to date, Xist RNA decoration of the single X chromosome was found to occur in a proportion of cells. This has been taken to indicate that the counting process is being triggered. In transgenic line L412, which contains two copies of wild-type YAC PA-2, or L117, which contains multiple copies of YAC PA-3 F1n, the X chromosome was found to be affected in 7 and 10%, respectively, of differentiated cells with Xist RNA domains (20). We set out to determine whether ΔDXPas34 transgenes were still capable of inducing inactivation of the X chromosome. Simultaneous RNA and DNA FISH with either Xist (λ 510)-plus-pYAC4 or Xist (λ 510)-plus-BAC X probe combinations was performed on differentiated ES cells to establish whether Xist RNA domains were produced by the unique X chromosome in the multicopy lines LF1 and LP5. An X chromosome-derived Xist RNA domain was never observed in cells differentiated for 7 days (n > 500), suggesting that the deletion of DXPas34 in YAC transgenes may lead to skewed inactivation in favor of the transgene.
DISCUSSION
In this study we provide functional evidence that the DXPas34 locus, situated 15 kb 3′ to the Xist gene, is a regulator of Xist expression and propose that it may play a key role in controlling the initiation of X inactivation. We also provide evidence that widespread antisense transcription across a large region including Xist and DXPas34 appears to be intimately linked to the function of DXPas34.
The DXPas34 locus was deleted in the context of two large YACs, which were then used as transgenes in male ES cells. Transgenes containing the Xist gene and an intact DXPas34 locus have previously been found to show correct Xist expression prior to differentiation (20, 30, 31). In differentiated cells, such transgenes can lead to cis inactivation and counting when present as multicopy arrays (20, 31). We were therefore able to address a number of questions with respect to the role of the DXPas34 locus in regulating Xist expression and X inactivation by using our ΔDXPas34 transgenic lines. First, we assessed the effect of the deletion on Xist expression in undifferentiated ES cells. A striking phenotype was observed in ΔDXPas34 transgenic ES cell lines: transcription, both sense and antisense, at the transgenic Xist locus was found to be abolished, as far as could be detected by RNA FISH, in all cases. Second, we examined the capacity of ΔDXPas34 transgenes to express Xist RNA following in vitro differentiation. We found that Xist RNA expression and chromosome coating in cis occurred efficiently at multicopy ΔDXPas34 transgene loci. Neither the expression of Xist nor that of the antisense transcripts prior to differentiation is therefore a requirement for Xist expression following differentiation. Third, we assessed the ability of multicopy ΔDXPas34 loci to induce inactivation of the endogenous X chromosome (counting) upon differentiation. No evidence for this was found in the ΔDXPas34 cell line. Thus, deletion of DXPas34 may also lead to nonrandom inactivation.
Our results suggest that the DXPas34 locus might contribute to at least three activities which may or may not be linked: regulation of Xist expression and of antisense transcription prior to inactivation, and the capacity to induce random inactivation upon differentiation.
DXPas34 may be a developmentally regulated enhancer or LCR.
The location of DXPas34 3′ to Xist and the disruption of transcriptional activity observed when it is deleted are both reminiscent of features associated with enhancers (34). Given the ΔDXPas34 phenotype, DXPas34 could be an enhancer of the unstable Xist transcription found in undifferentiated ES cells. Since both sense and antisense transcription is disrupted in ΔDXPas34 mutants, DXPas34 could be an enhancer affecting the transcriptional activity of a region of at least 50 kb. Examples of other long-range enhancers include the locus control region (LCR), which contains several such elements that enhance transcription across a region of more than 60 kb (see reference 24 for a review). It is interesting that low-level intergenic transcription, similar to that found across the Xist-DXPas34 region and beyond, has also been found across the LCR and β-globin cluster (1). The significance of such low levels of transcription is currently unclear. On the one hand, transcription might be important in, for example, defining chromatin domains of activity which ensure that genes can be correctly activated by the appropriate developmental or tissue-specific factors. On the other hand, such transcription could simply be symptomatic of an “open” chromatin conformation, mediated by loci such as LCRs.
A number of experiments could be undertaken to assess further the LCR or enhancer-like properties of DXPas34. First, such elements should enable high-level expression of linked reporter genes in transient-transfection assays. Second, such elements should be able to function in an orientation-independent manner (see reference 34 for a review). Inversion of DXPas34 should therefore not alter its ability to enhance Xist and antisense expression. Third, LCRs (but not enhancers) have the capacity to overcome chromosomal position effects on gene expression in a transgenic context. There is already some evidence that DXPas34 may have this property, since short cosmid transgenes, which contain Xist but lack the DXPas34 locus, show very variable levels of Xist expression between lines (23). Finally, the DXPas34 locus would be predicted to contain ES cell-specific DNase I-hypersensitive sites and corresponding binding sites for developmentally regulated factors.
DXPas34 is associated with low-level antisense transcription across a large region including Xist.
The existence of a single, ES cell-specific, antisense transcript to Xist (Tsix), which initiates close to the DXPas34 locus, has recently been proposed (28). In our study, we provide evidence for a much more complex transcription pattern in the region covering Xist and DXPas34. Complexity was manifested in several ways. First, antisense transcription was detected across a much larger region than that reported by Lee et al. (28) and continuous antisense transcripts were found several kilobases 5′ of the proposed 5′ end of Tsix. This is probably because we used strand-specific RT-PCR (which appears to be more sensitive than random primed RT-PCR) to test the whole 50-kb region and not just the Xist-DXPas34 region (28). Second, different levels of transcription were found, with transcription in the Xist-DXPas34 region being more readily detected than in flanking regions in undifferentiated ES cells. Unlike Lee et al. (28), we also found evidence of low-level antisense transcription in differentiated ES cells and adult tissues. A third level of complexity concerns the kinetics of transcription in this region at the time of X-inactivation initiation. We found DXPas34 transcripts to be down-regulated on the X chromosome being inactivated, but down-regulation did not precede the accumulation of Xist RNA; rather, it coincided with and sometimes even followed the accumulation of Xist RNA. This contrasts with the results of Lee et al., who reported that with probes within (rather than 3′ to) Xist, antisense down-regulation was observed just prior to up-regulation of Xist on the future inactive X (28). This difference could be due to a slight up-regulation of the sense Xist transcript at the onset of X inactivation, which might impede efficient antisense transcript elongation within Xist but not 3′ to it. Alternatively, antisense transcription within Xist and at the DXPas34 locus might be independently regulated. This would be consistent with our finding that there are two domains of low-level antisense transcription in adult somatic cells and differentiated ES cells, one centered over Xist and the other centered over DXPas34. Taken together, our results could be reconciled with those of Lee et al. (28) if DXPas34 did not simply represent the 5′ end of a single “Tsix” transcript but if, instead, several heterogeneous antisense transcripts existed, initiating at different sites in a large region 3′ to Xist. DXPas34 could be a central regulatory element of this generalized antisense transcription. To address this, we are investigating the distribution of transcription initiation sites in this large region.
A role for this antisense transcription in regulating Xist expression and X inactivation initiation by inhibiting high-levels of Xist expression prior to differentiation has been proposed (28). Such inhibition might occur either via transcriptional interference or via transcript stability, since formation of sense-antisense RNA duplexes has been suggested to trigger specific degradation mechanisms in other organisms (RNA interference) (17). If either of the above were true, abolition of antisense transcripts, as seen in our ΔDXPas34 lines, would be predicted to lead to higher levels of Xist transcription. However, no sign of Xist transcription was detectable in our ΔDXPas34 lines. Given this finding, if antisense transcription did regulate Xist expression, a complex scenario would have to be envisaged, with DXPas34 playing two roles with opposite effects in undifferentiated ES cells: as an enhancer of Xist expression and as a regulator of antisense transcription, which itself represses Xist expression. A simpler explanation would be that the low-level antisense transcription associated with the presence of DXPas34 is just a by-product of the chromatin conformation needed to mediate the enhancer-like functions of this sequence.
Imprinted DXPas34 transcription during early development.
DXPas34 was originally identified as a result of its unusual differential methylation profile (13). Differential methylation between alleles is also a hallmark of imprinted genes (see reference 38 for a review). The expression of DXPas34 during preimplantation development may well be indicative of a role for this element in the regulation of imprinted X inactivation and/or the transition from imprinted to random X inactivation. Although the significance of the transcription itself remains to be defined, as discussed above, this transcription can nevertheless be used as a marker for DXPas34 activity. DXPas34 transcripts are detected on only the maternal, not the paternal, X chromosome in preimplantation embryos, and the maternal Xist allele is correspondingly underexpressed. It is not yet known whether low-level maternal Xist expression at this stage of embryogenesis is due to transcript instability and/or to repression of transcription. The fact that expression is seen in only a proportion of cells suggests that the latter is certainly true. In cells where maternal Xist expression is observed, DXPas34 might be acting as an enhancer of unstable, P0-derived Xist transcription, as discussed above in the context of ES cells, preparing the way for random X inactivation in embryonic cells. Alternatively, DXPas34 could play a totally different role in imprinted X inactivation, for example in preventing high-level Xist expression from the maternal X chromosome. Clearly, analysis of the Xist expression phenotype of paternally or maternally inherited ΔDXPas34 transgenes during embryogenesis will be critical to address this, and chimeric animals are being created to this end.
Comparison of the ΔDXPas34 phenotype with other mutants.
When the phenotype of the DXPas34 deletion is considered in the light of other deletion mutants and transgenics generated in this region, it becomes apparent that the Xic is likely to consist of a very complex series of regulatory sequences. For example, a 35-kb cosmid Xist transgene which does not contain DXPas34 and yet is capable of expressing Xist prior to differentiation would appear to suggest that DXPas34 is not an essential regulatory element of Xist (23, 26). However, the variability in Xist expression levels between cell lines carrying such short transgenes suggests that they are susceptible to position effects (see above) and may have integrated within regions permissive to transgene expression (as would be predicted, given that they were subjected to drug selection). Alternatively, since the cosmid contains only 5 kb of sequence 3′ to Xist, it may lack downstream repressor elements which, in the absence of DXPas34, would constitutively inhibit Xist expression in the context of larger transgenes. In another study, deletion of 65 kb of sequence 3′ to Xist and including DXPas34 leads to a phenotype similar but not identical to the one we have observed. The Xist RNA signal in this case was reduced but not absent in undifferentiated ES cells (12). However, given the large size of the deletion, a number of different regulatory elements may have been affected in this case. Furthermore, the juxtaposition of the 3′ end of an actively transcribed gene (Brx) to the 3′ end of Xist at the deleted locus may actually reconstitute the wild-type situation of 3′ antisense transcription to some extent. In this context, it would be of interest for both the cosmid transgene and the 65-kb deletion mutant to determine whether antisense transcription is present or absent. This could address the question whether Xist expression prior to differentiation can be uncoupled from antisense transcription.
The sequences potentially regulating choice and counting in X inactivation also appear to be diverse. Both the DXPas34 mutant reported here and the 65-kb deletion of Clerc and Avner (12) result in nonrandom inactivation of the deleted allele, suggesting the presence of elements involved in choice and counting in this region. On the other hand, the genetically defined Xce locus, which affects the random nature of X inactivation (10), has been shown to be genetically distinct from Xist and DXPas34, lying distal to them (37a, 43). Additional elements potentially affecting choice and counting have been reported to lie within the Xist gene itself (32). Furthermore, the region upstream of Xist may also contain elements involved in counting, since a 120-kb region 5′ to Xist has recently been shown to be enriched for the hyperacetylated form of histone H4 in female but not in male ES cells or in female ES cells carrying a Xist deletion on one of the two X chromosomes (35). Indeed, the data we have presented here suggests that elements lying 3′ to Xist, such as the DXPas34 locus, might influence the choice of X chromosome to be inactivated via their effects on transcription initiating upstream of Xist. The challenge is now to define whether DXPas34 controls Xist expression through a direct mechanism, for example as an enhancer which binds specific transcription factors, or indirectly, by inducing wide-range chromatin conformational changes which could affect Xist promoter accessibility.
ACKNOWLEDGMENTS
We thank C. Fairhead for the gift of YAC mutagenesis plasmids and helpful advice, N. Brockdorff for the gift of Xist P0 plasmids and primers, C. Morey and P. Clerc for providing strand-specific Xist RNA FISH probes, and V. Colot for critical reading of the manuscript.
This work was supported by grants from the Association Française contre les Myopathies and the Association de Recherche sur le Cancer and from the Ministère de l’Enseignement Supérieur et de la Recherche to E.D.
REFERENCES
- 1.Ashe H L, Monks J, Wijgerde M, Fraser P, Proudfoot N J. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avner P. X chromosome inactivation: Xce and the candidate region for the X inactivation centre. In: Russo E, Riggs A, Martienssen R, editors. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 249–265. [Google Scholar]
- 3.Avner P, Prissette M, Arnaud D, Courtier B, Cecchi C, Heard E. Molecular correlates of the murine Xce locus. Genet Res. 1998;72:217–224. doi: 10.1017/s0016672398003516. [DOI] [PubMed] [Google Scholar]
- 4.Beddington R S P. Isolation, culture and manipulation of postimplantation mouse embryos. Oxford, England: IRL Press; 1987. [Google Scholar]
- 5.Borsani B, Tonlorenzi R, Simmler M-C, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzni D, Lawrence C, Willard H F, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 6.Brockdorff N, Ashworth A, Kay G F, McCabe V M, Norris D P, Cooper P J, Swift S, Rastan S. The product of the mouse Xist gene is a 15kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 7.Brown C J, Ballabio A, Rupert J L, Lafrenière R G, Grompe M, Tonlorenzi R, Willard H F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 8.Brown C J, Hendrich B D, Rupert J L, Lafrenière R G, Xing Y, Lawrence R G, Willard H F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 9.Camus A, Kress C, Babinet C, Barra J. Unexpected behavior of a gene trap vector comprising a fusion between the Sh ble and the lacZ genes. Mol Reprod Dev. 1996;45:255–263. doi: 10.1002/(SICI)1098-2795(199611)45:3<255::AID-MRD1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 10.Cattanach B M, Williams C E. Evidence of non-random X-chromosome activity in the mouse. Genet Res. 1972;19:229–240. doi: 10.1017/s001667230001449x. [DOI] [PubMed] [Google Scholar]
- 11.Clemson C M, McNeil J A, Willard H F, Lawrence J B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clerc P, Avner P. Role of the region 3′ to Xist exon 6 in the counting process of X-chromosome inactivation. Nat Genet. 1998;19:249–253. doi: 10.1038/924. [DOI] [PubMed] [Google Scholar]
- 13.Courtier B, Heard E, Avner P. Xce haplotypes show modified methylation in a region of the active X chromosome lying 3′ to Xist. Proc Natl Acad Sci USA. 1995;92:3531–3535. doi: 10.1073/pnas.92.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debrand E, Heard E, Avner P. Cloning and localization of the murine Xpct gene: evidence for complex rearrangements during the evolution of the region around the Xist gene. Genomics. 1998;48:296–303. doi: 10.1006/geno.1997.5173. [DOI] [PubMed] [Google Scholar]
- 15.Fairhead C, Heard E, Arnaud D, Avner P, Dujon B. Insertion of unique sites into YAC arms for rapid physical analysis following YAC transfer into mammalian cells. Nucleic Acids Res. 1995;23:4011–4012. doi: 10.1093/nar/23.19.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairhead C, Llorente B, Denis F, Soler M, Dujon B. New vectors for combinational deletions in yeast chromosomes and for gap-repair cloning using ‘split-maker’ recombination. Yeast. 1996;12:1439–1457. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1439::AID-YEA37%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 17.Grant S R. Dissecting the mechanisms of posttranscriptional gene silencing: divide and conquer. Cell. 1999;96:303–306. doi: 10.1016/s0092-8674(00)80541-3. [DOI] [PubMed] [Google Scholar]
- 18.Heard E, Avner P, Rothstein R. Creation of a deletion series of mouse YACs covering a 500 kb region around Xist. Nucleic Acids Res. 1994;22:1830–1837. doi: 10.1093/nar/22.10.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heard E, Kress C, Mongelard F, Courtier B, Rougeulle C, Ashworth A, Vourc’h C, Babinet C, Avner P. Transgenic mice carrying an Xist-containing YAC. Hum Mol Genet. 1996;5:441–450. doi: 10.1093/hmg/5.4.441. [DOI] [PubMed] [Google Scholar]
- 20.Heard E, Mongelard F, Arnaud D, Avner P. Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol. 1999;19:3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heard E, Simmler M C, Larin Z, Rougeulle C, Courtier B, Lehrach H, Avner P. Physical mapping and YAC contig analysis of the region surrounding Xist on the mouse X chromosome. Genomics. 1993;15:559–569. doi: 10.1006/geno.1993.1108. [DOI] [PubMed] [Google Scholar]
- 22.Heard H, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 23.Herzing L B K, Romer J T, Horn J M, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 24.Higgs D R. Do LCRs open chromatin domains? Cell. 1998;95:299–302. doi: 10.1016/s0092-8674(00)81761-4. [DOI] [PubMed] [Google Scholar]
- 25.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 26.Johnston C M, Nesterova T B, Formstone E J, Newall A E, Duthie S M, Sheardown S A, Brockdorff N. Developmentally regulated Xist promoter switch mediates initiation of X inactivation. Cell. 1998;94:809–817. doi: 10.1016/s0092-8674(00)81739-0. [DOI] [PubMed] [Google Scholar]
- 27.Kay G F, Penny G D, Patel D, Ashworth A, Brockdorff N, Rastan S. Expression of Xist during mouse development suggests a role in initiation of X chromosome inactivation. Cell. 1993;72:171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- 28.Lee J T, Davidow L S, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 29.Lee J T, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–278. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 30.Lee J T, Lu N, Han Y. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc Natl Acad Sci USA. 1999;96:3836–3841. doi: 10.1073/pnas.96.7.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J T, Strauss W M, Dausman J A, Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 32.Marahrens Y, Loring J, Jaenisch R. Role of the Xist gene in X chromosome choosing. Cell. 1998;92:657–664. doi: 10.1016/s0092-8674(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 33.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 34.Marriott S J, Brady J N. Enhancer function in viral and cellular gene regulation. Biochim Biophys Acta. 1989;989:97–110. doi: 10.1016/0304-419x(89)90037-1. [DOI] [PubMed] [Google Scholar]
- 35.O’Neill L P, Keohane A M, Lavender J S, McKabe V, Heard E, Avner P, Brockdorff N, Turner B. A developmental switch in H4 acetylation upstream of Xist plays a role in X chromosome inactivation. EMBO J. 1999;18:2897–2907. doi: 10.1093/emboj/18.10.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 37.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. The Xist gene is required in cis for X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 37a.Prissette, M., et al. Unpublished data.
- 38.Reik W, Walter J. Imprinting mechanisms in mammals. Curr Opin Genet Dev. 1998;8:154–164. doi: 10.1016/s0959-437x(98)80136-6. [DOI] [PubMed] [Google Scholar]
- 39.Robertson E J. Embryo-derived stem cell lines. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Oxford, England: IRL Press; 1987. pp. 71–112. [Google Scholar]
- 40.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 41.Rougeulle C, Colleaux L, Dujon B, Avner P. Generation and characterisation of an ordered lambda clone array for the 460 kb region surrounding the murine Xist sequence. Mamm Genome. 1994;5:416–423. doi: 10.1007/BF00357001. [DOI] [PubMed] [Google Scholar]
- 42.Sheardown S A, Duthie S M, Johnston C M, Newall A E T, Formstone E J, Arkell R M, Nesterova T B, Alghisi G C, Rastan S, Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- 43.Simmler M C, Cattanach B M, Rasberry C, Rougeulle C, Avner P. Mapping the murine Xce locus with (CA)n repeats. Mamm Genome. 1993;4:523–530. doi: 10.1007/BF00364788. [DOI] [PubMed] [Google Scholar]
- 44.Simmler M C, Cunningham D B, Clerc P, Vermat T, Caudron B, Cruaud C, Pawlak A, Szpirer C, Weissenbach J, Claverie J M, Avner P. A 94kb genomic sequence 3′ to the murine Xist gene reveals an AT rich region containing a new testis specific gene Tsx. Hum Mol Genet. 1996;11:1713–1726. doi: 10.1093/hmg/5.11.1713. [DOI] [PubMed] [Google Scholar]